Abstract
Nucleotide sequences of all eight RNA segments of 10 human H3N2 influenza viruses isolated during a 5-year period from 1993 to 1997 were determined and analyzed phylogenetically in order to define the evolutionary pathways of all genes in a parallel fashion. It was evident that the hemagglutinin and neuraminidase genes of these viruses evolved essentially in a single lineage and that amino acid changes accumulated sequentially with respect to time. In contrast, amino acid differences in the internal proteins were erratic and did not accumulate over time. Parallel analysis of the phylogenetic patterns of all genes revealed that the evolutionary pathways of the six internal genes were not linked to the surface glycoproteins. Genes coding for the basic polymerase-1, nucleoprotein, and matrix proteins of 1997 isolates were closest phylogenetically to those of earlier isolates of 1993 and 1994. Furthermore, all six internal genes of four viruses isolated in the 1995 epidemic season consistently divided into two distinct branch clusters, and two 1995 isolates contained PB2 genes apparently originating from those of viruses before 1993. It was apparent that the lack of correlation between the topologies of the phylogenetic trees of the genes coding for the surface glycoproteins and internal proteins was a reflection of genetic reassortment among human H3N2 viruses. This is the first evidence demonstrating the occurrence of genetic reassortment involving the internal genes of human H3N2 viruses. Furthermore, internal protein variability coincided with marked increases in the activity of H3N2 viruses in 1995 and 1997.
The surface hemagglutinin (HA) glycoprotein of influenza viruses is the major target for neutralizing antibodies, and point mutations in the potential antigenic domains of this protein are thought to allow viruses to evade established immune antibodies in the human population. Some influenza seasons are more severe than others, and this is thought to be a reflection of the degree of antigenic change in the HA protein of newly appearing antigenic variants from those of the former strain. Viruses with antigenically drifted HA proteins thus have a selective advantage in becoming the subsequent epidemic strain. Analyses of epidemic influenza virus isolates, therefore, have chiefly focused on antigenic characterization of the HA glycoprotein in order to detect new variants of each epidemic strain for the recommendation of vaccine strains in each season.
Although there have been a number of reports on the characterization of human H3N2 influenza viruses, it was recently reported that detection of a new H3N2 antigenic variant is often associated with the rapid disappearance of the former variant (14, 31). For example, during the 1992-1993 epidemic season in Japan and England an A/Beijing/352/89-like H3N2 variant that circulated early in the season was displaced by an A/Beijing/32/92-like variant (14, 31) during the latter part of the same season. Other reports, however, have demonstrated cocirculation of phylogenetically distinct H3N2 viruses in the same season (12, 34, 35).
It has been reported repeatedly that the virulence and growth of influenza viruses are influenced by changes in the internal proteins. The matrix (M1) protein is a multifunctional protein which contributes to the control of virulence, growth, (15, 52, 62, 63) and host specificity of influenza viruses (10, 33). In experiments with single gene reassortants of influenza viruses, it was shown that changes in the NP, basic polymerase-2 (PB2), and M1 proteins were involved in host restriction in monkeys (33), while attenuation of human viruses was confirmed in human volunteers by changes in the NP, nonstructural-1 (NS1), PB1, and PB2 proteins (10). Considering this evidence, the severity of an influenza epidemic season may be influenced not only by variability in the surface glycoproteins (HA and NA) but also by differences in the internal proteins (PB2, PB1, PA, NP, M1, M2, NS1, and NS2) of circulating influenza viruses. In this study, parallel evolutionary analyses of all eight gene segments and amino acid comparisons of all 10 viral proteins of 10 human H3N2 epidemic strains isolated over a 5-year period from 1993 to 1997 in Japan was done in order to provide a complete profile of protein variability, as well as the evolutionary patterns of all gene segments of these viruses. Also, phylogenetic and amino acid sequence data were compared with seasonal epidemiological data, including the total number of influenza-like illnesses and the numbers of virus isolations in Japan.
MATERIALS AND METHODS
Viruses.
The viruses used in this study and their abbreviations are summarized in Table 1. Abbreviations for Japanese strains isolated between 1993 and 1997 reflect the epidemic season in which the strain was isolated and not necessarily the year of isolation. For example, isolate A/Aichi/69/94, which was isolated during the influenza season of 1994-1995, was assigned the abbreviation Aic95 in order to more easily distinguish this virus from viruses of the 1993-1994 season, such as A/Akita/1/94 (Aki94). Viruses whose genes were sequenced in this report were propagated in 11-day-old embryonated chicken eggs (E) or MDCK-cell monolayers (M) as follows: A/Sichuan/2/87 (E), A/Hokkaido/20/89 (E), A/Beijing/352/89 (E), A/Washington/15/91 (E), A/Beijing/32/92 (E), A/Hebei/12/93 (E), A/Tianjin/33/92 (E), A/Kitakyushu/159/93 (E), A/Akita/1/94 (E), A/Tochigi/44/95 (M), A/Shiga/20/95 (E), A/Akita/1/95 (E), A/Wuhan/359/95 (E), A/Miyagi/29/95 (M), A/Fukushima/114/96 (E), A/Niigata/137/96 (M), A/Fukushima/140/96 (E), and A/Shiga/25/97 (M).
TABLE 1.
Sequence data and abbreviations of H3N2 isolates used in this study
| Strain | Abbreviation | Sequence referencesa for:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Segment 1 PB2 gene | Segment 2 PB1 gene | Segment 3 PA gene | Segment 4 HA gene | Segment 5 NP gene | Segment 6 NA gene | Segment 7 M gene | Segment 8 NS gene | ||
| A/Aichi/2/68 | Aic68 | M63515 | —b | M63515 | M34829 | ||||
| A/NT/60/68 | NT68 | J02140 | J02138 | J02139 | J02136 | ||||
| A/Beijing/1/68 | Bei68 | L07340 | |||||||
| A/Hong Kong/8/68 | HK68 | U42630 | |||||||
| A/Memphis/1/71 | Mem71 | L07341 | |||||||
| A/Hong Kong/107/71 | HK71 | U42631 | |||||||
| A/Udorn/307/72 | Udn72 | M91712 | D00051 | J02167 | J02169 | ||||
| A/Dunedin/4/73 | Dun73 | M74899 | |||||||
| A/PortChalmers/73 | PC73 | X08092 | |||||||
| A/Beijing/39/75 | Bei75 | L07358 | |||||||
| A/Victoria/3/75 | Vic75 | J02173 | |||||||
| A/Alaska/6/77 | Ala77 | K01332 | |||||||
| A/Texas/1/77 | Tex77 | D00602 | |||||||
| A/Guangdong/38/77 | Gua77 | L07359 | |||||||
| A/Bangkok/1/79 | Ban79 | K01150 | K01140 | ||||||
| A/Memphis/5/80 | Mem80 | L07360 | |||||||
| A/Shanghai/31/80 | Sha80 | L07361 | |||||||
| A/HongKong/5/83 | HK83 | M22577 | |||||||
| A/Yamagata/96/85 | Yam85 | NAc | |||||||
| A/Houston/24269/85 | Hou85 | M17699 | |||||||
| A/Mississippi/1/85 | Mis85 | U42632 | |||||||
| A/Memphis/14/85 | Mem85 | L07364 | |||||||
| A/Memphis/6/86 | Mem86 | L07365 | |||||||
| A/Los Angeles/2/87 | Los87 | U62543 | |||||||
| A/Guangdong/9/87 | Gua87 | L07366 | |||||||
| A/Sichuan/2/87 | Sic87 | D21173 | D30671*a | ||||||
| A/Shanghai/11/87 | Sha87 | U42633 | |||||||
| A/Memphis/8/88 | Mem88 | M36040 | M259361 | L07370 | M63516 | ||||
| A/England/427/88 | Eng88 | U42634 | |||||||
| A/Guangdong/38/89 | Gua89 | L07373 | |||||||
| A/Guangdong/39/89 | Gua89 | L18995 | |||||||
| A/Guizhou/54/89 | Gui89 | D49963 | |||||||
| A/Hokkaido/20/89 | Hok89 | D49960 | D30672* | ||||||
| A/Beijing/352/89 | Bei89 | D49961 | L07353 | U42635 | D30670* | ||||
| A/Shanghai/6/90 | Sha690 | L07357 | |||||||
| A/Shanghai/24/90 | Sha2490 | U42636 | |||||||
| A/Memphis/1/90 | Mem90 | L07355 | |||||||
| A/Bangkok/144/90 | Ban90 | D49966 | |||||||
| A/Shiga/2/91 | Shi91 | D49967 | |||||||
| A/Brazil/2/91 | Bra91 | D30662 | |||||||
| A/Washington/15/91 | Was91 | D30663 | U42637 | D30673* | |||||
| A/Beijing/32/92 | Bei92 | Z46392 | U42770 | D30667* | |||||
| A/Yokohama/83/92 | Yok92 | D30665 | |||||||
| A/Tianjin/33/92 | Tia92 | D30664 | D30674* | ||||||
| A/Sapporo/304/92 | Sap92 | U42776 | |||||||
| A/Netherlands/938/92 | Net92 | U42775 | |||||||
| A/Harbin/15/92 | Har92 | U42773 | |||||||
| A/Nanchang/3332/93 | Nan93 | U42779 | |||||||
| A/Washington/26/93 | Was93 | U43420 | |||||||
| A/Louisiana/4/93 | Lou93 | U43417 | |||||||
| A/Hebei/12/93 | Heb93 | D30668 | U65561* | ||||||
| A/Shangdong/9/93 | Shd93 | Z46417 | U43419 | ||||||
| A/Kitakyushu/159/93 | Kit93 | AF037412* | AF037418* | AF037424* | D30669 | AF038254* | AF038260* | U65562* | D30676* |
| A/Guangdong/24/93 | Gua2493 | U42777 | |||||||
| A/Guangdong/25/93 | Gua2593 | U42778 | |||||||
| A/Bangkok/122/94 | Ban94 | U43426 | |||||||
| A/Hebei/41/94 | Heb94 | U48441 | |||||||
| A/Guangdong/28/94 | Gua94 | U48442 | |||||||
| A/Sendai/c384/94 | Sen94 | U48440 | |||||||
| A/Singapore/7/94 | Sin94 | U43422 | |||||||
| A/NewYork/16/94 | NY94 | U43424 | |||||||
| A/Vermont/3/94 | Ver94 | U43423 | |||||||
| A/Akita/1/94 | Aki94 | U71132* | U71128* | U71136* | U48443 | U71144* | U71140* | U65564* | U65670* |
| A/Johannesburg/33/94 | Joh94 | U43425 | |||||||
| A/Tochigi/44/95 | Toc95 | AF037413* | AF037419* | AF037425* | U65554* | AF038255* | AF038261* | U65569* | AF038275* |
| A/Aichi/69/94 | Aic95 | U48446 | |||||||
| A/Shiga/20/95 | Shi95 | U71134* | U71130* | U71138* | U65557* | U71146* | U71142* | U65573* | U65673* |
| A/Akita/1/95 | Aki95 | U71133* | U71129* | U71137* | U48444 | U71145* | U71141* | U65574* | U65671* |
| A/Alaska/10/95 | Ala95 | U51245 | |||||||
| A/Wuhan/359/95 | Wuh95 | AF038268* | U51246 | ||||||
| A/Miyagi/29/95 | Miy95 | U71135* | U71131* | U71139* | U65560* | U71147* | U71143* | U65577* | U65674* |
| A/Fukushima/114/96 | Fuk96 | AF037414* | AF037421* | AF037427* | AF038266* | AF038257* | AF038265* | AF038272* | AF038277* |
| A/Niigata/137/96 | Nii96 | AF037415* | AF037420* | AF037426* | AF038267* | AF038256* | AF038262* | AF038271* | AF038276* |
| A/Fukushima/140/96 | Fuk97 | AF037416* | AF037422* | AF037428* | AF038269* | AF038258* | AF038263* | AF038273* | AF038278* |
| A/Shiga/25/97 | Shi97 | AF037417* | AF037423* | AF037429* | AF038270* | AF038259* | AF038264* | AF038274* | AF038279* |
∗, Sequenced in this report.
—, Sequence not available.
NA, accession number not available (Nakajima et al. [35]).
RNA extraction and nucleotide sequencing.
RNA from 100 μl of virus sample was purified as described previously (9) and suspended in 50 μl of RNase-free distilled water. Whenever possible, samples were taken directly from virus stocks received from municipal and prefectural centers for hygiene without being propagated further in our laboratory. Reverse transcription (RT)-PCR was performed by using a slightly modified protocol of a commercial kit (RT-PCR kit with avian myeloblastosis virus [AMV]; version 2.1; Takara). Briefly, first-strand cDNA synthesis was done by mixing 9.5 μl of RNA with 1 μl of “influenza A universal RT” primer (3′-AGCAAAAGCAGG-5′) (20 μM) and 9.5 μl of RT reaction mixture (4 μl of 25 mM MgCl2, 2 μl of 10× RT-PCR buffer, 2 μl of 10 mM dNTP, 1 μl of AMV reverse transcriptase, 0.5 μl of RNase inhibitor). This mixture was incubated at 30°C for 10 min, at 42°C for 60 min, and finally at 95°C for 5 min. Resultant cDNA was then used in all subsequent PCR amplifications of overlapping cassettes covering the complete protein coding domain of each gene. Nucleotide sequences for all oligonucleotide primers used for PCR amplification are available from the authors upon request. RT-PCR products were purified and sequenced directly as described previously (48) on an ABI377 autosequencer (Perkin-Elmer). All nucleotide sequence data reported here will appear in the GSDB, DDBJ, EMBL, and NCBI nucleotide sequence databases under the accession numbers listed in Table 1. Accession numbers for previously reported sequence data used in this report for the PB2 (21, 24, 29, 45), PB1 (4, 22, 25), PA (5), HA (14, 35, 37), NP (1, 6, 18, 50), NA (2, 32, 53, 59), M (23, 28, 43, 60, 64), and NS (7, 27, 42) genes are also summarized in Table 1.
Phylogenetic analyses.
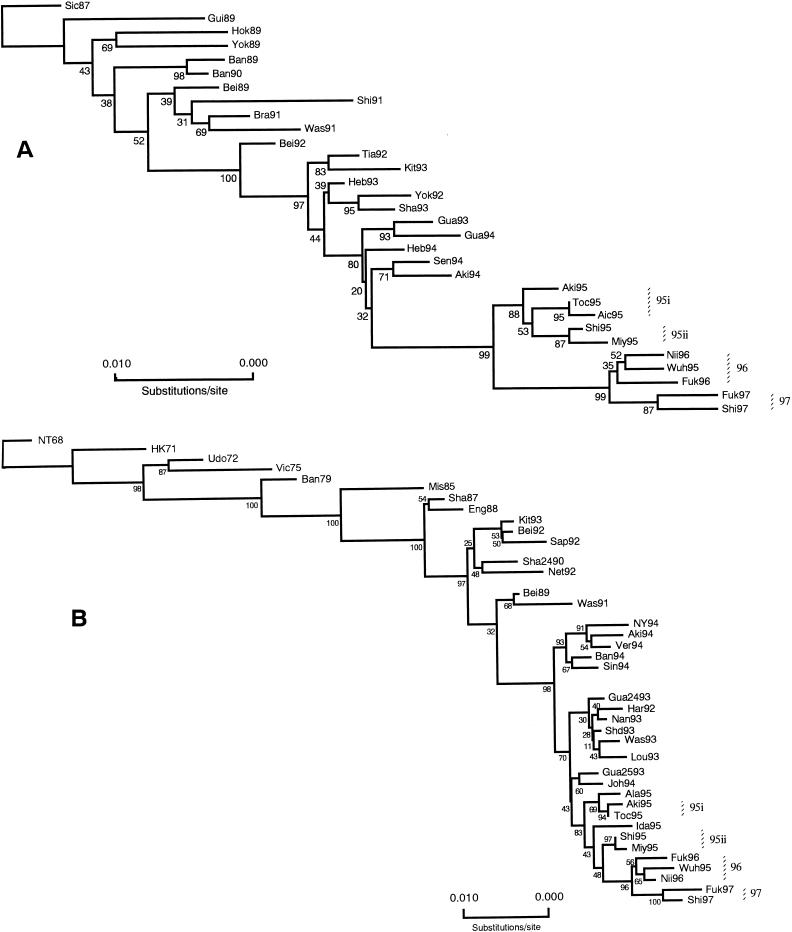
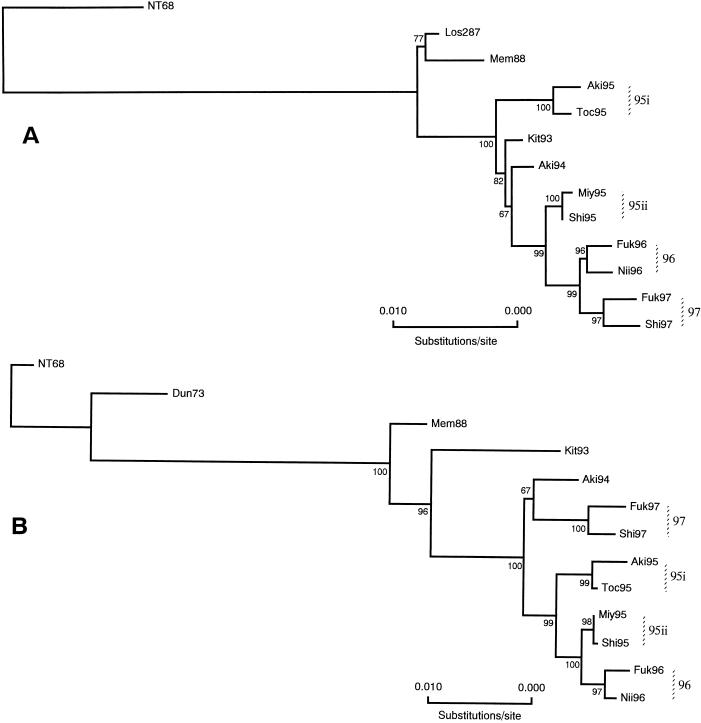
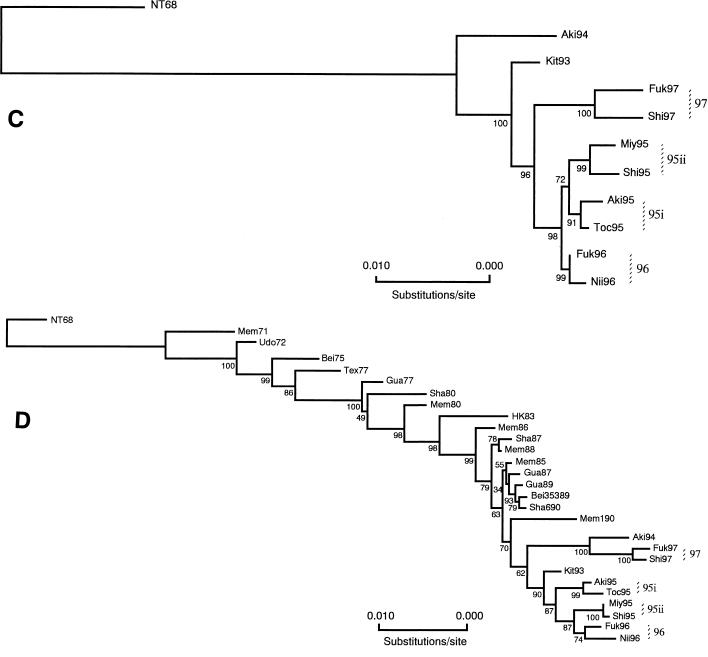
Phylogenetic analyses of the complete protein coding regions of all RNA segments were done using the neighbor-joining (NJ) method (20, 47). Nucleotide distance matrices were estimated by the six-parameter method (19) based on the number of total nucleotide substitutions, and evolutionary trees for the HA (Fig. 1A), NA (Fig. 1B), PB2 (Fig. 2A), PB1 (Fig. 2B), PA (Fig. 2C), NP (Fig. 2C), M (Fig. 3A), and NS (Fig. 3B) genes were constructed (20). To evaluate the robustness of the trees, the probabilities of the internal branches were determined by 500 bootstrap replications (16).
FIG. 1.
Evolutionary trees based on the total number of nucleotide substitutions of the HA1 domain of the HA (A) and NA (B) genes of human H3N2 influenza viruses constructed by NJ analysis. Numbers at the nodes indicate confidence levels of bootstrap analysis with 500 replications as a value of percent.
FIG. 2.
Evolutionary trees of the PB2 (A), PB1 (B), PA (C), and NP (D) genes of human H3N2 influenza viruses based on the total number of nucleotide substitutions in the protein coding regions constructed by NJ analysis. Numbers at the nodes indicate confidence levels of bootstrap analysis with 500 replications as a percentage value.
FIG. 3.
Evolutionary trees of the M (A) and NS (B) genes of human H3N2 influenza viruses based on the total number of nucleotide substitutions in the complete protein coding regions constructed by NJ analysis. Numbers at the nodes indicate confidence levels of bootstrap analysis with 500 replications as a percentage value.
RESULTS
Amino acid differences in the HA and NA proteins.
As shown in Table 2, amino acid changes in the HA and NA glycoproteins accumulated in a sequential fashion over time. Indeed, when compared to that of Kit93, 10 amino acid differences in the HA1 protein of 1995 isolates were also found in those of 1997 isolates (Fuk97 and Shi97). Previously uncharacterized substitutions were observed in proposed antigenic sites A and B, as well as the receptor binding domain of the HA protein (55, 56). Even though a novel change at position 226 of the receptor binding domain of the HA protein in Japanese and Chinese isolates of the 1994-1995 influenza season was reported (31), this residue was found to have changed again from isoleucine to valine in viruses of 1996 and 1997. Changes from leucine to glutamine at position 226 have been reported to be involved in determining binding specificity to host cell receptors (36, 41, 46); however, the effect of isoleucine or valine at this critical site have not been determined and warrant further study.
TABLE 2.
Amino acid variation observed in human H3N2 viruses isolated from 1993 to 1997 in Japana
| Protein | PB2 aa at position:
|
PB1 aa at position:
|
PA aa at position:
|
||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 82 | 107 | 221 | 318 | 539 | 553 | 559 | 570 | 697 | 14 | 56 | 83 | 97 | 163 | 179 | 216 | 382 | 439 | 517 | 622 | 212 | 250 | 256 | 275 | 276 | 312 | 341 | 342 | 349 | 350 | 376 | 409 | 432 | 437 | 548 | 557 | 712 | |
| Kit93 | S | T | A | R | I | V | T | I | L | A | R | D | E | N | I | S | S | D | I | Q | R | S | K | D | C | R | V | L | E | T | P | N | V | Y | M | M | T |
| Aki94 | M | K | G | R | L | I | |||||||||||||||||||||||||||||||
| Aki95 | M | V | K | G | R | I | H | L | K | ||||||||||||||||||||||||||||
| Toc95 | M | V | K | D | G | R | I | H | K | ||||||||||||||||||||||||||||
| Miy95 | K | M | K | D | G | R | Q | M | Q | I | K | ||||||||||||||||||||||||||
| Shi95 | M | K | G | R | S | M | I | I | K | ||||||||||||||||||||||||||||
| Fuk96 | T | M | I | K | N | S | N | V | R | I | |||||||||||||||||||||||||||
| Nii96 | M | I | K | S | V | V | R | I | |||||||||||||||||||||||||||||
| Fuk97 | N | S | I | N | M | I | K | G | R | F | K | N | I | ||||||||||||||||||||||||
| Shi97 | L | M | I | K | G | G | R | H | K | K | N | I | |||||||||||||||||||||||||
| Protein | HA (HA1 domain) aa at position:
|
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 27 | 34 | 111 | 114 | 133 | 135 | 142 | 156 | 157 | 158 | 164 | 167 | 172 | 183 | 196 | 197 | 214 | 219 | 226 | 248 | 262 | 275 | 276 | 278 | 279 | |
| Kit93 | G | K | I | I | D | D | G | G | K | S | E | L | S | G | H | V | Q | T | S | Q | T | N | G | T | S | F |
| Aki94 | R | K | L | T | R | I | F | N | S | |||||||||||||||||
| Aki95 | T | G | K | L | T | D | I | I | I | N | N | S | ||||||||||||||
| Toc95 | T | G | K | L | T | D | I | I | D | N | N | S | ||||||||||||||
| Miy95 | T | G | K | L | T | D | I | I | D | N | N | S | ||||||||||||||
| Shi95 | T | G | K | L | T | D | I | I | D | N | N | S | ||||||||||||||
| Fuk96 | R | V | T | G | T | L | T | D | I | V | S | N | N | S | ||||||||||||
| Nii96 | T | G | N | T | L | T | D | I | V | S | D | N | N | S | ||||||||||||
| Fuk97 | N | S | N | T | R | L | T | D | L | A | I | V | S | K | N | S | ||||||||||
| Shi97 | N | S | N | T | R | Q | L | k | Q | T | D | I | V | S | N | N | S | |||||||||
| Protein | NP aa at position:
|
NA aa at position:
|
||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18 | 65 | 127 | 197 | 239 | 259 | 290 | 375 | 384 | 451 | 24 | 43 | 47 | 51 | 52 | 54 | 65 | 71 | 82 | 127 | 136 | 141 | 151 | 212 | 221 | 248 | 262 | 285 | 336 | 338 | 339 | 342 | 344 | 346 | 369 | 381 | 385 | 400 | 418 | 435 | |
| Kit93 | E | R | D | I | M | S | N | E | R | A | M | S | N | I | P | Q | I | T | V | G | Q | N | D | V | N | E | V | P | Y | Q | N | N | K | S | E | G | T | S | I | K |
| Aki94 | K | E | V | L | G | G | N | S | M | L | E | A | I | R | K | K | R | E | ||||||||||||||||||||||
| Aki95 | D | N | M | L | E | A | I | L | E | G | K | K | R | V | E | |||||||||||||||||||||||||
| Toc95 | D | N | M | L | E | A | I | L | E | G | K | K | R | V | E | |||||||||||||||||||||||||
| Miy95 | D | T | V | N | M | L | E | A | R | G | I | L | E | G | K | K | R | E | ||||||||||||||||||||||
| Shi95 | D | T | N | M | L | E | A | G | I | L | E | G | K | K | R | E | ||||||||||||||||||||||||
| Fuk96 | D | T | N | M | L | E | I | A | D | G | A | K | I | H | L | E | G | K | K | R | E | |||||||||||||||||||
| Nii96 | D | T | N | M | L | E | A | D | K | G | I | S | H | L | E | G | K | K | R | E | ||||||||||||||||||||
| Fuk97 | D | K | E | V | V | L | D | G | G | N | M | L | G | A | D | K | K | G | I | H | L | D | E | G | K | E | K | R | E | |||||||||||
| Shi97 | D | K | E | V | V | L | D | G | G | N | M | L | E | M | A | D | K | G | I | H | L | E | G | K | E | K | R | E | ||||||||||||
| Protein | M1 aa at position:
|
M2 aa at position:
|
NS1 aa at position:
|
NS2 aa at position:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 219 | 227 | 230 | 16 | 21 | 43 | 57 | 33 | 66 | 68 | 71 | 101 | 112 | 129 | 144 | 164 | 209 | 212 | 228 | 26 | |
| Kit93 | I | A | K | G | D | L | H | L | E | I | E | N | E | M | L | S | N | P | S | E |
| Aki94 | V | K | P | |||||||||||||||||
| Aki95 | E | G | Y | I | K | D | P | |||||||||||||
| Toc95 | E | G | F | Y | K | D | P | |||||||||||||
| Miy95 | K | I | P | |||||||||||||||||
| Shi95 | I | P | V | |||||||||||||||||
| Fuk96 | I | P | ||||||||||||||||||
| Nii96 | I | P | ||||||||||||||||||
| Fuk97 | V | T | R | I | P | D | S | |||||||||||||
| Shi97 | V | T | R | Q | P | P | ||||||||||||||
Predicted amino acid (aa) differences in the proteins of human H3N2 viruses isolated from 1993 to 1997. Only locations where differences were observed are indicated. Differences located in the proposed antigenic sights and of the HA and NA proteins are underlined with a single line. Amino acid differences in the proposed receptor binding domain of the HA protein, as well as the functional domain of the NA protein, are double underlined.
As with the HA, amino acid differences in the NA proteins were maintained in those of later viruses. Substitutions were observed throughout the NA protein, although most changes were located in the functional globular region. Amino acid differences were also revealed in locations which have been implicated as antigenic sites of the NA protein (11) at position 336 (Y to H) in viruses of 1996 and 1997 as well as at residue 342 (N to D) in Fuk97. One change was also revealed in the substrate-binding pocket at position 151 (D to G), although this change was observed in only one isolate, Fuk96.
Amino acid differences in the internal M1, M2, NS1, and NS2 proteins.
Analysis of the M genes revealed that the M1 proteins of influenza viruses were highly conserved, supporting the observations by Ito et al. (23). Indeed, amino acid substitutions were only observed in the most recent isolates of 1997 (Fuk97 and Shi97), which contained three amino acid substitutions at positions 219 (I to V), 227 (A to T), and 230 (K to R). Also, it was of interest to reveal that, although most M2 proteins of viruses isolated from the 1993–1997 period were completely conserved, those of the 1995 isolates showed as many as four amino acid changes (Table 2) at positions 16 (G to E) and 21 (D to G) in the external amino domain, position 57 (H to Y) in the internal carboxyl domain, and position 43 (L to F) in the functional transmembrane domain.
Twelve amino acid substitutions were observed in the NS1 proteins of viruses isolated from 1994 to 1997, although most changes were restricted to a single virus or to viruses of a particular season. Only one substitution, at position 164 (S to P), was shown in all viruses from 1994 to 1997 when compared to that of Kit93. In contrast to the high variability of the NS1 proteins, the NS2 proteins of these viruses were highly conserved. Only one difference was observed in the NS2 protein of Shi95, at position 26 (E to V).
Amino acid differences in the proteins of the PB2, PB1, PA, and NP proteins of the RNP complex.
Analysis of amino acid sequence changes in the PB2, PB1, and PA proteins of recent Japanese viruses found only a low number of amino acid changes in these proteins, changes which were often not maintained in other isolates. Of the nine amino acid substitutions observed in the PB2 proteins of viruses from 1993 to 1997, only two were maintained, at position 570 (I to M) in all viruses isolated after 1994 and at position 697 (L to I) in isolates of 1996 and 1997. Similarly, only 2 of 11 amino acid substitutions among the PB1 proteins were conserved, at positions 56 (R to K) and 622 (Q to R), in all of the viruses isolated after 1994. However, with the exception of Fuk96 and Nii96, one amino acid difference was revealed in all of the isolates, at position 216 (S to G). Remaining substitutions in the PB1 protein were restricted to one isolate or to viruses of a single season. The PA protein had slightly higher variability, with 17 differences observed between isolates, although 9 of them were erratic and 5 were shared only among viruses of their respective epidemic seasons. Only two changes, at positions 432 (V to I) and 712 (T to K), were maintained in isolates of 1995 and 1996, although these disappeared in viruses of 1997 which contained changes at residues 312 (R to K), 350 (T to N), and 557 (M to I).
The pattern of amino acid differences in the NP proteins is worth notice from an evolutionary point of view. For example, all six amino acid changes observed in the NP proteins of Aki94 also existed in those of the 1997 viruses (Fuk97 and Shi97) but were not found in those of the 1995-1996 viruses. Also, a change at residue 451 (A to T) in viruses isolated in 1995 and 1996 (Miy95, Shi95, Fuk96, and Nii96) was not maintained in viruses of 1997. Two additional amino acid differences were found in isolates of 1997 at positions 18 (E to D) and 239 (M to V) which were not observed in those of other isolates.
Evolutionary analysis of the HA and NA genes.
Phylogenetic profiles of the HA gene (Fig. 1A) were consistent with those of previous reports (14, 31) and showed that the HA gene had evolved in a sequential fashion, with isolates of each season forming distinct clades. The HA genes of viruses of 1995 formed a clade which could be further divided into two branch clusters (BC) represented by 95i (Aki95, Toc95, and Aic95) and 95ii (Shi95 and Miy95), respectively, with a bootstrap value (BSV) of 87. Although 1995 viruses were divided phylogenetically into two branch clusters, variability among the HA genes of these isolates was a reflection of silent nucleotide mutations in the HA1 domain. For instance, although Miy95 and Toc95 were on different branch clusters, their HA1 proteins were identical to that of Toc95 (Table 2). Also, the 1996 viruses Fuk96 and Nii96 formed a branch cluster together with the vaccine strain, Wuh95 (BC 96) (BSV 35), while Fuk97 and Shi97 (BC 97) (BSV 87), were located on the newest lineage.
Like the HA gene, the NA genes of H3N2 isolates were shown to evolve in an essentially linear fashion (Fig. 1B). Analysis of the NA gene in this report showed the evolutionary patterns of the NA genes of the most recent viruses of 1996 and 1997 to have divided into two clades including Wuh95, Fuk96, and Nii96 (BC 96) (BSV 56) and Fuk97 and Shi97 (BC 97) (BSV 100). However, in contrast to the HA gene, cocirculation of distinct NA genes in the same epidemic season was observed, a finding supporting the phylogenetic variability observed by Xu et al. (59). Viruses of 1995 were revealed to be located in two distinct branch clusters, including 95i (Aki95 and Toc95) (BSV 94) and 95ii (Miy95 and Shi95) (BSV 97).
Evolutionary analyses of the PB2, PB1, PA, and NP genes.
Results of parallel phylogenetic analyses of the genes coding for the proteins of the RNP complex were particularly noteworthy. The PB2 genes (Fig. 2A) of viruses isolated from 1993 to 1997 were found to evolve generally in a sequential fashion, as isolates of 1993 (Kit93), 1994 (Aki94), 1995 (BC 95ii), 1996 (BC 96), and 1997 (BC 97) were on the same lineage. However, two viruses of 1995 (BC 95i) (BSV 100) formed a branch cluster which was distinguished from other viruses and apparently diverged before 1993. In the case of the PB1 gene (Fig. 2B), bootstrap analysis determined that isolates of 1997 (BC 97) (BSV 100) were most similar genetically to that of an earlier isolate of 1994 (Aki94) (BSV 67), whereas those of other isolates of 1995 and 1996 formed a separate lineage which was further divided into three branch clusters containing those of 1995 (BC 95i and 95ii) (BSV 99 and 98, respectively) and 1996 isolates (BC 96) (BSV 97). Construction of the evolutionary tree for the PA genes (Fig. 2C) revealed yet another distinct topology which showed little correlation with the chronology of the virus isolates, although the probabilities of the internal branches of the tree were very high. Viruses of 1995 (BC 95i and 95ii) (BSV 91 and 99, respectively) and 1996 (BC 96) (BSV 99) formed clades distinct from the viruses of 1997 (BC 97) (BSV 100), which appeared to have diverged sometime earlier. Also, the evolutionary position of a 1994 virus (Aki94) indicated that this gene branched off prior to that of a 1993 virus (Kit93) (BSV 100).
As shown in Fig. 2D, results of phylogenetic analysis of the NP genes of many human H3N2 viruses showed that these genes had evolved essentially as a single lineage, supporting observations by Shu et al. (50). However, an examination of the NP genes of viruses isolated from 1993 to 1997 sequenced in this study demonstrated that those of 1997 (BC 97) (BSV 100) viruses formed a distinct lineage with that of a 1994 isolate (Aki94) (BSV 100) that evidently had diverged prior to 1993. Viruses of 1995 and 1996, which appeared to have derived from the NP genes similar to 1993 viruses, evolved into three clades distinguishing the viruses of 1995 (BC 95i) (BSV 99) and (BC 95ii) (BSV 100) and those of 1996 (BC 96) (BSV 74).
Evolutionary analysis of the M and NS genes.
Even though the M genes of human H3N2 viruses were highly conserved, the present study revealed distinct phylogenetic differences (Fig. 3A). The M genes of the 95i branch (BSV 98) were distantly related to other viruses of 1995 (BC 95ii) (BSV 78), while the M genes of two 1996 isolates were also different from one another. Most interestingly, it was clearly shown that the M genes of viruses of 1997 (BC 97) (BSV 99) were distinguished from those of other recent Japanese viruses and showed the highest degree of genetic similarity to that of a Chinese isolate of 1993, Heb93 (BSV 95).
The results of an evolutionary analysis of the NS genes of recent isolates revealed nonlinear evolutionary pathways (Fig. 3B). The phylogenetic relationships among 1996 and 1997 viruses could not be elucidated, since the BSVs were relatively low, allowing for other possible branching patterns. However, it was apparent that viruses of the 95ii clade (BSV 96) were distinct from other 1995 viruses of the 95i clade (BSV 57), which were more similar to that of Aki94 (BSV 63).
Influenza virus activity in Japan from 1993 to 1997.
From year to year, the level of human morbidity and mortality attributed to influenza virus activity may vary considerably. In order to estimate the levels of influenza virus activity in Japan, various types of data are collected annually from local hospitals, clinics, and schools, as well as from prefectural and municipal institutes of hygiene. The numbers of reported influenza-like illnesses (ILIs) and the numbers of virus isolates of A/H3N2, A/H1N1, and B influenza virus are good indices in the estimation of morbidity due to influenza (Table 3). Also, these indices allow the estimation of the yearly relative morbidity in humans caused by each influenza virus. As shown in Table 3, the numbers of ILIs reported in the five influenza seasons from 1993 to 1997 varied considerably, as did the numbers of isolates of each influenza virus (A/H1N1, A/H3N2, and B). By calculation of the relative morbidity due to A/H3N2 viruses for each season, it was revealed that A/H3N2 activity was very high in the 1993, 1995, and 1997 seasons, with morbidity cases of 304,665, 361,831, and 229,490, respectively. These values are in sharp contrast to the values determined for the 1994 and 1996 seasons of 65,192 and 11,245 cases, respectively.
TABLE 3.
Estimation of relative morbidity due to influenza A (H3N2) viruses from 1993 to 1997 in Japan
| Epidemic seasona | No. of ILIsb | No. of influenza virus isolatesc
|
Relative morbidity due to H3N2 influenza virusd | |||
|---|---|---|---|---|---|---|
| A/H1N1 | A/H3N2 | B | Total | |||
| 1993 (10/92–10/93) | 877,520 | 0 | 1,278 | 2,403 | 3,681 | 304,665 |
| 1994 (10/93–10/94) | 76,461 | 0 | 457 | 79 | 536 | 65,192 |
| 1995 (10/94–10/95) | 735,175 | 88 | 1,917 | 1,890 | 3,895 | 361,831 |
| 1996 (10/95–10/96) | 176,260 | 3,475 | 237 | 3 | 3,715 | 11,245 |
| 1997 (10/96–10/97) | 317,305 | 0 | 3,217 | 1,231 | 4,448 | 229,490 |
The epidemic season in Japan begins in October and continues until October of the next year, as indicated in parenthesis.
Numbers of ILIs represent the number of influenza-like illnesses reported by elementary, junior high, and high schools to the Ministry of Health and Welfare, Tokyo, Japan.
Influenza virus isolates represent the numbers of viruses isolated by prefectural and municipal institutes of hygiene as reported to the National Institute of Infectious Diseases, Tokyo, Japan.
Relative morbidity due to H3N2 influenza virus was determined as follows: relative morbidity = ILI × (number of H3N2 isolates/total number of isolates).
DISCUSSION
Influenza viruses contain a segmented genome consisting of eight minus-sense RNA segments which code for 10 known viral proteins that are capable of reassortment during coinfection of a single host with two influenza viruses. Indeed, reassortment between human influenza viruses of different subtypes and between human and swine influenza viruses has been demonstrated repeatedly (3, 37–40, 51, 61), and it has been suggested that swine may serve as an intermediate host in which reassortment between human, swine, and avian influenza viruses may occur to give rise to new pandemic strains (3, 8, 26, 37, 44, 51, 54, 58). Also, a natural H1N2 reassortant containing the HA of recent human H1N1 viruses and the NA of recent human H3N2 viruses has been isolated from humans in China (30). With this evidence, it may be expected that reassortment between cocirculating human influenza viruses of the same subtype occurs. Indeed, recent phylogenetic divergence of the NA gene was suggested to be the result of genetic reassortment between recent human viruses (59). However, through evolutionary analysis of the internal NS gene, it was proposed that reassortment among cocirculating viruses appears not to occur very often and, therefore, it has been suggested that fixation of mutations in the genes coding for the internal proteins is dependent on immune pressure on the HA protein, effectively linking the evolution of the internal genes of influenza viruses to the HA gene (7, 17). Although this may indeed often be the case, parallel analyses of the genes coding for the surface glycoproteins and internal proteins have never been reported. A scarcity of sequence data of the internal genes of human H3N2 viruses has made it very difficult to analyze the phylogenetic patterns of these genes in a parallel manner. Also, available sequence data is of different viruses, isolated in different years and in different parts of the world, making an accurate comparison of the evolutionary patterns of these genes very difficult. The determination in this study of the phylogenetic pathways of all eight genes of 10 recent influenza viruses revealed that the gene segments coding for the internal proteins of these viruses are evolving in a more-independent manner than was previously speculated and were not linked to the evolution of the HA gene.
Although the HA proteins of four viruses isolated in the epidemic season of 1995 were almost identical, differences were observed in the PB1, PA, NP, NA, M2, and NS1 proteins which distinguished these viruses into two pairs. With the exception of the HA gene, the differences in the proteins of 1995 viruses were further supported by evolutionary analysis, indicating that the genes of 1995 isolates consistently diverged into two distinct branch clusters (95i and 95ii) with bootstrap probabilities of 95 to 100. It was, therefore, understood that considerable variability existed among cocirculating viruses in the same epidemic season which was not apparent through analysis of the HA protein alone. Also, it appeared that distinct RNA segment constellations of 95i and 95ii viruses may have been established through reassortment among cocirculating H3N2 viruses.
As summarized in Table 4, evolutionary patterns determined for each of the eight genes of human H3N2 viruses isolated from 1993 to 1997 indicated that reassortment between cocirculating human influenza viruses apparently occurred during this 5-year period. Two isolates of 1995, Aki95 and Toc95, contained PB2 and NP genes which were genetically more similar to those of a 1993 virus than to those of other viruses isolated in the same epidemic season. Japanese viruses of 1997 contained HA, NA, and PB2 genes that appeared to have originated from those of the previous variant of 1996, while the genes coding for the internal PB1, PA, NP, and M proteins apparently derived from earlier viruses of the 1993-1994 season. Most notably, the evolutionary patterns of the internal genes clearly demonstrated with bootstrap probabilities of 99 to 100 that the PB1 and NP genes of 1997 viruses were most similar to those of a 1994 isolate, whereas the M genes were distinct from all Japanese isolates from 1993 to 1996 and were instead most similar to that of a Chinese isolate of 1993. Even though the topology of the phylogenetic tree for the PA gene did not correlate well with the dates of the isolates, the PA genes of the 1997 isolates were shown to have diverged from a virus other than those of 1995 or 1996. Although it was reported that the NS genes of human H3N2 viruses evolves in a rapid and sequential fashion (7, 17), our analyses of the NS genes of recent H3N2 viruses revealed a nonlinear pattern of evolution and amino acid substitutions which seldom survived longer than one season. The nonlinear evolutionary pattern of the NS genes created difficulty in the determination of the origins of the NS genes of recent viruses, since significant probabilities for the internal branches could not be calculated. Nevertheless, the evidence suggested that distinct RNA constellations appeared to be a result of genetic recombination between cocirculating human H3N2 viruses and that genetic exchange may or may not accompany antigenic drift of the HA protein.
TABLE 4.
Proposed derivative strains of RNA segments of recent human H3N2 viruses
| Evolutionary clade (isolates) | RNA segment no. (protein) | Phylogenetically closest virus or branch clustera |
|---|---|---|
| 95i (A/Akita/1/95 and A/Tochigi/44/95) | 1 (PB2) | Kit93 |
| 2 (PB1) | 95ii | |
| 3 (PA) | 95ii | |
| 4 (HA) | 95ii | |
| 5 (NP) | Kit93 | |
| 6 (NA) | Ala95 | |
| 7 (M) | Nii96 | |
| 8 (NS) | ND | |
| 95ii (A/Shiga/20/95 and A/Miyagi/29/95) | 1 (PB2) | Aki94 |
| 2 (PB1) | 95i | |
| 3 (PA) | 95i | |
| 4 (HA) | 95i | |
| 5 (NP) | 96 | |
| 6 (NA) | Ida95 | |
| 7 (M) | Nii96 | |
| 8 (NS) | ND | |
| 96 (A/Fukushima/114/96 and A/Niigata/137/96) | 1 (PB2) | 95ii |
| 2 (PB1) | 95ii | |
| 3 (PA) | 95i | |
| 4 (HA) | Wuh95 | |
| 5 (NP) | 95ii | |
| 6 (NA) | Wuh96 | |
| 7 (M) | 95i | |
| 8 (NS) | ND | |
| 97 (A/Fukushima/140/96 and A/Shiga/25/97) | 1 (PB2) | 96 |
| 2 (PB1) | Aki94 | |
| 3 (PA) | Kit93 | |
| 4 (HA) | Fuk96 | |
| 5 (NP) | Aki94 | |
| 6 (NA) | 96 | |
| 7 (M) | Heb93 | |
| 8 (NS) | ND |
ND, the phylogenetically closest virus could not be accurately determined.
It has been suspected for some time that new epidemic variants of H3N2 influenza virus originated from China (49, 54), since antigenically variable viruses may circulate for some time in China before becoming epidemic. This was apparent when A/Beijing/32/92-like viruses were isolated as early as 1990 in China but did not become epidemic until 1993 (13, 14, 31, 57). A/Beijing/32/92-like viruses, therefore, appear to have circulated for at least 3 years in China before becoming the predominant epidemic strain globally. It is unclear why a particular strain will suddenly emerge after circulating for years in China, although it is generally thought that this is because the HA protein has not undergone sufficient antigenic change to effectively evade established immunity in the human population. In the five epidemic seasons in Japan investigated in this report (1993 to 1997), the relative morbidity due to H3N2 viruses in the 1993, 1995, and 1997 seasons was considerably higher than in the 1994 or 1996 seasons. High levels of morbidity due to H3N2 virus activity in 1995 and 1997 coincided with observed variability in the internal proteins, which was apparently the result of genetic reassortment. The results of this study provide evidence that strongly suggests for the first time that genetic exchange among cocirculating H3N2 influenza virus strains involving gene segments coding for the internal proteins occurs naturally in the human population and that this mechanism of genetic reassortment may be important in virus evolution and pathogenicity. In addition to antigenic drift of the HA protein, emergence of new epidemic H3N2 strains may be influenced by the establishment of a suitable RNA segment constellation through a combination of genetic mutation and reassortment between cocirculating viruses. This study establishes the importance of analyzing the entire genome of human influenza viruses when studying new epidemic strains. Changes in the internal proteins, as well as antigenic variability in the surface glycoproteins, should be considered when analyzing and predicting newly emerging influenza viruses in humans.
ACKNOWLEDGMENTS
The authors thank T. Gojobori for suggestions about the evolutionary calculations used in this report.
This work was supported by grants from the Japanese Ministry of Health and Welfare.
REFERENCES
- 1.Altmueller A, Fitch W M, Scholtissek C. Biological and genetic evolution of the nucleoprotein gene of human influenza A viruses. J Gen Virol. 1989;70:2111–2119. doi: 10.1099/0022-1317-70-8-2111. [DOI] [PubMed] [Google Scholar]
- 2.Bentley D R, Brownlee G G. Sequence of the N2 neuraminidase from influenza virus A/NT/60/68. Nucleic Acids Res. 1982;10:5033–5042. doi: 10.1093/nar/10.16.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bikour M H, Frost E H, Deslandes S, Talbot B, Weber J M, Elazhary Y. Recent H3N2 swine influenza virus with haemagglutinin and nucleoprotein genes similar to 1975 human strains. J Gen Virol. 1995;76:697–703. doi: 10.1099/0022-1317-76-3-697. [DOI] [PubMed] [Google Scholar]
- 4.Bishop D H L, Huddleston J A, Brownlee G G. The complete sequence of RNA segment 2 of influenza A/NT/60/68 and its encoded P1 protein. Nucleic Acids Res. 1982;10:1335–1343. doi: 10.1093/nar/10.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop D H L, Jones K L, Huddleston J A, Brownlee G G. Influenza A virus evolution: complete sequences of influenza A/NT/60/68 RNA segment 3 and its predicted acidic P polypeptide compared with those of influenza A/PR/8/34. Virology. 1982;120:481–489. doi: 10.1016/0042-6822(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 6.Buckler-White A J, Murphy B R. Nucleotide sequence analysis of the nucleoprotein gene of an avian and a human influenza virus strain identifies two classes of nucleoproteins. Virology. 1986;155:345–355. doi: 10.1016/0042-6822(86)90198-4. [DOI] [PubMed] [Google Scholar]
- 7.Buonagurio D A, Nakada S, Parvin J D, Krystal M, Palese P, Fitch W M. Evolution of human influenza A viruses over 50 years: rapid, uniform rate of change in the NS gene. Science. 1986;232:980–982. doi: 10.1126/science.2939560. [DOI] [PubMed] [Google Scholar]
- 8.Castrucci M R, Donatelli I, Sidoli L, Barigazzi G, Kawaoka Y, Webster R G. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology. 1993;193:503–506. doi: 10.1006/viro.1993.1155. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Clements M L, Subbarao E K, Fries L F, Karron R A, London W T, Murphy B R. Use of single-gene reassortant viruses to study the role of avian influenza A virus genes in attenuation of wild-type human influenza A virus for squirrel monkeys and adult human volunteers. J Clin Microbiol. 1992;30:655–662. doi: 10.1128/jcm.30.3.655-662.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colman P, Varghese J, Laver W. Structure of the catalytic and antigenic sites in influenza virus neuraminidase. Nature. 1983;303:41–44. doi: 10.1038/303041a0. [DOI] [PubMed] [Google Scholar]
- 12.Cox N J, Bender C A. The molecular epidemiology of influenza viruses. Semin Virol. 1995;6:359–370. [Google Scholar]
- 13.de Jong J C, Bestebroer T M, Bijlsma K, Verweij C. Preliminary report on influenza viruses of seasons 1992 and 1992/93, second version. Amsterdam, The Netherlands: National Institute of Public Health and Environmental Protection; 1993. [Google Scholar]
- 14.Ellis J S, Chakraverty P, Clewley J P. Genetic and antigenic variation in the haemagglutinin of recently circulating human influenza A (H3N2) viruses in the United Kingdom. Arch Virol. 1995;140:1889–1904. doi: 10.1007/BF01322680. [DOI] [PubMed] [Google Scholar]
- 15.Enami K, Qiao Y, Fukuda R, Enami M. An influenza virus temperature-sensitive mutant defective in the nuclear-cytoplasmic transport of the negative-sense viral RNAs. Virology. 1993;194:822–827. doi: 10.1006/viro.1993.1324. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 17.Fitch W M, Leiter J M, Li X Q, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci USA. 1991;88:4270–4274. doi: 10.1073/pnas.88.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gammelin M, Mandler J, Scholtissek C. Two subtypes of nucleoproteins (NP) of influenza A viruses. Virology. 1989;170:71–80. doi: 10.1016/0042-6822(89)90353-x. [DOI] [PubMed] [Google Scholar]
- 19.Gojobori T, Ishii K, Nei M. Estimation of average number of nucleotide substitutions when the rate of substitution varies with nucleotide. J Mol Evol. 1982;18:414–423. doi: 10.1007/BF01840889. [DOI] [PubMed] [Google Scholar]
- 20.Gojobori T, Moriyama E N, Kimura M. Molecular clock of viral evolution, and the neutral theory. Proc Natl Acad Sci USA. 1990;87:10015–10018. doi: 10.1073/pnas.87.24.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorman O T, Donis R O, Kawaoka Y, Webster R G. Evolution of influenza A virus PB2 genes: implications for evolution of the ribonucleoprotein complex and origin of human influenza A virus. J Virol. 1990;64:4893–4902. doi: 10.1128/jvi.64.10.4893-4902.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herlocher, M. L., M. W. Shaw, and H. F. Maassab. 1991. Unpublished data.
- 23.Ito T, Gorman O T, Kawaoka Y, Bean W J, Webster R G. Evolutionary analysis of the influenza A virus M gene with comparison of the M1 and M2 proteins. J Virol. 1991;65:5491–5498. doi: 10.1128/jvi.65.10.5491-5498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones K L, Huddleston J A, Brownlee G G. The sequence of RNA segment 1 of influenza virus A/NT/60/68 and its comparison with the corresponding segment of strains A/PR/8/34 and A/WSN/33. Nucleic Acids Res. 1983;11:1555–1566. doi: 10.1093/nar/11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaoka Y, Krauss S, Webster R G. Avian to human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge K F, Kawaoka Y, Webster R G. Potential for transmission of avian influenza viruses to pigs. J Gen Virol. 1994;75:2183–2188. doi: 10.1099/0022-1317-75-9-2183. [DOI] [PubMed] [Google Scholar]
- 27.Krystal M, Buonagurio D, Young J F, Palese P. Sequential mutations in the NS genes of influenza virus field strains. J Virol. 1983;45:547–554. doi: 10.1128/jvi.45.2.547-554.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamb R A, Lai C J. Conservation of the influenza virus membrane protein (M1) amino acid sequence and an open reading frame of RNA segment 7 encoding a second protein (M2) in H1N1 and H3N2 strains. Virology. 1981;112:746–751. doi: 10.1016/0042-6822(81)90319-6. [DOI] [PubMed] [Google Scholar]
- 29.Lawson C M, Subbarao E K, Murphy B R. Nucleotide sequence changes in the polymerase basic protein 2 gene of temperature-sensitive mutants of influenza A virus. Virology. 1992;191:506–510. doi: 10.1016/0042-6822(92)90221-a. . (Erratum, 195:302.) [DOI] [PubMed] [Google Scholar]
- 30.Li X S, Zhao C Y, Gao H M, Zhang Y Q, Ishida M, Kanegae Y, Endo A, Nerome R, Omoe K, Nerome K. Origin and evolutionary characteristics of antigenic reassortant influenza A (H1N2) viruses isolated from man in China. J Gen Virol. 1992;73:1329–1337. doi: 10.1099/0022-1317-73-6-1329. [DOI] [PubMed] [Google Scholar]
- 31.Lindstrom S L, Sugita S, Endo A, Ishida M, Huang P, Xi S H, Nerome K. Evolutionary characterization of H3N2 influenza viruses: novel changes in the receptor binding domain. Arch Virol. 1996;141:1349–1355. doi: 10.1007/BF01718836. [DOI] [PubMed] [Google Scholar]
- 32.Martinez C, Del Rio L, Portela A, Domingo E, Ortin J. Evolution of the influenza virus neuraminidase gene during drift of the N2 subtype. Virology. 1983;130:539–545. doi: 10.1016/0042-6822(83)90108-3. [DOI] [PubMed] [Google Scholar]
- 33.Murphy B R, Buckler-White A J, London W T, Snyder M H. Characterization of the M protein and nucleoprotein genes of an avian influenza A virus which are involved in host range restriction in monkeys. Vaccine. 1989;7:557–561. doi: 10.1016/0264-410x(89)90283-1. [DOI] [PubMed] [Google Scholar]
- 34.Nakajima S, Nishikawa F, Nakamura K, Nakajima K. Comparison of the HA genes of type B influenza viruses in herald waves and later epidemic seasons. Epidemiol Infect. 1992;109:559–568. doi: 10.1017/s0950268800050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakajima S, Takeuchi Y, Nakajima K. Location on the evolutionary tree of influenza H3 haemagglutinin genes of Japanese strains isolated during 1985-6 season. Epidemiol Infect. 1988;100:301–310. doi: 10.1017/s0950268800067431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson J, Couceiro S S, Paulson J C, Baum L G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium: the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 37.Nerome K, Kanegae Y, Shortridge K F, Sugita S, Ishida M. Genetic analysis of porcine H3N2 viruses originating in southern China. J Gen Virol. 1995;76:613–624. doi: 10.1099/0022-1317-76-3-613. [DOI] [PubMed] [Google Scholar]
- 38.Nerome K, Kanegae Y, Yoshioka Y, Itamura S, Ishida M, Gojobori T, Oya A. Evolutionary pathways of N2 neuraminidases of swine and human influenza A viruses: origin of the neuraminidase genes of two reassortants (H1N2) isolated from pigs. J Gen Virol. 1991;72:693–698. doi: 10.1099/0022-1317-72-3-693. [DOI] [PubMed] [Google Scholar]
- 39.Nerome K, Sakamoto S, Yano N, Yamamoto T, Kobayashi S, Webster R G, Oya A. Antigenic characteristics and genome composition of a naturally occurring recombinant influenza virus isolated from a pig in Japan. J Gen Virol. 1983;64:2611–2620. doi: 10.1099/0022-1317-64-12-2611. [DOI] [PubMed] [Google Scholar]
- 40.Nerome K, Yoshioka Y, Sakamoto S, Yasuhara H, Oya A. Characteristics of a swine recombinant influenza virus isolated in 1980: recombination between swine and the earliest Hong Kong (H3N2) viruses. Vaccine. 1985;3:267–273. doi: 10.1016/0264-410x(85)90120-3. [DOI] [PubMed] [Google Scholar]
- 41.Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 42.Odagiri T, Tobita K. Mutation in NS2, a nonstructural protein of influenza A virus, extragenically causes aberrant replication and expression of the PA gene and leads to generation of defective interfering particles. Proc Natl Acad Sci USA. 1990;87:5988–5992. doi: 10.1073/pnas.87.15.5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ortin J, Martinez C, Del Rio L, Davila M, Lopez-Galindez C, Villanueva N, Domingo E. Evolution of the nucleotide sequence of influenza virus RNA segment 7 during drift of the H3N2 subtype. Gene. 1983;23:233–239. doi: 10.1016/0378-1119(83)90055-0. [DOI] [PubMed] [Google Scholar]
- 44.Ouchi A, Nerome K, Kanegae Y, Ishida M, Nerome R, Hayashi K, Hashimoto T, Kaji M, Kaji Y, Inaba Y. Large outbreak of swine influenza in southern Japan caused by reassortant (H1N2) influenza viruses: its epizootic background and characterization of the causative viruses. J Gen Virol. 1996;77:1751–1759. doi: 10.1099/0022-1317-77-8-1751. [DOI] [PubMed] [Google Scholar]
- 45.Parkin N T, Chiu P, Coelingh K L. Temperature-sensitive mutants of influenza A virus generated by reverse genetics and clustered charged to alanine mutagenesis. Virus Res. 1996;46:31–44. doi: 10.1016/s0168-1702(96)01373-1. [DOI] [PubMed] [Google Scholar]
- 46.Rogers G N, Paulson J C, Daniels R S, Skehel J J, Wilson I A, Wiley D C. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 47.Saitou M, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 48.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitions. Proc Natl Acad Sci USA. 1977;14:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shortridge K F, Stuart-Harris C H. An influenza epicentre? Lancet. 1982;ii:812–813. doi: 10.1016/s0140-6736(82)92693-9. [DOI] [PubMed] [Google Scholar]
- 50.Shu L L, Bean W J, Webster R G. Analysis of the evolution and variation of the human influenza A virus nucleoprotein gene from 1933 to 1990. J Virol. 1993;67:2723–2729. doi: 10.1128/jvi.67.5.2723-2729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shu L L, Lin Y P, Wright S M, Shortridge K F, Webster R G. Evidence for interspecies transmission and reassortment of influenza A viruses in pigs in southern China. Virology. 1994;202:825–833. doi: 10.1006/viro.1994.1404. [DOI] [PubMed] [Google Scholar]
- 52.Smeenk C A, Brown E G. The influenza virus variant A/FM/1/47-MA possesses single amino acid replacements in the hemagglutinin, controlling virulence, and in the matrix protein, controlling virulence as well as growth. J Virol. 1994;68:530–534. doi: 10.1128/jvi.68.1.530-534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Rompuy L, Jou W M, Huylebroeck D, Fiers W. Complete nucleotide sequence of a human influenza neuraminidase gene of subtype N2 (A/Victoria/3/75) J Mol Biol. 1982;161:1–11. doi: 10.1016/0022-2836(82)90274-1. [DOI] [PubMed] [Google Scholar]
- 54.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiley D C, Wilson I A, Skehel J J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 56.Wilson I A, Skehel J J, Wiley D C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 57.World Health Organization. Weekly epidemiological record. Vol. 68. Geneva, Switzerland: World Health Organization; 1993. pp. 57–64. [Google Scholar]
- 58.Wright S M, Kawaoka Y, Sharp G B, Senne D A, Webster R G. Interspecies transmission and reassortment of influenza A viruses in pigs and turkeys in the United States. Am J Epidemiol. 1992;136:488–497. doi: 10.1093/oxfordjournals.aje.a116522. [DOI] [PubMed] [Google Scholar]
- 59.Xu X, Cox N J, Bender C A, Regnery H L, Shaw M W. Genetic variation in neuraminidase genes of influenza A (H3N2) viruses. Virology. 1996;224:175–183. doi: 10.1006/viro.1996.0519. [DOI] [PubMed] [Google Scholar]
- 60.Xu X, Kilbourne E D, Hall H E, Cox N J. Nonimmunoselected intrastrain genetic variation detected in pairs of high-yielding influenza A (H3N2) vaccine and parental viruses. J Infect Dis. 1994;170:1432–1438. doi: 10.1093/infdis/170.6.1432. [DOI] [PubMed] [Google Scholar]
- 61.Xu X, Rocha E P, Regenery H L, Kendal A P, Cox N J. Genetic and antigenic analyses of influenza A (H1N1) viruses, 1986–1991. Virus Res. 1993;28:37–55. doi: 10.1016/0168-1702(93)90088-5. [DOI] [PubMed] [Google Scholar]
- 62.Yasuda J, Bucher D J, Ishihama A. Growth control of influenza A virus by M1 protein: analysis of transfectant viruses carrying the chimeric M gene. J Virol. 1994;68:8141–8146. doi: 10.1128/jvi.68.12.8141-8146.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yasuda J, Toyoda T, Nakayama M, Ishihama A. Regulatory effects of matrix protein variations on influenza virus growth. Arch Virol. 1993;133:283–294. doi: 10.1007/BF01313769. [DOI] [PubMed] [Google Scholar]
- 64.Zebedee S L, Richardson C D, Lamb R A. Characterization of the influenza virus M2 integral membrane protein and expression at the infected-cell surface from cloned cDNA. J Virol. 1985;56:502–511. doi: 10.1128/jvi.56.2.502-511.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]