Abstract
Retinoblastoma protein (Rb) is a key regulator of cellular proliferation, controlling entry into G1/S in the cell cycle, largely through its action in binding the cellular transcription factor E2F, which activates genes important in DNA synthesis. Small DNA tumor viruses encode gene products which can functionally inactivate Rb, promoting cellular proliferation and viral DNA synthesis. In this study, the Epstein-Barr virus (EBV) immediate-early lytic gene product, BRLF1 (R), is shown to bind Rb in vivo, shortly after induction of the viral lytic cycle in EBV-infected Akata cells. Furthermore, the temporal kinetics of R-Rb interaction correlate with displacement of E2F1 from Rb. Mapping of the domains required for the interaction of R and Rb proteins reveals that R binds specifically to the N terminus of Rb, outside the Rb pocket, and that the first 200 amino acids of R are required for this interaction. The interaction of R and Rb may initiate cell cycle progression and facilitate viral DNA synthesis during lytic replication.
Epstein-Barr virus (EBV), a lymphotrophic human gammaherpesvirus, is the causative agent of infectious mononucleosis and is associated with several lymphoid malignancies, including Burkitt’s lymphoma (BL), Hodgkin’s disease, and lymphoproliferative diseases in immunocompromised persons (reviewed in reference 38). EBV also appears to be the primary agent in the epithelial cancer nasopharyngeal carcinoma (54).
Infection of primary B cells is predominantly latent, with only a subset of viral genes being expressed. Following infection, cells rapidly enter a proliferative phase and eventually become immortalized. Six latency-associated genes are required for the immortalization process, the Epstein-Barr nuclear antigens (EBNA1, -2, -3A, -3C, and -LP) and latent membrane protein 1. The viral genome in latently infected cells is maintained as a circular episome which is replicated by the host polymerase (41).
The mechanism of cell immortalization driven by EBV is not fully understood but appears to differ from that of the small DNA tumor viruses. Adenovirus (55, 78, 80), papillomavirus (19, 45, 77), and simian virus 40 (17, 45, 55) all encode viral oncoproteins which functionally inactivate the tumor suppressor gene products, p53 and Rb (43, 46, 74). Inactivation of Rb indirectly stimulates cellular proliferation (79), while coordinate inactivation of p53 prevents induction of the apoptotic pathway (59). Coordinate inactivation of tumor suppressor genes by small DNA tumor viruses is thought to promote the induction of S phase in cells, which is necessary for viral DNA replication. The induction of proliferative signals and suppression of apoptotic signals can lead to unrestricted cellular proliferation and, in some cases, cell transformation. In contrast, transformation by EBV is characterized by latent infection; lytic viral DNA replication is not thought to occur in cells destined for immortalization, and no virus is produced in the immortalized cells.
The EBV latent gene product, EBNA3C (also called EBNA6), has been reported to bind Rb in vitro and enhance transformation of rat embryo fibroblasts by ras. EBNA3C also transactivated the B-myb promoter in an E2F-dependent manner, which suggests that EBNA3C can inactivate Rb, releasing free E2F (49). A second EBV gene product, EBNALP (EBNA5), is reported to bind p53 and Rb in vitro and to colocalize with Rb in the cell (64, 65). The functional significance of this interaction is unknown (35, 64). Both EBNA3C and EBNALP lack the LXCXE motif found in other viral proteins which bind the Rb pocket. It is possible that either of these latency genes can contribute to cellular transformation if it is able to bind Rb in vivo; however, viral DNA synthesis, which occurs in productively infected cells, would not be affected.
In contrast to latent infection in B cells, EBV infection of epithelial cells is usually productive and results in cell lysis. Immunosuppression, however, may trigger reactivation of the virus in latently infected B cells, which leads to productive infection (48). This switch to a replicative pattern of viral gene expression can be mimicked by treating latently infected cells with phorbol ester (16, 84) or immunoglobulin G (IgG) antibody (66). Upon reactivation, the two key EBV immediate-early (IE) lytic genes, BZLF1 and BRLF1, are expressed (5, 68). These genes encode transactivators which activate viral and cellular promoters and lead to an ordered cascade of viral gene expression (6, 20–22, 29, 31, 37, 57). Expression of the BZLF1 gene product, Z (also called Zta or EB1), alone is sufficient to activate the EBV lytic cascade (12–14, 25, 69). Recent studies implicate Z in regulation of cellular proliferation. Z can bind p53 and inactivate p53-mediated transactivation functions in transient assays (83), and expression of Z in some epithelial tumor cell lines causes G0/G1 arrest in a p53-dependent manner. Expression of Z in these cells also results in upregulation of the cyclin-dependent kinase inhibitors p21 and p27, which causes accumulation of the hypophosphorylated form of Rb (8).
Like Z, the BRLF1 gene product, R (Rta), is a transactivator and can act alone or in tandem with Z to activate viral and cellular promoters (11, 12, 15, 24, 28, 31, 37, 44, 53). Recently, it has been shown that BRLF1 alone can initiate activation of lytic gene expression in latently infected epithelial cell lines (81). The 605-amino-acid (aa) R protein contains two transactivation domains and an N-terminal DNA-binding and dimerization domain (29, 44). R can activate promoters both by binding a specific DNA sequence, an R-responsive element (29, 44), and indirectly, possibly by activating or recruiting cellular transcription factors (42, 81, 82). Promoters activated in the latter manner include BRLF1 itself (57, 82), the human immunodeficiency virus long terminal repeat (52), and c-myc (26). Recent data show that the promoter for the EBV DNA polymerase, which is responsible for lytic viral DNA replication, is also activated by R indirectly through the transcription factors USF and E2F (42).
The Rb protein suppresses cellular proliferation by associating with and suppressing the activity of cellular transcription factors, in particular the E2F family of proteins, which activate genes important in cellular DNA replication (2, 47). Active, hypophosphorylated Rb can sequester E2F, preventing entry of cells into S phase. Phosphorylation of Rb by cyclin-dependent kinase complexes inactivates Rb and releases E2F, which subsequently activates promoters for genes involved in DNA synthesis (1, 40). Viral oncoproteins encoded by the small DNA tumor viruses can bind to underphosphorylated Rb and displace E2F, driving cellular proliferation (reviewed in reference 75).
We hypothesized that since R can indirectly activate some promoters by recruitment or activation of other transcription factors, including E2F, R might interact with the Rb protein and displace associated transcription factors. In this study, we show that R specifically binds Rb protein at early times following viral reactivation. The interaction is limited to early times following induction and correlates temporally with release of E2F1 from Rb.
MATERIALS AND METHODS
Plasmids.
Plasmid pGEX2T, containing the glutathione S-transferase (GST) open reading frame, was purchased from Pharmacia. GSTEBNA2 was prepared by PCR amplification of the EBNA2 open reading frame from DNA prepared from EBV-infected B95-8 cells, using the amplimers 5′ AACGCTGAGATCTATGCCTACATTCTATCTTGCG 3′ and 5′ AATCAAAGATCTTGATTACTGGATGGAGGGGCGAG 3′. The PCR product was cloned into the BamHI site of pGEX2T. Plasmid GSTE2F was a gift from Joe Nevins. GST5′Rb (previously called HURB; aa 1 to 380) and the GST mutant plasmids Δ39-89, Δ89-140, Δ126-166, Δ249-309, Δ309-343, and Δ343-383 were generous gifts from Jonathan Horowitz and have been described previously (60, 61). GST3′Rb (aa 377 to 928) has been described elsewhere (27). Plasmid pGEXR (53) and the R mutant plasmids used for in vitro translation, Rt201, Rt356, Rt356i81, Rt515, RΔ2-22, and RΔ81-184 (44), were generous gifts from Evelyne Manet and Alain Sergeant and have been previously described. Plasmid RcDNA, used for in vitro translation, was a gift from Shannon Kenney. pRBd5′ was a gift from R. Weinberg; pBSKglobE2F, a gift from Ed Harlow, contains E2F1 cDNA (nucleotides 130 to 1456) in pBSKglob.
Cell culture.
DG75 is an EBV-negative BL cell line (4); Akata is an EBV-infected type 1 lymphoblastoid line (66, 67). Akata and DG75 cells were maintained in RPMI 1640 with 10% fetal bovine serum (GIBCO-BRL, Gaithersburg, Md.) and antibiotics at 37°C in 5% CO2. To induce the latently infected Akata cell line, cells were treated with 0.1 mg of anti-human IgG (Sigma, St. Louis, Mo.) per ml. Samples were harvested at various times following induction.
In vitro translation.
Plasmids were transcribed and translated in vitro in the presence of [35S]methionine, using the TnT system (Promega, Madison, Wis.) according to the manufacturer’s specifications and with the appropriate RNA polymerase (SP6 or T7; Promega).
Bacterial expression of GST fusion proteins.
GST plasmids were transformed into Escherichia coli BL21, and expression was induced by 0.1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG). The samples were incubated at 37°C for 2 h with shaking, then harvested, and centrifuged for 30 min at 5,000 rpm. The bacterial pellet was resuspended in phosphate-buffered saline solution (PBS) to 1/10 the original culture volume. For binding to beads, the bacteria were lysed by freezing and thawing, followed by sonication on ice. After centrifugation, 20 to 800 μl of supernatant fluid was incubated with 25 μl of glutathione-Sepharose beads (Pharmacia, Uppsala, Sweden) in a total volume of 1 ml in PBS for 30 min at 25°C. The beads were washed in buffer (20 mM HEPES, 150 mM KCl, 1% Nonidet P-40, 1 mM dithiothreitol), then boiled in Laemmli buffer, and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The amount of GST fusion protein was determined by Coomassie blue staining of the gel.
GST protein precipitation.
Fusion protein was loaded onto beads as described above and then incubated with 35S-labeled reticulocyte lysate in 500 μl of buffer (above) for 90 min at 4°C. The beads were washed with buffer, then resuspended in Laemmli buffer, and boiled. The samples were resolved by SDS-PAGE. The gel was fixed in 25% methanol–7% acetic acid for 30 min, then incubated in 1 M sodium salicylate for an additional 30 min, and subjected to autoradiography.
Immunoprecipitation.
Cells were harvested, washed in PBS, resuspended in ELB+ buffer (0.25 M NaCl, 0.1% Nonidet P-40, 0.05 M HEPES [pH 7.0], 0.001 M phenylmethylsulfonyl fluoride, 0.005 M EDTA, 0.5 mM dithiothreitol), and lysed by repeated freezing and thawing. Total protein content was determined with a Bio-Rad (Richmond, Calif.) protein assay kit. For immunoprecipitations, 200 to 800 μg of cellular lysate was incubated with antibody in a volume of 1 ml (ELB+ buffer) on ice for 1 h. The samples were incubated for an additional 3 h at 4°C with shaking. Protein A-Sepharose (50 μl) (Pharmacia) was added, and the incubation continued for an additional 60 min. The beads were pelleted and washed four times with ELB+ buffer, then resuspended in Laemmli buffer, and boiled.
Immunoblot analysis.
Samples in Laemmli buffer were resolved by SDS-PAGE (6 to 12% polyacrylamide) and then transferred to an Immobilon membrane (Millipore, Bedford, Mass.). The membrane was blocked with 5% dry milk powder in TBST (0.9% NaCl, 0.02 M Tris-HCl [pH 7.5], 0.05% Tween 20). The membrane was incubated in primary antibody in blocking solution for 60 min, washed three times with TBST, and then incubated in secondary antibody (horseradish peroxidase-conjugated anti-mouse or anti-rabbit Ig; Amersham, Buckinghamshire, England) for 60 min. The membrane was washed three times with TBST and then developed in enhanced chemiluminescence reagents according to the protocol of the manufacturer (Amersham).
Antibodies.
The Rb-specific antibody, C-15X, and the E2F1 antibodies, KH95 and sc193, were from Santa Cruz Biotechnology (Santa Cruz, Calif.). The Rb antibody used for immunoprecipitation, AB-1, was from Oncogene Science (Cambridge, Mass.). The R monoclonal antibody, 8C12, was a generous gift from Alain Sergeant. The R2 antibody is an affinity-purified polyclonal serum raised against the R peptide corresponding to R aa 1 to 20 (CMRPKKDGLEDFLRLTPEIKK). The peptide was synthesized in the protein chemistry facility at the University of North Carolina, Chapel Hill. Immunization of rabbits and antibody purification were performed by Immunodynamics (La Jolla, Calif.).
Phosphatase assay.
Twenty micrograms of protein was incubated in buffer with 2,000 U of lambda phosphatase for 3 h at 30°C in a total volume of 50 μl according to the protocol of the manufacturer (New England Biolabs). Control samples containing no phosphatase were incubated under the same conditions. The samples were separated by SDS-PAGE as described above.
RESULTS
R binds Rb in vitro.
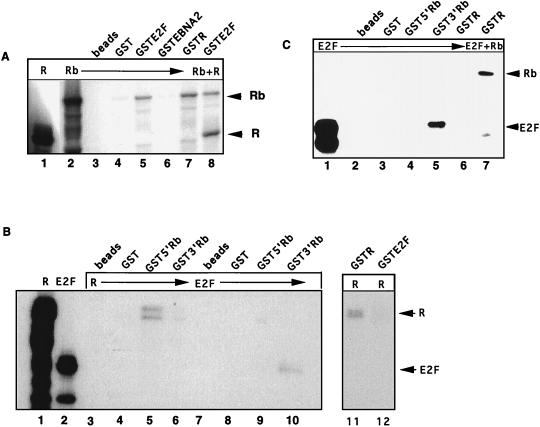
Since the EBV IE protein Z can bind and functionally inactivate p53 (81), we investigated whether the second EBV IE protein, R, could interact with the Rb protein. Radiolabeled R, Rb, and E2F proteins were expressed in reticulocyte lysates and then incubated with equivalent amounts of bacterially expressed GST fusion proteins, as judged by Coomassie staining. As expected, the positive control, GSTE2F (Fig. 1A, lane 5), specifically precipitated radiolabeled Rb, as did the viral fusion protein, GSTR (lane 7). Rb did not interact with beads alone, GST-loaded beads (lanes 3 and 4), or GSTEBNA2, an EBV latency protein used as a negative control (lane 6). Surprisingly, GSTE2F could simultaneously precipitate cotranslated R and Rb proteins (lane 8). This was unexpected, as most proteins such as E2F that interact with Rb specifically bind its pocket region, and the presence of another Rb-binding protein, such as a viral oncoprotein, usually results in displacement of E2F (75). These results indicated that R may bind Rb outside the E2F-binding pocket region or that R may interact with E2F directly.
FIG. 1.
EBV R protein binds Rb in vitro. Bacterially expressed GST fusion proteins linked to glutathione-Sepharose beads (Pharmacia) were incubated with 35S-labeled reticulocyte lysate programmed with Rb, R, or E2F DNA for 1 h at 4°C. (A) 35S-labeled Rb (lane 2) was incubated with beads alone (lane 3), GST-loaded beads (lane 4), or GSTEBNA2 as a negative controls (lane 6). GSTE2F (lane 5) and GSTR (lane 7) bound in vitro-labeled Rb. GSTE2F was incubated with cotranslated R and Rb protein (lane 8). (B) Radiolabeled R or E2F protein was incubated with GST proteins linked to Glutathione beads. Lanes 3 and 7, beads alone; lanes 4 and 8, GST; lanes 5 and 9, GST5′Rb (aa 1 to 380); lanes 6 and 10, GST3′Rb (aa 377 to 928); lane 11, GSTR; lane 12, GSTE2F. Direct loading of in vitro-labeled R and E2F proteins is shown in lanes 1 and 2, respectively. (C) GST fusion proteins were incubated with 35S-labeled E2F-1 protein. Lane 1, directly loaded E2F protein; lanes 2 to 4, E2F incubated with beads alone, GST-loaded beads, and GST5′Rb, respectively. GST3′Rb precipitated E2F (lane 4), but GSTR did not (lane 5). GSTR incubated with equal amounts of Rb and E2F proteins is shown in lane 7.
To address this question and to test whether the reciprocal interaction of R and Rb could be demonstrated, GST5′Rb (aa 1 to 380) and GST3′Rb (aa 377 to 928) were tested for interaction with radiolabeled R or E2F protein. Radiolabeled R protein was able to form dimers with GSTR (Fig. 1B, lane 11) and was precipitated by 5′Rb, but not 3′Rb, which contains the pocket region (lanes 5 and 6). R did not interact with beads or GST alone (lanes 3 and 4) and did not directly bind E2F (lane 12). As R and E2F bind different regions of Rb, this result is in agreement with the observation that Rb can interact simultaneously with R and E2F. More importantly, lanes 9 and 10 show that the GST-Rb constructs performed as expected: E2F was precipitated only by 3′Rb, which contains the pocket domain, and not by 5′Rb (lanes 9 and 10). An R doublet precipitated by Rb (lane 5) may be due to posttranslational modification of R or to a truncated form resulting from internal initiation or to partial protein degradation.
To confirm that R could not interact directly with E2F, GSTR was incubated with radiolabeled E2F protein. E2F protein was precipitated by the positive control, GST3′Rb (Fig. 1C, lane 5), but not by GSTR (lane 6). E2F was not precipitated by beads alone, GST, or GST5′Rb (lanes 2 to 4). GSTR precipitated Rb protein alone from a mixture of radiolabeled E2F and Rb (lane 7), indicating that the GSTR protein was indeed functional. An additional band which binds to GSTR is believed to be a degradation product of Rb, as it migrates more rapidly than E2F. The results in Fig. 1C, lane 7, differed from those in Fig. 1A, lane 8, where a tripartite complex of R, Rb, and E2F was observed. These differences may be due to the fact that the reticulocyte lysates were cotranslated in the earlier experiment (Fig. 1A) but not in Fig. 1C.
Taken together, these data indicate that R can bind Rb in vitro and that binding of R occurs outside the Rb pocket. The interaction is probably not mediated by E2F, as R did not interact directly with E2F, but there is potential for the formation of a complex containing R, Rb, and E2F proteins.
The EBV R protein coprecipitates Rb from induced Akata cells.
To determine whether the proteins interacted in vivo, coimmunoprecipitations were performed with BL cells. Akata is a latently EBV-infected BL line which expresses IgG; viral replication can be reactivated by cross-linking of the surface Ig with antibody. Induction of viral lytic cycle genes is rapid and synchronous and occurs in 50 to 75% of the cells (66, 68).
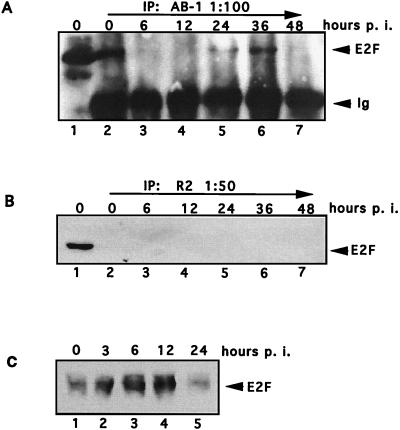
Samples were collected from Akata cells following IgG induction, and protein lysates were prepared. Protein (600 μg) was immunoprecipitated with R2 antibody, which recognizes R protein; the complexes were separated by SDS-PAGE and then immunoblotted with antibody C-15X, a polyclonal serum which recognizes all forms of the Rb protein (Santa Cruz). R specifically coprecipitated Rb at 6 and 12 h postinduction (Fig. 2A, lanes 5 and 6). R2 antibody did not precipitate Rb protein from uninduced cells, which do not express R (lane 4), nor did R2 precipitate Rb from radiolabeled reticulocyte lysates (lane 2). The interaction of R and Rb was limited to early times postinduction; it was not detected by 24 h postinduction (Fig. 2A, lanes 7 to 9).
FIG. 2.
EBV R protein precipitates Rb in induced Akata cells. EBV-infected Akata cells were treated with 0.1 mg of anti-IgG (Sigma) per ml, and samples were harvested at various times postinduction (hours p.i.). (A) Akata protein lysate samples were immunoprecipitated with R2 antiserum (1:100) and then probed for Rb with antibody C-15X (1:5,000; Santa Cruz). Shown are Western blots of in vitro-translated Rb protein (lane 1) and uninduced Akata protein (lane 3) and immunoprecipitation (IP) of in vitro-translated Rb (lane 2) and Akata samples induced for 0, 6, 12, 24, 36, or 48 h (lanes 4 to 9). (B and C) Western blots of induced and uninduced Akata protein samples probed with antibody 8C12 (1:10) (B) or Rb-specific antibody C-15X (C).
Western blot analyses of lysates from the same time course were performed to detect R and Rb protein directly. As expected, R protein was not detected in uninduced Akata cells but was readily detectable 6 h postinduction (Fig. 2B). Rb protein levels remained constant in both induced and uninduced Akata cells (Fig. 2C). Clearly, the interaction of R and Rb at early times is specific, and the lack of complex formation at later times is not limited by availability of the two proteins. It is possible that posttranslational modification of the protein(s) or availability of a tethering protein contributes to regulating R-Rb interaction.
Rb-E2F1 complex is disrupted by induction of the lytic cycle in EBV-infected Akata cells.
To determine whether E2F1, which binds hypophosphorylated Rb, was associated with Rb in Akata cells following induction, Akata protein lysates (the same samples as used in Fig. 2) were immunoprecipitated with Rb-specific antibody AB-1, and the complexes were probed for E2F1 protein. E2F1 was bound to Rb in uninduced Akata cells (Fig. 3A, lane 2) but was released from Rb at 6 and 12 h postinduction (lanes 3 and 4). The complexes reassociated by 24 h (lane 5). Therefore, the kinetics of E2F release from Rb paralleled those of R and Rb association. Rb-E2F complex dissociates again at 48 h (lane 7) independent of R binding, and this may reflect cell cycle-associated events such as Rb phosphorylation.
FIG. 3.
Association of Rb and E2F in EBV-infected Akata cells. Akata cell protein samples were collected at various times after anti-IgG induction (hours p. i.). (A) A 200-μg aliquot of lysate was immunoprecipitated (IP) with antibody to Rb protein (AB-1; Oncogene Science). The complexes were resolved by SDS-PAGE and probed for E2F1 protein (KH95; Santa Cruz). Lane 1, total E2F1 protein in 50 μg of Akata lysate; lanes 2 to 7, immunoprecipitations with Rb-specific antibody. (B) Akata protein samples were immunoprecipitated with R2 antibody and then immunoblotted with antibody for E2F1 protein (lanes 2 to 7). Direct loading of uninduced Akata lysate probed for E2F1 protein is shown in lane 1. (C) Akata lysate samples (100 μg) were immunoblotted with E2F1 antibody sc193 (1:100; Santa Cruz).
The in vitro data suggested that a complex of R, Rb, and E2F proteins might form in some circumstances. To determine whether R might directly bind E2F in vivo, or if R-Rb association might include E2F, Akata protein from induced cells was immunoprecipitated with the R-specific antibody R2, and the complexes were probed with antibody to E2F1. Figure 3B, lane 1, shows E2F1 protein directly in uninduced Akata cell lysate. E2F protein was not coprecipitated with R in any of the induced Akata lysate samples (lanes 2 to 7). These data demonstrate that R does not bind E2F1 directly and confirm that R-Rb association is not dependent on E2F.
Next, total E2F protein in Akata lysates was determined by Western blot analysis. Akata cells were harvested at 0, 3, 6, 12, and 24 h following IgG cross-linking, and whole-cell lysates were prepared. Immunoblot analysis was performed with antibody sc193 (1:100) (Fig. 3C). E2F levels appear to increase at early times following induction of the lytic cycle (Fig. 3C). Therefore, the disappearance of the E2F-Rb complex at 6 and 12 h postinduction, seen in Fig. 3A, is not due to a decrease in E2F protein levels.
Phosphorylation of R and Rb following induction of the lytic cycle.
Since Rb is a phosphoprotein and R may be phosphorylated, we investigated the phosphorylation status of R and Rb following induction of the lytic cycle in Akata cells. Akata cells were induced by IgG cross-linking as described above, samples were harvested following post induction at the times indicated, and protein lysates were prepared. To distinguish differently migrating forms of the proteins, we separated protein lysate samples by SDS-PAGE at conditions selected to enhance separation of the desired protein and then performed immunoblot analysis. For detection of Rb, 25 μg of lysate was separated on a 7% polyacrylamide gel. The protein was transferred to an Immobilon membrane and probed with antibody C-15X. R was detected with antibody 8C12 (1:100) following separation of 200 μg of protein with a 10% polyacrylamide gel. Multiple forms of both proteins were detected. At least two forms of Rb were detected in uninduced Akata cells (Fig. 4A, lane 1). At 3 h postinduction, virtually all Rb protein was underphosphorylated (lane 2). A more slowly migrating species of Rb started to accumulate at 6 h postinduction (lane 3) and became the predominant form of Rb at later times (lanes 4 to 8). Presumably Rb-binding proteins would preferentially interact with Rb at early times after induction.
FIG. 4.
Phosphorylation of R and Rb following lytic induction in Akata cells. Total protein lysate samples from induced Akata cells at various times postinduction (hours p.i.) were separated by SDS-PAGE and then probed for Rb (A) or R (B) protein. (C) A 20-μg aliquot of each lysate was incubated with lambda phosphatase (New England Biolabs) for the times indicated at 30°C prior to immunoblot analysis.
A rapidly migrating form of R was detected at 3 and 6 h postinduction (Fig. 4B, lanes 2 and 3). A second, more slowly migrating species was seen at later times following induction (lanes 4 to 8). To confirm that the differentially migrating forms of R and Rb were due to phosphorylation, we performed a phosphatase assay. Twenty micrograms of total lysate was incubated with 2,000 U of lambda phosphatase as described, followed by electrophoresis and immunoblot analysis. When samples were incubated in the presence of phosphatase, both R and Rb proteins were reduced to a single rapidly migrating species (Fig. 4C), indicating that the more slowly migrating species of the proteins were phosphorylated derivatives of each protein.
R aa 1 to 201 are required for Rb interaction.
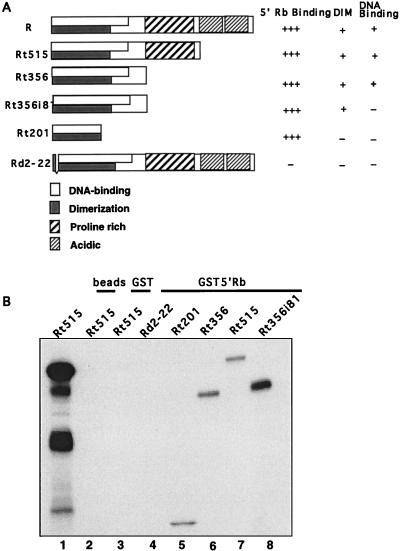
Next we determined which region of the R protein specifically interacted with Rb. For mapping studies, a series of mutant R constructs which contain C-terminal or internal deletions (44) were translated and radiolabeled with [35S]methionine in reticulocyte lysates, using T7 RNA polymerase (R and RΔ2-22) or SP6 RNA polymerase (Rt201, Rt356, Rt356i81, Rt515, and RΔ81-184) (Fig. 5A). The proteins were analyzed by gel electrophoresis followed by autoradiography to confirm that each of the translated proteins was of the expected molecular weight (data not shown). Equivalent amounts of the radiolabeled proteins as judged by autoradiography were precipitated with GST5′Rb fusion protein. Figure 5B shows that all of the C-terminally truncated R mutants interacted in vitro with 5′Rb (lanes 5 to 8), but the internal deletion mutant, RΔ2-22 (lane 4), did not bind 5′Rb. Another internal deletion mutant, RΔ81-184, also failed to coprecipitate with Rb (data not shown).
FIG. 5.
Structures and functions of R mutants. (A) Structures of R mutants used in mapping studies (generous gifts from Evelyne Manet). DNA binding and dimerization were described previously (44). (B) R plasmids were transcribed and translated in vitro in the presence of [35S]methionine, using the TnT system (Promega). Radiolabeled R protein was incubated with glutathione beads (lane 2), GST-beads (lane 3), or GST5′Rb (aa 1 to 380; lanes 4 to 8) for 90 min at 4°C. The samples were washed and separated by electrophoresis as described in the text.
These R mutants were previously used to map functional domains in the R protein. Manet et al. (44) localized the dimerization domain of R to aa 1 to 232, within the DNA-binding domain (aa 1 to 280). It was also shown that mutant Rt201, as well as the two deletion mutants, RtΔ2-22 and RtΔ81-184, were all deficient in both dimerization and DNA-binding capacity (44). Therefore, dimerization of R is not necessary for the interaction with Rb, as mutants Rt201 and Rt356i81 can precipitate the Rb protein.
Mapping the regions of the Rb protein that interact with R.
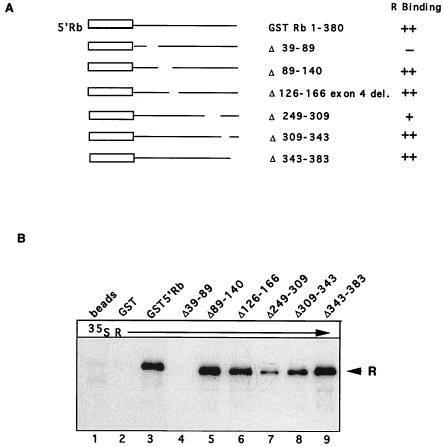
Results from Fig. 1 show that R specifically binds to the 5′ end of Rb, outside the Rb-binding pocket, perhaps because R does not contain an LXCXE motif previously found in the other proteins that bind the Rb pocket (46). However, possible functions for the N-terminal region of Rb have recently been uncovered (reviewed in reference 76). Therefore, the region of Rb necessary for interaction with R was mapped more closely. A series of GST5′Rb constructs which contained internal deletions (a generous gift from J. Horowitz) are shown in Fig. 6A (61). Equivalent amounts of bacterially expressed mutant proteins, as judged by Coomassie blue staining (data not shown), were incubated with radiolabeled, in vitro-translated R protein. As shown in Fig. 6B, deletion of two separate regions in Rb interfered with binding of R. Deletion of aa 39 to 89 (lane 4) completely disrupted interaction with R, and deletion of aa 249 to 309 significantly reduced the amount of R binding (lane 7). When the same experiment was repeated with radiolabeled Rt201 instead of full-length R, the results were similar (data not shown). These data indicate that aa 39 to 89 in Rb are critical for interaction with R. Another deletion, from aa 249 to 309, decreased R binding, perhaps because of conformational changes caused by the deletion.
FIG. 6.
Mapping the domains in Rb required for interaction with R protein. (A) Structures of GST-Rb constructs used for this experiment, which were generous gifts from Jonathan Horowitz and have been described previously (61). (B) Lysates from bacterially expressed GST fusion proteins were prepared and then bound to glutathione beads as described in the text. In vitro-translated 35S-labeled R protein was incubated with beads, GST, or a GSTRb construct as indicated. After the binding reaction, the complexes were separated by SDS-PAGE and visualized by autoradiography as described in the text.
DISCUSSION
The Rb and p53 gene products were identified by their function as tumor suppressor proteins, but their effects are more encompassing; it is now known that these proteins are critical for normal cellular proliferation. It is not surprising, then, that these proteins may be targeted not only during cellular transformation but also during viral infection to circumvent normal controls on cellular proliferation and establish an environment that favors viral DNA replication. This may be a common theme not only among the small DNA tumor viruses but also among other DNA viruses. Human cytomegalovirus encodes a gene product, IE2, which binds Rb and inactivates its E2F-mediated growth-suppressive function (27, 58), and infection of cells with this virus leads to a rapid burst in DNA, RNA, and protein synthesis (23, 62, 63, 70). Infection of cells with herpes simplex virus type 1 leads to an increase in S-phase E2F complexes and free E2F (30).
Previous data from our laboratory suggested that Rb may be a target for the R protein. R activation of the promoter for the EBV pol gene has been mapped to elements which bind the transcription factors USF and E2F; mutation at either site reduced R-mediated activation of pol (42). Additional promoters are activated by R in an indirect manner (26, 52, 57, 82). Taken together, these results indicate that R may activate some promoters by activation of cellular transcription factors. One mechanism by which indirect activation of promoters could occur is through interaction of R and Rb proteins and release of transcription factors bound to Rb.
The IE gene product, R, specifically binds Rb both in vitro and in vivo. The interaction of R and Rb occurs shortly after induction of the viral lytic cycle in Akata cells treated with IgG antibody and is limited to early times following induction. Beyond 24 h postinduction no interaction could be detected even though levels of R and Rb remained constant for 48 h, which indicates that the interaction may be regulated posttranslationally, possibly by phosphorylation. Rb activity is regulated by phosphorylation (71), and R may also be a target for phosphorylation by cellular kinases (51). We show here that R is indeed a phosphoprotein and that the status of both R and Rb phosphorylation varies following induction of the lytic cycle. Multiple forms of Rb are detected in uninduced cells, but hypophosphorylated forms of Rb accumulate 3 h postinduction, and Rb becomes progressively more phosphorylated at later times after induction (Fig. 4A and C). Therefore, it appears that induction may trigger a rapid, transient dephosphorylation of Rb. The mechanism underlying Rb dephosphorylation and the subsequent phosphorylation seen at later times remain to be investigated. Similarly, R appears to be underphosphorylated or not phosphorylated when it is first detected after induction; at later times, a mixture of phosphorylated and unphosphorylated forms is detected (Fig. 4B and C). Therefore, the strongest interaction of R and Rb proteins occurs at early times following induction, when both proteins are predominantly underphosphorylated or not phosphorylated. The disruption of R and Rb complexes could be due to increasing phosphorylation of R or Rb or both. It has not yet been determined precisely which phosphorylated form of Rb can bind R, but it is likely that R binds underphosphorylated Rb, as all proteins to date which bind Rb preferentially bind the underphosphorylated form of the protein (76). These data are exciting, as they indicate that EBV specifically targets Rb early in the lytic cycle.
E2F1 is bound to Rb in latently infected Akata cells. This finding suggests that at least some of the Rb protein is hypophosphorylated in these cells. This result contrasts with data obtained for lymphoblastoid cell lines, where Rb is known to be hyperphosphorylated. The result probably reflects differences between type 1 (Akata) and type 3 (lymphoblastoid cell lines) latent viral gene expression (3, 7).
The release of E2F from Rb during viral reactivation correlates temporally with the kinetics of R-Rb interaction. It would be tempting to speculate that R binding to Rb results in the displacement of E2F, which could then activate both viral and cellular E2F-responsive promoters, linking R to proliferation and/or cell cycle progression. However, the data suggest that interaction of R and Rb may have additional functional consequences.
The in vitro data indicate that R, Rb, and E2F may form a tripartite complex under some circumstances (Fig. 1A and C), whereas in vivo, E2F displacement from Rb clearly correlates with R binding to Rb. A complex of all three proteins formed only when R and Rb were cotranslated before addition of E2F (Fig. 1A). Therefore, it is likely that the conformation of Rb protein is changed with each binding partner, and this change may determine whether additional proteins can then bind Rb. The cellular context, i.e., other proteins binding to Rb, could determine whether binding of R might displace E2F. Other factors may play a role in regulating Rb protein interactions; since R is a phosphoprotein, it is possible that the phosphorylation status of R determines whether E2F is released. R has not been shown to have any kinase activity and thus is unlikely to play a direct role in Rb phosphorylation, but it is possible that R recruits other proteins which play a role in E2F displacement.
In addition, R binds specifically to the N-terminal region of Rb, outside the E2F-binding pocket. The deletion of aa 39 to 89, which completely abolished R binding, has been linked to several functions of Rb. This region was shown to contribute to growth suppression activity; it was essential for generation of flat cells in an SAOS2 cell proliferation assay (50). Data suggest that this region may bind the Sp1 inhibitor (9, 39, 72, 73). Deletion of this region completely abolished the ability of Rb to activate promoters for the human insulin receptor (56) and insulin-like growth factor II (39) genes through Sp1. This same region lies within a putative structural domain important for association of Rb with the p84 nuclear protein and subnuclear localization of Rb (18). Rb mutant protein deleted in either region, aa 39 to 89 or aa 249 to 309, was impaired in both biological activity and the ability to become hyperphosphorylated (50). It is possible that R binding specifically affects some of these functions. Both of these deletions map outside sequences that are conserved among the Rb family members. Therefore, it is unlikely that R will bind p107 or p130, but this possibility needs to be tested directly. Recently, it has been shown that the amino terminus of Rb binds and negatively regulates MCM7, a component of DNA-licensing factor, a complex required for initiation of replication (32). It is possible that this factor is needed for viral DNA replication and is displaced from Rb by binding of R.
Two regions in Rb, aa 39 to 89 and aa 249 to 309, may interact with R, but it is unclear at this point whether either region is directly bound by R. R may interact with both regions of Rb, possibly by a primary interaction at one site followed by contacts at a second site. This type of interaction occurs in binding of other proteins to Rb (10, 33, 34, 36). The deletion of aa 39 to 89 is associated with loss of many functions of the protein and may result in a conformational change in Rb folding which precludes R binding. Likewise, the decrease in R binding noted with the deletion of aa 249 to 309 may also be due to alteration in Rb protein folding which masks the R interaction domain.
Rb binds within the first 200 aa of the R protein, in the DNA-binding and dimerization domains. However, neither dimerization nor DNA binding of R is necessary for its interaction with Rb, as the R mutant Rt201, which cannot form dimers or bind DNA, also binds Rb (44). This finding does not, however, exclude the possibility that R dimers can bind Rb. It may be difficult to map this region further by using deletion mutants of R, as internal deletions within the dimerization region would severely disrupt protein confirmation. Neither of the two deletion mutants that we tested, RΔ2-22 and RΔ81-184, bound Rb, and further mapping studies would probably require point mutations in this region of R.
It is possible that R is a multifunctional protein that plays a larger role in viral DNA replication than was previously suspected. R, alone or in tandem with Z, can activate viral promoters to regulate viral gene expression during productive infection. R may also help regulate the cellular environment to promote viral replication. R can activate c-myc, a cellular gene important in proliferation (26). R also activates the EBV gene BHRF1, a viral homolog of the cellular antiapoptotic gene bcl-2 (28). We show here that R can interact with Rb; therefore, R is likely to have a direct effect on cellular proliferation during the initial stages of viral reactivation.
In addition, R binds to the Rb protein in a region that has been linked to additional functions of Rb, and this result suggests that interaction of R and Rb may have additional functional consequences beyond relief of Rb-mediated repression of E2F. R, therefore, may work in tandem with the other major IE EBV protein, Z, which binds p53, to control cellular environment during EBV lytic infection.
ACKNOWLEDGMENTS
This work was made possible through the generosity of several investigators who supplied constructs and reagents: E. Harlow, J. Horowitz, S. Kenney, E. Manet, J. Nevins, A. Sergeant, and R. Weinberg. We thank J. Horowitz for helpful advice and discussions, and we thank J. Horowitz, S. Kenney, and N. Raab-Traub for critical review of the manuscript.
This investigation was supported by NCI grant CA-19014.
REFERENCES
- 1.Adams P D, Kaelin W G. Transcriptional control by E2F. Semin Cancer Biol. 1995;6:99–108. doi: 10.1006/scbi.1995.0013. [DOI] [PubMed] [Google Scholar]
- 2.Almasan A, Yin Y, Kelly R E, Lee E Y, Bradley A, Li W, Bertino J R, Wahl G M. Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F-responsive genes, and apoptosis. Proc Natl Acad Sci USA. 1995;92:5436–5440. doi: 10.1073/pnas.92.12.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arvanitakis L, Yaseen N, Sharma S. Latent membrane protein-1 induces cyclin D2 expression, pRb hyperphosphorylation, and loss of TGF-B1-mediated growth inhibition in EBV-positive B cells. J Immunol. 1995;155:1047–1055. [PubMed] [Google Scholar]
- 4.Ben-Bassat H, Goldblum N, Mitrani S, Goldblum T, Yoffey J M, Cohen M M, Bentwich Z, Ramot B, Klein E, Klein G. Establishment in continuous culture of a new type of lymphocyte from a ‘Burkitt like’ malignant lymphoma (line DG-75) Int J Cancer. 1977;19:27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- 5.Biggen M, Bodescot M, Perricaudet M, Farrell P. Epstein-Barr virus gene expression in P3HR1-superinfected Raji cells. J Virol. 1987;61:3120–3122. doi: 10.1128/jvi.61.10.3120-3132.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buisson M, Manet E, Trescol-Biemont M-C, Gruffat H, Durand B, Sergeant A. The Epstein-Barr virus (EBV) early protein EB2 is a posttranscriptional activator expressed under the control of EBV transcription factors EB1 and R. J Virol. 1989;63:5276–5284. doi: 10.1128/jvi.63.12.5276-5284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cannell E J, Farrell P J, Sinclair A J. Epstein-Barr virus exploits the normal cell pathway to regulate Rb activity during the immortalisation of primary B-cells. Oncogene. 1996;13:413–421. [PubMed] [Google Scholar]
- 8.Cayrol C, Flemington E K. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L I, Nishinaka T, Kwan K, Kitabayashi I, Yokoyama K, Fu Y-H F, Grunwald S, Chiu R. The retinoblastoma gene product RB stimulates Sp1-mediated transcription by liberating Sp1 from a negative regulator. Mol Cell Biol. 1994;14:4380–4389. doi: 10.1128/mcb.14.7.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen P-L, Riley D J, Chen-Kiang S, Lee W-H. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chevallier-Greco A, Gruffat H, Manet E, Calender A, Sergeant A. The Epstein-Barr virus (EBV) DR enhancer contains two functionally different domains: domain A is constitutive, and cell specific, domain B is transactivated by the EBV early protein, R. J Virol. 1989;63:615–623. doi: 10.1128/jvi.63.2.615-623.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein-Barr virus (EBV) transactivators, EB1 and EB2, are required to activate transcription from an early promoter. EMBO J. 1986;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Countryman J, Jensen H, Seibel R, Wolf H, Miller G. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J Virol. 1987;61:2521–2528. doi: 10.1128/jvi.61.12.3672-3679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpes virus after gene transfer with a small clones subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox M, Leahy J, Hardwick J M. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J Virol. 1990;64:313–321. doi: 10.1128/jvi.64.1.313-321.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta A K, Feighny R J, Pagano J S. Induction of Epstein-Barr virus-associated DNA polymerase by 12-O-tetradecanoyl-phorbol-13-acetate. J Biol Chem. 1980;255:5120–5125. [PubMed] [Google Scholar]
- 17.DeCaprio J A, Ludlow J W, Figge J, Shew J-Y, Huang C-M, Lee W-H, Marsillo E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 18.Durfee T, Mancini M A, Jones D, Elledge S J, Lee W-H. The amino-terminal region of the retinoblastoma gene product binds a novel nuclear matrix protein that co-localizes to centers for RNA processing. J Cell Biol. 1994;127:609–622. doi: 10.1083/jcb.127.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyson N, Howley P M, Munger L, Harlow E. The human papillomavirus-16E7 oncoprotein is able to bind the retinoblastoma gene product. Science. 1989;243:943–947. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 20.Farrell P J, Rowe D T, Rooney C M, Kouzarides T. Epstein-Barr virus BZLF1 transactivator specifically binds to a consensus sequence AP-1 site and is related to c-fos. EMBO J. 1989;8:127–133. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemington E, Speck S H. Epstein-Barr virus BZLF1 transactivator induces the promoter of a cellular cognate gene, c-fos. J Virol. 1990;64:4549–4552. doi: 10.1128/jvi.64.9.4549-4552.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flemington E K, Goldfield A E, Speck S H. Efficient transcription of the Epstein-Barr virus immediate-early BZLF1 and BRLF1 genes requires protein synthesis. J Virol. 1991;65:7073–7077. doi: 10.1128/jvi.65.12.7073-7077.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furukawa T, Tanaka S, Plotkin S A. Stimulation of macromolecular synthesis in guinea pig cells by human CMV. Proc Soc Exp Biol Med. 1975;148:211–214. doi: 10.3181/00379727-148-38508. [DOI] [PubMed] [Google Scholar]
- 24.Giot J-F, Mikaelian I, Buisson M, Manet E, Joab I, Nicholas J-C, Sergeant A. Transcriptional interference between the EBV transcription factors EB1 and R: both DNA-binding and activation domains of EB1 are required. Nucleic Acids Res. 1991;19:1251–1258. doi: 10.1093/nar/19.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grogan E J, Jenson J, Countryman J, Heston L, Gradoville L, Miller G. Transfection of a rearranged viral DNA fragment WZhet, stably converts latent Epstein-Barr virus infection to productive infection in lymphoid cells. Proc Natl Acad Sci USA. 1987;84:1332–1336. doi: 10.1073/pnas.84.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutsch D E, Marcu D B, Kenney S C. The Epstein-Barr virus BRLF1 gene product transactivates the murine and human c-myc promoters. Cell Mol Biol. 1994;40:747–760. [PubMed] [Google Scholar]
- 27.Hagemeier C, Caswell R, Hayhurst G, Sinclair J H, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardwick J M, Lieberman P M, Hayward S D. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol. 1988;62:2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardwick J M, Tse L, Applegren N, Nicholas J, A V M. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilton M J, Mounghane D, McLean T, Contractor N V, O’Neil J, Carpenter K, Bachenheimer S L. Induction by herpes simplex virus of free and heteromeric forms of E2F transcription factor. Virology. 1995;213:624–638. doi: 10.1006/viro.1995.0034. [DOI] [PubMed] [Google Scholar]
- 31.Holley-Guthrie E A, Quinlivan E B, Mar E-C, S K. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen D (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J Virol. 1990;64:3753–3759. doi: 10.1128/jvi.64.8.3753-3759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horowitz, J. Personal communication.
- 33.Hu Q J, Dyson N, Harlow E. The regions of the retinoblastoma protein needed for binding to adenovirus E1A or SV40 large T antigen are common sites for mutations. EMBO J. 1990;9:1147–1155. doi: 10.1002/j.1460-2075.1990.tb08221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang S, Wang N P, Tseng B Y, W.-H. L, Lee E Y-H P. Two distinct and frequently mutated regions of retinoblastoma protein are required for binding to SV40 T antigen. EMBO J. 1990;9:1815–1822. doi: 10.1002/j.1460-2075.1990.tb08306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inman G J, Farrell P J. Epstein-Barr virus EBNA-LP and transcription regulation properties of pRB, p107 and p53 in transfection assays. J Gen Virol. 1995;79:2141–2149. doi: 10.1099/0022-1317-76-9-2141. [DOI] [PubMed] [Google Scholar]
- 36.Kaelin W G, Ewen M E, Livingston D M. Definition of the minimal simian virus 40 large T antigen- and adenovirus E1A-binding domain in the retinoblastoma gene product. Mol Cell Biol. 1990;10:3761–3769. doi: 10.1128/mcb.10.7.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenney S, Holley-Guthrie E, Mar E-C, Smith M. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J Virol. 1989;63:3878–3883. doi: 10.1128/jvi.63.9.3878-3883.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Fields virology. 2nd ed. New York, N.Y: Raven Press; 1993. pp. 1889–1920. [Google Scholar]
- 39.Kim S J, Onwuta U S, Lee Y I, Li R, Botchan M R, Robbins P D. The retinoblastoma gene product regulates Sp1-mediated transcription. Mol Cell Biol. 1992;12:2455–2463. doi: 10.1128/mcb.12.6.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam E W F, La Thangue N B. DP and E2F proteins: coordinating transcription with cell cycle progression. Curr Opin Cell Biol. 1994;6:859–866. doi: 10.1016/0955-0674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 41.Liebowitz D, Kieff E. Epstein-Barr virus. In: Roizman B, Whitely R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press; 1993. pp. 107–172. [Google Scholar]
- 42.Liu C, Sista N D, Pagano J S. Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J Virol. 1996;70:2545–2555. doi: 10.1128/jvi.70.4.2545-2555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludlow J W. Interactions between SV40 large-tumor antigen and the growth supressor proteins pRB and p53. FASEB J. 1993;7:866–871. doi: 10.1096/fasebj.7.10.8344486. [DOI] [PubMed] [Google Scholar]
- 44.Manet E, Rigolet A, Gruffat H, Giot J-F, Sergeant A. Domains of the Epstein-Barr virus (EBV) transcription factor R required for dimerization, DNA binding and activation. Nucleic Acids Res. 1991;19:2661–2667. doi: 10.1093/nar/19.10.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mietz J A, Unger T, Huibregtse J M, Howley P M. The transcriptional transactivation function of wild type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moran E. Interaction of adenoviral proteins with pRb and p53. FASEB J. 1993;7:880–885. doi: 10.1096/fasebj.7.10.8344487. [DOI] [PubMed] [Google Scholar]
- 47.Nevins J R. A link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 48.Pagano J S. Epstein-Barr virus. In: Kelley N, editor. Textbook of internal medicine. 2nd ed. Philadelphia, Pa: Lippincott; 1991. pp. 1499–1503. [Google Scholar]
- 49.Parker G A, Crook T, Bain M, Sara E A, Farrell P J, Allday M J. Epstein-Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene. 1996;13:2541–2549. [PubMed] [Google Scholar]
- 50.Qian Y, Luckey C, Horton L, Esser M, Templeton D J. Biological function of the retinoblastoma protein requires distinct domains for hyperphosphorylation and transcription factor binding. Mol Cell Biol. 1992;12:5363–5372. doi: 10.1128/mcb.12.12.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlivan, E. B. Unpublished data.
- 52.Quinlivan E B, Holley-Guthrie E, Mar E C, Smith M S, Kenney S. The Epstein-Barr virus BRLF1 immediate-early gene product transactivates the human immunodeficiency virus type 1 long terminal repeat by a mechanism which is enhancer independent. J Virol. 1990;64:1817–1820. doi: 10.1128/jvi.64.4.1817-1820.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quinlivan E B, Holley-Guthrie E A, Norris M, Gutsch D, Bachenheimer S L, Kenney S C. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter BMRF1. Nucleic Acids Res. 1993;21:1999–2007. doi: 10.1093/nar/21.8.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raab-Traub N. Epstein-Barr virus and nasopharyngeal carcinoma. Semin Cancer Biol. 1992;3:297–307. [PubMed] [Google Scholar]
- 55.Sarnow P, Ho Y S, Williams J, Levine A. Adenovirus E1B-58 kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54kd cellular protein. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 56.Shen W-J, Kim H S, Tsai S Y. Stimulation of human insulin receptor gene expression by retinoblastoma gene product. J Biol Chem. 1995;270:20525–20529. doi: 10.1074/jbc.270.35.20525. [DOI] [PubMed] [Google Scholar]
- 57.Sinclair A J, Brimmell M, Shanahan F, Farrell P J. Pathways of activation of the Epstein-Barr virus productive cycle. J Virol. 1991;65:2237–2244. doi: 10.1128/jvi.65.5.2237-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sommer M H, Scully A L, Spector D H. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stellar H. Mechanisms and genes of cellular suicide. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 60.Sterner J M, Murata Y, Kim H G, Kennett S B, Templeton D J, Horowitz J M. Characterization of cellular proteins that form complexes with the amino-terminus of the retinoblastoma (Rb) protein: a novel cell-cycle regulated kinase associates with Rb in G2/M phases. J Biol Chem. 1995;270:9281–9288. doi: 10.1074/jbc.270.16.9281. [DOI] [PubMed] [Google Scholar]
- 61.Sterner J M, Tao Y, Kennett S B, Kim H G, Horowitz J M. The amino terminus of the retinoblastoma (Rb) protein associates with a cyclin-dependent kinase-like kinase via Rb amino acids required for growth suppression. Cell Growth Differ. 1996;7:53–64. [PubMed] [Google Scholar]
- 62.Stinski M F. Synthesis of proteins and glycoproteins in cells infected with human cytomegalovirus. J Virol. 1977;23:751–767. doi: 10.1128/jvi.23.3.751-767.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.St. Jeor S C, Albrecht T B, Funk F D, Rapp F. Stimulation of cellular DNA synthesis by human cytomegalovirus. J Virol. 1974;13:353–362. doi: 10.1128/jvi.13.2.353-362.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szekely L, Pokrovskaja K, Jiang W-Q, Selivanova G, Lowbeer M, Ringertz N, Wiman K G, Klein G. Resting B-cells, EBV-infected B-blasts and established lymphoblastoid cell lines differ in their RB, p53 and EBNA-5 expression patterns. Oncogene. 1995;13:1413–1421. [PubMed] [Google Scholar]
- 65.Szekely L, Selivanova G, Magnusson K P, Klein G, Wiman K G. EBNA-5, an Epstein-Barr virus-encoded nuclear antigen, binds to the retinoblastoma and p53 proteins. Proc Natl Acad Sci USA. 1993;90:5455–5459. doi: 10.1073/pnas.90.12.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takada K. Cross-linking of cell surface immunoglobulins induces Epstein-Barr virus in Burkitt’s lymphoma lines. Int J Cancer. 1984;33:27–32. doi: 10.1002/ijc.2910330106. [DOI] [PubMed] [Google Scholar]
- 67.Takada K, Horinouchi K, Ono Y, Aya T, Osato M, Takahashi M, Hayasaka S. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes. 1991;5:147–156. doi: 10.1007/BF00571929. [DOI] [PubMed] [Google Scholar]
- 68.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takada K, Shimuzu N, Sakuma S, Keating A. Transactivation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV BamHI Z DNA fragment. J Virol. 1986;57:1016–1022. doi: 10.1128/jvi.57.3.1016-1022.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanaka S, Furukawa T, Plotkin S A. Human cytomegalovirus stimulates host cell RNA synthesis. J Virol. 1975;15:297–304. doi: 10.1128/jvi.15.2.297-304.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taya Y. RB kinases and RB-binding proteins: new points of view. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 72.Udvadia A J, Rogers K T, Higgins P D R, Murata Y, Martin K H, Humphrey P A, Horowitz J M. Sp-1 binds promoter elements regulated by the RB protein and Sp-1-mediated transcription is stimulated by RB coexpression. Proc Natl Acad Sci USA. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Udvadia A J, Templeton D J, Horowitz J M. Functional interactions between the retinoblastoma (Rb) protein and Sp-family members: superactivation by Rb requires amino acids necessary for growth suppression. Proc Natl Acad Sci USA. 1995;92:3953–3957. doi: 10.1073/pnas.92.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vousden K H. Interaction of human papillomavirus transforming proteins with the products of tumor suppressor genes. FASEB J. 1993;7:872–879. doi: 10.1096/fasebj.7.10.8393818. [DOI] [PubMed] [Google Scholar]
- 75.Vousden K H. Regulation of the cell cycle by viral oncoproteins. Semin Cancer Biol. 1995;6:109–116. doi: 10.1006/scbi.1995.0014. [DOI] [PubMed] [Google Scholar]
- 76.Wang J Y J, Knudsen E S, Welch P J. The retinoblastoma tumor suppressor protein. Adv Cancer Res. 1994;64:25–85. doi: 10.1016/s0065-230x(08)60834-9. [DOI] [PubMed] [Google Scholar]
- 77.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 78.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature (London) 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 79.Wiman K G. The retinoblastoma gene: role in cell cycle control and cell differentiation. FASEB J. 1993;7:841–845. doi: 10.1096/fasebj.7.10.8393817. [DOI] [PubMed] [Google Scholar]
- 80.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early region 1B protein. Nature (London) 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 81.Zalani S, Holley-Guthrie E, Kenney S. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci USA. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zalani S, Holley-Guthrie E A, Gutsch D E, Kenney S C. The Epstein-Barr virus immediate-early promoter BRLF1 can be activated by the cellular Sp1 transcription factor. J Virol. 1992;66:7282–7289. doi: 10.1128/jvi.66.12.7282-7292.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Q, Gutsch D, Kenney S. Functional and physical interaction between p53 and BZLF1: implication for Epstein-Barr virus latency. Mol Cell Biol. 1994;14:1929–1938. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.zur Hausen H, O’Neill F J, Freese U K, Hecker E. Persisting oncogenic herpesviruses induced by the tumor promoter TPA. Nature (London) 1978;272:373–375. doi: 10.1038/272373a0. [DOI] [PubMed] [Google Scholar]