Abstract
Most common genetic variants associated with disease are located in non-coding regions of the genome. One mechanism by which they function is through altering transcription factor (TF) binding. In this study, we explore how genetic variation is connected to differences in the regulatory landscape of livers from C57BL/6J and 129S1/SvImJ mice fed either chow or a high-fat diet. To identify sites where regulatory variation affects TF binding and nearby gene expression, we employed an integrative analysis of H3K27ac ChIP-seq (active enhancers), ATAC-seq (chromatin accessibility) and RNA-seq (gene expression). We show that, across all these assays, the genetically driven (i.e. strain-specific) differences in the regulatory landscape are more pronounced than those modified by diet. Most notably, our analysis revealed that differentially accessible regions (DARs, N = 29635, FDR < 0.01 and fold change > 50%) are almost always strain-specific and enriched with genetic variation. Moreover, proximal DARs are highly correlated with differentially expressed genes. We also show that TF binding is affected by genetic variation, which we validate experimentally using ChIP-seq for TCF7L2 and CTCF. This study provides detailed insights into how non-coding genetic variation alters the gene regulatory landscape, and demonstrates how this can be used to study the regulatory variation influencing TF binding.
Graphical Abstract
Graphical Abstract.
Introduction
The large majority of single nucleotide polymorphisms (SNPs) identified in genome wide association studies (GWAS) are located in non-coding regions of the human genome. A recent study examined 90361 obesity-trait related SNPs from 72 GWAS and demonstrated that over 90% were in the non-coding regions of the genome (1). Understanding the functional consequences of disease-associated SNPs, however, remains incomplete. A primary mechanism through which non-coding regulatory SNPs (rSNPs) may influence disease is by modifying transcription factor (TF) binding and target gene expression (2). However, characterizing the TF binding sites affected by genetic variation in humans is challenging due to the scarcity of available datasets, and the complexities associated with collecting and analysing novel human samples.
Inbred mouse strains offer an interesting comparative genetics platform to study the effects of genetic variation on gene regulation, as continued inbreeding has led to a near-complete lack of heterozygosity. For example, two commonly used strains, C57Bl/6J (B6) and 129S1/SvImJ (129), differ genetically by 5.2 million homozygous SNPs. These two strains also differ in their predisposition to diet-induced obesity and insulin resistance, with B6 mice demonstrating a greater susceptibility to insulin resistance compared to 129 mice (3,4). Despite this, previous studies using inbred mouse strains have focused primarily on the diet-induced epigenetic landscape, frequently quantitated by examining chromatin accessibility and H3K27 acetylation (H3K27ac), and have not directly explored the interplay between gene regulation and genetic variation (5,6). Some studies, using 129 and B6 mice and their close relatives, have connected genes and obesity-related QTLs in an effort to explain the phenotype under study, but these studies similarly did not dissect the mechanisms of how genetic variation affects transcriptional regulation (7,8).
Chromatin accessibility and enhancer activity can be used to identify candidate rSNPs (9). Advances in the interpretation of chromatin accessibility data from Assay for Transposase-Accessible Chromatin sequencing (ATAC-seq) have made this approach feasible (10–12). Another marker for transcriptional activity is the H3K27ac histone modification, which enables the segregation of active, poised and inactive enhancers. This facilitates the direct detection of TF binding sites actively regulating gene expression (13,14). Although the complexity of the epigenetic signals that underlies the enhancer landscape goes beyond chromatin accessibility and H3K27ac, recent approaches utilizing integrative analysis of these epigenetic markers have nonetheless been successful in identifying rSNPs (9,15). However, these studies have not explored how perturbation, including diet, modulate genetically determined enhancer function and TF binding.
Even though sequence conservation per se can be considered poor between mice and human, recent studies in liver and pancreatic islets have shown that conserved regulatory regions are enriched for metabolic GWAS SNPs (2,16). Moreover, TFs and their target genes have conserved functions between mice and humans, which enables the detection of functionally related rSNPs, even in the absence of direct sequence homologs (17,18). A recent study by Soccio et al. explored the genetic determinants of PPARγ binding in the white adipose tissue of 129 and B6 mice (19). Using an approach based on chromatin immunoprecipitation sequencing (ChIP-seq), they discovered several strain-selective motif-altering rSNPs that affect PPARγ-mediated gene regulation. However, they did not explore how chromatin modifications were affected at the strain-specific regulatory sites.
Studies on the relationship of TF binding, epigenetic landscape, and gene regulation for well characterized liver TFs have recently been performed (20). However, many other TFs connected to metabolic diseases have not been studied in this regard. For example, transcription factor-7-like 2 (TCF7L2), a known metabolic regulator in liver that is part of the WNT signalling cascade offers an interesting target for studying metabolism-related diseases. Impaired TCF7L2 function has been connected to dysregulation of glucose and lipid metabolism, and type 2 diabetes, in both mice and human (21–23). In addition, several studies on the metabolic role of TCF7L2 in the liver have been performed and its relationship with other liver-enriched TFs has been explored, making it an interesting TF to study further (23–26). However, studies on the genetic determinants of TCF7L2 binding in this context are lacking. In addition, since TCF7L2 co-factors in liver have been previously identified, it provides an interesting platform to study how genetic variation can effect TF binding through altered co-factor binding (27).
In this study, we sought to identify and mechanistically detail mouse candidate rSNPs in liver using an integrative approach using (i) ATAC-seq for assessing chromatin accessibility, (ii) H3K27ac ChIP-seq for detecting active enhancers, (iii) RNA-sequencing (RNA-seq) for measuring gene expression, (iv) CCCTC-binding factor (CTCF) ChIP-seq to validate the (in)active enhancer detection and, as an example case study, (v) TCF7L2 ChIP-seq for identifying regulatory sites for a metabolically relevant TF. To enhance the significance of our findings to common metabolic diseases beyond genetics, we used inbred B6 and 129 mice that were fed with either chow or high-fat diet (HFD). Finally, we mapped our mouse candidate rSNPs to the human genome using available GWAS and eQTL databases. The results of these experiments reveal that the most widespread differences across all assays were observed between the mouse strains. This was especially clear for both chromatin accessibility and TCF7L2 and CTCF binding, where we observed a near total absence of diet-induced changes. The strong enrichment of genetic variation to the sites of chromatin-level differences highlights the importance of a comprehensive genomics view into the gene regulatory landscape when seeking for the functional explanation to genetic findings like those of GWAS.
Materials and methods
Mouse experiments
Mice were acquired from Jackson Laboratories (Boston, USA), housed at the Lab Animal Centre of the University of Eastern Finland, and experimented on under a permit from the local Animal Experiment Board (ESAVI-2015-002081). Male C57BL/6J and 129S1/SvImJ mice were fed either a chow (Teklad 2016, Envigo) or HFD (TD.88137, Harlan) from 9 weeks of age for 8 weeks. At 17 weeks of age, mice were euthanized by CO2 for harvesting tissues. The livers were weighted, the outer edge of the left lateral (the biggest) lobe cut to smaller pieces, snap-frozen in liquid N2 and stored in −80°C. Liver samples of chow-fed first filial generation (F1) hybrid mice that were a cross between C57BL/6J females and 129S1/SvImJ males (B6129SF1/J, cat. 101043) were purchased from The Jackson Laboratory, Boston, USA.
RNA extraction
Total RNA from mouse liver tissue was extracted using the miRNeasy Mini Kit (QIAGEN) and QIAzol Lysis Reagent (QIAGEN) according to the manufacturer's protocol. Extracted RNA was treated with DNase using RNase-Free DNase (QIAGEN). RNA quality was assessed using an Agilent Bioanalyzer with RNA 6000 Nano kit. All samples had RIN score ≥7.
Library preparation and RNA-sequencing
RNA-seq libraries were prepared using 800 ng of total RNA. First, ribosomal RNA was depleted using the NEBNext rRNA Depletion Kit (New England BioLabs). Libraries were prepared using a NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England BioLabs) following the manufacturer's instructions. The library yield was quantified with Qubit DNA High Sensitivity assay (Invitrogen) and quality control was performed on the Agilent Bioanalyzer with DNA 1000 kit (Agilent). The indexed libraries were pooled, and single-end sequenced on the NextSeq 500 (Illumina) platform with 75 bp-reads.
Nuclei extraction, library preparation and sequencing for ATAC-seq
Libraries were generated using ∼20 mg of liver tissue following Omni-ATAC protocol (28) with minor adjustments. Nuclei were isolated using a Dounce homogenizer and 1× homogenization buffer (320 mM sucrose, 0.1 mM EDTA, 0.1% NP40, 5 mM CaCl2, 3 mM Mg(Ac)2, 10 mM Tris pH 7.8, 1× protease inhibitors (Roche, cOmplete), and 1 mM β-mercaptoethanol). After tissue homogenization, nuclei were recovered by OptiPrep/iodixanol (Sigma-Aldrich) density gradient centrifugation. For transposition reaction 50 000 nuclei were resuspended in transposition mix (25 μl 2 × TD buffer, 2.5 μl transposase, 16.5 μl PBS, 0.5 μl 1% digitonin, 0.5 μl 10% Tween-20, 5 μl H2O). After the transposition reaction, DNA was purified with DNA Clean & Concentrator-5 Kit (Zymo Research). Ten microliters of purified transposed DNA were amplified for 9–11 cycles (predetermined) using 25μl of 2 × Ultra II Q5 Master Mix (New England BioLabs) and 1 μl of both amplification primers (1.25 μM final concentration) (29). For clean-up and to remove primer dimers and large (>1000 bp) fragments from the libraries, SPRIselect beads (Beckman Coulter) were used. Library yield was quantified using NEBNext Library Quant Kit for Illumina (New England Biolabs) and quality assessed with the Agilent Bioanalyzer and High Sensitivity DNA kit (Agilent). ATAC-seq libraries were sequenced on the NextSeq 500 (Illumina) platform with paired-end 75 bp-reads.
ChIPmentation-based ChIP-seq analysis of CTCF and TCF7L2 binding, and H3K27 acetylation
To perform ChIP-Seq on mouse livers, ∼40 mg of frozen tissue was Dounce homogenized with the loose pestle A in 1 × PBS with 1 × protease inhibitors (cOmplete, Roche). Tissue suspension was centrifuged at 2000 rpm for 5 min and the pellet was resuspended in 1 ml PBS containing 1% formaldehyde for 10 min and quenched with glycine 150 mM for 5 min at room temperature. The cross-linked samples were washed twice with ice-cold 1 × PBS with 1 × protease inhibitors. Cells were lysed by pelleting and diluting twice in ice-cold SDS lysis buffer (50 mM Tris–Cl, 0.5% SDS, and 10 mM EDTA, 1 × protease inhibitor) and incubating for 10 min at 4°C with gentle mixing. The cell lysate was then Dounce homogenized with the tight pestle B in 1 ml ice-cold of SDS lysis buffer and the homogenate was filtered through a 100 μm cell strainer. The samples were split into 3 × 300 μl aliquots in 1.5 sonication tubes and incubated on ice for 10 min. Chromatin was sheared with Bioruptor Plus sonicator (Diagenode) for 35 cycles at high setting. Each cycle was 30s ON, 30s OFF and after each 10 cycles the sonicator was allowed to cool down for 5 min. To neutralize SDS, Triton-X-100 was added to final concentration of 1% along with 50 × protease inhibitors (final 1×). All the aliquots were combined and centrifuged at 13000 rpm for 20 min. Supernatant with sheared chromatin was collected. An aliquot of 100 μl was taken for assessment of chromatin shearing and a 10 μ aliquot was taken for preparation of input control. Rest of the sample was divided to 190 μl aliquots for immunoprecipitation.
ChIP and library preparation were performed using the recently described ChIPmentation protocol (30) with minor modifications. For ChIP, 2 μg of rabbit polyclonal anti-CTCF antibody (Diagenode, #C15410210), 5μl of rabbit monoclonal anti-TCF4/TCF7L2 antibody (Cell Signaling, C48H11) or 2 μg of Anti-Histone H3 (acetyl K27) antibody (Abcam, #4729) was added to 50 μl Protein G-coupled Dynabeads (Thermo Fisher Scientific) in 1 × PBS with 0.5% bovine serum albumin (BSA) and rotated at 40 rpm for 4 h at 4°C. Antibody-coated Dynabeads were washed three times with PBS with 0.5% BSA and then mixed with 190 μl chromatin samples in 1.5 ml tubes. The samples were rotated at 40 rpm overnight at 4°C. Immunoprecipitated chromatin was washed with 150 μl of low-salt buffer (50 mM Tris-Cl, 150 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, and 1 mM EDTA), high-salt buffer (50 mM Tris-Cl, 500 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, and 1 mM EDTA) and LiCl buffer (10 mM Tris–Cl, 250 mM LiCl, 0.5% IGEPAL CA-630, 0.5% sodium deoxycholate, and 1 mM EDTA), followed by two washes with TE buffer (10 mM Tris–Cl and 1 mM EDTA) and two washes with ice-cold Tris–Cl pH 8. Immunoprecipitated bead-bound chromatin was resuspended in 30 μl of 2 × TD buffer and 1 μl of transposase (Nextera, Illumina) for tagmentation. Samples were incubated at 37°C for 10 min followed by two washes with low-salt buffer. Bead-bound tagmented chromatin was diluted in 23 μl of water and 25 μl of 2 × Ultra II Q5 Master Mix (New England BioLabs, M0544S) and 1 μl of both amplification primers (29) were added. For adapter extension and reverse cross-linking, the libraries were incubated as follows: 72°C 5 min (adapter extension); 95°C 5 min (reverse cross-linking); followed by 11 cycles of 98°C 10s, 63°C 30s and 72°C 3 min. After PCR amplification, double-sided purification was performed using SPRIselect beads (Beckman Coulter). To prepare input controls, 2 μl of 50 mM MgCl2 was added to 10 μl sonicated lysate to neutralize the EDTA in the SDS lysis buffer. Tagmentation and amplification was done as described above. The library yield was quantified with Qubit DNA High Sensitivity assay (Invitrogen) and quality control was performed on the Agilent Bioanalyzer with DNA 1000 kit (Agilent). ChIP-seq libraries were sequenced on the NextSeq 500 (Illumina) with single-end 75 bp-reads.
ChIP-qPCR
To perform ChIP-qPCR on mouse livers ∼100–150mg of frozen tissue from chow-fed mice was Dounce homogenized with a tight pestle B in Farham Lysis Buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% IGEPAL) and then cross-linked with 1% formaldehyde in Farham Lysis Buffer for 10 min at RT while rotating. This reaction was stopped by adding 125 mM glycine at RT. Homogenized tissue was then collected in ice-cold 1x PBS with 1x protease inhibitors and subsequently centrifuged, and resuspended in RIPA buffer (1 × PBS, 1% IGEPAL, 0.5% sodium deoxycholate, 0.1% SDS), supplemented with 1× protease inhibitors. Chromatin was sheared with Bioruptor Plus sonicator (Diagenode, #UCD-300) at 4°C for 40 cycles at high setting to yield 200–400 bp DNA fragments.
For ChIP 10 μg of mouse anti-HNF4α antibody (K9218, ab41898; Abcam) was added to the sonicated chromatin in RIPA buffer and rotated at 40 rpm overnight at 4°C. After 16 h magnetic protein G Dynabeads (Invitrogen, 10004D) in 1× PBS with 0.5% BSA were added to the antibody bound chromatin and rotated at 40 rpm overnight at 4°C. Immunoprecipitated chromatin was washed five times in LiCl IP wash buffer [100 mM Tris (pH 7.5), 500 mM LiCl, 1% (w/v) IGEPAL, 1% (w/v) sodium deoxycholate] and twice in TE buffer [1 mM EDTA, 10 mM Tris–HCl (pH 8.1)]. Bead bound chromatin was eluted in 200 μl elution buffer (0.1 M NaHCO3 and 10% SDS) by incubation at 65°C for 1 hour. The proteins were decrosslinked by adding Proteinase K (New England BioLabs, #T2002) and incubating at 65°C for 4 h and DNA purified using the Monarch PCR & DNA Cleanup Kit (New England BioLabs, #T1030), and eluting in 50 μl of Monarch DNA Elution Buffer. qPCRs were carried out as individual biological replicates for each of the immunoprecipitated and input DNA samples. qPCR analysis was carried out with LightCycler 480 SYBR Green I Master (Roche Diagnostics Gmbh). Results were calculated using the formula 100 × 2DCp× E, where DCp= Cp (Input) – Cp (ChIP) and E=Percentage of Input over Total Chromatin as a factor. Control region for the B6-up regions was selected as intronic non-enhancer regions within Cenpl region and for 129-up the selected control region was a region with accessible chromatin in both strains near Egf.
Pseudogenome generation
Pseudogenome and transcriptome for 129 mice was created using g2gtools (v0.2.9, https://Github.com/churchill-lab/g2gtools) with SNPs and InDels from Mouse Genomes Project version 5 (MGPv5), GRCm38 genome and Gencode M17 gene annotation.
RNA-seq preprocessing
Raw reads were trimmed with Trimmomatic (v0.36) (31). Bowtie (v2.2.3) (32) was used to identify reads that align to known possible contaminants, which were then discarded. Reads were then aligned using STAR (v2.5.4b) (33) with manual two-pass alignment: on the first pass the reads were aligned to strain-specifically indexed genome and transcriptome, after which the splice junctions were collected and used on the second pass alignment with the ‘–sjdbFileChrStartEnd’ parameter along with the 2nd pass index generated with the splice junctions. Other STAR parameters were selected from a study reporting optimal alignment parameters (–outFilterMultimapNmax 100 –outFilterMismatchNmax 33 –seedSearchStartLmax 12 –alignSJoverhangMin 15 –outFilterMatchNminOverLread 0 –outFilterScoreMinOverLread 0.3 –outFilterType BySJout) (34). Read counts for genes were generated using STAR’s ‘-quantMode GeneCounts’ option.
RNA-seq analysis
Differentially expressed genes (DEGs) were detected using DESeq2 (v1.26.0) (35) between groups of 5 mice for 129 on chow, 6 mice for B6 on chow and 129 on HFD, and 7 mice for B6 on HFD. Genes without any reads in any sample were excluded from further analysis. PCA was performed using DESeq2. WGCNA, to obtain co-expression modules, and GSEA for the module-associated genes, were performed using CEMiTool (v1.12.0) (36). Optimal cut-offs for module merging (Pearson correlation > 0.95) and gene filtering (P-value < 0.25) were calculated using centralized enrichment score (CES) as described in Russo et al, 2018. Counts were normalized before performing network analysis using variance-stabilizing transformation with DESeq2. Topological overlap matrix and networks were constructed as signed. Over-representation analysis was ran using PANTHER™ GO slim (version 17.0) annotations (37). BisqueRNA (v1.05) was used to estimate cell type ratios using FACS-based mouse liver scRNA-seq dataset from Tabula Muris (38,39). Cell type specific genes were determined from the Tabula Muris dataset as genes that have the mean expression of ln(CPM + 1) >1 and are >5 × ln(CPM + 1) more expressed in one cell type compared to all other cell types. Cell type specific genes were enriched to modules using clusterProfiler's (v4.2.2) enricher function (40). The proActiv (v1.0.0) was used with default settings in R to identify actively transcribed TSSs, using both Major and Minor TSSs when connecting genes to chromatin features (41).
ATAC- and ChIP-seq pre-processing
Raw reads were trimmed with Trimmomatic (v0.36) and aligned to strain-specific genome using STAR (v2.5.4b) with ‘–alignIntronMax 1′ to turn off splice awareness and –alignEndsType ‘EndToEnd’ to prevent soft clipping. For ATAC-seq the reads were filtered for true pairs and MAPQ > 30 with Samtools (v1.9) (42). For ChIP-seq the reads were filtered for MAPQ > 20. For both, duplicates were removed after filtering using Picard (v2.8.13, https://Broadinstitute.github.io/picard/) MarkDuplicates. ATAC-seq peaks were called with HMMRATAC (v1.2.9) (12) using ‘–means 75200400600 –upper 20 –lower 10′. H3K27ac, CTCF and TCF7L2 ChIP-seq peaks were called with MACS2 (v2.2.7.1) using ‘-g mm -B –call-summits –keep-dup auto’, and ‘-q 0.05′ for H3K27ac, ‘-q 0.01′ for TCF7L2, and ‘-q 0.001′ for CTCF (43). After peak calling, summits were widened to peaks with Bedtools (v2.27.1) using ‘slop -b 75′ to prevent the summits of the 129 strain that overlapped the B6 deletions to be lost during the coordinate conversion (44). Consensus peak sets for ATAC-seq and TCF7L2 ChIP-seq were formed from the peak centres using kernel density estimation -based approach adapted from Tuoresmäki et al. (45). The consensus peak set for CTCF was created by merging the narrowPeaks from MACS2 using Bedtools merge. For H3K27ac, we used ChIP-R (v1.2.0) to identify group-wise the regions marked by H3K27ac (46). The consensus peak set for H3K27ac was created by merging the group-specific regions using Bedtools merge. We also generated harmonized H3K27ac summits using kernel density estimation -based approach and selected the summits overlapping group-specific H3K27ac regions for valley-based active nucleosome free region (NFR) detection. NFRs between two H3K27ac summits (±1500 bp) were defined as active. Peaks were filtered for the ENCODE blacklisted regions (47). For both ATAC and ChIP-seq, peaks and filtered BAM-files for the 129-strain were shifted to B6 coordinates using the g2gtools ‘convert’ function.
ATAC- and ChIP-seq differential analysis
Differentially accessible regions (DARs), differentially histone-acetylated regions (DHARs) and TCF7L2 differentially bound regions (DBRs) were detected with csaw (v1.28.0) adapting from Reske et al. (10). DARs were detected between groups of 5 mice for 129 on chow, 6 mice for B6 on chow and 129 on HFD, and 7 mice for B6 on HFD), DHARs in 4 mice per group, and TCF7L2 DBRs in 2 mice per group. Peaks with log2(CPM) > –3 were selected for analysis. Normalization was done using local regression fit. To focus on the Tn5 cut sites in read counting, ATAC-seq reads were shortened to 1 bp tags at 5′-ends. For ChIP-seq data, csaw function ‘correlateReads’ was used to calculate cross-correlation coefficients between read positions, and average fragment lengths were determined from the maximum cross-correlation coefficient. Reads were extended to the average fragment length before counting (48).
Transcription factor binding motif analysis
TF motif enrichment for known motifs was done using GimmeMotifs (v16.1.fix_rfe) with ‘gimme motifs’ using directly determined binding motifs from CIS-BP v2.0 collection and mm10 (B6) genome (49,50). Motif hits for TCF7L2 were obtained using TCF7L2 binding motifs from the CIS-BP collection with the motifmatchr R-package (v1.16.0) using 1e-4 as significance cut-off (51). Motif alterations for the same motif set were analyzed using motifbreakR (v2.0.0) with ‘filterp = TRUE, threshold = 1e-4, method = ‘ic’, legacy.score = FALSE’ and MGPv5 SNPs and InDels (52). Comparative motif analyses for gene co-expression module associated active NFRs and TCF7L2 overlap classes were performed using GimmeMotifs maelstrom. For TCF7L2 overlap analysis, the peaks were filtered of DBRs and peaks overlapping DARs. Additionally, the CIS-BP collection was filtered for TCF7L2 and highly similar motifs using universalmotif (v1.8.0) (53). First, the motifs were filtered for average information score >0.75 and then compared using Weighted Pearson correlation coefficient (WPCC) with normalized scores. Motifs with WPCC >0.7 to any of the TCF7L2 motifs in CIS-BP collection were discarded from the analysis. Motif clustering and alignment for the DBR-enriched altered motifs were done using universalmotif commands ‘motif_tree’, with arguments ‘method = ‘EUCL’, tryRC = FALSE’, and ‘view_motifs’ with arguments ‘tryRC = FALSE, method = ‘ALLR_LL’’. Venn diagrams for DBRs containing motif altering variants were plotted with nVennR (54).
Online resources
Known TF binding loci for mouse and human (Q-value < 1 × 10−5) were obtained from ChIP-Atlas Peak Browser (https://chip-atlas.org/peak_browser) (55). For mouse ‘Cell type: Liver’ and for human ‘Cell type Class: Liver’ were used for filtering liver specific TF binding sites. For mouse QTL phenotype enrichment analysis, lists of mouse QTL alleles, genotypes, mammalian phenotype annotation, and genetic markers were obtained from MGI database (https://www.informatics.jax.org/downloads/reports/index.html) (56).
Statistical analysis and visualization
Statistical analyses were performed with R software v3.6.1 (57) except for the Fisher's tests, and chipenrich (v2.18.0) (58) enrichment analyses, which were performed in R v4.1.0. For TCF7L2 peaks, chipenrich was run with default settings and mm10 genome. Chipenrich analyses for active DARs, DHARs and TCF7L2 DBRs were performed using ‘method = ‘polyenrich’, genome = ‘mm10’, max_geneset_size = 10000’. Confidence intervals for chipenrich results were calculated using mgcv.helper R package (https://github.com/samclifford/mgcv.helper). All plots were generated in R v4.1.0. Heatmaps were plotted using ComplexHeatmap (v2.1.0) and EnrichedHeatmap (v1.24.0) (59,60). Correlation analyses were done using Spearman rank correlation with logCPM normalized counts. For H3K27ac vs. NFR correlation, signal at consensus regions overlapping the valley summits of active NFRs were used. P-values shown as <2.2 × 10−16 exceed the limit of the precision of the calculation.
ATAC-seq pre-processing and statistical analysis of the F1-cross samples
To detect sites presenting allelic imbalance, we used ASEReadCounter*-pipeline together with Qllelic (v0.3.2) (61) statistical analysis tool. 129xB6 reference genome was created using 129 SNPs from MGPv5. To turn off splice-awareness, STAR parameter ‘–alignIntronMax 1′ was added to the pipeline. Allelic reads were counted using our ATAC-seq consensus peak set. For Qllelic, we selected sites that had >5 reads for statistical testing and used FDR P-value correction instead of Bonferroni correction.
Transferring mouse rSNP candidates to the human context
We used liftOver (v1.18.0) for DARs to transfer mouse genomic coordinates to their syntenic sequences in the human genome (hg38) (62). GWAS SNPs were acquired from the EBI catalog using gwascat (v2.18.0) R package (query on 4.6.2021) and significant variant-gene pairs for liver eGenes were acquired from GTEx Analysis V8 release (Single-Tissue cis-QTL Data) (63–65). Motif alteration analysis for human SNPs was performed using motifbreakR as described earlier except using directly determined human motifs from CIS-BP v2.0 (50,52).
Results
Genetics is the stronger driver of differences in the liver transcriptome than diet
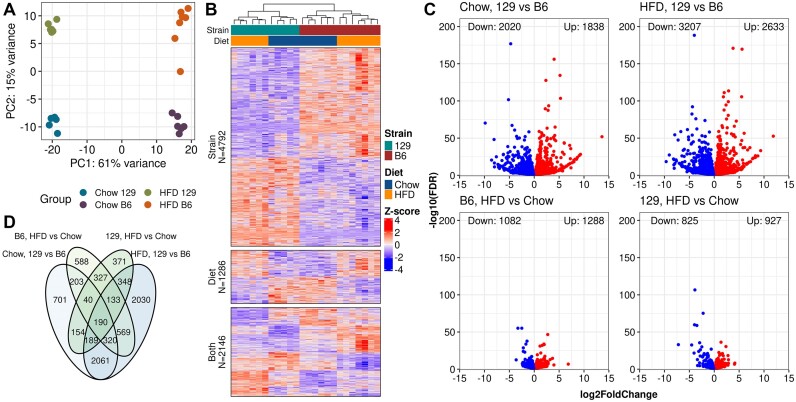
High-fat feeding increased the body mass more in B6 (chow: 28.4 ± 3.2 g vs HFD: 39.1 ± 4.3 g, T-test P-value = 1.3 × 10−4) than in 129 mice (chow: 25.9 ± 1.2 g versus HFD: 27.6.1 ± 3.1 g, T-test P-value = 0.022) (Supplementary Figure SF1A). Furthermore, concomitant differences were also evident in the liver weights, indicative of greater steatosis in B6 mice fed the HFD (Supplementary Figure SF1B) (5). We generated RNA-seq data (N = 5–7 per group), Supplementary Table S1: Table 1) to identify DEGs (FDR < 0.05) between the strains and diets (Supplementary Table S2: Tables 1-4). The main segregating factor between samples was the strain rather than the diet (Figure 1A). Out of the 8224 DEGs, more than half (4792, 58.3%) were strain-specific whereas only 15.6% (1286) were diet-specific (Figure 1B). The largest number of DEGs for any comparison (5840) was observed between the strains on HFD. The strain-DEGs also displayed greater fold changes compared to diet-DEGs in either strain (Figure 1C). A large proportion (2061, 43.0%) of the strain-DEGs were common in the two diet comparisons (Figure 1D). Panther GO term enrichment analysis revealed that the strain-specific DEGs were enriched in a variety of immune-related processes whereas the diet-specific DEGs were strongly enriched to lipid metabolism and other processes related to liver metabolism (Supplementary Figure SF1C, D). Fittingly, also genes that were both diet and strain-DEGs were mostly enriched in processes related to metabolism (Supplementary Figure SF1E).
Figure 1.
Most of the differences in gene expression are observed between the strains. (A) Principal component analysis on genes with any reads (PC, principal component). (B) Heatmap of Z-scored VST-normalized gene expression counts for the DEGs observed in at least one comparison. Row splits are determined by the comparisons in which the DEG was observed (Strain: Only in strain, Diet: Only in diet, and Both: Both in diet and strain). Columns are clustered using Pearson correlation with complete linkage and rows are clustered by Pearson correlation within the splits. (C) Volcano plots for the DEGs by comparisons. (D) Venn diagram of DEG counts.
Weighted gene correlation network analysis reveals differences in metabolic gene regulation between B6 and 129 mice
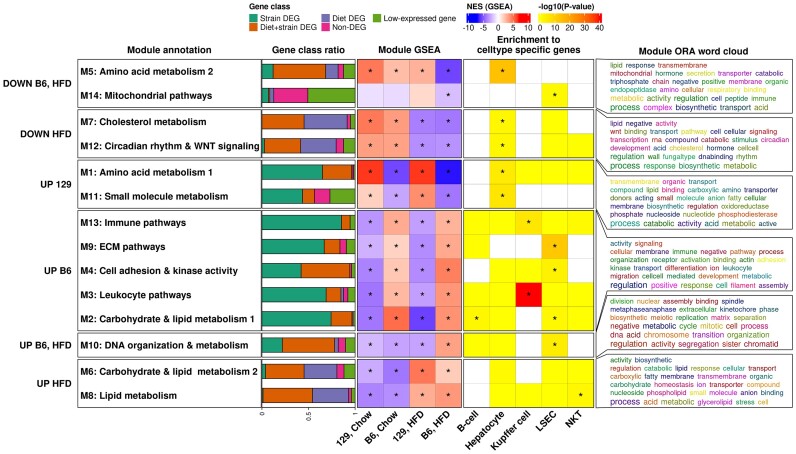
To shed more light on the biological processes underlying the observed differences in gene expression, we performed weighted gene correlation network analysis (WGCNA, parameter selection shown in Supplementary Figure SF2A) using CEMiTool (36). The analysis identified 14 co-expression modules (M1-14 which were significantly (FDR < 0.05) enriched to at least one GO Panther pathway in an over-representation analysis (Figure 2, Supplementary Figure SF2B). Gene set enrichment analysis (GSEA) revealed that several modules were differentially enriched between both the strains and the diets (Figure 2 panel: Module GSEA). Co-expression modules up-regulated in 129 mice were related to amino acid (M1) and small molecule metabolism (M11). In contrast, modules that were up-regulated in B6 mice were enriched for pathways related to carbohydrate and lipid metabolism (M2), extracellular matrix (M4 and 9) and the immune system (M3 and 13). There were also modules which were specifically diet-responsive in B6, including metabolic pathways related to amino acid metabolism (M5) and DNA organization and metabolism (M10) (Figure 2 panel: Module ORA word cloud, Supplementary Figure SF2B).
Figure 2.
Gene co-expression analysis reveals differences between the strains in metabolic gene co-expression modules. Module annotation: Module annotation determined by hand from the results of over representation analysis. Gene class ratio: Fractions of genes in different DEG-classes per module (Strain: Only in strain, Diet: Only in diet and Diet + strain: Both in diet and strain, Non-DEG: expressed non-DEG gene with zFPKM > –3, Low-expressed gene: gene that passed the unsupervised filter of CEMiTools, but is not classified expressed in other analyses). Module GSEA: Gene set enrichment analysis results for the modules. NES, normalized enrichment score. Enrichment to cell type specific genes: ORA results for module-associated genes in cell type specific gene sets. Module ORA word cloud: Word cloud annotation for the results of over representation analysis of GO terms. *P-value < 0.05.
Given that some modules were clearly related to the immune system, we explored the cell type compositions of our bulk liver RNA-seq samples using BisqueRNA deconvolution and the cell type specific marker genes derived from the Tabula Muris data (38,39). This analysis suggested that in all samples the most frequent cell type was the hepatocyte, representing about 50% of all cells, while the rest were composed mostly of liver sinusoidal endothelial cells (LSECs, about 25–30%) and Kupffer macrophages (about 10%). Interestingly, the B6 livers had a higher proportion of Kupffer macrophages than the 129 livers, and a concomitantly lower proportion of hepatocytes (Supplementary Figure SF2C). Enrichment analysis of the cell type marker gene sets in network modules showed that the Kupffer cell markers were highly enriched in the immune-related networks (M3 and 13). In contrast, hepatocyte markers tended to be enriched in the metabolic networks (e.g. M1 and 5), and the LSEC markers e.g. in the cell adhesion (M10) and ECM (M4) networks (Figure 2 panel: Enrichment to cell type specific genes). Based on these data, it is likely that the higher immune-related gene expression in B6 livers is due to their higher relative Kupffer macrophage content rather than the transcriptional activation of these genes in hepatocytes.
Joint analysis of chromatin accessibility and H3K27ac ChIP-seq identifies differentially active regulatory regions between B6 and 129 mice
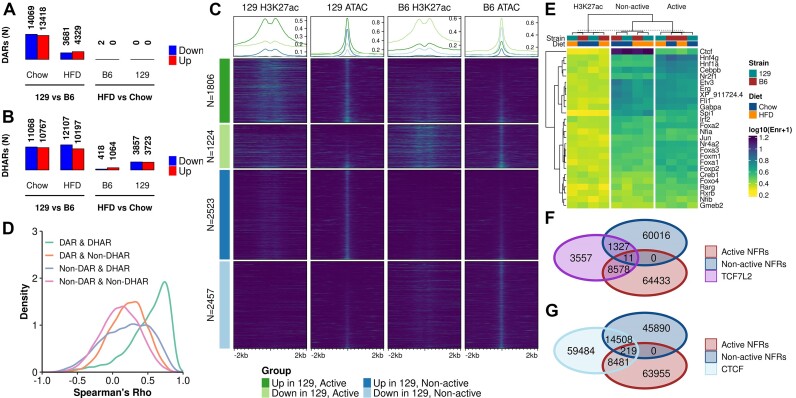
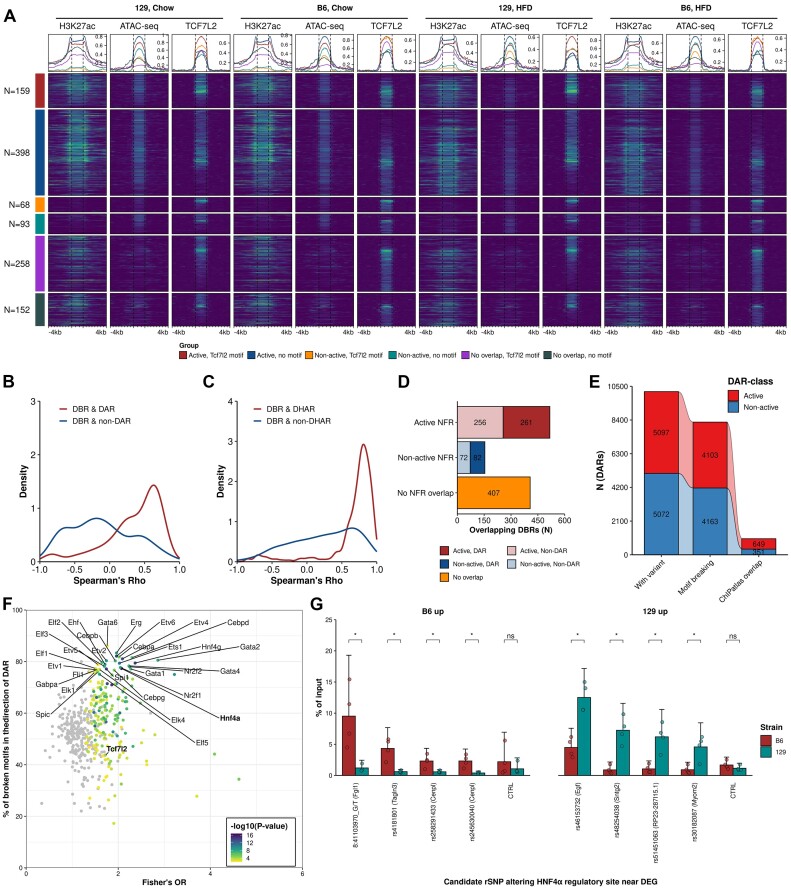
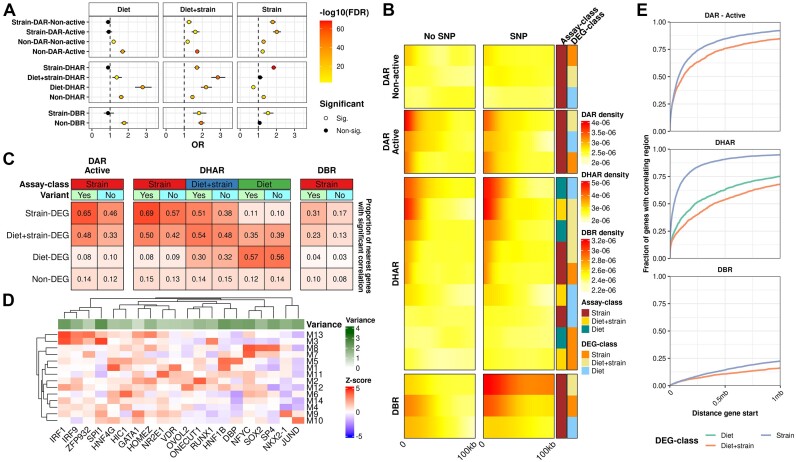
To gain insight into the regulatory chromatin landscape in the livers of B6 and 129 mice on chow and HFD, we generated ATAC-seq data (N = 5–7 per group) (Supplementary Table S1: Table 2). We called ATAC-seq peaks to represent NFRs which, among the cross-nucleosomal fragments, were prevalent in our data (Supplementary Figure SF3A). Across all samples, we identified a consensus set of 135044 non-overlapping NFRs ranging from 150–721bp that localized preferentially in the promoters and the non-coding genome (Supplementary Figure SF3B). For the identification of NFRs that represent DARs between the experimental groups, we opted to use the recently suggested stringent cut-offs (FDR < 0.01 and fold change > 50%) (10,11). In stark contrast with the RNA-seq results, there were almost no diet-induced NFRs in either strain while there was a plethora of DARs observed between the strains (strain-DARs): 8010 on HFD and 27487 on chow (Figure 3A, Supplementary Figure SF3C). Even with the more lenient cut-offs, only a few diet-induced DARs were observed in any comparison (Supplementary Table S3). Since it has been suggested that the NFRs flanked on both sides by the H3K27ac histone modification mark regions that participate actively in transcriptional regulation (9), we generated H3K27ac ChIP-seq data (4 mice per group) (Supplementary Table S1: Table 3) to be integrated with our ATAC-seq data. First, we identified a consensus set of 121119 genome regions marked by H3K27ac across all samples, which were mostly localized in the promoters and the non-coding genome (Supplementary Figure SF3D). The analysis of DHARs (FDR < 0.05) revealed that while the diet induced some changes (1482 DHARs for B6, 7580 for 129), mostly they were observed between the strains (22304 DHARs on HFD, 21835 on chow) (Figure 3B, Supplementary Figure SF3E).
Figure 3.
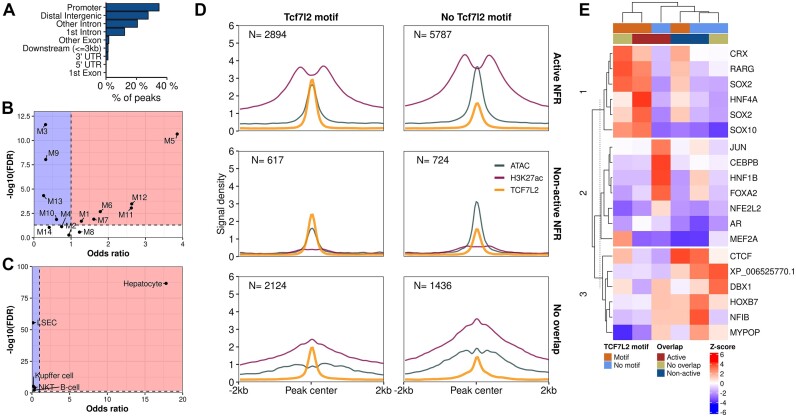
Differences in enhancer activity and accessibility between strains are dominant over diet-induced effects. (A) DAR (FDR < 0.001 and fold change > 50%) and (B) DHAR (FDR < 0.05) counts in the indicated comparisons. (C) Enrichment heatmap of ATAC-seq and H3K27ac signals at DARs observed between 129 and B6 mice on HFD. Rows are grouped to up and down differences between the strains at active and non-active regions. (D) Density plot of correlations between active NFRs (DARs and non-DARs) and the classes of histone acetylated regions of the H3K27ac valley summits (DHARs and non-DHARs). (E) Motif enrichment for group-wise active and non-active NFRs and H3K27Ac peaks. Heatmap columns and rows are clustered by Euclidean distance and columns are split using K-means clustering. Values shown are for the most significant motif for a given TF. Motifs for heatmap were selected by group-wise filtering of redundant motifs. (F, G) Venn diagrams of TCF7L2 (F) and CTCF (G) bound regions overlapping active and non-active NFRs.
Upon integration of the ATAC-seq and H3K27ac ChIP-seq data, 73646 (55%) of all NFRs were determined as active in at least one of our experimental groups. Of the active NFRs, 34761 (54%) were shared by all groups (Supplementary Figure SF3F). Most of the strain-DARs observed in either HFD or chow were in non-active regions (71% and 69%, respectively), highlighting the importance of identifying the active NFRs that represent active enhancers (Figure 3C, Supplementary Figure SF3G). In addition, ATAC- and H3K27ac signals were strongly positively correlated at the DARs that overlapped DHARs, highlighting the connection between strain-specific chromatin accessibility and enhancer activity (Figure 3D). Our integrative approach was further validated by the finding that active NFRs were, compared to the non-active NFRs or H3K27ac-only sites, more enriched for the motifs of well-known hepatic regulators, such as HNF4α, and enhancer binding proteins belonging to the CEBP-family (66). The inactive NFRs were most notably highly enriched for the motif of CTCF which is a known transcriptional insulator (Figure 3E) (67). ChIP-seq assays of individual TFs further validated the differences observed in the motif enrichment analysis. We chose TCF7L2 (2 mice per group) instead of a transcription factor arising from the enrichment analysis due to its’ prevalent role in the regulation of metabolic genes (22,23). We also performed ChIP-seq for CTCF (2 mice per group) to confirm its’ binding at non-active NFRs. TCF7L2 binding sites (N = 13571) primarily (63.7%) overlapped active NFRs (Figure 3F). In contrast, majority (71.9%) of CTCF binding was outside of both active and non-active NFRs. However, 24.0% of non-active NFRs were bound by CTCF, which is 1.7 times that of active NFRs (14.2%) (Figure 3G).
TCF7L2 binds primarily to NFRs classified as active enhancers hosting a binding motif
For a closer evaluation of how TF binding relates to the identified active NFRs, we performed additional analyses using the TCF7L2 ChIP-seq data (Supplementary Table S1: Table 4). Compared to NFRs and H3K27ac, TCF7L2 binding was even more prominently focused on promoters and intergenic regions (Figure 4A). Mapping TCF7L2 binding sites to the nearest TSSs of module-associated genes revealed that TCF7L2 binding was depleted at immune-associated modules M3 and 13 as well as LSEC specific gene enriched M9. Interestingly, TCF7L2 bound regions were enriched to metabolism and hepatocyte associated-related modules such as M5 and WNT-signalling associated M12 (Figure 4B). Additionally, the TCF7L2-bound regions were highly enriched to the loci of hepatocyte-specific genes confirming the previously documented role of TCF7L2 in the liver metabolism (Figure 4C) (22–24).
Figure 4.
Effects of location, chromatin state, and motif co-occupancy on overall TCF7L2 binding. (A) Enrichment of TCF7L2 binding sites to genomic features. Enrichment of TCF7L2-bound regions at the loci of (B) module-wise and (C) cell type -specific gene sets. (D) Mean signals of TCF7L2 ChIP-seq, ATAC-seq and H3K27ac ChIP-seq in different overlap classes of NFRs and TCF7L2 binding sites with and without a TCF7L2 motif. (E) Heatmap of motif enrichment in different TCF7L2 classes. Column splits are done using K-means clustering with the number of clusters determined using gap-statistic with 1-SE rule. Rows and columns are clustered using Euclidean distance.
As our approach in identifying TF binding sites that are altered by genetic variation is reliant on TF binding motifs, we scanned the active/non-active NFRs overlapping and non-overlapping TCF7L2 binding sites for three direct-evidence TCF7L2 binding motifs from the CISBP database. In all NFR overlap categories, the presence of a TCF7L2 binding motif associated with higher TCF7L2 signal density (Figure 4D). Interestingly, the dependency of TCF7L2 binding on the presence of TCF7L2 motif was the highest at non-overlapping sites (2124 of 3560 sites, 60%), second highest at non-active NFRs (617 of 1341 sites, 46%), and the lowest at active NFRs (2894 of 8681 sites, 33%). We also explored the differences in motif enrichment across the three different classes of TCF7L2 bound regions with and without TCF7L2 motif and observed three distinct motif clusters that defined the classes (Figure 4E). Even though we excluded the motifs of TCF7L2 and its’ close relatives from this enrichment analysis, Cluster 1, which defined all classes with a priori TCF7L2 motif, became populated by binding motifs belonging to the SOX-family TFs such as SOX10, as well as HNF4α, which all bear some similarity to the TCF7L2 motif used in the classification (50). The most important difference in motif enrichment patterns, however, was observed in Cluster 2, where the TCF7L2 peaks with no motif overlapping active NFRs were highly enriched with motifs for FOXA2, a known TCF7L2 co-binding factor, and CEBPB (27); this cluster may indeed identify TFs that assist TCF7L2 binding at sites that lack a clear TCF7L2 binding motif. Finally, as expected, Cluster 3 that defined TCF7L2 binding at non-active NFRs was enriched for CTCF motifs. These findings show that an in silico analysis based on a simple binary classification of active NFRs, while highly effective, appears to miss some of the true binding events.
Differentially accessible regions are enriched with genetic variation and connected with metabolism associated traits
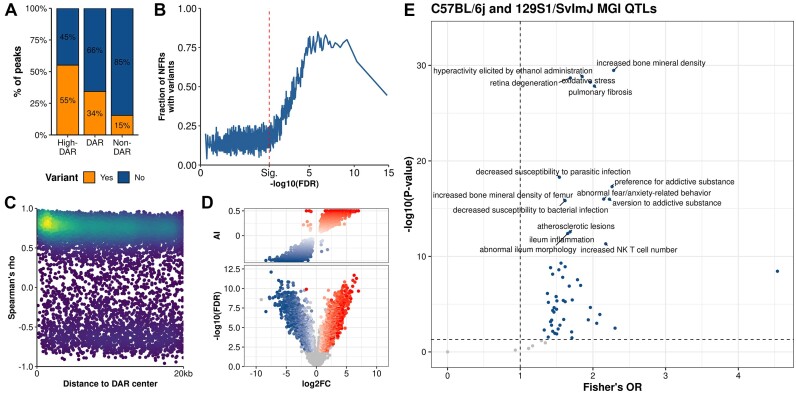
Since differences in chromatin accessibility were only observed between the strains, we combined the strain-DARs for both diets for further analysis of the role of genetic differences in differential accessibility. Compared to non-DARs, DARs overlapped genetic variation (MGPv5 SNPs, insertions and deletions) more often (Figure 5A; the breakdown of overlap counts per diet to variant class is given in Supplementary Figure SF4A). This was especially evident for high-confidence DARs (FDR < 0.001 and fold change > 50%) which overlapped with variants 3.7 times more often than did non-DARs (55 versus 15%, Fisher's Exact test P-value < 2.2 × 10−16). Furthermore, the fraction of DARs containing variants was higher at higher statistical significance levels suggesting a strong link between chromatin accessibility and genetic variation (Figure 5B). In addition, the most significant DARs (absolute fold-change > 50% and FDR < 10−5) more often had multiple variants (>3) compared to less significant DARs (Supplementary Figure SF4B). Since we observed differential accessibility to be closely connected to genetic variation, we hypothesized that the DARs without variants might not represent independent differences in accessibility, but rather reflect the presence of nearby DARs with variants. Of the DARs without SNPs 4564 (23.4%) were found within 20 kb of a DAR with variant(s) and 2791 (14.3% of the DARs without SNPs) had at least one DAR with SNP that presented strong positive correlation within this window (Spearman's rho > 0.7, P-value < 0.05) (Figure 5C; Supplementary Figure SF4C). In summary, genetic variation strongly affects chromatin accessibility directly but also propagates equidirectional effects onto the nearby chromatin landscape.
Figure 5.
Differentially accessible regions are enriched for genetic variants. (A) The proportions of combined high-confidence DARs (High-DAR, FDR < 0.001, fold change > 50%), DARs (FDR < 0.01, fold change > 50%), and non-DARs with (orange) and without (blue) overlapping genetic variants. (B) The fraction of ATAC-seq peaks containing at least one genetic variant (y-axis) in windows of 100 NFRs ordered by the significance of difference (x-axis). FDR is for the more statistically significant comparison (129 versus B6 on either chow or HFD). (C) Scatter plot displaying correlations of ATAC signal between DARs with SNPs and their neighbouring DARs without SNPs in ±20 kb windows around the NFR centres. Colouring scale indicates count density (from dark blue for low density to yellow for high density). (D) Overlaps of DARs and sites exhibiting AI. The upper plot presents the distribution of AI by logarithmic fold change (log2FC) for the more significant DAR of chow and HFD comparisons (x-axis). The lower volcano plot shows the log2FC (x-axis) and -log10(FDR) (y-axis) of sites from the strain-wise ATAC-seq differential accessibility analysis that overlap an F1 ATAC-seq site that passed the AI analysis. Sites with significant AI are shown, B6-dominant sites in white-to-blue and 129- dominant sites in white-to-red colour scale. AI: allelic imbalance. (E) Enrichment (Fisher's test) of DARs with variants to MGI QTL alleles associated with a phenotype in B6 and 129 QTLs, compared to non-DARs without variants.
To validate that it is genetics that underlies differential accessibility, we performed ATAC-seq on liver samples from chow-fed male 129xB6 F1-mice (N = 4). Within our consensus set of NFRs, 3688 exhibited allelic imbalance (AI) (Supplementary Figure SF4D), most of which (N = 2890, 78%) overlapped a strain-DAR (Supplementary Figure SF4E). Most notably, the strain whose allele exhibited dominance in AI was also almost always the strain for which the DAR was ‘up’ (Figure 5D). It should be noted that at comparable sequencing depths, AI analysis is less sensitive than DAR detection, which explains why many of our smaller DARs did not exhibit statistically significant AI. Secondly, the AI-analysis pipeline we used, or any other to our knowledge, cannot currently incorporate InDels into the analysis, which leads to the exclusion of those variant-overlapping DARs that only have InDels (6.6%). In summary, these results confirm the very strong association of differential chromatin accessibility and genetic variation.
After establishing the strong connection between genetic variation and chromatin accessibility, we explored the association of our DARs to the QTL-associated phenotypes obtained from the MGI database (56). Interestingly, DARs with variants were enriched (P-value < 0.05, compared to NFRs with variants using Fisher's test) to QTLs associated with metabolism and immune response, including the top hit for increased body weight (OR = 2.31, P-value = 2.58 × 10−274) (Supplementary Figure SF4F). Since these QTLs are derived from several different strains and crosses, we also explored the QTLs identified specifically from 129 and B6 strains. The results differed from the above ‘all MGI QTLs’ analysis, among the top hits being for oxidative stress (OR = 1.96, P-value = 5.62 × 10−29) and increased bone mineral density (OR = 2.03, P-value = 3.40 × 10−30) (Figure 5E). These findings could indicate that additional information on how genetics relates to phenotypes, including obesity, in these strains is there to be discovered.
Variants in binding motifs are predictive of differential TCF7L2 and CTCF binding
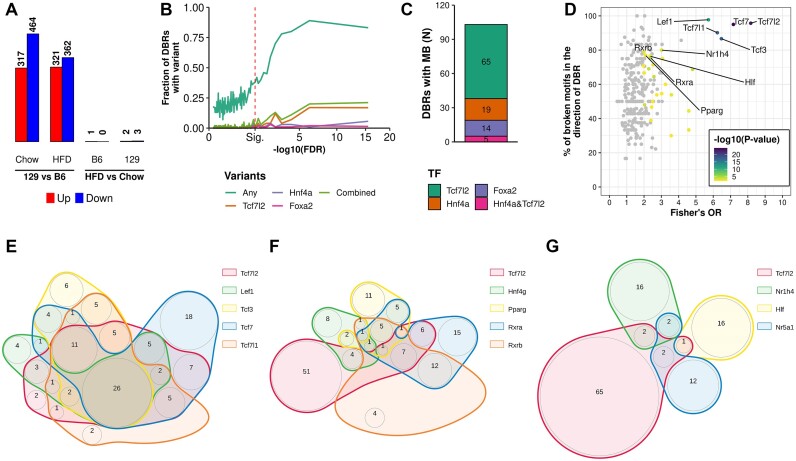
To directly evaluate the association between genetic variation and TF binding, we used TCF7L2 ChIP-seq data for B6 and 129 livers. Like DARs, almost all 1078 differentially bound regions (DBRs) for TCF7L2 were observed only between the strains (Figure 6A). Similarly, also the TCF7L2 strain-DBRs were enriched for variants, although only 17% of even the most significant DBRs (FDR < 6.5 × 10−7) contained any TCF7L2 motif-altering variants (Figure 6B). Even with the inclusion of altered motifs for known hepatic TCF7L2 co-binding factors, HNF4α and FOXA2 (27), only 24% of the highest-significance DBRs contained any of the included motif-altering variants. Genetic variants affecting binding motifs for TCF7L2 and HNF4α only had a minor overlap with the five TCF7L2 binding variants also affecting an HNF4α motif, suggesting that despite the motif similarity, the candidate rSNPs for these two TFs are mostly specific (Figure 6C).
Figure 6.
Effects of variants on strain-specific TF and TCF7L2 binding. (A) TCF7L2 DBR (FDR < 0.05) counts by comparison. (B) The fraction of TCF7L2 binding sites containing at least one genetic variant (‘Any’) or at least one motif-altering variant for TCF7L2 or a known co-binder for TCF7L2 in windows of 100 binding sites (y-axis) ordered by the significance of difference (x-axis). The displayed FDR-value is the highest per peak of the two strain comparisons (129 vs B6 on either chow or HFD). Combined: at least one of the TFs (TCF7L2, HNF4α and/or FOXA2) has an altered motif. (C) Counts for altered binding motifs for TCF7L2 and known co-binders in DBRs. (D) Comparison of variant-altered TF binding motifs between DBRs and non-DBRs using Fisher's test. TFs with P-value < 0.01 are coloured, and those with >80% of altered motifs with collateral motif score to DBR log2FC and P-value <0.01 are labelled. Venn diagrams of TCF7L2 peaks with altered binding for (E) TCF7L2 and TCF7L2-like motifs and (F, G) motifs not related to TCF7L2, significantly overrepresented in the Fisher's enrichment analysis.
Among all the 13571 TCF7L2 peaks, the variant-altered TCF7L2 motif frequently co-occurred with differential binding (67.3%; 70 DBRs among 104 peaks with variant-altered motif). Of note, for CTCF, for which the strain-specificity and motif-altering variant frequency of DBRs were very similar to those of TCF7L2 (Supplementary Figure SF5A, B), also the frequency of variant-altered motif co-occurrence with differential binding was similar (70.7%; total peaks: 82823, 1132 DBRs among 1602 peaks with variant-altered motif). When an alteration at any motif was allowed, the co-occurrence with DBRs was much less frequent: 22.1% for TCF7L2 and 32.0% for CTCF. Due to the fundamental functional differences between TCF7L2 and CTCF, these data suggest that the potential of TF-specific motif-altering variants to predict differential TF binding on ChIP-seq data is high.
Since altered binding motifs for the known co-binding TFs for TCF7L2 were only present in a fraction of DBRs, we explored more broadly the commonly occurring altered TF motifs in the DBRs. Unsurprisingly, the highest enrichment in DBRs over non-DBRs was observed for the binding motif for TCF7L2 and motifs highly similar to it (Figure 6D, Supplementary Figure SF6A). There was a large overlap between DBRs hosting altered TCF7L2 and TCF7L2-like motifs, and some instances where the DBR only hosted an altered TCF7L2-like motif (Figure 6E). There were also other altered motifs enriched to the DBRs (Fisher's test P-value < 0.01 and > 80% of altered motifs with collateral scores, i.e. higher motif score in the strain with higher TCF7L2 binding): HNF4γ, PPARγ, RXRα, RXRβ, NR5A1, NR1H4 and HLF. Even though the motifs of these TFs shared similarity with the TCF7L2 binding motif (Supplementary Figure SF6B), the sets of DBRs with altered motifs for these TFs were largely independent of the DBRs with altered TCF7L2 and TCF7L2-like motifs (Figure 6F-G). To further consolidate the view on altered motifs and TCF7L2 DBRs, we explored the concordance of the SNP-driven change in the motif score and the log2FC of TCF7L2 DBRs. Unsurprisingly, the motif for TCF7L2 itself showed the highest concordance, but also the other motifs arising from the enrichment analysis showed statistically significant positive correlation (Spearman's P-value < 0.05, Supplementary Figure SF6C). In some contrast, for CTCF DBRs the only enriched motifs where those for CTCF itself and its paralog CTCFL (Supplementary Figure SF7A). Collectively, these observations suggest that while the binding of TFs can also be affected by genetic variation at similar and possible co-binder TF motifs, the binding of TFs is mainly mediated by their own, specific binding motif.
Differential TCF7L2 binding is linked to differences in chromatin landscape
We next explored how the differential TCF7L2 binding is reflected onto the chromatin landscape. Among the TCF7L2 DBRs, 517 (48%) overlapped active NFRs, out of which approximately half (50.5%) were DARs. However, a large proportion of the DBRs overlapped neither active nor inactive NFR (37.8%) (Figure 7A). Similar to the pattern already seen with all TCF7L2 peaks (Figure 4D), these non-overlapping DBRs more often had a motif for TCF7L2 than the DBRs overlapping active NFRs (2.2-fold, 62.9% versus 28.5%) (Figure 7A). In addition, the DBRs very often correlated positively with overlapping chromosome accessibility at DARs as well as histone acetylation at DHARs (Figure 7B, C).
Figure 7.
Chromatin landscape of genetically determined transcription factor binding. (A) Signals of TCF7L2 ChIP-seq, ATAC-seq and H3K27ac ChIP-seq at NFRs with or without a TCF7L2 motif overlapping or not overlapping DBRs. Density plots of correlations between TCF7L2 DBRs and the classes of (B) active NFRs (DARs and non-DARs) and (C) H3K27ac regions (DHARs and non-DHARs). (D) Counts of TCF7L2 DBRs in different overlap categories. Three DBRs overlapping adjacent active and non-active NFRs were counted as active (1 active DAR and 2 active NFR). (E) Alluvial plot of DAR counts across study phases. (F) Comparison of variant-altered TF binding motifs between DARs and non-DARs using Fisher's test. TFs with P-value < 0.01 are coloured, and those with > 80% of altered motifs with collateral motif score to DBR log2FC and P-value < 0.01 are labelled. (G) Results from HNF4α ChIP-qPCR of altered HNF4 binding sites in DARs with ChIP-Atlas overlap. *, Mann–Whitney U-test P-value < 0.05; ns, non-significant.
Although the overall overlap between DBRs and DARs was relatively poor, we nevertheless evaluated the predictive potential of using motif-altering variants with differential chromatin accessibility data instead of differential binding data since ATAC-seq data is often easier to generate than TF-specific ChIP-seq data. In these analyses we used as the ground truth TCF7L2 and CTCF DBRs which are well predicted by respective motif alterations. Surprisingly, only 8.73% of all NFRs, or 10.71% of all DARs, with TCF7L2 motif-altering variants overlapped a TCF7L2 DBR (Table 1). When the NFR and DAR sets were further divided to active and non-active subset, the overlaps in active sets were clearly higher than in non-active sets, reaching 13.85% for active DARs. These results were contrasted by much higher overlaps between accessible chromatin with altered motif and CTCF DBRs (up to 58.5% for non-active DARs; Table 1), although in line with CTCF’s tendency to bind non-active NFRs, here the DBRs at non-active NFRs and DARs were better predicted than those at active NFRs and DARs. In summary, although candidate rSNPs clearly are identifiable based only on chromatin accessibility data, especially if integrated with histone mark data, the view remains incomplete in the absence of direct TF binding data in the form of e.g. ChIP-seq data.
Table 1.
Altered binding motifs overlapping TCF7L2 and CTCF DBRs across different region sets
| Motif-DBR overlapa | ||||
|---|---|---|---|---|
| Region set | TCF7L2 | CTCF | ||
| Altered Motif | TF-specific motif | NFR | 18/206 (8.74%) | 170/495 (34.34%) |
| Non-active NFR | 3/86 (3.49%) | 122/270 (45.19%) | ||
| Active NFR | 15/120 (12.50%) | 48/225 (21.33%) | ||
| DAR | 12/112 (10.71%) | 99/210 (47.14%) | ||
| Non-active DAR | 3/47 (6.38%) | 76/130 (58.46%) | ||
| Active DAR | 9/65 (13.85%) | 23/80 (28.75%) | ||
| TCF7L2 peak | 70/104 (67.31%) | 1132/1602 (70.66%) | ||
| Any motif | NFR | 315/20160 (1.56%) | 1370/20160 (6.80%) | |
| Non-active NFR | 65/9323 (0.70%) | 889/9323 (9.54%) | ||
| Active NFR | 250/10837 (2.31%) | 481/10 837 (4.44%) | ||
| DAR | 199/8266 (2.41%) | 642/8266 (7.77%) | ||
| Non-active DAR | 44/4163 (1.06%) | 400/4163 (9.61%) | ||
| Active DAR | 155/4103 (3.78%) | 242/4103 (5.90%) | ||
| TCF7L2 peak | 544/2461 (22.10%) | 6306/19734 (31.96%) | ||
aGiven as: altered motifs overlapping DBR/altered motifs in peaks (%).
We finally evaluated whether ChIP-seq data from public sources could allow the detection of variants altering TF binding. To identify potential rSNPs and their affected TFs, we first performed motifbreakR analysis for the active DARs with variants using the CIS-BP database motif collection (50,52). The analysis identified 7345 variants, which altered motifs for 461 TFs in 4103 of the 5097 active DARs with variants (Figure 7E). Among the most enriched altered motifs were those for HNF4α and other TFs shown to exhibit pioneer activity like ETV2 and CEBPA (68,69). Also TCF7L2 was enriched, although most of the altered TCF7L2 motifs were not in the direction of DAR (Figure 7F). We then evaluated these findings using mouse liver ChIP-seq data from ChIP-Atlas for the available TFs (55). Score changes at the 1352 motifs overlapping a DAR and a TF binding site from ChIP-Atlas (Supplementary Table S4) displayed stronger positive correlation with the log2FC of the overlapping DAR (Spearman's rho 0.49, P-value < 2.2 × 10−16) (Supplementary Figure SF7B), compared to all motif score changes observed at any DAR (Spearman's rho 0.15, P-value < 2.2 × 10−16) (Supplementary Figure SF7C). This suggests that ChIP-seq data from public sources might identify rSNPs using active DARs as a marker for altered TF binding. As an additional validation, we performed HNF4α ChIP-qPCR (N = 4, per strain) for eight sites that were predicted to have altered HNF4α binding by the motifbreakR analysis and overlapped active DARs that express allelic imbalance and locate near strain-DEGs (Supplementary Figure SF7D). All eight sites displayed high concordance with the predicted outcome, i.e. there was more HNF4α binding in the strain that had a stronger binding motif and higher accessibility (Figure 7G).
Differential accessibility and histone acetylation are predictive of nearest gene expression
Next, we aimed to identify how chromatin accessibility at NFRs, histone acetylation, and TF binding by TCF7L2 were associated to gene expression. To that end, active NFRs, H3K27ac regions and TCF7L2 bound regions were assigned to all expressed genes within ±1Mb, except for regions at active gene promoters which were only assigned to that gene. We then performed an enrichment analysis to the nearest DEGs across the different assays using chipenrich (Figure 8A), on the background of expressed genes. Active non-DARs were enriched to all DEG classes likely due to the known, general association of active chromatin accessibility and gene expression. On the other hand, strain-DARs were enriched especially to strain-DEGs and, as expected, not enriched to diet-DEGs. Additionally, active NFRs, both within DAR and non-DAR, showed higher enrichment to different DEG-categories compared to the respective non-active class, further solidifying the added benefit of the categorization of NFRs. The same pattern was observed with a more basic peak density approach: Active DARs populated the proximal regions of the strain and ‘diet + strain’-DEGs more densely than the non-active DARs (Figure 8B). DHARs also displayed an expected pattern as each class of DHARs was most enriched to the corresponding class of DEGs (Figure 8A). Strain-DBRs and non-DBRs had a largely similar pattern than their active DAR counterparts. The enrichment of non-DBRs to diet-DEGs and ‘diet + strain’-DEGs may reflect the role of TCF7L2 in metabolic pathways affected by the HFD (Figure 8A). Interestingly, the density of DBRs near strain-DEGs was observed to be much higher for those DBRs that contained variants compared to those that had no variants (Figure 8B).
Figure 8.
Relationships between DEGs and DARs, variants and TF binding motifs. (A) chipenrich enrichment analysis results for different assays. Non-DEGs were included in the background. (B) Densities of the distances between peaks of the different classes and their nearest DEG of every class. (C) Heatmap of the fractions of DAR, DHAR and DBR peaks with significant correlation (Spearman's P-value < 0.05) with the nearest gene in each DEG-class. (D) Heatmap of motif enrichment in the nearest active NFRs of module-associated genes. Rows and columns are clustered using Euclidean distance. Only the most relevant TFs (Z-score > 3 in at least one module) are shown. (E) Cumulative distribution plots for the absolute distance between the active DAR (top panel), DHAR (middle panel) and DBR (bottom panel) peaks and the nearest DEG of the same class. Y-axis depicts the proportion out of all the DEGs in the class. (B, C) Assay-class: Differential analysis annotation for NFRs, H3K27ac and DBRs. Strain: different between strains only, Diet + strain: difference observed in both inter-strain and intra-strain diet comparisons, and Diet: different only between diets.
Based on our findings that TCF7L2 preferentially binds at active NFRs (Figure 4D) and active DARs show enrichment and proximity to DEGs (Figure 8A, B), we focused on active NFRs to assess how the regions identified by the different assays correlate with the expression of their nearest DEGs (Figure 8C). Strain-DARs most often correlated (Spearman's P-value < 0.05) with the expression of their nearest strain-DEGs, especially when the DAR hosted a variant (with variant: 65.4%, without: 46.3%). The mixed-effect ‘diet + strain’-DEGs correlated less often (with variant: 48.2%, without: 33.2%) whereas the diet-DEGs and expressed non-DEGs rarely correlated. DHARs and DEGs correlated the most within the corresponding class. TCF7L2 DBRs more rarely correlated with their nearest DEGs: only 31.0% of strain-DBRs with variant correlated with the nearest strain-DEGs (Figure 8C).
Since co-expression is thought to represent co-regulation, we performed motif enrichment analysis with the closest active NFRs of module-associated genes to identify potential direct regulators and found that the modules clustered mostly according to their likely originating cell type. Immune modules M3 and 13 were more enriched for motifs for known immune-related TFs like the interferon regulatory factors (IRF) (70), while hepatocyte modules M1 and M5 were more enriched for e.g. HNF4γ (M1) and HNF1β (M1 & 5) (Figure 8D).
Finally, we assessed the degree of differential gene expression that could be explained by the different datasets. We linked every DEG to their closest (within 1Mb) correlating active-DAR, DHAR and DBR of the same class (e.g. diet-DEG to diet-DHAR). Additionally, DARs and DBRs were also linked to ‘diet + strain’-DEGs. Strain comparisons were highlighted in this approach (Figure 8E). Correlating strain-DHARs were found in the ±1 Mb neighbourhoods of 95% of the strain-DEGs, which was the highest percentage among all the assays. In addition, 92% of strain-DEGs had co-correlating active DARs in the neighbourhood. As expected, among all DEGs fewer genes had a co-correlating DBR in the neighbourhood, reflecting the inherently smaller target gene set of a single TF (Figure 8E). Altogether, these results show that especially the inter-strain differences in the chromatin landscape underlie differential gene expression.
Linking rSNP candidates to DEGs reveals both known and novel regulatory interactions between TFs and genes
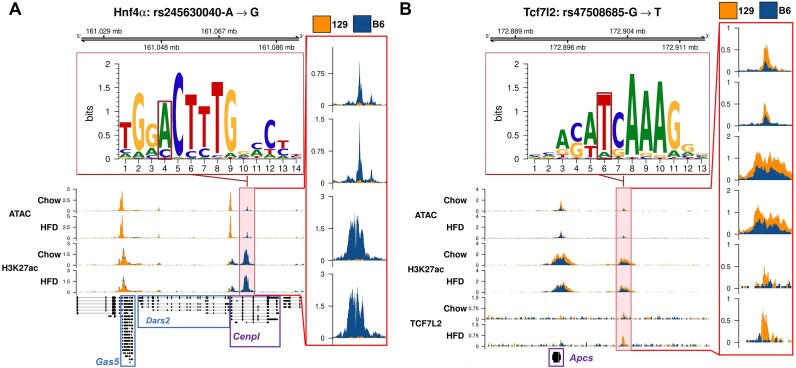
After observing that differential accessibility at active NFRs is strongly linked to differential gene expression and that the closest active NFRs often contain relevant TF binding motifs, we aimed to identify possible targets of that regulatory variation. We linked active DARs with motif breaking variants to their closest correlating (Spearman's P-value < 0.05) DEG of either ‘strain’ or ‘diet + strain’-class (Supplementary Table S5A). To establish whether there were known TF-gene interaction pairs within these connections, we used the TRRUSTv2 TF-gene interaction database (71). There were 23 known interactions of TFs and target genes overlapping our data. In addition, based on ChIP-Atlas data, 694 of our candidate rSNPs overlapped with a binding site of the corresponding TF. For example, an active B6-up DAR upstream of Cenpl overlapped an HNF4α binding site from ChIP-Atlas and hosted rs45630040 whose G allele of 129 mice presents a weaker binding motif for HNF4α (Figure 9A). Additionally, this locus also hosted rs258291433 whose T allele in 129 mice also weakens the binding motif for HNF4α. Cenpl was also more expressed in B6 mice, strengthening the case of HNF4α-driven regulation of Cenpl (Supplementary Figure SF8A). The Cenpl locus also hosted two other B6-upregulated DEGs (Gas5 and Dars2), suggesting a broader local regulation mediated through this enhancer region, and affected by the two rSNPs. Roles of both Cenpl and Dars2 in liver have not been explored outside of hepatocellular carcinoma but Gas5 has been shown to play a critical role in non-alcoholic fatty liver disease (72–74).
Figure 9.
Example loci of candidate rSNPs for known hepatic regulators and their target genes. (A) Candidate rSNP locus where rs245630040 alters a known binding motif for HNF4α in an active DAR overlapping HNF4α from ChIP-Atlas that correlates with Cenpl expression. Additional correlating DEGs in the neighbourhood include Dars2 and Gas5. (B) A DBR-DAR near the Apcs gene, which hosts rs47508685 that alters the binding motif for TCF7L2. (A, B) Gene names of inferred target DEGs are shown and their transcripts boxed in purple. Blue boxes indicate a down-regulated DEG which was not directly inferred as the target by the analysis. The insets on the right display zoomed views into the indicated data tracks of the main plot highlighted in pale red background.
We also overlapped altered TCF7L2 motifs in active DARs with TCF7L2 DBRs at DEG loci and identified eight overlapping candidate TCF7L2 rSNPs (Supplementary Table S5B). At seven of these SNPs, the difference in allele score was to the same direction as the change in TCF7L2 binding and in six of the sites the DAR fold change was in the same direction as the DBR fold change. Interestingly, half of the genes presented positive correlation, potentially supporting the proposed dual role of TCF7L2 as both transcriptional activator and repressor (75). An example of a likely TCF7L2 rSNP is rs47508685 that is located 8.4kb upstream of the 129-up DEG Apcs and whose T allele of 129 mice creates a TCF7L2 binding motif in a 129-up strain-DAR that overlaps a 129-up TCF7L2 DBR (Figure 9B, Supplementary Figure SF8A). Interestingly, Apcs has been previously recognized as a candidate gene for body fat percentage related QTL in an F2-cross of 129 and B6 mice (8). Additionally, the protein product encoded by this gene, serum amyloid P, has been recently shown to be of great importance in reducing steatosis and inflammation in mice. Apcs has also been recognized as a possible inhibitor of obesity-induced effects in humans (76).
Additionally, we linked TCF7L2 DBRs with motif altering variants to their closest correlating DEG of either ‘strain’ or ‘diet + strain’-class to identify additional candidate rSNPs not found within active DARs (Supplementary Table S5C). This analysis yielded 57 SNPs in 54 DBRs within 1Mb of DEGs that hosted a TCF7L2 altering variant. Of the DBRs, 14 overlapped with DARs where in six the motif altering variant was observed within the TCF7L2 DBR and not the active DAR. One interesting candidate rSNP, rs238427830, creating a stronger binding motif in 129, was found 17 kb upstream of Ttc39b, which is a 129-up and HFD-down ‘diet + strain’-DEG suggesting that TCF7L2 may directly regulate Ttc39b (Supplementary Figure SF8A, B). Interestingly, Ttc39b has been suggested to participate in the regulation of lipid homeostasis through trans-activating LXRβ mediated signalling cascades (77).
Human eQTL SNPs of the candidate rSNP locus gene orthologues are connected to liver-specific metabolic functions
Lastly, we explored the possibility to infer potential mechanisms of action for human rSNPs based on our candidate mouse rSNPs. First, we lifted the active DARs overlapping SNPs that broke any motif to the human genome using liftOver, then identified all human SNPs in these syntenic regions from the EBI GWAS catalogue, and finally used motifbreakR to identify the human TF motifs targeted by these SNPs (52,62,64). This approach yielded 10 human SNPs for the 12 mouse SNPs in 10 active DARs which altered the motif for the same TF as in mouse (Supplementary Table S6A). Interestingly, these SNPs were associated with various metabolic traits. For example, rs36991149, overlapping an active DAR, and its human counterpart rs2070895 both altered the binding motif of PAX7. In humans, rs2070895 is associated with several GWAS traits related to cholesterol metabolism.
For the second approach, since TFs and their target gene sets are well conserved between mice and humans (17,18), we paired the TFs with genetically altered motifs in active DARs to their nearest genes and mapped these TF-gene pairs to human liver eQTL data (65). The analysis yielded 1280 eQTL SNPs in the neighbourhoods of 137 genes that altered the motif of the same TF than in mouse and that locate near an orthologous human gene, 53 of which overlapped with the corresponding human TF binding site from ChIP-Atlas (Supplementary Table S6B). It should be noted here that the overlap between the GTEx eQTL SNPs and ChIP-Atlas tracks is poor: only 13.2% (53923 out of 407285) of the liver eQTL SNPs overlap with any liver associated ChIP-Atlas track for TFs. We also queried the eQTL SNP hits from the EBI GWAS catalogue and identified GWAS annotation for 113 of them (Supplementary Table S6C). Interestingly, also these were often related to metabolism. For example, the human rs3809898 that alters an RARγ binding site associated with TNKS, linked to low density lipoprotein cholesterol levels, has a mouse counterpart rs30303466 that overlaps an active DAR and both locates near the mouse orthologue Tnks and alters the binding motif for RARγ.
Out of our 1280 eQTL SNP hits, 44 overlapped a TCF7L2 DBR in mouse but none altered the binding motif for TCF7L2 (Supplementary Table S6D). However, four mouse variants (rs245402086, rs223056549, rs235169540 and rs579006641) altered the binding motifs for FOXM1 and FOXJ3 which are relatives of FOXA2, a known liver co-binding factor of TCF7L2 (27). These variants had eight human eQTL SNP counterparts also altering one of either FOXM1 or FOXJ3 binding motifs. Finally, we checked whether TCF7L2 DBRs with TCF7L2 altering variants could be found near eGenes with linked TCF7L2 motif-altering variant. We identified one eQTL SNP with TCF7L2-altering motif found near RAPGEF5 (Supplementary Table S6E). Interestingly, Rapgef5 had an upstream TCF7L2 DBR where an insertion creates a weaker binding motif in 129 mice, coinciding with weaker TCF7L2 binding in 129 mice. In summary, using a strategy based on functional conservation of TF-gene pairs, our analysis short-listed several hundred human SNPs, including many metabolic GWAS hits, as rSNP candidates for further exploration and functional validation.
Discussion
Even though B6 and 129 mice have been commonly used to study diet-induced obesity, the genetically driven differences in their gene regulatory networks are not well understood (4). Here we present our findings on how genetic variation, as observed in the livers of B6 and 129 mice, is linked to differences in the chromatin landscape and how these relate to the regulation of gene expression. We also propose functionally likely causal rSNPs based on an integrative analysis of genome-wide assays of gene expression and epigenetic markers and evaluate the outcomes by using TCF7L2 binding as an example.
As revealed by both cell type specific gene enrichment analysis of module-associated genes and, to a minor extent, TF motif enrichment analysis of their neighbouring active regulatory regions, differences in the cell-type composition of the two mouse strains affect some of our analyses. Based on our data, the analyses prone to this kind of an effect are those that rely on the correlation of signals, such as WGCNA, in which the genes co-expressed in their specific cell types appear to drive the co-expression module generation. Another analysis affected by cell type specific signals was the motif enrichment analysis for active NFRs neighbouring expressed genes. These findings indicate that studying the effects of genetic variation and TF binding would greatly benefit from single cell -based approaches. These studies could additionally provide interesting prospects for studying the differing obesity predisposition in 129 and B6 mice. For example, higher proportion of Kupffer cells in the B6 livers may explain the HFD-induced insulin resistance phenotype observed in these mice because Kupffer cell activation is known to contribute to hepatic insulin resistance (4,78).
The integration of chromatin accessibility and active enhancer marker H3K27ac data allows for the segregation of functionally meaningful subsets of enhancers (9). Building on that, we report here that classifying NFRs as active and non-active refines the identification of TF binding sites hosting binding motifs for relevant TFs. In addition, based on the enrichment of TF binding motifs, the NFRs classified as active outperform the H3K27ac-only approach in accurately identifying TF binding sites. However, it should be noted that even though the binary classification we used here was shown to be effective, it may not fully address the complexity of enhancer types. Indeed, a recent publication by Sahu et al. describes four types of enhancers which can present both accessible chromatin and H3K27ac or neither (15). These enhancers are very often marked by a signal from self-transcribing active regulatory region sequencing (STARR-seq), which could be a robust addition for similar analyses in the future. In addition, they describe a class of chromatin-dependent enhancers where CTCF binding co-locates with accessible chromatin and H3K27ac, validating the small overlap of CTCF binding sites and active NFRs seen in our analysis.
TF binding, as exemplified by our TCF7L2 ChIP-seq data, can also occur at inaccessible chromatin if the TF has pioneer activity (20). In our study, TCF7L2 frequently bound to chromatin that was inaccessible but still marked by H3K27ac. As TCF7L2 has been recently shown to have a dual role as both a transcriptional activator and repressor, this observation could be indicative of transcriptional repression or highlighting a mechanism for pioneer activity by TCF7L2 (79). Moreover, we observed that there were differences in the motif enrichment patterns across the TCF7L2 overlap classes, hinting further towards the possibility of different functional roles that depend, site by site, on the local chromatin context. These findings underline the importance of also including specific TF ChIP-seq data when searching for, or validating, high-confidence rSNP candidates.
The differences we observed across all the genome-wide assays were much more pronounced between the strains compared to the effects of HFD. In fact, chromatin accessibility measured by ATAC-seq was non-responsive to diet in both B6 and 129 mice, similar to a previous report on B6 mice and using DNAse-seq (6). The observed high occurrence of genetic variants in DARs, and their validation by the allelic imbalance analysis of the F1 livers, identifies genetics as the key determinant of chromatin accessibility. In addition, the high positive correlation of the DARs without SNPs in the close proximity of DARs with SNPs highlights a cis-acting role of chromatin accessibility and genetic variation. These observations align with previous reports on the heritability of chromatin accessibility (80). Nevertheless, some of the DARs without variants had no nearby DARs with variants. Similar incomplete overlap of strain-specific sites and local variants has been previously described (17). This, together with the incomplete overlap of TCF7L2 DBRs with active DARs, highlights the need for future studies on the degree at which the differences in chromatin accessibility are mediated by e.g. pre-established enhancers during liver development, and the degree at which they are related to the pioneer activity of TFs in the mature liver (6,20,81).
We assessed how well altered TCF7L2 and CTCF binding motifs can be used to predict differential binding events in NFRs, DARs and with TF-specific ChIP-seq derived binding sites. To no surprise, TF specific ChIP-seq predicted rSNP candidates better than chromatin accessibility data. In addition, while altered CTCF binding motifs provided quite good results in predicting CTCF DBRs, most of the CTCF binding located outside accessible chromatin, leaving multiple rSNPs outside the DAR-based analysis. Based on our findings, TF-specific ChIP-seq is a necessity at the very least for the validation of rSNP candidates. This was further supported by the improved correlation between the DAR fold changes and altered motif scores for altered motifs that overlap their corresponding TF binding sites from the ChIP-Atlas database and HNF4α ChIP-qPCR (55). Indeed, an interesting future study could be to assess how well public data, such as the ChIP-Atlas data, aligns with in silico identified rSNP candidates. In addition, an interesting avenue for future studies would be to compare genome-wide the induced accessibilities at genetically altered binding sites between e.g. TCF7L2, a hepatocyte-enriched, functionally diverse TF, and the TFs with pioneer activity like HNF, ETV/ETS or CEBP family-related TFs that were overrepresented in the DARs (20,68,69,75). This would help to identify the TFs that modify the liver accessibility landscape and characterize how this reflects to the observed phenotype since pioneer factors have been shown to be the prominent modifiers of local accessibility (20,82).
Like ATAC-seq, also altered TCF7L2 binding displayed a strong connection with genetic variation and minimal diet-induced effects. A large majority of the genetic effects, however, appear unspecific for a given TF, since only approximately 15% of the most significant TCF7L2 DBRs contained variants that specifically altered the binding motif for TCF7L2. This is in line with a previous observation by Soccio et al. who reported that only 20% of the strain-selective PPARγ-bound regions in white adipose tissue contained motif altering variants (19). One possible explanation for the apparently unspecific genetic effects could be that they target co-binders of the TF and thereby indirectly also affect the measured binding the TF itself. However, based on our enrichment analysis of altered binding motifs in TCF7L2 DBRs, it would seem that TF specific binding motifs are most predictive of the TF’s binding. In addition, differences between TFs in this regard are present as can be seen by comparing the enrichment results of altered binding motifs of TCF7L2 and CTCF DBRs. The high specificity of altered binding motifs in CTCF DBRs could be due to its solitary role as a chromatin domain regulator whereas TCF7L2 is known to have multiple co-binding TFs (27). Nevertheless, since altered binding motifs explain only a minority of the strain-specific binding of any TF, additional strategies are still needed to characterize the mechanisms behind the strain-selective TF binding events.
We also explored how our individual assays relate to the observed differences in gene expression. Inter-strain differences were observed to be the most concordant. This was especially true for DARs which, when compared to other assays, more often correlated with their nearest strain-DEG. In addition, for all assays the regions with variants presented higher likelihood of correlation compared to regions without variants. However, it is important to note that not all DEGs had DARs and DHARs in their neighbourhood. This is in line with a recent study by Zhang et al. where the depletion of H3K27ac signal at enhancers had only minimal effects on gene expression (83). However, in our analysis, correlating strain-DHARs were present within 1Mb of almost all strain-DEGs. Overall, the strong concordance between inter-strain enhancer accessibility and activity to gene expression, compared to the diet-induced differences, suggests that genetically determined enhancer activation is a major driver of the gene regulatory landscape in 129 and B6 mice. However, even though assigning the nearest gene to each genomic region has been widely used, and also provided well-aligned results in our study setting, the use of methods like chromatin conformation capture that directly link regulatory regions to target genes could assist in identifying targets of regulatory variation (84). For example, as seen with our TCF7L2 data, only a fraction of DEGs were observed to be correlating within 1Mb of DBRs. This might suggest a more distal action of regulation. Inferring distal regulatory connections in silico would greatly benefit from chromatin architecture data. Alternatively, together with the high enrichment of TCF7L2 binding sites to hepatocyte-specific genes, this may mean that TCF7L2 has fairly few target genes. Yet another possibility is that TCF7L2 binds only is certain cell types and if so, likely hepatocytes. This could also explain why the presence of genetically affected motifs in DARs or NFRs predicted TCF7L2 DBRs less well than CTCF DBRs; a ubiquitous binder like CTCF would tend to bind in chromatin that is accessible in many cell types whereas cell type specific binding might focus on chromatin only accessible in some cell types, which could additionally limit the detectability of cell type specific NFRs because in a bulk tissue sample their ATAC signal becomes ‘diluted" by the many other cell types.
Previous QTL studies of the 129 and B6 strains have described genomic regions related to the phenotypic differences observed also in our mice (7,8). For example, a study on 129P3/J and C57BL/6ByJ mice, close relatives to our 129 and B6 strains, respectively, identified QTLs driving dietary obesity (7). Although these studies used SNP arrays that are low-resolution by modern standards, several QTLs overlapped our DARs with variants. This warrants a closer look, using e.g. higher-density genotyping arrays, onto the genetic variation that relates to metabolism in these mice. In addition, we describe an interesting binding site hosting motif-altering variant for TCF7L2 near Apcs, a gene that has been documented as a candidate gene for the genetic obesity predisposition in B6 mice by multiple studies (8,85), and the human orthologue of which is reported to have anti-obesity properties (76). Since the Apcs knockout studies, to our knowledge, have only been performed on persistent lung inflammation and fibrosis, further investigation on the role of Apcs in obesity, liver metabolism, and regulation by TCF7L2, seem warranted (86). It should be noted that even though we present large-scale association of DARs and genetic variation that involves a multitude of variants, it remains a remote possibility that much of what we see is secondary to the effects by just a few driver variants.
Sequence conservation at open chromatin between human and mouse is reportedly 10–20% (87). Thus, it was no surprise that chromatin accessibility proved a poor inter-species mapping strategy between candidate mouse rSNPs and the human GWAS hits. However, our mouse DARs with candidate rSNPs near DEGs hosted many variants that mapped to human liver eQTL SNP-gene pairs in which the SNP affected the binding motif for the same TF as in mice. Moreover, these candidate ‘shared eQTLs’ also included several GWAS hits relevant for the metabolic functions of the liver, and even for the observed phenotypic differences between the mouse strains. Interestingly, one of the themes that arose from both sequence conservation and eQTL analysis was bone mineral density (BMD). In addition, BMD was one of the top hits from the QTL enrichment analysis of DARs with variants. Interestingly, BMD has been connected to circulating LDL and HDL cholesterol levels (88,89), but the causal relationships between the two phenomena are still largely unclear. Based on our findings, and supported by previous QTL studies, 129 and B6 mice could provide an interesting platform for studying the genetic determinants of BMD and blood cholesterol that could then be translated to the human setting. One limiting factor in such studies would be the presently low overlap of the human ChIP-Atlas data and the human SNP, which speaks for the need of liver ChIP-seq data for additional TFs.
In conclusion, differences in chromatin accessibility in 129 and B6 mice are enriched for genetic variation and associated with differences observed in the hepatic gene regulatory landscape. Both H3K27ac at the DARs, and expression of the nearest DEGs of the DARs often correlate with chromatin accessibility. The extent by which the observed differences in the chromatin accessibility are due to developmentally pre-established enhancers and how this relates to strain-specific TF binding events needs further clarification. We also show that while active DARs can be used to identify candidate rSNPs, better results in the identification of genetically determined binding sites are achieved using TF specific ChIP-seq. The data presented in this study offers several leads for investigating potentially causal liver rSNPs that are shared between mice and humans. Additionally, we provide targets of interest in identifying genetic determinants of the obesity predisposition in B6 mice. Our study also highlights TCF7L2 as an interesting, possibly hepatocyte-specific candidate for further studies into the transcriptional regulation of hepatic metabolism.
Supplementary Material
Acknowledgements
Analyses were performed on the high-performance computing cluster provided by UEF Bioinformatics Center, University of Eastern Finland, Finland. Sequencing was performed using the facilities of the Genome Center of the University of Eastern Finland. We thank assist. prof. Luke Norton, UT Health San Antonio, USA, for constructive comments.
Contributor Information
Juho Mononen, Institute of Biomedicine, Faculty of Health Sciences, University of Eastern Finland, Kuopio FI-70211, Finland.
Mari Taipale, A.I. Virtanen Institute, Faculty of Health Sciences, University of Eastern Finland, Kuopio FI-70211, Finland.
Marjo Malinen, Department of Environmental and Biological Sciences, Faculty of Science and Forestry, University of Eastern Finland, Joensuu FI- 80101, Finland; Department of Forestry and Environmental Engineering, South-Eastern Finland University of Applied Sciences, Kouvola FI-45100, Finland.
Bharadwaja Velidendla, Institute of Biomedicine, Faculty of Health Sciences, University of Eastern Finland, Kuopio FI-70211, Finland.
Einari Niskanen, Institute of Biomedicine, Faculty of Health Sciences, University of Eastern Finland, Kuopio FI-70211, Finland.
Anna-Liisa Levonen, A.I. Virtanen Institute, Faculty of Health Sciences, University of Eastern Finland, Kuopio FI-70211, Finland.
Anna-Kaisa Ruotsalainen, A.I. Virtanen Institute, Faculty of Health Sciences, University of Eastern Finland, Kuopio FI-70211, Finland.
Sami Heikkinen, Institute of Biomedicine, Faculty of Health Sciences, University of Eastern Finland, Kuopio FI-70211, Finland.
Data availability
The datasets supporting the conclusions of this article are available in the NCBI GEO repository, under identifier GSE196942. The bioinformatics scripts used in this work are available in Zenodo at https://doi.org/10.5281/zenodo.10355100. The ChIP-qPCR primers are available on request.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Academy of Finland [308875]; Finnish Diabetes Research Foundation (1-year grant). Funding for open access charge: Institute of Biomedicine, University of Eastern Finland.
Conflict of interest statement. None declared.
References
- 1. Yengo L., Sidorenko J., Kemper K.E., Zheng Z., Wood A.R., Weedon M.N., Frayling T.M., Hirschhorn J., Yang J., Visscher P.M.. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 2018; 27:3641–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yue F., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D., Shen Y.et al.. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014; 515:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Almind K., Kahn C.R.. Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes. 2004; 53:3274–3285. [DOI] [PubMed] [Google Scholar]
- 4. Chu D.-T., Malinowska E., Jura M., Kozak L.P.. C57BL/6J mice as a polygenic developmental model of diet-induced obesity. Physiol. Rep. 2017; 5:e13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Siersbæk M.S., Ditzel N., Hejbøl E.K., Præstholm S.M., Markussen L.K., Avolio F., Li L., Lehtonen L., Hansen A.K., Schrøder H.D.et al.. C57BL/6J substrain differences in response to high-fat diet intervention. Sci. Rep. 2020; 10:14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siersbæk M., Varticovski L., Yang S., Baek S., Nielsen R., Mandrup S., Hager G.L., Chung J.H., Grøntved L.. High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss. Sci. Rep. 2017; 7:40220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin C., Theodorides M.L., McDaniel A.H., Tordoff M.G., Zhang Q., Li X., Bosak N., Bachmanov A.A., Reed D.R.. QTL analysis of dietary obesity in C57BL/6byj X 129P3/J F2 mice: diet- and sex-dependent effects. PLoS One. 2013; 8:e68776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Su Z., Korstanje R., Tsaih S., Paigen B.. Candidate genes for obesity revealed from a C57BL/6J x129S1/SvImJ intercross. Int. J. Obes. (Lond). 2008; 32:1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu H., Duncan K., Helverson A., Kumari P., Mumm C., Xiao Y., Carlson J.C., Darbellay F., Visel A., Leslie E.et al.. Analysis of zebrafish periderm enhancers facilitates identification of a regulatory variant near human KRT8/18. Elife. 2020; 9:e51325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reske J.J., Wilson M.R., Chandler R.L.. ATAC-seq normalization method can significantly affect differential accessibility analysis and interpretation. Epigenetics Chromatin. 2020; 13:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gontarz P., Fu S., Xing X., Liu S., Miao B., Bazylianska V., Sharma A., Madden P., Cates K., Yoo A.et al.. Comparison of differential accessibility analysis strategies for ATAC-seq data. Sci. Rep. 2020; 10:10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tarbell E.D., Liu T.. HMMRATAC: a hidden Markov ModeleR for ATAC-seq. Nucleic Acids Res. 2019; 47:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A.et al.. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Herrera-Uribe J., Liu H., Byrne K.A., Bond Z.F., Loving C.L., Tuggle C.K.. Changes in H3K27ac at gene regulatory regions in porcine alveolar macrophages following LPS or PolyIC exposure. Front. Genet. 2020; 11:817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sahu B., Hartonen T., Pihlajamaa P., Wei B., Dave K., Zhu F., Kaasinen E., Lidschreiber K., Lidschreiber M., Daub C.O.et al.. Sequence determinants of human gene regulatory elements. Nat. Genet. 2022; 54:283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keller M.P., Rabaglia M.E., Schueler K.L., Stapleton D.S., Gatti D.M., Vincent M., Mitok K.A., Wang Z., Ishimura T., Simonett S.P.et al.. Gene loci associated with insulin secretion in islets from nondiabetic mice. J. Clin. Invest. 2019; 129:4419–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Link V.M., Duttke S.H., Chun H.B., Holtman I.R., Westin E., Hoeksema M.A., Abe Y., Skola D., Romanoski C.E., Tao J.et al.. Analysis of genetically diverse macrophages reveals local and domain-wide mechanisms that control transcription factor binding and function. Cell. 2018; 173:1796–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J.et al.. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015; 518:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Soccio R.E., Chen E.R., Rajapurkar S.R., Safabakhsh P., Marinis J.M., Dispirito J.R., Emmett M.J., Briggs E.R., Fang B., Everett L.J.et al.. Genetic variation determines pparγ function and anti-diabetic drug response In vivo. Cell. 2015; 162:33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hansen J.L., Cohen B.A.. A quantitative metric of pioneer activity reveals that HNF4A has stronger in vivo pioneer activity than FOXA1. Genome Biol. 2022; 23:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen X., Ayala I., Shannon C., Fourcaudot M., Acharya N.K., Jenkinson C.P., Heikkinen S., Norton L.. The diabetes gene and wnt pathway effector TCF7L2 regulates adipocyte development and function. Diabetes. 2018; 67:554–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Norton L., Chen X., Fourcaudot M., Acharya N.K., DeFronzo R.A., Heikkinen S.. The mechanisms of genome-wide target gene regulation by TCF7L2 in liver cells. Nucleic Acids. Res. 2014; 42:13646–13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oh K.-J., Park J., Kim S.S., Oh H., Choi C.S., Koo S.-H.. TCF7L2 Modulates glucose homeostasis by regulating CREB- and FoxO1-dependent transcriptional pathway in the liver. PLoS Genet. 2012; 8:e1002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee D.S., An T.H., Kim H., Jung E., Kim G., Oh S.Y., Kim J.S., Chun H.J., Jung J., Lee E.-W.et al.. Tcf7l2 in hepatocytes regulates de novo lipogenesis in diet-induced non-alcoholic fatty liver disease in mice. Diabetologia. 2023; 66:931–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boj S.F., van Es J.H., Huch M., Li V.S.W., José A., Hatzis P., Mokry M., Haegebarth A., van den Born M., Chambon P.et al.. Diabetes risk gene and wnt effector Tcf7l2/TCF4 controls hepatic response to perinatal and adult metabolic demand. Cell. 2012; 151:1595–1607. [DOI] [PubMed] [Google Scholar]
- 26. Neve B., Le Bacquer O., Caron S., Huyvaert M., Leloire A., Poulain-Godefroy O., Lecoeur C., Pattou F., Staels B., Froguel P.. Alternative human liver transcripts of TCF7L2 bind to the gluconeogenesis regulator HNF4α at the protein level. Diabetologia. 2014; 57:785–796. [DOI] [PubMed] [Google Scholar]
- 27. Frietze S., Wang R., Yao L., Tak Y.G., Ye Z., Gaddis M., Witt H., Farnham P.J., Jin V.X.. Cell type-specific binding patterns reveal that TCF7L2 can be tethered to the genome by association with GATA3. Genome Biol. 2012; 13:R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Corces M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B.et al.. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 2017; 14:959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J.. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013; 10:1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gustafsson C., Paepe A.D., Schmidl C., Månsson R.. High-throughput ChIPmentation: freely scalable, single day ChIPseq data generation from very low cell-numbers. BMC Genomics. 2019; 20:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langmead B., Wilks C., Antonescu V., Charles R.. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics. 2019; 35:421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baruzzo G., Hayer K.E., Kim E.J., Di Camillo B., FitzGerald G.A., Grant G.R.. Simulation-based comprehensive benchmarking of RNA-seq aligners. Nat. Methods. 2017; 14:135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Russo P.S.T., Ferreira G.R., Cardozo L.E., Bürger M.C., Arias-Carrasco R., Maruyama S.R., Hirata T.D.C., Lima D.S., Passos F.M., Fukutani K.F.et al.. CEMiTool: a bioconductor package for performing comprehensive modular co-expression analyses. BMC Bioinformatics. 2018; 19:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D.. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019; 47:D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jew B., Alvarez M., Rahmani E., Miao Z., Ko A., Garske K.M., Sul J.H., Pietiläinen K.H., Pajukanta P., Halperin E.. Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat. Commun. 2020; 11:1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tabula Muris Consortium Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018; 562:367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu G., Wang L.-G., Han Y., He Q.-Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics (Larchmont, N.Y.). 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Demircioğlu D., Cukuroglu E., Kindermans M., Nandi T., Calabrese C., Fonseca N.A., Kahles A., Lehmann K.-V., Stegle O., Brazma A.et al.. A pan-cancer transcriptome analysis reveals pervasive regulation through alternative promoters. Cell. 2019; 178:1465–1477. [DOI] [PubMed] [Google Scholar]
- 42. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.1000 Genome Project Data Processing Subgroup . The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-seq (MACS). Genome. Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tuoresmäki P., Väisänen S., Neme A., Heikkinen S., Carlberg C.. Patterns of genome-wide VDR locations. PLoS One. 2014; 9:e96105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Newell R., Pienaar R., Balderson B., Piper M., Essebier A., Bodén M.. ChIP-R: assembling reproducible sets of ChIP-seq and ATAC-seq peaks from multiple replicates. Genomics. 2021; 113:1855–1866. [DOI] [PubMed] [Google Scholar]
- 47. Amemiya H.M., Kundaje A., Boyle A.P.. The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 2019; 9:9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lun A.T.L., Smyth G.K.. csaw: a bioconductor package for differential binding analysis of ChIP-seq data using sliding windows. Nucleic Acids Res. 2016; 44:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heeringen S.J.v., Veenstra G.J.C. GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinformatics. 2011; 27:270–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P., Najafabadi H.S., Lambert S.A., Mann I., Cook K.et al.. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014; 158:1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schep A. 2022; motifmatchr: Fast Motif Matching in R.
- 52. Coetzee S.G., Coetzee G.A., Hazelett D.J.. motifbreakR: an R/bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics. 2015; 31:3847–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tremblay B. 2022; universalmotif: Import, Modify, and Export Motifs with R. 1.12.3.
- 54. Pérez-Silva J.G., Araujo-Voces M., Quesada V.. nVenn: generalized, quasi-proportional Venn and Euler diagrams. Bioinformatics. 2018; 34:2322–2324. [DOI] [PubMed] [Google Scholar]
- 55. Zou Z., Ohta T., Miura F., Oki S.. ChIP-Atlas 2021 update: a data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and bisulfite-seq data. Nucleic Acids Res. 2022; 50:W175–W182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Smith C.L., Eppig J.T.. The Mammalian Phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009; 1:390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. R Core Team R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing. 2021; Vienna, Austria. [Google Scholar]
- 58. Lee C.T., Cavalcante R.G., Lee C., Qin T., Patil S., Wang S., Tsai Z.T.Y., Boyle A.P., Sartor M.A.. Poly-Enrich: count-based methods for gene set enrichment testing with genomic regions. NAR. Genom. Bioinform. 2020; 2:lqaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gu Z., Eils R., Schlesner M.. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016; 32:2847–2849. [DOI] [PubMed] [Google Scholar]
- 60. Gu Z., Eils R., Schlesner M., Ishaque N.. EnrichedHeatmap: an R/bioconductor package for comprehensive visualization of genomic signal associations. BMC Genomics. 2018; 19:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mendelevich A., Vinogradova S., Gupta S., Mironov A.A., Sunyaev S.R., Gimelbrant A.A.. Replicate sequencing libraries are important for quantification of allelic imbalance. Nat. Commun. 2021; 12:3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maintainer B.P. 2021; liftOver: changing genomic coordinate systems with rtracklayer. 1.18.0.
- 63. Carey V. 2021; gwascat: representing and modeling data in the EMBL-EBI GWAS catalog. 2.26.0.
- 64. Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E.et al.. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019; 47:D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. The Genotype-Tissue Expression Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020; 369:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guzman-Lepe J., Cervantes-Alvarez E., Collin de l’Hortet A., Wang Y., Mars W.M., Oda Y., Bekki Y., Shimokawa M., Wang H., Yoshizumi T.et al.. Liver-enriched transcription factor expression relates to chronic hepatic failure in humans. Hepatol. Commun. 2018; 2:582–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jia Z., Li J., Ge X., Wu Y., Guo Y., Wu Q.. Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genome. Biol. 2020; 21:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Koyano-Nakagawa N., Gong W., Das S., Theisen J.W.M., Swanholm T.B., Van Ly D., Dsouza N., Singh B.N., Kawakami H., Young S.et al.. Etv2 regulates enhancer chromatin status to initiate Shh expression in the limb bud. Nat. Commun. 2022; 13:4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nurminen V., Neme A., Seuter S., Carlberg C.. Modulation of vitamin D signaling by the pioneer factor CEBPA. Biochim. Biophys. Acta. Gene. Regul. Mech. 2019; 1862:96–106. [DOI] [PubMed] [Google Scholar]
- 70. Zhao G.-N., Jiang D.-S., Li H.. Interferon regulatory factors: at the crossroads of immunity, metabolism, and disease. Biochim. Biophys. Acta. Mol. Basis. Dis. 2015; 1852:365–378. [DOI] [PubMed] [Google Scholar]
- 71. Han H., Cho J.-W., Lee S., Yun A., Kim H., Bae D., Yang S., Kim C.Y., Lee M., Kim E.et al.. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids. Res. 2018; 46:D380–D386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cui Z., Xiao L., Chen F., Wang J., Lin H., Li D., Wu Z.. High mRNA expression of CENPL and its significance in prognosis of hepatocellular carcinoma patients. Dis. Markers. 2021; 2021:9971799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Qin X., Li C., Guo T., Chen J., Wang H.-T., Wang Y.-T., Xiao Y.-S., Li J., Liu P., Liu Z.-S.et al.. Upregulation of DARS2 by HBV promotes hepatocarcinogenesis through the miR-30e-5p/MAPK/NFAT5 pathway. J. Exp. Clin. Cancer Res. 2017; 36:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chen T., Meng Y., Zhou Z., Li H., Wan L., Kang A., Guo W., Ren K., Song X., Chen Y.et al.. GAS5 protects against nonalcoholic fatty liver disease via miR-28a-5p/MARCH7/NLRP3 axis-mediated pyroptosis. Cell Death Differ. 2023; 30:1829–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hrckulak D., Kolar M., Strnad H., Korinek V.. TCF/LEF transcription factors: an update from the internet resources. Cancers. 2016; 8:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pilling D., Cox N., Thomson M.A., Karhadkar T.R., Gomer R.H.. Serum amyloid P and a dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin ligand inhibit high-fat diet-induced adipose tissue and liver inflammation and steatosis in mice. Am. J. Pathol. 2019; 189:2400–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hsieh J., Koseki M., Molusky M.M., Yakushiji E., Ichi I., Westerterp M., Iqbal J., Chan R.B., Abramowicz S., Tascau L.et al.. TTC39B deficiency stabilizes LXR reducing both atherosclerosis and steatohepatitis. Nature. 2016; 535:303–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lanthier N., Molendi-Coste O., Horsmans Y., Rooijen N.v., Cani P.D., Leclercq I.A. Kupffer cell activation is a causal factor for hepatic insulin resistance. Am. J. Physiol. Gastrointest. Liver Physiol. 2010; 298:G107–G116. [DOI] [PubMed] [Google Scholar]
- 79. Guo Q., Kim A., Li B., Ransick A., Bugacov H., Chen X., Lindström N., Brown A., Oxburgh L., Ren B.et al.. A β-catenin-driven switch in TCF/LEF transcription factor binding to DNA target sites promotes commitment of mammalian nephron progenitor cells. Elife. 2021; 10:e64444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gate R.E., Cheng C.S., Aiden A.P., Siba A., Tabaka M., Lituiev D., Machol I., Gordon M.G., Subramaniam M., Shamim M.et al.. Genetic determinants of co-accessible chromatin regions in activated T cells across humans. Nat. Genet. 2018; 50:1140–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Samstein R.M., Arvey A., Josefowicz S.Z., Peng X., Reynolds A., Sandstrom R., Neph S., Sabo P., Kim J.M., Liao W.et al.. Foxp3 Exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012; 151:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hammelman J., Krismer K., Banerjee B., Gifford D.K., Sherwood R.I.. Identification of determinants of differential chromatin accessibility through a massively parallel genome-integrated reporter assay. Genome Res. 2020; 30:1468–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang T., Zhang Z., Dong Q., Xiong J., Zhu B.. Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biol. 2020; 21:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Martin P., McGovern A., Orozco G., Duffus K., Yarwood A., Schoenfelder S., Cooper N.J., Barton A., Wallace C., Fraser P.et al.. Capture hi-C reveals novel candidate genes and complex long-range interactions with related autoimmune risk loci. Nat. Commun. 2015; 6:10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Li J., Lu Z., Wang Q., Su Z., Bao Y., Shi W.. Characterization of Bglu3, a mouse fasting glucose locus, and identification of Apcs as an underlying candidate gene. Physiol. Genomics. 2012; 44:345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pilling D., Gomer R.H.. Persistent lung inflammation and fibrosis in serum amyloid P component (APCs-/-) knockout mice. PLoS One. 2014; 9:e93730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vierstra J., Rynes E., Sandstrom R., Zhang M., Canfield T., Hansen R.S., Stehling-Sun S., Sabo P.J., Byron R., Humbert R.et al.. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science (New York, N.Y.). 2014; 346:1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ackert-Bicknell C.L. HDL cholesterol and bone mineral density: is there a genetic link?. Bone. 2012; 50:525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Makovey J., Chen J.S., Hayward C., Williams F.M.K., Sambrook P.N.. Association between serum cholesterol and bone mineral density. Bone. 2009; 44:208–213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are available in the NCBI GEO repository, under identifier GSE196942. The bioinformatics scripts used in this work are available in Zenodo at https://doi.org/10.5281/zenodo.10355100. The ChIP-qPCR primers are available on request.