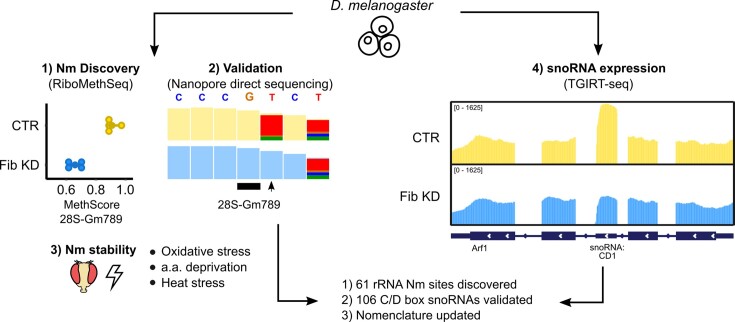
Abstract
During their maturation, ribosomal RNAs (rRNAs) are decorated by hundreds of chemical modifications that participate in proper folding of rRNA secondary structures and therefore in ribosomal function. Along with pseudouridine, methylation of the 2′-hydroxyl ribose moiety (Nm) is the most abundant modification of rRNAs. The majority of Nm modifications in eukaryotes are placed by Fibrillarin, a conserved methyltransferase belonging to a ribonucleoprotein complex guided by C/D box small nucleolar RNAs (C/D box snoRNAs). These modifications impact interactions between rRNAs, tRNAs and mRNAs, and some are known to fine tune translation rates and efficiency. In this study, we built the first comprehensive map of Nm sites in Drosophila melanogaster rRNAs using two complementary approaches (RiboMethSeq and Nanopore direct RNA sequencing) and identified their corresponding C/D box snoRNAs by whole-transcriptome sequencing. We de novo identified 61 Nm sites, from which 55 are supported by both sequencing methods, we validated the expression of 106 C/D box snoRNAs and we predicted new or alternative rRNA Nm targets for 31 of them. Comparison of methylation level upon different stresses show only slight but specific variations, indicating that this modification is relatively stable in D. melanogaster. This study paves the way to investigate the impact of snoRNA-mediated 2′-O-methylation on translation and proteostasis in a whole organism.
Graphical Abstract
Graphical Abstract.
Introduction
In eukaryotic cells, the path leading to a functional ribosome is orchestrated by approximately 80 ribosomal proteins, 4 ribosomal RNAs (rRNAs) and hundreds of non-ribosomal proteins (1). The 28S, 18S and 5.8S rRNAs are transcribed as one single pre-rRNA molecule from a variable number of gene copies that cluster in nucleoli (2–4). Folding and assembly of the mature rRNAs begins as soon as they are processed in a highly timely and complex way. The correct folding of rRNA is a sine qua non for the catalytic activity of ribosomes. This process relies on small nucleolar ribonucleoprotein complexes (snoRNPs) that bring distant nucleotides in three-dimensional proximity but also covalently modify rRNAs (5). The most abundant rRNA modifications are 2′-O-methylation (Nm) and pseudouridine (Ψ). The presence of Nm enhances nucleotide stacking and therefore dictates the flexibility of secondary structures of the rRNA, which influences the core function of the ribosome (6). Previous studies reported that both Nm and Ψ modifications concentrate around functionally crucial domains such as the Peptidyl Transferase Centre (PTC), the A- and the P-sites, the Decoding Centre (DC) as well as the inter-subunit bridge (7,8).
In the case of Nm, the vast majority of site-specific modifications are deposited by a single snoRNP composed of one small nucleolar RNA (snoRNA) of the C/D box family, two copies of fibrillarin, which carries the catalytic methyltransferase activity, and NH2L1 and a single copy of NOP58 and NOP56 (Fib, hoip, nop5 and nop56, respectively in D. melanogaster) (9). Cryo-EM structures have revealed how these proteins form a cavity that bind simultaneously rRNAs and snoRNAs (10). SnoRNAs are encoded as independent genes or within introns of coding and non-coding genes, with a predominance of the latest in higher eukaryotes (11). For intronic snoRNAs the major maturation step and assembly with combining proteins occurs before snoRNAs are spliced out (12,13). As their name suggests, C/D box snoRNAs are usually short RNAs (<100 nt) that possess one box C [RUGAUGA] and one box D [CUGA] motif that are separated by one weaker copy of the same motifs, called boxes C’ and D’ (14). The C and D-box sequences, which are located in close proximity to the 5′ and 3′ ends, respectively, interact and form a kink-turn structure that is recognized by the snoRNP complex (15). According to the strength of the motif, the boxes C’ and D’ located in the middle of snoRNAs will also fold into a kink turn and form a short hairpin. Most importantly, the 10 to 21 nt-sequences upstream of the boxes D/D’ can each contain an AntiSense Element (ASE) which is complementary to different rRNA segments (16). Canonical C/D box snoRNAs follow the D + 5 rule, meaning that they guide Nm deposition on the rRNA site that faces the 5th nucleotide upstream of the boxes D/D’ (17). The presence of two ASEs gives the possibility to some C/D box snoRNAs to guide Nm on two distinct targets. In some organisms such as yeast, the same ASE can hybridize to more than one rRNA target, resulting into 42 snoRNAs for a total of 54 rRNA Nm sites (18). On the other hand, in higher eukaryotes such as humans, where there are twice as more Nm sites in rRNA (n = 109), canonical C/D box snoRNAs are proportionally more numerous (n = 273) than in yeast where often a single snoRNA targets multiple Nm sites (19,20). Despite having identified Nm sites in multiple organisms, the matching between the sites and their corresponding snoRNA has not yet been systematically addressed, and results mainly from predictions.
The alteration of snoRNP members or of single C/D box snoRNAs can lead to a broad spectrum of translational defects. For example, knock-down (KD) of fibrillarin in human cell lines leads to a decrease in rRNA levels and reduced translation efficiency of transcripts that contain IRES-dependent translation initiation sequences (8,21). Also, mutation of D243 in the catalytic domain of nop1, the yeast ortholog of Fib, leads to a significant reduction of ribosome levels and a relative increase in free 40S subunits (7). Knocking out a single C/D box snoRNA is sufficient to shift the codon usage bias of ribosomes (22). Thus, there is increasing evidence that rRNA modifications can fine-tune translation, yet their impact at the physiological level still remains poorly understood.
In Drosophila melanogaster, Nm sites have only been predicted based on their complementarity to snoRNA ASEs, which themselves have been partly annotated based on motif predictions from genomic data (23). Various methods have been developed to detect and quantify Nm-modified sites (24,25). In this study, we use both RiboMethSeq, a nucleotide-resolution sequencing method that employs chemical probing coupled to next generation sequencing, and direct RNA nanopore sequencing (DRS), a long-read sequencing technology that can sequence native RNA molecules, in which modified sites can be identified based on alterations in the current intensity when the RNA molecules are translocated through the nanopores (26). As for C/D box snoRNA quantification, we obtained the full transcriptome of different tissues using TGIRT, a highly processive reverse transcriptase (RT) enzyme that reads through RNA modifications and complex secondary structures. Putting all techniques together, we built a comprehensive map of 61 rRNA Nm sites in D. melanogaster, most of which matched with at least one of the 106 expressed C/D box snoRNAs. Our results show that Nm sites on rRNA are relatively stable upon environmental stresses, yet specific changes were observed depending on the type of stress, suggesting specific adaptations. Our work opens the path for investigating the role of individual snoRNAs in multicellular organisms.
Materials and methods
Drosophila stocks and dissection
D. melanogaster CantonS reared at 25°C and 65% humidity were used for all in vivo assays. Mated females were chosen for all assays as Fibrillarin expression is higher based on modENCODE. Approximately 30 heads were collected for each replicate. For the oxidative stress assay, we extracted RNA from 30 individuals fed for 8 days with 5% sucrose and 0.1% nipagin in the control group and additionally 2.5 nM Paraquat (Sigma 856177) in the treated group. Flies submitted to nutrient stress were either fed on complete medium (7 g of yeast, 10 g of sucrose, 2g of agar and 1.5 ml of 10% Nipagin for 100 ml of H2O) or low nutrient medium medium (2.5 g of sucrose, 2g of agar and 1.5 ml of 10% nipagin for 100 ml of H2O) for 10 days (27). Flies submitted to heat stress were grown for 8 days at 29, 25 or 18°C on complete medium and in darkness (28).
Cell culture, RNA interference and transfection
D. melanogaster S2R+ cells were grown in Schneider's medium (Gibco) supplemented with 10% FBS (Sigma) and 1% penicillin–streptomycin (Sigma). For RNA interference (RNAi) experiments, PCR templates for the dsRNA were prepared using T7 megascript Kit (NEB). dsRNA against bacterial β-galactosidase gene (lacZ) was used as a control for all RNA interference (RNAi) experiments (T7-Fib-F: 5′-ACTTCTTACTGCTTGGGCG; T7-Fib-R: 5′-ACCAATGGCGAGAAGATTG; T7-CG8939-F: 5′-GAAAGACGCGCAAGGATAAG; T7-CG8939-R: 5′- TTGTCACGGAAATCATTGGA). S2R+ cells were seeded at the density of 106 cells/ml in serum-free medium and 7.5 μg of dsRNA was added to 106 cells. After 6 h of cell starvation, serum supplemented medium was added to the cells. dsRNA treatment was repeated after 48 and 96 h and cells were collected 24 h after the last treatment. Effectene (Qiagen) was used to transfect vector constructs in all overexpression experiments following the manufacturer's protocol.
RNA isolation and RT-PCR
Total RNA from S2R+ cells and Canton-S tissues was isolated with Trizol (Invitrogen) and treated with DNase I treatment (New England Biolabs) for all assays in this study. 250 ng of DNAsed RNA were retro-transcribed using random hexamer primers with the M-MLV-RT enzyme (Promega). cDNA were amplified with the GoTaq qPCR Master Mix (Promega) and the fluorescence was measured on a QuantStudio 6 Flex System (Applied Biosystems). Relative expression was calculated with the ΔΔCt method (qRT-Fib-F: 5′-GCCATTGGTCTCAACGGAG; qRT-Fib-R: 5′-GAGGGAGTGTTCATTGCGC; qRT-CG8939-F: 5′-GCTCAGAGTCATCCGAATCC; qRT-CG8939-R: CGAACCTTCTTGGCATTTGT).
Detection of 2′-O-methylation by RiboMethSeq (RMS)
RiboMethSeq (RMS) analysis was performed as previously described (29). Briefly, 150 ng of total RNA were subjected to alkaline hydrolysis for 16 min at 96°C followed by ethanol precipitation. The extremities of RNA fragments were end-repaired and converted to libraries using NEBNext Small RNA library preparation kit according to the manufacturer's recommendations. Libraries were quantified and multiplexed and subjected for high-throughput sequencing on a NextSeq2000 instrument with a 50 bp single read mode.
After removal of adapter sequences by trimmomatic v0.39, reads <40nt were selected and mapped to the D. melanogaster rRNA sequences FBtr0346885 (28S), FBtr0346878 (18S) and FBtr0346887 (5.8S) using bowtie2 in End-to-End mode (30,31). Reads’ extremities (5′-ends and 3′-ends) were counted and the different RiboMethSeq scores were calculated. A combination of ScoreMean >0.92 and ScoreA >0.5. ScoreC (MethScore) was used for quantification of the methylation level. Conserved Nm sites between species were done based on the alignment of rRNA sequences with Clustal 2.1 (Supplementary Table S1) (32).
Quantification of RNA modifications using LC-MS/MS
RNA from S2R+, HEK293 and S288c cells was enzymatically digested using benzonase (Santa Cruz Biotech) and nuclease P1 (Sigma) in 10 mM ammonium acetate pH 6.0 and 1 mM MgCl2 at 40°C for 1 h, added ammonium bicarbonate to 50 mM, phosphodiesterase I and alkaline phosphatase (Sigma), and incubated further at 37°C for 1 h. Digested samples were precipitated with 3 volumes of acetonitrile and supernatants were lyophilized and dissolved in a solution of stable isotope labelled internal standards (I.S., see mass list) for LC-MS/MS analysis. An Agilent 1290 Infinity II UHPLC system with an ZORBAX RRHD Eclipse Plus C18 150 × 2.1 mm (1.8 μm) column protected with an ZORBAX RRHD Eclipse Plus C18 5 × 2.1 mm (1.8 μm) guard (Agilent) was used for chromatographic separation. The mobile phase consisted of A: water and B: methanol (both added 0.1% formic acid) at 0.22 ml/min, starting with 5% B for 0.5 min followed by 2.5 min of 5–20% B, 3.5 min of 20–95% B, and 4 min re-equilibration with 5% B. Mass spectrometric detection was performed using an Agilent 6495 Triple Quadrupole system monitoring the mass transitions 268.1–136.1 (A), 284.1–152.1 (G), 244.1–112.1 (C), 245.1–113.1 (U), 282.1–136.1 (Am), 298.1–152.1 (Gm), 258.1–112.1 (Cm), 259.1–113.1 (Um), 285.1/153.1 (d3-m6A, I.S), 301.1/152.1 (d3-Gm, I.S.), 261.1/112.1 (d3-Cm), 264.1/127.1 (13C5-m5U, I.S.), 273.1/136.1 (13C5-A, I.S.), 246.1/114.1 (d2-C, I.S.) in positive electrospray ionization mode.
Library preparation for direct RNA nanopore sequencing (DRS)
Total RNA extracted from untreated and Fib knockdown samples (Fib_KD) in biological duplicates was DNase-treated with Turbo DNase (Thermofisher, AM2238) for 10′ at 37°C. Subsequently, 1000 ng of each sample was polyadenylated with E. coli PolyA Polymerase (NEB #M0276L) for 15 min at 37°C. 200 ng of poly(A)-tailed RNA was ligated to pre-annealed custom barcoded adaptors, following previously published protocols (33). Ligated RNA was prepared for direct RNA sequencing using the SQK-RNA002 kit following the ONT Direct RNA Sequencing protocol version DRS_9080_v2_revI_14Aug2019, with minor changes to allow for sample multiplexing. Briefly, for each sample, barcoded oligonucleotides A and B were mixed in annealing buffer (0.01 M Tris–Cl pH 7.5, 0.05M NaCl) to a final concentration of 1.4 μM each in a total volume of 75 μl. The mixture was then incubated at 94°C for 5 min and slowly cooled down (–0.1°C/s) to room temperature. Then, 200 ng of total RNA were ligated to the pre-annealed custom RT adaptors (33) using concentrated T4 DNA Ligase (NEB-M0202T). Ligated RNA was reverse transcribed using Maxima H Minus RT (Thermo Scientific, EP0752) at 60°C for 30 min, without the heat inactivation step. The products were purified using 1.8× Agencourt RNAClean XP beads (Fisher Scientific-NC0068576) and washed with 70% freshly prepared ethanol. 50 ng of reverse transcribed RNA from each reaction were pooled together, and the RNA:DNA hybrid was ligated to the RMX adapter. The mix was purified using 1× Agencourt RNAClean XP beads, washing with Wash Buffer (WSB) twice. The sample was then eluted in Elution Buffer (EB) and mixed with RNA Running Buffer (RRB) prior to loading onto a primed R9.4.1 flowcell. The samples were run on MinION sequencing devices.
Direct RNA sequencing data pre-processing
Raw fast5 files were processed with the Master of Pores pipeline (version 1.5, (34), https://github.com/biocorecrg/master_of_pores). Fast5 files were demultiplexed with DeePlexiCon using default parameters (33) and basecalled with Guppy basecaller v4.0. (https://nanoporetech.com). Reads were mapped to D. melanogaster rRNA sequences, obtained from Ensembl: FBtr0346885 (28S), FBtr0346878 (18S), FBtr0346887 (5.8S) and FBtr0086426 (5S). Mapping was performed using minimap2 v2.17 (https://github.com/lh3/minimap2) with ‘-ax splice -k 14 -uf’ options.
Analysis of RNA modifications in direct RNA sequencing data
Basecalling features (base quality, mismatch frequency, insertion frequency and deletion frequency) of each 5-mer were extracted from 2 replicates of untreated and Fib KD S2R+ reads using Epinano (35), version 1.2 (https://github.com/enovoa/EpiNano). Then, each position at the centre of the 5-mer was assigned a score that consisted of the difference between the scaled sum of the aforementioned frequencies in the untreated and in the Fib KD sample, as previously described (36). Then, we calculated the median of scores for each rRNA transcript and each replicate. For de novo discovery of Nm sites, nucleotides with a score greater than 3× the median score of all positions in the same transcript in both replicates or 5× the median in one of the replicates were further kept. Given that Nm modifications affects the score over multiple subsequent nucleotides – not just the modified site –, we merged nucleotides passing the previous criterion across four consecutive nucleotides into ‘regions’. Regions of ≤6 nts were expanded to 7-mer windows and regions ≥10 nts were split into two 7-mer windows to maximise the occurrence of only 1 Nm site per window. Regions overlapping with the 28S break and 5S rRNA were excluded. For the validation of RMS Nm sites by DRS, we extracted 7-mer windows around each confident RMS site and we kept regions with a score greater than 3x the median score in both replicates or 5× the median in one of the replicates. Integrative Genomics Viewer (IGV) version 2.8.13 was used to visualise mismatches and Epinano scores.
Preparation of TGIRT-Seq libraries
Total RNA from S2R+ cells and Canton-S tissues was isolated with Trizol (Invitrogen) and 3 μg were DNAsed with DNase I treatment (New England Biolabs). RNA was directly ribodepleted with the riboPOOL kit (siTOOLs) that was completed with oligos hybridising to 5S and 2S rRNAs following the manufacturer's instructions. The quality of RNA and ribodepletion was assessed on an Agilent 2100 Bioanalyzer. The purified ribodepleted RNA samples were fragmented for 3 min at 94°C in Magnesium RNA Fragmentation Module (New England Biolabs) and 3′ ends were dephosphorylated with T4 polynucleotide kinase (Lucigen). Between 50 and 75 ng were retrotranscribed in cDNA with 1.5 μl of template-switching TGIRT enzyme. The remaining library was prepared exactly as in Boivin and Deschamps-Francoeur et al (37). After 12–13 cycles of PCR and a 1.4x cleanup with Ampure XP beads (Bekman-Coulter), the profile of TGIRT libraries were evaluated once more by Bioanalyzer (± 250 bp average). Finally, the libraries were sequenced on an Illumina Next-seq 500 platform (2 × 150) yielding between 9.5 and 19.9 million paired-end reads.
TGIRT-seq data analysis
Adapters and low quality reads were trimmed from fastq files with cutadapt (version 3.4, (38)). rRNA sequences were filtered out with bowtie2 (-p 20 -L 15 -k 20 –fr –end-to-end) losing up to 4% of reads (31). The remaining reads were mapped with STAR as described in (39) (version 2.7.8a, (40)). Finally the reads were counted and annotated with a corrected annotation of Drosophila_melanogaster. BDGP6.32.103.gtf with CoCo (cc -c both -p -s 1), a pipeline that is efficient in attributing reads to genes that are nested in others, such as snoRNAs (41). Counts, counts per million (CPM) and transcripts per million (TPM) were taken from the CoCo output. We considered a snoRNA was expressed if at least 20 reads were measured in at least one TGIRT-seq library. Analysis of differentially expressed genes was done with DeSeq2 (42) and plots were drawn in R using basic functions and ggplot2 (43) as well as Inkscape (version 1.0.1).
Screening of canonical C/D box snoRNAs with snoScan
To validate the C/D box features and to predict Nm targets of the 143 snoRNAs (biotype: snoRNA and ncRNA), we used both human yeast and mammalian probabilistic search models of snoScan 1.0 (44). All publications used for mapping snoRNAs in D. melanogaster are listed on Supplementary Table S2. Briefly, 142 snoRNA fasta files with corrected coordinates were compared against the same rRNA sequences used for the RiboMethSeq. Target predictions of expressed snoRNAs matching experimentally identified Nm sites were considered as highly confident. When the snoRNA was expressed but matched an unknown target or no target at all, it was classified as orphan. The target predictions of snoRNAs for which no expression was detected in the tested tissues were considered as lowly confident.
Nomenclature of Drosophila melanogaster C/D box snoRNAs
Because of the heterogeneity and obsolescence of the current nomenclature of C/D box snoRNAs in D. melanogaster, we decided in collaboration with FlyBase to propose an updated nomenclature. First, 27 genes lacking any evidence for expression that were erroneously curated from a personal communication to FlyBase in 2006 (under the ID FBrf0199239) have been deleted in FlyBase (since release FB2023_05). We then renamed the remaining 120 C/D box snoRNA genes with expression evidence using the following syntax: (i) a ‘snoRNA:CD’ prefix, followed by (ii) an incremental number reflecting their order of discovery/publication, and where necessary (iii) a letter to distinguish between copies of the same gene (Supplementary Tables S2 and S6). This new system provides a simple, scalable consistency to D. melanogaster snoRNA nomenclature, similar to that used by the HGNC for human snoRNA genes (Seal et al., 2020), and avoids overloading the gene symbol with information about genomic location, orthology or RNA target(s) that is prone to revision or going out-of-date.
Results
Identification of Nm sites on Drosophila melanogaster rRNA
Different rRNA annotations are available in Drosophila melanogaster including multiple ones from the Ensembl 103 assembly, one GenBank reference that was used for previous snoRNA predictions, and a PDB reference that was used for 3D representations. To facilitate the navigation between previously predicted and novel identified Nm positions (see below), we aligned annotated rRNA sequences with Sanger-sequenced data from both embryonic cell line S2R+ cells and wildtype Canton-S flies (Supplementary Table S1 and Supplementary Figure S1A). The most similar sequences were FBtr0346885 (28S), FBtr0346878 (18S), FBtr0346887 (5.8S) and FBtr0086426 (5S) from the Ensembl assembly. Consistent with previous observations (45), we confirmed that the 28S rRNA has a ’hidden break’ based on the lower read coverage that spans nucleotides 1814 to 1858, but also based on the presence on the electrogram of two RNA peaks just below 2 kb that reflects a mixture of 18S and two similarly sized fragments of 28S (Supplementary Figure S1B, C). This break is also present in 5.8S, which is in fact shorter in D. melanogaster (123 nts) in comparison to human or yeast (157 and 158 nts, respectively) (46).
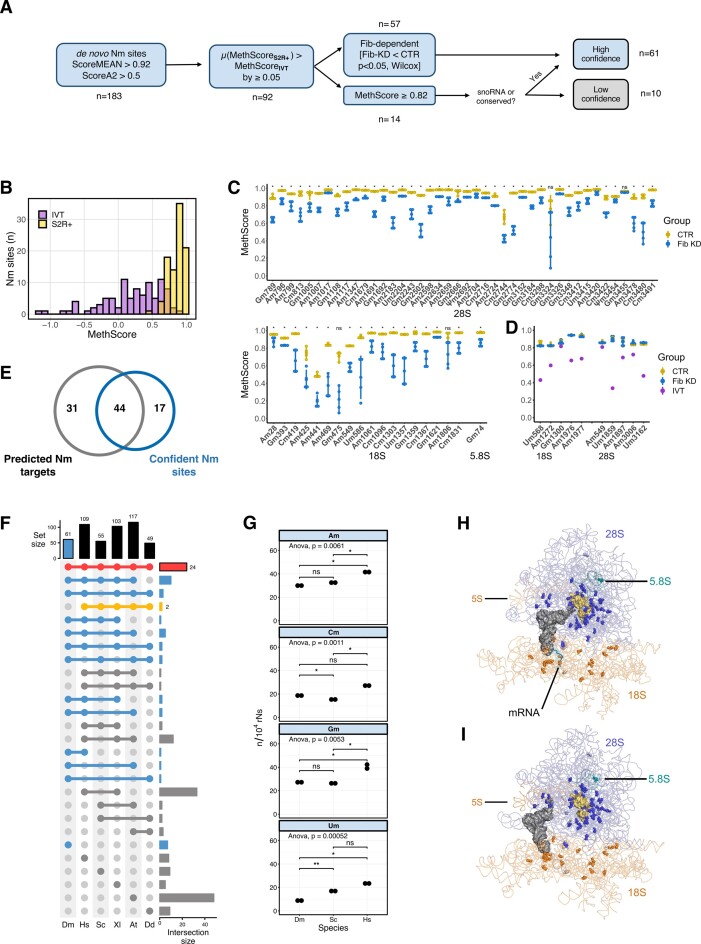
To map Nm sites onto the rRNA molecules, we generated RiboMethSeq (RMS) libraries from total RNA extracted from S2R+ cells under control (CTR) and Fibrillarin knockdown (Fib-KD) conditions. It is important to note that only transient and partial KD of Fib can be achieved, since this protein is essential for snoRNP maturation and rRNA processing. The construction of RMS libraries is based on the increased resistance to alkaline pH of the phosphodiester bond between the 2′-O-methylated nucleotide and the one following it. As this method is prone to false positives related to RNA structure, sequence and ligation biases (29), we applied different filters in order to ensure a robust and conservative calling of Nm sites (Figure 1A). First, we used a combination of score MEAN >0.92 and scoreA2 >0.5 as in previous studies (47) and identified 183 putative Nm sites by their protection against alkaline cleavage. A MethScore, which is a semi-quantitative parameter that reflects the fraction of sites that are methylated, was then computed for each position. In parallel, we extracted the MethScores from data obtained from in vitro transcribed rRNAs (IVTs), which are not methylated and can therefore serve as negative controls. We then filtered candidate Nm sites that had an average MethScore in CTR (n = 4) that was higher by 0.05 in comparison to the MethScore observed for IVT rRNAs. This filtering reduced the number of putative Nm sites to 92 (Figure 1A, B, Supplementary Table S3). The third filter was based on the significant drop of MethScore that was observed after knocking-down Fib (ΔMeth score ≥ 0.05, P-value < 0.05, Wilcoxon test). With this criterion, 57 candidate Nm sites out of 92 previously retained candidates had a significantly lower MethScore in Fib-KD conditions and thus were ranked as highly confident sites (Figure 1A, C and Supplementary Table S3). Interestingly, the differential MethScore was as low as 0.03 for 28S-Am1017 and as high as 0.49 for 18S-Gm475, underlining the very heterogenous response of individual Nm level to the reduction of Fib-KD, probably reflecting differential stability and/or activity of snoRNP complexes. Similar behaviour was also observed for human cell lines upon fibrillarin KD where only a fraction of rRNA sites showed substantial decrease in methylation level (8,21).
Figure 1.
(A) Data processing performed for the identification of Nm sites in D. melanogaster using RiboMeth-seq. (B) Distribution of MethScores for S2R+ cells (yellow) and of in vitro transcribed rRNAs (purple). (C) MethScore for each highly confident Nm site in 28S, 18S and 5.8S rRNAs in control (yellow) and Fibrillarin knockdown (blue) S2R+ cells. Asterisks indicate the significance of loss of methylation (Standard deviation bars, n = 4, Wilcoxon test, * P< 0.05 and NS P> 0.05). (D) MethScore for low confidence Nm sites on 18S and 28S rRNAS in control (yellow) or Fibrillarin knockdown (blue) S2R+ cells and in IVT rRNAs (purple). (E) Overlap between the 61 confident Nm sites and the Nm candidates predicted from snoRNA complementarity in previous literature (summarised in Supplementary Table S2). (F) Upset plot of Nm sites that are conserved across different model organisms (Dm: D. melanogaster; Hs: H.sapiens; Sc: S. cerevisiae; Xl: X. laevis; At: A. thaliana; Dd: D. discoideum). Nm sites found in Dm are highlighted in blue, Nm sites conserved in all species are in red and those that are absent in Dm are highlighted in yellow. (G) Number of Am, Cm, Gm and Um normalized over 10000 non modified bases for D. melanogaster (Dm), H. sapiens (Hs) and S. cerevisiae (Sc). Measurement was done by mass spectrometry (ANOVA test for all samples, Wilcoxon test for paired comparison) (H, I) Three-dimensional location of Nm sites in two elongating conformations of ribosomes loaded with a tRNA in the P-site and an mRNA in the decoding centre (testis polysome, PDB 6XU7, reported resolution 4.9 Å) (H) and a tRNA in the E-site (embryos ribosome, PDB 4v6w, reported resolution 6 Å) (I). The main chain of Nm sites is shown as blue (28S), cyan (5.8S) and orange (18S) spheres and the bases are shown as sticks. The nucleotides facing the Peptidyl Transferase Centre are shown in yellow.
The remaining population of candidates (35 sites, 92–57 = 35) did not show Fib-KD dependent reduction in MethScore values (Figure 1A). From these, 14 had a MethScore level ≥0.82, a cut-off that was set in a way that the first Nm site above the cut-off was predicted to have a snoRNA match. Since the majority of rRNA Nm methylation events (with the exception of 28S-Gm3455, see below) are snoRNA-dependent in all studied eukaryotic species, the existence of known or predicted C/D-box snoRNA guide complementary to the site can be used as an additional criterion for validation, even in the absence of a clear Fib-dependence. Out of 14 retained sites, 3 had a complementary snoRNA guide and 1 revealed to be highly conserved in other eukaryotic species. These 4 candidates were thus ranked along with the previous high confidence Nm sites (57 + 3 + 1 = 61). The conserved Nm site with no snoRNA match is 28S-Gm3455 that is predicted to be deposited by CG8939, the orthologue of the stand-alone methyltransferases FTSJ3 in human and Spb1 in yeast (48). Curiously, the knockdown of CG8939 led only to a minor drop of 0.03 at the expected 28S-Gm3455 in comparison to a 0.07 drop at the Nm site located one nucleotide upstream (Supplementary Figures S2A and S2B). The ten remaining Nm candidate sites with no associated snoRNA or conservation match were ranked as low confident (Figure 1D). For those, the only supporting evidence is high MethScores indicating protection against alkaline cleavage and lower protection observed in IVT. It is not excluded that these could be targets of non-canonical snoRNA guides or yet uncharacterised guide-independent methyltransferases. Alternatively, these low confidence sites could be indicators of other RNA modifications that could contribute to alkaline resistance of RNA. Lastly, biases due to robust secondary structure or in adapter ligation can also create underrepresented fragments in the RMS analysis.
In total, 61 highly confident Nm sites were identified by this successive filtering, with 42 in 28S, 18 in 18S and 1 Nm site in 5.8S, while 5S and 2S rRNAs had no detectable Nm modification (Table 1). Only 44 of these highly confident sites overlapped with the 75 Nm sites predicted so far in the literature, essentially based on snoRNA complementarity (Figure 1E). We nevertheless checked the MethScore of the remaining 31 predicted sites in the pre-filtered data and none had a value of MethScore above 0.56 or was Fib-dependent. As shown by our expression data (see below), these predicted snoRNAs are not even expressed, suggesting that these Nm sites were inaccurately predicted. Among the 61 highly confident sites, 53 are present in at least one of the five model organisms for which Nm sites have been mapped and 24 are universally conserved in all five species (Figure 1F, Supplementary Table S4). In contrast, D. melanogaster lacks 2 Nm sites that were found to be conserved in all the other studied eukaryotic species. As the number of rRNA Nm sites in humans is almost double of that observed in flies (Supplementary Figure S1D), we used this information to see if we could detect the same ratio in bulk measurements obtained by mass spectrometry as a mean to check whether we may have missed some Nm sites. Assuming that methylation is close to 100% for most sites in all organisms and even though the total length of human rRNAs is 15% longer than fly ones, we could observe that the normalized number of Am, Gm and Um was significantly higher in human cells than in fly cells (Wilcoxon test, Figure 1G). This trend is in line with the ratio of modified bases in each species and particularly visible for Um, which exceeds a 2-fold-change (20 and 6 sites respectively for human and fly) (Supplementary Figure S1D). In yeast, the normalized number of Nm sites was similar for Am and Gm, lower for Cm and higher for Um (Figure 1G, Supplementary Table S4). Altogether, the comparative ranking of Nm counts between species suggests that our criteria to identify Nm sites are specific enough without hindering sensitivity.
Table 1.
Nm sites in Drosophila melanogaster rRNAs identified by RiboMeth-seq
| Target rRNA | Nm site† | Conservation§ | Gene id (FlyBase) | Proposed gene name |
|---|---|---|---|---|
| 18S | Am28N | DmHYAXDd | FBgn0063388 | snoRNA:CD5a |
| FBgn0063387 | snoRNA:CD5b | |||
| FBgn0063386 | snoRNA:CD5c | |||
| Gm393N | DmHAX | FBgn0065058 | snoRNA:CD21 | |
| FBgn0082931 | snoRNA:CD53a | |||
| Cm419N | DmHYAX | FBgn0086533 | snoRNA:CD35 | |
| Am425N | DmHYX | FBgn0086056 | snoRNA:CD23 | |
| Am441N | DmHYAXDd | FBgn0263489 | snoRNA:CD78 | |
| Am469N | DmHAX | FBgn0086041 | snoRNA:CD36 | |
| Gm475N | DmDd | FBgn0263851 | snoRNA:CD68 | |
| Am549N | DmHYAX | FBgn0261973 | snoRNA:CD63a | |
| FBgn0261974 | snoRNA:CD63b | |||
| Um586N | DmHYAXDd | FBgn0060291 | snoRNA:CD22b | |
| FBgn0082926 | snoRNA:CD46 | |||
| Am1061N | DmHYAX | FBgn0082952 | snoRNA:CD24 | |
| Cm1096N | DmA | FBgn0086055 | snoRNA:CD37 | |
| Cm1303N | DmHA | FBgn0086061 | snoRNA:CD62 | |
| Um1357N | DmHYAXDd | FBgn0082949 | snoRNA:CD38a | |
| FBgn0082947 | snoRNA:CD38c | |||
| FBgn0082948 | snoRNA:CD38b | |||
| FBgn0286758 | snoRNA:CD39c | |||
| Gm1359N | DmHYAXDd | FBgn0086053 | snoRNA:CD39a | |
| FBgn0086054 | snoRNA:CD39b | |||
| FBgn0286758 | snoRNA:CD39c | |||
| Cm1367N | Dm | FBgn0082951 | snoRNA:CD40 | |
| Gm1621N | DmHYAXDd | FBgn0063389 | snoRNA:CD8 | |
| Am1806N | DmHX | FBgn0086074 | snoRNA:CD41 | |
| Cm1831N | DmHYAXDd | FBgn0065053 | snoRNA:CD15 | |
| 28S | Gm789N | DmH | FBgn0020518 | snoRNA:CD1 |
| Am796N | DmHXDd | FBgn0086067 | snoRNA:CD43 | |
| Am799N | DmHYAXDd | FBgn0086078 | snoRNA:CD25b | |
| FBgn0086079 | snoRNA:CD25a | |||
| FBgn0086067 | snoRNA:CD43 | |||
| Cm813N | DmHYAX | FBgn0086062 | snoRNA:CD26b | |
| FBgn0086063 | snoRNA:CD26a | |||
| Gm1005N | DmHYAX | FBgn0063392 | snoRNA:CD9a | |
| FBgn0065047 | snoRNA:CD11b | |||
| FBgn0065048 | snoRNA:CD11a | |||
| FBgn0086042 | snoRNA:CD44 | |||
| Am1007N | DmHYAX | FBgn0082943 | snoRNA:CD27a | |
| FBgn0082942 | snoRNA:CD27b | |||
| Am1017N | DmHYAX | FBgn0082941 | snoRNA:CD45 | |
| Gm1108N | DmHYAX | FBgn0082935 | snoRNA:CD28a | |
| FBgn0082934 | snoRNA:CD28b | |||
| FBgn0082933 | snoRNA:CD28c | |||
| FBgn0082932 | snoRNA:CD28d | |||
| Am1117N | Dm | FBgn0082935 | snoRNA:CD28a | |
| FBgn0082934 | snoRNA:CD28b | |||
| FBgn0082933 | snoRNA:CD28c | |||
| FBgn0082932 | snoRNA:CD28d | |||
| Am1347N | DmHYAXDd | FBgn0015543 | snoRNA:CD16 | |
| Cm1679N | DmHYAXDd | FBgn0086068 | snoRNA:CD29 | |
| Am1691N | DmHYAXDd | FBgn0063377 | snoRNA:CD4a | |
| FBgn0063376 | snoRNA:CD4b | |||
| FBgn0086068 | snoRNA:CD29 | |||
| Gm1692N | DmHYAXDd | FBgn0063377 | snoRNA:CD4a | |
| FBgn0063376 | snoRNA:CD4b | |||
| Am2183N | DmHAX | FBgn0086039 | snoRNA:CD48 | |
| Um2204N | DmHYAXDd | FBgn0260002 | snoRNA:CD30b | |
| FBgn0086051 | snoRNA:CD30a | |||
| Gm2243N | DmHX | FBgn0086028 | snoRNA:CD49 | |
| Gm2502N | DmHA | snoRNA not found | ||
| Am2598N | DmHYAX | FBgn0065071 | snoRNA:CD18 | |
| Am2634 | DmHYAXDd | FBgn0065066 | snoRNA:CD14 | |
| Am2659N | DmHYAXDd | FBgn0086023 | snoRNA:CD51a | |
| FBgn0086024 | snoRNA:CD51b | |||
| FBgn0086025 | snoRNA:CD51c | |||
| Gm2666N | DmHYAXDd | FBgn0025881 | snoRNA:CD7 | |
| Ψm2692N | DmHXDd | FBgn0086603 | snoRNA:CD20 | |
| Am2704N | DmHAXDd | FBgn0082944 | snoRNA:CD52c | |
| FBgn0082945 | snoRNA:CD52b | |||
| FBgn0082946 | snoRNA:CD52a | |||
| Cm2716 | DmHYAXDd | FBgn0082938 | snoRNA:CD31a | |
| FBgn0082937 | snoRNA:CD31b | |||
| FBgn0082936 | snoRNA:CD31c | |||
| Am2724N | Dm | FBgn0082938 | snoRNA:CD31a | |
| FBgn0082937 | snoRNA:CD31b | |||
| FBgn0082936 | snoRNA:CD31c | |||
| Cm2744N | DmHAX | FBgn0065073 | snoRNA:CD13 | |
| FBgn0263467 | snoRNA:CD67 | |||
| Gm2774N | DmHYAXDd | FBgn0082931 | snoRNA:CD53a | |
| FBgn0082930 | snoRNA:CD53b | |||
| FBgn0082929 | snoRNA:CD53c | |||
| Gm3152N | DmHYAXDd | FBgn0063381 | snoRNA:CD2a | |
| FBgn0063380 | snoRNA:CD2b | |||
| FBgn0063379 | snoRNA:CD2c | |||
| FBgn0063378 | snoRNA:CD2d | |||
| Gm3184 | DmHAXDd | FBgn0086064 | snoRNA:CD32b | |
| FBgn0086065 | snoRNA:CD32a | |||
| Cm3298N | Dm | FBgn0082940 | snoRNA:CD54a | |
| FBgn0082939 | snoRNA:CD54b | |||
| Gm3324N | DmYADd | FBgn0086075 | snoRNA:CD55 | |
| Gm3326N | DmHYAX | FBgn0082928 | snoRNA:CD33a | |
| FBgn0082927 | snoRNA:CD33b | |||
| FBgn0086075 | snoRNA:CD55 | |||
| Gm3348N | DmHYAXDd | FBgn0063375 | snoRNA:CD6a | |
| FBgn0063374 | snoRNA:CD6b | |||
| FBgn0063373 | snoRNA:CD6c | |||
| Cm3412N | DmHAXDd | FBgn0060292 | snoRNA:CD22a | |
| FBgn0060291 | snoRNA:CD22b | |||
| FBgn0065070 | snoRNA:CD19 | |||
| Um3415N | Dm | FBgn0060292 | snoRNA:CD22a | |
| FBgn0060291 | snoRNA:CD22b | |||
| FBgn0086048 | snoRNA:CD56b | |||
| FBgn0086049 | snoRNA:CD56a | |||
| Am3420N | Dm | FBgn0086057 | snoRNA:CD47 | |
| Cm3422N | Dm | FBgn0086069 | snoRNA:CD57 | |
| Ψm3454 | DmHYAXDd | snoRNA not found (CG8939-dependent) | ||
| Gm3455 | DmHYAXDd | snoRNA not found (SPB1 in yeast) | ||
| Am3478N | DmHYAXDd | FBgn0063385 | snoRNA:CD3a | |
| FBgn0063384 | snoRNA:CD3b | |||
| FBgn0063383 | snoRNA:CD3c | |||
| FBgn0063382 | snoRNA:CD3d | |||
| Cm3480N | DmYADd | FBgn0063385 | snoRNA:CD3a | |
| FBgn0063384 | snoRNA:CD3b | |||
| FBgn0063383 | snoRNA:CD3c | |||
| FBgn0063382 | snoRNA:CD3d | |||
| Cm3491N | DmHYAXDd | FBgn0083988 | snoRNA:CD58a | |
| FBgn0083989 | snoRNA:CD58b | |||
| 5.8S | Gm74 | DmHAX | FBgn0086066 | snoRNA:CD34 |
†Ensembl 104 28S/FBtr0346885; 18S/FBtr0346878; 5.8S/FBtr0346887.
§Dm: D. melanogaster; H: H. sapiens; S: S. cerevisiae; X: X. laevis; A: A. thaliana; Dd: D. discoideum.
NNm site validated with Nanopore.
Consistent with previous work, we found that most Nm sites in rRNA were highly methylated (56/61) (Figure 1C, Table 1) (8,49). The 5 Nm sites that were partially methylated (MethScore <0.80) were still Fib-dependent (Figure 1C). Intriguingly, three of them were located back-to-back from position 425 to 475 in the 18S rRNA. Cryo-EM structures revealed that these sites are less protected by ribosomal proteins (50,51). In line with observations for other organisms, Nm sites on the large ribosomal subunit (LSU) are mainly clustered in the immediate surrounding of the PTC and the peptide exit tunnel (PET) (Figure 1H–I and Supplementary Figure S3A for 2D-projection). In addition, about 1/3 of 18S Nm modifications are gathered around the decoding centre (DC) while the other 2/3 are distributed across the 18S rRNA (Supplementary Figure S3B for 2D-projection). As for the only Nm site of 5.8S rRNA, it is found on the wall of the PET. The existence of cryo-EM snapshots containing a tRNA in the E site (51) (Figure 1I) revealed a so far undescribed cluster of 4 Nm sites on the 28S rRNA that are in proximity to the amino acid attachment site of tRNAs – two of them, Gm3326 and Cm3298 being as close as 3.5 Å to the closest tRNA atom. The examination of the Nm sites in the context of the secondary structure showed that they are primarily located at the base of helices and in small bulges or loops and are systematically absent from expansion segments, which are usually species-specific (Supplementary Figures S3A and S3B).
Orthogonal validation of Nm sites using nanopore sequencing
Since the presence of other modifications (e.g. pseudouridine) as well as the secondary structure of rRNAs can occult the detection of Nm sites and/or lead to false positives candidates in RMS analysis (29) we sought to use an alternative approach to identify and validate RMS-identified Nm sites. Specifically, we employed direct RNA nanopore sequencing (DRS), which has been shown to detect Nm sites on the basis of an increased base-calling error in the 5-mer window containing each Nm-modified site (36).
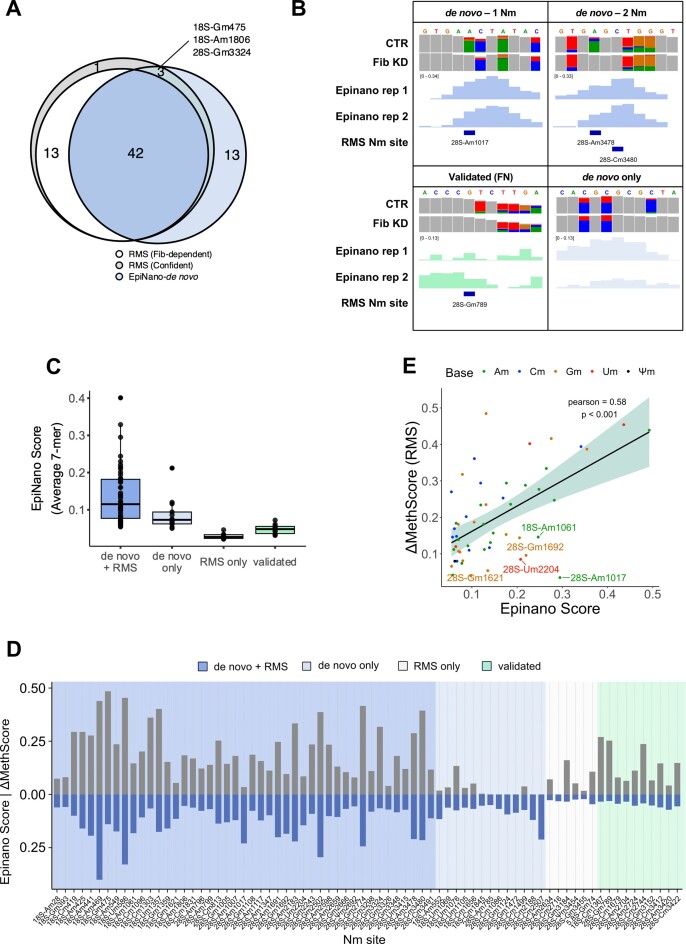
Briefly, two independent replicates of Fib-KD and two CTR samples that were used for RMS were also sequenced using DRS. The de novo Nm discovery approach (described in the Materials and methods section) identified 53 regions (7-mers) as significantly altered when comparing Fib-KD and CTR samples. Of these regions, 40 overlapped with a total of 42 Fib-dependent and 3 non-dependent Nm sites identified using RMS (Figure 2A, Table 1). Nevertheless, some of them were part of a region overlapping with two RMS sites, suggesting that the signal from a single Nm site can mislead the detection of a neighbouring Nm site, due to the spreading of the Nm signal across several nucleotides (36). On a similar note, the longer the ‘regions’ were before re-sizing, the more likely it was that they overlapped with two consecutive Nm sites (Supplementary Figure S4A). For example, our de novo Nm detection pipeline could not identify that a given region contained 2 modified sites in the following cases: 28S-Am3478 and Cm 3480, or Gm3324 and Gm3326, which are in short distance from one another (Figure 2B).
Figure 2.
(A) High confidence RMS Nm sites (grey), RMS Fibrillarin-dependent Nm sites (white, ΔmethScore ≥ 0.05) that overlap de novo nanopore sites (blue). (B) IGV tracks of representative examples of de novo and validation analysis by DRS. Positions with base-calling allele error frequencies greater than 20% are coloured; grey represents match to reference. (C) Distribution of Epinano score of 7-mer regions based on whether they were discovered de novo or validated and whether they overlapped with RMS Nm sites. (D) Differential methylation between Fib KD and CTR S2R+ cells obtained with RiboMethSeq (ΔMethScore, n = 4, blue) and with Nanopore (Epinano score, n = 2, turquoise) for high confidence Nm sites (n = 61). (E) Pearson correlation of ΔMethScore and Epinano score for overlapping Nm sites. Nm sites are coloured by base and those with significantly higher Epinano score are labelled with their coordinates.
The 13 regions that were discovered by the de novo DRS but not the RMS analysis were in average shorter and had lower Epinano score in comparison to overlapping regions (Figure 2A–C) (35). We therefore used different approaches to evaluate their validity. Five sites were eliminated as they were also picked in two independent negative controls (CG8939 and CG11447 KDs, data not shown). Three other regions were also discarded as they were generated artificially by splitting one large region that overlapped a single RMS site. In total, only five regions remained after this additional filtering, without an obvious explanation, as none of them overlapped with Nm sites found in other organisms (Supplementary Figure S4B) and no matching snoRNA could be predicted. They could result from the alteration of the current provoked by other factors such as 2D rRNA structure that can be indirectly affected by distal Nm sites (although the rRNA molecules were linearized during the library preparation to minimize the effect of RNA structure in the nanopore signal). Further work will be necessary to determine whether these regions do indeed encompass real Nm sites.
Finally, we examined the capacity of DRS to validate known RMS sites. For this, 7-mer windows around each confident RMS site were extracted and kept when the Epinano score was 3-fold greater than the background (see Methods). This way, 10 Nm sites could be validated along with the 45 de novo ones, corresponding to a total of 55 Fib-dependent RMS sites (95%) (Figure 2C and D, Table 1). We noted that the sites captured in the validation, but not in the de novo approach, were often missed because of the score being altered in shifted windows, or because the score was extremely low in only one of the two replicates, but not the other one as it was the case for 28S-Gm789 (Figure 2B and Supplementary Table S5). Finally, as expected, the 3 Nm sites that were Fib-independent in the RMS dataset and missed by the de novo approach were also missed by the validation approach (Figure 2A). Overall, RMS ΔMethScores and Epinano scores correlated significantly (Pearson r = 0.58, P< 0.001), although the amplitude of RMS ΔMethScores was larger than that of Epinano scores (Figure 2D and E). We further noticed that nanopore was more sensitive to differential methylation on Am sites in comparison to RMS, which had no visible bias towards any base (Figure 2E, Supplementary Figures S4C and S4D). In conclusion, while 77% of RMS-identified Nm sites were de novo discovered with nanopore sequencing, 95% of Fib-dependent RMS-identified sites could be validated with nanopore, indicating that the nanopore approach is a bona fide validation tool of Nm modifications on rRNA.
Validation of C/D box snoRNA expression and prediction of Nm targets
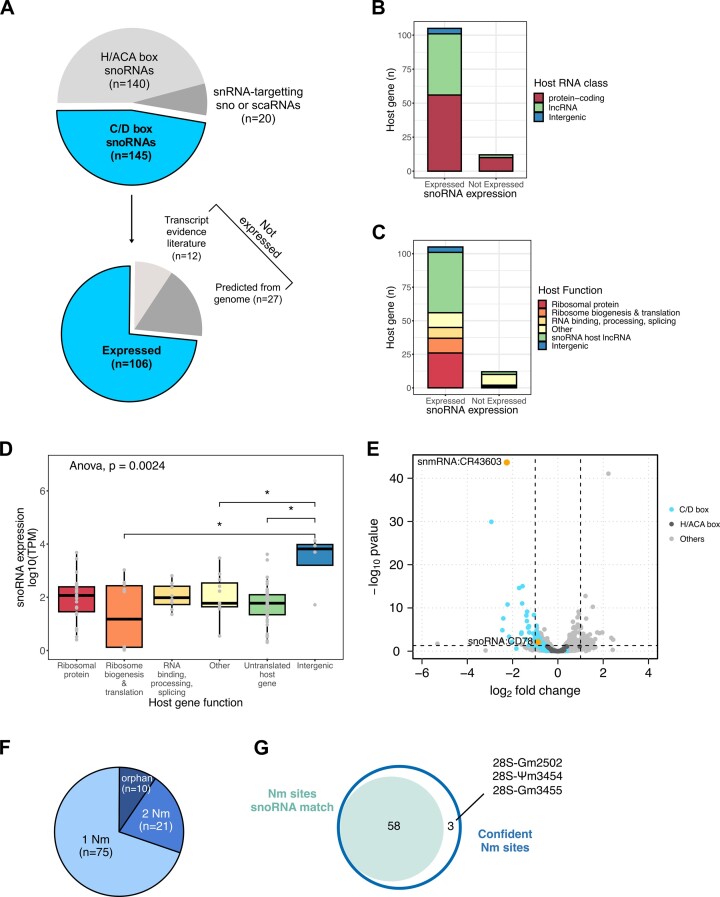
So far, predictions from expression and genomic data from our and previous works have led to the annotation of 145 transcripts as canonical C/D box snoRNAs targeting rRNAs in D. melanogaster (Figure 3A up, and Supplementary Table S2). However current polyA-enriched libraries miss out on many snoRNAs, likely due to the complex structure (Supplementary Figure S5A). To validate snoRNA expression in a comprehensive manner, we sequenced the whole transcriptome of S2R+ cells, ovaries and heads of wild-type flies using TGIRT-seq—a rRNA-depleted library preparation that covers all RNA species and revealed that snoRNAs is the second more abundant RNA class after tRNAs (Supplementary Figures S5B and S5C) (37,39). We first noticed a discrepancy between coverage and coordinates reaching up to 63 nucleotides across all tissues (Supplementary Figure S5D). Indeed, canonical snoRNAs have a characteristic ‘square’ coverage as end-to-end reads span the snoRNA gene which facilitates the identification of gene extremities (Supplementary Figure S5D). Therefore, we redefined genomic coordinates for expressed C/D box snoRNAs based on the read coverage and proceeded to downstream analyses (Supplementary Table S6).
Figure 3.
(A) Distribution of transcripts labelled as ’biotype:snoRNA’ in the Ensembl 104 assembly (upper pie chart). Number of C/D box snoRNAs that are expressed in S2R+, heads and ovaries cells (lower pie chart). The non-expressed C/D box snoRNAs are further separated based on expression evidence in literature (Supplementary Table S2). Number of expressed and non-expressed snoRNAs according to the RNA class (B) and biological function (C) of their host gene. (D) Expression levels of snoRNAs in S2R+ cells in function of the biological function of their host gene (Data points in grey correspond to the mean of three samples. Pairwise comparisons, Wilcoxon test, * P< 0.05). (E) Volcano plot of whole-transcriptome changes following Fib KD. C/D box and H/ACA snoRNAs and the remaining ±10k expressed transcripts are highlighted in turquoise, dark grey and light grey respectively. Two Fib-dependent lncRNAs are highlighted in orange. The horizontal and vertical dashed lines illustrate respectively the FDR = 0.05 and |logFC| > 1 (DeSeq2). (F) Number of rRNA Nm targets predicted for each expressed C/D box snoRNA. (G) Number of high confident Nm sites with at least one snoRNA match.
In total, 106 out of the 145 annotated snoRNAs were expressed in the above-mentioned tissues (Figure 3A up, Supplementary Figure S5C), including one novel snoRNA candidate predicted in this work (Supplementary Figures S5E and S5F). Among the remaining 39 non-expressed snoRNAs, 27 were annotated in 2006 based exclusively on sequence predictions and without validated gene expression data in modENCODE (Figure 3A down, Supplementary Figure S5A, Supplementary Tables S2 and S6). Following a re-evaluation by FlyBase, those specific 27 genes were removed in the release FB2023_05.
We then examined different features of the 106 expressed and the 12 non-expressed genes that may correlate or explain expression differences between and within these two groups. We focused particularly on the expressed group where TPMs span 1 to 5-digit numbers (Supplementary Figure S5C). In terms of genomic location, all but 5 C/D box snoRNAs are encoded within introns of other genes. These intergenic snoRNAs were more expressed than other snoRNAs in all three tissues (Figure 3B–D, Supplementary Figure S5A). Similarly to humans, the remaining C/D box snoRNAs are expressed in a narrow majority within protein-coding genes (53%) and lncRNAs (42%) (Figure 3B) (52). Non-expressed genes were mostly encoded in genes with miscellaneous functions. Host genes, including those hosting non-expressed snoRNAs, were systematically expressed apart from Uhg7, lncRNA:CR31647 and ect. LncRNAs are represented by seven different genes all starting with the prefix Uhg that stands for ‘U snoRNA host gene’ (Supplementary Table S6). The most represented class of protein-coding host genes coded for ribosomal proteins, followed by ribosome biogenesis and translation factors then RNA binding and processing factors (Figure 3C). Host gene expression levels mildly correlated with their embedded snoRNAs but this relation was lost when snoRNA expression levels were considered for individual host gene functions (Figure 3D, Supplementary Figures S6A and S6B). While the existence of isoforms or the C and D box conservation scores had no impact on snoRNA expression levels, snoRNAs targeting 28S were significantly more expressed than orphans or those targeting 18S. In parallel, those targeting 2 Nm sites tended to be less expressed, particularly in head tissue (Supplementary Figure S6D). In summary, among the features we examined, only gene location and target rRNA were associated to different expression levels in all three tissues included in this work.
In line with previous work, we found that the depletion of Fib led to a significant downregulation of 40 C/D box snoRNAs (FDR < 0.05) and to a visible downregulation of most of the remaining expressed C/D box snoRNA transcripts (Figure 3E). In contrast, H/ACA snoRNAs that are often co-expressed in neighbouring introns, remained unaffected and are globally more highly expressed than C/D box snoRNAs (Figure 3E). Interestingly, two uncharacterised ncRNAs, snmRNA:CR43603 and lncRNA:CR43604, were also downregulated. This lncRNA is covered by two distinct blocks of reads (Supplementary Figure S5E). The C/D boxes and the ASE are identified in the second block. By screening for snoRNA features in these downregulated genes, we found that the lncRNA CR43604 is predicted to target 18S-Am441, a Nm site that is conserved and partially methylated in all other model organisms, for which no canonical snoRNA was predicted so far (Supplementary Figure S5F).
The final step was to match each snoRNA to a target nucleotide in rRNAs. For this purpose various prediction tools have been developed over the years such as snoScan (44). Briefly, we favoured the prediction with the higher score (detailed pipeline in Supplementary Figure S7 and Supplementary Methods). When multiple rRNA positions were predicted for a given snoRNA, the secondary prediction was also kept if it matched a consecutive confident Nm site. As a result, we de novo predicted that most of the expressed C/D box snoRNAs (96/106) can target at least one confident Nm site (Table 1 and Supplementary Table S6). From the 12 non-expressed snoRNAs, only 3 were assigned with a Nm prediction (Supplementary Table S6). In total we predicted a new primary or secondary target for 32 snoRNAs and re-assigned 5 of them to a new canonical target (Supplementary Table S6). Furthermore, 74 matched to only one confident Nm site, 10 remained unmatched (‘orphan’) while 22 snoRNAs were predicted to target at least 2 Nm sites on rRNAs (Figure 3F). In sum, the combination of the RMS and of TGIRT-seq data led to the prediction of 33 new snoRNA:rRNA matches (Supplementary Table S6) as well as the attribution of at least one snoRNA to 58 out of the 61 Nm sites (Figure 3G).
rRNA Nm levels are relatively stable during stress conditions
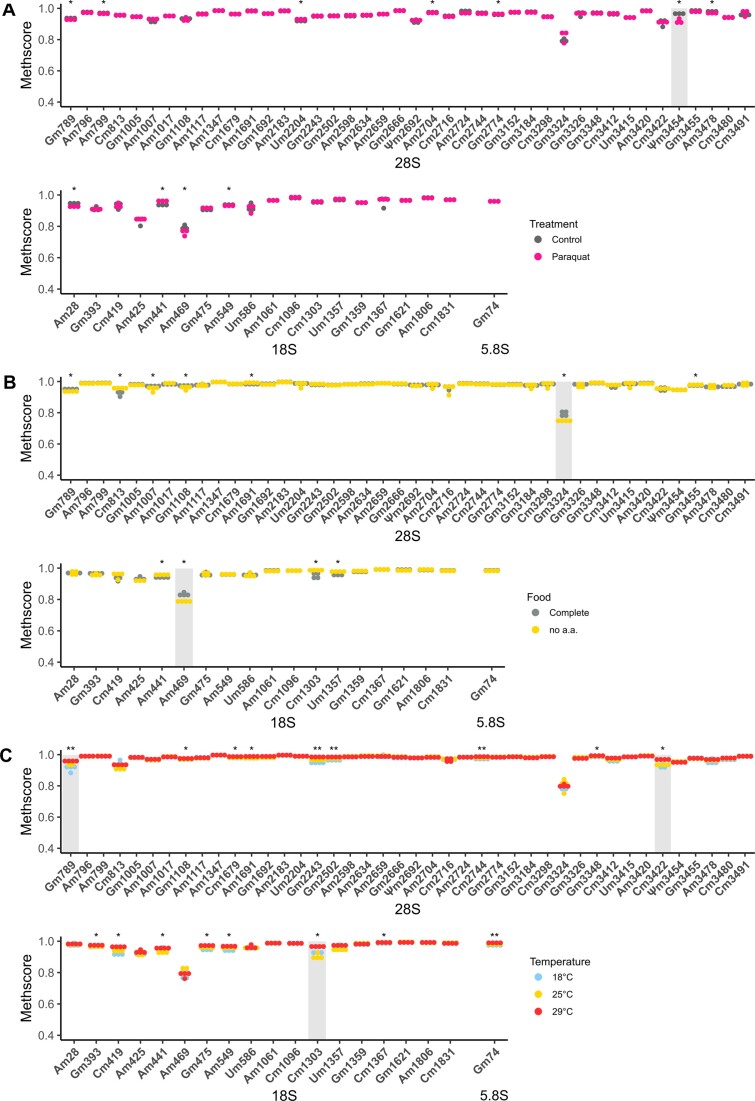
RNA modifications can be dynamically regulated upon different stress and developmental conditions. Although several reports have shown that rRNA Nm modifications are variable in different cell types and developmental stages, no study has quantified Nm levels in response to long environmental stresses (19,53,54). Having built a list of confident Nm sites we therefore sought to explore whether this variability can be observed in D. melanogaster subjected to different stresses. We selected environmental stresses that affect ribosome composition or abundance and we first challenged female flies to paraquat, a well-known herbicide inducing oxidative stress in cells (55). Flies were fed on medium containing paraquat up to 8 days, to allow enough time for turnover of existing ribosomes (56). Total RNA from heads was isolated and submitted to RMS. We first noticed that all Nm sites identified in S2R+ cells were found in wildtype Canton-S flies. Overall, we found very little variation on Nm levels in paraquat treated flies versus control conditions (Figure 4A) as almost no Nm site varied more than 0.02 in MethScore value. Only Ψm3454 in 28S rRNA showed a mild reduction of 0.05 in MethScore. Thus, we concluded from this experiment that oxidative stress conditions have negligible impact on the establishment and maintenance of Nm on rRNA in D. melanogaster.
Figure 4.
Nm levels in rRNAs from heads of female Canton-S individuals that were exposed (A) to paraquat over 8 days (n = 3); (B) that were fed with complete food medium or amino-acid-depleted and low sugar diet over 10 days (n = 4) and (C) that were exposed to 18°C, 25°C or 29°C over 8 days (n = 4). Asterisks indicate the significance of methylation changes for each Nm site (Kruskal–Wallis test, ** P< 0.01, * P< 0.05 and NS P> 0.05). Nm sites with an average ΔMethscore ≥0.05 are highlighted in grey.
We next wondered whether this absence of variability also holds true during other stress conditions. We fed adult females on low nutrient food (no amino acid and 0.25X sugar) for 10 days and monitored the Nm levels on total RNA isolated from heads (27). Again, Nm levels were very stable, despite numerous sites showing a significant difference in nutrient-deprived flies in comparison to controls (Figure 4B). Nevertheless, two sites 28S-Gm3324 and 18S-Am469 were both hypomethylated by 0.05 in the nutrient-deprived group. Lastly, we challenged the flies to different temperatures and repeated the RMS assay. This time, 18 sites showed differential Nm methylation (Figure 4C). While this number was higher compared to the other stresses, the extent of variation was again relatively mild. Among these 18 sites 28S-Gm789, 28S-Cm3422 and 18S-Cm1303 were differentially methylated by at least 0.05. Interestingly flies grown at 29°C had systematically higher Nm levels while those grown at 18°C tended to have lower Nm levels.
In conclusion, these experiments indicate that Nm on rRNA is quite stable and relatively insensitive to environmental changes. In addition, it appears that the little variations are distinct depending of the stress that was applied.
Discussion
Along with pseudouridine, Nm is the most abundant chemical modification found on rRNAs. Together with post-translational modifications and paralogs of ribosomal proteins, rRNA modifications contribute to the emerging field of ribosome heterogeneity (57,58). Several studies show the importance of individual Nm modifications on translation in mammalian cell models as well as on fitness in yeast and archaea (22,59–61). Nevertheless, much less is known on how differences in Nm between tissues or environmental conditions shape the proteome and therefore the identity of a cell. Drosophila melanogaster provides many benefits for studying the role of snoRNAs and of Nm sites on ribosomes because a) it is a multicellular organism and b) has a relatively short lifespan both of which allow to study snoRNA expression and Nm levels in a spatiotemporal manner; c) it presents limited snoRNA gene duplications in comparison to mammals, which makes snoRNA knockouts functional (52) and finally d) a multitude of genetic tools exist for spatiotemporal re-activation of silenced genes.
To fully profit of these aspects, we started by mapping Nm sites on rRNA using two orthogonal techniques: RiboMethSeq and Nanopore DRS. This yielded a list of 61 Nm sites, 50 of which are also conserved in human (Figure 1F). In parallel, we sequenced snoRNAs using TGIRT-seq, a customized NGS-based library particularly fit for small and complex small RNAs (Figure 3A–E) (41). This data allowed us to faithfully validate expressed snoRNAs, to curate their coordinates, to uniformly predict rRNA targets and to flag snoRNAs that come from likely wrong annotations. The set of confident C/D box snoRNAs were renamed to resemble the human nomenclature (Table 1 and Supplementary Table S6). Finally, we saw that Nm levels are overall very stable in response to environmental stresses with a few exceptions (Figure 4A–C).
First, we applied a series of filters to RMS data to identify 61 Nm sites in D. melanogaster. These briefly consisted into filtering MethScores that are higher than a non-methylated control, up-ranking conserved sites, matching snoRNAs and being responsive to Fib (Figure 1A). This was completed by the validation of a large majority of sites by Nanopore. Although we found that the potency of Nanopore DRS for de novo discovery of Nm sites was limited, we showed nevertheless that Nanopore was an excellent tool for validating RMS hits. We believe our overall conservative approach is yet sensitive enough since the measurements of bulk Am, Cm, Gm, and Um meet the expected Nm:Ntotal ratio for three different species (Figure 1G, Supplementary Figure S1D). Surprisingly, the Nm levels could not be explained by the abundance of corresponding snoRNA, neither in steady nor in Fib KD conditions. In a steady state of S2R+ culture, Nm sites are in majority hypermethylated while snoRNA levels are highly variable (Figure 1C, Supplementary Figure S8A). Following Fib KD, the decrease of both Nm levels and matching snoRNA levels also did not correlate (Supplementary Figure S8B). At last, the initial level of snoRNA abundance could not explain the Nm drop either (Supplementary Figure S8C). We tried to address more parameters that could explain this heterogeneous response to Fib KD, such as the conservation of target Nm sites, their secondary structure context, or the order in which they are methylated during ribogenesis, but none of them gave a clear answer. Although the order of Nm deposition has been divided into early (co-transcriptional) and later times (nucleolar) in yeast, the precise order of deposition remains approximate (62). As the knowledge of function of individual Nm sites per se is very limited (22,59), our map of Nm sites in D. melanogaster combined to that of other model organisms is setting the base for future functional studies (Supplementary Table S4).
C/D box snoRNAs expression levels are also heterogeneous both across transcripts (4-digit difference between lowest and highest expressed snoRNA) and within each transcript in response to Fib KD (from no difference up to 2 fold-change). It has been recently shown in humans that the expression of the host gene per se is an indicator of snoRNA expression and we also observe this in D. melanogaster (52). Consistent with humans we also observe no linear correlation between host gene and snoRNA expression levels. Although snoRNAs are co-transcribed in the same transcript as their host gene, they have in most cases a much higher abundance than that of its host gene. Therefore, snoRNAs seem to have higher stability than their hosts, which might be explained by innate features or by their function(s). This might be the case for instance for snoRNAs targeting 28S rRNA which are in average more abundant than those targeting 18S rRNA (Supplementary Figure S6C). However, once snoRNAs are segregated by the number of Nm targets, it becomes visible that (a) 2-Nm-targeting snoRNAs mostly associate with 28S rRNA, (b) they are less abundant than 1-Nm targeting snoRNAs while (c) 1-Nm-targeting snoRNAs that target 18S rRNA are less expressed than those targeting 28S rRNA (Supplementary Figure S6D). We do not have a simple explanation for these observations at the moment. One hypothesis is that different RNP may associate with snoRNA on the 18S versus 28S, which would differently affect their stability. The same would be true for 1-Nm versus 2 Nm targeting snoRNAs. An alternative hypothesis is that these snoRNAs could be more often involved in non-canonical functions (e.g. mRNA splicing, miRNA and piRNA precursor, Nm on tRNAs), requiring therefore a higher abundance of transcripts (63–66). More experiments would be required to test these possibilities.
The only way currently to study the function of individual Nm sites is through the depletion or knock-out of their corresponding snoRNAs. While both are feasible in D. melanogaster, the annotation of C/D box snoRNAs and their rRNA targets were outdated. We therefore curated the coordinates of expressed snoRNAs and improved considerably rRNA target predictions in a traceable manner. We used only snoScan to predict canonical rRNA targets, yet we find it to be the most complete tool as it covers the limitations of other tools: snoReport looks for C/D box motifs in stringent positions and structure constraints, PLEXY requires knowing aforehand the location and sequence of D/D’ boxes and snoGlobe seeks only for anti-sense element complementarity, but not for C/D boxes (67–69). Here, we propose bona fide predictions for 106 C/D box snoRNAs, a quarter of which were predicted to be an exclusive match for 27 individual Nm sites. This feature is particularly interesting for generating single-snoRNA mutants and studying the demethylation of single Nm sites. The 12 snoRNA detected in previous work but not ours are perhaps not expressed in the tissues and developmental stages we examined or not covered enough by our sequencing (Figure 3A). As for the remaining non-expressed 27 genes, there is no evidence of expression in the literature and they have now been withdrawn from the current FlyBase annotation (Figure 3A). Finally, our de novo search for non-annotated snoRNAs yielded only one candidate, snoRNA:CD78, that could be a lncRNA-derived snoRNA (70). We cannot exclude that future tools or different parameters could reveal new C/D box snoRNAs in D. melanogaster.
The canonical role of snoRNAs is guiding chemical modifications on rRNAs. Based on this, about two-thirds of C/D box snoRNAs in D. melanogaster were named after the main rRNA target-nucleotide, the annotation of which has been updated since the last snoRNA naming. Because there is growing evidence that snoRNAs are involved in the modification of other RNAs and mechanisms (64,68,71) but also because we generated more consistent rRNA target predictions, we saw the need to propose a harmonised and less rRNA-centered nomenclature. The latter is free of any association to specific targets and only reveals the class it belongs to through the prefix ‘snoRNA:CD’. As this prefix is followed by a numeric-based index, it can easily be extended in the future by any new snoRNA gene or isoform (Table 1). In addition, we did not wish to pick the same index as human for conserved snoRNAs as there is no strict 1-to-1 match for all snoRNAs. Instead, our numbering of D. melanogaster snoRNAs follows their order of publication. Both the new nomenclature and revised coordinates will be integrated into FlyBase.
As part of other modulable features that contribute to ribosome heterogeneity, rRNA modifications are also thought to shape the translatome (72). This specialisation has been observed during development and to a lesser extent in response to environmental cues (73,74). rRNA modifications are perhaps not surprisingly resistant to the latter (36,75). Given they are mostly deposited during ribosome biogenesis and that their half-life is in the order of several hours in bacteria and 3–5 days in mammalian cells, a certain degree of ribosomal turnover must occur to observe changes (BioNumbers 108025, 108023, 110053) (56,76,77). In addition, there are no enzyme known to erase rRNA modifications. Based on literature, we selected environmental stresses that affect ribosome composition or abundance. So far, heat is the only stress known to affect rRNA modifications, particularly in T. kodakarensis where the number of acetylated cytidines increases dramatically when these archaea are grown in 85°C instead of 55°C (61). Other stresses such as acute nutrient deprivation or oxidative stress can affect RNA modifications indirectly by reducing the number of ribosomes or damaging their integrity, respectively (78,79). With these examples in mind, we tested oxidative, nutrient and heat stresses for 8 to 10 days, a duration that is sufficient for significant renewal of the ribosome pool. The mild difference observed in 6 Nm sites across all stresses could not be linked neither to a change of the corresponding snoRNA expression, nor to the expression of the stand-alone CG8939 (data not shown). This very limited variation of Nm levels indicate that Nm modification machinery is very resistant to ambient stresses in D. melanogaster. Yet the significant variation of certain Nm sites we observed might serve as a functional response to adapt to the environment.
In conclusion, our work opens the door to studying the role of rRNA Nm sites at the whole organism level. With a confident map of Nm sites and a better characterized set of matching snoRNAs, it will become feasible to individually manipulate Nm sites and study their function in specific tissues and developmental stages.
Supplementary Material
Acknowledgements
We thank members of the Roignant, Motorin and Novoa labs for helpful discussion as well as Nuria Bosch Guiteras for providing us with S. cerevisiae strain S288c (https://www.yeastgenome.org/strain/s288c). Special thanks are also given to Ryan Nottingham from the Lambowitz Lab for his precious advice on the TGIRT-seq protocol. We acknowledge the support of the MEIC to the EMBL partnership, Centro de Excelencia Severo Ochoa and CERCA Programme/Generalitat de Catalunya.
Contributor Information
Athena Sklias, Center for Integrative Genomics, Faculty of Biology and Medicine, University of Lausanne, 1015 Lausanne, Switzerland.
Sonia Cruciani, Center For Genomic Regulation (CRG), The Barcelona Institute of Science and Technology, Dr Aiguader 88, 08003 Barcelona, Spain.
Virginie Marchand, Université de Lorraine, CNRS, INSERM, Epitranscriptomics and RNA sequencing (EpiRNA-Seq) Core Facility (UAR2008/US40 IBSLor) and UMR7365 IMoPA, Nancy, France.
Mariangela Spagnuolo, Institute of Pharmaceutical and Biomedical Sciences, Johannes Gutenberg-University Mainz, Staudingerweg 5, 55128 Mainz, Germany.
Guillaume Lavergne, Center for Integrative Genomics, Faculty of Biology and Medicine, University of Lausanne, 1015 Lausanne, Switzerland.
Valérie Bourguignon, Université de Lorraine, CNRS, INSERM, Epitranscriptomics and RNA sequencing (EpiRNA-Seq) Core Facility (UAR2008/US40 IBSLor) and UMR7365 IMoPA, Nancy, France.
Alessandro Brambilla, Proteomics and Modomics Experimental Core (PROMEC), Norwegian University of Science and Technology and the Central Norway Regional Health Authority, Trondheim, Norway.
René Dreos, Center for Integrative Genomics, Faculty of Biology and Medicine, University of Lausanne, 1015 Lausanne, Switzerland.
Steven J Marygold, FlyBase, Department of Physiology, Development and Neuroscience, University of Cambridge, Downing Street, Cambridge, United Kingdom.
Eva Maria Novoa, Center For Genomic Regulation (CRG), The Barcelona Institute of Science and Technology, Dr Aiguader 88, 08003 Barcelona, Spain; University Pompeu Fabra (UPF), Dr Aiguader 88, 08003 Barcelona, Spain.
Yuri Motorin, Université de Lorraine, CNRS, INSERM, Epitranscriptomics and RNA sequencing (EpiRNA-Seq) Core Facility (UAR2008/US40 IBSLor) and UMR7365 IMoPA, Nancy, France.
Jean-Yves Roignant, Center for Integrative Genomics, Faculty of Biology and Medicine, University of Lausanne, 1015 Lausanne, Switzerland; Institute of Pharmaceutical and Biomedical Sciences, Johannes Gutenberg-University Mainz, Staudingerweg 5, 55128 Mainz, Germany.
Data availability
Basecalled FAST5 of nanopore direct RNA sequencing runs have been deposited to ENA, under accession code PRJEB45722. Predicted rRNA modifications in D. melanogaster (based on homology to known human rRNA modifications), EpiNano RNA modification scores, as well as the code to perform EpiNano score analysis per k-mer and de novo discovery of Nm sites, can be found in Zenodo (https://zenodo.org/record/8271283). TGIRT-seq fastq files and read counts obtained with CoCo are publicly available in NCBI’s Gene Expression Omnibus (GEO) and are accessible under GEO Series accession number GSE233853 and GSE241399. RiboMethSeq fastq files and score tables are accessible under GSE241400.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Research in the laboratory of J.Y.R. is supported by the University of Lausanne, the Swiss National Science Foundation [310030_197906]; Deutsche Forschungsgemeinschaft [RO 4681/9-1, RO 4681/12-1, RO 4681/13-1, TRR319 RMaP]; Y.M. was supported by FRCR EpiARN from Grand Est Région, France. EMN was supported by funds from the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) [PID2021-128193NB-100 to E.M.N.]; European Research Council [ERC-StG-2021 No 101042103 to E.M.N.]; S.C. was supported by ‘la Caixa’ InPhINIT PhD fellowship [LCF/BQ/DI19/11730036]; Centro de Excelencia Severo Ochoa funding.
Conflict of interest statement. None declared.
References
- 1. Klinge S., Woolford J.L.. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019; 20:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu K.L., Nelson J.O., Watase G.J., Warsinger-Pepe N., Yamashita Y.M.. Transgenerational dynamics of rDNA copy number in Drosophila male germline stem cells. eLife. 2018; 7:e32421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miller O.L., Beatty B.R.. Visualization of nucleolar genes. Science. 1969; 164:955–957. [DOI] [PubMed] [Google Scholar]
- 4. Mullineux S.-T., Lafontaine D.L.J.. Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand?. Biochimie. 2012; 94:1521–1532. [DOI] [PubMed] [Google Scholar]
- 5. Rodgers M.L., Woodson S.A.. A Roadmap for rRNA Folding and Assembly during Transcription. Trends in Biochemical Sciences. 2021; 46:889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Natchiar S.K., Myasnikov A.G., Hazemann I., Klaholz B.P.. Visualizing the role of 2’-OH rRNA methylations in the human ribosome structure. Biomolecules. 2018; 8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao Y., Rai J., Yu H., Li H.. CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations. Structure. 2022; 30:983–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharma S., Marchand V., Motorin Y., Lafontaine D.L.J.J.. Identification of sites of 2′-O-methylation vulnerability in human ribosomal RNAs by systematic mapping. Sci. Rep. 2017; 7:11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jansen R.P., Hurt E.C., Kern H., Lehtonen H., Carmo-Fonseca M., Lapeyre B., Tollervey D.. Evolutionary conservation of the human nucleolar protein fibrillarin and its functional expression in yeast. J. Cell Biol. 1991; 113:715–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lapinaite A., Simon B., Skjaerven L., Rakwalska-Bange M., Gabel F., Carlomagno T.. The structure of the box C/D enzyme reveals regulation of RNA methylation. Nature. 2013; 502:519–523. [DOI] [PubMed] [Google Scholar]
- 11. Kufel J., Grzechnik P.. Small nucleolar RNAs tell a different tale. Trends Genet. 2019; 35:104–117. [DOI] [PubMed] [Google Scholar]
- 12. Talross G.J.S., Deryusheva S., Gall J.G.. Stable lariats bearing a snoRNA (slb-snoRNA) in eukaryotic cells: a level of regulation for guide RNAs. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2114156118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grzechnik P., Szczepaniak S.A., Dhir S., Pastucha A., Parslow H., Matuszek Z., Mischo H.E., Kufel J., Proudfoot N.J.. Nuclear fate of yeast snoRNA is determined by co-transcriptional Rnt1 cleavage. Nat. Commun. 2018; 9:1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tollervey D., Kiss T.. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 1997; 9:337–342. [DOI] [PubMed] [Google Scholar]
- 15. Rothé B., Manival X., Rolland N., Charron C., Senty-Ségault V., Branlant C., Charpentier B.. Implication of the box C/D snoRNP assembly factor Rsa1p in U3 snoRNP assembly. Nucleic Acids Res. 2017; 45:7455–7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang Z., Lin J., Ye K.. Box C/D guide RNAs recognize a maximum of 10 nt of substrates. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:10878–10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kiss-László Z., Henry Y., Bachellerie J.-P., Caizergues-Ferrer M., Kiss T.. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996; 85:1077–1088. [DOI] [PubMed] [Google Scholar]
- 18. Sloan K.E., Warda A.S., Sharma S., Entian K.D., Lafontaine D.L.J., Bohnsack M.T.. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017; 14:1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Motorin Y., Quinternet M., Rhalloussi W., Marchand V.. Constitutive and variable 2’-O-methylation (Nm) in human ribosomal RNA. RNA Biol. 2021; 18:88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bergeron D., Paraqindes H., Fafard-Couture É., Deschamps-Francoeur G., Faucher-Giguère L., Bouchard-Bourelle P., Abou Elela S., Catez F., Marcel V., Scott M.S.. snoDB 2.0: an enhanced interactive database, specializing in human snoRNAs. Nucleic Acids Res. 2023; 51:D291–D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Erales J., Marchand V., Panthu B., Gillot S., Belin S., Ghayad S.E., Garcia M., Laforêts F., Marcel V., Baudin-Baillieu A.et al.. Evidence for rRNA 2′-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:12934–12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jansson M.D., Häfner S.J., Altinel K., Tehler D., Krogh N., Jakobsen E., Andersen J.V., Andersen K.L., Schoof E.M., Ménard P.et al.. Regulation of translation by site-specific ribosomal RNA methylation. Nat. Struct. Mol. Biol. 2021; 28:889–899. [DOI] [PubMed] [Google Scholar]
- 23. He F., James A., Raje H., Ghaffari H., DiMario P.. Deletion of Drosophila Nopp140 induces subcellular ribosomopathies. Chromosoma. 2015; 124:191–208. [DOI] [PubMed] [Google Scholar]
- 24. Motorin Y., Marchand V.. Detection and analysis of RNA ribose 2′-O-methylations: challenges and solutions. Genes (Basel). 2018; 9:642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krogh N., Nielsen H.. Sequencing-based methods for detection and quantitation of ribose methylations in RNA. Methods. 2019; 156:5–15. [DOI] [PubMed] [Google Scholar]
- 26. Garalde D.R., Snell E.A., Jachimowicz D., Sipos B., Lloyd J.H., Bruce M., Pantic N., Admassu T., James P., Warland A.et al.. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods. 2018; 15:201–206. [DOI] [PubMed] [Google Scholar]
- 27. Doroszuk A., Jonker M.J., Pul N., Breit T.M., Zwaan B.J.. Transcriptome analysis of a long-lived natural Drosophila variant: a prominent role of stress- and reproduction-genes in lifespan extension. Bmc Genomics [Electronic Resource]. 2012; 13:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klepsatel P., Gáliková M., Xu Y., Kühnlein R.P.. Thermal stress depletes energy reserves in Drosophila. Sci. Rep. 2016; 6:33667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marchand V., Blanloeil-Oillo F., Helm M., Motorin Y.. Illumina-based RiboMethSeq approach for mapping of 2′-O-me residues in RNA. Nucleic Acids Res. 2016; 44:e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J.et al.. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011; 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith M.A., Ersavas T., Ferguson J.M., Liu H., Lucas M.C., Begik O., Bojarski L., Barton K., Novoa E.M.. Molecular barcoding of native RNAs using nanopore sequencing and deep learning. Genome Res. 2020; 30:1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cozzuto L., Liu H., Pryszcz L.P., Pulido T.H., Delgado-Tejedor A., Ponomarenko J., Novoa E.M.. MasterOfPores: a workflow for the analysis of Oxford nanopore direct RNA sequencing datasets. Front. Genet. 2020; 11:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu H., Begik O., Lucas M.C., Ramirez J.M., Mason C.E., Wiener D., Schwartz S., Mattick J.S., Smith M.A., Novoa E.M.. Accurate detection of m6A RNA modifications in native RNA sequences. Nat. Commun. 2019; 10:4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Begik O., Lucas M.C., Pryszcz L.P., Ramirez J.M., Medina R., Milenkovic I., Cruciani S., Liu H., Vieira H.G.S., Sas-Chen A.et al.. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat. Biotechnol. 2021; 39:1278–1291. [DOI] [PubMed] [Google Scholar]
- 37. Boivin V., Deschamps-Francoeur G., Couture S., Nottingham R.M., Bouchard-Bourelle P., Lambowitz A.M., Scott M.S., Abou-Elela S.. Simultaneous sequencing of coding and noncoding RNA reveals a human transcriptome dominated by a small number of highly expressed noncoding genes. 2018; 24:950–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10–12. [Google Scholar]
- 39. Boivin V., Reulet G., Boisvert O., Couture S., Elela S.A., Scott M.S.. Reducing the structure bias of RNA-seq reveals a large number of non-annotated non-coding RNA. Nucleic Acids Res. 2020; 48:2271–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deschamps-Francoeur G., Boivin V., Abou Elela S., Scott M.S.. CoCo: rNA-seq read assignment correction for nested genes and multimapped reads. Bioinformatics. 2019; 35:5039–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2016; NY: Springer-Verlag. [Google Scholar]
- 44. Lowe T.M., Eddy S.R.. A computational screen for methylation guide snoRNAs in yeast. Science. 1999; 283:1168–1171. [DOI] [PubMed] [Google Scholar]
- 45. Natsidis P., Schiffer P.H., Salvador-Martínez I., Telford M.J.. Computational discovery of hidden breaks in 28S ribosomal RNAs across eukaryotes and consequences for RNA integrity numbers. Sci. Rep. 2019; 9:19477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pavlakis G., Wurst R., Vournakis J.. Russell T.R., Brew K., Faber H., Schultz J.. Identification of Drosophila 2S rRNA as the 3′-part of 5.8S rRNA. From Gene to Protein: Information Transfer in Normal and Abnormal Cells. 1979; Academic Press; 617. [Google Scholar]
- 47. Azevedo-Favory J., Gaspin C., Ayadi L., Montacié C., Marchand V., Jobet E., Rompais M., Carapito C., Motorin Y., Sáez-Vásquez J.. Mapping rRNA 2’-O-methylations and identification of C/D snoRNAs in Arabidopsis thaliana plants. RNA Biol. 2021; 18:1760–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Simabuco F.M., Morello L.G., Aragão A.Z.B., Paes Leme A.F., Zanchin N.I.T.. Proteomic characterization of the Human FTSJ3 preribosomal complexes. J. Proteome Res. 2012; 11:3112–3126. [DOI] [PubMed] [Google Scholar]
- 49. Khoshnevis S., Dreggors-Walker R.E., Marchand V., Motorin Y., Ghalei H.. Ribosomal RNA 2′-O-methylations regulate translation by impacting ribosome dynamics. Proc. Natl. Acad. Sci. U.S.A. 2022; 119:e2117334119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hopes T., Norris K., Agapiou M., McCarthy C.G.P., Lewis P.A., O’Connell M.J., Fontana J., Aspden J.L. Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching. Nucleic Acids Res. 2022; 50:2240–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Anger A.M., Armache J.-P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R.. Structures of the human and Drosophila 80S ribosome. Nature. 2013; 497:80–85. [DOI] [PubMed] [Google Scholar]
- 52. Fafard-Couture É., Jacques P.-É., Scott M.S.. Motif conservation, stability, and host gene expression are the main drivers of snoRNA expression across vertebrates. Genome Res. 2023; 33:540–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramachandran S., Krogh N., Jørgensen T.E., Johansen S.D., Nielsen H., Babiak I.. The shift from early to late types of ribosomes in zebrafish development involves changes at a subset of rRNA 2′-O-me sites. RNA. 2020; 26:1919–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Häfner S.J., Jansson M.D., Altinel K., Kraushar M.L., Andersen K.L., Abay-Nørgaard C., Fontenas M., Sørensen D.M., Gay D.M., Arendrup F.S.et al.. Ribosomal RNA 2’-O-methylation dynamics impact cell fate decisions. Developmental Cell. 2022; 58:1593–1609. [DOI] [PubMed] [Google Scholar]
- 55. Shcherbik N., Pestov D.G.. The impact of oxidative stress on ribosomes: from injury to regulation. Cells. 2019; 8:1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Milo R., Jorgensen P., Moran U., Weber G., Springer M.. BioNumbers–the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010; 38:D750–D753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Georgeson J., Schwartz S.. The ribosome epitranscriptome: inert—Or a platform for functional plasticity?. RNA. 2021; 27:1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Milenkovic I., Santos Vieira H.G., Lucas M.C., Ruiz-Orera J., Patone G., Kesteven S., Wu J., Feneley M., Espadas G., Sabidó E.et al.. Dynamic interplay between RPL3- and RPL3L-containing ribosomes modulates mitochondrial activity in the mammalian heart. Nucleic Acids Res. 2023; 51:5301–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liang X.H., Liu Q., Fournier M.J.. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009; 15:1716–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Esguerra J., Warringer J., Blomberg A.. Functional importance of individual rRNA 2′-O-ribose methylations revealed by high-resolution phenotyping. RNA. 2008; 14:649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sas-Chen A., Thomas J.M., Matzov D., Taoka M., Nance K.D., Nir R., Bryson K.M., Shachar R., Liman G.L.S., Burkhart B.W.et al.. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature. 2020; 583:638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Birkedal U., Christensen-Dalsgaard M., Krogh N., Sabarinathan R., Gorodkin J., Nielsen H.. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. 2015; 54:451–455. [DOI] [PubMed] [Google Scholar]
- 63. Bergeron D., Faucher-Giguère L., Emmerichs A.-K., Choquet K., Song K.S., Deschamps-Francoeur G., Fafard-Couture É., Rivera A., Couture S., Churchman L.S.et al.. Intronic small nucleolar RNAs regulate host gene splicing through base pairing with their adjacent intronic sequences. Genome Biol. 2023; 24:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bratkovič T., Božič J., Rogelj B.. Functional diversity of small nucleolar RNAs. Nucleic Acids Res. 2019; 48:1627–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Falaleeva M., Welden J.R., Duncan M.J., Stamm S.. C/D-box snoRNAs form methylating and non-methylating ribonucleoprotein complexes: old dogs show new tricks. Bioessays. 2017; 39: 10.1002/bies.201600264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang M., Li K., Bai J., Van Damme R., Zhang W., Alba M., Stiles B.L., Chen J.-F., Lu Z.. A snoRNA–tRNA modification network governs codon-biased cellular states. Proc. Natl. Acad. Sci. U.S.A. 2023; 120:e2312126120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. de Araujo Oliveira J.V., Costa F., Backofen R., Stadler P.F., Machado Telles Walter M.E., Hertel J.. SnoReport 2.0: new features and a refined Support Vector machine to improve snoRNA identification. BMC Bioinf. 2016; 17:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Deschamps-Francoeur G., Couture S., Abou-Elela S., Scott M.S.. The snoGloBe interaction predictor reveals a broad spectrum of C/D snoRNA RNA targets. Nucleic Acids Res. 2022; 50:6067–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kehr S., Bartschat S., Stadler P.F., Tafer H.. PLEXY: efficient target prediction for box C/D snoRNAs. Bioinformatics. 2011; 27:279–280. [DOI] [PubMed] [Google Scholar]
- 70. Shi J., Zhou T., Chen Q.. Exploring the expanding universe of small RNAs. Nat. Cell Biol. 2022; 24:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Han C., Sun L.-Y., Luo X.-Q., Pan Q., Sun Y.-M., Zeng Z.-C., Chen T.-Q., Huang W., Fang K., Wang W.-T.et al.. Chromatin-associated orphan snoRNA regulates DNA damage-mediated differentiation via a non-canonical complex. Cell Rep. 2022; 38:110421. [DOI] [PubMed] [Google Scholar]
- 72. Genuth N.R., Barna M.. Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat. Rev. Genet. 2018; 19:431–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hebras J., Krogh N., Marty V., Nielsen H., Cavaillé J.. Developmental changes of rRNA ribose methylations in the mouse. RNA Biol. 2020; 17:150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Delhermite J., Tafforeau L., Sharma S., Marchand V., Wacheul L., Lattuca R., Desiderio S., Motorin Y., Bellefroid E., Lafontaine D.L.J.J.. Systematic mapping of rRNA 2’-O methylation during frog development and involvement of the methyltransferase fibrillarin in eye and craniofacial development in Xenopus laevis. PLoS Genet. 2022; 18:e1010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bailey A.D., Talkish J., Ding H., Igel H., Duran A., Mantripragada S., Paten B., Ares M.. Concerted modification of nucleotides at functional centers of the ribosome revealed by single-molecule RNA modification profiling. eLife. 2022; 11:e76562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Defoiche J., Zhang Y., Lagneaux L., Pettengell R., Hegedus A., Willems L., Macallan D.C.. Measurement of ribosomal RNA turnover In vivo by use of deuterium-labeled glucose. Clin. Chem. 2009; 55:1824–1833. [DOI] [PubMed] [Google Scholar]
- 77. Nikolov E.N., Dabeva M.D., Nikolov T.K.. Turnover of ribosomes in regenerating rat liver. Int. J. Biochem. 1983; 15:1255–1260. [DOI] [PubMed] [Google Scholar]
- 78. An H., Ordureau A., Körner M., Paulo J.A., Harper J.W.. Systematic quantitative analysis of ribosome inventory during nutrient stress. Nature. 2020; 583:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shedlovskiy D., Zinskie J.A., Gardner E., Pestov D.G., Shcherbik N.. Endonucleolytic cleavage in the expansion segment 7 of 25S rRNA is an early marker of low-level oxidative stress in yeast. J. Biol. Chem. 2017; 292:18469–18485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Basecalled FAST5 of nanopore direct RNA sequencing runs have been deposited to ENA, under accession code PRJEB45722. Predicted rRNA modifications in D. melanogaster (based on homology to known human rRNA modifications), EpiNano RNA modification scores, as well as the code to perform EpiNano score analysis per k-mer and de novo discovery of Nm sites, can be found in Zenodo (https://zenodo.org/record/8271283). TGIRT-seq fastq files and read counts obtained with CoCo are publicly available in NCBI’s Gene Expression Omnibus (GEO) and are accessible under GEO Series accession number GSE233853 and GSE241399. RiboMethSeq fastq files and score tables are accessible under GSE241400.