Abstract
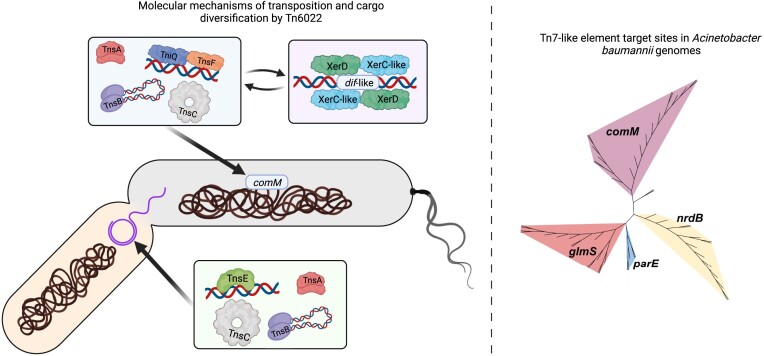
Mobile genetic elements play an important role in the acquisition of antibiotic and biocide resistance, especially through the formation of resistance islands in bacterial chromosomes. We analyzed the contribution of Tn7-like transposons to island formation and diversification in the nosocomial pathogen Acinetobacter baumannii and identified four separate families that recognize different integration sites. One integration site is within the comM gene and coincides with the previously described Tn6022 elements suggested to account for the AbaR resistance island. We established Tn6022 in a heterologous E. coli host and confirmed basic features of transposition into the comM attachment site and the use of a novel transposition protein. By analyzing population features within Tn6022 elements we identified two potential novel transposon-encoded diversification mechanisms with this dynamic genetic island. The activities of these diversification features were confirmed in E. coli. One was a novel natural gain-of-activity allele that could function to broaden transposition targeting. The second was a transposon-encoded hybrid dif-like site that parasitizes the host dimer chromosome resolution system to function with its own tyrosine recombinase. This work establishes a highly active Tn7-like transposon that harnesses novel features allowing the spread and diversification of genetic islands in pathogenic bacteria.
Graphical Abstract
Graphical Abstract.
Introduction
Transposons are mobile genetic elements capable of moving between positions in a genome. Tn7 and related transposons are notable for the control they display over target site selection allowing integration into fixed safe sites in the chromosome (1). A type of transposons related to the original Tn7 element, called Tn6022 elements, encode the same core Tn7 functional domains and have been suggested to account for the important AbaR resistance island in Acinetobacter baumnannii (2–4). Prototypic Tn7 uses five proteins to catalyze transposition into different types of target sites, TnsA, TnsB, TnsC, TnsD(TniQ) and TnsE (1). TnsA and TnsB form the heteromeric transposase that moves the element to a new site using a cut-and-paste transposition mechanism (5,6). Tn7 is activated for high frequency transposition when a preferred target site sequence is recognized by the protein TnsD, which contains a TniQ domain (7). TnsC communicates transposition signals between the TnsD(TniQ) protein and the TnsA + TnsB transposase. Diverse Tn7-like elements use the same core domain activities (8,9) but have evolved to recognize different types of target sites that maximize the dispersal of the element while minimizing the chance of inactivating important host genes. The capacity of Tn7-like elements to integrate into conserved positions in bacterial genomes called attachment sites (att sites) allows the formation of genetic islands using a transposition mechanism (10). In most cases, a specific DNA sequence within an essential gene is recognized but transposition is directed outside of the coding region thereby preserving gene functionality. A precise distance between the sequence recognized through TniQ-domain proteins and the point of insertion comes from a ring or filament of a fixed number of TnsC molecules that positions the transposase to integrate the element (11–13). TniQ domain containing proteins have naturally fused to different DNA binding domains or co-opted CRISPR-Cas systems to recognize different essential genes, such as glmS, ffs, parE, guaC, rsmJ and tRNA genes among others (9,14–17). Interestingly, Tn6022 elements are found within the coding sequence of the comM gene. Integration at this position has been noted for some time as inactivating the gene and forming the important AbaR resistance island in A. baumannii (for Acinetobacter baumanniiResistance) before the connection to the transposition mechanism was suggested (18,19).
Tn7-like elements are also known to be important vehicles for spreading genes that are beneficial to the host and element but are not directly related to transposition called cargo genes (20,21). When a transposon inserts into the chromosome, cargo genes under regular selection can persist longer than the transposon itself. For example, the transposition genes and end sequences can become inactivated and degrade, but if the cargo they delivered is under selection, genetic information can persist to form a fitness, pathogenicity, or resistance island (10). In these cases, these genetic islands can form by a transposition-based process but have little or no evidence of the element that allowed their formation. The diversity of cargo and novel configurations of elements related to Tn6022 has been noted by many groups, but the molecular underpinnings of this diversity have remained enigmatic (22–24).
We bioinformatically assessed Tn7-like elements in A. baumannii genome sequences to understand the role these elements play in the formation of resistance islands. This analysis revealed the previously described types of Tn7-like elements targeting the glmS and comM att sites and two types not described previously in A. baumannii targeting parE and nrdB att sites. We found that a specific group of near identical elements within the Tn6022 elements targeting the comM gene was responsible for almost all the diversity of antibiotic resistance in A. baumannii. We established a novel molecular mechanism for Tn7-like transposition involving an accessory protein that functions with TniQ. Two additional genetic features were discovered: a naturally occurring gain-of-activity allele and a novel tyrosine recombinase system. Both of these systems were tested in heterologous Escherichia coli, demonstrating their ability to generate diversity through novel genetic mechanisms.
Materials and methods
Identification of TniQ and tyrosine site-specific recombinase homologs in A. baumannii genomes
Annotated nucleotide (19,282) and protein (67,128,995) FASTA files (Genebank and Refseq) of Acintobacter baumannii genomes (17,469) were downloaded from National Center for Biotechnology Information (NCBI) FTP site (retrieved March 2023). Profile HMMs associated with TniQ (PF06527) downloaded from the European Bioinformatics Institute (EMBL-EBI) Pfam database, were used for detecting homologs by using hmmsearch (HMMER3) with default parameters (25,26). The search generated 2440 hits. A comprehensive non-redundant TniQ (56 protein) sequences was generated by using CD-HIT (https://www.weizhongli-lab.org/cd-hit/) with sequence identity threshold 0.95 (27). These TniQ protein sequences were aligned with the MUSCLE algorithm by using the Neighbor-joining clustering method (maximum number of iterations: 500) with Geneious Prime (v2022.2.1) (28). The aligned sequences were then used to build an approximately-maximum-likelihood phylogeny using RAxML algorithm (29). The tree was displayed with iTOL(v5) (30).
An iterative jackhmmer search (n = 5) was performed against all annotated protein (67,128,995) sequences (Genebank and Refseq) of A. baumannii genomes (17,469) by using host XerC (ABO13046.2) and XerD (EXS72628.1) proteins as the query. Following every cycle of this search, the hit sequences were aligned, and a profile hidden Markov model (HMM) was built. This profile HMM was then used as a new query in the next search cycle. This iterative procedure allows identification of distantly related site-specific tyrosine recombinase homologues of the original query. The resulting sequences (165,907) were then clustered by using CD-HIT (https://www.weizhongli-lab.org/cd-hit/) with sequence identity threshold 0.65, and the representatives (232) of the clusters were aligned as described above. The aligned sequences were then used to build an approximately-maximum-likelihood phylogeny using FastTree algorithm in Geneious Prime (v2022.2.1) (31). The tree was displayed with iTOL (v5) (30).
To determine the structural similarity of the XerC proteins, the structures of XerC-like(Tn6022), XerC (A. baumannii) and XerC (E. coli) were obtained using Alphafold2 (32). The structure comparisons and RMSD values were obtained in ChimeraX using the command Matchmaker (33).
Analyzing insertions in closed genomes of A. baumannii
The coordinates for the glmS and comM att sites were obtained through BLAST searches. To determine transposon insertions within comM we computed the distance between two points (40 bp) that flank the known insertion point for AbaR. To determine insertions downstream of glmS, we computed the distance between glmS and the downstream gene murI. The comM output size of 108 bps or less was considered an empty comM site, the glmS output size of less than 722 bp was considered and empty glmS att site (Supplementary Tables S1 and S2).
To analyze the att sites in A. baumannii the accession of the genomes and coordinates, corresponding nucleotide sequences were extracted from NCBI. A command line software tool Prokka (v 1.14.5) was used for annotation. Genbank format files (.gb) generated by Prokka were visualized with Geneious prime (v2022.2.1) (34). To assess the relatedness of the elements the TnsA and TnsB protein sequences were concatenated and aligned with the MUSCLE algorithm by using the neighbor-joining clustering method (maximum number of iterations: 500) with Geneious Prime (v2022.2.1) (28). The aligned sequences were used to build an approximately-maximum-likelihood phylogeny using RAxML algorithm (29). The tree was displayed with iTOL(v5) (30). WebLogo (v2.8.2) was used for displaying the TnsB binding sites (35).
The A. baumannii similarity tree for closed genomes was constructed with 203 genomes based on seven housekeeping genes (fusA, gltA, groEL, pyrG, recA, rplB, rpoB). Briefly, these seven genes were used to search the closed/complete genomes by BLASTN, and all significant hits were collected. A customized python code was used to remove the duplicates (two or more hits per genome), collect the genome records only with seven genes, and concatenate all sequences for each genome into a single FASTA file (Code available at https://data.mendeley.com/datasets/5nc43tsxg3/1). The nucleotide sequences were aligned with the MUSCLE algorithm by using default parameters (28). Neighbor-joining algorithm in Geneious Prime (v2022.2.1) was used to build a similarity tree and the tree was displayed with iTOL (v5) (30).
To compare among Tn6022 elements (Supplement Figure S2), TBLASTX was performed to compare genomes having different elements. The genomic regions were visualized with EasyFig V2.2.6 and sequence similarities are shown with gradient (grey to black; between 70% and 100%) straight lines (36).
Growth conditions
Escherichia coli strains (Table 1) were grown in lysogeny broth (LB) or on solid LB agar supplemented with the following concentrations of antibiotics and supplements where appropriate unless noted otherwise: 100 μg/ml carbenicillin, (carb), 100 ug/ml trimethoprim, (tmp),10 μg/ml gentamicin, (gen), 30 μg/ml chloramphenicol, (cam), 8 μg/ml tetracycline, (tet), 50 μg/ml kanamycin, (kan), 50 μg/ml spectinomycin, (spec), 20 μg/ml nalidixic acid, (nal), 100 μg/ml rifampicin, (rif), 50 μg/ml X-gal, 0.2% glucose, 0.02% rhamnose, 0.02% arabinose and 0.1mM IPTG.
Table 1.
Bacterial strains used in the study
| Strain | Genotype | Source |
|---|---|---|
| BW25141 | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), Δ(phoB-phoR)580, λ-, galU95, ΔuidA3::pir+, recA1, endA9(del-ins)::FRT, rph-1, Δ(rhaD-rhaB)568, hsdR514 | (72) |
| BW27783 | F−, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, Δ(araH-araF)570(::FRT), ΔaraEp-532::FRT, φPcp8-araE535, rph-1, Δ(rhaD-rhaB)568, hsdR514 | (73) |
| CW51 | F−, ara−, arg-, Δ(lac-pro)XIII, nalR, rifR, recA56 | (74) |
| JY12 | BW27783 attTn7::R-tet-oriR6k-dif-FRT-L-R-kan-oriR10k-dif-FRT-L | This work |
| AC158 | BW27783 attTn7::mini-Tn7(KanR), F ΔfinO-fxsA::glmS- SpecR | This work |
| AC159 | BW27783 attTn7::mini-Tn7(KanR), F ΔfinO-fxsA::lacZ- SpecR | This work |
| AC199 | BW27783 attTn7::mini-Tn7-mini-Tn6022(KanR), F ΔfinO-fxsA:: comM::mini-Tn6022(dhfr)-SpecR | This work |
| AC220 | BW27783 attTn7::mini-Tn7::mini-Tn6022(KanR), F EMG2 ΔfinO-fxsA::comM-SpecR | This work |
Strain and plasmid construction
Transposition and site-specific recombination tester strains were all constructed in E. coli strain BW27783 using standard techniques (Table 1) (37). For monitoring transposition using a mating-out assay a Tn6022 mini-element was situated in the E. coli attTn7 site as described (38). The Tn6022 mini-element consisted of 149 bp from the left end and 155 bp from the right end of the Tn6022 element flanking a gene for kan resistance. For monitoring site-specific recombination, Tn7 mini-elements were produced with genes encoding for kan or tet resistance flanked by the 144 bp from the left end and 70 bp from the right end of Tn7. They also contained the frt site recognized by FLP recombinase and the candidate dif-like site associated with Tn6022 elements in A. baumannii. The elements were constructed as tandem insertions into the attTn7 site using lambda red recombination as described previously (39).
Candidate transposition or site-specific recombination genes were cloned into expression vectors using gene fragments that were ordered or cloned from PCR products amplified from bacterial strains. Final constructs were sequenced verified. Sequences for all vectors are provided in the Supplementary Data Table S1. A minimal conjugal F plasmid was constructed from the original F plasmid in strain EMG2 (CGSC#4401). The minimal F plasmid with the comM target (F-‘comM’) has a 600 bp fragment of comM (‘comM’), 300 bp from each side of the AbaR pathogenicity island. The immune F ‘comM’ was made by recombining an immobile mini-Tn6022 element into the exact site normally used in the comM gene. This element had the same left and right ends as the KanR element but was immobile based on changes at the terminal base pairs in the element (TGT/ACA to ACT/TGA) (40) and was TmpR because it has the dhfr gene from prototypic Tn7 (WJK06983.1).
Mating-out transposition assay
In the assay, movement of a mini-transposon (KanR) from the chromosome to a conjugal F plasmid is monitored with different targets, the comM or glmS att site or lacZ as a control (15). Candidate transposition proteins are expressed from plasmid vectors. Following induction of transposition proteins, the frequency of transposition is determined by mating the F plasmids into recipient cells and determining the percentage of cells with and without the transposon marker. In the assay a single colony of the appropriate donor strain was grown overnight at 37°C in 5 ml of LB with the appropriate antibiotics to maintain the plasmids and glucose to prevent expression of the transposition proteins. Overnight cultures were washed to remove the glucose and sub-cultured 1:50 in fresh LB supplemented with antibiotics and arabinose, rhamnose, or IPTG added as noted to induce expression of the candidate transposition proteins. During induction donor strains were grown with aeration at 37°C to an OD600 of 0.6 (∼2 h). To assess the amount of transposition that occurred in the donor strain, F plasmids were mated into mid-log recipient cells (CW51) at a ratio of 1:10 (donor: recipient) at 37°C for 1.5 h on a culture wheel turning at the lowest setting. Transconjugants were selected following serial dilution by plating on solid media with and without kanamycin to determine the percentage that contained the KanR mini-element integrated into the F plasmid (LB with spec, rif, and nal, with and without kan). Where noted, the percentage of transposition was determined by patching 200 transconjugants from the LB strep + nal + rif plate to the same media with and without kan.
Mapping insertions
For insertions verified by Sanger DNA sequencing, isolates were obtained for colony PCR from the mating-out assay plates to check for insertions in comM using a pair of primers that flank ‘comM’ on the F plasmid. After verifying the correct size, PCR products were purified and sent for Sanger DNA sequencing. For insertions verified by Illumina NGS sequencing, colonies were washed from the surface of LB kan + spec mating-out assay plates and resuspended in LB. Total genomic DNA was extracted from the resuspended cells and sent for paired end Illumina sequencing through SeqCenter, LLC (seqcenter.com). Tn6022 left and right ends were identified in the returned FASTA files and their coordinates were plotted to the length of the F plasmid. All analysis of NGS data was done using custom Python and R scripts (Data and code available at https://data.mendeley.com/datasets/5nc43tsxg3/1).
Site-specific recombination assays
Competent JY12 cells were transformed with different combinations of pBAD322 (empty vector, pBAD-FLP or pBAD-XerC-like) and pBCKS+ (empty vector or pBCKS-XerD) plasmids and plated in LB-agar containing kan (10 ug/ml), tet (8 ug/ml), cam (30 g/ml), carb (100 μg/ml) and glucose (0.2%). Overnight cultures of single colonies were grown at 37°C using the same selective media. The next day, cells were diluted 5000 times into induction media containing LB cam (30 μg/ml), carb (100 μg/ml), Arabinose (0.2%) and IPTG (0.1 mM) and grown at 37°C for 5 h. To monitor site-specific recombination by loss of the KanR gene, after 5 h of growth, the cells were diluted 105–106 times, plated into LB-agar containing 0.2% Glucose and grown overnight. The next day, 200 single colonies from each plate were patched into LB agar containing kan (50 μg/ml). To monitor site-specific recombination by capture of the resolved circular product, after 5 h of growth, the cells were collected, and the plasmid DNA extracted. The plasmids were transformed into BW25141 strain and plated into LB-agar containing kan (50 μg/ml).
Results
Major types of Tn7-like elements in Acinetobacter baumannii
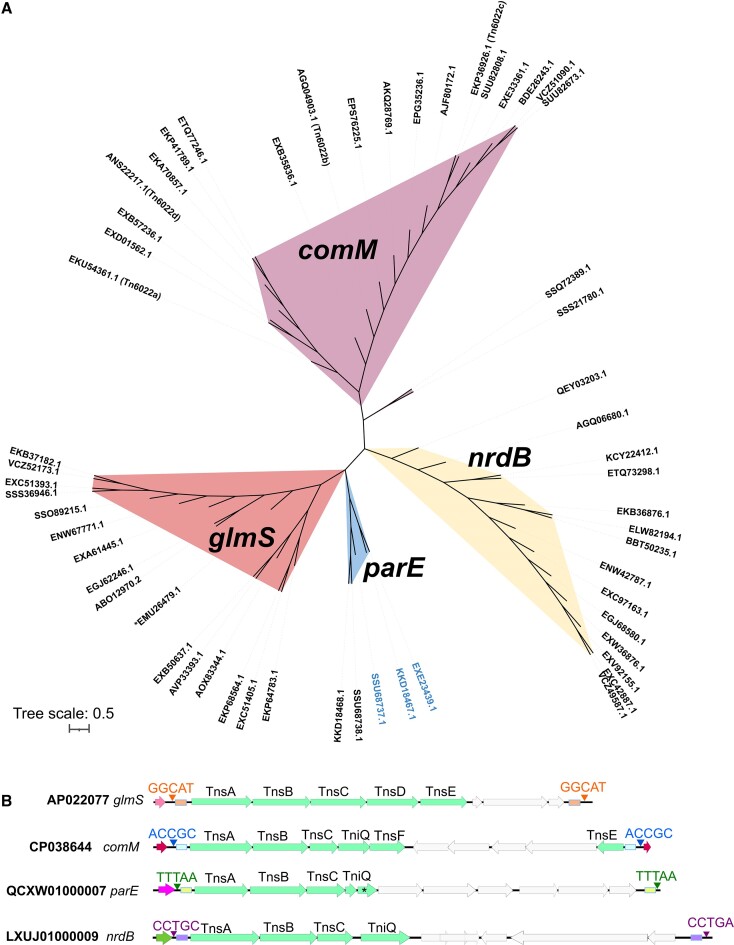
To analyze the role of Tn7-like elements in generating genetic diversity in A. baumannii we searched annotated open reading frames in 17469 A. baumannii genomes from NCBI. Searching for TniQ-domain containing proteins revealed 2440 unique amino acid sequences which were compiled into 56 groups with proteins showing 95% identity. A similarity tree constructed from representative TniQs established four types of Tn7-like elements based on att site usage (Figure 1A). Direct examination of the other transposition proteins associated with each TniQ indicated the two major types of Tn7-like elements previously noted in A. baumannii targeting the glmS and comM attachment sites (9) (see below). Among the glmS targeting elements included the original well-studied prototypic Tn7 element (indicated in Figure 1A) (41). We also identified two types of Tn7-like elements that were not previously described in A. baumannii. One small branch of elements integrates at an att site downstream of the parE gene encoding one subunit of topoisomerase IV, which was previously identified as an att site in Acinetobacter ursingii and other gammaproteobacteria (15,42). In addition to the ∼486 amino acid TniQ protein that targeted the parE att site, they also contained a smaller ∼170 amino acid TniQ protein of unknown targeting specificity (Figure 1B). The fourth type of Tn7-like element based on TniQ similarity likely targets the gene encoding ribonucleotide-diphosphate reductase subunit beta, nrdB. While the elements in this branch from A. baumannii were typically degraded at their end sequences, homologs of the TniQ protein in Acinetobacter nosocomialis strain NCTC 8102 (CP029351) and Guyparkeria sp. SB14A (SWAW01000013) integrated downstream of the nrdB gene in these genomes.
Figure 1.
Diverse TniQ proteins in Acinetobacter baumannii genomes recognize different attachment sites. (A) TniQ (PF06527) was queried against 17469 Acintobacter baumannii genomes using HMM to identify putative homologs. An approximately-maximum-likelihood similarity tree was built using 56 non-redundant TniQ protein sequences and displayed with iTOL(v6). Four distinct clades were predicted to utilize four major classes of att sites (comM, glmS, parE, and nrdB) based on the genomic neighbourhood, which are indicated by color above. Two groups of TniQ proteins are found in the genomes which have parE att sites. The accessions numbers colored blue in the tree carry a large TniQ protein (∼486aa). (B) Four unique types of Tn7-like elements were found within Acinetobacter baumannii genomes that use distinct att sites. Core transposition proteins are indicated with the genes colored green. Accession numbers of four examples that recognize different att sites are indicated to the left. Transposon ends (small rectangles) and the five base pair target site duplication are shown with different colors. The prototypic Tn7 group is indicated (*).
Analysis of Tn7-like elements in closed A. baumannii genomes
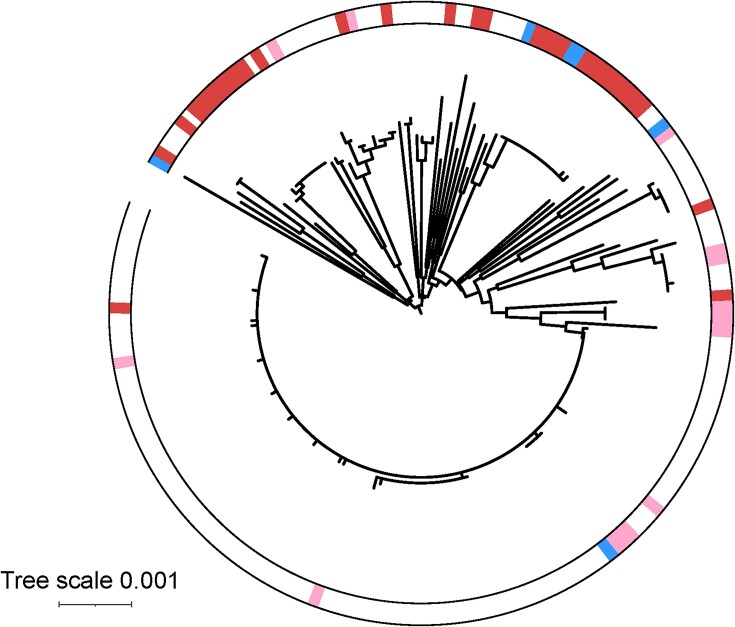
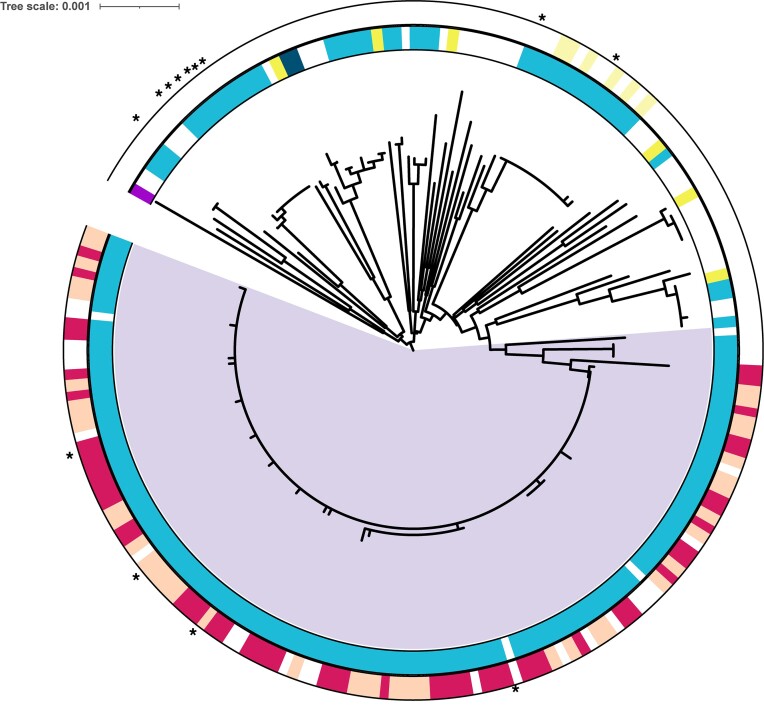
To assess the role of Tn7-like elements in genomic diversification we obtained 203 closed A. baumannii genomes and constructed a similarity tree based on seven housekeeping genes (fusA, gltA, groEL, pyrG, recA, rplB, rpoB) (Figures 2 and 3). Having a closed genome, meaning one assembled into a single circular DNA molecule, allowed us to understand genome rearrangements that could be related to Tn7-like integration events. We then searched the genomes for glmS and comM targeting elements by screening for insertion events within the expected att sites (Supplemental Tables S1 and S2). We found that 54/203 (24%) of the genomes contained evidence of a Tn7-like insertion event downstream of glmS, but less than half of these 21/203 (∼9%) are predicted to be functional transposons based on possessing essential transposition genes and complete left and right transposon end sequences (Figure 2). The majority of complete A. baumannii genomes had evidence for comM inserting elements, 165/203 (81%) (Figure 3). We note that many of the genomes used in our analysis are from outbreaks of antibiotic resistant strains that are likely clonal. It was of interest that strains that had near-identical housekeeping genes still displayed diverse transposon configurations (Figures 2 and 3). None of the parE targeting elements were found in the set of 203 closed genomes. Manual examination of the elements predicted to target the nrdB site suggest that all of the A. baumannii genomes have remnants of an ancestral insertion downstream of nrdB with some maintaining the tns encoding genes, but the transposons ends were difficult to recognize preventing a meaningful analysis of the cargo they originally transported.
Figure 2.
Distribution of integration events in the glmS att site in Acinetobacter baumanii genomes. A similarity tree was constructed with 203 closed Acintobacter baumannii genomes based on seven housekeeping genes (fusA, gltA, groEL, pyrG, recA, rplB, rpoB). The presence of Tn7-like elements that are potentially active or inactive is denoted by a pink or red fill respectively. Genomes that harbor at least one active and one inactive Tn7-like transposon are denoted with a blue fill.
Figure 3.
Families of Tn6022 elements in the comM att site in Acinetobacter baumanii genomes. A similarity tree was constructed with 203 closed Acintobacter baumannii genomes based on seven housekeeping genes (fusA, gltA, groEL, pyrG, recA, rplB, rpoB). Inner ring markings represent the presence of one of the four identified clades of Tn6022 elements. The A, B, C and D groups are represented by yellow, dark blue, light blue, and purple, respectively. The outer ring indicates presence or absence of the XerC-like tyrosine recombinase and/or TnsC’-‘TnsF (features only found in the branch indicated in purple). Genomes with a Tn6022 element that has TnsC’-‘TnsF is indicated in magenta. The presence of both TnsC’-‘TnsF and the XerC-like tyrosine recombinase is indicated with beige. Yellow indicates an uncharacterized integrase tyrosine recombinase. Asterisk marks indicate genomes that contained scrambled elements whose sizes could not be assessed (Supplemental Table S1).
To determine the relationship between the glmS and comM inserting elements identified in the closed genomes, a sequence of TnsA and TnsB from each identified element were concatenated, aligned, and graphed as a similarity tree using RAxML (Supplementary Figure S1). Predicted active elements inserted downstream of glmS all encode TnsABCDE genes related to prototypic Tn7, forming seven clades of transposons (Supplementary Table S1). Some of the antibiotic resistance genes found with these elements were beta-lactam and aminoglycoside resistance in the ∼46 kb clade Tn7g elements, anguibactin and an annotated multi-drug resistance gene in the ∼55 kb clade Tn7c elements and an annotated multi-drug resistance gene in the 24 kb Tn7f element. Two branches had genome islands formed by Tn7 elements. One of these had the 8 kb island and had a more recent insertion of a 42 kb Tn7e element (Supplementary Figure S1). Genetic islands varied from 1.9 to 25.2 kb and intact elements from 13 to 58 kb (Supplementary Tables S1 and S2). The average size of the insertion downstream of glmS was 15.8 kb.
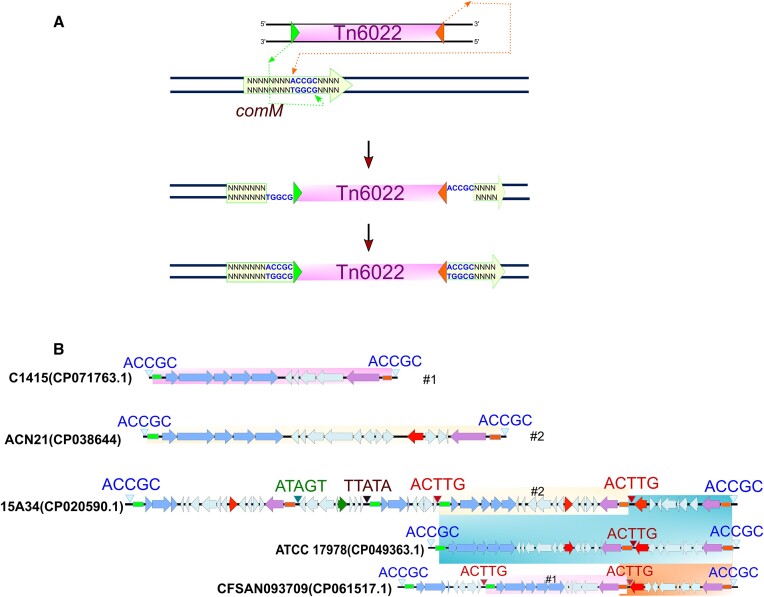
The comM targeting elements clustered into four clades that we refer to as Tn6022 elements based on the TnsA(Orf1), TnsB(TniA), TnsC(TniB), TniQ(Orf2), Orf3(TniE or TnsF), and TnsE(Orf4) encoding genes (3,4,9,20) (Figures 1B and Supplementary Figure S1). One subgroup of Tn6022 elements is predicted to be nonfunctional and degraded based on unrecognizable ends and interrupted genes. While the three other subgroups of Tn6022 elements are likely functional based on predicted full-length genes, one subgroup was by far more dominant that the others suggesting it had features that allow an evolutionary advantage compared to the others within the selected isolates (see below) (Figure 3). Thirteen of the Tn6022 elements gave a calculated size of > 230kb (indicated by asterisk in Figure 3), but manual examination revealed one or more inversions at the element ends which prevent a straightforward characterization of their sizes (Supplementary Tables S1). Insertion elements are common in many Tn6022 elements that can provide homology for inversion events scrambling the element. Excluding these elements showed lengths from 9.3 through 89 kb with and average size of 28 kb (Supplementary Tables S1 and S2).
Tn6022 had properties that were different than predicted from the literature for prototypic Tn7. Prototypic Tn7 precisely directs transposition into the glmS att site at a specific distance from the glmS gene (11). Subsequent insertions into the site are prevented by a process called target immunity. Target immunity acts on the inability of the transposition complex to be assembled through TnsC once an insertion is already integrated into the site with a process involving TnsB and its binding sites across the ends of the element (43–45). The process of target immunity was shown to occur with other Tn7-like elements, like the CRISPR-Cas associated transposons (46–48). This was at odds with the finding that there are multiple examples with Tn6022 in A. baumannii genomes that suggest that target immunity is weak or can be overwhelmed (4) (Figure 4 and Supplementary Figure S2).
Figure 4.
Tn6022 configurations in Acinetobacter baumannii. (A) Illustration of cut and paste transposition employed Tn7 and related elements (B) Multiple transposition and recombination events seemingly account for the diversification of Tn6022 elements inserted within comM. The top example illustrates a typically acquired Tn6022c element (shaded pink) through transposition in A. baumannii strain C1415 (Accession #: CP071763.1). In the middle is A. baumannii strain ACN21 (Accession #: CP038644) which appears to have a second Tn6022c (shaded light yellow) element with different cargo (Red = antibiotic resistance blaOXA and tetR) genes. Multiple events of transposition and recombination are observed in A. baumannii strain 15A34, which encode the TnsC-TnsF fusion (not highlighted), as well as a putative XerC-like tyrosine recombinase (green filled arrow). Multiple insertions can be detected by the presence of complementary TSDs (text shown in blue and red) in A. baumannii strain CFSAN093709 (Accession #: CP061517.1). The Tn6022c elements (shaded blue) in A. baumannii strain ATCC17978 (Accession #: CP049363.1) appear to be generated through a deletion event from CFSAN093709 or 15A34. TnsABC, TniQ and TnsF are shown with blue filled arrows, and TnsE is shown as a purple filled arrow. Cargo genes are represented by cyan filled arrows. Unique TSDs are shown with triangles, and element RE are green rectangles and LE are orange rectangles.
The process of transposition results in certain characteristic features, like small direct repeats that flank the element called the target site duplication (TSD). The TSD forms as a consequence of the staggered joining events to the target DNA during transposition that result in flanking single stranded gaps outside the elements ends (Figure 4A). When these gaps are filled by repair a TSD results, which in the case of Tn7 and Tn7-like elements is five bp long. The sequence of the TSD can be used to understand which ends of the element integrated at the same time with the same transposition event. For example, insertion of a second element (ACTTG TSD) was found within another element (ACCGC TSD) (Figure 4). Additionally, many of the elements appear to have captured a 6.2 kb fragment of host DNA sequence using a mechanism akin to composite transposons that involves transposition proximal to an existing element (Figure 4) (4,22). The TSD from the original transposition events that captured this fragment can also be surmised, ATAGT and TTATA. We noticed that these instances of local and internal insertions only occurred across one branch of closely related A. baumannii strains in our set of closed genomes (In purple in Figure 3). While the transposon end sequences and most of the proteins had the identical sequence to other Tn6022 elements within the subgroup, we noted genetic features that were unique to this branch (see below).
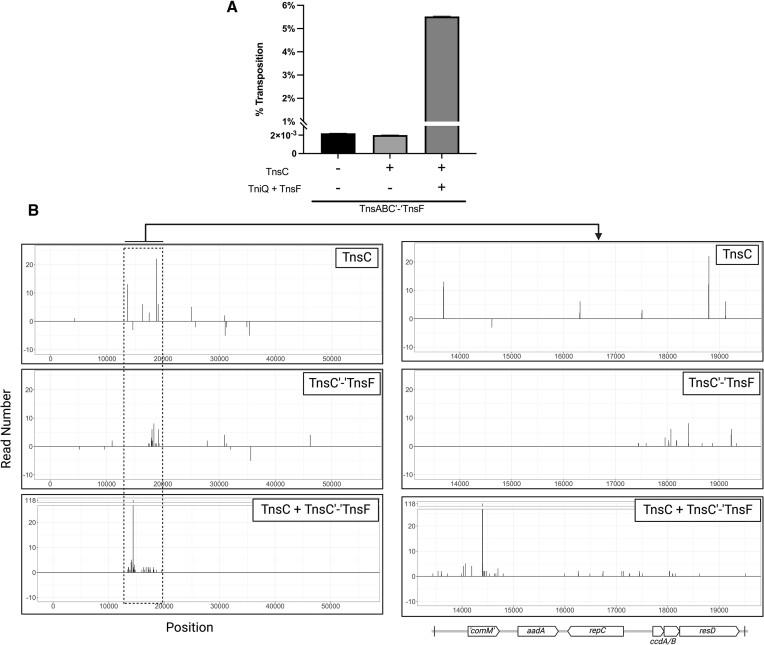
Tn6022 transposition into comM could be established in a heterologous E. coli host
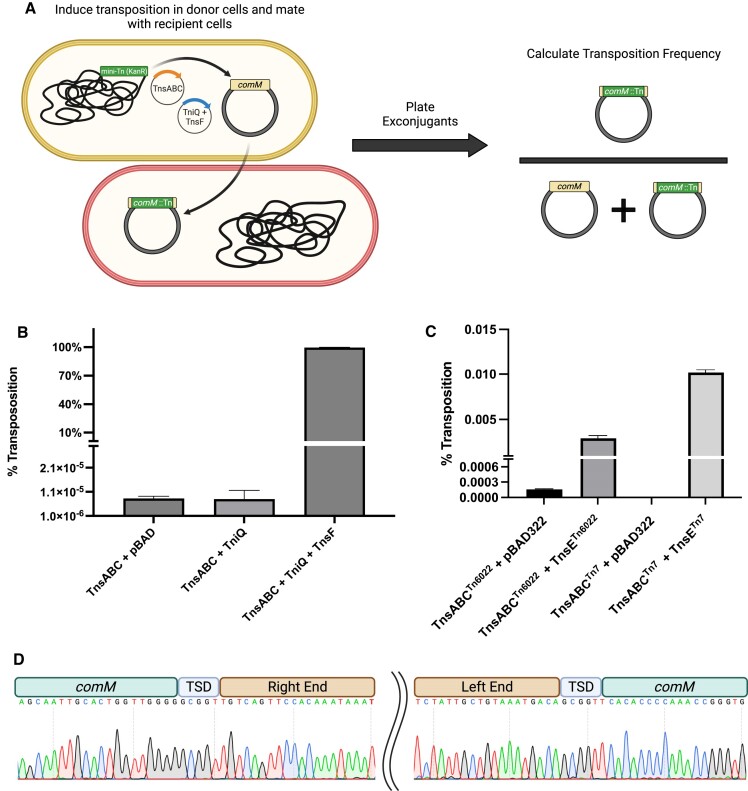
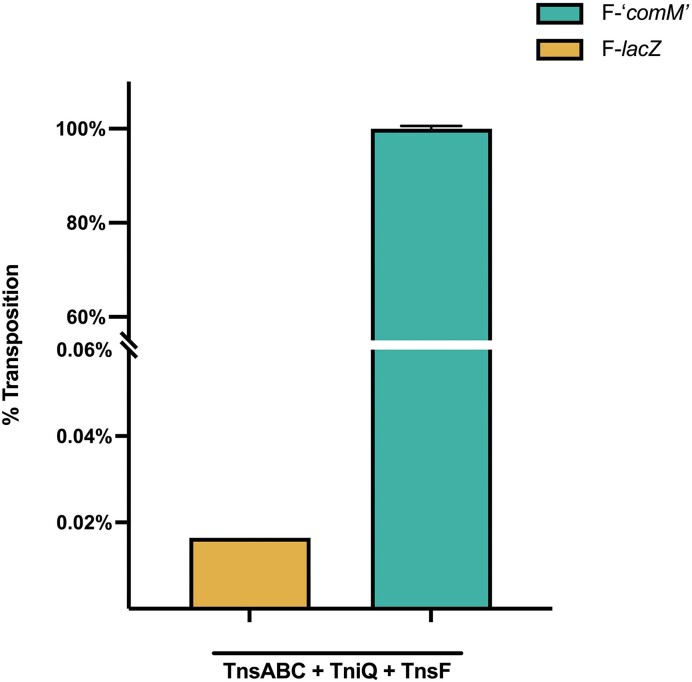
Transposition with Tn6022 elements has not been analyzed at the molecular level. We sought to establish Tn6022 transposition in the heterologous E. coli host as described previously using a mating-out assay (15) (Figure 5A). A mini-element containing the predicted left and right transposon end sequences flanking a kanamycin resistance gene was situated in a neutral position in the E. coli chromosome. A 600bp region of the comM gene was cloned into a derivative of the F plasmid to be used as a target that was capable of self-mobilization via conjugation. The putative Tn6022 transposition genes from the most active clade integrated into the genome of A. baumannii strain ATCC17978 were cloned into E. coli expression vectors. Using the mating-out assay, transposition can be monitored following induction of the candidate genes by mating the F plasmid into a new bacterial host. The frequency of transposition is calculated by enumerating F plasmids with and without the genetic marker encoded on the mini-element. We found that unlike other Tn7-like transposon systems, the TnsA, TnsB, TnsC and a TniQ family protein was not sufficient to establish high frequency transposition into the comM att site (15–17,42) (Figure 5B). We noted that Tn6022 elements within comM universally contained orf3, a gene encoding a 481 amino acid protein, forming an operon with tniQ, which was recently renamed tnsF (see discussion). TnsF showed regions of homology to a XerH-family tyrosine recombinase, these regions were recently suggested to be repurposed for DNA binding and to interact with TniQ (49). Expressing TnsF along with TnsABC + TniQ reconstituted robust transposition in E. coli at essentially 100% at full induction levels (0.02% Arabinose and 0.1mM IPTG) (Figure 5B). Sanger sequencing confirmed that transposition occurred with the same target site duplication and in the same orientation always found in the natural A. baumannii hosts (Figure 5C). High levels of Tn6022 transposition required the 600bp sequence from comM as only a low level of transposition was detected when the mating-out assay was performed with an F plasmid without this sequence (Figure 6). We predict that the TnsF protein provides the DNA binding specificity to the TniQ domain protein, similar to how DNA binding specificity is provided by effector complexes in the RNA guided transposons (16,17,47,48).
Figure 5.
TnsA, TnsB, TnsC, TniQ and a new open reading frame TnsF are required for comM att targeting in the heterologous E. coli host. (A) Schematic representing the general steps of the mating-out assay protocol. A mini-Tn6022 element harboring kanamycin resistance is integrated into the attTn7 site on the E. coli chromosome in donor cells (yellow) which also harbor a conjugal plasmid with a 600 bp portion of comM on it. Following a period of induction to produce transposition proteins, donor cells are mated with a population of counter-selectable recipient cells (red). Recipient cells are plated on selective media to calculate the number of exconjugants that received a conjugal plasmid with an integrated mini-element vs. total exconjugants to determine transposition frequency. (B, C) Percent transposition as measured by the mate-out assay using different combinations of Tn6022 transposition proteins in the presence of a conjugal plasmid harboring a 600 bp region of comM (‘comM’). (D) Sanger sequencing demonstrates precise ‘comM’ targeting which exactly recapitulates insertions found in nature. A DNA chromatogram demonstrating Sanger sequencing of Tn6022 insertions within ‘comM’ obtained from recipient cell colonies that were mated with donors expressing TnsABC + TniQ + TnsF. 10 isolates were screened for insertions via PCR using a pair of ‘comM’ flanking primers before sequencing. In all 10 isolates the transposon ends are clearly distinguished interrupting comM along with the hallmark target-site duplication (TSD) which occurs as a result of transposition. All experiments were performed with 0.02% arabinose induction to express the full length TniQ + TnsF operon, and 0.1mM IPTG to express the full length TnsABC operon. TnsE experiments were performed with 0.0002% arabinose to express TnsE. All experiments were performed in triplicate with error bars representing the standard error of the mean. Created with BioRender.com.
Figure 6.
Transposition with TniQ and TnsF in the absence of ‘comM’ is severely reduced. Percent transposition as measured by the mating-out assay using strains expressing TnsABC + TniQ + TnsF in the presence or absence of a conjugal plasmid harboring a 600 bp region of comM. All experiments were performed in triplicate with error bars representing the standard error of the mean. F-lacZ sample error bars are plotted but not visible.
Transposition with TnsABC alone gave a low but detectable level of transposition above the vector control, about four orders of magnitude below the level found with TnsABC + TniQ + TnsF transposition into comM (Figures 5B). To characterize the targeting specificity of TnsABC + TniQ + TnsF directed transposition, 24 isolates from samples expressing these proteins were screened via PCR and the predicted change in size indicated that all occurred within the 600bp sequence from comM in the F plasmid target. Sanger sequencing of ten examples revealed a 5bp target site duplication that is characteristic of transposition and in all cases exactly recapitulated the precise targeting found in genome sequences from clinical A. baumannii samples (Figure 5C). Isolates expressing only TnsABC had no insertions within comM when 24 isolates were examined by PCR. Sequencing by NGS confirmed TnsABC alone were capable of producing transposition resulting in a TSD (See below).
We directly compared the TnsABC + TnsD mediated transposition frequency of prototypic Tn7 into the glmS att site with Tn6022 transposition into the comM att site using the mating-out assay. We reduced the transposition protein expression time to allow a broader range for comparison and found that the Tn6022 transposition frequency was somewhat higher than the frequency found with prototypic Tn7 (Figure 7). In both cases checking ten examples showed the precise targeting found in sequenced genomes. The high efficiency of these systems supports the bioinformatic finding that these elements are pervasive in A. baumannii.
Figure 7.
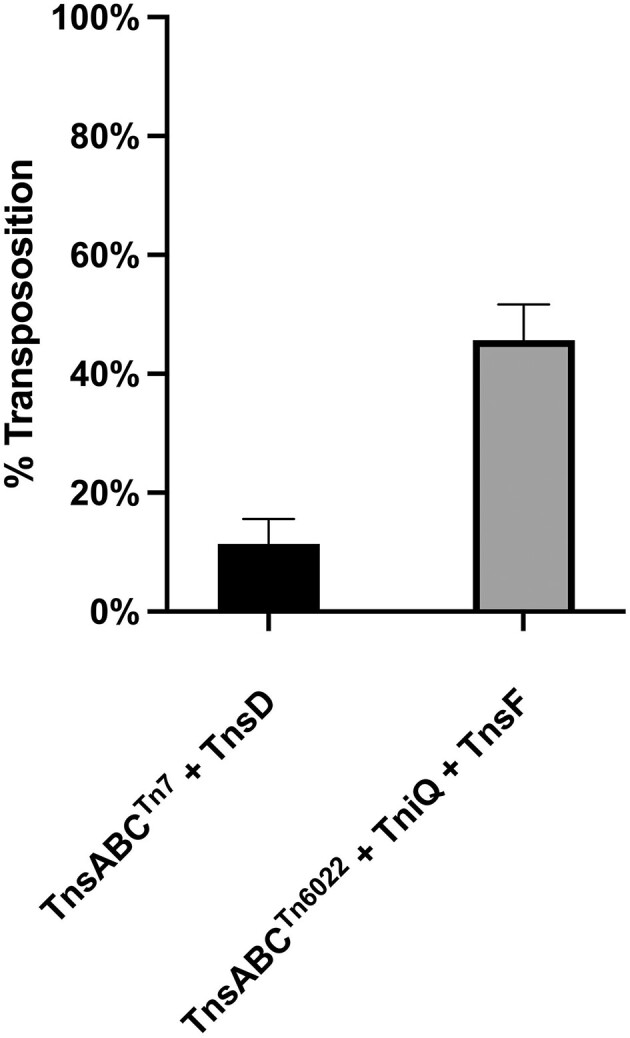
Transposition frequency comparison between Tn7 and Tn6022. Percent transposition measured and compared by the mating-out assay using prototypic Tn7 (TnsABC + TnsD) and Tn6022 transposition proteins (TnsABC + TniQ + TnsF) in the presence of a conjugal plasmid harboring a region of glmS (Tn7 experiments) or a 600 bp region of comM (Tn6022 experiments). The percentage of transposition events was calculated by patching 200 transconjugants to determine the percentage that had the transposon genetic marker (KanR). All experiments were performed in triplicate with 0.02% arabinose induction and 0.1mM IPTG for 90 min. All experiments were performed in triplicate with error bars representing the standard error of the mean.
Prototypic Tn7 has a pathway of transposition that preferentially targets molecular features associated with DNA processing found during plasmid conjugation with the target selecting protein TnsE (50). TnsABC + TnsE preferentially directs transposition into DNA sequences that are undergoing plasmid conjugation or DNA repair (51–54). At one end of the element distal from the other transposition genes, Tn6022 transposons encode a distant homolog of TnsE found in Tn7 (Figure 1) (20). Initial experiments with TnsABC + TnsE from this element didn’t indicate transposition (Figure 5B). However, monitoring transposition found with overnight induction revealed TnsE-dependent transposition with the version found in Tn6022 elements (Figure 5C). Ilumina sequencing confirmed bone fide TnsABC + TnsE transposition occurred into the F plasmid with the Tn6022 system. In the context of this assay transposition events occurred randomly across the F plasmid with TnsE-mediated transposition in the Tn7 and Tn6022 systems (Supplemental Figure S3).
Tn6022 elements display target immunity, strongly discouraging a second insertion from occurring into an att site where an insertion already resides
Our bioinformatics suggested that target immunity may not be present with Tn6022 because we identified multiple cases where internal transposition occurs within or proximal to elements (Figure 4 and Supplementary Figure S2). However, this seemed to only occur in one branch of elements (Figure 3). Our experiments examining Tn6022 transposition in E. coli suggested target immunity was occurring as only a single insertion was found in our mating-out experiments, even in cases where we could show ∼100% transposition. To directly test for target immunity in Tn6022 elements we constructed an immobilized mini-Tn6022 element into our F plasmid derivative within the 600 bp ‘comM’ sequence. This element had the TnsB-binding sites normally found with the element, but the TGT/ACA terminal base pairs were mutated to ACT/TGA (40). We found a drastic decrease in transposition monitored with the mating-out assay when a mini-Tn6022 element was already found in the comM att site. Transposition was reduced from ∼100% with comM to ∼2 × 10−5% when a mini-Tn6022 element resided in the comM site (Figure 8). The minimal transposition found with TnsABC only was also sensitive to target immunity, but in this case the reduction was only about 50-fold (Figure 8).
Figure 8.
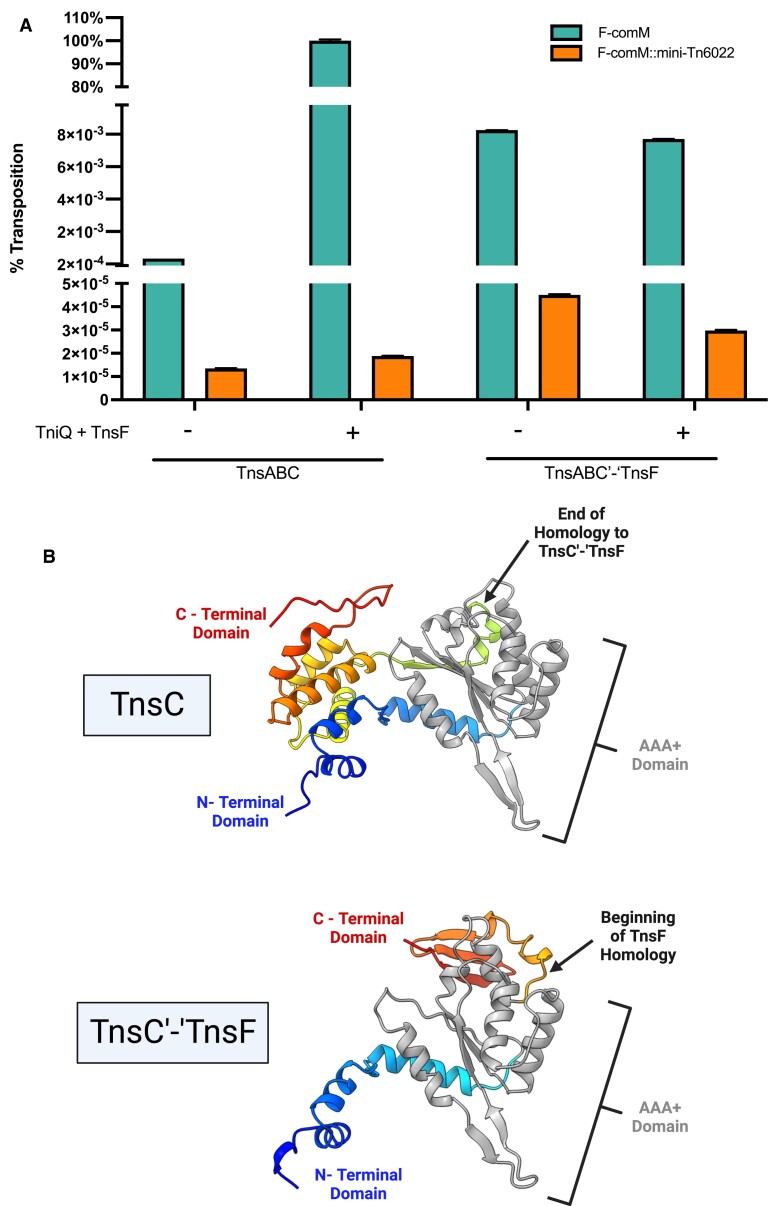
A naturally occurring TnsC and TnsF fusion is capable of promoting transposition and Tn6022 insertions are repressed by target immunity. (A) Percent transposition as measured by the mateing-out assay using different combinations of Tn6022 transposition proteins. Samples in green harbor a conjugal plasmid with a 600 bp region of comM, and samples in orange have an immobile Tn6022 mini-element inserted into the expected target site within ‘comM’. All experiments were performed in triplicate with error bars representing the standard error of the mean. (B) Structures of both TnsC and TnsC’-‘TnsF were predicted using AlphaFold2 for comparison. The N-terminal and C-terminal amino acids are shown in blue and red respectively, with the core AAA + domain shown in grey. The beginning of sequence from TnsC that is missing from TnsC’-‘TnsF is marked with an arrow in the top panel, and the beginning of homology between TnsC’-‘TnsF and TnsF is marked with an arrow in the bottom panel. All experiments were performed with 0.02% arabinose induction to express the full length TniQ + TnsF operon, and 0.1 mM IPTG to express the full length TnsABC or TnsABC’-‘TnsF operon. All experiments were performed in triplicate with error bars representing the standard error of the mean. Created with BioRender.com.
A natural TnsC’-‘TnsF fusion protein acts as a gain-of-function allele altering basic features of transposition
We found that Tn6022 displayed strong target immunity when tested in E. coli. However, there were multiple examples in A. baumannii genomes that suggest that target immunity can be overwhelmed (4) (Figure 4). Given that these examples were with the exact element tested in our experiments, we looked for other features in these elements that might help account for the internal or proximal Tn6022 insertions. Tn6022 elements in the branch showing the highest amount of rearrangement frequently had a TnsC protein that was fused to TnsF, removing the intervening tniQ gene (Figure 3, outer ring). The 212 amino acid TnsC’-‘TnsF fusion protein maintained the first 167 amino acids from TnsC, including the AAA+ region of the protein and had only the C-terminal 46 amino acids from TnsF (Figure 8B). Despite missing around half of TnsC and almost all of TnsF the fusion protein was capable of catalyzing transposition with TnsA + TnsB. Transposition with TnsABC’-‘TnsF occurred at a frequency about ten-fold higher than found with TnsABC. However, TnsABC’-‘TnsF transposition was no higher when TniQ + TnsF were co-expressed indicating that the TnsC’-‘TnsF fusion could no longer interact with the comM targeting system, the TniQ + TnsF proteins (Figure 8A). This suggests that loss of a region of either TnsC or Orf3 renders the TnsC’-‘TnsF fusion incapable of effectively communicating with the target site selection proteins. Transposition with TnsABC’-‘TnsF was sensitive to target immunity, showing about a 200-fold decrease in transposition into the comM site that already contained a copy of the element (Figure 8A). Target immunity in the prototypic Tn7 element involves the ATPase activity of TnsC (55–57). If the same is true for immunity in the Tn6022 elements it would suggest that the ATPase function of TnsC’-‘TnsF fusion protein is still functional even with only 167 amino acids of the original TnsC protein. In the prototypic Tn7 system the TnsC region that interacts with TnsA and TnsB is found at the very C-terminus of the protein (58,59). Given that the TnsC’-‘TnsF fusion allows transposition in the Tn6022 system indicates that interaction with the TnsAB transposase must occur by a fundamentally different mechanism in this system (See Discussion).
Many of the elements with the TnsC’-‘TnsF fusion allele are present in composite elements that have full length versions of all the Tn6022 proteins including TniQ and TnsF (Figure 4). We found a low level of transposition with TnsAB and the TnsC’-‘TnsF fusion protein (TnsABC’-‘TnsF) in a mating-out assay, although transposition with the TnsC fusion occurred at about 10-times the level found with TnsC wild type without TniQ + TnsF (Figure 8A). We tested for synergistic effects of the TnsC’-‘TnsF fusion proteins with the full-length proteins from the comM inserting Tn6022 elements in E. coli to better understand the mechanisms forming diverse Tn6022 elements found in clinical settings. To assess the capacity of TnsC’-‘TnsF to interact with full-length TnsC we designed a mating-out assay where full-length TnsC was provided on an inducible vector and co-expressed in cells with TnsABC’-‘TnsF. When full-length TnsC was co-expressed with TnsC’-‘TnsF without TniQ + TnsF, transposition rates were not different than what was observed with either TnsABC or TnsABC’-‘TnsF alone (Figure 8A). However, when TniQ and full-length TnsF were also expressed, transposition rates were substantially boosted to ∼5% transposition in the mating-out assay (Figure 9A). To understand the interplay between wild type TnsC and the TnsC’-‘TnsF fusion and the response to TniQ + TnsF we mapped insertions using Illumina sequencing (Figure 9B). As expected based on the results above, the gene fragment from comM was not a target with the low transposition found with TnsABC or with TnsABC’-‘TnsF (Figure 9B). There did seem to be a regional preference for insertions around the toxin gene ccdB and the tyrosine recombinase gene resD with TnsABC’-‘TnsF insertions (See magnified panels in Figure 9B). When wild type TnsC, TnsC’-‘TnsF and TniQ + TnsF were expressed we observed a different pattern of insertions. While there was a strong peak of reads at the exact base pair position expected for comM directed insertions, there was also a regional bias that was found around this site (Figure 9B). The finding that insertions shifted with co-expression of TnsC’-‘TnsF and TnsC suggests that TnsC’-‘TnsF is capable of a functional interaction with TnsC and disrupts the precise targeting of these elements (see Discussion).
Figure 9.
TnsC’-‘TnsF fusion produces a dominant negative phenotype when co-expressed with full length TnsC. (A) Percent transposition as measured by the mating-out assay using strains expressing various combinations of Tn6022 transposition proteins. In all samples TnsA, TnsB and TnsC’-‘TnsF were all expressed in their native operon from a lactose inducible promoter. TnsC was separately cloned into a second expression vector under the control of a rhamnose inducible promoter. Experiments were performed in triplicate and error bars are reported as the standard error of the mean. (B) Illumina sequencing was done on total genomic DNA extracted from colonies isolated from mate-out assay recipient cells. Transposon left and right ends were identified and the number of reads that corresponded to each identified end were plotted on the x-axis, with bp position of F-‘comM’ plotted on the y-axis. Negative values represent reverse orientation insertions. All reactions were performed with 0.02% arabinose induction to express the full length TniQ + TnsF operon, 0.02% rhamnose to express the TnsC’-‘TnsF fusion, and 0.1mM IPTG to express the full length TnsABC operon. Created with BioRender.com.
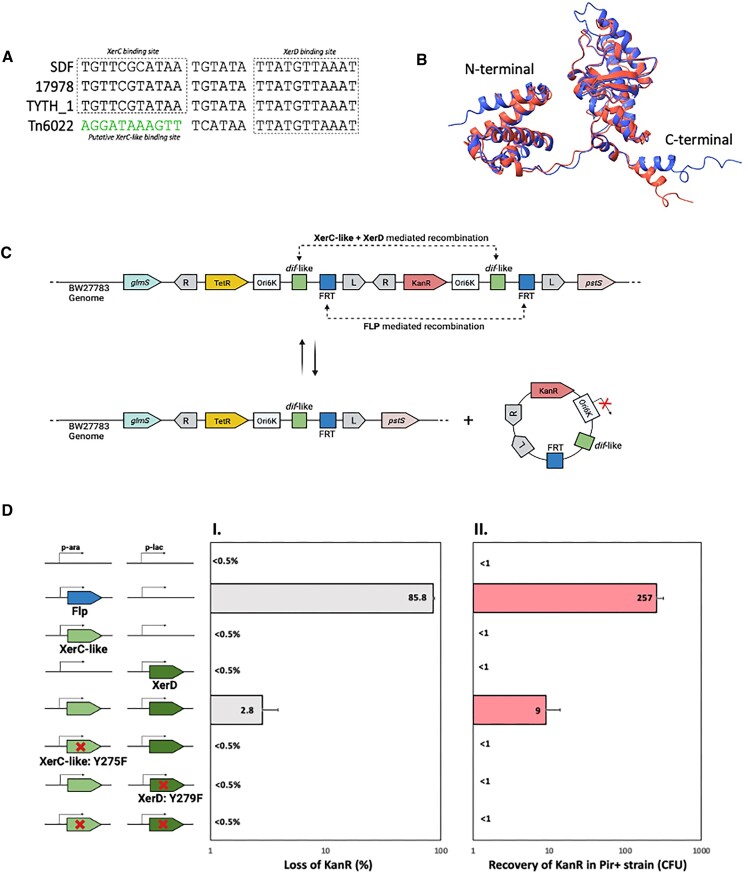
A XerC-like protein allows recombination at a dif-like site found on mobile elements with A. baumannii XerD
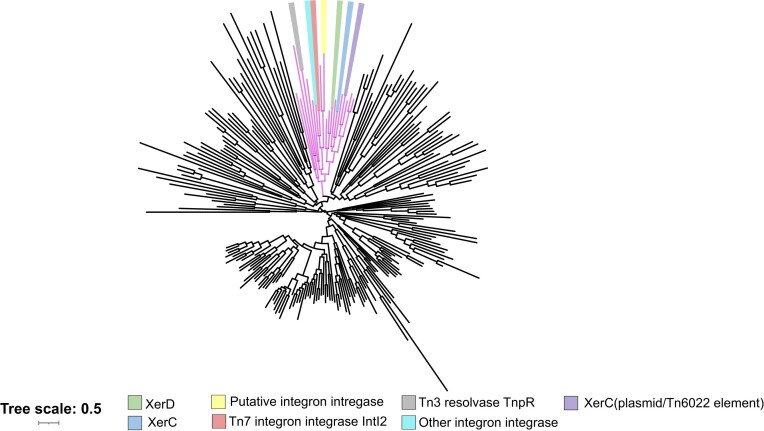
A genetic feature that was commonly associated with the most diverse branch of Tn6022 elements was a homolog of the XerC protein (we call XerC-like) and a downstream dif-like site (Figures 4 and 10). The XerC and XerD proteins are tyrosine recombinases that act at a specific dif site they recognize in the region where DNA replication terminates in circular chromosomes. Recombination with XerC and XerD at the dif site allows dimer chromosomes to be resolved at the time and location of cell division via an association with the chromosome translocation protein FtsK (60). The dif-like site had a perfect match to the half site recognized by XerD in A. baumannii (Figure 10A). We suspected the other half site of the dif-like site was recognized by the XerC-like protein associated with the Tn6022 elements (Figure 10A). The XerC/dif-like system was located in a region captured from the host between two Tn6022 elements (Figures 2 and 4) (4). Host and mobile element-encoded Xer systems have previously been implicated as important in A. baumannii, but to our knowledge this Xer system has never been described (61,62). A bioinformatic search for tyrosine recombinases across the 17 469 available A. baumanni genome sequences found them to be common and diverse (Figure 11). The XerC-like protein associated with Tn6022 elements branched with the host XerC and XerD proteins (Figure 11) while the alignment of the transposon encoded XerC-like with the host XerC indicated only 37.9% identity (Query coverage 95%). Comparison between the A. baumannii host XerC and Tn6022 associated XerC-like proteins with AlphaFold2 predicts the proteins are structurally similar despite differences in amino acid sequence (Figure 10B). Alignment of the XerC and XerD proteins in E. coli, A. baumannii, and Tn6022 shows they all maintain the important amino acids for resolution (Supplemental Figure S4). A similarity tree indicates that this protein is more closely related to Xer proteins that act at the terminus than tyrosine recombinases associated with integron cassette systems (Figure 11).
Figure 10.
Evaluation of the site-specific recombination mediated by Tn6022 XerC-like and A. baumannii host XerD at Tn6022 dif-like site in E. coli. (A) Alignment of the dif sites from three different strains of A. baumannii (SDF, 17978 and TYTH) and the dif-like site found associated with Tn6022. The putative XerC-like binding site is represented in green. (B) Comparison of the tridimensional structures of XerC protein from A. baumannii (red) and XerC-like protein found in Tn6022 (blue). The structures were obtained with Alphafold2. (C) The N- and C-terminal domains are marked. Schematic representation of the experimental design used to evaluate the site-specific recombination at dif-like site mediated by XerC-like protein in partnership with A. baumannii XerD. FLP recombinase and its cognate frt site were used as positive control. Two consecutive mini elements containing dif-like and frt sites, the origin of replication OriR6K and an antibiotic resistance gene were cloned between glmS and pstS genes in the genome of E. coli strain BW27783. The element closer to glmS carries a Tetracycline resistance gene (TetR) while the element closer to pstS gene carries a Kanamycin resistance gene (KanR). If the recombination occurs in either dif-like or frt site, a circular product unable to replicate in the BW27783 background is excised from the genome and the cells will lose KanR. (D) Recombination mediated by XerC-like and XerD proteins tested by two different methods. The five different conditions schematically represented (empty vectors, FLP only, XerC only, XerD only, XerC + XerD) were used in each method. (DI) After inducing protein expression, single colonies were streaked into LB-agar containing Kanamycin. The graph represents the percentage of colonies unable to grow in the presence of Kanamycin, which indicates they have lost KanR due to a recombination event in either dif-like or FRT site. Two hundred (200) colonies were tested for each condition. (DII) After inducing protein expression, plasmid DNA was extracted, transformed into BW25141 (encoding the π protein) and plated onto LB-agar containing Kanamycin. The graph represents the number of transformants that acquired the KanR gene. All experiments were performed in triplicate with error bars representing the standard error of the mean. Created with BioRender.com.
Figure 11.
Tyrosine recombinases in Acinetobacter baumannii genomes. An iterative search was performed against all annotated proteins (67 128 995) of Acinetobacter baumannii genomes (17 469). The representative hits (232) were aligned with the MUSCLE algorithm. Aligned sequences were then used to build an approximately-maximum-likelihood phylogeny using the FastTree algorithm and displayed with iTOL(v6). Branch nodes with host XerC (light blue highlight) and XerD (light green highlight) are included with the major group of well-studied tyrosine recombinases from mobile genetic elements (magenta branches). Others branches of tyrosine recombinases are represented as black.
We tested the functionality of the Xer-like system associated with Tn6022 elements in E. coli. Presumably such a system would facilitate recombination between elements in the chromosome and on plasmids and between elements in the chromosome. For example, we find that the same segment of DNA encoding the XerC-like protein and dif-like site in Tn6022 elements is also found on a A. baumannii conjugal virulence plasmid pIOMTU433 without the Tn6022 element (Accession AP014650). We were especially interested in testing if recombination with the site could allow excision between adjacent elements to form new variants of the transposon, a process that would expand diversity generation. We assembled miniTn7 elements in the glmS attachment site in E. coli with different genetic markers (Figure 10C). Included in the elements were the dif-like site from the transposons and plasmids found in A. baumannii. As a positive control we also included the frt site recognized by the Flp recombinase when expressed in the cell (63–65). These constructs also contain a conditional R6K plasmid origin of replication (66). Typical strains of E. coli used in the laboratory will not replicate the R6K plasmid origin, but circular DNAs can be selected in strains that encode the π protein on the chromosome (i.e. BW25141 (39,67,68)).
Expressing the Flp recombinase allowed the expected loss of the intervening KanR genetic marker at a high efficiency, ∼86% (Figure 10D). The excised product could be captured, and sequence confirmed by isolating DNA and transforming into a strain expressing the π protein required for replication of the circular element with the conditional replicon. We were also able to confirm loss of the intervening KanR genetic marker when the XerC-like and XerD(A.b.) proteins were expressed at the same time (Figure 10D). Expressing each of these components alone did not allow recombination. Consistent with these findings we could only capture the E. coli excised circular product in the permissive host for conditional plasmid replication from cells expressing both the XerC-like and XerD (Figure 10D). To confirm that the resolution activity of the XerC-like protein was needed for excision of a circular molecule, we repeated the experiment with versions missing the critical tyrosine residues. The Xer-like(Y275F) and XerD(Y279F) mutants were incapable of resolution (Figure 10D).
Discussion
Tn7 elements display a high level of control over target site selection with targeting pathways recognizing a conserved site in the chromosome and a second pathway recognizing mobile plasmids. We screened for Tn7-like elements bioinformatically across A. baumannii genomes to understand how these elements can contribute to the formation of resistance islands in this nosocomial pathogen that is known to be highly antibiotic resistant. Four families of Tn7-like elements were identified that target att site at glmS, comM, parE, and nrdB (Figure 1). To understand the contribution of Tn7-like elements we examined a subset of A. baumannii sequences with closed genomes. While insertions at the glmS and especially the comM site were common, one specific version of the Tn6022 element found to insert within comM dominated among these isolates (Figures 2 and 3). To understand factors that contributed to this transposon's success we tested transposon components in the heterologous E. coli host. Tn6022 transposition required an extra protein, not used in the prototypic Tn7 system and allowed robust (100%) transposition recapitulating the exact targeting and orientation control found in A. baumannii (Figure 5). The most dynamic Tn6022 elements had candidate systems for boosting diversification which we could confirm in E. coli (Figure 4 and Supplemental Figure S2). One was a novel natural gain-of-activity allele that could function alone or with the wild type gene product to broaden transposition targeting (Figure 9). The second was a transposon-captured hybrid dif-like site that parasitizes the host dimer chromosome resolution system to function with its own tyrosine recombinase (Figure 10).
Prototypic Tn7 has been studied for decades, but over the last ten years the widespread nature and diversity of Tn7-like elements is becoming clearer (4,9,17,20,49,69,70). In almost all cases Tn7-like elements have evolved to recognize a conserved integration site in the bacterial chromosome and a second pathway that targets features of mobile elements capable of cell-to-cell transfer, usually conjugal plasmids. Tn6022 provides the first example of an accessory target site selection protein, TnsF, like the CRISPR-Cas effector complexes recruited on multiple occasions. Very recently, another group made the same finding calling the Orf3 protein TnsF, a protein also used to direct other types of mobile elements to integrate into the comM gene (49) (Figure 5B). We have used the TnsF designation in the current work but note that this is unfortunately not the same gene as a TnsF encoding gene in a Tn6230 element from a previous publication (4). Similar to their finding, we also found targeting to the E. coli comM equivalent gene at the same exact position (49). Other Tn7-like elements have evolved to recognize comM and other competency genes as an integration site using crRNAs indicating that programed inactivation of natural transformation may be beneficial for the transposon (17).
We identified a natural gain-of-function allele that fuses TnsC to a small section of the element encoded TnsF protein, removing half of TnsC. This fusion protein is functional for untargeted transposition indicating that the ability to recruit TnsA and TnsB is different in the Tn6022 elements than with Tn7 (Figure 8). Previous work with TnsC from prototypic Tn7 indicates that it interacts with TnsA and TnsB using features at the C-terminal end of TnsC (58) a region missing from TnsC from Tn6022. The TnsC’-‘TnsF fusion protein is not capable of being targeted to comM with the TniQ + TnsF proteins in our assays suggesting these interaction regions reside in the second half of the TnsC protein (Figure 8). We found genetic evidence for a functional interaction between wild type TnsC and the TnsC’-‘TnsF fusion protein (Figure 9). Co-expression of wild type TnsC and TnsC’-‘TnsF were found to display an intermediate frequency of transposition from the rate found with each protein individually with the rest of the transposition components. The pattern of transposition events also differed with co-expression of TnsC and TnsC’-‘TnsF showing a regional bias broadly around the comM sequence (Figure 9B). Further research will be needed to understand the molecular interactions between TnsC and TnsC’-‘TnsF driving this novel pattern of integration. The presence of both TnsC and TnsC’-‘TnsF is associated with examples of internally directed transposition events that could insertionally inactivate the element but would also offer the ability to form new transposon combinations (Figures 3, 4, and Supplemental Figure S2). As described previously, cycles of addition by transposition and loss by deletion should allow accelerated diversification (4). The elements with multiple left and right ends would also allow various sub-combinations of elements to leave the locus as a new stable element. For example, diversity could be generated in this structure, but elements leaving without the diversity features would again have the attributes of the wild type system (Supplementary Figure S2). We suggest that this system is what leads to the success of this Tn6022 element in A. baumannii. We further suggest that there may be other examples of a similar process with other mobile elements that allow an element to sample diversification but ultimately lock in strict control.
We identified a XerC-like protein in a captured segment found in some of the Tn6022 elements (Figures 3 and 4). The XerC-like protein was associated with a dif-like site that would be recognized by host XerD proteins but was only functional in our assay when the XerD protein from A. baumannii was co-expressed (Figure 10). This suggests that features of the A. baumannii XerD protein may be important for activating this system as the native E. coli XerD protein does not support this activity. This is surprising given it recognizes the same sequence and that the A. baumannii and E. coli XerD proteins are structurally similar based on AlphaFold2 predications (Figure 10B). The tyrosine recombinase is found associated with Tn6022 elements common in clinical strains and on plasmids with and without Tn6022 elements. Recombination mediated at the dif-like could allow multiple benefits for generating diversity between transposons and plasmids. In the case of multiple local insertion events, recombination at the mobile element encoded dif-like site would theoretically also allow diversification by loss of the intervening sequence or transposition with the circular form of the element that was removed. XerC + XerD dimer resolution systems acting at the dif site in the chromosome require an interaction with the host FtsK protein which helps target its activity specifically at the point of cell division. Other mobile elements that utilize Xer recombination systems need accessory host proteins like PepA and ArgR or their own accessory protein like XafT found in satellite phage TLCΦ to activate XerCD recombination independent of FtsK (71). Future work will be needed to understand the mechanistic underpinnings of the system we identified here from A. baumannii.
Tn7 and Tn7-like elements are known for their precise control over target site selection and target immunity. These characteristics are expected to help maintain and spread the element but would also put constrains on generating new elements with novel cargo. The mechanisms indicated here would allow diversity to be generated in these elements that could contribute to the formation of new elements and a mechanism for capturing new cargo. Segregation away from the diversity generating systems would allow novel elements to be borne out of these systems, benefiting from the temporary diversification but allowed to revert to tight control when the element returns to the native genetic context of transposition genes. We expect that new forms of Tn7-like elements and additional diversification systems will continue to be found that help explain the outsized role these transposition systems have in antibiotic and biocide resistance in pathogenic bacteria.
Supplementary Material
Acknowledgements
Work in the Peters lab was supported by NIH R01 GM129118 and R21 AI148941.
Author contributions: B.J.K., J.C. and S.S. helped design and carried out the bioinformatics, M.T.P., K.A.E., S.S., J.T., L.M.C. and A.C. helped design and did the transposition experiments, M.T.P., Y.J. and L.C.M. helped design and did the tyrosine recombinase experiments, J.E.P. helped design and oversaw all aspects of the research and wrote the paper that was edited and approved by all authors.
Contributor Information
Alberto Correa, III, Department of Microbiology, Cornell University, Ithaca, NY, USA.
Saadlee Shehreen, Department of Microbiology, Cornell University, Ithaca, NY, USA.
Laura Chacon Machado, Department of Microbiology, Cornell University, Ithaca, NY, USA.
Jordan Thesier, Department of Microbiology, Cornell University, Ithaca, NY, USA.
Lille M Cunic, Department of Microbiology, Cornell University, Ithaca, NY, USA.
Michael T Petassi, Department of Microbiology, Cornell University, Ithaca, NY, USA.
Joshua Chu, Department of Microbiology, Cornell University, Ithaca, NY, USA.
Bennett J Kapili, Department of Microbiology, Cornell University, Ithaca, NY, USA.
Yu Jia, College of Life Sciences and Engineering Research Center of Bioreactor and Pharmaceutical Development (Ministry of Education), Jilin Agricultural University, Changchun City, Jilin Province, China.
Kevin A England, Department of Microbiology, Cornell University, Ithaca, NY, USA.
Joseph E Peters, Department of Microbiology, Cornell University, Ithaca, NY, USA.
Data availability
The data underlying this article are available in the article, in its online supplementary material, and online. https://data.mendeley.com/datasets/5nc43tsxg3/1.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institutes of Health [R01 GM129118 and R21 AI148941]. Funding for open access charge: National Institutes of Health [R01 GM129118].
Conflict of interest statement. None declared.
References
- 1. Peters J.E Tn7. Microbiol. Spectr. 2014; 10.1128/microbiolspec.MDNA3-0010-2014. [DOI] [PubMed] [Google Scholar]
- 2. Rose A. TnAbaR1: a novel Tn7-related transposon in Acinetobacter baumannii that contributes to the accumulation and dissemination of large repertoires of resistance genes. Biosci. Horiz. 2010; 3:40–48. [Google Scholar]
- 3. Hamidian M., Hall R.M.. AbaR4 replaces AbaR3 in a carbapenem-resistant Acinetobacter baumannii isolate belonging to global clone 1 from an Australian hospital. J. Antimicrob. Chemother. 2011; 66:2484–2491. [DOI] [PubMed] [Google Scholar]
- 4. Peters J.E., Fricker A.D., Kapili B.J., Petassi M.T.. Heteromeric transposase elements: generators of genomic islands across diverse bacteria. Mol. Microbiol. 2014; 93:1084–1092. [DOI] [PubMed] [Google Scholar]
- 5. Sarnovsky R., May E.W., Craig N.L.. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996; 15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- 6. May E.W., Craig N.L.. Switching from cut-and-paste to replicative Tn7 transposition. Science. 1996; 272:401–404. [DOI] [PubMed] [Google Scholar]
- 7. Bainton R.J., Kubo K.M., Feng J.N., Craig N.L.. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993; 72:931–943. [DOI] [PubMed] [Google Scholar]
- 8. Peters J.E. Targeted transposition with Tn7 elements: safe sites, mobile plasmids, CRISPR/Cas and beyond. Mol. Microbiol. 2019; 112:1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peters J.E., Makarova K.S., Shmakov S., Koonin E.V.. Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc. Natl. Acad. Sci. 2017; 114:E7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parks A.R., Peters J.E.. Transposon Tn7 is widespread in diverse bacteria and forms genomic islands. J. Bacteriol. 2007; 189:2170–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen Y., Gomez-Blanco J., Petassi M.T., Peters J.E., Ortega J., Guarne A.. Structural basis for DNA targeting by the Tn7 transposon. Nat. Struct. Mol. Biol. 2022; 29:143–151. [DOI] [PubMed] [Google Scholar]
- 12. Park J., Tsai A., Rizo A., Truong V., Wellner T., Schargel R., Kellogg E.. Structures of the holo CRISPR RNA-guided transposon integration complex. Nature. 2022; 613:775–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffmann F.T., Kim M., Beh L.Y., Wang J., Vo P.L.H., Gelsinger D.R., George J.T., Acree C., Mohabir J.T., Fernandez I.S.et al.. Selective TnsC recruitment enhances the fidelity of RNA-guided transposition. Nature. 2022; 609:384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McKown R.L., Orle K.A., Chen T., Craig N.L.. Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J. Bacteriol. 1988; 170:352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petassi M.T., Hsieh S.C., Peters J.E.. Guide RNA categorization enables target site choice in Tn7-CRISPR-cas transposons. Cell. 2020; 183:1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Saito M., Ladha A., Strecker J., Faure G., Neumann E., Altae-Tran H., Macrae R.K., Zhang F.. Dual modes of CRISPR-associated transposon homing. Cell. 2021; 184:2441–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hsieh S.C., Peters J.E.. Discovery and characterization of novel type I-D CRISPR-guided transposons identified among diverse Tn7-like elements in cyanobacteria. Nucleic Acids Res. 2022; 51:765–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Post V., White P.A., Hall R.M.. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 2010; 65:1162–1170. [DOI] [PubMed] [Google Scholar]
- 19. Godeux A.S., Svedholm E., Lupo A., Haenni M., Venner S., Laaberki M.H., Charpentier X.. Scarless removal of large resistance island AbaR results in antibiotic susceptibility and increased natural transformability in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2020; 64:e00951-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benler S., Faure G., Altae-Tran H., Shmakov S., Zheng F., Koonin E.. Cargo genes of Tn7-like transposons comprise an enormous diversity of defense systems, mobile genetic elements, and antibiotic resistance genes. mBio. 2021; 12:e0293821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parks A.R., Peters J.E.. Tn7 elements: engendering diversity from chromosomes to episomes. Plasmid. 2009; 61:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bi D., Zheng J., Xie R., Zhu Y., Wei R., Ou H.Y., Wei Q., Qin H.. Comparative analysis of AbaR-type genomic islands reveals distinct patterns of genetic features in elements with different backbones. mSphere. 2020; 5:e00349-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bi D., Xie R., Zheng J., Yang H., Zhu X., Ou H.Y., Wei Q.. Large-scale identification of AbaR-type genomic islands in Acinetobacter baumannii reveals diverse insertion sites and clonal lineage-specific antimicrobial resistance gene profiles. Antimicrob. Agents Chemother. 2019; 63:e02526-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holt K., Kenyon J.J., Hamidian M., Schultz M.B., Pickard D.J., Dougan G., Hall R.. Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb Genom. 2016; 2:e000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A.et al.. The Pfam protein families database in 2019. Nucleic Acids Res. 2019; 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Potter S.C., Luciani A., Eddy S.R., Park Y., Lopez R., Finn R.D.. HMMER web server: 2018 update. Nucleic Acids Res. 2018; 46:W200–W204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang Y., Niu B., Gao Y., Fu L., Li W.. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010; 26:680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004; 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014; 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Letunic I., Bork P.. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021; 49:W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Price M.N., Dehal P.S., Arkin A.P.. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010; 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A.et al.. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E.. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021; 30:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014; 30:2068–2069. [DOI] [PubMed] [Google Scholar]
- 35. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sullivan M.J., Petty N.K., Beatson S.A.. Easyfig: a genome comparison visualizer. Bioinformatics. 2011; 27:1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peters J.E. Methods for General and Molecular Microbiology, Third Edition. 2007; American Society of Microbiology. [Google Scholar]
- 38. Sibley M.H., Raleigh E.A.. A versatile element for gene addition in bacterial chromosomes. Nucleic. Acids. Res. 2012; 40:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peters J.E. Reddy C.A., Beveridge T.J., Breznak J.A., Marzluf G.A., Schmidt T.M., Snyder L.R.. Gene transfer - Gram-negative bacteria, Methods for General and Molecular Microbiology. 2007; 3rd edn.Washington D. C.ASM Press; 735–755. [Google Scholar]
- 40. Gary P.A., Biery M.C., Bainton R.J., Craig N.L.. Multiple DNA processing reactions underlie Tn7 transposition. J. Mol. Biol. 1996; 257:301–316. [DOI] [PubMed] [Google Scholar]
- 41. Lichtenstein C., Brenner S.. Unique insertion site of Tn7 in E. coli chromosome. Nature. 1982; 297:601–603. [DOI] [PubMed] [Google Scholar]
- 42. Klompe S.E., Jaber N., Beh L.Y., Mohabir J.T., Bernheim A., Sternberg S.H.. Evolutionary and mechanistic diversity of Type I-F CRISPR-associated transposons. Mol. Cell. 2022; 82:616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Skelding Z., Queen-Baker J., Craig N.L.. Alternative interactions between the Tn7 transposase and the Tn7 target DNA binding protein regulate target immunity and transposition. EMBO J. 2003; 22:5904–5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hauer B., Shapiro J.A.. Control of Tn7 transposition. Mol. Gen. Genet. 1984; 194:149–158. [DOI] [PubMed] [Google Scholar]
- 45. Arciszewska L.K., Drake D., Craig N.L.. Transposon Tn7 cis-acting sequences in transposition and transposition immunity. J. Mol. Biol. 1989; 207:35–52. [DOI] [PubMed] [Google Scholar]
- 46. Vo P.L.H., Ronda C., Klompe S.E., Chen E.E., Acree C., Wang H.H., Sternberg S.H.. CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat. Biotechnol. 2020; 39:480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Klompe S.E., Vo P.L.H., Halpin-Healy T.S., Sternberg S.H.. Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature. 2019; 571:219–225. [DOI] [PubMed] [Google Scholar]
- 48. Strecker J., Ladha A., Gardner Z., Schmid-Burgk J.L., Makarova K.S., Koonin E.V., Zhang F.. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019; 365:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Faure G., Saito M., Benler S., Peng I., Wolf Y.I., Strecker J., Altae-Tran H., Neumann E., Li D., Makarova K.S.et al.. Modularity and diversity of target selectors in Tn7 transposons. Mol. Cell. 2023; 83:2122–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wolkow C.A., DeBoy R.T., Craig N.L.. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 1996; 10:2145–2157. [DOI] [PubMed] [Google Scholar]
- 51. Shi Q., Straus M.R., Caron J.J., Wang H., Chung Y.S., Guarne A., Peters J.E.. Conformational toggling controls target site choice for the heteromeric transposase element Tn7. Nucleic Acids Res. 2015; 43:10734–10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shi Q., Parks A.R., Potter B.D., Safir I.J., Luo Y., Forster B.M., Peters J.E.. DNA damage differentially activates regional chromosomal loci for Tn7 transposition in Escherichia coli. Genetics. 2008; 179:1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Peters J.E., Craig N.L.. Tn7 recognizes target structures associated with DNA replication using the DNA binding protein TnsE. Genes Dev. 2001; 15:737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peters J.E., Craig N.L.. Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. Mol. Cell. 2000; 6:573–582. [DOI] [PubMed] [Google Scholar]
- 55. Stellwagen A.E. Analysis of gain of function mutants of an ATP-dependent regulator of Tn7 transposition. J. Mol. Biol. 2001; 305:633–642. [DOI] [PubMed] [Google Scholar]
- 56. Stellwagen A.E., Craig N.L.. Mobile DNA elements: controlling transposition with ATP-dependent molecular switches. Trends Biochem. Sci. 1998; 23:486–490. [DOI] [PubMed] [Google Scholar]
- 57. Stellwagen A., Craig N.L.. Gain-of-function mutations in TnsC, an ATP-dependent transposition protein which activates the bacterial transposon Tn7. Genetics. 1997; 145:573–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Choi K.Y., Li Y., Sarnovsky R., Craig N.L.. Direct interaction between the TnsA and TnsB subunits controls the heteromeric Tn7 transposase. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E2038–E2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ronning D.R., Li Y., Perez Z.N., Ross P.D., Hickman A.B., Craig N.L., Dyda F.. The carboxy-terminal portion of TnsC activates the Tn7 transposase through a specific interaction with TnsA. EMBO J. 2004; 23:2972–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Carnoy C., Roten C.A.. The dif/Xer recombination systems in proteobacteria. PLoS One. 2009; 4:e6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cameranesi M.M., Moran-Barrio J., Limansky A.S., Repizo G.D., Viale A.M.. Site-specific recombination at XerC/D sites mediates the formation and resolution of plasmid Co-integrates carrying a bla(OXA-58)- and TnaphA6-resistance module in Acinetobacter baumannii. Front. Microbiol. 2018; 9:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Harmer C.J., Pong C.H., Hall R.M.. Insertion sequences related to ISAjo2 target pdif and dif sites and belong to a new IS family, the IS1202 family. Microb Genom. 2023; 9:mgen000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cherepanov P.P., Wackernagel W.. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995; 158:9–14. [DOI] [PubMed] [Google Scholar]
- 64. Guzman L.-M., Belin D., Carson M.J., Beckwith J.. Tight regulation, modulation, and highlevel expression by vectors containing the arabinose PBAD promoter. J Bact. 1995; 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cronan J.E. A family of arabinose-inducible Escherichia coli expression vectors having pBR322 copy control. Plasmid. 2006; 55:152–157. [DOI] [PubMed] [Google Scholar]
- 66. Kolter R., Inuzuka M., Helinski D.R.. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978; 15:1199–1208. [DOI] [PubMed] [Google Scholar]
- 67. Metcalf W.W., Jiang W., Daniels L.L., Kim S.K., Haldimann A., Wanner B.L.. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996; 35:1–13. [DOI] [PubMed] [Google Scholar]
- 68. Metcalf W.W., Jiang W., Wanner B.L.. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene. 1994; 138:1–7. [DOI] [PubMed] [Google Scholar]
- 69. Park J.U., Petassi M.T., Hsieh S.C., Mehrotra E., Schuler G., Budhathoki J., Truong V.H., Thyme S.B., Ke A., Kellogg E.H.et al.. Multiple adaptations underly co-option of a CRISPR surveillance complex for RNA-guided DNA transposition. Mol. Cell. 2023; 83:1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hsieh S.-C., Peters J.E.. Tn7-CRISPR-Cas12K elements manage pathway choice using truncated repeat-spacer units to target tRNA attachment sites. 2021; bioRxiv doi:06 February 2021, preprint: not peer reviewed 10.1101/2021.02.06.429022. [DOI]
- 71. Miele S., Provan J.I., Vergne J., Possoz C., Ochsenbein F., Barre F.X.. The Xer activation factor of TLCPhi expands the possibilities for Xer recombination. Nucleic Acids Res. 2022; 50:6368–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Khlebnikov A., Datsenko K., Skaug T., Wanner B., Keasling J.. Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology. 2002; 147:3241–3247. [DOI] [PubMed] [Google Scholar]
- 74. Waddell C.S., Craig N.L.. Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes Dev. 1988; 2:137–149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article, in its online supplementary material, and online. https://data.mendeley.com/datasets/5nc43tsxg3/1.