Abstract
Retrovirus genomes contain a sequence at the 5′ end which directs their packaging into virions. In Rous sarcoma virus, previous studies have identified important segments of the packaging signal, Ψ, and support elements of a secondary-structure prediction. To further characterize this sequence, we used an in vivo selection strategy to test large collections of mutants. We generated pools of full-length viral DNA molecules with short stretches of random sequence in Ψ and transfected each pool into avian cells. Resulting infectious virus was allowed to spread by multiple passages, so that sequences could compete and the best could be selected. This method provides information on the kinds of sequences allowed, as well as those that are most fit. Several predicted stem-loop structures in Ψ were tested. A stem at the base of element O3 was highly favored; only sequences which maintained base pairing were selected. Two other stems, at the base and in the middle of element L3, were not conserved: neither base pairing nor sequence was maintained. A single mutation, G213U, was seen upstream of the randomized region in all selected L3 stem mutants; we interpret this to mean that it compensates for the defects in L3. Randomized mutations adjacent to G213 maintained the wild-type base composition but not its sequence. The kissing-loop sequence at end of L3, postulated to function in genome dimerization, was not required for infectivity but was selected for over time. Finally, a deletion of L3 was constructed and found to be poorly infectious.
The packaging of the RNA genome into retrovirus particles is a specific and efficient process. The nature of packaging is only partially understood, but cis- and trans-acting factors have been identified. Expression of the Gag protein alone is sufficient for virus assembly and is also sufficient for encapsidation of the genome (29). An RNA packaging signal, called Ψ or E, has been found in the 5′ end of the genome of many retroviruses (reviewed in references 5 and 9). The core of Ψ is generally located between the primer binding site and the start of gag, although other sequences can contribute to packaging efficiency. The segment of RNA encompassing Ψ is predicted to be highly structured. RNA stem-loops have been implicated in packaging for a number of retroviruses (7, 20, 23, 28, 42).
Two of the best-studied packaging signals are in murine leukemia virus (MLV) and human immunodeficiency virus type 1 (HIV-1). MLV has been particularly well studied because of its utility as a vector in gene therapy. The MLV packaging signal has been mapped to a 350-nucleotide (nt) sequence in the 5′ untranslated region (UTR), downstream of the splice donor (22). In vitro and in vivo studies show that four stem-loop structures in the RNA are necessary and sufficient for packaging, although sequences downstream improve the efficiency (27, 28). In HIV-1, Ψ is less well defined. As in MLV, a sequence in the 5′ UTR is important for packaging. Several studies indicate a role for four stem-loops located between the 5′ splice donor and the beginning of gag (reviewed in reference 5). A sequence of 46 nt which includes the third stem-loop was shown to be sufficient for packaging of a reporter gene into virus-like particles in a vaccinia virus expression system (16). Deletion and mutation of the other stem-loops cause packaging defects in a variety of in vivo expression and in vitro assay systems (4, 6, 8, 25, 30), indicating their importance. However, other sequences that have been reported to affect packaging include (i) the transactivation-responsive element (TAR) and other 5′ sequences and (ii) the Rev-responsive element (25). Furthermore, the function of the stem-loops appears to be context dependent: the addition of certain reporter genes inhibits packaging (5, 24); and the position of the 46-nt packaging sequence was found to be critical for its function, presumably because surrounding sequences affected its ability to fold into a stem-loop (16). The complete packaging signal may be composed of several widely separated sequences, and its secondary and tertiary structure are almost certainly important for its recognition by the assembling virus.
In Rous sarcoma virus (RSV), the RNA packaging signal has been mapped to the 5′ UTR (19, 21). A 160-nt sequence, located between U5 and the start of gag, is sufficient to direct packaging of a reporter gene (2). RNA secondary-structure prediction (14) yielded a model of the 5′ UTR consisting of several stem-loop elements. One element, O3, was shown to be essential: mutations which disrupted base pairing of the stem at the base of this element reduced packaging of a reporter gene 100-fold, while compensatory mutations which restored base pairing rescued packaging to nearly half of wild-type levels (2, 20).
In RSV, Ψ overlaps sequences that are predicted to have other functions in virus replication. Wild-type viruses have dimeric genomes. The packaging signal overlaps a putative dimerization initiation sequence in RSV (13); a similar signal is found in one of the stem-loops of the HIV-1 Ψ (4). The relationship of packaging and dimerization is unclear (10, 31, 33). The 5′ UTR of RSV also contains three small open reading frames (ORFs) which serve to regulate Gag protein translation (10, 11, 15, 18, 33). Therefore, in studying the effect of mutations in Ψ, one must consider the effects on dimerization and translation also.
Our approach to the study of packaging was inspired by the RNA in vitro selection procedure (12, 17, 37). This method, which takes advantage of the fact that RNA has both informational and functional capacity, begins with a pool of RNA molecules of random sequence. A selection pressure, such as requiring the RNA to bind to a protein, is applied. The bound molecules are isolated, reverse transcribed, and amplified. This creates a new pool, which can be transcribed back into RNA and selected again. After multiple rounds, the best sequences are highly enriched in the pool. Our method, in vivo selection, uses the natural replication cycle of the virus in place of the in vitro steps of selection, reverse transcription, and amplification. By building a randomized stretch into a proviral clone and transfecting the resulting mixture of DNA into cells, one generates a population of viruses which can evolve over time. This method was first used by Berkhout and Klaver to examine the bulge and loop of the HIV transactivation-responsive region sequence (3); they randomized three nucleotides at a time. The study presented here extends the method, using much longer random stretches and multiple random stretches to test larger secondary-structure elements.
The in vivo selection technique has allowed us to map more of the RSV packaging signal. One predicted stem was shown to be essential, while two other stems were disrupted with no reduction in virus infectivity. A predicted dimerization initiation signal was shown to be favored but not strictly required.
MATERIALS AND METHODS
Plasmids.
All nucleotide sequences and numbers in this report refer to the RNA sequence of the Schmidt-Ruppin A (SR-A) strain of RSV (36) (Genbank accession no. L29198). Plasmid RCAS.bpΔA/B was the generous gift of Stephen Hughes. This plasmid, a member of the RCAS series, contains an infectious RSV DNA sequence in a pBR322 vector, without src or selectable genes (40). Except for part of the pol gene, RCAS.bpΔA/B has the SR-A sequence. We constructed a derivative of this plasmid, called RCAS.SE, which contains a unique SpeI site corresponding to nt 205 in the viral RNA and an EagI site at nt 325. A second RCAS derivative was made in which nt 205 to 325 are replaced with a nonviral sequence. This plasmid, called RCAS.stuffer, was made by replacing the small SpeI-EagI fragment of RCAS.SE with a 2.1-kb SpeI-EagI fragment from YEP24 (New England BioLabs).
Construction of randomized pools.
The wild-type RSV sequence from nt 205 to 325 was replaced by mutagenic cassettes. The sequences of the cassettes are shown in Fig. 2. For L3 Base, L3 Mid, L3 KL, and O3 Mid, oligonucleotides corresponding to sense-strand nt 193 to 331 in the viral RNA (RCAS.SE sequence) were synthesized by the Cornell BioResource Facility. Within each oligonucleotide, one or two stretches of sequence, 6 to 14 nt in length, were replaced with random sequence (25% each base at each position). The oligonucleotides were made double stranded by PCR to facilitate cloning; the sequences of the primers are presented in Table 1. PCR was performed with one of the upstream primers SpeI top, SpeI long, and SpeI G/T and downstream primer EagI bottom or EagI bottom2. For O3 Base, a pair of oligonucleotides was synthesized by Genosys, Inc.; one corresponded to sense-strand nt 193 to 263 and contained random nucleotides at positions 228 to 237; the other corresponded to the antisense-strand nt 331 to 247. This pair was mixed for PCR with primers SpeI top and EagI bottom. In all cases, the PCR mixtures contained 100 ng of each long oligonucleotide, 375 ng of each primer, 1 μl of Taq polymerase (Boehringer Mannheim Corporation [BMB]), 1.5 mM MgCl2, 10 mM Tris (pH 8.3), 50 mM KCl, and 0.2 mM each deoxynucleoside triphosphate (dNTP). PCR products were purified by phenol extraction and ethanol precipitation, or by using a High-Pure kit (BMB), and then digested with SpeI and EagI. Cut DNA was purified by phenol extraction and ethanol precipitation prior to ligation.
FIG. 2.
Sequences of viral RNA from the SR-A strain of RSV and from RCAS.SE-derived virus from nt 194 to 332 (top two lines) and sequences of the five mutagenic cassettes used in the randomization experiments. N, randomized position; ., base that matches the SR-A sequence.
TABLE 1.
Primers used in this study
| Primer name | Sequence |
|---|---|
| 52 | GTT GAT TCC CTG ACG ACT ACG AG |
| 619 | GCC TTC AAT GCC CCC AAA ACC A |
| 844 | CCG CAA TGA TAG CAG GAT GTG |
| 5′EcoRI | GCC ATT TGA GAA TTC ACC ACA TTG |
| EagI bottom | TCG TCG GCC GCA CTC T |
| EagI bottom2 | TCG TCG GCC GCA CTC TCC GT |
| SpeI G/T | TTC GAT GAC ACT AGT GGA GTG GGC |
| SpeI long | TTC GAT GAC ACT AGT GGA GGG GGC |
| SpeI top | TTC GAT GAC ACT AGT GG |
To generate vector for mass cloning without contamination by wild-type Ψ sequences, plasmid RCAS.stuffer was used. Ten micrograms of DNA was digested with SpeI and EagI. Due to the large size of the plasmid backbone (11 kb), we were unable to recover sufficient DNA for cloning by the standard procedures of separation on an agarose gel and extraction of the relevant band. Therefore, the fragments were separated on a 5 to 20% sucrose gradient in a buffer consisting of 10 mM Tris (pH 8), 1 mM EDTA, and 50 mM NaCl. Centrifugation was at 50,000 rpm for 5 h in an SW60 rotor (256,000 × g). Fractions were collected; those containing the 11-kb backbone and no insert were recovered, and the DNA was concentrated by isopropanol precipitation. For the ligation, 400 ng of backbone and 1 ng of insert were mixed with 500 U of T4 DNA ligase (New England Biolabs or BMB). Forty nanograms of the ligation mix was used to transform Escherichia coli DH5α by electroporation (BioRad Gene Pulser II). After electroporation, 1 ml of SOC medium (32) was added, and the cells were incubated for 30 min at 37°C with shaking at 225 rpm. Ten microliters of cells was plated on ampicillin-containing agar; the resulting colonies were counted to estimate the number of transformants. The remaining cells were added to 100 ml of Luria broth containing 100 μg of ampicillin per ml and grown overnight at 37°C with shaking at 225 rpm. Plasmid DNA was purified by standard techniques (32, 39).
Cells and virus.
Turkey embryo fibroblasts (TEFs) were the generous gift of Rebecca Craven. These cells do not contain endogenous sequences capable of recombining with RSV (41). Early-passage cells (from between 6 and 15 passages after the initial preparation) were used in all experiments. TEFs were grown in low-glucose Dulbecco’s modified Eagle medium (DMEM) supplemented with 1% vitamins, 20 mM l-glutamine, 100 U of penicillin G and 100 μg of streptomycin sulfate per ml, 2% heat-inactivated chick serum (Gibco BRL), and 10% fetal calf serum (Gemini). Cells were kept in 5% CO2 at 39°C. The chick serum was found to contain avian retrovirus particles, which could be detected in nested PCRs; therefore, the chick serum was omitted from the medium when virus was to be collected for RNA analysis.
Infections were carried out by overnight incubation of TEFs with virus in complete medium. Transfections were performed by the DEAE-dextran method. For a 100-mm-diameter plate, 10 μg of plasmid was added to 1.5 ml of Tris-buffered saline containing 100 μg of DEAE-dextran. Cells were washed twice with phosphate-buffered saline and twice with Tris-buffered saline, and then the DNA was added. The plates were incubated at 39°C and rocked every 15 min for 60 to 90 min. The DNA was aspirated, and the cells were shocked with 4 ml of dimethyl sulfoxide 27% in serum-free DMEM. After 4 min, the cells were washed with medium and fed with complete DMEM. To estimate the transfection efficiency, one plate was transfected with the reporter plasmid pHBAPr-1-βgal, which contains the lacZ gene driven by the β-actin promoter (26), in parallel with each experimental transfection; histochemical staining was performed to allow counting of the transfected cells (1). We found that 1 × 105 to 5 × 105 cells were stained per 100-mm-diameter plate.
The presence of virus was monitored by an exogenous reverse transcriptase (RT) assay as described previously (34). Medium from transfected or infected plates was filtered through a 0.45-μm-pore-size filter. Virus was collected from 2.5 ml of medium by centrifugation through a 15% sucrose cushion at 200,000 × g for 18 min at 4°C. The pellet was resuspended in 30 μl of 1× VSB (25 mM Tris [pH 7.5], 50 mM NaCl, 1 mM EDTA). Of this, 5 μl of virus was added to 20 μl of a reaction cocktail for final concentrations of 50 mM Tris (pH 8), 20 mM dithiothreitol, 0.05% Nonidet P-40, 6 mM MgCl2, 60 mM NaCl, 5 μg of oligo(dT) and 10 μg of poly(A) per ml, 10 μM cold dTTP, and 8 μCi of [α-32P]dTTP (3,000 Ci/mmol; Amersham). The reaction mixtures were incubated for 2 h at 37°C. Five microliters of each reaction mixture was spotted on DE81 paper (Whatman), washed three times in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), rinsed in H2O and then in 95% ethanol, dried, and quantitated by Cerenkov counting in a scintillation counter.
Analysis of viral RNA.
Virus was collected as described above. The pellets were resuspended and lysed in 100 μl of VLB (100 mM NaCl, 50 mM Tris [pH 7.5], 10 mM EDTA, 0.5% sodium dodecyl sulfate) with 10 μg of yeast tRNA and 10 μg of proteinase K. RNA was purified by phenol extraction and ethanol precipitation. Alternatively, pelleted virus was resuspended in 10 mM Tris (pH 8)–20 mM NaCl, and the RNA was purified by using a QIamp kit (Qiagen). To generate cDNA for sequencing, RT-PCR was performed. cDNA synthesis reactions were performed in a 20-μl volume containing the RNA recovered from 0.8 ml of medium, 1 μl of avian myeloblastosis virus RT (BMB), 800 ng of primer 844, 0.8 mM each dNTP, 0.5 μl of RNase inhibitor (PRIME), 50 mM Tris (pH 8.5), 8 mM MgCl2, 30 mM KCl, and 1 mM dithiothreitol. RNA and primer 844 were heated to 80°C for 5 min and chilled on ice to remove secondary structure; all components except the enzyme were mixed and incubated for 10 min to allow annealing of the primer to the RNA. Reactions were warmed to 42°C, and enzyme was added. The reaction mixtures were incubated at 42°C for 90 min. This was followed by PCR using all of the cDNA synthesis reaction mixture, to which was added 375 ng each of primers 844 and 5′EcoRI and 1.3 μl of Taq polymerase (BMB), in a total volume of 100 μl with final conditions of 18 mM Tris (pH 8.3), 46 mM KCl, and 2.8 mM MgCl2. No additional dNTPs were added. When insufficient product resulted, nested PCR was performed with the product of the first reaction as a template, with primers 52 and 619. PCR products were purified by using a SpinBind column (FMC) and sequenced with the Gibco BRL double-stranded DNA cycle sequencing system or by the BioResource Center, Cornell University, on an Applied Biosystems ABI Prism 377. Alternatively, the RT-PCR products were subcloned into pBluescript KSII+ (Stratagene), using the EcoRI site introduced at nt 10 by primer 5′EcoRI and an XhoI site at nt 644, and individual constructs were sequenced.
Sequence analysis.
Secondary-structure analysis was performed by using mfold (43, 38) on the server located at http://www.ibc.wustl.edu/∼zuker/rna/form1.cgi. An alignment of the first 400 nt of viral RNA sequence from different avian sarcoma and leukosis virus strains was performed with MegAlign. The strains and, in parentheses, their GenBank accession numbers are CT10 (Y00302), FSV (J02194), HBI (M11784), HPRS-103 (Z46390), MC29 (J02247), PR2257T (X51863), Prague B[LA23] (J02339), Prague B[LA23 mutant A] (M31387), Prague C (J02342), Prague C [duck adapted] (X51860), Schmidt-Ruppin A (L29198), SRA-V (U41731), Schmidt-Ruppin D (D10652), UR2 (M10455), and Y73 (J02027).
RESULTS
In vivo selection.
In vivo selection is based on the production of a large pool of viral genomes containing one or more stretches of randomized sequence. When such a pool of randomized genomes is transfected into cells, any genome capable of Gag expression will direct virion production. However, many of the genomes will have defective packaging signals, and thus many of the virions will not contain viral RNA. Only those which do carry genomes are infectious; therefore, when cell-free virus is used to infect fresh cells, only viruses with functional packaging signals will spread. After many passages, those with better packaging signals will come to dominate the population.
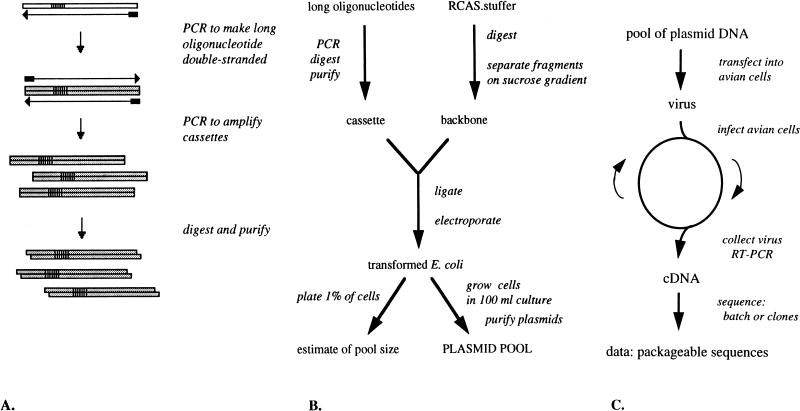
Generating the pool of mutant genomes is a multistep process (Fig. 1B). We began with a full-length, infectious clone of RSV, plasmid RCAS.bpΔA/B (40), which contains the complete SR-A provirus, with some pol sequence from strain BH, on a pBR322 backbone. We modified this construct by introducing unique SpeI and EagI restriction sites flanking the core of Ψ. The resulting plasmid, RCAS.SE, was as infectious as the parental virus (data not shown). A further construct, RCAS.stuffer, was made by replacing the SpeI-EagI fragment in RCAS.SE with an irrelevant 2.1-kb fragment from a yeast plasmid. RCAS.stuffer, rather than RCAS.SE, was digested and used for all of the cloning steps, in order to avoid contaminating the pools with wild-type sequence arising from incompletely digested plasmid. As expected, this construct was not infectious (data not shown). To purify large quantities of the RCAS backbone, we sedimented the digested DNA through a 5 to 20% sucrose gradient, collected fractions that contained the backbone but not the stuffer insert, and after concentration used this DNA in ligation reactions. For each experiment, the 120-nt SpeI-EagI fragment corresponding to Ψ was replaced with a mutagenic cassette containing the random stretches of 6 to 14 nt (25% of each nucleotide [Fig. 2 and 3]). The cassettes were created with long oligonucleotides that were amplified to make them double stranded (Fig. 1A). After digestion with the two restriction enzymes, the cassettes were ligated into the RCAS backbone. E. coli cells were transformed with the ligation mixture by electroporation and then grown overnight in batch culture in presence of ampicillin. Plasmid DNA was purified from the cultures and then transfected into TEFs to initiate an infection.
FIG. 1.
In vivo selection: experimental procedures and cloning. (A) Production of mutagenic cassette. A long oligonucleotide (white box) is synthesized with a viral sequence and a stretch of randomized sequence (striped segment). PCR is performed to first make the DNA double stranded and then amplify it. The resulting cassette is digested with restriction enzymes and purified. (B) Cloning of plasmid pool. The cassette produced is ligated into an infectious clone of the virus, in this case an RCAS plasmid. E. coli is transformed with the ligation reaction and grown as batch culture; a small aliquot is used to quantitate the number of transformants. Plasmids are purified from the batch culture. (C) In vivo selection experiment. The pool of plasmids is transfected into cultured cells. The resulting virus is passaged many times. After one or more passages, the virus is collected and its genome is recovered, amplified, and sequenced.
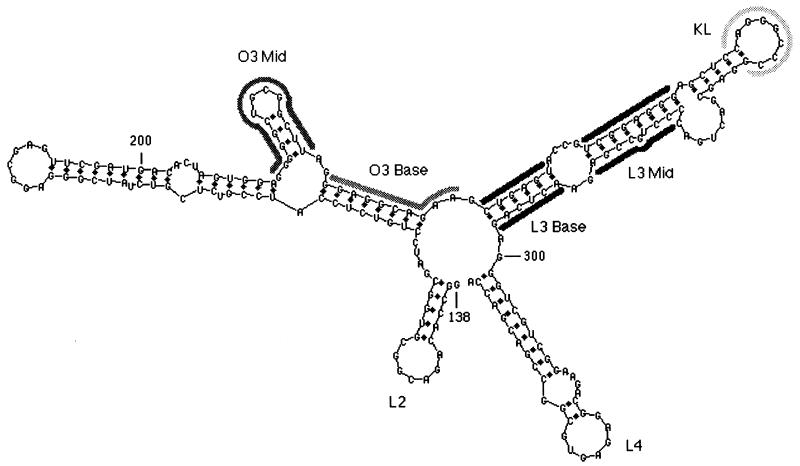
FIG. 3.
Secondary-structure prediction for Ψ RNA. The computer program mfold was used to predict the structure of nt 138 to 335, a sequence which includes the minimal packaging region MΨ (2). The elements L2, O3, L3, and L4 are shown (14). Thick gray lines indicate the sequences that were randomized in the different experiments of this study.
The virus produced by the transfected cells was used to infect fresh TEFs, and after a period of cell growth the virus in the medium again was collected and passaged to new cells for up to seven rounds of infection, representing up to 56 days of culture (Fig. 1C). Virus shed into the medium was monitored by standard exogenous RT assays (34). After one or more rounds of infection, the virus was collected and its RNA purified and subjected to RT-PCR to generate cDNA. cDNAs were subcloned, and the individual clones were sequenced; alternatively, if only a single sequence was used for transfection, then the cDNA was sequenced directly.
To assess the number of sequences in each random pool, we performed two types of controls. The number of E. coli transformants, i.e., the pool size, was estimated by plating 1% of the cells after electroporation onto selective medium and counting the resulting colonies. Pool sizes typically ranged from 1 × 105 to 5 × 105. Up to ca. 5 × 105 TEFs could be transfected on one plate, and thus most or all of the plasmid pool could be sampled in one transfection. For the L3 KL pool with seven randomized nucleotides (see below), that is enough to ensure that all possible sequences were sampled (47 = 1.6 × 104 possible sequences). Other pools had more randomized positions, such that the total number of possible sequences was much larger than the pool size; the largest was 17 nt, for a total of 1.7 × 1010 possible sequences. In such experiments, only a small fraction of the possible sequences could be sampled.
In addition to pool size, the extent of randomness in each pool was investigated by sequencing of the relevant stretch of DNA in plasmid preps of four to eight randomly picked E. coli transformants. We calculated the frequency of each base averaged over all positions in the randomized stretch. For most experiments, this was reasonably close to 25% for each nucleotide. However, as noted below, the L3 Base and L3 Mid pools showed a distinct nucleotide bias, with many more purines than pyrimidines at all positions. This bias can be attributed to an error in the oligonucleotide synthesis.
O3 Base.
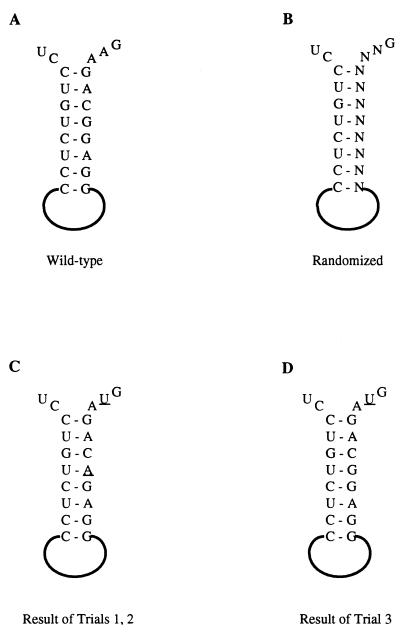
To test the method, we first examined a sequence whose importance was known. Knight et al. (20) showed in a packaging assay that a base-paired stem, here called O3 Base, was critical. Point mutations on one side of the stem abolished packaging, while mutants with compensatory changes which restored base pairing had 40% of the wild-type activity in the assay. We tested this stem in the in vivo selection system by randomizing 10 nucleotides (nt 227 to 236), of which the first 8 are one side of the stem and 2 are predicted to be unpaired (Fig. 4A and B). We constructed a pool containing 5 × 105 transformants, slightly less than the theoretical complexity for 10 random nt, 106, and hence not all sequences would be expected to be represented. In two independent experiments after one cycle of infection, a single sequence was recovered (Fig. 4C). It contains only two changes relative to the wild-type sequence: G232A and A236U. The G-to-A change preserves base pairing in the predicted stem, and A236 is predicted to be unpaired. In a third experiment, a different mutant, containing only the A236U mutation, was recovered (Fig. 4D). The lack of recovery of the wild-type sequence presumably is due to its absence in the pool. We interpret these results to strongly support the hypothesis that base pairing in O3 Base is critical for infectivity and to validate this experimental strategy as a method to analyze the packaging signal.
FIG. 4.
Randomization of O3 Base. (A) Diagram of the O3 element and adjacent sequences. The nucleotides of the O3 Base stem are shown. (B) Sequence randomized in the O3 Base experiment. Ten positions, shown as N, were substituted with mixed nucleotides. The DNA pool containing the randomized sequences was transfected into TEFs. The resulting virus was used to infect a fresh culture. Virus from infected cells was collected, and its RNA was subjected to RT-PCR and sequencing. (C) Results from two experiments. The recovered virus showed the same single sequence in each trial, with only two changes relative to the wild-type sequence (underlined bases). (D) Result from a third trial with the same pool. One position (underlined) differed from the wild type.
L3 Base and L3 Mid.
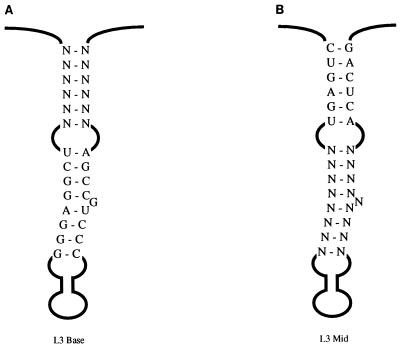
The predicted L3 element (nt 239 to 298) is a 60-nt sequence downstream of O3 and containing three base-paired stems, two internal loops, and a 7-nt loop at the end (Fig. 3). The stems are highly conserved across the 15 viral strains for which sequence information is available (see Materials and Methods). For example, L3 Base shows a single change in two strains, the 5′ half of L3 Mid shows a single change in one strain, and the 3′ half of L3 Mid shows variability in two nucleotides, one of which is predicted to bulge out of the stem. In contrast, several of the predicted internal loops of L3 show multiple changes. To address the importance of the L3 element, we first constructed two viral DNA pools with randomized sequences corresponding to L3 Base (nt 238 to 243 and 292 to 297, total of 12 nt) and to L3 Mid (nt 248 to 255 and 281 to 289, total of 17 nt) (Fig. 5). In each experiment, both sides of each stem are randomized; we would expect that if the stems are required, only sequences that allowed base pairing of the two halves would be recovered. Due to a bias in the oligonucleotide synthesis, which was discovered after completion of the selections, the input sequences had an excess of purines. For example, L3 Mid had 17 randomized positions; seven clones from the pool were sequenced, for a total of 119 relevant nucleotides. The composition was 52% G, 30% A, 9.4% T, and 8.6% C. This bias is an artifact that must be considered in interpretation of the results.
FIG. 5.
Randomization of L3 Base (A) and L3 Mid (B). Diagrams of the L3 element are shown. N indicates a randomized position.
The L3 Mid pool was transfected into TEFs, and the resulting virus was used for three consecutive infections of fresh TEF cultures. Viral RNA was recovered after the third passage and subjected to RT-PCR; the products were subcloned into pBluescript, and individual clones were sequenced. The results were striking. We recovered many different mutant sequences, all highly enriched in G and A due to the input bias. All of the sequences were very different from the wild-type sequence (Table 2). Furthermore, examination of the two halves of each sequence revealed that the mutants did not maintain base pairing: no stem can form at this position in any of the mutants. We conclude from these results that neither the primary sequence nor the predicted secondary structure is essential for viral infectivity. The fact that no single sequence was recovered more than twice indicates that no one sequence had a large selective advantage over the others.
TABLE 2.
L3 Mid mutants
| Construct | Sequence
|
|
|---|---|---|
| 5′ half | 3′ half | |
| Wild type | UCGGAGGG | CCCUGCCGA |
| Mutant | UUAGGAAG | GAAGGGAGA (2a) |
| UGUCAGCG | GGGGAGGAA | |
| UUAGGAAG | GAACGGAGA | |
| GGAUAACG | GAGAGAAGG | |
| GGAUAGCG | GAGAGAGGG | |
| GAAGGACG | GAGGAGUAG | |
| GGAUAGCG | GAGAGAGGG | |
| GAAUGACG | GAGGAGUAG | |
| GAAUGACG | GAGGAGUAA | |
Number of clones with indicated sequence.
In a parallel experiment, the L3 Base pool was analyzed in the same way, with virus being passaged seven times. Virus was recovered after passages 4 and 7. As in the case of the L3 Mid experiment, the L3 Base sequences recovered were varied, purine rich, and very different from the wild-type sequence. They also did not base pair (Table 3). Several contained a single base deletion in the 3′ half. Two sequences were recovered multiple times. One was found in two clones from passage 4 and two clones from passage 7, while the other was found in six clones from passage 4 and two from passage 7. This may be an artifact of the PCR amplification, or it may indicate that those sequences were overrepresented in the virus population; however, the fact that the number of clones with these sequences did not increase from passages 4 to 7 indicates that they do not have a strong selective advantage over the other sequences. These data clearly show that the predicted stems are dispensable for infectivity. Furthermore, the large excess of purines was not detrimental at these positions, despite the dissimilarity of this base composition from the wild type.
TABLE 3.
L3 Base mutants
| Construct | Sequence
|
|
|---|---|---|
| 5′ half | 3′ half | |
| Wild type | CTGAGT | ACTCAG |
| Mutant | GAGGGA | AAGAGA (8a) |
| AAGGA- | GGAAGG (4) | |
| AAGGA- | GGAAAG | |
| AAGGA- | GGAAGG | |
| AGGAA- | GAGAGA | |
| AAGGGA | GGAAGA | |
| AAGGGA | GGACGA | |
| AAGGGA | GGAAGG | |
| GAGGGA | GAGGGA | |
| GAGGGA | GGAAGG | |
| GGUAGG | GAGAGG | |
Number of clones with indicated sequence.
Sequence analysis revealed that a spontaneous point mutation had been selected in all 31 clones of L3 Mid and L3 Base mutants. Nucleotide 213, which is in the O3 element, was changed from G to U. Secondary-structure predictions for this change in the context of the wild-type sequence showed only a minor alteration, the extension of a small hairpin by 1 bp. In an attempt to understand these results, we generated several recombinant viruses of defined sequence. A virus in which the G213U mutation was placed in the wild-type context replicated normally (data not shown). Four constructs which contained single recovered L3 Base or L3 Mid mutant sequences were made (Fig. 6A). Construct BR4-5 corresponds to a particular sequence recovered from passage 4 of the L3 Base virus pool, including the mutations in the stem and the G213U change. The derivative BR4-5 U/G has the same stem sequence but contains the wild-type G at position 213. Likewise, constructs M2 and M2 U/G contain the stem sequences from a third-round L3 Mid virus, with and without G213U, respectively. These defined DNAs were transfected into TEFs, and equal amounts (by RT assay) of the resulting viruses were used for infections. The two viruses with the U at position 213 replicated normally, as assessed by RT assays of infected cultures. The two viruses with the G at position 213 were tested twice. In one experiment, both showed very low levels of virus production; in a second, both replicated with delayed kinetics and then reached wild-type levels, suggesting that a reversion event had occurred (data not shown). These experiments support the idea that the single nucleotide change had a very strong effect on replication.
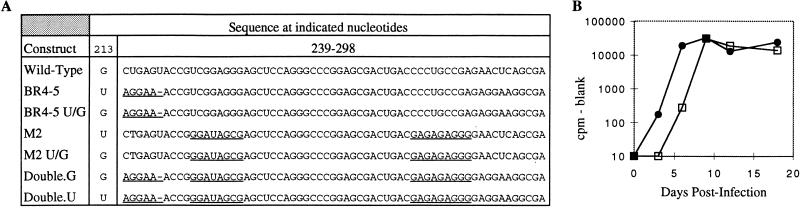
FIG. 6.
Recombinant L3 Stem mutants. (A) Sequences of nt 213 and 239 to 298 in the wild-type virus and indicated mutants. Mutated bases are underlined. (B) Kinetics of virus spread. Equal amounts of virus particles produced by transfected cells were used to infect TEFs. Virus production was monitored by RT assay. Each time point represents virus from a confluent 10-cm-diameter plate; the medium was on the plate for 24 h prior to collection. •, mutant Double.U; □, mutant Double.G.
The fact that individual mutants from the L3 Base and L3 Mid selected pools could replicate normally indicates that each stem alone is dispensable. We considered the possibility that the virus could replicate with one of the two stems intact, but not if both stems were disordered. To address this possibility, we made a recombinant which contained one selected sequence from each pool (Fig. 6B). In this viral RNA, neither stem should be able to form. We constructed two versions of this viral DNA, one with the wild-type G213 (called Double.G) and one with G213U (called Double.U). Again, equal amounts of virus produced by transfected cells were used for infections. We found that Double.U, like the wild-type virus, could replicate when both stems were disrupted. Double.G showed delayed kinetics but reached wild-type levels after several days (Fig. 6B), again suggesting a reversion event. Seven days after the Double.G infection, the virus was recovered and the relevant portion of the RNA was amplified by RT-PCR. Sequencing showed that it had indeed reverted to G213U. These data all strongly support the idea that the point mutation is critical when the L3 structure is lost.
O3 Mid.
The recurring selection of the spontaneous G213U mutation would appear to be an important clue about the structure and function of Ψ. Some but not all secondary-structure predictions show a small stem-loop from nt 214 to 225 in the wild-type sequence, and the G213U mutation is predicted to lengthen the stem by 1 bp. To better understand the structural features surrounding nt 213, we randomized nt 212 to 225 and analyzed the recovered virus in the same manner as described above. The pool size was estimated at 1.6 × 105, much smaller than the theoretical complexity of 3 × 108 for 14 nt. Hence, only about 1 in 1,000 of the possible sequences would be sampled in this experiment. Virus was recovered after the first and third rounds of infection, and cDNAs were subcloned and sequenced (Table 4).
TABLE 4.
O3 Mid mutants
| Clone | Sequence of nt 212–225a | ORF 3 translation | No. of clones with sequence after:
|
|
|---|---|---|---|---|
| Round 1 | Round 3 | |||
| SR-A | GGGGGCUGCGGCUU | MTLVEGAAA | ||
| SE | GGGGGCUGCGGCUU | MTLVEGAAA | ||
| G213U | GUGGGCUGCGGCUU | MTLVEWAAA | ||
| Mutant | GUGGCCGGGGGGUG | MTLVEWPGGEGGRS | 5 | 8 |
| GUGCCCCGCGGGA | MTLVECPAGKGGRS | 5 | ||
| GUGGCGUGUUGGUU | MTLVEWRVG | 3 | 2 | |
| GAGUGGGCGCGCUC | MTLVESGRAQGGRS | 1 | 1 | |
| GUGGCCGGGGGGUU | MTLVEWPGG | 2 | ||
| GUGGGGGAGUCCUC | MTLVEWGSPOGGRS | 1 | ||
| GUGGGGGAGUCCUC | MTLVEWGSPOGGRS | 1 | ||
| GUGGCCGGGUGGUA | MTLVEWPGGKGGRS | 1 | ||
| GUGGCCGGGGGGUG | MTLVEWPGGEGGRS | 1 | ||
| GUGGGCCGCGGCAA | MTLVEWAAAKGGRS | 1 | ||
| GUGGCUGUGGGGUU | MTLVEWLWG | 1 | ||
Underlined nucleotides and amino acids differ from the SR-A sequence.
The sequence characteristics for the two rounds were similar. Several sequences were observed more than once, indicating that the population of virus had limited diversity. The wild-type sequence did not appear in these clones, as expected given the sampling size, but the observed sequences share some general properties with the wild-type sequence. All are rich in G bases and have few A bases. All but one can be modeled as a stem-loop containing at least two base pairs. Nucleotides 212 and 214 are invariably G, the wild-type nucleotide. Position 213 is U in all but one sequence (it is A in clone O3MR1-14); no clones have the wild-type G. Thus, in the context of these mutations, as in the L3 stem mutations, a U at that position is highly favored. No other positions are conserved relative to the wild-type sequence. One example of base covariation is evident: nucleotide 216 is a G and 223 is a C in SR-A and all other known strains. These bases were seen in about one-third of our recovered sequences, but in the remaining clones, a C at 213 and a G at 223 were seen. This covariation indicates that a base pair between these nucleotides is highly selected. According to secondary-structure predictions, all but two of the sequences can form a hairpin which includes this base pair. We also found that nt 225 could be any base. This position is the first nucleotide of the UAG stop codon of ORF 3 in the wild-type virus. The fact that the other three bases are allowed, despite ruining the stop codon, agrees with the previous finding that a U225G mutant showed wild-type infectivity and packaging, presumably due to the presence of an in-frame stop codon 15 nt downstream of the mutation (11). In addition, many of the mutants had altered amino acid sequences for the peptide encoded by ORF 3; this also is consistent with previous studies which concluded that the amino acid sequences of the ORFs are not important (15, 18). In summary, these results indicate that the sequence of nt 212 to 225 need not be conserved, although the local structure may be important; however, the data do not allow us to explain the strong selection for the G213U mutation. We can only speculate as to the effect of G213U on viral replication. It is possible that this mutation stabilizes the predicted small hairpin structure, or all of O3. Perhaps the stem is usually stabilized by the presence nearby of L3, so that in the absence of L3, the additional stability imparted by G213U is needed. The mutation might also prevent alternative, incorrect foldings with downstream sequences by stabilizing the stem.
Kissing loop.
We next examined the loop at the end of element L3. In the SR-A strain of RSV the loop has the sequence AGGGCCC, while in other strains the loop contains other 6-nt, GC-rich palindromes. The loop, called a kissing loop, has been shown to promote dimerization of RNAs in vitro (13). A kissing loop contains a palindromic sequence that can base pair with the corresponding loop of an identical monomer. Several studies suggest that a kissing loop initiates genome dimerization in HIV-1 (5). Based on the in vitro data, a similar role was proposed for the loop in RSV (13). To test this hypothesis, we randomized the 7 nt of the loop. The pool size was estimated at 5 × 105, greater than the theoretical complexity of 1.6 × 104, and thus all possible sequences of the seven 7-nt stretch should have been sampled in this experiment. We recovered and sequenced viral genomes after two and five passages. The sequences of the 24 clones recovered after passage 2 were clearly biased toward palindromic symmetry (Table 5). Nine sequences, of which one was repeated once, contained 6-nt palindromes and five contained 4-nt palindromes. The remaining nine sequences were not palindromic and thus could not serve as kissing loops. By contrast, the recovered sequences after five passages were all palindromes (Table 5). Six of 24 clones contained the wild-type sequence. Several other GC-rich palindromes appeared multiple times. The fact that several sequences were seen more than once in this small sample indicates that they are highly selected. Three of the 24, including one of the wild-type clones, also contained the G213U mutation. We interpret these data to mean that a palindromic sequence in the loop is favored but not absolutely required for replication.
TABLE 5.
KL mutants
| Sequencea
| ||||
|---|---|---|---|---|
| Wild type | Round 2
|
Round 5 | ||
| 6-nt palindrome | 4-nt palindrome | No palindrome | ||
| AGGGCCC | UGGCGCC (2b) | AGCUGCA | GAGGAGG | UGGCGCC(9) |
| AGCGCGC | AGGUUGA | GCAGGGA | AGGGCCC(7c) | |
| UACGCGU | GGGUUGU | AUGGUGA | CGGGCCC(4d) | |
| AAAGUUU | GUGUCAA | GGGUGGU | UGGGCCC(3) | |
| GGUGCCA | UGGGGCU | UCGUCUA | ACCGCGG | |
| GUGUGUC | GGAGCGA | |||
| CGGCUGC | AUCUUUG | |||
| GACGUCGc | GCGAAUC | |||
| UAAGUAG | ||||
Palindromes are underlined.
Number of clones with indicated sequence.
One clone also contained the G213U mutation.
Two clones contained the G213U mutation.
ΔL3.
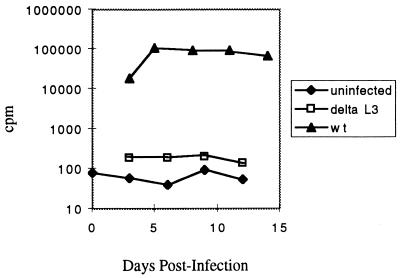
Based on the findings that the predicted stems and loop of L3 can be greatly altered, we chose to test a complete deletion of L3. We constructed a mutant called ΔL3, in which the entire L3 element is cleanly deleted. This mutant was transfected into TEFs, and the resulting virus used to infect fresh cells. The infections were followed for up to 13 days by RT assay. While cultures infected with wild-type virus contained detectable RT activity within 3 to 5 days and had sustained high levels after 5 to 7 days, ΔL3 virus levels increased slowly and never exceeded 5% of wild-type levels. The infectivity experiment was repeated four times, and in each case the RT activity was 20- to 300-fold lower than that of a fully infected wild-type culture; a representative experiment is shown in Fig. 7. The virus was recovered from one of the infected cultures, its RNA was subjected to RT-PCR, and the cloned sequence was examined. It had maintained the deletion and, unlike the L3 Base and L3 Mid mutants, did not contain G213U or any other mutations in the leader. These results are consistent with the hypothesis that L3 is not required for but does contribute significantly to packaging specificity or to other viral functions.
FIG. 7.
Kinetics of ΔL3 virus replication. Equal amounts of virus particles produced by transfected cells were used to infect TEFs. Virus production was monitored by RT assay. Each time point represents virus from a confluent 10-cm-diameter plate; the medium was on the plate for 24 h prior to collection. wt, wild type.
DISCUSSION
In this study, we have used an in vivo selection scheme to examine short sequences and predicted RNA secondary structures in the RSV packaging signal. We were able to select functional sequences from pools of virus with up to 17 randomized nt. Some experiments confirmed the importance of known elements; others showed the nonessential nature of several conserved sequences.
Our first experiment involved randomization of 10 nt including 8 nt of the stem O3 Base. The input material was a large and diverse sequence pool; the output, selected virus, was very restricted—only two different sequences were recovered. The sequences were very similar to the wild-type sequences and were selected after just one round of infection. The strong selection for the stem structure points to its importance for infectivity, in agreement with the data of Knight et al. (20) and Banks et al. (2). The results gave us confidence in the validity of the technique for examining the Ψ sequence and for the possibility of selecting functional sequences out of pools of this size and complexity.
In contrast to the strong selection seen in the O3 Base experiment, we found no selection for sequence or predicted structure in L3 Base and L3 Mid. The recovered sequences after many rounds of infection were unlike the wild-type sequence in both sequence and base composition and could not base pair to form stems. A recombinant virus with both stems disrupted replicated like the wild type, a fact which strengthens our conclusion: the predicted stems are dispensable for infectivity. This result is surprising, considering the high sequence conservation of the stems in natural strains of the virus. However, the data do agree with the finding of Katz et al. (19) that a deletion of 22 nt in L3 does not abrogate infectivity.
An unexpected result of the L3 Base and Mid experiments was the discovery of the spontaneous mutation, G213U. This single nucleotide change appears able to rescue infectivity of the L3 stem mutants: one recombinant virus, constructed with both stems disrupted and the G at position 213, replicated poorly and then reverted to U at position 213. This kind of rapid genetic change is common in retroviruses due to the high error rate of RT. In an attempt to understand the phenotype of this mutation, we probed the sequence and local structure around it. We randomized nt 212 to 225. The recovered sequences, although different from the natural sequence, had a base composition very close to that of the wild type. Positions 212 and 214 were always G, as in the wild type, and nearly all recovered clones had G213U, implying that this change may compensate for nearby mutations as well as the changes in L3. One pair of bases covaried, making a putative GC or CG base pair. All of the recovered sequences can be modeled as short stem-loops; we interpret this to mean that the local structure is likely to be important. Several sequences were found multiple times among the recovered clones, indicating that the virus population had a limited diversity and that some of the sequences may be more fit than others. This experiment did not answer the question of why G213U is selected in the presence of L3 stem mutants; however, it did reveal some of the sequence requirements at that location.
The apparent dispensability of the stems led us to question related function proposed for L3, dimerization. L3 was shown to act as a kissing-loop dimer initiation signal in vitro (13). A kissing loop is a loop containing a palindromic sequence, which can base pair with the loop of a second identical molecule. Fossé et al. (13) made transcripts in vitro, corresponding to nt 1 to 626 of RSV, which dimerize spontaneously. They found that an oligonucleotide which is complementary to nt 258 to 274 interfered with the dimerization reaction and that deletion of nt 207 to 270 reduced dimerization. This led to the suggestion that L3 contains a kissing-loop structure. We decided to test this hypothesis in vivo by randomizing the 7-nt loop at the end of L3. A variety of sequences were recovered. Over five rounds of infection, we observed a dramatic change in the virus population. After two rounds, the majority of sequences did not contain palindromes, although more were observed (42%) than would be expected by chance (5%); after five rounds, all contained palindromes. Compared to the case of O3 Base, in which the stem was selected after only one round, these results suggest a weak but significant selection pressure for a palindrome. From these experiments, it is impossible to distinguish whether the advantage imparted by the palindrome is due to an effect on dimerization, packaging or both; this question awaits further study, using more specific assays for those functions. The assertion that a palindromic loop in the predicted dimer initiation sequence is not essential is supported by a recent study on HIV-1. It was found that recombination between genomes with different kissing-loop sequences occurred as efficiently as recombination between identical genomes (35). Recombination depends on the copackaging of two genomes into a single virion; this result indicates that copackaging can occur in the absence of a kissing-loop interaction.
Given the data on the various parts of L3 (the dispensability of the stems and the loose requirement for a palindromic loop), we questioned whether the L3 element was needed at all. The ΔL3 mutant, missing all 60 nt of L3, was constructed to address that issue. This mutant replicated, but did so quite poorly. The severity of the defect shows that L3 has some function in replication; the fact that it replicates at all, however, means that the function is not required or can be carried out at least partially by other sequences. A quantitative assessment of packaging and other functions might sort out which functions are affected in this mutant.
We have shown that replication-competent sequences can be recovered from pools containing up to 14 random nts in a row, or two stretches totaling 17 random positions. Sample size may be a limitation when a sequence has stringent requirements. For example, in the O3 Base experiment, strong selection for the stem resulted in only two sequences being selected out of roughly 105. If a longer stretch had been randomized, probably no virus at all would have been recovered due to the sampling problem. For the other experiments, the selection was not as stringent, and we were able to recover fit molecules when sampling a tiny fraction of the possible sequences. Stretches even longer than those assayed here should be workable in this system, if the requirements are not very strict for the sequence in question.
Overall, our results demonstrate the power of in vivo selection as a tool for studying packaging and other viral RNA functions. One advantage of the method over the more traditional site-directed mutagenesis approach is that vast numbers of mutants are tested at one time. One can simultaneously discover which of sequences are allowed and which are not. Furthermore, one can follow the evolution of a virus population over time, distinguishing weak and strong selection pressures. A feature of the method which is both an advantage and a disadvantage is the fact that the readout of the assay is infectivity, while the actual selection may occur at the level of one or more viral functions. Complementing the in vivo procedures with assays for specific steps of replication would allow a more thorough investigation. In sum, in vivo selection has proved to be a useful approach for the study of packaging and should be applicable to many other retroviral RNA sequences of limited complexity.
ACKNOWLEDGMENTS
This work was supported by grant CA20081 from the USPHS.
We thank Maxine Linial, Michael Sakalian, Lance Stewart, and Steinar Johansen for many helpful conversations and suggestions. Thanks are due to Stephen Hughes and Rebecca Craven for reagents.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. I. New York, N.Y: John Wiley & Sons; 1998. [Google Scholar]
- 2.Banks J B, Yeo A, Green K, Cepeda F, Linial M. A minimal avian retroviral packaging sequence has a complex structure. J Virol. 1998;72:6190–6194. doi: 10.1128/jvi.72.7.6190-6194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkhout B, Klaver B. In vivo selection of randomly mutated retroviral genomes. Nucleic Acids Res. 1993;21:5020–5024. doi: 10.1093/nar/21.22.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout B, Van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 6.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 7.Burns C C, Moser M, Banks J, Alderete J P, Overbaugh J. Identification and deletion of sequences required for feline leukemia virus RNA packaging and construction of a high-titer feline leukemia virus packaging cell line. Virology. 1996;222:14–20. doi: 10.1006/viro.1996.0393. [DOI] [PubMed] [Google Scholar]
- 8.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffin J M, editor. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 10.Donze O, Damay P, Spahr P F. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donze O, Spahr P-F. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11:3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellington A D, Szostak J W. In vitro selection of RNA molecules that bind specific ligands. Nature (London) 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 13.Fossé P, Motte N, Roumier A, Gabus C, Muriaux D, Darlix J L, Paoletti J. A short autocomplementary sequence plays an essential role in avian sarcoma-leukosis virus RNA dimerization. Biochemistry. 1996;35:16601–16609. doi: 10.1021/bi9613786. [DOI] [PubMed] [Google Scholar]
- 14.Hackett P B, Dalton M W, Johnson D P, Petersen R B. Phylogenetic and physical analysis of the 5′ leader RNA sequences of avian retroviruses. Nucleic Acids Res. 1991;19:6929–6934. doi: 10.1093/nar/19.24.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hackett P B, Petersen R B, Hensel C H, Albericio F, I. G S, Palmenberg A C, Barany G. Synthesis in vitro of a seven amino acid peptide encoded in the leader RNA of Rous sarcoma virus. J Mol Biol. 1986;190:45–57. doi: 10.1016/0022-2836(86)90074-4. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi T, Shioda T, Iwakura Y, Shibuta H. RNA packaging signal of human immunodeficiency virus type 1. Virology. 1992;188:590–599. doi: 10.1016/0042-6822(92)90513-o. [DOI] [PubMed] [Google Scholar]
- 17.Joyce G F. Amplification, mutation and selection of catalytic RNA. Gene. 1989;82:83–87. doi: 10.1016/0378-1119(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 18.Katz R A, Cullen B R, Malavarca R, Skalka A M. Role of the avian retrovirus mRNA leader in expression: evidence for novel translational control. Mol Cell Biol. 1986;6:378–379. doi: 10.1128/mcb.6.2.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz R A, Terry R W, Skalka A M. A conserved cis-acting sequence in the 5′ leader of avian sarcoma virus RNA is required for packaging. J Virol. 1986;59:163–167. doi: 10.1128/jvi.59.1.163-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knight J B, Si Z H, Stoltzfus C M. A base-paired structure in the avian sarcoma virus 5′ leader is required for efficient encapsidation of RNA. J Virol. 1994;68:4493–4502. doi: 10.1128/jvi.68.7.4493-4502.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linial M, Medeiros E, Hayward W S. An avian oncovirus mutant (SE21Q1b) deficient in genomic RNA: biological and biochemical characterization. Cell. 1978;15:1371–1381. doi: 10.1016/0092-8674(78)90062-4. [DOI] [PubMed] [Google Scholar]
- 22.Mann R, Mulligan M C, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 23.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–5058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride M S, Schwartz M D, Panganiban A T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortlock D, Keller E B, Ziegra C J, Suter M M. High efficiency transfection of monkey kidney COS-1 cells. J Tissue Culture Methods. 1994;15:76–180. [Google Scholar]
- 27.Mougel M, Barklis E. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J Virol. 1997;71:8061–8065. doi: 10.1128/jvi.71.10.8061-8065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mougel M, Zhang Y, Barklis E. cis-active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J Virol. 1996;70:5043–5050. doi: 10.1128/jvi.70.8.5043-5050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oertle S, Spahr P-F. Role of the Gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J Virol. 1990;64:5757–5763. doi: 10.1128/jvi.64.12.5757-5763.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paillart J C, Berthoux L, Ottmann M, Darlix J L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rein A. Retroviral RNA packaging: a review. Arch Virol. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Sonstegard T S, Hackett P B. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J Virol. 1996;70:6542–6552. [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart L, Schatz G, Vogt V M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.St. Louis D C, Gotte D, Sanders-Buell E, Ritchey D W, Salminen M O, Carr J K, McCutchan F E. Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection in vitro. J Virol. 1998;72:3991–3998. doi: 10.1128/jvi.72.5.3991-3998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanstrom R, Varmus H E, Bishop J M. Nucleotide sequence of the 5′ noncoding region and part of the gag gene of Rous sarcoma virus. J Virol. 1982;41:535–541. doi: 10.1128/jvi.41.2.535-541.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 38.Walter A E, Turner D H, Kim J, Lyttle M H, Mueller P, Mathews D H, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L-F, Voysey R, Yu M. Simplified large-scale alkaline lysis preparation of plasmid DNA with minimal use of phenol. BioTechniques. 1994;17:26–28. [PubMed] [Google Scholar]
- 40.Whitcomb J M, Ortiz-Conde B A, Hughes S H. Replication of avian leukosis viruses with mutations at the primer binding site: use of alternative tRNAs as primers. J Virol. 1995;69:6228–6238. doi: 10.1128/jvi.69.10.6228-6238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang S, Temin H M. A double hairpin structure is necessary for the efficient encapsidation of spleen necrosis virus retroviral RNA. EMBO J. 1994;13:713–726. doi: 10.1002/j.1460-2075.1994.tb06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]