Abstract
Occupied between ~10,300 and 9300 years ago, the Pre-Pottery Neolithic site of Aşıklı Höyük in Central Anatolia went through early phases of sheep domestication. Analysis of 629 mitochondrial genomes from this and numerous sites in Anatolia, southwest Asia, Europe, and Africa produced a phylogenetic tree with excessive coalescences (nodes) around the Neolithic, a potential signature of a domestication bottleneck. This is consistent with archeological evidence of sheep management at Aşıklı Höyük which transitioned from residential stabling to open pasturing over a millennium of site occupation. However, unexpectedly, we detected high genetic diversity throughout Aşıklı Höyük’s occupation rather than a bottleneck. Instead, we detected a tenfold demographic bottleneck later in the Neolithic, which caused the fixation of mitochondrial haplogroup B in southwestern Anatolia. The mitochondrial genetic makeup that emerged was carried from the core region of early Neolithic sheep management into Europe and dominates the matrilineal diversity of both its ancient and the billion-strong modern sheep populations.
Analyses of 629 sheep mitogenomes detected no early domestication bottleneck but a Late Neolithic bottleneck in Anatolia.
INTRODUCTION
The establishment of Neolithic sedentary societies in southwest Asia was associated with the development of farming practices between 10 thousand and 12 thousand calibrated years before the present (ka cal BP). Those practices included the cultivation of cereals and legumes and the management of sheep, goats, cattle, and pigs, which ultimately resulted in their domestication (1–9). Crop-livestock subsistence strategies started gaining ground around 10.5 ka cal BP in the northern “Fertile Crescent,” and by 9.5 ka cal BP, this mode of subsistence had replaced the foraging lifestyle in parts of southwest Asia and Cyprus (2, 10–14). Archaeobotanical and zooarcheological analyses showed that these millennia-long practices of crop cultivation and ungulate management led to phenotypic changes in both plants and animals (2–4, 15–18). To understand these changes, it is often necessary to integrate the evidence of multiple sites and millennia (3, 19). However, only a few Pre-Pottery Neolithic sites preserved a sufficiently long occupation history and representative faunal assemblages to track morphological, biometric, and demographic changes related to early livestock management at a single location. The list of such exceptional sites in Anatolia includes Çayönü, Cafer Höyük, and Nevalı Çori in Southeastern Anatolia and Aşıklı Höyük in Central Anatolia (1, 3, 20).
Aşıklı Höyük is situated on the bank of the Melendiz River (21, 22). Here, large numbers of caprine bones (i.e., sheep and goats) have been excavated from occupational phases spanning over a thousand years, between ~10.3 and 9.3 ka cal BP. The importance of small livestock management at the site was such that, during the thousand-years occupation, the composition of animal remains identified as sheep and goat increased from ~50% to 87% (table S1) (8, 23, 24). Analyses of this extraordinary assemblage provided a unique glimpse into early strategies of sheep management. This includes mortality curves that are indicative of the culling of juvenile males, which in turn reflects exploitation for meat (23), and spatial patterns of skeletal distribution that imply that slaughtering took place near the living quarters (18). Further analyses of intra-articular joint pathologies suggested restricted mobility close to the village, including residential stabling (25), which led to the accumulation of dung and urine salts in the sediments (26, 27).
By 9.7 ka cal BP, however, sheep management strategies apparently shifted toward extensive herding. Evidence for this includes a decrease in urine salt and dung concentrations in residential areas (26, 27), an increase in carcass size (table S2), an improvement in joint health implying greater mobility (table S3) (25), and shifts in phytolith and stable isotope profiles that imply more extensive grazing (18, 28, 29). Together, the evidence obtained at Aşıklı Höyük demonstrates that sheep management in early Neolithic communities was a dynamic process of learning by doing (8, 18, 25).
Although management strategies at Aşıklı Höyük likely affected the phenotype of sheep populations, it is not clear whether they initiated evolutionary changes that ultimately led to the strong genetic differentiation between wild and domestic populations that we observe today. A common assumption is that capture and spatial isolation of a subset of a wild population induced a “domestication bottleneck,” provoking the general reduction of genetic diversity evident in modern domestic sheep populations (30).
Here, to address whether the initial management of sheep at Aşıklı Höyük caused shifts in their genetic makeup, we analyzed 629 whole mitogenomes sourced from 15 countries, including 62 from Aşıklı Höyük, spanning a period of over 10,000 years. This allowed us to infer the mitochondrial phylogeography and maternal demographic history of Anatolian and European sheep, and the contribution of the Aşıklı Höyük community to the formation of the Neolithic package dispersing across north and southwestern Anatolia between 10.0 ka and 8.0 ka cal BP, and subsequently into Europe.
RESULTS
Mitogenome dataset
We investigated 171 ancient samples from 7 sites in Anatolia (n = 102), 8 localities in the Levant and Caucasus (n = 24), and 18 localities in Europe (n = 45). Radiocarbon dating of associated charcoal and plant annuals placed our ancient samples in the Neolithic, Chalcolithic, Bronze Age, Iron Age, Middle Ages, and post-Medieval periods. In addition, we used data mining to retrieve two historic sequences (York, UK, 0.247 ka cal BP—the year 1776 AD—) from the Sequence Read Archive (see Materials and Methods).
For modern sheep, our study generated 147 mitogenomes and retrieved 164 mitogenomes by data mining from the Sequence Read Archive. Furthermore, we sourced 136 mitogenomes from published studies (30–40) and 9 directly from GenBank (acc. numbers in table S5). The complete dataset, which included mitogenomes from 24 modern mouflons (Ovis gmelini) from Iran, and 1 urial (Ovis vignei), encompasses 173 ancient and 456 modern sheep mitogenomes for a total of 629 individuals (Fig. 1 and tables S4 and S5).
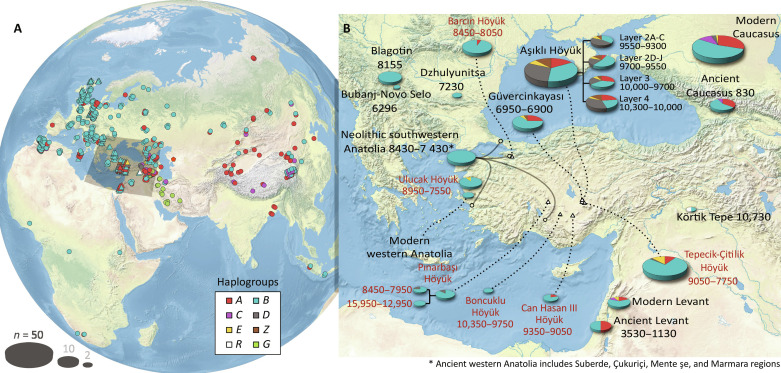
Fig. 1. Temporal and geographical distributions of haplogroup frequencies.
(A) Worldwide (jittered) location of all 629 samples analyzed in this study. (B) The map shows details of Anatolia and surrounding regions with haplogroup frequencies by archeological site, except for Neolithic southwestern Anatolia, where the grouping is at the regional scale (notice the tentacles out of the pie chart indicating the included locations, named below the map). Text in red indicates data reported in previous studies (42, 43), wherein haplogroups A to E were assigned using five diagnostic single-nucleotide polymorphism (SNP) markers of a 144–base pair (bp) control region fragment. Pie chart areas are proportional to their sample sizes (key, bottom left). The numbers accompanying location names refer to their estimated age range (calibrated years before the present) based on the site’s stratigraphy, material culture, and radiocarbon dating of appropriate materials including charcoal and annual plants.
Haplogroups distribution and diversity/neutrality indexes
In our dataset, we identified previously reported domestic sheep haplogroups A to E, as well as an unknown haplogroup that we labeled as Z (Fig. 2). In addition, we labeled modern mouflon (O. gmelini) haplogroups collectively as G. In modern sheep, the major mitochondrial haplogroups show a marked structure. In western herds populating Europe and North Africa, haplogroup B predominated (frequencies = 0.87 and 0.95, respectively). However, haplogroup B appeared less prominently in the Caucasus (frequency = 0.54) and in the Levant (frequency = 0.63) and exhibited a lower frequency than A in eastern Asia [frequency (B) = 0.36; frequency (A) = 0.5], where it coexisted with haplogroups C, D, and E that were less common and mostly endemic (Fig. 1 and Table 1).
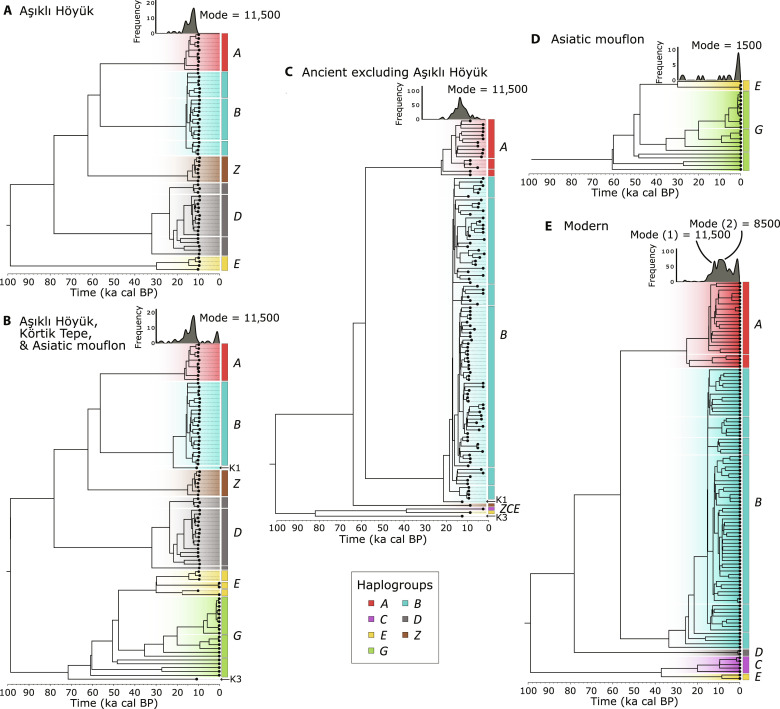
Fig. 2. Subtrees from the phylogenetic inference.
This figure shows five subtrees of the full phylogenetic tree (figs. S3 to S7) to facilitate interpretation: (A) subtree of all 62 Aşıklı Höyük samples; (B) subtree of samples from Aşıklı Höyük, Körtik Tepe, and Asiatic mouflons (O. gmelini); (C) subtree of all ancient samples except those from Aşıklı Höyük; (D) subtree of Asiatic mouflons; and (E) subtree from present day samples (randomly subsampled with proportional representation of haplogroups). K1 and K3 indicate the mouflons from Körtik Tepe. Graphs above the trees correspond to smoothed histograms of inferred coalescent times (nodes). Their horizontal scale (times) is the same as that of the trees. The tree of modern domestic sheep exhibits a first peak of coalescences at 11.5 ka cal BP and a second peak in the tree of modern sheep around 8.5 ka cal BP. These are absent in the wild sheep sample.
Table 1. Mitogenomic haplogroup frequencies.
Individual samples match the sample codes in table S5. Temporal tests of haplogroup frequencies used the groups and frequencies shown here, except for modern Anatolia, Körtik Tepe, and Asiatic mouflon, that were not subject to such analyses due to sample size. The table shows the absolute frequencies of mitochondrial haplogroups A, B, C, D, E, Z, G, and unassigned (R).
| Sample code | Sample origin | Mean age (ka cal BP) | Haplogroup | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | Z | R | G | Total | |||
| aEU | Ancient Europe | 4.11 | 1 | 46 | 47 | ||||||
| aSWAN | Neolithic southwestern Anatolia | 7.72 | 18 | 18 | |||||||
| aCC | Ancient Caucasus | 0.83 | 5 | 8 | 1 | 14 | |||||
| aLEV | Ancient Levant | 2.72 | 5 | 5 | 10 | ||||||
| aAH | Asikli Höyük (all layers) | 9.83 | 10 | 22 | 19 | 4 | 7 | 62* | |||
| aAH1 | Asikli Höyük levels 2A-C | 9.43 | 4 | 5 | 1 | 1 | 11 | ||||
| aAH2 | Asikli Höyük levels 2D-J | 9.63 | 1 | 8 | 3 | 1 | 2 | 15 | |||
| aAH3 | Asikli Höyük level 3 | 9.85 | 4 | 5 | 3 | 1 | 1 | 14 | |||
| aAH4 | Asikli Höyük level 4 | 10.15 | 5 | 5 | 8 | 1 | 3 | 22 | |||
| aGK | Chalcolithic Güvercinkayası | 6.93 | 5 | 13 | 1 | 1 | 20 | ||||
| aKT | Körtik Tepe | 10.73 | 1 | 1 | 2 | ||||||
| mAFR | Modern Africa | 0.0 | 3 | 89 | 2 | 94 | |||||
| mEU | Modern Europe | 0.0 | 23 | 154 | 177 | ||||||
| mWAN | Modern western Anatolia | 0.0 | 3 | 3 | |||||||
| mCC | Modern Caucasus | 0.0 | 18 | 32 | 6 | 2 | 1 | 59 | |||
| mLEV | Modern Levant | 0.0 | 2 | 7 | 1 | 1 | 11 | ||||
| mEASIA | Modern eastern Asia | 0.0 | 44 | 32 | 11 | 87 | |||||
| mOO | Asiatic mouflon | 0.0 | 3 | 21 | 24 | ||||||
| OUT | Urial | 0.0 | 1 | 1 | |||||||
| Total | 629 | ||||||||||
*Includes all four layers below.
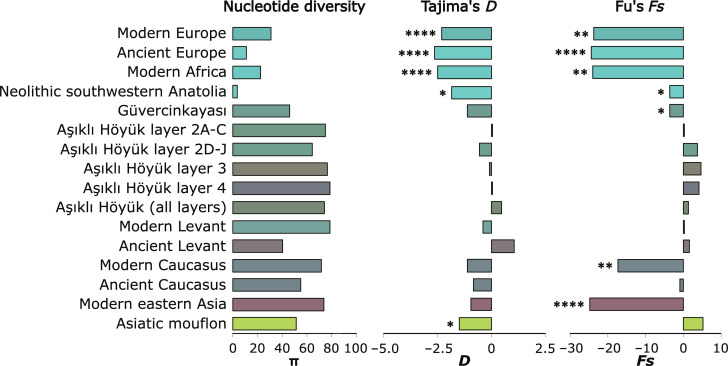
A spatial pattern compatible with the haplogroups distribution appeared in the nucleotide diversity as well as in the neutrality indexes and tests (Fig. 3). The samples from modern Africa, modern and ancient Europe, and Neolithic southwestern Anatolia, which had haplogroup B at high frequencies, show a depleted nucleotide diversity (0.0004 to 0.0022) and values of Tajima’s D and Fu’s Fs statistics that were negative, large, and significant (Tajima’s D = −2.63 to −1.84; Fu’s Fs = −23.8 to −to 3.65; P < 0.00001 to 0.015 and 0.002 0.048, respectively). In contrast, all post-Neolithic samples (including modern) from Anatolia, Caucasus, Levant, and eastern Asia displayed markedly higher nucleotide diversities (0.0033 to 0.0053), higher nonsignificant Tajima’s D values (−1.121 to 1.048; P = 0.113 to 0.894), and low and significant Fu’s Fs values (−24.80 to −3.653; P ≤ 0.0001 to 0.712). More notably, Aşıklı Höyük displayed the highest nucleotide diversity (0.0053), a positive nonsignificant Tajima’s D value (0.453; P = 0.762), and a positive nonsignificant value of Fu’s Fs (1.283; P = 0.722). In addition, the subsequent levels of Aşıklı Höyük did not show significant differences among them for these statistics and tests (Fig. 3).
Fig. 3. Population genetics statistics.
Estimations of nucleotide diversity, Tajima’s D, and Fu’s Fs. The asterisks show the significance level of the associated statistical test on the statistics (* = 0.01 to 0.05, ** = 0.001 to 0.01, *** = 0.0001 to 0.001, and **** = 0.00001 to 0.0001). Significant values of Tajima’s and Fu’s neutrality tests could indicate past demographic bottlenecks. The bars’ colors correspond to the sample frequency-weighted average of the haplogroup’s colors shown in Figs. 1 and 2. These statistics suggest a bottleneck in the ancestry line of the European populations and possibly Neolithic southwestern Anatolia, while the colors reflect the haplogroup differences between eastern and western samples (notice the gray tones of Aşıklı Höyük samples).
Tests of temporal change in haplogroup frequencies
When we tested the temporal changes in haplogroup frequencies, we found, in general, significant differences between, but not within, groups. The western group included Africa, Europe (modern and ancient), and Neolithic southwestern Anatolia, while the eastern group included Neolithic Aşıklı Höyük, Chalcolithic Güvercinkayası, Caucasus, Levant, and eastern Asia. This pattern was true when the tests were performed with effective population sizes (Ne) between 104 and 106 (we refrained from writing P values here because of the numerous combinations of Ne used in these analyses; see table S6 for the exact values and fig. S2 for a graphical summary of these results). The whole pattern of significance makes sense of the patterns of diversity/neutrality indexes and leads to an important observation that the divergence between eastern and western groups seems to have originated between central and southwestern Anatolia, during the Neolithic (fig. S2).
In addition, the temporal tests detected no significant changes in haplogroup frequencies across Aşıklı Höyük’s occupation layers (P = 0.28 to 0.97; fig. S2 and table S6).
Phylogenetic analyses and demographic inference on coalescent times
Our phylogenetic analysis recovered clades corresponding to the major domestic sheep haplogroups A to E, as well as an unknown haplogroup that we labeled as Z, in addition to the modern mouflon haplogroups (O. gmelini) collectively labeled as G (Fig. 2 and figs. S3 to S7). Notably, within haplogroups A to E and Z, we found a high density of coalescent events (nodes) between 12.0 ka and 7.6 ka cal BP (50% high-density interval), peaking around 11.5 ka cal BP.
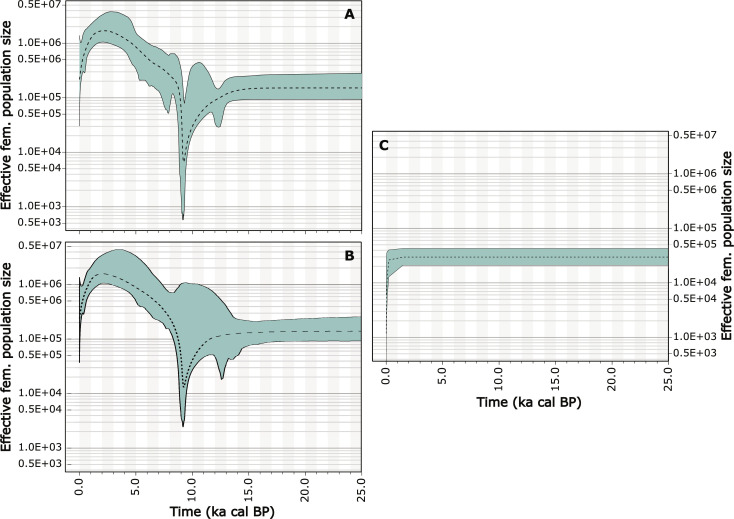
We tested if this concentration of coalescences was an artifact of sampling by comparatively inferring the demographic histories of European-Anatolian samples and mouflon samples based solely on coalescent times (nodes ages) and sample ages. For this, we used an in-house developed method that takes advantage of the statistical technique known as Markov chain Monte Carlo (MCMC) (see text S8). Although at low resolution, the inferred demographic history of European-Anatolian sheep was compatible with the demographic history inferred by extended Bayesian skyline plots that were co-estimated with the phylogenetic reconstruction in the software BEAST. Both the MCMC procedure and the skyline plots suggest the occurrence of at least one major bottleneck that reached its peak between 8.0 ka and 10.0 ka cal BP, and a long-term demographic expansion thereafter, except for the last millennium that is characterized by a decline (Fig. 4 and fig. S8).
Fig. 4. Extended Bayesian skyline plots.
The charts show demographic histories inferred with the samples of (A) modern and ancient Europe, the Neolithic sites of Aşıklı Höyük, Körtik Tepe, and southwestern Anatolian (including Suberde, Çukuriçi, Menteşe, and Marmara regions); (B) the same samples minus Neolithic southwestern Anatolian sites; and (C) modern Asiatic mouflon, plus two Körtik Tepe and one Aşıklı Höyük samples for time calibration. The dotted lines indicate the median values, while the gray areas represent the 95% highest probability density (HPD) regions. The plots in (A) and (B) show a strong population bottleneck peaking around 9.0 ka to 9.5 ka cal BP followed by a significant population expansion (consider the HPD areas). This may be a compendium of the domestication process, a demographic constriction postdating site occupation at Aşıklı Höyük, and a historical expansion of flocks of sheep into Europe.
Simulation-based inference by approximate Bayesian computation
After integrating the results of the different analyses, we constructed some hypotheses regarding the evolution of domestic sheep in Eurasia. To test them, we used the simulations-based statistical technique known as approximate Bayesian computation (ABC). The specific goal was to infer a temporal population structure relating to European and Anatolian sampled populations and to estimate key parameters such as Ne. We split this inference into four phases with each phase targeting a simple inferential goal to prevent a number of technical problems such as high dimensionality and overparametrization. Figure 4A and figs. S9 to S11 show schematic representations of the models used in each phase.
The first phase of analyses by ABC targeted a demographic inference at Aşıklı Höyük. It showed that a model with two demographic phases (fig. S9B) was moderately better supported than a non-changing model (fig. S9A and table S9) (Bayes factor = 1.7 to 3.8). The parameter estimation of such a model supported a major increase in the sheep population throughout site occupation, although posterior distributions showed a large variance (fig. S12). Furthermore, an analysis of pseudo-observed datasets suggested that this analysis did not lack statistical power to detect large bottlenecks, as our ABC methodology could detect an eightfold bottleneck at Aşıklı Höyük around 90% of the time with very low stringency criteria (Bayes factor > 1.0) (fig. S13).
The second phase of ABC analyses expanded the focus of interest to the spatiotemporal structure of the sheep populations in Neolithic-Chalcolithic Anatolia. Specifically, the analyses aimed to pick the best among eight alternative scenarios (fig. S10). The best support was divided between two models. In both models, the populations of Neolithic southwestern Anatolia and Güvercinkayası were sisters, but in one model they descended directly from Aşıklı Höyük (model likelihood = 0.110 to 0.171), while in the other, they descended from an independent ancestral population (model likelihood = 0.159 to 0.188) (fig. S10 and table S11).
The third phase of ABC analyses aimed to select the best scenario of the ancestry of the European sheep population with respect to the populations of Neolithic-Chalcolithic Anatolia. In the best-supported scenario, Neolithic southwestern Anatolia was the ancestor of European sheep (Bayes factor = 1.4 to 342.0) (fig. S11 and table S13). This result was expected, though, considering the high prevalence of haplogroup B in both the European and Neolithic western Anatolian samples (frequencies = 0.979 and 0.870 in ancient and modern Europe; and = 1.0 in Neolithic western Anatolia) (see Fig. 1).
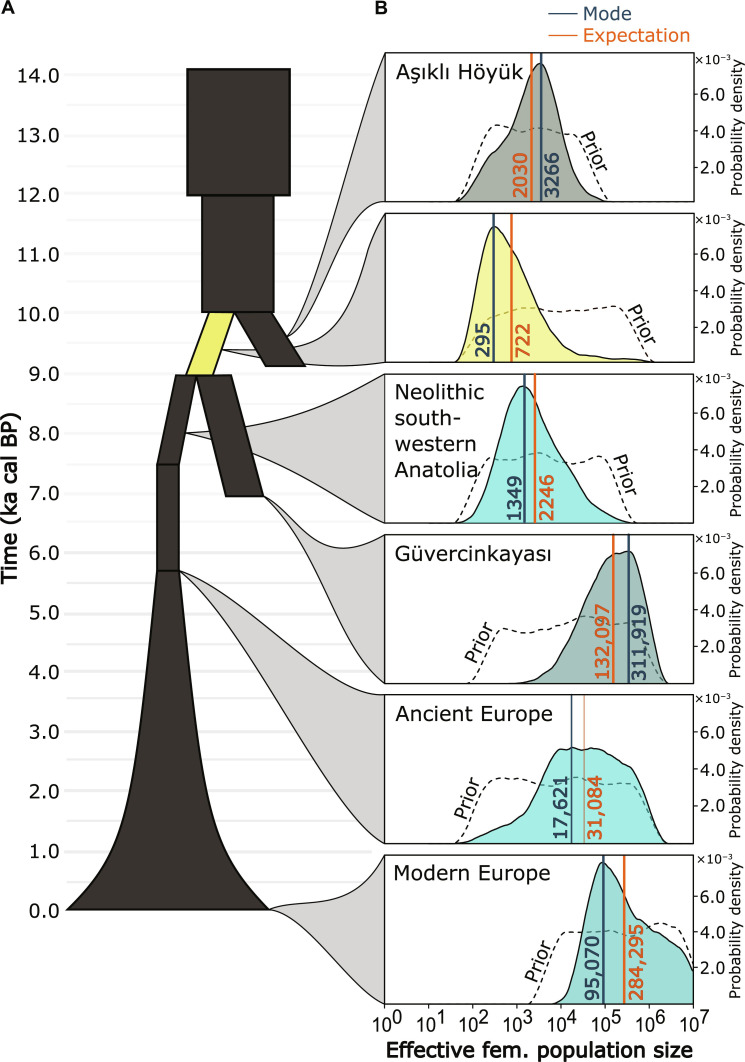
In the last phase of ABC analyses, we estimated with improved precision the Ne values of sampled and unsampled populations of a simulation design that was constructed upon the selected models of the second and third phases. In such a model, modern and ancient European sheep descend from herds populating Neolithic southwestern Anatolia, which in turn descended (together with Güvercinkayası) from a population that branched off Aşıklı Höyük’s stem population at a variable time between the Epipaleolithic and Aşıklı Höyük’s abandonment (see fig. S11). The punctual estimations for Ne in Aşıklı Höyük were in the few thousand (mode = 2030), but the value estimated for the population that followed (i.e., the ancestor of Neolithic southwestern Anatolia and Güvercinkayası) dropped one order of magnitude (mode = 295). The estimations of Ne for Neolithic western Anatolia were ~1.0 × 103 to 2.0 × 103, while for Güvercinkayası they were ~2.0 × 105 to 3.0 × 105. Expectedly, the population of sheep increased from the Anatolian Neolithic to the European Neolithic, and from there to the modern European sheep population (Fig. 5).
Fig. 5. Modern and ancient Ne values inferred by ABC.
(A) Schematic representation of the model used in phase four of the simulation-based inference by ABC. Rectangles and trapezoids represent constant-size populations, while the inverted-funnel shape at the bottom represents a population with exponential growth (distributions corresponding to initial and final Ne values); (B) posterior probability distributions (in color) of Ne. The color matches the average haplogroup color of the corresponding samples (as in Fig. 3), except for the ancestral population (in yellow) of Neolithic southwestern Anatolia and Chalcolithic Güvercinkayası. Notice the small value of the central estimations of Ne in such a population as compared to the sheep population of Aşıklı Höyük.
DISCUSSION
Haplogroups as potential markers of independent domestication
The phylogeographic structure that we observed in modern mitochondrial sheep haplogroups, where haplogroup B predominates in western populations and haplogroup A does so in eastern populations, is consistent with various reports [e.g., (33, 41–43)]. Such structure has been explained by means of multiple domestication origins (33, 44–47), the domestication of different wild populations or species (33), introgression (41), and lineage sorting-and-gene drift (33). Although multiple domestication origins are still being invoked to explain haplogroup structure (44–47), its popularity was most certainly influenced a few decades ago by the discovery of multiple domestication origins in pigs and cattle (33). However, sheep domestication centers outside southwest Asia have never been identified (48). Furthermore, goats, also believed to have had a unique domestication, exhibited a phylogeographic structure similar to the one in sheep before the post-Neolithic rise of the modern dominant haplogroup (49). In cattle, the two independently domesticated species (Bos taurus and Bos indicus) display a mitochondrial differentiation that is much higher than the observed among sheep populations, and their phylogeographic structure is heavily influenced by post-Neolithic introgression (50). Further criticisms of the multiple domestication hypothesis in sheep point out that the hypothesis equals haplogroups with populations while ignoring the diversity of the ancestral wild populations (48). In this regard, we show here that early Neolithic Aşıklı Höyük from Central Anatolia had haplogroups A, B, D, E, and unreported Z at high frequencies (0.161, 0.355, 0.306, 0.065, and 0.113, respectively) which is broadly consistent with what is reported in Neolithic sheep (see fig. S14) (41–43). These findings most definitely refute multiple domestication origins as an explanation for the modern distribution of these haplogroups.
Regarding another hypothesis, available evidence disregards the contribution of species other than Asiatic mouflon (O. gmelini) to the domestic sheep lineage, despite ample documentation of introgression among wild sheep species (33, 41, 48). It is possible, however, that domestic sheep descend from several mouflon populations with some degree of differentiation (see discussion below). Regarding lineage sorting and gene drift, we believe that they have some etiological value that is, however, conferred by further anthropogenic and demographic causes. When put in context, our evidence suggests that the near-fixation of haplogroup A in eastern Eurasian and haplogroup B in Europe and Africa emerged from Neolithic human-mediated migrations and demographic bottlenecks.
Links between an increased density of coalescences and sheep domestication
In our phylogenetic reconstruction, we found a very high density of coalescent events (nodes) between 12.0 ka and 7.6 ka cal BP (peaking around 11.5 ka cal BP) within haplogroups A to E and Z. Two types of analyses, our in-house developed MCMC procedure and the extended Bayesian skyline plots, suggest that the observed pattern of concentrated coalescences is not due to a sampling bias but rather associated with demographic constriction. Furthermore, Asiatic mouflons did not present a concentration of coalescences nor bottlenecks in the demographic histories inferred by the skyline plots or our MCMC procedure (Fig. 3 and fig. S8), suggesting that the demographic process responsible for it may well be unique to managed rather than to wild sheep. One more clue regarding the origin of the concentration of coalescences is the fact that it coincides with the early phases of sheep management in southwest Asia (2, 19). A similar coalescence pattern (between 11.0 ka and 9.5 ka cal BP) also appeared in ancient domestic goats (51) and is consistent with an inferred Neolithic bottleneck in cattle (52). Therefore, it seems that the concentration of coalescent events in sheep relates to founder events associated with the species’ early management potentially because of multiple capture events and breeding in captivity, briefly a domestication bottleneck.
That said, we acknowledge our limited statistical power to detect a comparable phenomenon in wild sheep due to the lack of samples of pre-domestic Anatolian sheep. Thus, we cannot completely discard the possibility that the concentration of coalescences was, at least partially, due to pre-domestication events in mouflons. Further studies, especially those incorporating ancient mouflon specimens and whole genome sequencing data, could confirm or reject that such a pattern resulted from initial capture and human management of sheep, subsequent demographic changes, or demographic changes caused by non-anthropogenic factors, such as the climate conditions of the Younger Dryas cold spell.
Impact of sheep management on microevolutionary processes at Aşıklı Höyük
One exciting possibility that our dataset allowed us to test is the hypothesis that management practices at Aşıklı Höyük caused the bottleneck suggested by the coalescence patterns in domestic sheep. However, nucleotide diversity, Tajima’s D, and Fu’s Fs statistics did not show significant changes across layers and levels (Fig. 3) and neither did the temporal tests on haplogroup frequencies (P = 0.28 to 0.97; fig. S2 and table S6). More powerful simulation–based analyses better supported a two-phased model with an increase rather than a decrease in population size (table S9). Although statistical support for this was moderate, a population increase is in line with zooarcheological evidence (8, 23, 24). Overall, our findings indicate that 1000 years of local management did not alter sheep mitogenomic diversity. Although our findings only refer to female ancestry, they nonetheless contradict the commonly held narrative that a genetic bottleneck accompanied early centuries of human management of livestock.
The constancy of diversity and neutrality indexes and haplogroups’ composition across archeological levels, combined with the presence in the phylogenetic reconstruction of sequences that coalesce during the site’s occupation (see Fig. 2 and figs. S3 to S7), suggests breeding continuity in the sheep of Aşıklı Höyük. The development of breeding practices could help explain the decline in the relative importance of game observed in the human diet during site occupation (18, 23, 25). However, the deep phylogenetic branching of some individuals at Aşıklı Höyük and the fact that some of these cluster with modern mouflons in haplogroups E may be a consequence of human exploitation of non–locally bred individuals. Those individuals can be compared to two Körtik Tepe individuals (Fig. 2, B and C, labeled K1 and K3) that coalesce, one at the base of haplogroup B and one to a deep branch that is paraphyletic to all mouflon and sheep individuals. The Körtik Tepe individuals, dated to 11.7 ka cal BP, are considered wild based on their archeological context, morphology, and dating (53). The possibility that the entire sheep population of Aşıklı Höyük was hunted or captured and raised [see, e.g., (8)] could explain the high diversity—and lack of bottleneck—at the site but is at odds with the mitogenomic relatedness that we observe and with the archeological evidence pointing toward dense stabling. Thus, we hypothesize that a minority of individuals at Aşıklı Höyük are likely wild sheep, which is in agreement with archeological evidence of hunted individuals that are distinguishable from the local stock, both in Aşıklı Höyük and in related sites (8, 23).
The simultaneous presence of presumably wild and managed individuals in Aşıklı Höyük brings up an obvious question: Why? Although speculative, the presence of wild individuals may be explained by re-stocking that was used to compensate for losses caused by abortions and lamb mortality (54). It is also possible that some individuals at Aşıklı Höyük were exchanged with other communities of sheep keepers or that the founder herd of managed individuals was relatively large and diverse in geographic origin, possibly coming from somewhere else. A geographical translocation was the case for a “new” glume wheat that was coevally cultivated at Cafer Höyük, Aşıklı Höyük, and Boncuklu. This variety was not endemic to Central Anatolia but transferred long-distance from the Upper Euphrates valley (7, 55, 56). Since in this part of Anatolia, early cereal cultivation went along with sheep and goat management (2, 19, 20), a transfer of small stock on the hoof must be taken into consideration.
Beyond Aşıklı Höyük
We found, in the phylogenetic reconstruction, multiple mitogenomes from modern and ancient Anatolian and European sheep that coalesced with mitogenomes from Aşıklı Höyük within 2000 years of the site’s occupation. This suggests that the genetic makeup of modern sheep populations retains a legacy from early Central Anatolia. As Central Anatolia participated in extensive trade networks since the Epipaleolithic (57), we assume that this legacy stemmed from an ancestral sheep metapopulation that included Aşıklı Höyük.
In contrast to Aşıklı Höyük, we found haplogroup B dominating later western Anatolian sites, Chalcolithic Güvercinkayası, and reported Anatolian sites dated between the Epipaleolithic and the Late Neolithic (41–43). Outside Anatolia, the same haplogroup appeared quasi-fixed in ancient and modern European and African sheep but became less dominant in the Levant and the Caucasus regions and turned out to be less prevalent than haplogroup A in modern sheep from central and eastern Asia. The spatial and temporal pattern of diversity and neutrality statistics, complemented and refined by the results of the temporal tests, suggests that the near fixation of haplogroup B in western Eurasia and A in central and eastern Asia resulted from demographic processes that postdate the occupation of Aşıklı Höyük yet predate the introduction of sheep to Europe (~8.5 ka cal BP) and central (~7.0 ka cal BP) and eastern Asia (~5.0 ka cal BP). More specifically, a bottleneck.
We tracked the origin of this putative bottleneck with several rounds of simulations-based analyses. These analyses revealed an approximately tenfold reduction in Ne in the ancestral sheep population from which the populations in Neolithic southwestern Anatolia and Chalcolithic Güvercinkayası putatively descended (Fig. 5). This conspicuous bottleneck most likely occurred after Aşıklı Höyük had already been abandoned and coincided with the expansion of the Neolithic way of life beyond its formative zone. From the archeological record, the Neolithic expansion beyond Central Anatolia into western Anatolia and further (north-)west occurred by an inland route, via the Lake District, as well as through maritime expansion following the southern Anatolian coast, as has also been suggested recently (58–60). The migration of humans and their flocks was neither a single event nor a linear phenomenon, but a complex process involving various groups, which has recently been confirmed by the palaeogenomics of Neolithic Anatolian farmers (61). We therefore hypothesize that the demographic constraint observed in sheep could be the result of serial founder events that took place as sheep husbandry dispersed into (north-)western Anatolia.
Our analyses provide a definitive answer to previous reports that hinted at a possible gradient of increasing diversity and decreasing frequency of haplogroup B toward central and Eastern Anatolia and toward the Neolithic [e.g. (41, 42)] but failed to track its origin. Our results indicate that the rise of haplogroup B was not the result of founder events that co-occurred with the introduction of sheep into Europe but rather related to a prior demographic process when sheep husbandry gained a foothold beyond Central Anatolia.
Our ABC analyses, MCMC-estimated historical demography, and Skyline Plots also support a substantial population expansion (~10×) in European sheep over the past ~5000 years (Figs. 3 and 4 and fig. S8). This demographic process can conceivably be the result of post-Neolithic human expansion, and an increase in genetic diversity related to the dispersal of wool sheep lineages originating in southwest Asia and beyond (62, 63).
MATERIALS AND METHODS
Experimental design
To evaluate the mitogenomic diversity and the possible presence of a domestication bottleneck during the early stages of sheep management, we used a worldwide sample of mitogenomic sequences of sheep that was exceptionally dense in Neolithic southwest Asia and Europe. We computed population genetics statistics and applied multiple statistical analyses, including a phylogenetic reconstruction and simulation-based analyses by ABC. To better understand the numerous analyses of our study, consider that they pertain to three spatiotemporal scales. The first one focuses on Aşıklı Höyük, an aceramic Neolithic site in Central Anatolia with a long occupation span; at the next scale, analyses focused on Neolithic-Chalcolithic sites in Anatolia, and at the largest scale, our analyses involved all ancient and modern sites in Anatolia and Europe.
With respect to Aşıklı Höyük, we compared four subsequent archeological occupation layers that experienced substantial changes in sheep management. We first estimated and compared gene and nucleotide diversities, and the neutrality statistics Fs of Fu and D of Tajima (along with the associated statistical tests). Next, we tested the temporal changes in the haplogroup frequencies across layers, and afterward, we carried out simulation-based analyses by ABC for testing a set of possible scenarios of demographic change (or lack of it) across one millennium of site occupation while estimating their Ne.
In a subsequent phase of analyses, we inferred the relationships between Aşıklı Höyük and the other ancient Anatolian samples in our sheep dataset, namely, Neolithic southwestern Anatolia and Chalcolithic Güvercinkayası. For this stage, we also compared distinct scenarios and inferred Ne by means of simulations and ABC.
With the results of the previous analyses, we moved forward to study the relationship between European domestic sheep and their Anatolian ancestors. As in previous phases of analysis, we used simulation-based analyses by ABC to select the best statistically supported scenario among a suite of alternatives. With the selected scenario we carried out a second simulation-based inference (by ABC) to estimate Ne for all populations of the selected scenario, including Aşıklı Höyük, Neolithic southwestern Anatolia, Chalcolithic Güvercinkayası, and ancient and modern Europe.
Sampling
Samples were obtained from three sources (see tables S4 and S5 for details and table S14 for contact information).
1) De novo sequences obtained from direct sampling of both live specimens and ancient remains. The set of ancient samples resulted from the study of >500 bone and tooth specimens from archeological sites and museum collections in southwest Asia and Europe, and subsequent processing through targeted capture and next-generation sequencing.
2) Sequences retrieved from public repositories of high throughput sequencing data by means of data mining techniques (see text S7).
3) Published mitogenomes. We gathered mitogenomes associated with published studies or directly from GenBank. Published data from archeological sheep originated from sites in Bulgaria, Georgia, Germany, Ireland, Israel, Malta, Serbia, Türkiye, and the United Kingdom. Details of the samples including age, geographical origin, and haplogroup are shown in table S5.
DNA extraction and sequencing
We processed modern and ancient samples for DNA extraction, polymerase chain reaction, mitochondrial DNA (mtDNA) bait capture of sequencing libraries, and sequencing, in separate facilities, following established protocols (see texts S1 to S6). We processed ancient DNA samples in clean room facilities designed to deal with ancient and highly degraded materials both at the University of Munich (LMU) and Trinity College Dublin. Sequencing took place at the Gene Center, LMU, and the TrinSeq facility, Trinity College Dublin. We aligned sequences and generated consensus FASTA files as described in texts S4 to S6. The final database of 629 mitogenomes was initially subject to five packages of statistical analyses.
Haplogroup identification and population genetics indexes
After alignment, we identified haplogroups A to E by using the published sequences in our dataset as a reference. These haplogroups displayed split times older than 30 ka cal BP. We called Z a new haplogroup constituting a deep monophyletic clade that was paraphyletic to haplogroups A and B. Another unreported mouflon-exclusive haplogroup was named G. This haplogroup is polyphyletic, though this was not problematic since it was not used for statistical inference. We noticed that haplogroup A contains two individuals that diverged so deep that it would even challenge their haplogroup calling. However, we kept their classification as A, as their impact on the statistical analysis would be negligible or null.
After assigning haplogroups to all samples, we grouped all samples into 12 groups based on temporal and geographic closeness and computed haplogroup frequencies for them. In addition, we subdivided the sample from Aşıklı Höyük into four groups corresponding to different archeological occupation layers. Last, we used these 12 + 4 sample groups to estimate the average number of pairwise differences, nucleotide diversity, and gene diversity as well as to compute the neutrality tests, and the corresponding indexes, of Tajima (D), Fu (Fs), Ewens-Watterson, and Chakraborty. We computed diversity and neutrality statistics and tests with the software Arlequin v3.5.2.2 (64) and MEGA X v10.0.5 (65).
Tests of temporal change in haplogroup frequencies
To discard gene drift and sampling error as the causes behind the observed differences in haplogroups, we applied temporal tests of allele frequencies, here applied to haplogroup frequencies. We applied these tests among all sample pairs that had different ages. The rationale is that a significant result with these tests would suggest the presence of evolutionary forces, other than gene drift under a constant population size. Those forces could be selection or gene flow but also intense demographic changes. We applied simultaneously four tests for every comparison: a conventional contingency-table chi-squared test, the test of Waples (66), and a Bayesian test that includes the estimation of a temporal version of the statistic FST (67). The Waples and the Bayesian tests are statistically more suited and accurate for testing temporal changes in genetic diversity but require knowledge of Ne values. To bypass this problem, we tested five values of Ne for each comparison (102, 103, 104, and 105). For these analyses, it was necessary to group temporally some samples that displayed different ages but were geographically and chronologically close (see Table 1 and table S6). Because of their large number, we presented the comparisons in two groups: only ancient Anatolian samples, and comparisons among Anatolian samples and other samples (see fig. S1).
Phylogenetic analyses
We reconstructed a temporally calibrated phylogenetic tree, taking advantage of our high coverage time-stamped mitogenomes, and inference of historical demography by means of an extended Bayesian skyline plot associated with the phylogenetic inference (68). We used all modern and ancient mitochondrial sequences reported here, including a urial specimen (O. vignei) as an outgroup. We partitioned the sequences in three codon positions (all protein-coding genes), tRNAs, rRNAs, and noncoding regions, and used the HKY85 + G as a substitution model, following the result of the model selection tool implemented in MEGA X v10.0.5 (65). For the phylogenetic inference, we used the Bayesian method with trees of the posterior set sampled by MCMC, as implemented in the software BEAST v2.6.6 (68). We used the software BEAUti (69) to create the input files. We carried out a pre-run of 100 million generations to obtain operator diagnostics for improving convergence, and two final parallel runs with 25% burn-in and an overall of one billion generations each. The full run allowed most parameters to reach the minimum effective sample size of 200. Mixing was assessed using the software Tracer (70).
To infer the extended Bayesian skyline plot (71, 72), we ran three separate runs: the first one using samples from modern and ancient Europe and Neolithic sites Aşıklı Höyük and Körtiktepe, the second one with the previously described set plus samples from Neolithic southwestern Anatolia (Suberde, Çukuriçi, Menteşe, and Marmara regions), and the third one using only samples of modern mouflons.
Demographic inference on coalescent times
In our phylogenetic reconstruction, we found a very high density of nodes between 12.0 ka and 7.6 ka cal BP, inside the clades corresponding to haplogroups A to E (excluding mouflons). To test whether this high density of coalescences is the result of a founder event or an artifact of having numerous ancient samples with broadly contemporaneous ages, we applied a Monte Carlo statistical procedure to the set of coalescent times and samples ages: the first ones obtained from the BEAST-estimated nodes’ ages and second ones from the radiocarbon- and stratigraphy-estimated samples ages. This procedure, developed in-house, takes advantage of the known mathematical relationship between demographic history and inter-coalescent times to sample the demographic history of Anatolian and European sheep by means of MCMC. The inference should produce an approximately constant demographic history if the increased density of coalescence events was solely due to sampling bias (see details in text S8).
Simulation-based inference by approximate Bayesian computation
The latest and most extensive package of analyses consisted in a series of simulation-based analyses performed by ABC (73). ABC analyses were performed in four phases, each with a specific inferential goal. They were the following:
1) To test and estimate the changes in Ne between the subsequent excavation layers of Aşıklı Höyük. These analyses included an analysis that used pseudo-observed datasets to estimate the statistical power that our methodology had to detect a bottleneck around Aşıklı Höyük.
2) To select the best model of ancestry relationships among the three sampled ancient Anatolian populations, namely, Neolithic Aşıklı Höyük, Neolithic southwestern Anatolia, and Chalcolithic Güvercinkayası.
3) To select the best ancestry model for the European population out of the ancient Anatolian populations.
4) To estimate with improved precision the population sizes in all involved populations, from Aşıklı Höyük to modern Europe and thus confirm or discard the presence of a domestication bottleneck.
All analyses included optimization and inferential steps, and the fit of the summary statistics was controlled by visual inspection on the predictive distribution of the summary statistics (figs. S15 and S16). All analyses were also replicated with different sets of summary statistics, as well as with a procedure based on random forests, a type of ABC inference assisted by machine learning (see text S9 and tables S7, S8, S10, and S12 for details on the simulation-based inference by ABC).
Ethics statement
All work involving living animals was conducted according to the national and international guidelines for animal welfare. Blood samples were collected with owner consent during routine examinations under good veterinary practice. Sampling was approved by the regional government of Upper Bavaria (55.2-1-54-2532.0-47-2016).
Acknowledgments
This study benefited from input by A. Galik, L. Gourichon, H. Hemmer, C. Malone, F. McCormick, T. O’Connor, J. Quade, A. Scheu, G. Tsartsidou, and A. Zeeb-Lanz. D.O. is thankful to J. Vuković for the permission to sample the Blagotin sheep specimens, while J.Bul. thanks A. Bulatović for the permission to sample the Bubanj sheep.
Funding: This work was supported by the Deutsche Forschungsgemeinschaft (DFG), project no. 165831460 (to J.P.); European Research Council under the European Union’s Horizon 2020 research and innovation program, grant agreement 885729-AncestralWeave and 295729-CodeX (to D.G.B.); Science Foundation Ireland grant number 21/PATH-S/9515 (to K.G.D.); Government of Ireland Postdoctoral Fellowship GOIPD/2020/605 (to V.E.M.); and U.S. NSF Division of Ocean Sciences Postdoctoral Fellowship NSF-OCE-PRF #2126500 (to J.T.A.).
Author contributions: Conceptualization: J.P., D.G.B., and I.Me. Data curation: E.S.-C., S.K., S.Ge., I.Me., and A.J.H. Formal analysis: E.S.-C., I.Me., A.J.H., E.A.D., and A.T.L. Funding acquisition: J.P., I.Me., and D.G.B. Investigation: I.W., S.Ge., K.G.D., V.E.M., A.J.H., V.M., G.L., N.A.Z., E.G., S.K., B.A., B.H., C.Ç., D.A., D.G., D.O., E.W.S., I.Ma., J.Bul., J.M., J.Bur., L.A., L.B., L.K.H., M.M., M.P.P., N.C., N.S., P.Z., S.Gü., S.S., V.Ö., G.D., H.B., J.P., J.T.A., M.C.S., M.Ö., M.U., N.D.M., N.P., and S.M.M. Methodology: S.K. Project administration: J.P., D.G.B., and I.Me. Resources: G.L., N.A.Z., E.G., B.A., B.H., C.Ç., D.A., D.G., D.O., E.W.S., I.Ma., J.Bul., J.M., J.Bur., L.A., L.B., L.K.H., M.M., M.P.P., N.C., N.S., P.Z., S.Gü., S.S., and V.Ö. Software: E.S.-C. Supervision: J.P., I.Me., and D.G.B. Validation: E.S.-C. and S.K. Visualization: E.S.-C., A.J.H., and I.Me. Writing—original draft: E.S.-C., I.Me., L.F., D.G.B., and J.P. Writing—review and editing: E.S.-C., I.Me., L.F., D.G.B., J.P., J.T.A., B.A., M.C.S., M.Ö., J.Bur., K.G.D., and V.E.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: For inquiries regarding the sheep samples newly reported in this study, see the contact information of the corresponding authors and the samples associated with them in table S14. The code for carrying out coalescent simulations and post-simulation analyses by ABC corresponds to an update of the software BaySICS. It is available at https://doi.org/10.5281/zenodo.10210930. The code for performing the MCMC procedure for inferring a demographic history out of coalescent times is available at https://doi.org/10.5281/zenodo.10392203. All sequencing data, including newly generated mitogenomes are available at https://doi.org/10.6084/m9.figshare.24552790. Their accession numbers and, in due case, the associated published articles are indicated in table S5. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Texts S1 to S10
Figs. S1 to S16
Tables S1 to S14
References
REFERENCES AND NOTES
- 1.H. Hongo, R. H. Meadow, Pig exploitation at Neolithic Çayönü Tepesi southeastern Anatolia, in Ancestors for the Pigs: Pigs in Prehistory, M. Nelson, Ed. (MASCA Research Papers in Science and Archaeology 15, Museum of Archaeology and Anthropology, University of Pennsylvania, Philadelphia 1998), pp. 77–89. [Google Scholar]
- 2.Peters J., von den Driesch A., Helmer D., Segui M. S., Early animal husbandry in the Northern Levant. Paléorient 25, 27–48 (1999). [Google Scholar]
- 3.J. Peters, A. von den Driesch, D. Helmer, in The Upper Euphrates-Tigris Basin: Cradle of agro-pastoralism?, in The First Steps of Animal Domestication, J.-D.Vigne, J. Peters, D. Helmer, Eds. (Oxbow Books, 2005), pp. 96–124. [Google Scholar]
- 4.Tanno K.-I., Willcox G., How fast was wild wheat domesticated? Science 311, 1886 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Asouti E., Fuller D. Q., A contextual approach to the emergence of agriculture in Southwest Asia: Reconstructing early neolithic plant-food production. Curr. Anthropol. 54, 299–345 (2013). [Google Scholar]
- 6.Kabukcu C., Asouti E., Pöllath N., Peters J., Karul N., Pathways to plant domestication in Southeast Anatolia based on new data from aceramic Neolithic Gusir Höyük. Sci. Rep. 11, 2112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baird D., Fairbairn A., Jenkins E., Martin L., Middleton C., Pearson J., Asouti E., Edwards Y., Kabukcu C., Mustafaoğlu G., Russell N., Bar-Yosef O., Jacobsen G., Wu X., Baker A., Elliott S., Agricultural origins on the Anatolian plateau. Proc. Natl. Acad. Sci. U.S.A 115, E3077–E3086 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stiner M. C., Munro N. D., Buitenhuis H., Duru G., Özbaşaran M., An endemic pathway to sheep and goat domestication at Aşıklı Höyük (Central Anatolia, Turkey). Proc. Natl. Acad. Sci. U.S.A 119, e2110930119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riehl S., Zeidi M., Conard N. J., Emergence of agriculture in the foothills of the Zagros mountains of Iran. Science 341, 65–67 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Conolly J., Colledge S., Dobney K., Vigne J.-D., Peters J., Stopp B., Manning K., Shennan S., Meta-analysis of zooarchaeological data from SW Asia and SE Europe provides insight into the origins and spread of animal husbandry. J. Archaeol. Sci. 38, 538–545 (2011). [Google Scholar]
- 11.Arbuckle B. S., Kansa S. W., Kansa E., Orton D., Çakırlar C., Gourichon L., Atici L., Galik A., Marciniak A., Mulville J., Buitenhuis H., Carruthers D., De Cupere B., Demirergi A., Frame S., Helmer D., Martin L., Peters J., Pöllath N., Pawłowska K., Russell N., Twiss K., Würtenberger D., Data sharing reveals complexity in the westward spread of domestic animals across Neolithic Turkey. PLOS ONE 9, e99845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vigne J.-D., Carrère I., Briois F., Guilaine J., The early process of mammal domestication in the Near East: New evidence from the Pre-Neolithic and Pre-Pottery Neolithic in Cyprus. Curr. Anthropol. 52, S255–S271 (2011). [Google Scholar]
- 13.J.-D. Vigne, L. Gourichon, D. Helmer, L. Martin, J. Peters, in Quaternary of the Levant. Environments, Climate Change, and Humans, E. Yehouda, B.-Y. Ofer, Eds. (Cambridge Univ. Press, 2017), pp. 753–760. [Google Scholar]
- 14.Gourichon L., Horwitz L. K., An inter-regional comparison of animal domestication in the northern and southern Levant. Food Hist. 19, 53–78 (2021). [Google Scholar]
- 15.Fuller D. Q., Van Etten J., Manning K., Castillo C., Kingwell-Banham E., Weisskopf A., Qin L., Sato Y.-I., Hijmans R. J., The contribution of rice agriculture and livestock pastoralism to prehistoric methane levels:An archaeological assessment. Holocene 21, 743–759 (2011). [Google Scholar]
- 16.H.-P. Uerpmann, Probleme der Neolithisierung des Mittelmeeraums (Beihefte zum Tübinger Atlas des Vordern Orients; Ludwing Reichert, 1979). [Google Scholar]
- 17.Ervynck A., Dobney K., Hongo H., Meadow R., Born Free? New evidence for the status of “Sus scrofa” at Neolithic Çayönü Tepesi (Southeastern Anatolia, Turkey). Paléorient 27, 47–73 (2001). [Google Scholar]
- 18.J. Peters, F. Neuberger, I. Wiechmann, M. Zimmermann, M. Balasse, N. Pöllath, Shaping the sheep: Human management and decision-making at Aşıklı Höyük, Central Anatolia, in The Early Settlement at Aşıklı Höyük. Essays in Honor of Ufuk Esin, M. Özbaşaran, G. Duru, M. Stiner, Eds. (Ege Yayınları, 2018), pp. 325–344. [Google Scholar]
- 19.J. Peters, B. S. Arbuckle, N. Pöllath, Subsistence and beyond: Animals in Neolithic Anatolia, in The Neolithic in Turkey. 10500–5200 BC: Environment, Settlement, Flora, Fauna, Dating, Symbols of belief, with Views from North, South, East, and West, M. Özdoğan, N. Başgelen, P. Kuniholm, Eds. (Archaeology & Art Publications, 2014), pp. 135–203. [Google Scholar]
- 20.D. Helmer, Révision de la faune de Cafer Höyük (Malatya, Turquie). Apports des méthodes de l’analyse de mélanges et the l’analyse de Kernel à la mise en évidence de la domestication, in Archaeozoology of the Near East VIII, E. Vila, L. Gourichon, A. M. Choyke, H. Buitenhuis, Eds. (Travaux de la Maison de l’Orient et de la Méditerranée 49, 2008), pp. 169–196. [Google Scholar]
- 21.Özbaşaran M., Re-starting at Aşıklı. Anatolia antiqua. Eski Anadolu 19, 27–37 (2011). [Google Scholar]
- 22.M. Özbaşaran, Asikli Höyük, in The Neolithic in Turkey. New Excavations and New Research: Central Turkey, M. Özdoğan, N. Başgelen, P. Kuniholm, Eds. (Archaeology & Art Publications, 2012), pp. 135–158. [Google Scholar]
- 23.Stiner M. C., Buitenhuis H., Duru G., Kuhn S. L., Mentzer S. M., Munro N. D., Pöllath N., Quade J., Tsartsidou G., Özbaşaran M., A forager–herder trade-off, from broad-spectrum hunting to sheep management at Aşıklı Höyük, Turkey. Proc. Natl. Acad. Sci. U.S.A 111, 8404–8409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.H. Buitenhuis, J. Peters, N. Pollath, M. C. Stiner, N. D. Munro, O. Sarıtaş, The faunal remains from levels 3 and 2 of Aşıklı Höyük: Evidence for emerging management practices, in The Early Settlement at Aşıklı Höyük. Essays in Honor of Ufuk Esin, M. Özbaşaran, G. Duru, M. C. Stiner, Eds. (Ege Yayınları, 2018), pp. 281–323. [Google Scholar]
- 25.Zimmermann M. I., Pöllath N., Özbaşaran M., Peters J., Joint health in free-ranging and confined small bovids—Implications for early stage caprine management. J. Archaeol. Sci. 92, 13–27 (2018). [Google Scholar]
- 26.S. M. Mentzer, Micromorphological analyses of anthropogenic materials and insight into tell formation processes at Aşıklı Höyük, in The Early Settlement at Aşıklı Höyük. Essays in Honor of Ufuk Esin, M. Özbaşaran, G. Duru, M. Stiner, Eds. (Ege Yayınları, 2018), pp. 105–128. [Google Scholar]
- 27.Abell J. T., Quade J., Duru G., Mentzer S. M., Stiner M. C., Uzdurum M., Özbaşaran M., Urine salts elucidate Early Neolithic animal management at Aşıklı Höyük, Turkey. Sci. Adv. 5, eaaw0038 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.F. Neuberger, M. Balasse, N. Pöllath, J. Peters, Diet of wild versus culturally controlled caprines in Early Neolithic Anatolia based on stable carbon isotope analysis of bone apatite, in Animals: Cultural Identifiers in Ancient Societies, J. Peters, G. McGlynn, V. Goebel, Eds. (Documenta Archaeobiologiae 15, M. Leidorf, 2019), pp. 251–260. [Google Scholar]
- 29.G. Tsartsidou, The microscopic record of Aşıklı Höyük: Phytolith analysis of material from the 2012–2016 field seasons, in The Early Settlement at Aşıklı Höyük. Essays in Honor of Ufuk Esin, M. Özbaşaran, G. Duru, M. Stiner, Eds. (Ege Yayınları, 2018), pp. 147–189. [Google Scholar]
- 30.Lv F.-H., Peng W.-F., Yang J., Zhao Y.-X., Li W.-R., Liu M.-J., Ma Y.-H., Zhao Q.-J., Yang G.-L., Wang F., Li J.-Q., Liu Y.-G., Shen Z.-Q., Zhao S.-G., Hehua E., Gorkhali N. A., Vahidi S. M. F., Muladno M., Naqvi A. N., Tabell J., Iso-Touru T., Bruford M. W., Kantanen J., Han J.-L., Li M.-H., Mitogenomic meta-analysis identifies two phases of migration in the history of eastern eurasian sheep. Mol. Biol. Evol. 32, 2515–2533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meadows J. R. S., Hiendleder S., Kijas J. W., Haplogroup relationships between domestic and wild sheep resolved using a mitogenome panel. Heredity 106, 700–706 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brahi O. H. D., Xiang H., Chen X., Farougou S., Zhao X., Mitogenome revealed multiple postdomestication genetic mixtures of West African sheep. J. An. Breed. Genet. 132, 399–405 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Hiendleder S., Mainz K., Plante Y., Lewalski H., Analysis of mitochondrial DNA indicates that domestic sheep are derived from two different ancestral maternal sources: No evidence for contributions from urial and argali sheep. J. Heredity 89, 113–120 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Burgstaller J. P., Schinogl P., Dinnyes A., Müller M., Steinborn R., Mitochondrial DNA heteroplasmy in ovine fetuses and sheep cloned by somatic cell nuclear transfer. BMC Dev. Biol. 7, 141 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lancioni H., Phylogenetic relationships of three italian merino-derived sheep breeds evaluated through a complete mitogenome analysis. PLOS ONE 8, e73712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X., Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat. Commun. 11, 2815 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mustafa S. I., Schwarzacher T., Heslop-Harrison J. S., Complete mitogenomes from Kurdistani sheep: Abundant centromeric nuclear copies representing diverse ancestors. Mitochondrial DNA Part A 29, 1180–1193 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Niu Y., Jin M., Li Y., Li P., Zhou J., Wang X., Petersen B., Huang X., Kou Q., Chen Y., Biallelic β-carotene oxygenase 2 knockout results in yellow fat in sheep via CRISPR/Cas9. An. Genet. 48, 242–244 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Reicher S., Seroussi E., Weller J. I., Rosov A., Gootwine E., Ovine mitochondrial DNA sequence variation and its association with production and reproduction traits within an Afec-Assaf flock1. J. An. Sci. 90, 2084–2091 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Yang J., Li W.-R., Lv F.-H., He S.-G., Tian S.-L., Peng W.-F., Sun Y.-W., Zhao Y.-X., Tu X.-L., Zhang M., Xie X.-L., Wang Y.-T., Li J.-Q., Liu Y.-G., Shen Z.-Q., Wang F., Liu G.-J., Lu H.-F., Kantanen J., Han J.-L., Li M.-H., Liu M.-J., Whole-genome sequencing of native sheep provides insights into rapid adaptations to extreme environments. Molec. Biol. Evol. 33, 2576–2592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Her C., Rezaei H.-R., Hughes S., Naderi S., Duffraisse M., Mashkour M., Naghash H.-R., Bălășescu A., Luikart G., Jordan S., Özüt D., Kence A., Bruford M. W., Tresset A., Vigne J.-D., Taberlet P., Hänni C., Pompanon F., Broad maternal geographic origin of domestic sheep in Anatolia and the Zagros. An. Genet. 53, 452–459 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Yurtman E., Özer O., Yüncü E., Dağtaş N. D., Koptekin D., Çakan Y. G., Özkan M., Akbaba A., Kaptan D., Atağ G., Vural K. B., Gündem C. Y., Martin L., Kılınç G. M., Ghalichi A., Açan S. C., Yaka R., Sağlıcan E., Lagerholm V. K., Krzewińska M., Günther T., Miranda P. M., Pişkin E., Şevketoğlu M., Bilgin C. C., Atakuman Ç., Erdal Y. S., Sürer E., Altınışık N. E., Lenstra J. A., Yorulmaz S., Abazari M. F., Hoseinzadeh J., Baird D., Bıçakçı E., Çevik Ö., Gerritsen F., Özbal R., Götherström A., Somel M., Togan İ., Özer F., Archaeogenetic analysis of Neolithic sheep from Anatolia suggests a complex demographic history since domestication. Commun. Biol. 4, 1279 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor W. T. T., Pruvost M., Posth C., Rendu W., Krajcarz M. T., Abdykanova A., Brancaleoni G., Spengler R., Hermes T., Schiavinato S., Hodgins G., Stahl R., Min J., Kyzy S. A., Fedorowicz S., Orlando L., Douka K., Krivoshapkin A., Jeong C., Warinner C., Shnaider S., Evidence for early dispersal of domestic sheep into Central Asia. Nat. Hum. Behav. 5, 1169–1179 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Meadows J. R. S., Cemal I., Karaca O., Gootwine E., Kijas J. W., Five ovine mitochondrial lineages identified from sheep breeds of the Near East. Genetics 175, 1371–1379 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamalakkannan R., Kumar S., Bhavana K., Prabhu V. R., Machado C. B., Singha H. S., Sureshgopi D., Vijay V., Nagarajan M., Evidence for independent domestication of sheep mtDNA lineage A in India and introduction of lineage B through Arabian sea route. Sci. Rep. 11, 19733 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedrosa S., Uzun M., Arranz J.-J., Gutiérrez-Gil B., Primitivo F. S., Bayón Y., Evidence of three maternal lineages in Near Eastern sheep supporting multiple domestication events. Proc. Biol. Sci. 272, 2211–2217 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh S., Kumar S. Jr., Kolte A. P., Kumar S., Extensive variation and sub-structuring in lineage A mtDNA in Indian sheep: Genetic evidence for domestication of sheep in India. PLOS ONE 8, e77858 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerbault P., Allaby R. G., Boivin N., Rudzinski A., Grimaldi I. M., Pires J. C., Vigueira C. C., Dobney K., Gremillion K. J., Barton L., Arroyo-Kalin M., Purugganan M. D., de Casas R. R., Bollongino R., Burger J., Fuller D. Q., Bradley D. G., Balding D. J., Richerson P. J., Gilbert M. T. P., Larson G., Thomas M. G., Storytelling and story testing in domestication. Proc. Natl Acad. Sci. U S A 111, 6159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colli L., Lancioni H., Cardinali I., Olivieri A., Capodiferro M. R., Pellecchia M., Rzepus M., Zamani W., Naderi S., Gandini F., Vahidi S. M. F., Agha S., Randi E., Battaglia V., Sardina M. T., Portolano B., Rezaei H. R., Lymberakis P., Boyer F., Coissac E., Pompanon F., Taberlet P., Marsan P. A., Achilli A., Whole mitochondrial genomes unveil the impact of domestication on goat matrilineal variability. BMC Genomics 16, 1115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cubric-Curik V., Novosel D., Brajkovic V., Stabelli O. R., Krebs S., Sölkner J., Šalamon D., Ristov S., Berger B., Trivizaki S., Bizelis I., Ferenčaković M., Rothammer S., Kunz E., Simčič M., Dovč P., Bunevski G., Bytyqi H., Marković B., Brka M., Kume K., Stojanović S., Nikolov V., Zinovieva N., Schönherz A. A., Guldbrandtsen B., Čačić M., Radović S., Miracle P., Vernesi C., Curik I., Medugorac I., Large-scale mitogenome sequencing reveals consecutive expansions of domestic taurine cattle and supports sporadic aurochs introgression. Evol. Appl. 15, 663–678 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daly K. G., Mattiangeli V., Hare A. J., Davoudi H., Fathi H., Doost S. B., Amiri S., Khazaeli R., Decruyenaere D., Nokandeh J., Richter T., Darabi H., Mortensen P., Pantos A., Yeomans L., Bangsgaard P., Mashkour M., Zeder M. A., Bradley D. G., Herded and hunted goat genomes from the dawn of domestication in the Zagros Mountains. Proc. Natl Acad. Sci. U S A 118, e2100901118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bollongino R., Burger J., Powell A., Mashkour M., Vigne J.-D., Thomas M. G., Modern taurine cattle descended from small number of Near-Eastern founders. Molec. Biol. Evol. 29, 2101–2104 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Emra S., Benz M., Siddiq A. B., Özkaya V., Adaptions in subsistence strategy to environment changes across the Younger Dryas - Early Holocene boundary at Körtiktepe Southeastern Turkey. Holocene 32, 390–413 (2022). [Google Scholar]
- 54.Pöllath N., R. García-González, S. Kevork, U. Mutze, M. I. Zimmermann, M. Özbaşaran, J. Peters, A non-linear prediction model for ageing foetal and neonatal sheep reveals basic issues in early neolithic husbandry. J. Archaeol. Sci. 130, 105344 (2021). [Google Scholar]
- 55.M. Ergun, M. Tengberg, G. Willcox, C. Douché, Plants of Aşıklı Höyük and changes through time: First archaeobotanical results from the 2010–14 excavation seasons, in The Early Settlement at Aşıklı Höyük. Essays in Honor of Ufuk Esin, M. Ösbaşaran, G. Duru, M. Stiner Eds. (Ege Yayınları, 2018), pp. 191–217. [Google Scholar]
- 56.Ulaş B., Fiorentino G., Recent attestations of “new” glume wheat in Turkey: a reassessment of its role in the reconstruction of Neolithic agriculture. Veget. Hist. Archaeob. 30, 685–701 (2021). [Google Scholar]
- 57.Marchi N., Winkelbach L., Schulz I., Brami M., Hofmanová Z., Blöcher J., Reyna-Blanco C. S., Diekmann Y., Thiéry A., Kapopoulou A., Link V., Piuz V., Kreutzer S., Figarska S. M., Ganiatsou E., Pukaj A., Struck T. J., Gutenkunst R. N., Karul N., Gerritsen F., Pechtl J., Peters J., Zeeb-Lanz A., Lenneis E., Teschler-Nicola M., Triantaphyllou S., Stefanović S., Papageorgopoulou C., Wegmann D., Burger J., Excoffier L., The genomic origins of the world’s first farmers. Cell 185, 1842–1859.e1818 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horejs B., Milić B., Ostmann F., Thanheiser U., Weninger B., Galik A., The Aegean in the early seventh Millennium BC: Maritime networks and colonization. J. World Prehist. 28, 289–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Özdoğan E., Current research and new evidence for the neolithization process in Western Turkey. Euro. J. Archaeol. 18, 33–59 (2015). [Google Scholar]
- 60.M. Özdoğan, in The Central/Western Anatolian Farming Frontier, M. Brami, B. Horejs, Eds. (Austrian Academy of Sciences, 2019), pp. 143–158. [Google Scholar]
- 61.Yaka R., Mapelli I., Kaptan D., Doğu A., Chyleński M., Erdal Ö. D., Koptekin D., Vural K. B., Bayliss A., Mazzucato C., Fer E., Çokoğlu S. S., Lagerholm V. K., Krzewińska M., Karamurat C., Gemici H. C., Sevkar A., Dağtaş N. D., Kılınç G. M., Adams D., Munters A. R., Sağlıcan E., Milella M., Schotsmans E. M. J., Yurtman E., Çetin M., Yorulmaz S., Altınışık N. E., Ghalichi A., Juras A., Bilgin C. C., Günther T., Storå J., Jakobsson M., de Kleijn M., Mustafaoğlu G., Fairbairn A., Pearson J., Togan İ., Kayacan N., Marciniak A., Larsen C. S., Hodder I., Atakuman Ç., Pilloud M., Sürer E., Gerritsen F., Özbal R., Baird D., Erdal Y. S., Duru G., Özbaşaran M., Haddow S. D., Knüsel C. J., Götherström A., Özer F., Somel M., Variable kinship patterns in Neolithic Anatolia revealed by ancient genomes. Curr. Biol. 31, 2455–2468 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.H. Greenfield, A reconsideration of the secondary products revolution in south-eastern Europe: On the origins and use of domestic animals for milk, wool, and traction in the central Balkans, in The Zooarchaeology of Fats, Oils, Milk and Dairying (Oxbow Books, 2005), pp. 14–31. [Google Scholar]
- 63.Becker C., The textile revolution. research into the origin and spread of wool production between the Near East and Central Europe. J. Ancient Stud. 6, 102–151 (2016). [Google Scholar]
- 64.Excoffier L., Lischer H. E. L., Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10, 564–567 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waples R. S., Temporal variation in allele frequencies: Testing the right hypothesis. Evolution 43, 1236–1251 (1989). [DOI] [PubMed] [Google Scholar]
- 67.Sandoval-Castellanos E., Testing temporal changes in allele frequencies: a simulation approach. Genet Res (Camb) 92, 309–320 (2010). [DOI] [PubMed] [Google Scholar]
- 68.Bouckaert R., Heled J., Kühnert D., Vaughan T., Wu C.-H., Xie D., Suchard M. A., Rambaut A., Drummond A. J., BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drummond A. J., Suchard M. A., Xie D., Rambaut A., Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molec. Biol. Evol. 29, 1969–1973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A., Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ho S. Y. W., Shapiro B., Skyline-plot methods for estimating demographic history from nucleotide sequences. Molec. Ecol. Resour. 11, 423–434 (2011). [DOI] [PubMed] [Google Scholar]
- 72.Heled J., Drummond A. J., Bayesian inference of population size history from multiple loci. BMC Evol. Biol. 8, 289 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beaumont M. A., Approximate Bayesian Computation in Evolution and Ecology. Ann. Rev. Ecol. Evol. Syst. 41, 379–406 (2010). [Google Scholar]
- 74.Rohland N., Reich D., Mallick S., Meyer M., Green R. E., Georgiadis N. J., Roca A. L., Hofreiter M., Genomic DNA Sequences from Mastodon and Woolly Mammoth Reveal Deep Speciation of Forest and Savanna Elephants. PLoS Biol. 8, e1000564 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rohland N., Hofreiter M., Comparison and optimization of ancient DNA extraction. Biotechniques 42, 343–352 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Dabney J., Knapp M., Glocke I., Gansauge M.-T., Weihmann A., Nickel B., Valdiosera C., García N., Pääbo S., Arsuaga J.-L., Meyer M., Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl Acad. Sci. U S A 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Korlević P., Gerber T., Gansauge M.-T., Hajdinjak M., Nagel S., Aximu-Petri A., Meyer M., Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. Biotechniques 59, 87–93 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Daly K. G., Delser P. M., Mullin V. E., Scheu A., Mattiangeli V., Teasdale M. D., Hare A. J., Burger J., Verdugo M. P., Collins M. J., Kehati R., Erek C. M., Bar-Oz G., Pompanon F., Cumer T., Çakırlar C., Mohaseb A. F., Decruyenaere D., Davoudi H., Çevik Ö., Rollefson G., Vigne J.-D., Khazaeli R., Fathi H., Doost S. B., Sorkhani R. R., Vahdati A. A., Sauer E. W., Kharanaghi H. A., Maziar S., Gasparian B., Pinhasi R., Martin L., Orton D., Arbuckle B. S., Benecke N., Manica A., Horwitz L. K., Mashkour M., Bradley D. G., Ancient goat genomes reveal mosaic domestication in the Fertile Crescent. Science 361, 85–88 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Meyer M., Kircher M., Gansauge M.-T., Li H., Racimo F., Mallick S., Schraiber J. G., Jay F., Prüfer K., de Filippo C., Sudmant P. H., Alkan C., Fu Q., Do R., Rohland N., Tandon A., Siebauer M., Green R. E., Bryc K., Briggs A. W., Stenzel U., Dabney J., Shendure J., Kitzman J., Hammer M. F., Shunkov M. V., Derevianko A. P., Patterson N., Andrés A. M., Eichler E. E., Slatkin M., Reich D., Kelso J., Pääbo S., A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gamba C., Jones E. R., Teasdale M. D., McLaughlin R. L., Gonzalez-Fortes G., Mattiangeli V., Domboróczki L., Kővári I., Pap I., Anders A., Whittle A., Dani J., Raczky P., Higham T. F. G., Hofreiter M., Bradley D. G., Pinhasi R., Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 5, 5257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gansauge M.-T., Meyer M., Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat. Protocols 8, 737–748 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Li H., Durbin R., Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koboldt D. C., Zhang Q., Larson D. E., Shen D., McLellan M. D., Lin L., Miller C. A., Mardis E. R., Ding L., Wilson R. K., VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.J. Hein, M. Schierup, C. Wiuf, Gene Genealogies, Variation and Evolution. A Primer in Coalescent Theory (Oxford Univ. Press, 2004), vol. 54. [Google Scholar]
- 85.Bertorelle G., Benazzo A., Mona S., ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Mol. Ecol. 19, 2609–2625 (2010). [DOI] [PubMed] [Google Scholar]
- 86.Robert C. P., Cornuet J. M., Marin J. M., Pillai N. S., Lack of confidence in approximate Bayesian computation model choice. Proc. Natl Acad. Sci. U S A 108, 15112–15117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pudlo P., Marin J.-M., Estoup A., Cornuet J.-M., Gautier M., Robert C. P., Reliable ABC model choice via random forests. Bioinformatics 32, 859–866 (2016). [DOI] [PubMed] [Google Scholar]
- 88.Sandoval-Castellanos E., Palkopoulou E., Dalen L., Back to BaySICS: A user-friendly program for Bayesian Statistical Inference from Coalescent Simulations. PLoS One 9, e98011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krauß R., Beginnings of the Neolithic in Southeast Europe: the Early Neolithic sequence and absolute dates from Džuljunica-Smărdeš (Bulgaria). Docum. Praehist. 41, 51–77 (2014). [Google Scholar]
- 90.De Groene D., Pigs and humans in Early Neolithic south-eastern Europe. Docum. Praehist. 45, 38–51 (2018). [Google Scholar]
- 91.M. Mashkour, R. Khazaeli, H. Fathi, S. Amiri, D. Decruyenaere, A. Mohaseb, H. Davoudi, S. Sheikhi, E. W. Sauer, in Sasanian Persia: Between Rome and the Steppes of Eurasia, E. Sauer, Ed. (Edinburgh Univ. Press, 2017), pp. 74-96. [Google Scholar]
- 92.Daly K., Arbuckle B. S., Rossi C., Mattiangeli V., Lawlor P. A., Mashkour M., Sauer E., Lesur J., Atici L., Erek C. M., Bradley D. G., A novel lineage of the Capra genus discovered in the Taurus Mountains of Turkey using ancient genomics. eLife 11, e82984 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.J. Oldenstein, KASTELL ALZEY Archäologische Untersuchungen im Spätrömischen Lager und Studien zur Grenzverteidigung im Mainzer Dukat (University of Mainz, 2009), vol. 1. [Google Scholar]
- 94.Boulestin B., Mass cannibalism in the linear pottery culture at Herxheim (Palatinate, Germany). Antiquity 83, 968–982 (2009). [Google Scholar]
- 95.N. Card, M. Edmonds, A. Mitchell, The Ness of Brodgar, as it Stands, M. Edmonds, A. Mitchell, Eds. (The Orcadian, 2020). [Google Scholar]
- 96.Card N., To cut a long story short: Formal chronological modelling for the Late Neolithic site of Ness of Brodgar Orkney. Europ. J. Archaeol. 21, 217–263 (2018). [Google Scholar]
- 97.D. Griffiths, J. Harrison, M. Athanson, Beside the Ocean: Coastal Landscapes at the Bay of Skaill, Marwick, and Birsay Bay, Orkney: Archaeological Research 2003-18 (Oxbow Books, 2019), p. 346. [Google Scholar]
- 98.N. M. Sharples, in From Machair to Mountains: Archaeological Survey and Excavation in South Uist, M. Parker Pearson, Ed. (Oxbow Books, 2012), vol. 4, pp. 215–258. [Google Scholar]
- 99.M. P. Pearson, J. Mulville, H. Smith, P. Marshall, Cladh Hallan: Roundhouses and the dead in the Hebridean Bronze Age and Iron Age, Part I: Stratigraphy, spatial organisation and chronology, in Sheffield Environmental and Archaeological Research Campaign in the Hebrides (Oxbow Books, 2021), vol. 8, p. 568. [Google Scholar]
- 100.N. Sharples, A Late Iron Age Farmstead in the Outer Hebrides. Excavations at Mound 1, Bornais, South Uist (Oxbow Books, 2012), p. 419. [Google Scholar]
- 101.M. P. Pearson, M. Brennand, J. Mulville, H. Smith, Cille Pheadair: A norse farmstead and pictish burial cairn in South Uist, in Sheffield Environmental and Archaeological Research Campaign in the Hebrides (Oxbow Books, 2018), vol. 7, p. 656. [Google Scholar]
- 102.A. J. Lawson, M. J. Allen, Potterne 1982-5: Animal Husbandry in Later Prehistoric Wiltshire (Trust for Wessex Archaeology, 2000). [Google Scholar]
- 103.B. W. Cunliffe, Danebury: Anatomy of an Iron Age Hillfort (Batsford, 1983). [Google Scholar]
- 104.P. Wallace, The archaeology of Viking Dublin, in The comparative history of urban origins in non-Roman Europe, H. B. Clarke, A. Simms, Eds. (Oxford 1985), BAR International Series, 255(1), pp. 103–145. [Google Scholar]
- 105.Stoddart S., Bonanno A., Gouder T., Malone C., Trump D., Cult in an island society: Prehistoric Malta in the Tarxien period. Cambridge Archaeological J. 3, 3–19 (1993). [Google Scholar]
- 106.H. Greenfield, T. Jongsma, in Space and Spatial Analysis in Archaeology, E. C. Robertson, J. D. Siebert, D. C. Fernandez, M. U. Zender, Eds. (University of Calgary Press, 2006), chap. 8, pp. 69–79. [Google Scholar]
- 107.Whittle A., Bartosiewicz L., Borić D., Pettitt P., Richards M. P., In the beginning : new radiocarbon dates for the Early Neolithic in northern Serbia and south-east Hungary. Antaeus. 25, 63–118 (2002). [Google Scholar]
- 108.Porčić M., Blagojević T., Pendić J., Stefanović S., The Neolithic demographic transition in the Central Balkans: Population dynamics reconstruction based on new radiocarbon evidence. Philosop. Trans. R. Soc. B: Biol. Sci. 376, 20190712 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.A. Bulatović, D. Milanović, Bubanj The Eneolithic and the Early Bronze Age Tell in Southeastern Serbia (Verlag der Österreichischen Akademie der Wissenschaften, 2020), vol. 90, p. 418. [Google Scholar]
- 110.Dothan T., Zukerman A., A preliminary study of the Mycenaean IIIC:1 pottery assemblages from Tel Miqne-Ekron and Ashdod. Bullet. Am. Schools Oriental Res. 333, 1 (2004). [Google Scholar]
- 111.L. K. Horwitz, E. Dahan. Animal husbandry practices during the historic periods, in Yoqne’am I. The Late Periods, A. Ben-Tor, M. Avissar, Y. Portugali, Eds. (Qedem Reports 3, 1996), pp. 246–255. [Google Scholar]
- 112.T. E. Levy, D. Alon, Shiqmim I: Studies Concerning Chalcolithic Societies in the Northern Negev Desert, Israel (1982-1984) (B.A.R., 1987). [Google Scholar]
- 113.B. Horejs, C. Schwall, Interaction as a stimulus?: Çukuriçi Höyük and the transition from the Late Chalcolithic to the Early Bronze Age in Western Anatolia, in Communities in Transition: The Circum-Aegean Area During the 5th and 4th Millennia BC, S. Dietz, F. Mavridis, Ž. Tankosić, T. Takaoğlu, Eds. (Oxbow Books, 2018), pp. 530–537. [Google Scholar]
- 114.Arbuckle B. S., Özkaya V., Animal exploitation at Körtik Tepe : An early Aceramic Neolithic site in southeastern Turkey. Paléorient 32, 113–136 (2006). [Google Scholar]
- 115.V. Özkaya, A. Coşkun, Körtik Tepe, in The Neolithic in Turkey. New Excavations and New Research. The Tigris Basin, M. Özdoğan, N. Başgelen, I. Kuniholm, Eds. (Archaeology and Art Publications, 2011), pp. 89–127. [Google Scholar]
- 116.Benz M., Coşkun A., Rössner C., Deckers K., Riehl S., Alt K. W., Özkaya V., First evidence of an Epipalaeolithic hunter-fisher-gatherer settlement at Körtik Tepe. Arkeometri Sonuçları Toplantısı 28, 65–78 (2013). [Google Scholar]
- 117.Arbuckle B. S., Revisiting Neolithic caprine exploitation at Suberde, Turkey. J. Field Archaeol. 33, 219–236 (2008). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Texts S1 to S10
Figs. S1 to S16
Tables S1 to S14
References