Abstract
Background
Little is known about prognostic factors of brain metastases (BM) from colorectal cancer (CRC). HER2 amplification/overexpression (HER2+) was previously described; its impact on prognosis remains uncertain.
Methods
In the translational study HEROES, extensive molecular analysis was performed on primary CRC (prCRC) and their matched resected BM by means of NGS comprehensive genomic profiling and HER2 status as assessed by immunohistochemical/ in situ hybridization. Count of tumour-infiltrating lymphocytes (TILs) was also performed. Primary objective: to describe the molecular landscape of paired BM/prCRC. Secondary objectives: to search for new prognostic biomarkers of outcome after BM resection: intracranial-only Progression-Free Survival (BM-iPFS), Progression-Free Survival (BM-PFS), and Overall Survival (BM-OS).
Results
Out of 22 patients having paired samples of prCRC and BM, HER2+ was found on 4 (18%) BM, 3 (75%) of which also HER2+ in matched prCRC. Lower tumour mutation burden (HR 3.08; 95%CI 1.06–8.93; p = 0.0386) and HER2-negative BM (HER2neg) (HR 7.75;95%CI 1.97–30.40; p = 0.0033) were associated with longer BM-iPFS; HER2neg BM (HR 3.44; 95%CI 1.03–11.53; p = 0.0449) and KRASmut BM (HR 0.31; 95%CI 0.12–0.80; p = 0.0153) conferred longer BM-PFS. Longer BM-OS was found in pts with TILs-enriched (≥1.6/HPF) BM (HR 0.11; 95%CI0.01–0.91; p = 0.0403).
Conclusions
This study shows HER2+ enrichment in both BM and their prCRC. TILs-enriched BM conferred better BM-OS.
Subject terms: Prognostic markers, Tumour biomarkers
Introduction
Brain metastases (BM) represent a rare event in colorectal cancer (CRC), but carry an extremely poor prognosis. Their incidence is estimated to be around 1 to 3% [1], with a trend to increase as a consequence of the prolonged CRC overall survival. The rarity of BM from CRC, together with the technical difficulties to access BM tissue, limits the feasibility of prospective studies and molecular characterization, resulting in scarce information about their development and prevention.
Clinical factors with prognostic value in patients with BM from CRC have been previously studied: a dedicated nomogram created by Pietrantonio et al., shows how age, Karnofsky performance status, site and number of BM can impact survival [2]. On the other hand, the advances in molecular diagnostics improved insight into molecular pathways and alterations underlying BM development. In analogy with breast cancer and other malignancies, enrichment in ERRB2 amplification/HER2 overexpression (HER2 + ) has been recently described in a cohort of patients with BM from gastrointestinal tumours [3]. HER2+ is rare in CRC, being detected in overall 1-2% of patients with CRC and no more than 5% of patients with RAS/BRAF wild-type disease [4–6]. In HER2-positive breast cancer, understanding tumour biology has been crucial to identify those patients at higher risk of BM development and therefore worthy of special surveillance, such as central nervous system periodic scan, for early diagnosis and treatment.
Another factor emerging from literature is the increased tendency to BM in CRC with BRAFV600E mutation [7]: the aggressiveness of this CRC subtype is well known, but conclusive data about BM can hardly be drawn due to the rarity of both BRAF-mutated metastatic CRC (mCRC) (8–12% of the total mCRC) [8] and BM.
Recently, deficiency in homologous recombination (HRD) and mismatch repair (MMRD) were also described in tissue from BM compared to matched primary CRC tumours in a cohort of 19 patients [9]: this finding opens interesting scenarios in the genomic profiling of BM from CRC.
Moving from such a background, we designed the present study in order to describe genomic landscape and clinical characteristics of patients with BM from CRC, with a special focus on HER2 expression.
Patients and methods
HEROES was a retrospective-prospective, translational study in which patients with resected BM from CRC and treated at our institution were enrolled to perform extensive molecular analysis of matched tissue from primary tumour and BM.
Criteria of inclusion were: i) histologically proven diagnosis of CRC, ii) diagnosis of BM, iii) availability of matched primary CRC tumour and BM tissue specimens. All the patients signed written informed consent; the protocol received approval from local Ethic Committee of Veneto Institute of Oncology IOV – IRCCS, and was conducted in accordance with the Declaration of Helsinki.
A dedicated database was created, in which both clinical and molecular characteristics were collected. Molecular characterization and tumour mutation burden (TMB) quantification were obtained by means of Next Generation Sequencing (NGS, FoundationOne CDx®). Furthermore, RAS/BRAF and Microsatellite Instability (MSI) were respectively assessed with MassArray (Myriapod Colon Status® kit) and immunohistochemistry (IHC) to validate NGS results. HER2 was assessed with IHC/in situ hybridization (ISH) using criteria previously reported in literature [10], and HER2 3+ overexpression or HER2 2+ overexpression with ISH-confirmed amplification were considered as HER2 HER2+. Tumour-infiltrating lymphocytes (TILs) were counted on hematoxylin-eosin-stained tissue sections; no immunohistochemical characterization was provided, so a quantitative-only assessment was conducted.
All the analyses were performed on matched primary tumour and BM tissue for each patient.
Primary objective of the study was to report the molecular landscape of paired tissues from primitive tumour and brain metastases, with special focus on HER2.
Secondary objectives were to search for new prognostic biomarkers in patients with resected brain metastases from CRC. Three survival endpoints were defined, all of them starting from BM resection: the first to the time of intracranial-only disease progression, death (any cause) or last follow-up, whichever occurred first (intracranial-only progression-free survival, BM-iPFS), the second to the disease progression in any site, death (any cause) or last follow up, whichever occurred first (progression free survival, BM-PFS), and the third to the time of death for any cause or at last follow up, whichever occurred first(overall survival, BM-OS). Canonical overall survival (OS), defined as the time from diagnosis of metastatic disease to death for any cause or last follow up, whichever occurred first., was also described.
Statistical design
Given the exploratory, descriptive nature of this study, no formal statistical hypothesis was generated. The median follow-up was calculated using the reverse Kaplan-Meier method, starting from BM resection. Fisher’s exact test was applied to evaluate the distribution of clinical and molecular characteristics. Survival endpoints (BM-iPFS, BM-PFS, BM-OS, OS) were described using the Kaplan-Meier method, and Cox proportional hazards regression model was applied to calculate the hazard ratios (HR); given the low sample size, no multivariate analysis nor interactions tests were feasible. All tests were two sided.
Cut-offs for TMB and TILs were set with ROC curves. All the statistical analyses were performed using R software v.4.2.3.
Results
Clinical characteristics of study population
Out of 101 pts with BM from CRC treated at our Institution from 1 January 2010 until 31 December 2021, 22 (10 males/12 females) underwent BM resection and were thus included in the analysis. Of them, 11 (50%) were retrospectively enrolled while the remaining 11 (50%) patients were prospectively enrolled starting from 1st March 2018. The large majority of the overall population was composed by patients aged ≥70 (18 out of 22, 82%), with left colon or rectal cancer (19 out of 22, 86%); furthermore, 17 (77%) of the patients enrolled had also lung metastases, while liver metastases were found in 11 (50%) patients. BM were metachronous (onset > 6 months after prCRC diagnosis) in most cases (68 vs 32%); at the time of brain surgery, the majority of patients had both intra and extracranial disease (64 vs 36%); just three out of 22 patients had more than a single BM at the time of surgery (Table 1). Of the 22 patients included in the analysis, 6 (27%) did not receive any systemic antitumoral treatment for mCRC, while 7 (32%) received three or more lines of treatment. Two patients received immunotherapy in experimental trials, one of them bearing MSI-high (MSI-H) mCRC; importantly, none of them received anti-HER2 treatments (Supplementary Table 1).
Table 1.
Clinical characteristics of study population.
| Patients N = 22 (%) | |
|---|---|
| Age at first tumour diagnosis, years | |
| Median (IQR) | 51 (47–65) |
| <70 | 4 (18) |
| ≥70 | 18 (82) |
| Sex | |
| Male | 10 (45) |
| Female | 12 (55) |
| ECOG PS at baseline | |
| 0 | 13 (59) |
| ≥1 | 9 (41) |
| Stage at diagnosis | |
| I-II-II | 11 (50) |
| IV | 11 (50) |
| Synchronous vs metachronous metastases | |
| Sync (<6 months) | 11 (50) |
| Meta (≥6 months) | 11 (50) |
| Primary tumour location | |
| Right | 3 (14) |
| Left | 9 (41) |
| Extraperitoneal rectum | 10 (45) |
| Primary tumour resection | |
| Yes | 17 (77) |
| No | 5 (23) |
| Liver metastases at any time | |
| Yes | 11 (50) |
| No | 11 (50) |
| Lung metastases at any time | |
| Yes | 17 (77) |
| No | 5 (23) |
| Lines of treatments received in total | |
| ≤3 | 15 (68) |
| >3 | 7 (32) |
| Age at brain metastases resection, years | |
| Median (IQR) | 58 (48–68) |
| <70 | 17 (77) |
| ≥70 | 5 (23) |
| ECOG PS at the time of brain metastases resection | |
| 0 | 8 (36) |
| ≥1 | 14 (64) |
| Brain metastases presentation | |
| Synchronous | 7 (32) |
| Metachronous | 15 (68) |
| Tumour burden at the time of brain surgery | |
| Intracranial-only disease | 8 (36) |
| Intra and extracranial disease | 14 (64) |
| Number of brain metastases at the time of brain surgery | |
| 1 | 19 (86) |
| >1 | 3 (14) |
Molecular and immunohistochemical landscape of matched CRC and BM
Molecular and immunohistochemical characteristics of BM were consistent with data reported in Literature: out of 22 analysed BM, HER2+ was documented in four (18%); three (14%) carried a BRAFV600E mutation; two (9%) displayed MSI-H: therefore, HER2, BRAFV600E and MSI-H enrichment in BM from CRC was confirmed.
Some heterogeneity between prCRC and corresponding BM was recognizable: out of four patients with HER2 + BM, only two displayed HER2+ also on matched prCRC; in the other 2 cases, HER2+ was acquired on BM, being not documented on corresponding prCRC. On the other hand, in one case HER2+ was lost from prCRC to coupled BM (Table 2). Therefore, in total 3 (14%) patients out of 22 had discordant HER2 status between BM and matched prCRC (p = 1.000).
Table 2.
Molecular and histological characteristics of matched BM and primitive CRC tissue.
| Primary tumour N = 22 (%) | Brain metastases N = 22 (%) | p-value | |
|---|---|---|---|
| HER2 | |||
| Ampl | 3 (14) | 4 (18) | 1.000 |
| Non ampl | 19 (86) | 18 (82) | |
| Not evaluable | 0 | 0 | |
| KRAS | |||
| WT | 5 (24) | 8 (36) | 0.310 |
| Mut | 16 (76) | 14 (64) | |
| Not evaluable | 1 | 0 | |
| NRAS | |||
| WT | 19 (95) | 21 (95) | 1.000 |
| Mut | 1 (5) | 1 (5) | |
| Not evaluable | 2 | 0 | |
| BRAF | |||
| WT | 19 (95) | 19 (86) | 0.608 |
| Mut | 1 (5) | 3 (14) | |
| Not evaluable | 2 | 0 | |
| MSI | |||
| MSS | 20 (91) | 20 (91) | 1.000 |
| MSI-H | 2 (9) | 2 (9) | |
| Not evaluable | 0 | 0 | |
| TMB | |||
| High ≥ 5 | 7 (44) | 10 (48) | 1.000 |
| Low < 5 | 9 (56) | 11 (52) | |
| Not evaluable | 6 | 1 | |
| TILs | |||
| High ≥ 1.6 | 8 (44) | 5 (25) | 0.307 |
| Low < 1.6 | 10 (56) | 15 (75) | |
| Not evaluable | 4 | 2 | |
| Grading | |||
| G1/G2 | 15 (71) | 5 (71) | 1.000 |
| G3/G4 | 6 (29) | 2 (29) | |
| Not evaluable | 1 | 15 | |
| TP53 | |||
| WT | 4 (20) | 5 (23) | 1.000 |
| Mut | 16 (80) | 17 (77) | |
| Not evaluable | 2 | 0 | |
| APC | |||
| WT | 4 (20) | 4 (18) | 1.000 |
| Mut | 16 (80) | 18 (89) | |
| Not evaluable | 2 | 0 | |
| PIK3CA | |||
| WT | 15 (75) | 20 (91) | 0.229 |
| Mut | 5 (25) | 2 (9) | |
| Not evaluable | 2 | 0 | |
Looking at the three patients with BRAF mutations on BM, in one case BRAFV600E mutation was documented on both prCRC and matched BM; in another patient, the mutation was detected on BM but not on the corresponding prCRC; for the third patient, BRAF on prCRC was not evaluable, so the comparison was not feasible.
KRAS mutations were more frequent than expected (16 out of 22, 76%) and were consistent between matched primary tumour and BM; only one patient out of 22 had a NRAS mutation (p. Q61R), which was found in both primary tumour and BM.
Other frequent mutations found by means of NGS in BM were observed in the TP53 (17 out of 22 BM, 77%), APC (18 out of 22 BM, 89%) and PIK3CA (2 out of 22 brain metastases, 9%) genes. The distribution of PIK3CA mutations was heterogeneous between coupled samples, being gained from primary tumour to BM in one patient and lost in other four cases (Table 2).
Median TMB value was 4 mut/Mb both on prCRC specimens (range 1 to 115 mut/Mb) and on BM (range 1 to 57 mut/Mb). Using ROC curves as formerly described, TMB was defined as high if ≥ 5.02 mut/Mb. Median TILs number was 2/HPF (range 1 to 5/HPF) on primary samples and 0.6/HPF (range 0 to 6/HPF) on BM. TILs were defined as high if ≥ 1.6/HPF. With such cut-offs, respectively 10 (48%) and 5 (25%) BM specimens had high TMB and TILs, being both consistent between matched primary tissue and BM.
Impact of clinical, molecular and immunohistochemical characteristics on survival
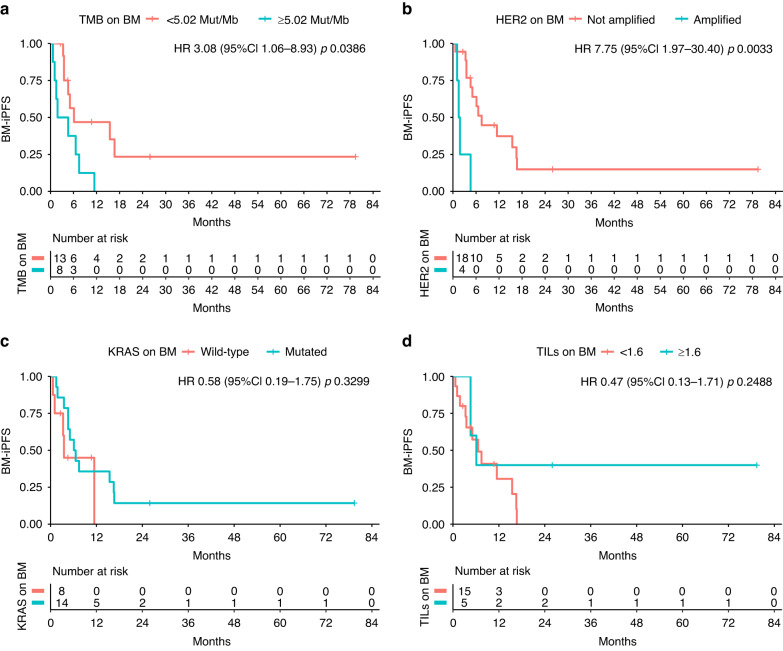
At a median follow-up of 45.89 months (95% CI 17.73 to 88.03), factors positively influencing BM-iPFS were low TMB (6.12 vs 3.23 months; HR 3.08; 95% CI 1.06 - 8.93; p value = 0.0386) and absence of HER2+ on brain metastases (7.47 vs 1.68 months; HR 7.75; 95% CI 1.97 −30.40; p value = 0.0033). On the contrary, KRAS mutations did not have a significant impact on BM-iPFS (HR 0.58; 95% CI 0.19–1.75; p value = 0.3299); as well, no differences in BM-iPFS were detected depending on TILs number on BM (HR 0.47; 95% CI 0.13–1.71; p value = 0.2488) (Fig. 1).
Fig. 1. BM-iPFS.
In our analyses, intracranial progression-free survival from BM (brain metastasis) resection (BM-iPFS) was improved in case of low TMB (Tumor Mutation Burden) (<5.02 Mut/Mb) (a) and absence of HER2 amplification (HER2+) (b) on BM; on the contrary, no correlation was observed with KRAS status (c) or TILs (Tumor Infiltrating Lymphocytes) on BM (d).
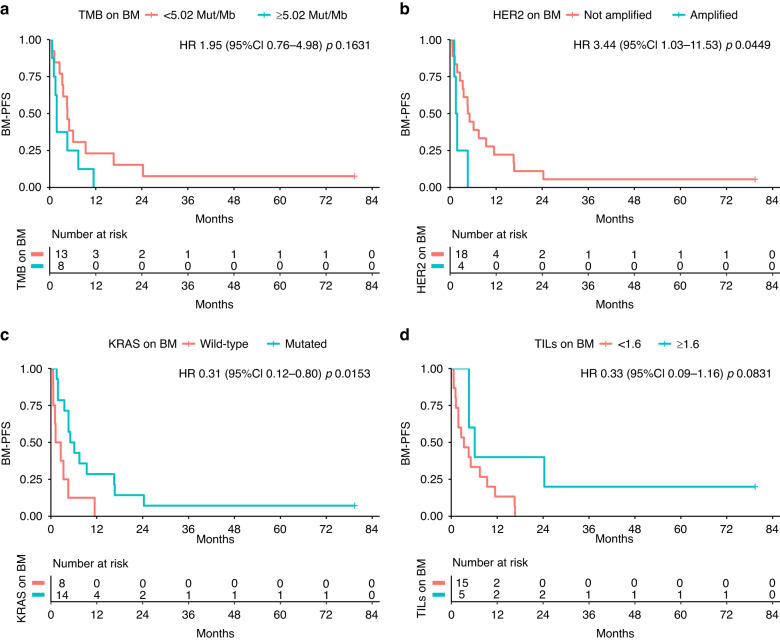
The absence of HER2+ on BM had also a positive impact on BM-PFS (4.84 vs 1.68 months; HR 3.44; 95%CI 1.03–11.53; p = 0.0449), as well as KRAS mutations (1.90 vs 5.60 months; 0.31; 95% CI 0.12–0.80; p value = 0.0153). On the contrary, TMB and TILs did not have a clear impact on BM-PFS (respectively HR 1.95; 95% CI 0.76–4.98; p value = 0.1631 and HR 0.33; 95% CI 0.09–1.16; p value = 0.0831) (Fig. 2).
Fig. 2. BM-PFS.
No correlation was confirmed between any-site progression free survival from brain metastasis (BM) resection (BM-PFS) and TMB (Tumor Mutation Burden) on BM (a); on the other hand, HER2 amplification (HER2+) (b) and KRAS mutations (c) on BM yielded better BM-PFS. A non-statistically significant advantage in BM-PFS was observed in case of TILS ≥ 1.6 on BM (d).
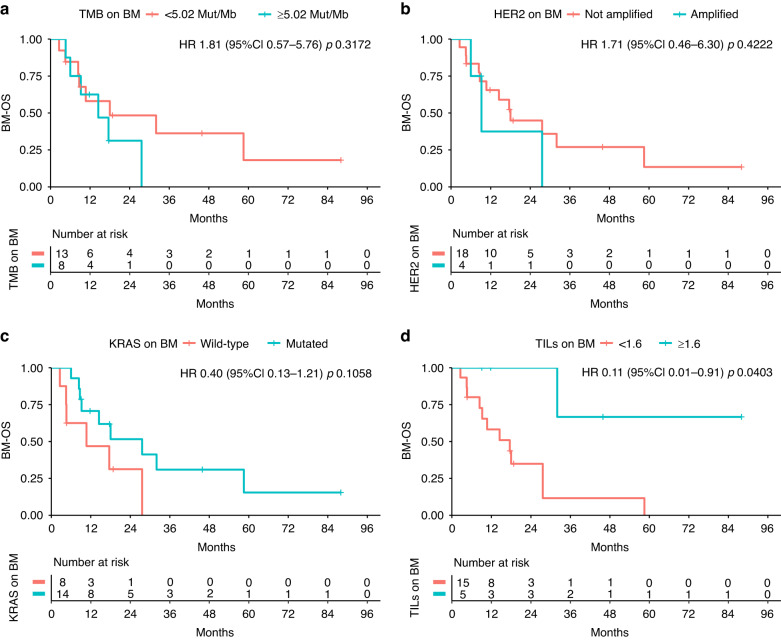
OS from the diagnosis of metastatic disease was 37.20 months (CI 95%: 21.30 - NA). Longer BM-OS was found in pts with higher TILs (≥1.6) on BM (p value = 0.0403), whereas TMB, HER2 status, and KRAS mutations had no clear effect (Fig. 3). As well, none of the other mutations individuated with NGS had an impact on survival (Fig. 4).
Fig. 3. BM-OS.
Looking at overall survival from brain metastasis (BM) resection (BM-OS), no correlation was demonstrated with TMB (Tumor Mutation Burden) (a), HER2 amplification (HER2+) (b) and KRAS mutations (c) on BM. Instead, statistically significant advantage was observed in case of TILS ≥ 1.6 on BM (d).
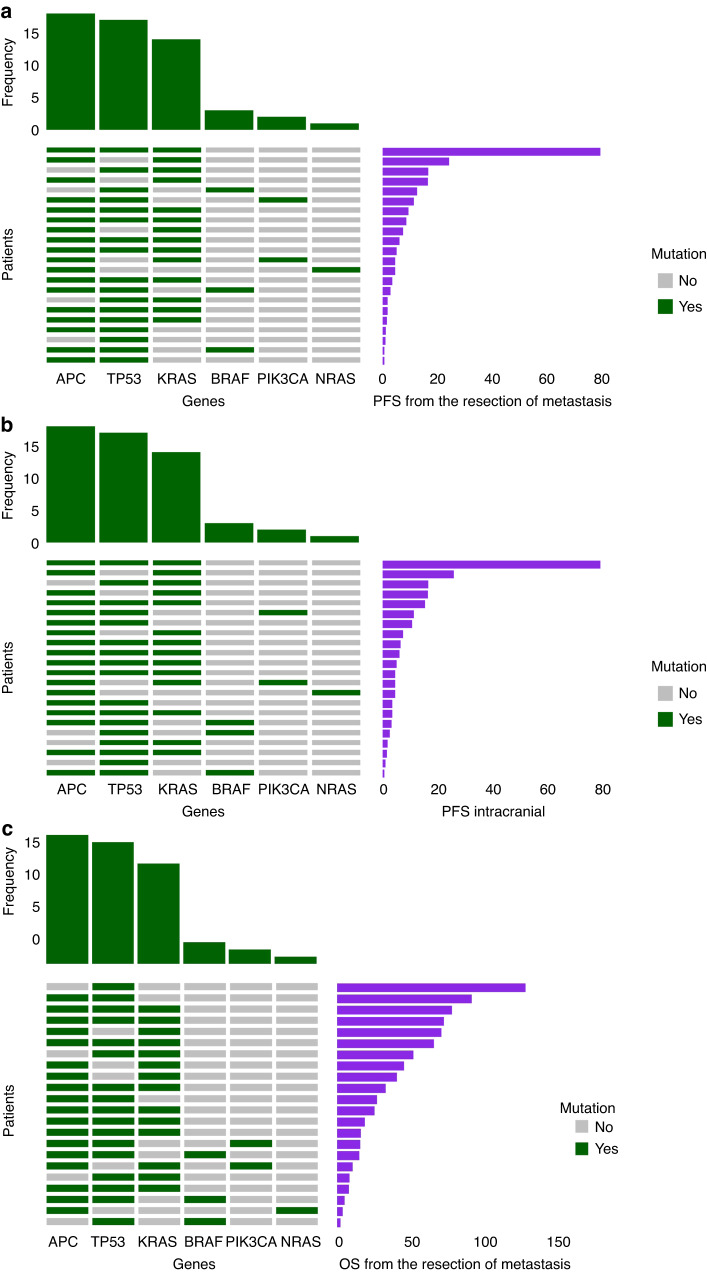
Fig. 4. Impact of single genes.
As depicted below, neither any-site (BM-PFS) (a) nor intracranial progression free survival (BM-iPFS) (b) and overall survival from brain metastasis (BM) resection (BM-OS) (c) were influenced by any of the molecular alterations detected with Next Generation Sequencing (NGS).
Discussion
In this study, extensive molecular landscape of BM from CRC was described, providing unique data on this poorly explored field of study. In our experience, both BM-iPFS and BM-PFS were positively influenced by absence of HER2 + . TMB < 5.02 mut/Mb conferred better BM-iPFS also; however, it must be pointed out that this is not a recognised cut-off, but was set as described above in order to better characterize the relatively small sample size of the present study. The presence of KRAS mutations was related to better BM-PFS only.
Considering these results, it might be argued that this study suggests a prognostic value for HER2, TMB and KRAS in patients with BM from CRC. Nevertheless, caution should be used in interpreting and generalizing our results. Indeed, apart from the small sample size, our cohort is also strictly selected from a clinical point of view: patients diagnosed with BM are deemed amenable to surgical resection only in case of optimal clinical conditions, small and/or single brain lesions with favourable localization; furthermore, it is more likely that these patients are referred to high volume and expertise surgical centres. Accordingly, our case series was composed of 22 individuals selected out of 101 patients with BM from CRC, because they were the only ones who were declared eligible for surgery. Thus, it would be more appropriate to infer that our results might guide in clinical management of patients amenable to neurosurgery for BM from CRC, and not to all of them.
The effect of HER2 on prognosis is intriguing and deserves special discussion. An interaction test between HER2 and other known strong prognostic factors such as BRAF or ECOG PS could provide further insight on their relative weight on prognosis; unfortunately, the sample size was not sufficient to proceed with further analyses. Nonetheless, the negative influence on BM-iPFS and BM-PFS conferred by HER2+ documented in our work could be explained by three main factors: negative impact of HER2+ per se on prognosis, absence of targeted therapies against HER2 at the time of treatment, and resistance conferred by HER2+ to anti-EGFR.
Resistance to anti-EGFR has been widely documented in patients with HER2 + CRC [11–13]; on the contrary, the prognostic role of HER2+ has not been well established in CRC; nevertheless, the tendency to worse outcomes in survival has been reported in patients with HER2 + CRC [14]. More recently, worse relapse-free survival (RFS) was described in patients with liver metastases from CRC displaying HER2 + . In our series, only three patients out of 22 (14%) received anti-EGFR and none of them received anti-HER2 therapy. According to the results of the HERACLES trial [6] and, more recently, to the results of the DESTINY-CRC01 trial [15], anti-HER2 agents could revert the negative impact on prognosis exerted by HER2 + . A special mention should be reserved to the intracranial activity of these drugs: in particular, trastuzumab is known to be unable to pass the blood-brain barrier. Of note, unexpectedly high rate of BM was observed in relationship with treatment-prolonged survival after trastuzumab and lapatinib in patients with HER2-amplified CRC [3]. Conversely, trastuzumab-deruxtecan showed remarkable intracranial activity in a dedicated phase II trial [16]; however, this study was conducted on patients with breast cancer, and no studies are currently available specifically addressing the activity of trastuzumab-deruxtecan on BM from CRC.
Putting our results into a purely clinical context, the lesson learnt from this experience was to always bear in mind the chance of BM development in patients with HER2 + CRC: thus, symptoms suggesting cerebral localizations should be carefully explored in this special population. Moreover, especially in case of long survivors, including cerebral imaging in routine follow-up might become reasonable if our data will be further confirmed.
Moving from the case of HER2, isolating specific molecular alterations potentially predictive of BM development could be intriguing: in this work, together with HER2, also KRAS, BRAFV600E, TP53 and APC mutations were observed in BM. On the other hand, we described how some of these alterations could be either lost or gained from prCRC to BM, suggesting an evolving molecular landscape from primary tumour to BM. Previous studies described intratumor heterogeneity of HER2 expression between prCRC and BM, documenting discordant HER2 status between primary tumour and matched metastases, both intra and extracranial: in these studies, the impact of HER2 discordance on prognosis is unclear [17, 18]; on the other hand, the indirect weight of the heterogeneity between prCRC and metastases on prognosis is intuitive, because molecular modifications could lead to increased therapeutic chances.
Safe access to brain tissue for molecular characterization would be useful to drive clinical choices, especially in the era of target therapy; given the well-known limits related to intracranial surgical procedures, liquid biopsy could be of special interest in these situations. Unfortunately, scarcity of circulating tumour DNA (ctDNA) has been documented in plasma in case of BM from solid tumours, as well as in case of central nervous system primary tumours [19]. Notwithstanding, cerebrospinal fluid (CSF) is emerging as a source of ctDNA from brain lesions: in fact, several actionable mutations have been identified in CSF-ctDNA, and there is some suggestion that CSF-ctDNA is more accurate than blood ctDNA in reproducing private molecular alterations of BM from breast cancer, being also detectable only in patients with BM, and decreasing accordingly to response to systemic treatments and/or intracranial surgery [20]. No specific data are available regarding CRC; however, these previous experiences could represent a valid starting point for further development.
A finding that is worth of special mention regards TMB: in our report, TMB < 5.02 mut/Mb was related to better BM-iPFS. This result must be interpreted with caution: high TMB has been related to better prognosis in both limited-stage CRC [21] and metastatic setting [22]; furthermore, predictive value of high TMB in case of therapy with immune checkpoint inhibitors has been demonstrated [23]. However, in our series only two patients received immune checkpoint inhibitors. Although FDA approval of pembrolizumab for patients with solid tumors with high TMB took as cutoff 10 mut/Mb, this threshold is still object of debate since it could be dependent on histology [24, 25] and on the assay employed to determine the TMB value itself. For example, in the study from Innocenti et al., positive prognostic value was attributed to high TMB in patients with metastatic MSS CRC; in this study, the threshold to define high TMB was 8 mut/Mb. Of note, in our cohort, the large majority of patients had left-sided CRC: therefore, a tendency to lower TMB was predictable.
Conclusion
Even with the limitation of small sample size, the HEROES study supports HER2+ enrichment in both prCRC and BM from CRC. Absence of HER2+ seems to confer better BM-iPFS and BM PFS in patients with resected BM from CRC. In the future, larger studies and new techniques like liquid biopsy could be important to better assess the molecular landscape evolution of BM from CRC in order to individuate new prognostic and predictive factors.
Supplementary information
Acknowledgements
The authors wish to say thank you to the patients and their family, as well as the sub-investigators, data manager and nurses.
Author contributions
Conceptualization: AAP, VA, MF, SL; methodology: AAP, MF; data collection and curation: VV, AAP; formal analysis: AAP, CDT; validation of results: all authors; original draft preparation: AAP; review and editing of the manuscript: VA, FB, MF, SL; supervision: FB, MF, SL; project administration and funding acquisition: FB, SL. All authors have read and agreed to the published this version of the manuscript.
Funding
This research was promoted by Veneto Institute of Oncology IOV – IRCCS and received no external funding. Matteo Fassan is supported by grants from the Italian Health Ministry/Veneto region research program NET-2016–02363853 and AIRC 5 per mille 2019 (ID. 22759 program). Furthermore, this research was funded by Italian Ministry of Health Ricerca Corrente.
Data availability
The data generated in this study are available within the article.
Competing interests
FB (Francesca Bergamo) reports: advisory/consulting role for Servier and Novartis; speakers’ fee from Eli-Lilly, MSD, EISAI, Bayer, BMS. SL reports: research funding (to Institution) from Amgen, Astellas, Astra Zeneca, Bayer, Bristol-Myers Squibb, Daichii Sankyo, Hutchinson, Incyte, Merck Serono, Mirati, MSD, Pfizer, Roche, Servier; personal honoraria as invited speaker from Amgen, Bristol-Myers Squibb, Incyte, GSK, Lilly, Merck Serono, MSD, Pierre-Fabre, Roche, Servier; participation in advisory board for Amgen, Astellas, Astra Zeneca, Bayer, Bristol-Myers Squibb, Daiichi-Sankyo, GSK, Incyte, Lilly, Merck Serono, MSD, Servier, Takeda. MF (Matteo Fassan) reported personal fees (as speaker bureau or advisor) from Roche, MSD, GSK, Amgen, Lilly, Novartis, Astellas Pharma, Pierre Fabre, Astra Zeneca and received research grants from Astellas Pharma, QED Therapeutics, Diaceutics, and Macrophage Pharma. All of the above-mentioned financial relationships were outside this work. All remaining authors have declared no conflicts of interest.
Ethics approval and consent to participate
This research received approval from the local Ethics Committee. Written informed consent and consent for publication were obtained from each patient to participate in the study.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Alessandra Anna Prete, Valentina Angerilli.
These authors jointly supervised this work: Matteo Fassan, Sara Lonardi.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-023-02569-4.
References
- 1.Mongan JP, Fadul CE, Cole BF, Zaki BI, Suriawinata AA, Ripple GH, et al. Brain metastases from colorectal cancer: risk factors, incidence, and the possible role of chemokines. Clin Colorectal Cancer. 2009;8:100–5. doi: 10.3816/CCC.2009.n.016. [DOI] [PubMed] [Google Scholar]
- 2.Pietrantonio F, Aprile G, Rimassa L, Franco P, Lonardi S, Cremolini C, et al. A new nomogram for estimating survival in patients with brain metastases secondary to colorectal cancer. Radiother Oncol. 2015;117:315–21. doi: 10.1016/j.radonc.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 3.Sartore-Bianchi A, Lonardi S, Aglietta M, Martino C, Ciardiello F, Marsoni S, et al. Central Nervous System as Possible Site of Relapse in ERBB2-Positive Metastatic Colorectal Cancer: Long-term Results of Treatment With Trastuzumab and Lapatinib. JAMA Oncol. 2020;6:927–9. doi: 10.1001/jamaoncol.2020.0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richman SD, Southward K, Chambers P, Cross D, Barrett J, Hemmings G, et al. HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials. J Pathol. 2016;238:562–70. doi: 10.1002/path.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kavuri SM, Jain N, Galimi F, Cottino F, Leto SM, Migliardi G, et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov. 2015;5:832–41. doi: 10.1158/2159-8290.CD-14-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:738–46. doi: 10.1016/S1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 7.Tran B, Kopetz S, Tie J, Gibbs P, Jiang ZQ, Lieu CH, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117:4623–32. doi: 10.1002/cncr.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 9.Sun J, Wang C, Zhang Y, Xu L, Fang W, Zhu Y, et al. Genomic signatures reveal DNA damage response deficiency in colorectal cancer brain metastases. Nat Comm. 2019;10:3190. doi: 10.1038/s41467-019-10987-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valtorta E, Martino C, Sartore-Bianchi A, Penaullt-Llorca F, Viale G, Risio M, et al. Assessment of a HER2 scoring system for colorectal cancer: results from a validation study. Mod Pathol. 2015;28:1481–91. doi: 10.1038/modpathol.2015.98. [DOI] [PubMed] [Google Scholar]
- 11.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–23. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 12.Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, Souglakos J, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med. 2011;3:99ra86. doi: 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sartore-Bianchi A, Amatu A, Porcu L, Ghezzi S, Lonardi S, Leone F, et al. HER2 Positivity Predicts Unresponsiveness to EGFR-Targeted Treatment in Metastatic Colorectal Cancer. Oncologist. 2019;24:1395–402. doi: 10.1634/theoncologist.2018-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang XY, Zheng ZX, Sun Y, Bai YH, Shi YF, Zhou LX, et al. Significance of HER2 protein expression and HER2 gene amplification in colorectal adenocarcinomas. World J Gastrointest Oncol. 2019;11:335–47. doi: 10.4251/wjgo.v11.i4.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshino T, Di Bartolomeo M, Raghav KPS, Masuishi T, Kawakami H, Yamaguchi K, et al. Trastuzumab deruxtecan (T-DXd; DS-8201) in patients (pts) with HER2-expressing metastatic colorectal cancer (mCRC): Final results from a phase 2, multicenter, open-label study (DESTINY-CRC01) J Clin Oncol. 2022;40:119–119. doi: 10.1200/JCO.2022.40.4_suppl.119. [DOI] [Google Scholar]
- 16.Bartsch R, Berghoff AS, Furtner J, Marhold M, Bergen ES, Roider-Schur S, et al. Trastuzumab deruxtecan in HER2-positive breast cancer with brain metastases: a single-arm, phase 2 trial. Nat Med. 2022;28:1840–7. doi: 10.1038/s41591-022-01935-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen PC, Yeh YM, Chu CT, Su PF, Chiu PH, Lin BW, et al. HER2 amplification in colorectal cancer with brain metastasis: A propensity score matching study. Eur J Cancer. 2023;181:62–69. doi: 10.1016/j.ejca.2022.12.019. [DOI] [PubMed] [Google Scholar]
- 18.Shan L, Lv Y, Bai B, Huang X, Zhu H. Variability in HER2 expression between primary colorectal cancer and corresponding metastases. J Cancer Res Clin Oncol. 2018;144:2275–81. doi: 10.1007/s00432-018-2744-z. [DOI] [PubMed] [Google Scholar]
- 19.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumour DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzpatrick A, Iravani M, Mills A, Childs L, Alaguthurai T, Clifford A, et al. Assessing CSF ctDNA to improve diagnostic accuracy and therapeutic monitoring in breast cancer leptomeningeal metastasis. Clin Cancer Res. 2022;28:1180–91. doi: 10.1158/1078-0432.CCR-21-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domingo E, Camps C, Kaisaki PJ, Parsons MJ, Mouradov D, Pentony MM, et al. Mutation burden and other molecular markers of prognosis in colorectal cancer treated with curative intent: results from the QUASAR 2 clinical trial and an Australian community-based series. Lancet Gastroenterol Hepatol. 2018;3:635–43. doi: 10.1016/S2468-1253(18)30117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Innocenti F, Ou FS, Qu X, Zemla TJ, Niedzwiecki D, Tam R, et al. Mutational analysis of patients with colorectal cancer in CALGB/SWOG 80405 identifies new roles of microsatellite instability and tumor mutational burden for patient outcome. J Clin Oncol. 2019;37:1217–27. doi: 10.1200/JCO.18.01798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020;21:1353–65. doi: 10.1016/S1470-2045(20)30445-9. [DOI] [PubMed] [Google Scholar]
- 24.Panda A, Betigeri A, Subramanian K, Ross JS, Pavlick DC, Ali S, et al. Identifying a clinically applicable mutational burden threshold as a potential biomarker of response to immune checkpoint therapy in solid tumors. JCO Precis Oncol. 2017;2017:PO.17.00146. doi: 10.1200/PO.17.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–6. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article.