Abstract
Nonstructural proteins encoded by measles virus (MV) include the V protein which is translated from an edited P mRNA. V protein is not associated with intracellular or released viral particles and has recently been found to be dispensable for MV propagation in cell culture (H. Schneider, K. Kaelin, and M. A. Billeter, Virology 227:314–322, 1997). Using recombinant MVs (strain Edmonston [ED]) genetically engineered to overexpress V protein (ED-V+) or to be deficient for V protein (ED-V−), we found that in the absence of V both MV-specific proteins and RNAs accumulated to levels higher than those in the parental MV molecular clone (ED-tag), whereas MV-specific gene expression was strongly attenuated in human U-87 glioblastomas cells after infection with ED-V+. The titers of virus released from these cells 48 h after infection with either V mutant virus were lower than those from cells infected with ED-tag. Similarly, significantly reduced titers of infectious virus were reisolated from lung tissue of cotton rats (Sigmodon hispidus) after intranasal infection with both editing mutants compared to titers isolated from ED-tag-infected animals. In cell culture, expression of V protein led to a redistribution of MV N protein in doubly transfected Cos-7 cells, indicating that these proteins form heterologous complexes. This interaction was further confirmed by using a two-hybrid approach with both proteins expressed as Gal4 or VP16 fusion products. Moreover, V protein efficiently competed complexes formed between MV N and P proteins. These findings indicate that V protein acts to balance accumulation of viral gene products in cell culture, and this may be dependent on its interaction with MV N protein. Furthermore, expression of V protein may contribute to viral pathogenicity in vivo.
As a member of the paramyxovirus subfamily of the Mononegavirales, measles virus (MV) has a nonsegmented RNA genome of negative polarity with six structural genes that are sequentially arranged. The entire coding region is flanked by the 3′ and 5′ promoter regions, and the individual genes are separated by conserved intergenic regions. With one exception, the MV gene products are encoded by genes that are functionally monocistronic: both those required for assembly (matrix [M], fusion [F], and hemagglutinin [H] proteins) and those required for genome replication and/or viral transcription (nucleocapsid [N] protein and polymerase or large protein [L]). Conversely, as in all paramyxoviruses, the gene encoding as the main product the phosphoprotein P, the polymerase cofactor, gives rise to additional proteins which are detectable in infected cells. The basic C protein (20 kDa) is a translation product of an overlapping reading frame (3), and the V protein (46 kDa) is translated from P mRNAs after insertion of a single G nucleotide at a specific editing site (5). Recently, a third protein, R (46 kDa), which requires ribosomal frameshifting for its expression, has been detected in infected cells at very low levels (17). Both MV C and V proteins are expressed at high levels but are found only in infected cells and not within the virion (10, 30). Although the reading frames for both C and V are highly conserved (1), the functions of these proteins are unknown. They are not required for propagation in cell culture, since recombinant MVs deficient for V or C function were successfully rescued from cloned cDNA and could be propagated in human 293, HeLa, and Vero cells as efficiently as the parental, nondefective strain (22, 25).
As an edited product of the P mRNA, V protein shares the 231 amino-terminal amino acid (aa) residues with the P protein (5). Within this region, casein kinase II phosphorylation sites have been mapped for P (8), and it is likely that these are also targeted by this enzyme in V, as the protein is also phosphorylated (30). Moreover, two interaction sites for N have been mapped within the P protein, one of which is localized at the amino terminus of the P protein (11). However, a direct interaction of MV V with N protein has not been confirmed (18). MV V protein, like all V proteins, has a highly conserved and cysteine-rich carboxy terminus that shows similarities to the zinc finger binding motifs of some DNA binding proteins and binds zinc ions (19).
The biological role of V proteins in paramyxovirus replication has not yet been clarified. For Sendai virus (SeV), binding of V protein to unassembled N molecules and V protein-dependent inhibition of genome replication, but not of transcription, have been shown in vitro (7, 12). Similar to MV, a recombinant SeV in which synthesis of V was abrogated by mutation of the editing site was efficiently rescued from cloned cDNA and propagated in cell culture; however, the kinetics and plaque morphology were slightly altered compared to those of the parental nondefective strain (15). Interestingly, accumulated levels of SeV-specific transcripts after infection of cell cultures as well as a reduced pathogenicity of V-deficient SeV in mice were also noted, indicating that V protein might act to balance viral gene expression in infected cells and in vivo. In the present study we present evidence that, similar to SeV, MV V protein is involved in the regulation of MV RNA and hence of protein accumulation in human glioblastoma U-87 cells. Moreover, MVs which either overexpress V or are deficient for V protein expression are less pathogenic as shown by experimental infection of cotton rats. As evidenced by transient-transfection studies, N-V complexes are likely to form in infected cells, and this may be associated with regulation of viral gene expression.
MATERIALS AND METHODS
Cells and viruses.
Human glioblastoma U-87 cells (ATCC) and Cos-7 cells were maintained in minimal essential medium (MEM) containing 10% heat-inactivated fetal calf serum (FCS). Vero cells were maintained in MEM containing 5% FCS, and U-937 and BJAB cells were maintained in RPMI 1640 containing 10% FCS. All cell lines were maintained in the presence of antibiotics. Recombinant MVs of strain Edmonston (ED) genetically engineered to overexpress V protein (ED-V+) or to be deficient for V protein (ED-V−) (mutant 3 or 1, respectively [25]) and the parental molecular clone (ED-tag [23]) were propagated on Vero cells to yield titers of 2 × 106 (for ED-tag and ED-V−) and 2 × 105 (ED-V+).
MV protein and RNA analysis.
After infection with recombinant MVs for the time intervals indicated, U-87 cells were labeled for 5 h with 150 μCi of TranS label (Amersham)/ml or were labeled after an overnight infection for 3 h, followed by chase periods of 8 or 16 h, subsequently lysed in RIPA detergent (150 mM NaCl, 10 mM Tris-HCl [pH 7.4], 1% Na-desoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 1 mM phenylmethylsulfonyl fluoride), and processed for immunoprecipitation following standard procedures. Immunocomplexes were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and MV-specific signals were quantified by phosphorimaging. For RNA analysis, total RNA was prepared from U-87 cells infected with ED-tag, ED-V−, or ED-V+ by using an RNeasy kit following the manufacturer’s protocol (Qiagen), separated on formaldehyde-agarose gels, blotted onto nitrocellulose filters, and analyzed using 32P-labeled RNA probes specific for the MV N, M, and H genes (4) or, for the control, with a 32P-labeled DNA probe specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Signals obtained were quantified by phosphorimaging.
Infection of cotton rats and virus titration.
Cotton rats (inbred strain COTTON/NIco) were obtained from Iffa Credo, Lyon, France. Animals were kept under controlled environmental conditions and used at the age of 6 to 8 weeks. For intranasal (i.n.) infection MV was given in phosphate-buffered saline (PBS) to ether-anesthetized cotton rats in a volume not exceeding 100 μl. Four (or five) days later, the animals were asphyxiated with CO2 and lungs were removed and weighed. Lung tissue was minced with scissors and dounced with a glass homogenizer. Serial 10-fold dilutions of homogenized lung tissue were assessed for the levels of infectious virus by using Vero cells. Plates (48 well) were scored microscopically for cytopathic effects after 7 days. The amount of virus in each inoculum was expressed as the quantity of virus that could infect 50% of the inoculated tissue culture monolayers.
Plasmid constructs.
pSG424, pVP16AASV19N, and the reporter gene construct G5BCAT were kindly supplied by A. K. Banerjee, Cleveland, Ohio. To obtain pGal-N, the MscI/XbaI fragment (covering, except for the first codon, the entire coding region of the MV ED N gene) was released from pEN (Bluescript-Vector) and ligated into the EcoRI (blunt ended by nuclease S1 trimming)/XbaI site of pSG424 downstream of and in-frame with the coding sequence for aa 1 to 147 of Gal4. The amino-terminal M was replaced by L by this procedure. The same fragment was ligated into a pGem vector with a reconstructed multiple cloning site between two SmaI restriction sites to yield pGem-MscI/XbaI-N. The SmaI fragment was then ligated into the EcoRV site of pVP16AASV19N upstream and in-frame with aa 398 to 479 of VP16 to yield pN-VP16. Peptides added to the amino terminus were AGL (replacing the initiator M), and the linker joining the N to the VP16 reading frame was LEPI.
The pGem-1 multiple cloning site was replaced by an oligonucleotide sequence containing restriction sites for HindIII-PstI-EcoRI-EcoRV-BamHI-NsiI-XbaI-EcoRI with the authentic 5′-terminal nucleotides of the P-V reading frame between the BamHI and NsiI sites to yield pGemP/Vstart. The NsiI/XbaI fragments were excised from pEP (containing the entire coding sequence of MV ED P) or pEV (containing the V gene) and ligated into pGemP/Vstart to yield pGemP/V. The EcoRI/XbaI fragment was excised from pGemP/V and ligated into pSG424, yielding pGal-P or pGal-V, respectively. The linker joining the Gal4 and P or V reading frames was EFDIGS. pP-VP16 was generated by ligation of the EcoRV fragment from pGemP into the EcoRV site of pVP16AASV19N. By this procedure, the 20 carboxy-terminal aa residues of P were lost. The amino-terminal-joining peptide was IGS. The generation of pSC-N, pSC-P, and pSC-V has been described previously (14).
Transfections, reporter gene assay, and immunofluorescent staining.
Transient transfections were performed by lipofection following the manufacturer’s protocol (Gibco-BRL). Routinely, 1 μg of each of the Gal4 and VP-16 fusion constructs was transfected along with 1 μg of pG5BCAT. For the competition experiments, competitor plasmids were cotransfected at the concentrations indicated. Cells were harvested 40 h after transfection and lysed by repeated freeze-thawing, and 10 μg of total protein extracts was used for a standard chloramphenicol acetyltransferase assay.
For the colocalization studies, Cos-7 cells were seeded onto coverslips and transfected with pSC-N (14) or pSC-V (10 μg each). After 40 h, cells were fixed and permeabilized (PBS–3.5% paraformaldehyde followed by treatment with PBS–0.25% Triton X-100) and then stained for MV antigens with a monoclonal anti-N antibody or a rabbit anti-V monospecific serum raised against the C-terminal domain. MV-specific protein expression was determined after infection with ED-tag, ED-V−, or ED-V+ at the time intervals indicated after fixation-permeabilization by using monoclonal antibodies directed against the MV N protein.
RESULTS
Effect of V protein expression on MV infection in cell cultures.
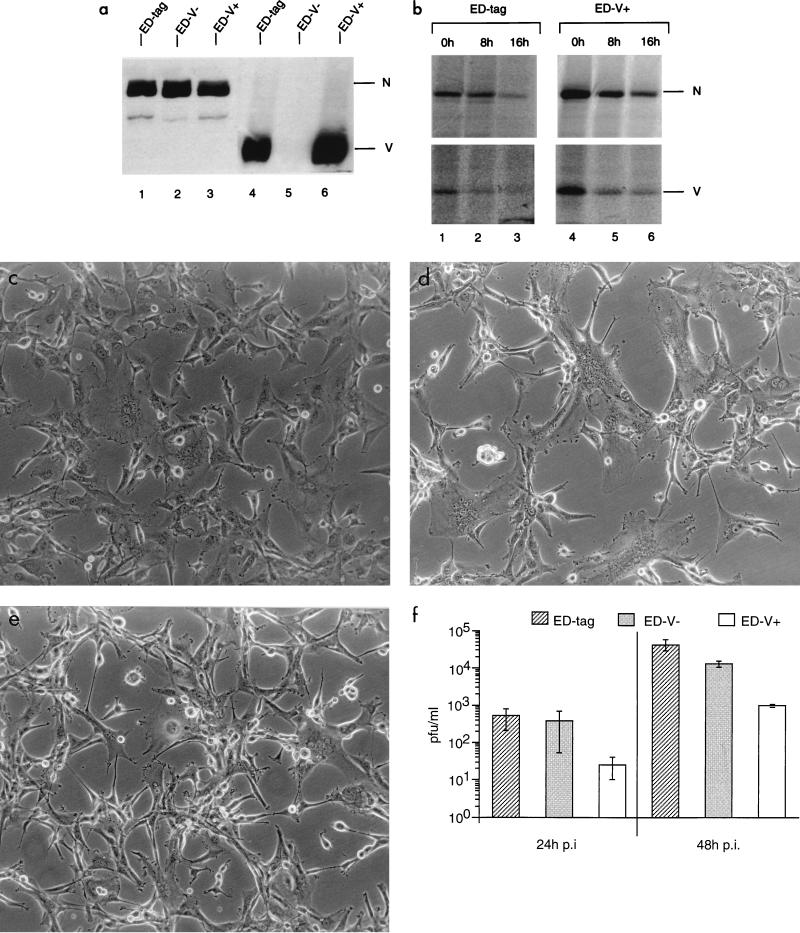
The role of V protein in MV replication was analyzed using a standard MV genome virus (ED-tag) rescued from cloned cDNA (23) and its derivatives in which V protein expression was abrogated by a single nucleotide exchange (ED-V−) or substantially enhanced by the addition of three nucleotides (ED-V+) within the original editing site (25). By the latter manipulation, an additional amino acid was introduced into the V and P protein reading frames. The phenotype of the mutant viruses with respect to the accumulation of V protein was determined by Western blot analyses after infection of U-87 glioblastoma cells (Fig. 1a). To determine whether the enhanced accumulation of V protein in U-87 cells infected with ED-V+ was based on a higher turnover rate of this protein in ED-tag-infected cells, U-87 cells were infected with ED-tag (multiplicity of infection [MOI] of 0.5) or ED-V+ (MOI of 5) (different MOIs were used for reasons outlined below) for 16 h, labeled for 3 h, and subsequently chased for 8 or 16 h, respectively (Fig. 1b). Based on the values obtained after the 8-h chase, the half-lives of the N and V proteins were found to be similar, if not shorter, in U-87 cells infected with ED-V+ (4.5 and 5.5 h, respectively) compared to those found for the parental ED-tag (7.5 h for both proteins). Thus, enhanced accumulation of V in ED-V+-infected cells was apparently not due to an abnormal stability of this protein.
FIG. 1.
Phenotypic characterizations, cytopathic effects, and titers of virus release of ED-tag, ED-V−, and Ed-V+ after infection of human glioblastoma U-87 cells (MOI of 0.1). (a) Cell extracts were harvested 40 h p.i. and the expression of N protein was adjusted to similar amounts as defined by Western blotting (lanes 1 to 3). Identical lysate amounts were then used to detect V protein expression (lanes 4 to 6). (b) U-87 cells were infected with ED-tag (MOI of 0.5) or ED-V+ (MOI of 5) for 16 h and labeled for 3 h and then were harvested immediately (lanes 1 and 4) or chased for 8, (lanes 2 and 5) or 16 h (lanes 3 and 6). A monoclonal MV N-specific antibody or a V-specific serum was used for immunoprecipitation, and the corresponding signals were quantified by phosphorimaging. (c to e) Syncytium formation in U-87 cells infected with ED-tag (c), ED-V− (d), or ED-V+ (e) 24 h p.i. (f) Supernatants of U-87 cells infected with ED-tag, ED-V−, or ED-V+ for 24 or 48 h were titrated in duplicate by standard plaque assays. Values shown are the means of three independent experiments.
Following infection of U-87 cells with ED-tag, ED-V−, or ED-V+ (MOI of 0.1), differences in the development of cytopathic effects were observed after 24 h. The formation of syncytia was markedly enhanced in U-87 cells infected with ED-V− (Fig. 1d) compared to that in cells infected with ED-tag (Fig. 1c), whereas cytopathic effects were less pronounced in cultures infected with ED-V+ (Fig. 1e). Similar results with respect to syncytium formation were obtained in lymphoblastoid B cells (BJAB) or monocytes (U-937) infected with the different viruses (not shown). Later in infection, U-87 cells infected with ED-V− disintegrated much earlier than those infected with ED-tag, while ED-V+-infected cultures survived substantially longer (not shown). Titers of infectious virus released from U-87 cells infected with ED-tag and ED-V− were similar 24 h after infection, whereas those after ED-V+ infection were consistently lower (Fig. 1f). After 48 h, MVs lacking or overexpressing V proteins yielded lower titers of infectious MV than the parental ED-tag (Fig. 1f).
MV-specific protein expression in U-87 cells infected with ED-tag, ED-V−, or ED-V+.
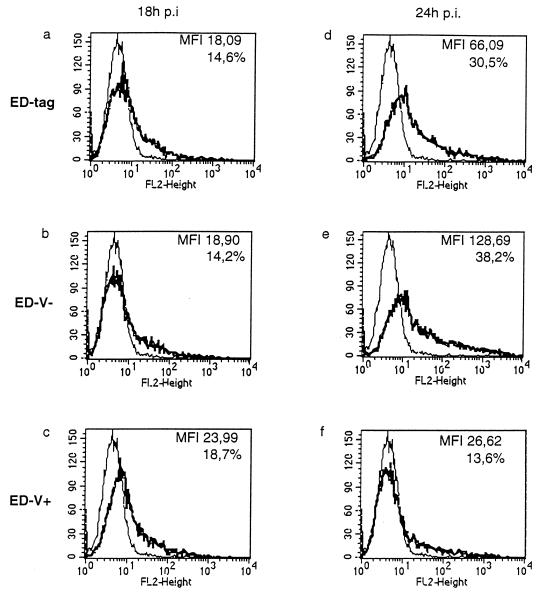
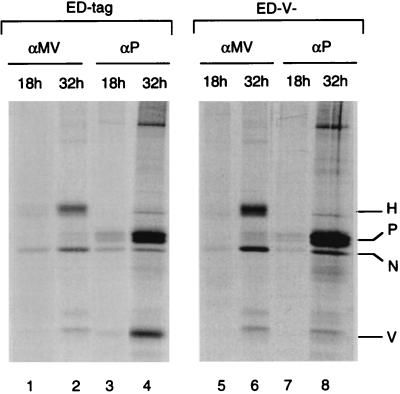
U-87 cells were infected with ED-tag, ED-V−, or ED-V+ (MOI of 0.1) and analyzed by indirect immunofluorescent staining for expression of the MV N protein after 18 h (Fig. 2a to c) or 24 h (Fig. 2d to f). No significant differences were observed after 18 h in the numbers of cells expressing N protein or in the intracellular accumulation levels (indicated by the mean fluorescent intensity) among the viruses. In contrast, at 24 h postinfection (p.i.) the numbers of infected cells were higher in U-87 cells infected with ED-tag or ED-V− (Fig. 2d and e) than in cells infected with ED-V+ (Fig. 2f). Moreover, the levels of N protein in ED-V−-infected U-87 cells was significantly increased (Fig. 2e), indicating that the absence or overexpression of V protein was associated with an augmented or restricted accumulation of virus-specific proteins, respectively. To assess MV protein synthesis, U-87 cells were metabolically labeled 13 or 27 h after infection with an MOI of 0.5 of ED-tag (Fig. 3, lanes 1 to 4) or ED-V− (Fig. 3, lanes 5 to 8), and virus-specific proteins were immunoprecipitated from cell lysates with a polyclonal anti-MV hyperimmune serum (Fig. 3, lanes 1, 2, 5, and 6) or a P-monospecific serum (Fig. 3, lanes 3, 4, 7, and 8). The P-specific antiserum also precipitated V protein in extracts from ED-tag-infected U-87 cells 32 h p.i. (Fig. 3, lane 4). As revealed by quantification, the increase in MV N and P protein synthesis after 32 h of infection was up to 10-fold higher in cells infected with ED-V− than in ED-tag-infected cells. The increase in H protein synthesis was about fivefold higher in the ED-V−-infected than in the ED-tag-infected cells. This suggested that the higher steady-state levels of MV-specific proteins observed by immunofluorescent staining (Fig. 2d to f) resulted from a higher rate of virus-specific protein synthesis.
FIG. 2.
Quantitative analysis of MV N protein expression in U-87 cells infected (MOI of 0.1) with ED-tag (a and d), ED-V− (b and e), or ED-V+ (c and f) for 18 (a to c) or 24 (d to f) h. In each panel, the mean fluorescence intensity (MFI) and the percentage of cells staining for N protein are indicated. As a negative control, uninfected U-87 cells were stained with the MV N-specific antibody.
FIG. 3.
MV-specific protein synthesis in U-87 cells infected at an MOI of 0.5 with ED-tag (lanes 1 to 4) or ED-V− (lanes 5 to 8) for 18 h (lanes 1, 3, 5, and 7) or 32 h (lanes 2, 4, 6, and 8) (each including a 5-h labeling period) was analyzed after precipitation of whole-cell lysates with MV hyperimmuneserum (lanes 1 and 2 and 5 and 6) or a P-specific antiserum (lanes 3 and 4 and 7 and 8). MV N-, P-, and H-specific signals were quantified by phosphorimaging.
Accumulation of MV-specific transcripts in U-87 cells infected with ED-tag, ED-V−, or ED-V+.
U-87 cells were infected with ED-tag, ED-V−, or ED-V+ (MOI of 0.1), and total cellular RNA was harvested 18 and 24 h (Fig. 4a) later and analyzed for the accumulation of MV-specific transcripts by using N (top)- and H (bottom)-specific RNA probes. RNA concentrations were controlled by using a GAPDH-specific probe (not shown). As early as 18 h p.i., N-specific transcripts accumulated to higher levels in cells infected with ED-V− than in cells infected with ED-tag and were barely detectable in U-87 cells infected with ED-V+ (H-specific signals could not be reliably quantified). Similarly, at 24 h p.i., the N-specific mRNA accumulated at significantly lower levels in ED-V+-infected cells and at higher levels in ED-V−-infected cells than in cells infected with ED-tag. As evidenced by quantification, H-specific signals were similar after infection with ED-tag or ED-V− after 24 h (Fig. 4a). Interestingly, the relative amount of the H-specific transcript was higher in ED-V− (N/H ratio of 7.3%) than in Ed-tag-infected cells (N/H ratio of 5.1%). H-specific signals were barely detectable in U-87 cells infected with ED-V+ and thus could not be reliably quantified. The same applied to signals for genomic transcripts of negative sense, which were always below our detection level. Signals corresponding to the antigenomic RNA could be detected only after 24 or 32 h after infection with ED-tag and ED-V− and were barely visible in ED-V+-infected U-87 cells (Fig. 4b).
FIG. 4.
(a) Northern blot analysis of total RNAs isolated from U-87 cells infected at an MOI of 0.1 with ED-tag, ED-V−, or ED-V+ for 18 or 24 h using MV N- and H-specific 32P-labeled riboprobes. Values obtained after quantification of the signals by phosphorimaging are indicated (in thousands). ni, not infected; nd, not detectable. (b) Total RNA from U-87 cells infected with ED-tag, ED-V−, or ED-V+ (MOI of 0.1) for 24 or 32 h was analyzed for expression of the MV antigenomic transcript by using a 32P-labeled N-specific riboprobe. Values obtained after phosphorimaging are indicated (in thousands). (c) U-87 cells were infected with ED-tag (MOI of 1), ED-V− (MOI of 0.5), or ED-V+ (MOI of 5), and total RNA was harvested after 18 h and analyzed for the accumulation of MV N-, M-, and H-specific mRNA as well as cellular GAPDH by using 32P-labeled riboprobes. Values for the signals are indicated (in thousands) after normalization for equal amounts of GAPDH (factors were 1.0 for ED-tag-, 0.48 for ED-V+-, and 0.89 for ED-V−-infected cells). For the individual infections, the N mRNA expression was set at 100%, and the values obtained for the corresponding M- or H-specific signals are indicated as percentages relative to N expression. The values were determined on the basis of counts and do not reflect copy numbers of the individual transcripts.
To determine whether there would be an effect on the transcriptional gradient, U-87 cells were infected with ED-tag (MOI of 1), ED-V− (MOI of 0.5), or ED-V+ (MOI of 5) (different MOIs were used to account for the differences in the overall accumulation levels of N-specific transcripts [Fig. 4a]) and the expression of the MV N-, M-, and H-specific monocistronic transcripts was analyzed after 18 h (Fig. 4c). Signals obtained were normalized to that of the corresponding loading control (GAPDH). As evidenced by the ratio of the counts obtained for the M- and H-specific signals to that of each corresponding N-specific signal, the gradient of mRNA expression appeared less polar after ED-V− infection compared to ED-tag infection, which again was less polar than that found after ED-V+ infection (Fig. 4c). The effect was most pronounced for the accumulation of H-specific transcripts, where the expression levels were about 50% higher in ED-V− (N/H ratio of 8.1%) and about 50% lower in ED-V+-infected U-87 cells (N/H ratio of 2.8%) than in cells infected with ED-tag (N/H ratio of 5.4%). These findings suggest that in the presence of high levels of V protein, not only is there an overall reduction of viral RNA synthesis but the slope of the gradient of MV-specific mRNAs is steeper than that for the parental ED-tag, whereas opposite effects were observed in the absence of V.
Recombinant MVs with altered V protein expression are attenuated in vivo.
Due to an intracellular block in MV replication, mice and rats (either transgenic for CD46 or their nontransgenic littermates [13, 21]) are not susceptible to experimental peripheral MV infection. We have recently adopted an experimental model in cotton rats (Sigmodon hispidus) in which MV grows to peak titers in lung tissue on days 4 and 5 after i.n. infection (20). Cotton rats were i.n. inoculated with 105 PFU of ED-tag, ED-V−, or ED-V+, and lung tissues were obtained for virus titration after 4 and 5 days. On day 4 p.i., lower titers of infectious virus were isolated from animals infected with either editing mutant than from those infected with the parental strain. On day 5 p.i., attempts to rescue infectious virus from animals infected with ED-V+ failed, and the yields of ED-V− were still lower than those obtained from animals infected with ED-tag (Fig. 5). Thus, as in tissue culture (Fig. 1e), both the absence and the overexpression of V protein are associated with a reduction of titers of infectious virus in vivo.
FIG. 5.
Titers of infectious virus (50% tissue culture infectious dose [TCID50]) per gram of lung tissue of cotton rats 4 and 5 days after i.n. infection with ED-tag, ED-V−, or ED-V+. Titers shown are means for each six animals and were determined by a standard plaque technique.
V protein forms complexes with N protein.
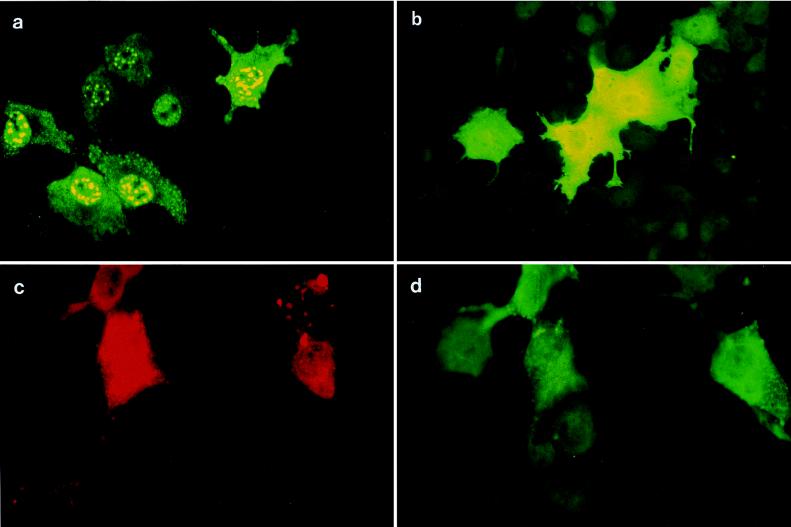
To study the interactions of MV ribonucleoprotein components and V, the subcellular distribution of N and V proteins was analyzed after transient transfection of pSC-N and pSC-V (in which the expression of N and V proteins was driven by a cytomegalovirus promoter) in Cos-7 cells. When expressed alone, N protein was found in large aggregates, mainly in the nucleus and to a lesser extent in the cytoplasm (Fig. 6a), whereas V protein was homogeneously distributed within the cell (Fig. 6b). Upon coexpression with V, N protein aggregates were significantly diminished and the protein localized mainly to the cytoplasm, while cells singly expressing N protein (but not V) maintained the nucleocapsidlike structures (Fig. 6c and d). Thus, redistribution of N protein upon coexpression with V in Cos-7 cells indicated physical interaction between these proteins. To confirm this, we fused the coding sequences of MV N, P and V proteins in-frame either carboxy terminally to the DNA-binding domain of Gal4 (to yield pGalN and pGalV) or amino terminally to the transactivating domain of VP16 (to yield pNVP16 and pPVP16) as schematically shown in Fig. 7a. The expression of the fusion proteins was controlled by immunofluorescent staining after transient transfection (not shown). U-87 cells were transfected with the fusion constructs along with the pG5BCAT reporter plasmid, where reporter gene activation could occur only upon reconstitution of the Gal4-VP16 hybrid transactivator. Transfection of either fusion construct alone (with or without the heterologous parental fusion vector, i.e., pGal4 or pAASVVP16) did not induce reporter gene activation, and neither did cotransfection of pGal4 and pAASVVP16 (not shown). As expected, high levels of reporter gene activation were observed after cotransfection of pGalN and pPVP16 (Fig. 7b). Cotransfection of pGalV with MV N fusion constructs confirmed the formation of N-V complexes (Fig. 7b). Only very weak interactions between P and V were reproducibly observed in this system (not shown). To test whether N-V complexes would also be formed in the presence of P protein, triple transfections were performed with two interacting partners expressed as fusion proteins (GalN, PVP16, or GalV) and the third protein expressed from a cytomegalovirus promoter (either pSC-V or pSC-P). As a control, an empty vector plasmid (pCMV) was used. While cotransfection of pCMV interfered with the formation of both N-P and N-V complexes only to a minor extent, N-P complexes were strongly sensitive to coexpression of V (from pSC-V) as were N-V complexes to coexpression of P protein (from pSC-P) (Fig. 7b). To gain insight into the relative affinities of V and P for N, the amount of competitor plasmid added to the N-P or N-V complex was titrated (Fig. 7c). The N-V complex (Fig. 7c) was equally sensitive to competition by V and P, whereas slightly higher amounts of pSC-V than pSC-P were required to interfere with N-P complex formation (Fig. 7c, upper).
FIG. 6.
Cos-7 cells were transfected with pSC-N (a), pSC-V (b), or pSC-N and pSC-V (c and d) and stained with monoclonal anti-N (a and c) or polyclonal anti-V (b and d) serum. In panels c and d the same field of a culture doubly transfected with pSC-N and pSC-V is shown in which V protein was detected by a fluorescein isothiocyanate-coupled secondary antibody and therefore, for N protein detection, a secondary antibody giving a red stain (coupled to phycoerythrin) had to be applied. Note that after double transfection, in cells not expressing V (panels c and d, upper right corners), N protein retained its punctate distribution.
FIG. 7.
(a) Schematic outline of the coding sequence of MV N, P, or V fused to the DNA-binding domain of Gal4 or the transactivating domain of VP16. (b) Plasmid constructs were transfected in triplicate assays either in doublets (GalN-PVP16 or GalV-NVP16) or together with 5 μg of nonspecific (pCMV) or specific (pSC-V or pSC-P) competitor plasmids into U-87 cells along with the indicator plasmid. (c) Titrations were performed after cotransfection of GalP-NVP16 or GalV-NVP16 with pSC-P or pSC-V (at the concentrations indicated) along with the reporter plasmid. For the results shown in panels b and c, lysates were harvested after 48 h and chloramphenicol acetyltransferase (CAT) activities were determined. Values are the means for two independent experiments.
DISCUSSION
The role of nonstructural proteins in the replication of viruses of the family Mononegavirales is still unknown. The fact that most of the paramyxoviruses edit their second reading frame suggests that these proteins are essential for viral replication in vivo. For MV, this assumption is further supported by the findings that the V reading frame reveals a high degree of sequence conservation (1), and is retained even in persistent infections (10). It is, however, also apparent that V protein function is dispensable for viral replication in cell culture, since MV editing mutants, regardless of whether they overexpress or lack V protein, have been successfully rescued from cloned cDNA and can be propagated normally (25). To further analyze the role of V protein, we compared the replication of a standard MV (ED) (rescued from cloned cDNA) and two editing variants. In ED-V+, a CCC stretch was inserted into the editing region (into the template strand) thus adding 1 aa to both the P and V proteins. To obey the rule of six, three nucleotides were deleted in the H to L intergenic region. The recombinant virus has been found to overexpress V protein (25) (Fig. 1a). As indicated by pulse-chase experiments, the high accumulation levels of V protein after ED-V+ infection were likely to result from an enhanced rate of V protein synthesis rather than an abnormal stability of this protein as previously suggested (25) (Fig. 1b). V protein expression in ED-V− has been abolished by the replacement of a uridine with a cytidine in the editing sequence (again in the template strand). As also noted for SeV (9, 15), abrogation of the editing process did not result in markedly enhanced accumulation levels of P protein (reference 25 and our own observations).
We now show that MV gene expression is affected in cell culture by the presence of V protein, as an editing mutant deficient for V expression accumulated viral mRNAs and proteins more rapidly than the parental strain while transcription of an MV overexpressing V was significantly attenuated in U-87 cells (Fig. 4a and c). In general, the effect of V overexpression (ED-V+) was found to be more pronounced than that observed in the absence of V. Since as a nonstructural protein V is synthesized only after primary transcription, and its expression peaks at 16 h p.i. (30), it is likely that secondary rather than primary MV RNA synthesis was affected. The effects of overexpression or lack of V protein on the replication of antigenomic RNA were similar to those on overall transcription. These data do not allow us to determine whether V-mediated control primarily affects genome replication (as revealed by the accumulation of the antigenomic RNA), which would lower the amount of template for transcription, or affects only transcription of monocistronic mRNAs, which, in turn, would lower the rate of genome replication. As, however, the accumulation of monocistronic mRNAs along the genome was apparently sensitive to the absence or overexpression of V, it is likely that transcription is directly affected (Fig. 4c). Observations made with SeV V− variants (15) suggest that the major effect of V would be on transcription, whereas in in vitro studies using mixed cell extracts trans-supplementation with V showed impaired replication but not transcription of SeV defective interfering particle genomes (7, 12). As predicted by the latter studies, the presence of the P protein N-terminal domain within V should enable its interaction with L in a manner similar to that of P protein (7), yet such an interaction has not been experimentally verified. On the other hand, SeV V protein has been shown to interact with N protein, however, not with N assembled into nucleocapsids, confirming that V does not interact with nucleocapsid structures (12).
When expressed alone, a large proportion of the MV N proteins localized to the nuclear compartment, predominantly as large aggregates (14), although we also observed some smaller aggregates in the cytoplasm (Fig. 6). Coexpression of V clearly was associated with a redistribution of N, indicating that the proteins interacted. This interpretation was further supported by our findings with a two-hybrid approach in U-87 cells which revealed that N-V complexes are formed and V protein even competed for N-P complexes (Fig. 7b and c). A domain located within aa 204 and 321 of P was found to be essential for retention of N in the cytoplasm (14), and this domain only partially overlaps with the V reading frame. Thus, the N-protein-interacting domain of V would have to localize between aa 204 and 231 or an additional domain located far more upstream at the amino terminus would have to be involved. In fact, studies with several paramyxoviruses and rhabdoviruses have confirmed that there are two binding sites on P for N, one at the amino terminus and the other at the carboxy terminus (2, 6, 24, 27). In transient-transfection assays, MV P proteins in which aa 2 to 204 were deleted were shown to be less efficient in retaining N protein in the cytoplasm than P proteins of full length (14), and more recently, a two-hybrid approach revealed that amino-terminal deletions of the P protein led to a dramatic loss of N protein interaction (11). These results together with our findings strongly suggest that for MV an amino-terminal domain of P as well as V protein can bind to N protein. As R protein also contains the amino-terminal domain of P and V, its interaction with N protein could be predicted. As this protein, however, is expressed only at very low levels (17) this interaction may not be of functional importance.
In contrast to these findings, MV V protein did not interact with N protein when expressed as glutathione S-transferase fusion protein or in a yeast two-hybrid system (18). The most likely explanation for this discrepancy is that different cell systems were used in these studies. Although not directly assayed, the effect of V on MV replication may also be dependent on the cell type. Whereas V-dependent differences in the accumulation levels of MV proteins and virus production were quite obvious in U-87 (Fig. 1f and 2), BJAB, and U-937 cells (not shown), these differences were less apparent in Vero cells where no differences in titers of infectious virus released were observed (25). Interestingly, a cell-type dependency for the V− phenotype of SeV was noted which has been tentatively ascribed to the interaction of V with cellular proteins that would modify its activity (15). In this respect, it is interesting to note that so far unidentified host cell proteins have been shown to interact with MV V protein (18).
Our data clearly show that replication of MV editing mutants was not only attenuated in cell culture but also after i.n. infection in cotton rats as indicated by the lower titers recovered from lung tissue (Fig. 5). Similar observations were recently made using an SeV in which V protein expression was completely abrogated by a mutation of the editing site, similar to our ED-V− variant (15). Both with single-cycle replication conditions and multiple rounds of replication, the SeV V− was found to grow slightly faster than the parental nondefective virus, and an altered plaque morphology and increased cytopathogenicity were also noted. Similar to that observed for ED-V−, an augmented viral protein synthesis, which was due to increased transcription of viral mRNAs, was observed after infection with SeV V−. Moreover, reinforced expression of a foreign gene introduced into an SeV V− has also been described (31). When inoculated into mice, the V− variant was much less pathogenic than the parental strain and did not spread in the epithelium, and viral titers obtained from the lungs were significantly reduced. Using another mutant encoding only the amino-terminal half and not the V-specific carboxy-terminal half of V (VΔC), the pathogenicity determinant of V protein was mapped to its carboxy terminus (16). It is, however, important to note that this particular mutant VΔC is largely homologous to a natural editing product of SeV, the W protein, and a corresponding protein is not present in MV (29). Thus, the presence or overexpression of the amino-terminal half of P might well have different effects on the pathogenicity of SeV and MV.
Both SeV and MV lacking V proteins are attenuated in vivo (Fig. 5) (15). As is evident from observations made in cell culture, an augmented synthesis of viral mRNAs and/or proteins might not be tolerated well by infected cells, as indicated by their enhanced cytopathogenicity. In vivo, this may favor rapid destruction of the host cell prior to efficient viral assembly and release. The attenuated phenotype of ED-V+ is most probably due to the strongly reduced levels of viral transcripts and proteins (Fig. 2 and 4), which may prevent efficient spread in vivo. Whether this attenuated intracellular gene expression might predispose the viruses overexpressing V to establish persistent infections remains to be investigated. It is, however, still possible that the nucleotide phase shift between the insert at the editing site and the deletion in the 3′-nontranslated H region may have an additional effect on the phenotype of the ED-V+.
Attenuation of MV transcription is a key feature of the primary interaction between MV and neural cells in vitro and of viral persistence in the central nervous system after both natural and experimental infection (for a review, see reference 26). So far, although V protein expression has been documented in cell cultures persistently infected with MV (3a, 10), its expression levels relative to other viral proteins have not been particularly addressed. Altered P mRNA editing has been described for a hamster neurotropic strain (HNT) of MV; however, a predominance of nonedited P transcripts has been found in both lytic and persistent infection (28). It will thus be interesting to investigate the pathogenic potential of MV editing mutants in experimental brain infections in rodents.
ACKNOWLEDGMENTS
We thank Bert Rima for helpful discussion and Ute Brinckmann for providing unpublished observations.
Financial support was provided by the Deutsche Forschungsgemeinschaft, the Bundesministerium für Forschung und Technologie, the Wellcome Trust, and the Schweizerische Nationalfonds.
REFERENCES
- 1.Baczko K, Pardowitz J, Rima B K, ter Meulen V. Constant and variable regions of measles virus proteins encoded by the nucleocapsid and phosphoprotein genes derived from lytic and persistent viruses. Virology. 1992;190:469–474. doi: 10.1016/0042-6822(92)91236-n. [DOI] [PubMed] [Google Scholar]
- 2.Barr J, Easton A J. Characterisation of the interaction between the nucleoprotein and phosphoprotein of pneumonia virus of mice. Virus Res. 1995;39:221–235. doi: 10.1016/0168-1702(95)00090-9. [DOI] [PubMed] [Google Scholar]
- 3.Bellini W J, Englund G, Arnheiter H, Rozenblatt S, Richardson C D. Measles virus P gene codes for two proteins. J Virol. 1985;53:908–919. doi: 10.1128/jvi.53.3.908-919.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Brinkmann, U. G. Unpublished observations.
- 4.Cattaneo R, Rebmann G, Schmid A, Baczko K, ter Meulen V, Billeter M A. Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 1987;6:681–687. doi: 10.1002/j.1460-2075.1987.tb04808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattaneo R, Kaelin K, Baczko K, Billeter M A. Measles virus editing provides an additional cysteine rich protein. Cell. 1989;56:759–764. doi: 10.1016/0092-8674(89)90679-x. [DOI] [PubMed] [Google Scholar]
- 6.Chenik M, Chebli K, Gaudin Y, Blondel D. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J Gen Virol. 1994;75:2889–2896. doi: 10.1099/0022-1317-75-11-2889. [DOI] [PubMed] [Google Scholar]
- 7.Curran J, Boeck R, Kolakofsky D. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 1991;10:3079–3085. doi: 10.1002/j.1460-2075.1991.tb07860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das T, Schuster A, Schneider-Schaulies S, Banerjee A K. Involvement of cellular casein kinase II in the phosphorylation of measles virus P protein: identification of phosphorylation sites. Virology. 1995;211:218–226. doi: 10.1006/viro.1995.1394. [DOI] [PubMed] [Google Scholar]
- 9.Delenda C, Hausmann S, Garcin D, Kolakofsky D. Normal cellular replication of Sendai virus without the trans-frame, nonstructural V protein. Virology. 1997;228:55–62. doi: 10.1006/viro.1996.8354. [DOI] [PubMed] [Google Scholar]
- 10.Gombart A F, Hirano A, Wong T C. Expression and properties of the V protein in acute measles virus and subacute sclerosing panencephalitis virus strains. Virus Res. 1992;25:63–78. doi: 10.1016/0168-1702(92)90100-n. [DOI] [PubMed] [Google Scholar]
- 11.Harty R N, Palese P. Measles virus phosphoprotein (P) requires the NH2- and COOH-terminal domains for interactions with the nucleoprotein (N) but only the COOH terminus for interactions with itself. J Gen Virol. 1995;76:2863–2867. doi: 10.1099/0022-1317-76-11-2863. [DOI] [PubMed] [Google Scholar]
- 12.Horikami S M, Smallwood S, Moyer S A. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology. 1996;222:383–390. doi: 10.1006/viro.1996.0435. [DOI] [PubMed] [Google Scholar]
- 13.Horvat B, Rivailler G, Varior-Krishnan G, Cardoso A, Gerlier D, Rabourdin-Combe C. Transgenic mice expressing human measles virus (MV) receptor CD46 provide cells exhibiting different permissivities to MV infection. J Virol. 1996;70:6673–6681. doi: 10.1128/jvi.70.10.6673-6681.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber M, Cattaneo R, Spielhofer P, Örvell C, Norrby E, Messerli M, Perriard J C, Billeter M A. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology. 1991;185:299–308. doi: 10.1016/0042-6822(91)90777-9. [DOI] [PubMed] [Google Scholar]
- 15.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 1997;16:578–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato A, Kitoyani K, Yoshida T, Shioda T, Nagai Y. Importance of the cysteine-rich carboxyl-terminal half of V protein for Sendai virus pathogenesis. J Virol. 1997;71:7266–7272. doi: 10.1128/jvi.71.10.7266-7272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liston P, Briedis D J. Ribosomal frameshifting during translation of measles virus P protein mRNA is capable of directing the synthesis of a unique protein. J Virol. 1995;69:6742–6750. doi: 10.1128/jvi.69.11.6742-6750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liston P, Diflumeri C, Briedis D J. Protein interactions entered into by measles virus P, V and C proteins. Virus Res. 1995;38:241–259. doi: 10.1016/0168-1702(95)00067-z. [DOI] [PubMed] [Google Scholar]
- 19.Liston P, Briedis D J. Measles virus V protein binds zinc. Virology. 1994;198:399–404. doi: 10.1006/viro.1994.1050. [DOI] [PubMed] [Google Scholar]
- 20.Niewiesk S, Eisenhuth I, Fooks A, Clegg J, Schnorr J J, Schneider-Schaulies S, ter Meulen V. Measles virus-induced immune suppression in the cotton rat (Sigmodon hispidus) model depends on viral glycoproteins. J Virol. 1997;71:7214–7219. doi: 10.1128/jvi.71.10.7214-7219.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niewiesk S, Schneider-Schaulies J, Ohnimus H, Jassoy C, Schneider-Schaulies S, Diamond L, Logan J S, ter Meulen V. CD46 expression does not overcome the intracellular block of measles virus replication in transgenic rats. J Virol. 1997;71:7969–7973. doi: 10.1128/jvi.71.10.7969-7973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radecke F, Billeter M A. The nonstructural C protein is not essential for multiplication of Edmonston B strain measles virus in cultured cells. Virology. 1996;217:418–421. doi: 10.1006/viro.1996.0134. [DOI] [PubMed] [Google Scholar]
- 23.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dötsch C, Christiansen G, Billeter M A. Rescue of measles viruses from cloned cDNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Randall R E, Bermingham A. NP:P and NP:V interactions of the paramyxovirus simian virus 5 examined using a novel protein:protein capture assay. Virology. 1996;224:121–129. doi: 10.1006/viro.1996.0513. [DOI] [PubMed] [Google Scholar]
- 25.Schneider H, Kaelin K, Billeter M A. Recombinant measles viruses defective for RNA editing and V protein synthesis are viable in culture. Virology. 1997;227:314–322. doi: 10.1006/viro.1996.8339. [DOI] [PubMed] [Google Scholar]
- 26.Schneider-Schaulies S, Schneider-Schaulies J, Dunster L M, ter Meulen V. Measles virus gene expression in neural cells. Curr Top Microbiol Immunol. 1995;191:101–115. doi: 10.1007/978-3-642-78621-1_7. [DOI] [PubMed] [Google Scholar]
- 27.Takacs A M, Das T, Banerjee A K. Mapping of interaction domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two-hybrid system. Proc Natl Acad Sci USA. 1993;90:10375–10379. doi: 10.1073/pnas.90.21.10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanchiere J A, Bellini W J, Moyer S A. Hypermutation of the phosphoprotein and altered mRNA editing in the hamster neurotropic strain of measles. Virology. 1995;207:555–561. doi: 10.1006/viro.1995.1116. [DOI] [PubMed] [Google Scholar]
- 29.Vidal S, Curran J, Kolakofsky D. A stuttering model for paramyxovirus P mRNA editing. EMBO J. 1990;9:2017–2022. doi: 10.1002/j.1460-2075.1990.tb08330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wardrop E A, Briedis D J. Characterization of V protein in measles virus-infected cells. J Virol. 1991;65:3421–3428. doi: 10.1128/jvi.65.7.3421-3428.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu D, Shioda T, Kato A, Hasan M K, Sakai Y, Nagai Y. Sendai virus-based expression of HIV-1 gp120: reinforcement by the V(−) version. Genes Cell. 1997;2:457–466. doi: 10.1046/j.1365-2443.1997.1340332.x. [DOI] [PubMed] [Google Scholar]