Abstract
JC polyomavirus (JCV), the causative agent of progressive multifocal leukoencephalopathy (PML), is ubiquitous in humans, infecting children asymptomatically and then persisting in the kidney. Renal JCV is not latent but replicates to excrete progeny in the urine. The renal-urinary JCV DNAs carry the archetype regulatory region that generates various rearranged regulatory regions occurring in JCVs derived from the brains of PML patients. Tissue cultures that support the efficient growth of archetype JCV have not been reported. We studied whether archetype JCV could replicate in COS-7 cells, simian cells transformed with an origin-defective mutant of simian virus 40 (SV40). Efficient JCV replication, as detected by a hemagglutination assay, was observed in cultures transfected with five of the six archetype DNAs. The progeny JCVs could be passaged to fresh COS-7 cells. However, when the parental cells of COS-7 not expressing T antigen were transfected with archetype JCV DNAs, no viral replication was detected, indicating that SV40 T antigen is essential for the growth of JCV in COS-7 cells. The archetype regulatory region was conserved during viral growth in COS-7 cells, although a small proportion of JCV DNAs underwent rearrangements outside the regulatory region. We then attempted to recover archetype JCV from urine by viral culture in COS-7 cells. Efficient JCV production was observed in COS-7 cells infected with five of the six JCV-positive urine samples examined. Thus, COS-7 cells should be of use not only for the production of archetype JCV on a large scale but also for the isolation of archetype JCV from urine.
JC virus (JCV) was first isolated by Padgett et al. (35) from the affected brain tissue of a patient with a fatal demyelinating disease of the central nervous system known as progressive multifocal leukoencephalopathy (PML). JCV is a member of the polyomavirus genus of the papovavirus family (37) and is closely related to simian virus 40 (SV40) and BK virus (BKV), the other polyomavirus infecting humans (14).
Serologic studies have shown that most humans are asymptomatically infected with JCV in childhood (33, 34). JCV which enters individuals as a primary infection is not completely cured by an immune response and persists in renal tissue throughout life, as shown by several types of evidence. In earlier studies, JCV was frequently detected in the urine of immunocompromised patients, whereas it was rarely detected in the urine of immunocompetent individuals (7). From this observation and the finding that JCV DNA was present in normal renal tissue (11), it was thought that JCV persists in renal tissue in a latent form. However, by using more sensitive methods, we showed that JCV DNA is frequently excreted in the urine of adults without obvious immunodeficiency (22, 23). In addition, we have recently demonstrated that renal JCV represents that which persisted after a primary infection (24).
We molecularly cloned urinary JCV DNAs from two healthy individuals and eight nonimmunocompromised patients (46). The basic structures of the regulatory regions of the cloned DNAs were identical, and the regulatory regions with this structure were thus designated the archetype. JCV DNAs in normal renal tissue also carry the archetype regulatory region (26, 42). Archetype JCV DNAs have been found in the urine of various individuals from around the world (1, 4, 13, 17, 28, 44–46). It was thus thought that archetype JCVs represent JCVs that circulate in the human population (46). (In normal hosts, JCV may also exist in sites other than renal tissue. Indeed, JCV has been recovered from lymphocytes of individuals without PML [reference 15 and references cited therein]. However, the structures of the JCV regulatory regions derived from lymphocytes have rarely been elucidated.)
Before the isolation of archetype JCV DNA from urine, it had been found that regulatory regions (designated PML type) from the brains of PML patients are hypervariable (26, 29, 31). It has been proposed that these PML-type JCVs were generated from archetype JCV strains in the course of persistence in patients (3, 9, 20, 21, 45, 46).
To our knowledge, tissue cultures that support the efficient growth of archetype JCV have not been reported. The lack of appropriate tissue cultures to propagate archetype JCV has greatly hampered biological studies of archetype JCV. Daniel et al. (12) recently reported that a low level of JCV replication, detectable by faint hybridization signals, was induced after the transfection of POJ cells with an archetype JCV DNA (POJ cells are primary human fetal glial cells transformed by an origin-defective mutant of JCV [27]). In this paper, we report that (i) various archetype JCV DNA clones can initiate efficient viral replication in COS-7 cells, which are simian cells constitutively expressing SV40 T antigen (16); (ii) the archetype regulatory region is conserved during the growth of JCV in COS-7 cells; and (iii) archetype JCV in urine can be isolated by using COS-7 cells.
MATERIALS AND METHODS
Cell lines.
COS-7 and CV-1 cells were obtained from the RIKEN Cell Bank (Tsukuba Science City, Japan). COS-7 is a derivative of CV-1 (a cell line established from the kidney of an African green monkey) that was transformed with an origin-defective mutant of SV40 (16). Dulbecco’s modified Eagle medium (DMEM) supplemented with kanamycin (50 μg/ml), amphotericin B (Fungizone) (0.6 μg/ml), and fetal bovine serum (FBS) (10%) was used as maintenance medium for both cell lines.
JCV DNA clones.
The origins of the recombinant JCV DNAs used in this study are shown in Table 1. The JCV DNAs that had been cloned at the BamHI site within the T-antigen gene were recloned at the EcoRI site within the VP1 gene. The recombinant plasmids were prepared by using the Wizard Midipreps DNA Purification System (Promega Corporation, Madison, Wis.) and then purified by centrifugation to equilibrium in CsCl-ethidium bromide gradients.
TABLE 1.
JCV DNA clones used in this study
| Clone(s) | Subtypea | Regulatory region | Origin | Reference |
|---|---|---|---|---|
| MY | MY | Archetype | Non-PML, urine | 46 |
| CY | CY | Archetype | Non-PML, urine | 46 |
| C1 | SC | Archetype | Non-PML, urine | 44 |
| SP-1 | EU | Archetype | Non-PML, urine | 17 |
| GH-1 to -4 | Af1 | Archetype | Non-PML, urine | 17 |
| ZA-1 | Af2 | Archetype | Non-PML, urine | 17 |
| Mad-1 | EU | PML type | PML, brain | 19 |
The classification is that of Sugimoto et al. (40).
Urine samples and their fractionation.
Urine samples were collected from six JCV-positive volunteers or patients without PML who participated in previous studies (23). These were U1 (male, 78 years old), U2 (male, 67 years old), U3 (male, 83 years old), U4 (male, 61 years old), U5 (male, 55 years old), and U6 (male, 79 years old).
The urine samples were fractionated by centrifugation as described previously (22) within the day when they were collected from the donors. Briefly, by centrifugation at 1,500 × g for 20 min at 4°C, urine was fractionated into the urinary sediment and supernatant, containing cell-associated and cell-free JCVs, respectively. The urinary sediment was resuspended in 1 ml of phosphate-buffered saline (PBS), sonicated at 200 W for 3 min, and sedimented at 1,500 × g for 10 min. The resultant supernatant was divided into aliquots and stored at −80°C to be used for the infection of COS-7 cells. The urinary supernatant was centrifuged at 100,000 × g for 3 h at 4°C. The pellet was resuspended in 1 ml of PBS, sonicated, and stored as described above.
Transfection.
The recombinant JCV DNA clones were excised with EcoRI, and linear JCV DNAs were recovered by phenol extraction followed by ethanol precipitation. Cells (1 × 105 to 1.4 × 105) were plated on a 35-mm-diameter dish and cultured overnight in the maintenance medium. Two microliters of Lipofectamine (Life Technologies, Gaithersburg, Md.) was diluted with 100 μl of DMEM, mixed with 100 μl of DMEM containing 1 μg of linear JCV DNA, and left standing at room temperature for 40 min (endonuclease-cleaved linear viral DNA can be recircularized after transfection [10]). After the addition of 800 μl of DMEM, the Lipofectamine-JCV DNA mixture was loaded onto cells grown on a 35-mm-diameter dish (see above). The cells were cultured at 37°C for 5 h, and the medium was then replaced with the maintenance medium. After the cells were incubated for 2 days, they were transferred to a 50-ml flask. The cells were then continuously cultured in the maintenance medium with transfer at a split ratio of 1:5 every 3 or 4 days.
Infection.
COS-7 cells (1.5 × 105) were plated on a 35-mm-diameter dish and cultured overnight in the maintenance medium. Two hundred microliters of a urine fraction containing either cell-associated or cell-free JCV (see above) was mixed with 400 μl of DMEM and added to cells on a 35-mm-diameter dish (see above). After adsorption for 2 h, the medium was replaced with the maintenance medium. The cells were incubated for 2 days and then transferred to a 50-ml flask. Subsequently, the cells were continuously cultured in the maintenance medium with transfer at a split ratio of 1:5 every 3 or 4 days.
The infection with JCV recovered from transfected COS-7 cells was performed as follows. Transfected cells grown on 100-mm-diameter dishes were washed once with PBS, resuspended in PBS, and lysed by sonication at 200 W for 3 min. The cell lysate was diluted with DMEM to a concentration of 1.6 hemagglutination (HA) units (HAU) per ml (see below), and 0.5 ml was used to infect cells. The procedures for infection and subsequent passage of cells were as described above.
To characterize the one-step growth of JCV in COS-7 cells, 5 × 105 COS-7 cells were plated on a 50-ml flask and cultured overnight in the maintenance medium. A virus stock derived from the urine-infected COS-7 cells was diluted with DMEM to concentrations of 1.2, 12, and 120 HAUs per ml, and 0.5 ml was used to infect the COS-7 cells as described above. After adsorption for 2 h, the cells were twice washed with DMEM and cultured in DMEM supplemented with 2% FBS. The cells received a medium change on the third day. On various days after infection, cells were harvested and processed for the assay of the output HA as described above.
HA assay.
Confluent cells in a 50-ml flask were washed once with PBS, resuspended in 1 ml of PBS, and lysed by sonication at 200 W for 3 min. The lysate was treated with 0.1 mg of neuraminidase (type V; Sigma Chemical Co., St. Louis, Mo.) per ml at 37°C overnight, incubated at 56°C for 30 min, and centrifuged at 2,100 × g for 5 min. The resultant supernatant was serially diluted with PBS and assayed for HA by using human type O erythrocytes as described previously (33). The HA titer was defined as the reciprocal of the greatest dilution of the virus suspension with which complete HA was observed.
Indirect immunofluorescence assay.
Infected cells were plated onto 13-mm-diameter coverslips. The medium was removed 3 or 4 days later, and the cells were washed with PBS and then fixed with acetone at room temperature for 10 min. The fixed cells were incubated for 1 h at 37°C with a 1:500 dilution, in PBS, of the rabbit antiserum JCAb1 (6), followed by incubation for 1 h at 37°C with fluorescein-conjugated goat immunoglobulin G fraction to rabbit immunoglobulin G (Cappel, Cochranville, Pa.). JCAb1 is an antibody raised against a peptide of JCV capsid protein (VP1) and lacks cross-reactivity to BKV as well as to SV40 (reference 6 and unpublished data).
Extraction of viral DNA.
Viral DNA was extracted from confluent cells on a 100-mm-diameter dish by the Hirt method (18). The final ethanol precipitate was dissolved in 50 μl of distilled water. Viral DNA in urine was extracted as described previously (22).
PCR.
JCV regulatory regions were amplified by using B1 and B3 as primers (39), and the VT-intergenic regions (IG regions) (8) were amplified by using P1 and P2 (25). (The IG region was previously established as a region of the JCV genome that contains abundant type-determining sites [8].) The total reaction volume of 50 μl contained 1.25 U of KOD polymerase (Toyobo Co., Osaka, Japan), 200 μM each deoxynucleoside triphosphate, 10 mM Tris-HCl (pH 8.0), 1 mM MgCl2, and 0.25 μM primers. The cycle profile was 94°C for 30 s, 50°C for 30 s, and 74°C for 60 s. The amplification of the viral sequences in COS-7 cells was performed with 20 cycles in a thermal sequencer (Iwaki Glass Co., Tokyo, Japan) with 100-times-diluted sample DNAs, whereas the amplification of urinary viral sequences was performed with 50 cycles and undiluted sample DNAs.
Cloning of PCR products.
The amplified fragments were digested with a combination of HindIII and PstI, which excises a fragment containing both the origin of replication and the regulatory region. The digested fragments were ligated to HindIII- and PstI-digested, alkaline phosphatase-treated pUC19 and used to transform competent Escherichia coli HB101 cells (Takara Shuzo Co., Kyoto, Japan).
Cloning of complete viral DNA.
The viral DNA extracted from cells was digested with BamHI, which cleaves the JCV DNA at a single site. The recovered DNA was ligated to BamHI-digested, alkaline phosphatase-treated pUC19 and was used to transform competent E. coli HB101 cells. Colonies containing recombinant plasmids were obtained by colony hybridization with JCV [32P]DNA as the probe (36).
Restriction fragment length polymorphism.
Restriction enzymes were obtained from Takara Shuzo Co. (HincII, HindIII, and PstI) and from Toyobo Co. (BamHI). Double digestion with HindIII and PstI was performed at 37°C in 50 mM Tris-HCl (pH 8.0)–10 mM MgCl2–50 mM NaCl supplemented with 0.1 mg of bovine serum albumin per ml. Double digestion with BamHI and HincII was performed at 37°C in 50 mM Tris-HCl (pH 7.5)–10 mM MgCl2–1 mM dithiothreitol–100 mM NaCl supplemented with 0.1 mg of bovine serum albumin per ml. Digests were analyzed by electrophoresis on 1.5% agarose gels stained with ethidium bromide.
Sequencing of the regulatory region.
Representative recombinant plasmids were prepared by using the Wizard Midipreps DNA Purification System (Promega) and sequenced by using an AutoRead sequencing kit and an ALFred DNA sequencer (Pharmacia-LKB, Uppsala, Sweden).
Sequencing of the complete JCV DNA.
A recombinant plasmid containing the complete JCV DNA was prepared as described above and sequenced by using an AutoCycle sequencing kit and an ALFred DNA sequencer (Pharmacia-LKB). The primers described by Agostini et al. (2) were used. The determined sequence was compared to the complete coding sequences of three JCV strains, 225 (L-254), 226 (L-264), and 228 (L-270) (GenBank accession numbers AF015530, AF015531, and AF015534, respectively) (5).
Nucleotide sequence accession numbers.
The DNA sequence data reported here have been deposited in the GSDB, DDBJ, EMBL, and NCBI nucleotide sequence databases with accession no. AB008765 (dp-1, regulatory region), AB008766 (U1, IG region), AB008767 (U2, IG region), AB0087688 (U4, IG region), AB008769 (U5, IG region), and AB008770 (U6, IG region).
RESULTS
Induction of efficient JCV replication by transfection of COS-7 cells with cloned archetype JCV DNAs.
On the basis of phylogenetic analyses, the archetype JCV DNAs in Europe, Africa, and Asia are classified into nine subtypes that occupy unique territories in the Old World (40). Clones (MY, CY, C1, SP-1, GH-1, and ZA-1) belonging to six subtypes (Table 1) and a PML-type clone (Mad-1) were excised from the vector and introduced into COS-7 cells by using Lipofectamine (an endonuclease-cleaved linear viral DNA can be recircularized after transfection [10]). The transfected cells were grown in the maintenance medium and serially transferred at a split ratio of 1:5 every 3 or 4 days. On day 10, cytopathology (rounding of the cells) started to appear in the cells transfected with all clones except GH-1. Cells were harvested on days 7, 14, 21, 26, 28, and 35 after transfection and assayed for HA as described in Materials and Methods. The data (Table 2) can be summarized as follows. In the archetype-transfected cultures, except those transfected with GH-1, HA began to be detected on day 14 or 21 and reached the highest titers on day 28. The HA titers decreased remarkably on day 35 in most of the transfected cells, probably because of the detachment of infected cells from the surfaces of the culture flasks.
TABLE 2.
Viral replication after transfection with various JCV DNA clones
| Clone | HA titer on day:
|
|||||
|---|---|---|---|---|---|---|
| 7 | 14 | 21 | 24 | 28 | 35 | |
| CY | <2 | 2 | 32 | 64 | 128 | 16 |
| MY | <2 | 4 | 64 | 64 | 256 | 32 |
| C1 | <2 | <2 | 2 | 16 | 128 | 32 |
| SP-1 | <2 | 2 | 16 | 32 | 128 | 16 |
| GH-1 | <2 | <2 | <2 | <2 | <2 | <2 |
| ZA-1 | <2 | <2 | 4 | 128 | 256 | 16 |
| Mad-1 | <2 | 2 | 32 | 32 | 32 | 4 |
In three independent experiments, COS-7 cells were transfected with the same set of JCV DNA clones as described above, and the transfected cells were processed as described above. We repeatedly detected the production of HA in all of the transfected cells except those transfected with GH-1, although the days when HA was first detected varied among the experiments. These results indicated that various archetype JCV DNAs can induce the efficient growth of JCV in COS-7 cells.
In cells transfected with Mad-1, HA appeared as early as in the archetype-transfected cells, but the HA titers in these cells remained at a medium level between days 21 and 28 (Table 2). This is probably because the detachment of infected cells occurred earlier in the Mad-1-transfected cells than in the archetype-transfected cells.
The GH-1-transfected cells continued to be cultured until 72 days, but they did not exhibit any detectable HA. Three other clones belonging to the Af1 subtype (GH-2, -3, and -4) (17, 40) were also used to transfect COS-7 cells, but we did not detect any viral growth in the transfected COS-7 cells.
The Af1 clones used have the normal replication origin and typical archetypal regulatory regions (17). We are currently examining what region of the Af1 genome is responsible for the lack of the capacity to induce viral growth in COS-7 cells.
Various transfected cultures on day 28 (Table 2), except for the GH-1-transfected cells, were lysed and used to infect fresh COS-7 cells (see Materials and Methods). The infected cells were cultured as described above. All infected cells produced an HA titer of 64 to 128 on day 7 after infection. Thus, the JCV produced after transfection with archetype JCV DNAs can be transferred to fresh COS-7 cells.
CV-1 cells (parental cells of COS-7) not expressing SV40 T antigen were similarly transfected with the same set of JCV DNAs as described above. On various days from 14 to 61 days after transfection, cells were harvested and assayed for HA, but no HA was detected on any day. By using PCR targeting the regulatory region, we also examined the replication of JCV in transfected CV-1 cells, but we detected no JCV replication (data not shown). Therefore, SV40 T antigen is essential for the growth of JCV in COS-7 cells.
Quantitative analysis of the JCV DNAs produced in COS-7 cells.
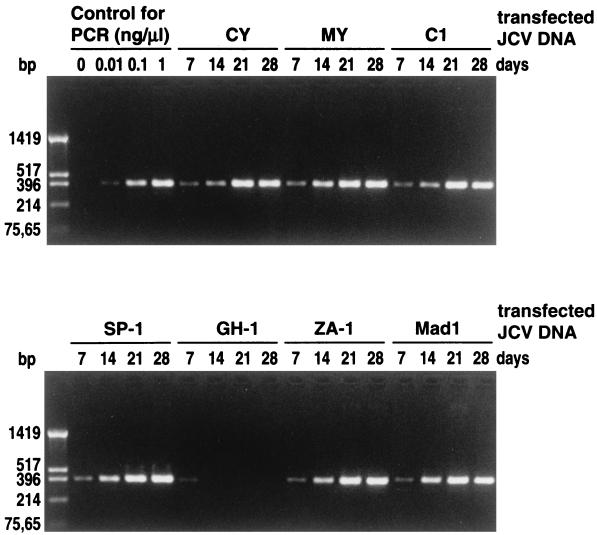
Viral DNAs extracted from cultures 7, 14, 21, or 28 days after transfection were diluted 100-fold and used for the amplification of the JCV regulatory region. Aliquots of the amplification mixtures were electrophoresed on an agarose gel (Fig. 1). The amounts of amplified fragments from all of the transfected cells except the GH-1-transfected cells increased with the incubation time. In the cells transfected with GH-1, a low level of amplification was seen on day 7, but the amplification ceased on subsequent days. These data indicated that the observed amplification on days 14 to 28 (Fig. 1) arose from progeny JCVs rather than from transfected JCV DNAs.
FIG. 1.
Detection of JCV regulatory regions from COS-7 cells transfected with various cloned JCV DNAs. COS-7 cells were transfected with cloned archetype JCV DNAs (CY, MY, C1, SP-1, GH-1, and ZA-1) and a cloned PML-type JCV DNA (Mad-1) and cultured for the indicated number of days. Viral DNAs extracted from the cells were diluted 100 times with water and used as the template to amplify the JCV regulatory region. Amplification was performed with 20 cycles under the conditions described in Materials and Methods. Serially diluted JCV DNA clone CY was used as the control for amplification. The products were resolved by electrophoresis on a 1.5% agarose gel stained with ethidium bromide. The sizes of the HinfI fragments of pUC19 are indicated to the left.
By comparing the densities of amplified fragments in the agarose gel for JCV DNAs from COS-7 cells and standard JCV DNAs (Fig. 1), the yields of JCV DNAs on day 28 were roughly estimated to be 5 to 10 μg per 25-cm2 flask for the transfected cells, except for those transfected with GH-1.
A quantitative discrepancy between the HA (Table 2) and PCR (Fig. 1) data was found. Thus, large amounts of amplified fragments were detected for some transfected cultures (e.g., the day 21 cells transfected with C1 and ZA-1) for which the HA titers were low. Although the reason for this is not clear, we speculate that JCV DNA replication may be enhanced in COS-7 cells transfected with C1 and ZA-1.
Structural analysis of the JCV DNAs that replicated in COS-7 cells.
The sizes of the amplified fragments appeared to be identical to that of the fragment amplified from cloned archetype JCV (CY) DNA (Fig. 1). To confirm this, the fragments amplified on day 28 were cloned into plasmid pUC19, inserts were excised from about 20 recombinant clones, and their sizes were analyzed by agarose gel electrophoresis (data not shown). All clones derived from the same transfected cells were judged to be identical in size. The sequencing of two or three representative clones confirmed that the nucleotide sequences of these clones were identical to those of the corresponding transfected JCV DNAs. We concluded that the archetype regulatory region is conserved during viral growth in COS-7 cells.
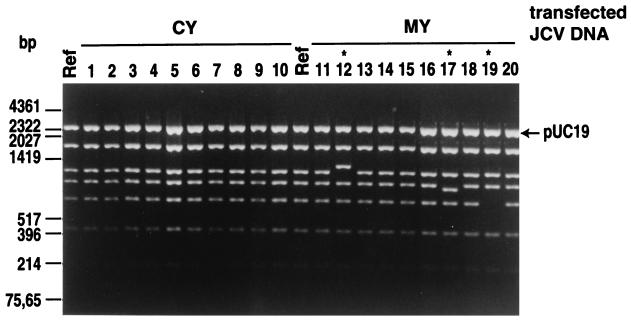
From the viral DNAs extracted from various transfected cells on day 28 (Table 2), complete JCV DNAs were cloned into pUC19 at the unique BamHI site. Sixteen to 23 recombinant clones for each sample were analyzed with a combination of BamHI and HincII (BamHI was used to excise the whole linear JCV DNA from the vector, while HincII was used to cut the linear JCV DNA into four or six fragments, depending on the subtypes [44]). Typical examples are shown in Fig. 2. A summary of the result of these analyses is given in Table 3. Four to 33% of the analyzed clones showed an increase or decrease in the sizes of certain fragments. All of these changes were mapped outside the regulatory region (data not shown).
FIG. 2.
Restriction analysis of JCV DNAs cloned from COS-7 cells transfected with various archetype JCV DNAs. COS-7 cells were transfected with cloned archetype JCV DNAs (CY, MY, C1, SP-1, GH-1, and ZA-1) and a cloned PML-type JCV DNA (Mad-1) and cultured for 28 days. Viral DNAs extracted from the cells were digested with BamHI, which cleaves the JCV DNA at a single site. The recovered DNAs were cloned into pUC19 as described in Materials and Methods. The recombinant plasmids obtained and the JCV DNAs (Ref) that were used for transfection were digested with a combination of BamHI and HincII. The digests were resolved by electrophoresis on 1.5% agarose gels stained with ethidium bromide. Results with representative clones derived from CY- and MY-transfected COS-7 cells are shown. The sizes of some HinfI fragments of pUC19 and some HindIII fragments of lambda DNA are indicated at the left.
TABLE 3.
Rearrangement of JCV DNAs during viral growth in COS-7 cellsa
| Transfected JCV DNA | No. of clones examined | No. (%) of rearranged clones
|
||
|---|---|---|---|---|
| Total | With shortened fragments | With elongated fragments | ||
| CY | 24 | 1 (4) | 1 (4) | 0 |
| MY | 24 | 5 (20) | 4 (17) | 1 (4) |
| C1 | 24 | 4 (17) | 3 (13) | 1 (4) |
| SP-1 | 24 | 8 (33) | 7 (30) | 1 (4) |
| ZA-1 | 24 | 6 (25) | 4 (17) | 2 (8) |
| Mad-1 | 23 | 1 (4) | 0 | 1 (4) |
Viral DNAs were extracted from transfected COS-7 cells on day 28 and cloned into pUC19 at the BamHI site. Recombinant clones were digested with BamHI and HincII and electrophoresed on 1.5% agarose gels. Changes in fragment size were detected by comparing the fragment pattern of each clone with that of the reference JCV DNA.
We determined the complete coding sequence of a JCV DNA clone (clone 11 in Fig. 2) derived from the MY-transfected cells. This clone apparently underwent no change throughout the genome (Fig. 2). The complete coding sequence of clone 11 was compared with the reported sequences of three JCV strains, 225, 226, and 228 (5) (MY, 225, 226, and 228 belong to the same JCV subtype [unpublished data]). We detected only a small number of nucleotide differences (0.5 to 0.6%) between clone 11 and the three JCV strains. Furthermore, we compared clone 11 with SV40; no distinct SV40 DNA segment was found in the complete DNA sequence of clone 11, although many short nucleotide stretches shared by JCV and SV40 (14) were detectable (data not shown).
From these data, it is clear that no recombination event between JCV and SV40 is involved in the efficient replication of the archetype JCV in COS-7 cells.
Recovery of JCV from urine by using COS-7 cells.
Urine samples (U1 to U6) were collected from JCV-positive volunteers or patients without PML who participated in previous studies (23). The urine samples were fractionated by centrifugation (22). Both cell-associated and cell-free JCVs were used to infect COS-7 cells. The infected COS-7 cells were allowed to grow with passages at a split ratio of 1:5 every 3 or 4 days. When a marked extension of the cytopathic effect (rounding of the cells) was observed in at least one culture, we started sampling for the HA assay from all cultures for each experiment. Thus, samples were taken on days 14, 21, and 28 when cell-associated JCVs were infected and on days 28, 35, and 42 when cell-free JCVs were infected. The cells were lysed and processed for the HA assay. As shown in Table 4, the cells infected with five of the six cell-associated JCVs yielded HA, whereas the cells infected with three of the six cell-free JCVs yielded HA. In addition, the cells infected with cell-associated JCVs produced HA much earlier than those infected with the corresponding cell-free JCVs.
TABLE 4.
Recovery of JCVs from urine samples by viral culture in COS-7 cellsa
| Urine donor | HA titer in cells infected with:
|
|||||
|---|---|---|---|---|---|---|
| Cell-associated JCV from urine
|
Cell-free JCV from urine
|
|||||
| Day 14 | Day 21 | Day 28 | Day 28 | Day 35 | Day 42 | |
| U1 | <2 | <2 | 64b | <2 | 64 | NDc |
| U2 | <2 | <2 | 64b | <2 | <2 | <2 |
| U3 | <2 | <2 | <2 | <2 | <2 | <2 |
| U4 | <2 | 32b | ND | <2 | 64 | ND |
| U5 | <2 | 2 | 128b | <2 | 2 | 64 |
| U6 | 32b | ND | ND | 64 | ND | ND |
JVCs were obtained by centrifugation as described in Materials and Methods.
Used for the extraction of viral DNA.
ND, not done.
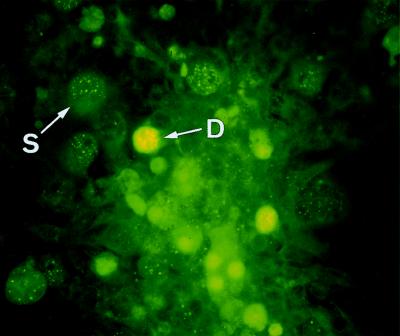
The related human polyomavirus BKV also shows HA activity (34) and is excreted in the urine of a small percentage of nonimmunocompromised individuals (22). We therefore performed an indirect immunofluorescence assay to examine whether the virus produced in COS-7 cells after the infection with JCV from urine would react with a rabbit antiserum specific to JCV (6). A typical result is shown in Fig. 3. There were two classes of fluorescence-positive cells; fluorescent granules were densely distributed in the nuclei in the one type and sparsely distributed in the other. The implications of the different manifestations of fluorescence are not clear, although both types of fluorescence appeared to represent the JCV multiplication in nuclei (fluorescence of any type was not observed in uninfected COS-7 cells [data not shown]). Cells with fluorescence were detected for all of the infected cultures examined except those infected with U3.
FIG. 3.
Immunofluorescence staining of COS-7 cells infected with urinary JCV with rabbit JCAb1 serum and fluorescein-labeled anti-rabbit immunoglobulin. JCAb1 is an antibody against a peptide of JCV capsid protein (VP1) without cross-reaction to BKV (6). Cells infected with JCV from the urinary sediment containing cell-associated JCVs were plated onto a 13-mm-diameter coverslip and processed for immunofluorescence microscopy on day 15 after infection. Densely (D) and sparsely (S) distributed fluorescences are indicated (see text). Magnification, ×200.
We amplified the 610-bp IG regions (8) from urinary JCV-infected COS-7 cells as well as from urine samples. These amplified IG regions were sequenced as described in Materials and Methods. Without exception, the IG sequences from the urine samples were identical to those from the COS-7 cells infected with JCV from the same urine samples, although there were variations in the IG sequences among the urine samples (Table 5) (Table 5 shows only nucleotides at positions where nucleotide differences were found among the urine samples). Therefore, the possibility that the viral growth after the infection with urine arose from contamination can be excluded.
TABLE 5.
Nucleotide variations in the IG region among urine samplesa
| Urine sample | Nucleotide at position:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2161 | 2209 | 2239 | 2251 | 2291 | 2317 | 2371 | 2416 | 2518 | 2593 | 2604 | 2606 | 2642 | 2645 | 2663 | 2712 | 2723 | |
| U1 | C | C | G | C | C | A | A | C | G | G | T | C | A | G | A | G | T |
| U2 | T | C | T | A | C | C | G | T | A | A | C | T | G | A | A | A | C |
| U4 | C | T | G | C | C | A | A | C | G | G | T | C | A | G | G | G | T |
| U5 | C | C | G | C | T | A | A | C | A | G | T | C | A | G | A | G | T |
| U6 | C | C | G | C | C | A | A | C | G | G | T | C | A | G | G | G | T |
The nucleotides shown are those at positions where differences were found among the urine samples. The nucleotide numbering is that of Frisque et al. (14). JCV isolated from each urine sample had the same IG sequence as that found in each urine sample.
Stability of the JCV regulatory region during the recovery of JCV from urine.
Viral DNAs extracted from infected cell cultures producing HA (Table 4) were used to amplify the JCV regulatory region. The amplified fragments were cloned into pUC19, and the resultant 11 or 12 clones were examined for the sizes of inserts by agarose gel electrophoresis. All clones examined except those from U2-infected cells appeared to have the same size as the archetype regulatory region (data not shown). The sequencing of two or three representative clones revealed that these clones had the archetype regulatory region. Therefore, the archetype regulatory region is conserved during the recovery of JCV from urine.
By contrast, although a majority of the 11 clones derived from the U2-infected cells seemed identical to the archetype (2 representative clones of these were sequenced and found to have the archetype sequence), 3 clones had a slightly but significantly elongated regulatory region. The sequencing of these clones revealed that they had the insertion of a single nucleotide (A) between nucleotides (nt) 180 and 181 (the nucleotide numbers are those of CY [46]) and the duplication of an 18-nt stretch (nt 201 to 218). This regulatory region was designated dp-1.
The dp-1 regulatory region may have been generated during the recovery of JCV in COS-7 cells. Alternatively, variant JCVs with these regulatory regions may have existed in the original urine sample and, together with archetype JCV, may have replicated in COS-7 cells. To distinguish between these possibilities, we amplified the JCV regulatory region from the U2 urine sample and cloned the amplified fragments into pUC19. One hundred forty resultant clones were screened for the presence of elongated regulatory regions. We detected two elongated clones, one of which was identical to dp-1. Therefore, we concluded that together with archetype JCV, a variant JCV with the dp-1 regulatory region was recovered in COS-7 cells. Since dp-1 occurred more frequently in the recovered JCVs (3 of 11) than in the original urine sample (1 of 140), JCV with dp-1 may have a higher growth capacity in COS-7 cells than archetype JCV.
One-step growth of archetype JCV in COS-7 cells.
We infected COS-7 cells with various amounts of JCV recovered from urine. The infected cells were cultured in DMEM with 2% FBS and harvested on various days for the assay of output HA. Typical results are shown in Table 6. The findings can be summarized as follows.
TABLE 6.
One-step growth of the archetype JCV in COS-7 cells
| Input JCV (HAU)a | HA titer on day:
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| 0.6 | <2 | <2 | <2 | 32 | 64 | 128 | 512 |
| 6 | 2 | 2 | 2 | 64 | 256 | 512 | NDb |
| 60 | 2 | 2 | 2 | 256 | 64 | ND | ND |
A JCV stock derived from the U4 urine sample (cell-associated fraction) was used for infection.
ND, not done.
(i) In the cultures infected with 6 and 60 HAU, we detected a low level of HA (HA titer of 2) on days 1 to 3, but this low HA titer probably represents residual input viruses.
(ii) A high HA titer appeared on day 4 in the cultures infected with a large amount of virus (60 HAU); the HA titer, however, decreased remarkably on day 5, probably because of the detachment of infected cells from the surfaces of the culture flasks. In addition, significant HA titers were first detected on day 4 in the cultures infected with smaller amounts of viruses (0.6 and 6 HAU). Thus, we concluded that the one-step growth of archetype JCVs in COS-7 cells takes nearly 4 days.
(iii) High yields of progeny viruses were also obtained by infection with smaller amounts (0.6 and 6 HAU) of viruses followed by longer cultivation of the infected cells.
DISCUSSION
We showed that (i) a large amount of progeny JCVs was produced after the transfection of COS-7 cells with various cloned archetype JCV DNAs, (ii) the expression of SV40 T antigen was essential for the production of progeny in COS-7 cells, (iii) the archetype regulatory region was conserved in the progeny DNAs, (iv) most of the progeny DNAs did not undergo rearrangements outside the regulatory region, and (v) the progeny were transferred to fresh COS-7 cells. From these findings, we conclude that COS-7 cells expressing SV40 T antigen can support the efficient replication of archetype JCV. The possibility that the observed viral replication represents that of recombinant viruses containing the SV40 regulatory region derived from the integrated SV40 genome can be excluded.
Our conclusion seems to be straightforward, but an explanation for the DNA changes observed outside the regulatory region may be required. Size changes of restriction fragments were detected in significant percentages (4 to 33%) of the progeny JCV DNAs. However, similar degrees of heterogeneity have been observed for JCV DNAs when PML-type JCVs were serially cultivated in primary human glial cells (30) and in a human neuroblastoma cell line (IMR-32) (32). Therefore, we consider that tissue culture-induced heterogeneity in the JCV DNA is an inevitable consequence of the genetic nature of JCV.
Daniel et al. (12) recently reported that an archetype JCV DNA induces a low level of replication of JCV in POJ cells. The replication of archetype JCV in POJ cells was detected by the continuous presence of faint hybridization signals during the incubation of transfected cells. Therefore, the replication of archetype JCV is far less efficient in POJ cells than in COS-7 cells. The COS-7 cell line was established by transforming a simian cell line (CV-1), derived from the kidney of an African green monkey, with an origin-defective mutant of SV40 (16). The POJ cell line was established by transforming human fetal glial cells with an origin-defective mutant of JCV (27). Thus, COS-7 cells constitutively express SV40 T antigen, while POJ cells constitutively express JCV T antigen. In addition, the parental cells of COS-7 and POJ differ in cell type and species. Some of these differences may be associated with the difference between these cell lines’ capacities to support the replication of archetype JCV. For example, since it has been reported that SV40 T antigen is more active than JCV T antigen in the replication of JCV DNA in vitro (12) as well as in vivo (38), JCV might have replicated more efficiently in COS-7 cells expressing SV40 T antigen.
It is remarkable that the archetype regulatory region is stable during the growth of JCV in COS-7 cells, since the archetype generates various PML-type regulatory regions by deletion and amplification during the persistence of JCV in patients (3, 9, 20, 21, 45, 46). This discrepancy may be explained by the fact that transfected or infected COS-7 cells were cultured for weeks in this study, but this period is much shorter than the persistence of archetype JCV in human hosts.
For the first time, we succeeded in recovering archetype JCV from urine by viral culture. We confirmed the identity between JCVs occurring in the original urine and those recovered in COS-7 cells. First, we found the identity of 610-bp IG sequences between the two JCVs, although they varied among urine samples. Thus, the possibility that viral growth after infection with urine arose from contamination can be excluded. Second, we showed that the archetype regulatory region was conserved during the recovery of JCV from urine. Although an elongated regulatory region (dp-1) was found in the recovered JCVs, this regulatory region was also present in the original urine sample. Thus, we concluded that the JCVs recovered by using COS-7 cells faithfully reflect those occurring in urine.
COS-7 cells supported the replication of Mad-1 (a strain with a PML-type regulatory region) (14) as well as JCV with the dp-1 regulatory region. These findings imply that JCV with various rearranged regulatory regions can replicate efficiently in COS-7 cells. Therefore, COS-7 cells may also be used for the isolation of PML-type JCVs from the brains of PML patients and for the isolation of new polyomaviruses from various primates.
Archetype JCV in urine has been detected as a distinct viral DNA (1, 4, 13, 17, 28, 44–46). However, since JCV DNA was recovered from the virion fraction, it was thought that JCV virions, rather than naked DNAs, are excreted in urine (22). In this study, we found that urinary JCV is indeed infectious. However, this infectivity of urinary JCV was dependent on the presence of SV40 T antigen synthesized from the integrated SV40 DNA. The present study demonstrated that urinary JCV can interact with receptors on the surface of cells and can penetrate into the cells. The reason for this is obvious; if JCV is not adsorbed to cells and does not penetrate into them, subsequent events in the viral replication cycle, including viral DNA replication regulated by T antigen, would not occur.
The life cycle of JCV in hosts very likely involves the following sequence of events: the entry of JCV into the body with multiplication at the site of entry, viremia with the transport of JCV to the target organ, and viral persistence in the target organ with urinary excretion of progeny. The details of each event remain to be elucidated. However, we recently demonstrated that the target of JCV persistence is the renal tissue (24). The present results further show that urinary JCV is the most likely source of infection. However, the natural cells for entry that can support JCV replication remain to be identified.
ACKNOWLEDGMENTS
We are grateful to Yoshiyuki Nagai and Souichi Nukuzuma for helpful discussions. We also thank Yukari Mitsunobu for excellent technical assistance.
This study was supported in part by grants from the Ministry of Education, Sciences, Sports and Culture, Japan; from the Ministry of Welfare, Japan; and from Kitasato University (grant-in-aid no. SAHS-B108-1997).
REFERENCES
- 1.Agostini H T, Brubaker G R, Shao J, Ryschkewitsch C F, Blattner W A, Stoner G L. BK virus and a new type of JC virus excreted by HIV-1-positive patients in rural Tanzania. Arch Virol. 1995;140:1919–1934. doi: 10.1007/BF01322682. [DOI] [PubMed] [Google Scholar]
- 2.Agostini H T, Ryschkewitsch C F, Brubaker G R, Shano J, Stoner G L. Five complete genomes of JC virus type 3 from Africans and African Americans. Arch Virol. 1997;142:637–655. doi: 10.1007/s007050050108. [DOI] [PubMed] [Google Scholar]
- 3.Agostini H T, Ryschkewitsch C F, Singer E J, Stoner G L. JC virus regulatory region rearrangements and genotypes in progressive multifocal leukoencephalopathy: two independent aspects of virus variation. J Gen Virol. 1997;78:659–664. doi: 10.1099/0022-1317-78-3-659. [DOI] [PubMed] [Google Scholar]
- 4.Agostini H T, Ryschkewitsch C F, Stoner G L. Genotype profile of human polyomavirus JC excreted in urine of immunocompetent individuals. J Clin Microbiol. 1996;34:159–164. doi: 10.1128/jcm.34.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agostini H T, Yanagihara R, Davis V, Ryschkewitsch C F, Stoner G L. Asian genotypes of JC virus in Native Americans and in a Pacific island population: markers of viral evolution and human migration. Proc Natl Acad Sci USA. 1997;94:14542–14546. doi: 10.1073/pnas.94.26.14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki N, Mori M, Kato K, Sakamoto Y, Noda K, Tajima M, Shimada H. Antibody against synthetic multiple antigens (MAP) of JC virus capsid protein (VP1) without cross reaction to BK virus: diagnostic tool for progressive multifocal leukoencephalopathy. Neuroscience. 1996;205:111–114. doi: 10.1016/0304-3940(96)12389-2. [DOI] [PubMed] [Google Scholar]
- 7.Arthur R R, Shah K V. Occurrence and significance of papovaviruses BK and JC in the urine. Prog Med Virol. 1989;36:42–61. [PubMed] [Google Scholar]
- 8.Ault G S, Stoner G L. Two major types of JC virus defined in progressive multifocal leukoencephalopathy brain by early and late coding region DNA sequences. J Gen Virol. 1992;73:2669–2678. doi: 10.1099/0022-1317-73-10-2669. [DOI] [PubMed] [Google Scholar]
- 9.Ault G S, Stoner G L. Human polyomavirus JC promoter/enhancer rearrangement patterns from progressive multifocal leukoencephalopathy brain are unique derivatives of a single archetypal structure. J Gen Virol. 1993;74:1499–1507. doi: 10.1099/0022-1317-74-8-1499. [DOI] [PubMed] [Google Scholar]
- 10.Carbon J, Shenk T E, Berg P. Biochemical procedure for production of small deletions in simian virus 40 DNA. Proc Natl Acad Sci USA. 1975;72:1392–1396. doi: 10.1073/pnas.72.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesters P M, Heritage J, McCance D J. Persistence of DNA sequences of BK virus and JC virus in normal human tissues and in diseased tissues. J Infect Dis. 1983;147:676–684. doi: 10.1093/infdis/147.4.676. [DOI] [PubMed] [Google Scholar]
- 12.Daniel A M, Swenson J J, Mayreddy R P R, Khalili K, Frisque R J. Sequence within the early and late promoters of archetype JC virus restrict viral DNA replication and infectivity. Virology. 1996;216:90–101. doi: 10.1006/viro.1996.0037. [DOI] [PubMed] [Google Scholar]
- 13.Flægstad T, Sundsfjord A, Arthur R R, Pedersen M, Traavik T, Subramani S. Amplification and sequencing of the control regions of BK and JC virus from human urine by polymerase chain reaction. Virology. 1991;180:553–560. doi: 10.1016/0042-6822(91)90069-n. [DOI] [PubMed] [Google Scholar]
- 14.Frisque R J, Bream G L, Cannella M T. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallia G L, Houff S A, Major E O, Khalili K. Review: JC virus infection of lymphocytes—revisited. J Infect Dis. 1997;176:1603–1609. doi: 10.1086/514161. [DOI] [PubMed] [Google Scholar]
- 16.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 17.Guo J, Kitamura T, Ebihara H, Sugimoto C, Kunitake T, Takehisa J, Na Y Q, Al-Ahdal M N, Hallin A, Kawabe K, Taguchi F, Yogo Y. Geographical distribution of the human polyomavirus JC virus types A and B and isolation of a new type from Ghana. J Gen Virol. 1996;77:919–927. doi: 10.1099/0022-1317-77-5-919. [DOI] [PubMed] [Google Scholar]
- 18.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 19.Howley P M, Rentier-Delrue F, Heilman C A, Law M-F, Chowdhury K, Israel M A, Takemoto K K. Cloned human polyomavirus JC DNA can transform human amnion cells. J Virol. 1980;36:878–882. doi: 10.1128/jvi.36.3.878-882.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iida T, Kitamura T, Guo J, Taguchi F, Aso Y, Nagashima K, Yogo Y. Origin of JC virus variants associated with progressive multifocal leukoencephalopathy. Proc Natl Acad Sci USA. 1993;90:5062–5065. doi: 10.1073/pnas.90.11.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato K, Guo J, Taguchi F, Daimaru O, Tajima M, Haibara H, Matsuda J, Sumiya M, Yogo Y. Phylogenetic comparison between archetypal and disease-associated JC virus isolates in Japan. Jpn J Med Sci Biol. 1994;47:167–178. doi: 10.7883/yoken1952.47.167. [DOI] [PubMed] [Google Scholar]
- 22.Kitamura T, Aso Y, Kuniyoshi N, Hara K, Yogo Y. High incidence of urinary JC virus excretion in nonimmunosuppressed older patients. J Infect Dis. 1990;161:1128–1133. doi: 10.1093/infdis/161.6.1128. [DOI] [PubMed] [Google Scholar]
- 23.Kitamura T, Kunitake T, Guo J, Tominaga T, Kawabe K, Yogo Y. Transmission of human polyomavirus JC virus occurs both within the family and outside the family. J Clin Microbiol. 1994;32:2359–2363. doi: 10.1128/jcm.32.10.2359-2363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamura T, Sugimoto C, Kato A, Ebihara H, Suzuki M, Taguchi F, Kawabe K, Yogo Y. Persistent JC virus (JCV) infection is demonstrated by continuous shedding of the same JCV strains. J Clin Microbiol. 1997;35:1255–1257. doi: 10.1128/jcm.35.5.1255-1257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunitake T, Kitamura T, Guo J, Taguchi F, Kawabe K, Yogo Y. Parent-to-child transmission is relatively common in the spread of the human polyomavirus JC virus. J Clin Microbiol. 1995;33:1448–1451. doi: 10.1128/jcm.33.6.1448-1451.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loeber G, Dörries K. DNA rearrangements in organ-specific variants of polyomavirus JC strain GS. J Virol. 1988;62:1730–1735. doi: 10.1128/jvi.62.5.1730-1735.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandl C, Walker D L, Frisque R J. Derivation and characterization of POJ cells, transformed fetal glial cells that retain their permissivity for JC virus. J Virol. 1987;61:755–763. doi: 10.1128/jvi.61.3.755-763.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markowitz R-B, Eaton B A, Kubik M F, Latorra D, McGregor J A, Dynan W S. BK virus and JC virus shed during pregnancy have predominantly archetypal regulatory regions. J Virol. 1991;65:4515–4519. doi: 10.1128/jvi.65.8.4515-4519.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin J D, King D M, Slauch J M, Frisque R J. Differences in regulatory sequences of naturally occurring JC virus variants. J Virol. 1985;53:306–311. doi: 10.1128/jvi.53.1.306-311.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin J D, Padgett B L, Walker D L. Characterization of tissue culture-induced heterogeneity in DNAs of independent isolates of JC virus. J Gen Virol. 1983;64:2271–2280. doi: 10.1099/0022-1317-64-10-2271. [DOI] [PubMed] [Google Scholar]
- 31.Matsuda M, Jona M, Yasui K, Nagashima K. Genetic characterization of JC virus Tokyo-1 strain, a variant oncogenic in rodents. Virus Res. 1987;7:159–168. doi: 10.1016/0168-1702(87)90077-3. [DOI] [PubMed] [Google Scholar]
- 32.Nukuzuma S, Yogo Y, Guo J, Nukuzuma C, Itoh S, Shinohara T, Nagashima K. Establishment and characterization of a carrier culture producing high titers of polyoma JC virus. J Med Virol. 1995;47:370–377. doi: 10.1002/jmv.1890470413. [DOI] [PubMed] [Google Scholar]
- 33.Padgett B L, Walker D L. Prevalence of antibodies in human sera against JC virus, an isolate from a case of progressive multifocal leukoencephalopathy. J Infect Dis. 1973;127:467–470. doi: 10.1093/infdis/127.4.467. [DOI] [PubMed] [Google Scholar]
- 34.Padgett B L, Walker D L. New human papovaviruses. Prog Med Virol. 1976;22:1–35. [PubMed] [Google Scholar]
- 35.Padgett B L, Walker D L, ZuRhein G M, Eckroade R J, Dessel B H. Cultivation of papova-like virus from human brain with progressive multifocal leukoencephalopathy. Lancet. 1971;i:1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Shah K V. Polyomaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Raven Publishers; 1995. pp. 2027–2043. [Google Scholar]
- 38.Sock E, Wegner M, Fortunato E A, Grummt F. Large T and sequence within the regulatory region of JC virus both contribute to features of JC virus DNA replication. Virology. 1993;197:537–548. doi: 10.1006/viro.1993.1627. [DOI] [PubMed] [Google Scholar]
- 39.Sugimoto C, Ito D, Tanaka K, Matsuda H, Saito H, Sakai H, Fujihara K, Itoyama Y, Yamada T, Kira J, Matsumoto R, Mori M, Nagashima K, Yogo Y. Amplification of JC virus regulatory DNA sequences from cerebrospinal fluid: diagnostic value for progressive multifocal leukoencephalopathy. Arch Virol. 1998;143:249–262. doi: 10.1007/s007050050284. [DOI] [PubMed] [Google Scholar]
- 40.Sugimoto C, Kitamura T, Guo J, Al-Ahdal M N, Shchelkunov S N, Otova B, Ondrejka P, Chollet J-Y, El-Safi S, Ettayebi M, Grésenguet G, Kocagöz T, Chaiyarasamee S, Thant K Z, Thein S, Moe K, Kobayashi N, Taguchi F, Yogo Y. Typing of urinary JC virus DNA offers a novel means to trace human migrations. Proc Natl Acad Sci USA. 1997;94:9191–9196. doi: 10.1073/pnas.94.17.9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi H, Yogo Y, Furuta Y, Takada A, Irie T, Kasai M, Sano K, Fujioka Y, Nagashima K. Molecular characterization of a JC virus (Sap-1) clone derived from a cerebellar form of progressive multifocal leukoencephalopathy. Acta Neuropathol. 1992;83:105–112. doi: 10.1007/BF00308469. [DOI] [PubMed] [Google Scholar]
- 42.Tominaga T, Yogo Y, Kitamura T, Aso Y. Persistence of archetypal JC virus DNA in normal renal tissue derived from tumor-bearing patients. Virology. 1992;186:736–741. doi: 10.1016/0042-6822(92)90040-v. [DOI] [PubMed] [Google Scholar]
- 43.Yogo Y, Guo J, Iida T, Satoh K, Taguchi F, Takahashi H, Hall W W, Nagashima K. Occurrence of multiple JC virus variants with distinctive regulatory sequences in the brain of a single patient with progressive multifocal leukoencephalopathy. Virus Genes. 1994;8:99–105. doi: 10.1007/BF01703608. [DOI] [PubMed] [Google Scholar]
- 44.Yogo Y, Iida T, Taguchi F, Kitamura T, Aso Y. Typing of human polyomavirus JC virus on the basis of restriction fragment length polymorphisms. J Clin Microbiol. 1991;29:2130–2138. doi: 10.1128/jcm.29.10.2130-2138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yogo Y, Kitamura T, Sugimoto C, Hara K, Iida T, Taguchi F, Tajima A, Kawabe K, Aso Y. Sequence rearrangement in JC virus DNAs molecularly cloned from immunosuppressed renal transplant patients. J Virol. 1991;65:2422–2428. doi: 10.1128/jvi.65.5.2422-2428.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yogo Y, Kitamura T, Sugimoto C, Ueki T, Aso Y, Hara K, Taguchi F. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J Virol. 1990;64:3139–3143. doi: 10.1128/jvi.64.6.3139-3143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]