Summary
The aim of this study was to analyze signaling pathways associated with differentially expressed messenger RNAs in people with restless legs syndrome (RLS). Seventeen RLS patients and 18 controls were enrolled. Coding RNA expression profiling of 12,857 gene transcripts by next-generation sequencing was performed. Enrichment analysis by pathfindR tool was carried-out, with p-adjusted ≤0.001 and fold-change ≥2.5. Nine main different network groups were significantly dysregulated in RLS: infections, inflammation, immunology, neurodegeneration, cancer, neurotransmission and biological, blood and metabolic mechanisms. Genetic predisposition plays a key role in RLS and evidence indicates its inflammatory nature; the high involvement of mainly neurotropic viruses and the TORCH complex might trigger inflammatory/immune reactions in genetically predisposed subjects and activate a series of biological pathways—especially IL-17, receptor potential channels, nuclear factor kappa-light-chain-enhancer of activated B cells, NOD-like receptor, mitogen-activated protein kinase, p53, mitophagy, and ferroptosis—involved in neurotransmitter mechanisms, synaptic plasticity, axon guidance, neurodegeneration, carcinogenesis, and metabolism.
Subject areas: Neurology, Bioinformatics, Transcriptomics
Graphical abstract
Highlights
-
•
This study analyzed pathways of differentially expressed mRNAs in people with RLS
-
•
Nine closely interconnected network groups emerged to be significantly dysregulated
-
•
Among them: infections, inflammation, immunology, neurodegeneration, and cancer
-
•
Infections could trigger inflammatory/immune reactions in predisposed subjects
Neurology; Bioinformatics; Transcriptomics
Introduction
Restless legs syndrome (RLS) is a sensory-motor disorder characterized by an urgent need to move the legs, often but not always accompanied or perceived as a consequence of unpleasant sensations, which begin or worsen during periods of rest or inactivity, especially in the evening or at night, partially or totally eliminated with movement. The symptoms are not attributable to another medical condition (such as myalgia, venous stasis, arthritis, edema, leg cramps, etc.).1
Several neuroimaging, neurophysiology, and basic research investigations have been conducted with the aim of better characterizing the disorder, overall highlighting an involvement of the mesolimbic and nigro-striatal dopaminergic pathways, with a thalamic metabolic involvement,2,3 of GABAergic and glutamatergic transmission, of the opioid system, iron metabolism, and CNS structures involved in the transmission of sensorimotor information.3,4 Several studies have also demonstrated a possible role of inflammatory mechanisms in the pathogenesis of RLS, with increased plasma levels of C-reactive protein (CRP) and a high neutrophil/lymphocyte ratio, although this is not yet well understood and further investigations are needed.5
Conventional treatment of RLS involves iron supplementation, dopamine agonists, calcium channel ligands, and opioids,6,7 as well as some non-pharmacological approaches.8 In children no drugs have been approved so far, but a good response to iron treatment has been demonstrated.9 However, especially dopamine agonists, can cause augmentation, characterized by a loss of therapeutic response and onset, paradoxically, of a more accentuated symptomatology, in body districts where it was previously absent and even during daylight hours.10
Therefore, it is important to conduct further studies for a better understanding of the etiopathogenesis of RLS in order to provide a stronger rationale for its management with drugs and ideas for new therapeutic options, possibly based on molecular evidence and with a precision medicine approach.11,12
To this end, genetics can provide an important help and very often persons affected by RLS have a positive family history. Having a first-degree relative with RLS increases the risk by 6-to-7 times13 and studies on twins have suggested that genetic factors contribute up to 70% in the risk of developing RLS.14 In addition, periodic leg movements during sleep (PLMS), which occur in most RLS patients, are also associated with some of the same risk polymorphisms as RLS, suggesting a shared genetic risk.15
Genome-wide association studies (GWASs) have in fact highlighted that the MEIS1, BTBD9, and PTPRD genes, which are implicated in dopaminergic transmission and iron metabolism, are involved in both RLS and PLMS.15 To date, GWAS studies on RLS have identified at least 13 different genes in the Northern European population, and recent studies have identified seven other major susceptibility loci. Their functions are yet to be defined, although they seem to be related to the neural development of limbs during embryogenesis.16 It seems that iron deficiency in the brain17 might represent a key biological driver in RLS, possibly due to low levels of peripheral iron or genetic factors, although the link between these conditions is still unclear.16 Additionally, it seems that the currently known variants represent only 20% of the genetic risk of RLS and, therefore, a large part of the heritability in this disorder remains to be discovered. In this perspective, new and innovative biotechnologies in the molecular field might provide a valid research support and additional pathophysiological information.14
Some studies have already been published on proteomics18,19,20,21,22 or other metabolomics23 in RLS, the results of which indicate genetic determinants only marginally overlapping with those indicated by genomics. In this perspective, considering that the transfer of gene information from DNA to metabolites involves gene methylation24 and messenger RNAs (mRNAs), transcriptome analysis may provide important and complementary information on RLS genetics and metabolomics. For this reason, the aim of this study was to analyze signaling pathways associated with differentially up- and downregulated mRNAs in people with RLS, in order to obtain information on any deregulated network playing a key role in its etiopathogenesis, thus also providing hints for new diagnostic therapeutic approaches.
Results
Main findings
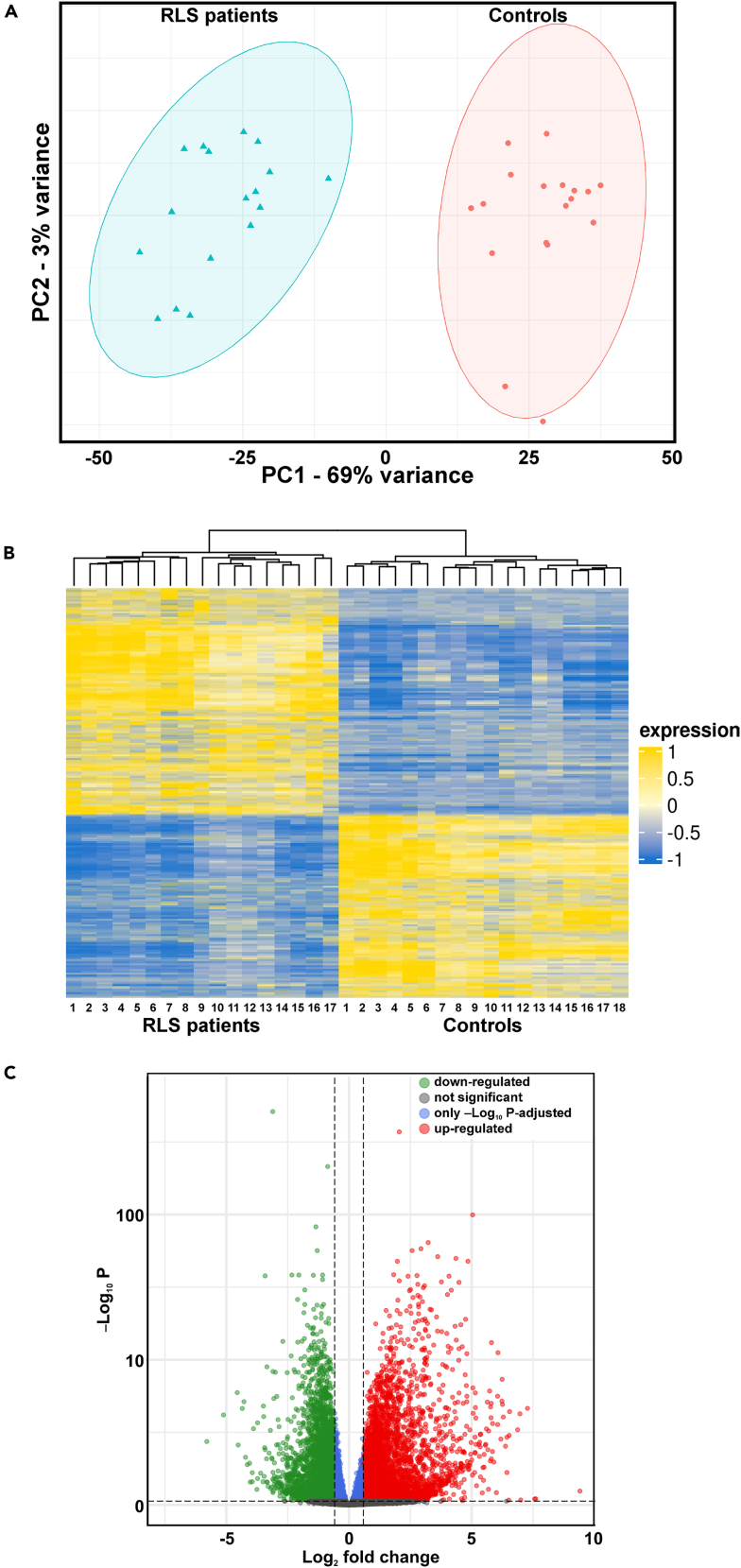
A whole transcriptome analysis was performed by next-generation sequencing in order to measure gene abundance, evaluate their possible deregulation, and identify novel transcriptome features in RLS patients. High throughput data sequences and high unique mapping rates of the samples indicated that the quantity and quality of the sequenced data were in accordance with the requirements meaning what 20–30 million reads per sample for large genomes and a mapping rate greater than or equal to 90%. Principal component analysis (PCA) revealed a clear clustering and showed complete separation between patients and controls (Figure 1A). To evaluate how sensitive the clustering performance was to sample size, we evaluated the distribution of the samples with respect to PC1 and PC2 and showed a multivariate normal distribution with a confidence of 95%.
Figure 1.
Main findings of the transcriptomics analysis
(A) Principal component analysis of transcripts found in RLS patients and controls showing the clustering f the two groups by using the first principal component (PC1) and the second (PC2).
(B) Heatmap of significant statistical differences of the mRNA expression profile found in RLS patients and controls.
(C) Volcano plot displays the distribution of differentially expressed transcripts by their fold change and p value. As also marked by the different colors, the most upregulated genes are toward the right, the most downregulated genes are toward the left, and the most statistically significant genes are on the top.
The expression level of each gene was calculated via feature-Counts by using common gene annotations (GenCode_Version37) with the whole annotated transcript. The obtained gene counts allowed to detect, in total 23,331 genes, expressed in at least 25% of RLS or control25 samples. In the present study, to investigate the global mRNA expression differences between the two groups, hierarchical clustering analyses were performed.
As shown in the heatmap (Figure 1B), significant statistical differences of the mRNA expression profile were found in RLS patients, compared to healthy controls. In detail, we identified 12,857 genes differentially regulated between the two groups (RLS vs. controls, padj ≤0.05). Among them, 4,932 were significantly (padj ≤0.05 and fold-change ≥1.5) upregulated (Table S1) and 3,971 (padj ≤0.05 and fold-change ≤–1.5) significantly downregulated in patients (Table S2).
The volcano plot displays the distribution of differentially expressed transcripts by their fold change and p value (Figure 1C). Among upregulated genes in RLS, in particular, four showed a fold-change greater than 100: HBG1, KDM5D, DDX3Y, and TXLNGY, with the top upregulated gene (HBG1) with fold-change 678. DM5D and DDX3Y genes showed fold-change close to 200, while other genes (such as RPS4Y1 fold-change 90, OLIG2 fold-change 76, and SHISA7 fold-change 47) were also significantly upregulated, although to a lesser extent. Conversely, among the top downregulated genes, five had fold-change less than −20: ZNF208, EREG, NR4A2, FOSB, and AREG; the latter, in particular, showed a fold-change of −55.
Functional and pathway enrichment analysis of differentially expressed genes
To elucidate the biological role of differentially expressed genes, 2,597 genes (padj ≤0.001 and |FC| ≥2.5) were subjected to gene ontology analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis to investigate biological functional variations between the two groups and to study possible pathways that might be potentially associated with RLS susceptibility.
First of all, we considered gene ontology categories to provide an in-depth perspective on the biological and molecular processes related to RLS.
Regarding gene ontology enrichment, we found downregulated and upregulated genes (Table S2) involved in several molecular functions (Table S2), biological processes (Table S3) and cellular components (Table S4). Interestingly among them, the upregulated HBA1, HBA2, HBB, PRDX5, PINK1, RARA, SPI1, ZBTB7B, SOX6 genes were found to be associated with the following molecular functions: (GO:0001217) DNA-binding transcription repressor activity, (GO:0004601) peroxidase activity binding, and (GO:0043422) protein kinase B binding.
Concerning the biological processes, we notably found the enrichment of (GO:0042744) hydrogen peroxide catabolism (GPX1, HBA1, HBA2, HBB), (GO:0015671) oxygen transport (HBA1, HBA2, HBB, HBD, HBG1, HBG2, HBZ), (GO:0042542) response to hydrogen peroxide (GPX1, HBA1, HBA2, HBB), (GO:0030218) erythrocyte differentiation (C18orf54, GATA1, TAL1, DYRK3, KLF1); while among cellular components, (GO:0005833) hemoglobin complex (HBA1, HBA2, HBB, HBD, HBG1, HBG2, HBZ) and (GO:0097386) glial cell projection (ABCA7) are interesting.
Moreover, in order to obtain an integrative view of the RLS-associated transcripts identified by next generatn sequencig (NGS), functional network analysis was performed by pathfindR tool as described in STAR Methods. In detail, to explore the pathways involved in the pathology we combined the analysis of pathfindR which use different database such as Reactome and BioCarta.
Enrichment analysis was performed by using the applying the p-adj threshold of 0.001 and |FC| ≥2.5. In this way we identified 65 statistically significant signaling pathways involving 139 terms. Table 1 shows the distribution of terms included in these pathways, divided by their involvement in a particular process and/or function and/organ (the total is higher than 139 because some terms could be associated to more than one process/function/organ). This table also shows the top five upregulated and downregulated genes among all terms included in each category.
Table 1.
Number of terms included in the 65 statistically significant pathways found by pathfindR, divided by their involvement in a particular process and/or function and/organ
| Process/Function/Organ | Number of terms | top 5 upregulated genes |
top 5 downregulated genes |
||||
|---|---|---|---|---|---|---|---|
| gene | frequency | percent | gene | frequency | percent | ||
| Infection | 27 | TNFRSF1A | 14 | 51.9 | NFKBIA | 20 | 74.1 |
| FADD | 13 | 48.1 | PIK3R1 | 17 | 63.0 | ||
| HLA-A | 8 | 29.6 | AKT3 | 16 | 59.3 | ||
| HLA-B | 8 | 29.6 | JUN | 15 | 55.6 | ||
| HLA-C | 8 | 29.6 | CXCL8 | 14 | 51.9 | ||
| Cancer | 27 | GADD45G | 12 | 42.9 | AKT3 | 24 | 85.7 |
| PTEN | 12 | 42.9 | PIK3R1 | 24 | 85.7 | ||
| TGFA | 8 | 30.0 | JUN | 12 | 42.9 | ||
| BCL2L1 | 7 | 25.0 | FOS | 9 | 32.1 | ||
| WNT6 | 5 | 17.9 | NFKBIA | 8 | 28.6 | ||
| Metabolism | 23 | CREB3L3 | 7 | 30.4 | AKT3 | 19 | 82.6 |
| CREB5 | 7 | 30.4 | PIK3R1 | 16 | 69.6 | ||
| TNFRSF1A | 7 | 30.4 | PLCB1 | 11 | 47.8 | ||
| RXRA | 6 | 26.1 | PLCB4 | 11 | 47.8 | ||
| MMP9 | 5 | 21.7 | FOS | 8 | 34.8 | ||
| PTEN | 5 | 21.7 | |||||
| Immunology | 18 | HLA-A | 5 | 27.8 | NFKBIA | 9 | 50.0 |
| HLA-B | 5 | 27.8 | PIK3R1 | 9 | 50.0 | ||
| HLA-C | 5 | 27.8 | JUN | 8 | 44.4 | ||
| CCL5 | 4 | 22.2 | AKT3 | 6 | 33.3 | ||
| FADD | 4 | 22.2 | FOS | 6 | 33.3 | ||
| IFNGR2 | 4 | 22.2 | |||||
| PYCARD | 4 | 22.2 | |||||
| Neurodegeneration | 17 | BCL2L1 | 7 | 41.2 | AKT3 | 9 | 52.9 |
| TNFRSF1A | 6 | 35.3 | PIK3R1 | 9 | 52.9 | ||
| KLC3 | 5 | 29.4 | JUN | 7 | 41.2 | ||
| MAP2K3 | 5 | 29.4 | NDUFV2 | 6 | 35.3 | ||
| TUBB2A | 5 | 29.4 | FOS | 5 | 29.4 | ||
| TUBB4A | 5 | 29.4 | |||||
| Inflammation | 14 | CXCL1 | 3 | 21.4 | PIK3R1 | 6 | 42.9 |
| NCF1 | 3 | 21.4 | NFKBIA | 5 | 35.7 | ||
| ACTN1 | 2 | 14.3 | AKT3 | 4 | 28.6 | ||
| CCL5 | 2 | 14.3 | CXCL8 | 4 | 28.6 | ||
| CXCL6 | 2 | 14.3 | PLCB1 | 4 | 28.6 | ||
| FADD | 2 | 14.3 | PLCB4 | 4 | 28.6 | ||
| Cellular processes | 9 | ACTN1 | 4 | 44.4 | AKT3 | 4 | 44.4 |
| VASP | 3 | 33.3 | PIK3R1 | 4 | 44.4 | ||
| WAS | 3 | 33.3 | DOCK1 | 3 | 33.3 | ||
| FCGR2A | 2 | 22.2 | EEA1 | 2 | 22.2 | ||
| FCGR3B | 2 | 22.2 | FN1 | 2 | 22.2 | ||
| Neurotransmission | 6 | CAMK2A | 5 | 83.3 | AKT3 | 3 | 50.0 |
| CAMK2B | 5 | 83.3 | FOS | 3 | 50.0 | ||
| CREB3L3 | 3 | 50.0 | PIK3R1 | 3 | 50.0 | ||
| CREB5 | 3 | 50.0 | PLCB1 | 3 | 50.0 | ||
| CACNA1A | 2 | 33.3 | PLCB4 | 3 | 50.0 | ||
| Blood | 5 | IFNGR2 | 3.0 | 60.0 | AKT3 | 4.0 | 80.0 |
| IL6R | 3.0 | 60.0 | PIK3R1 | 4.0 | 80.0 | ||
| CSF1 | 2.0 | 40.0 | TFRC | 2.0 | 40.0 | ||
| CSF3R | 2.0 | 40.0 | CNTF | 1.0 | 20.0 | ||
| EPOR | 2.0 | 40.0 | FLT3 | 1.0 | 20.0 | ||
| Hormonal | 2 | ||||||
| Pregnancy | 1 | ||||||
| Heart | 1 | ||||||
| Circadian | 1 | ||||||
| Addiction | 1 | ||||||
The top five upregulated and downregulated genes among all terms included in each category are also indicated.
Discussion
Because of the richness of the results obtained, we will focus our discussion on the analysis of the implication of the dysregulation of the pathways listed in Table 1 involving 5 or more terms. More details about the findings in each of these groups of networks are reported in Figures S1–S9.
Infections
We found an important involvement of gene networks involved in infections; in particular, 27 infection gene networks were found to be differently expressed in patients with RLS, of which 13 connected with viral infections and, the remaining with bacteria, parasites, and plasmodia. An important involvement was found, in terms of number of genes involved and greater regulation of gene expression, of measles, toxoplasmosis, shigellosis, herpesvirus associated with Kaposi’s sarcoma, human immunodeficiency virus 1 (HIV), and many other herpetic viruses: Epstein-Barr virus, human cytomegalovirus (CMV), human papillomavirus and, to a lesser extent, influenza A and COVID-19, hepatitis B, and carcinogenesis-related viruses (Figure S2).
Recently, a connection between RLS and long COVID-19 syndrome was found in women, the authors hypothesized that post-infectious mechanisms could contribute to the onset of RLS,26 while in another work a greater severity of RLS symptoms was found during the first phase of the COVID-19 pandemic.27 A connection between RLS and HIV infection is still being studied: while several authors reported this link,28,29 possibly mediated by interleukin (IL)-1β and IL-17α,30 with a possible damage to the basal ganglia by the virus,31 others do not agree.32,33
There are no data on a possible correlation between RLS and the other infectious agents found in our study (Figure S2), such as toxoplasmosis, rubella, and herpetic viruses; however, our findings might indicate the need to further assess their possible role.
It is important to note that an association between RLS and inflammatory/autoimmune diseases with a connection with herpetic viruses is often found: RLS has a prevalence of 13.3%–65.1% in patients with multiple sclerosis (MS),34 in which herpes viruses seem to play a pathogenetic role,35 especially the Epstein-Barr virus.36 Another very intriguing observation is the finding of the infectious agents of the TORCH complex: congenital infections of toxoplasmosis, others (syphilis, hepatitis B), rubella, CMV, and herpes simplex (other pathogens associated with congenital infections include HIV, parvovirus, and chickenpox virus).37 They are also all pathogens with neurotropism. An interesting study pointed out that pathogens of the TORCH complex have devastating effects on the self-renewal of neurons, also being able to influence the offspring, with long-term effects in the neuronal lineage—a paradigmatic example seems to be the neurodegeneration of Alzheimer’s disease (AD).38
A recent condition, called pseudo-TORCH, has been reported that involves symptoms of a TORCH infection in the absence of a real infection, based on genetic causes yet to be discovered.39 The association between RLS and pregnancy is well known, the latter favors the onset or worsens RLS, especially in women who have a family history of the disorder,40 which might explain our finding and that, in genetically predisposed subjects, triggering conditions (such as pregnancy) can favor a pseudo-TORCH symptomatology, in the absence of a real infection in progress (but because of possible previous TORCH infections in the parental line). After all, infectious agents of the TORCH complex can induce dysfunction of the basal ganglia, with dopaminergic dysfunction, movement disorders and even parkinsonism.31,41,42
Inflammation
A recent systematic review showed a significant trend toward higher levels of CRP and a higher neutrophil-to-lymphocyte ratio in patients with RLS, compared to controls; although the authors stressed the need for further studies, a possible role of inflammatory factors in the pathogenesis of the disorder was envisaged.5 In addition, research conducted in the field of proteomics in patients with RLS has also highlighted the involvement of inflammatory mechanisms, such as increased production of oxygen free radicals and activation of complement fractions),18,20 as well as proteins involved in the inflammatory response (Alpha-2 macroglobulin precursor, Alpha-2 HS glycoprotein).20 Finally, only one metabolomics study was conducted in patients with RLS in patients with peritoneal dialysis in which the authors underlined how the increase in oxidative stress (and therefore inflammation) could play a key role in the connection between RLS and kidney disorders chronic;43 however, metabolomics studies in idiopathic RLS are lacking.
Our findings support the involvement of inflammatory mechanisms in RLS, with as many as 14 different inflammatory networks involved (Figure S3); the involvement of infectious and immunological networks (Figures S1 and S3) provide additional support. Among the networks most implicated we found those of IL-17, chemokines, extracellular formation of neutrophil traps and, to a lesser extent, transendothelial migration pathways of leukocytes, regulation of the inflammatory mediator of transient receptor potential (TRP) channels, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and RIG-I-like receptors. T cells IL-17 cytokines are implicated in protective immunity against many pathogens and, if dysregulated, may promote immunopathology in the context of infection or autoimmunity; moreover, they are involved in cardiovascular and neurological diseases, such as atherosclerosis, MS, AD, and Parkinson’s disease (PD).44
One study has shown an involvement of this pathway in RLS in HIV patients;30 it has also been reported that IL-17 plays a role in the dopaminergic system. Implicated in RLS,45 by interrupting the blood-brain barrier and influencing dopamine synthesis. Moreover, dopamine has been shown to reduce the immune response mediated by Th17 cells.46 Importantly, several antidepressants, and selective serotonin reuptake inhibitors in particular, can induce or worsen RLS or PLMS45,47,48 and have an action on the IL-17 pathway. Also amisulpride, chlorpromazine, haloperidol, clozapine, and quetiapine have been associated with elevated levels of IL-17.46 In contrast, bupropion (the only antidepressant that does not worsen or induce RLS or PLMS49) reduces the inflammatory response mediated by IL-17, similarly to pramipexole, also used in RLS.46 Therefore, the involvement of this network in RLS could be responsible for the modulation of the dopaminergic pathways.
Regarding the role of chemokine networks, extracellular neutrophil trap formation, and transendothelial leukocyte migration, several studies have shown an increase in inflammatory markers and a higher neutrophil-lymphocyte ratio in RLS;5 a brain autopsy study showed gliotic microvascular infiltrates and T cells in patients with RLS,50 proving a probable involvement of IL-17.44
The regulation pathway of the inflammatory mediator of TRP channels is also involved in a wide range of neurodegenerative conditions: cancer, neuronal apoptosis, epilepsy, and pain.51,52 TRP channel receptors are more specifically involved in temperature nociception by GABAergic transmission.53 Therefore, their role in RLS could be connected with the sensory symptomatology of these patients, probably indicating an involvement of GABAergic circuits, as well as with the response to drugs modulating these receptors.45
Finally, both RIG-I-like receptors and NF-κB are involved in virus infections,54,55 confirming a possible role of viral infection in the pathogenesis of RLS; NF-κB is also involved in synaptic plasticity processes and is fundamental for memory consolidation at the level of the amygdala,55 a structure also involved in RLS.2
Immunology
RLS is associated with multiple autoimmune diseases, such as MS,34 has a 2.56-fold greater risk in patients with Hashimoto’s thyroiditis, compared to the general population56 and is also very frequent in psoriasis and rheumatoid arthritis,57,58 as well as in celiac and Chron’s diseases.59,60,61 Proteomic studies also highlight the involvement of complement fractions, demonstrating a possible role of the innate immune response in RLS.18,20
Our results demonstrate an involvement of 18 different networks involved in immune processes, with a very high expression of tumor necrosis factor (TNF) pathway, toll-like receptor pathway, nucleotide-binding and oligomerization domain (NOD-like pathway), Th 17 cell differentiation, and transendothelial leukocyte migration (Figure S4).
TNF is fundamental in the regulation of the immune system, especially in response to exposure to “non-self” elements such as bacteria or viruses, since it suggests that an external agent may be related to a dysregulation of this pathway in RLS, with a fold enrichment among the highest in our study. In this network we found, again, elements connected with the IL-17 pathway, Th 17 cell differentiation, as well as an involvement of toll-like receptors which, together with RIG-I-like receptors, are involved in virus infections54 and have a role in transendothelial migration of leukocytes, present in infections by external agents. These results support the finding of gliotic microvascular infiltrates and T cells in patients with RLS,50 adding important new hints for the understanding of its pathogenetic mechanisms.
A novel finding is the important role of the NOD-like receptor (NLR) pathway in RLS: we have in fact detected the second highest fold enrichment and the greatest gene expression compared to controls (Figure S4). NLR receptors are involved in defense against pathogen invasion and maintaining normal homeostasis; some NLRs form multiprotein complexes called inflammasomes producing inflammatory cytokines and inducing pyroptosis (highly inflammatory form of programmed cell death, in response to pathogens) through the proteolytic cascade.62 The interest on the implication of this pathway in RLS also lies in its modulatory role of neuropathic pain, via the extracellular signal-regulated kinase (ERK) signaling pathway;63 moreover, recent findings link the role of dopamine in modulating NLR-mediated neuroinflammation in PD and AD, with a correlation with neurodegeneration.64 However, it seems that dopamine triggers but does not activate the NLR inflammasome: this role belongs to the NF-κB, which increases or decreases cytokine production.65 In recent years, studies have assessed the role of inflammasome in PD, in which it might contribute to dopaminergic degeneration,66 with an important function of the STAT3 and NLR-related pathways in pyroptosis.67 We are not aware of any prior study of these aspects in RLS.
Neurodegeneration
We found a dysregulation of 17 networks involved in neurodegeneration, with the highest fold enrichment for the mechanisms of mitophagy, ferroptosis, and p53 signaling pathway, while the greatest gene expression was observed for the mitogen-activated protein kinase (MAPK) pathway (Figure S5).
Despite substantial clinical, neuroimaging, neuropathological, and genetic differences between them, several studies describe a higher prevalence of RLS in PD68 and amyotrohic lateral sclerosis (ALS).69 A meta-analysis has shown a correlation between RLS and cognitive deficits,70 although specific investigations on the subject are still few and very recent, needing further confirmation: an interesting 12-year retrospective cohort study suggests that RLS is associated with an increased risk of AD and vascular dementia.71
The mechanism of these interconnections is still largely unknown, but a possible key to understand it could be the involvement of ferroptosis both in RLS and in the aforementioned diseases, considering its possible involvement in substance abuse and neurodegenerative diseases (PD, ALS, and AD).72
Ferroptosis is a form of cell death dependent on iron and driven by lipid peroxidation, some of the molecules that regulate it are also involved in iron homeostasis. Excess iron is stored in ferritin heteropolymers, a redox-inactive form of iron, to protect cells and tissues from iron-mediated damage; autophagic degradation of ferritin increases labile iron levels and contributes to ferroptosis.73 RLS is associated to reduced ferritin levels in the cerebrospinal fluid,74 even when in comorbidity with PD, in which ferritin levels, but not sideremia or hemoglobin, are significantly correlated with RLS.75 In addition, guidelines for the treatment of RLS include the supplementation of iron, to ensure adequate levels of ferritin,9,74 even in the absence of pathological values of hemoglobin or iron.
These observations are important for future therapeutic perspectives, as ferroptosis inhibitors are expected to be promising treatments in PD and AD.76 Ferroptosis is also responsible for an increased production of reactive oxygen species with mitochondrial damage and neurodegeneration, it is no coincidence that there is an interconnection between ferroptosis and mitophagy, through a pathway involving NLR.77 Among pathways related to neurodegeneration, we found the highest fold enrichment for mitophagy (Figure S5); there are no other similar findings for RLS but only for PD, with a proposed interaction between mitophagy/autophagy, dopamine metabolism and adaptive immune system at the basis of its pathogenesis.78
Our findings also highlight an important role of MAPK signal transduction pathways in RLS; these contribute to the regulation of cell proliferation, including the p53 pathway. The loss of control of one of these pathways can evolve into processes of neurodegeneration or carcinogenesis.79 A recent study showed that Map2k5 (a member of the MAPK family) mutant mice have decreased dopaminergic cell survival and levels of tyrosine hydroxylase (an enzyme limiting dopaminergic synthesis and requiring iron), indicating its crucial role in the regulation of the dopaminergic system.80 In addition, we found a high fold enrichment both for the p53 signaling pathway (regulating apoptosis and connected to MAPK79), and for apoptosis itself. Recently, p53-mediated apoptotic cell death has been identified as the leading cause of dopaminergic neuron loss in PD, with a causal link of TNF-NF KappaB signaling in limiting cell survival;81 this is also in agreement with studies on the role of inflammasome in PD, which might contribute to dopaminergic degeneration by NF-κB.66
Therefore, the involvement in RLS of these different neurodegenerative pathways, closely interconnected with each other and with the inflammasome, could explain the association of RLS with other neurodegenerative diseases and with dopaminergic dysfunction common to all them, as well as the effectiveness of iron for RLS; indeed, a research hot topic is the relationship between inflammation and ferroptosis.82
A recent study of DNA methylation (DNAm) in RLS supports the notion of neurodevelopmental alterations in this pathology, but not neurodegeneration;83 however, it must be considered that DNAm algorithms are not markers of neurodegeneration, instead they provide a reliable estimate of an individual’s chronological age, also predicting the risk of age-related diseases and mortality and are influenced by multiple variables.84
Cancer
We also observed the dysregulation of 28 networks related to cancer, with the greatest combination of fold enrichment and gene expression on the MAPK signal pathway, the Erb signal pathway and that of the Rap 1 signal (both related to MAPK) and tumors associated with viruses, such as: Kaposi’s sarcoma associated with herpesvirus, viral carcinogenesis, human T cell leukemia virus 1 infection, acute myeloid leukemia, colorectal cancer, prostate cancer, and small-cell lung cancer (Figure S6).
In this group also, viral infections have an important role, probably being able to interfere with transcriptional mechanisms and therefore with some pathways involved in carcinogenesis. Among these, the main one is MAPK for which we have already underlined its involvement in the dopaminergic system regulation.79,80 Moreover, our finding of a possible important role of ferroptosis in RLS could also explain the implication of pathways involved in lung and colorectal cancers and acute myeloid leukemia, in which ferroptosis is involved.82
To date, no association has been found between cancer and RLS, except for a high prevalence of this disorder in patients with myeloma,85,86 while another study found a high prevalence of PLMS in breast cancer;87 clinical and epidemiological studies on the relationship between RLS and cancer are therefore needed to better understand our findings.
Neurotransmission
We also found several networks involved in neurotransmission: the highest expression was observed for cholinergic synapses, followed by the cyclic adenosine monophosphate (cAMP) pathway, dopaminergic synapses, and axon guidance (Figure S7).
The involvement of the dopaminergic system in RLS is known both from the literature45 and clinical evidence: the guidelines on the treatment of the disorder indicate dopamine agonists as drugs of first choice6 and neuroimaging studies confirm a role of the basal ganglia structures included in the mesolimbic dopaminergic pathway.2,3 studies in mouse models also confirm a role of dopaminergic receptors in RLS, concluding that the identification of causal links between genetic risk factors, altered organ functions and changes in molecular pathways in neural circuits may suggest new therapeutic options.88
The involvement of cholinergic synapses is predominant among our findings, the first direct evidence in RLS in humans, to our knowledge. Cases of patients with severe forms of RLS resistant to the common medications suggested by guidelines but responsive to nicotinic acetylcholine receptor agonists89 have been reported; on the other hand, the only antidepressant that does not induce symptoms of RLS or its worsening is bupropion (inhibitor of dopamine and norepinephrine reuptake), which modulates different types of cholinergic receptors and nicotinic acetylcholine receptors of neurons of the dorsal raphe nucleus and hippocampus.49 In addition, studies in mouse models have demonstrated an important role of the modulation of striatal cholinergic neurons in the pathogenesis of RLS.90 Considering that low levels of ACh (as during NREM sleep) induce slow and synchronous oscillations, while high concentrations of ACh (as during wakefulness and REM sleep) lead to fast and asynchronous oscillations,91 and taking into account that the release of ACh from the afferents of the basal forebrain influences attention, learning, and sensory processing (the initial stages of cortical sensory processing are under direct cholinergic control),92 our results allow us to better understand why both an impairment of attention70 and an important sensory component of symptoms occur in RLS.15 In addition, our data provide a further explanation why in RLS there is a significant instability of sleep architecture with hyperarousability,93,94 on which dopamine agonists have little efficacy.95,96,97
Again, in animal studies, it has been seen that adenosine striatal receptor heteromers, located in the cortico-striatal neuronal terminals, play a fundamental role in the release of acetylcholine and dopamine.98 A recent pathogenetic hypothesis of RLS is that a hypoadenosynergic state, induced by cerebral iron deficiency, provides the link between sensorimotor symptoms of RLS and hyperarousal.99 Adenosine plays a fundamental role in biochemical processes, such as energy transfer and signal transduction, through cAMP; our results show high fold enrichment and gene expression in the cAMP pathway, thus confirming a role of adenosine also in human RLS, as proposed by animal models.
Finally, we also found a dysregulation of the axon guidance pathway. Axon guidance is mediated by a variety of signals, such as netrines, semaphorins and cell adhesion molecules and is involved in cell migration and formation of neuronal connections.100 A recent meta-analysis of GWASs in individuals with RLS confirmed MEIS1 as the strongest genetic risk factor for this disorder, while genetic correlation analyses identified neurodevelopmental pathways: genes linked to axon guidance (associated with SEMA6D), synapse formation (NTNG1), and neuronal specification (HOXB and MYT1 cluster family).101 Our findings support this evidence and identify other genes involved in axon guidance, as well as in synaptic plasticity: we have detected an upregulation of OLIG2, involved in axon-guidance and important for the development of pre-thalamus and thalamocortical projections;102 as well as an upregulation of RPS4Y1 transcription. Both these genes are inhibitors of the STAT3 pathway103,104 and associated with an increased risk of autism spectrum disorder and pre-eclampsia,103 conditions frequently associated with RLS.40,105 In addition, the transcription factors STAT3, OLIG2, and NF-κB play a regulatory role in astrocyte activation and glial scar formation.104
We have already reported previously, about immune system networks, the role of the NLR pathway which, together with the STAT3 pathway, contributes to pyroptosis and dopaminergic degeneration;67 while in the inflammatory network we observed dysregulation in the expression of NF-κB, implicated in virus infections54,55 and in synaptic plasticity processes.55 It is therefore essential to note that our findings regarding the different networks are closely interconnected with each other, proving the complexity of the pathogenesis of RLS, to which numerous biological mechanisms seem to contribute, all equally important for the understanding of the disease.
Finally, we also found an upregulation of the SHISA7 gene, also involved in the regulation of synaptic plasticity, as well as in the binding between GABA and glutamate receptor,106,107 through a modulation of AMPA receptors, necessary for synaptic plasticity of the hippocampus and memory recall.108
Cellular processes
Regarding gene networks associated with cellular processes, we found that the main dysregulations involved Fe γ receptor-mediated phagocytosis, phagosome, autophagy, and endocytosis, as well as mechanisms of focal adhesion and regulation of cytoskeletal actin (Figure S8).
The involvement of phagocytosis mechanisms confirms the activation of biological mechanisms in response to an external agent, as already seen for inflammation, infection, and immunological networks. In particular, Fe γ receptor-mediated phagocytosis is implicated in iron metabolism.109 The role of focal adhesion mechanisms is interconnected with axon guidance and neuroplasticity mechanisms;100 most of the signaling pathways that control the neuronal migration process during cortical development and the axonal growth pattern converge to regulate the dynamics of the actin cytoskeleton,110 one of the routes most dysregulated among those assessed in this study.
Mouse models have shown that abnormalities in neuronal connectivity, of the cytoskeleton of the dendritic spines and of the cytoskeletal actin correlate with anomalies of the cortical-mesencephalic circuit because of frontal and striatal cortical hyperexcitability at the dopaminergic level; the main symptomatic manifestations of this anomaly is psychomotor agitation,111 which is one of the cardinal symptoms of RLS. Actin cytoskeleton is also involved in membrane pore functioning and neurotransmitter release mechanisms, including that of the dopamine precursor L-DOPA (L-3,4-dihydroxyphenylalanine);112 consequently, a malfunction of this pathway could be among the contributing causes of the dopaminergic alterations found in RLS.
Our findings highlight, therefore, a dysregulation of genes and mechanisms involved in neurogenesis and neuroplasticity, especially of crossroads structures in the limbic system and cortical thalamus projection. Considering that the human brain requires a long period for its development, growing evidence suggests that long-lasting neuroplasticity also confers risk for developmental disorders, possibly manifesting also later in life.113 We can, therefore, hypothesize that, during the cortical maturation of patients with RLS, there are genetically determined factors that modify the processes of neuroplasticity, leading to the creation of aberrant sensory-motor and associative areas, hence the symptoms reported by patients.
Blood
This network group includes two pathways with high fold enrichment and gene expression in RLS: osteoclast differentiation and hematopoietic cell line (Figure S9).
No study has ever directly found a role of the hematopoietic cell line in RLS, although in PD, often associated with RLS,75 accelerated hematopoietic cell mitosis has recently been proposed, with possible correlations with immune imbalances and motor and cognitive progression.114
In our study we found alterations in the genes involved in the hematopoietic lineage, precisely: oxygen transport (HBA1, HBA2, HBB, HBD, HBG1, HBG2, HBZ), erythrocyte differentiation (C18orf54, GATA1, TAL1, DYRK3, KLF1), and hemoglobin complex (HBA1, HBA2, HBB, HBD, HBG1, HBG2, HBZ), data that provide support of the alterations of iron metabolism in RLS and for the therapeutic response to iron of these patients.7,9
The role of the osteoclast differentiation pathway in RLS needs further studies; however, it is known that parathyroid hormone (PTH) stimulates the differentiation of osteoclasts and a recent review evaluated the interaction between vitamin D and calcium, phosphorus and PTH and their possible role in RLS. Vitamin D deficiency stimulates the production of PTH,115 possibly explaining the greater expression in osteoclast differentiation. In addition, a significant association between the vitamin D/PTH axis and leg restlessness in PD has recently been suggested, considering that PTH has a presumed role in nociceptive modulation, the authors proposed a possible interrelationship with RLS.116 In this regard, our results highlight a considerable involvement of the PTH synthesis/secretion/action pathway (Figure S9), supporting our hypothesis.
Metabolism
Considering the multiple networks involved in RLS and the complexity of their interactions, we can also understand the possible consequences at the level of different organs and systems and therefore metabolic processes. In fact, our results show that as many as 23 different metabolic pathways are dysregulated in RLS; among these, those with greater gene expression and fold enrichment are: lipid and atherosclerosis, insulin resistance, PI3K-Akt pathway, PTH synthesis/secretion/action pathway, estrogen signaling pathway, relaxin signaling pathway, and cardiomyocyte adrenergic signaling pathway (Figure S10).
One of the most important pathways is that of PTH synthesis/secretion/action, in accordance with our hypothesis described in the hematopoietic network about the possible function of the osteoclast differentiation pathway.
Also interesting is the involvement of numerous pathways concerning cardiovascular risk factors: lipids and atherosclerosis, insulin resistance, estrogen signal, cardiomyocyte adrenergic signal, and PI3K-Akt pathway, which seem to play a role in reperfusion mechanisms following ischemia.117 There is a large literature concerning the association between RLS and cardiovascular disease, however, with conflicting data and without a definitive explanation of the reason for this association.118,119 The proposed mechanisms are: alterations of the autonomic nervous system (ANS), glucose intolerance and reduced insulin sensitivity/diabetes, oxidative stress, inflammatory mechanisms, impaired endothelial functioning, and activation of the hypothalamic-pituitary axis or of the renin-angiotensin-aldosterone system.120
Our results therefore represent an important step to shed light on the cardiovascular risk in RLS, demonstrating a role of lipid metabolism, with consequent risk of atherosclerosis. Moreover, lipid metabolism is also involved in ferroptosis, in association with inflammatory mechanisms. Endothelial damage, also favoring atherosclerosis, can also occur as a consequence of activation of the IL-17 pathway and transendothelial migration of leukocytes, both related to atherosclerosis, hyperlipidemia, and myocardial infarction.121 The increased expression of the cardiomyocyte adrenergic signaling pathway could represent an indirect signal of a role of ANS in the pathogenesis of RLS, thus corroborating the clinical observations on the increased risk of heart rate alterations and arterial hypertension in RLS and PLMS.122,123,124
The role of relaxin in RLS is unknown, it may be related to pregnancy, although many other physiological roles have been identified for this hormone, such as cardiovascular and neuropeptide functions, and it has been implicated in fibromyalgia,125 a disorder frequently associated with RLS.126
Finally, we found an upregulation of the estrogen signaling pathway in RLS, in agreement with the higher prevalence of the disorder in women than in men,127 in endometriosis (stimulated by estrogens),128 in pregnancy especially during the third trimester,129 when an increase in estrogen occurs. On the other hand, estrogens protect against cardiovascular risk, which is known to be greater in men than in women until menopause;130 the fact that RLS involves both metabolic pathways that favor cardiovascular risk and protective pathways in this sense, could contribute to the conflicting epidemiological results on cardiovascular risk in RLS. Heterogeneous samples for sex and age might affect the results, considering also that the severity of RLS, as well as that of PLMS, vary according to age.131 However, a very interesting fact is that proteomic studies also support a role of cardiovascular factors in RLS: a recent study showed a significant increase in kininogen-1 and a reduction in alpha-1-antitrypsin, in relation to the severity of the disease, supporting a higher cardiovascular risk.22 Cardiovascular factors were found to be significant also in another proteomic analysis of patients with RLS.18
The increase in kininogen-1 was also found by another study, in addition to the overexpression of inflammatory and immunological factors (complement fractions, alpha-1B-glycoprotein, alpha-1-acid glycoprotein 1), as previously described also in the inflammatory and immunological networks, supporting the results of our study, as well as the concept that the pathogenesis of RLS is likely multifactorial and involves several biological mechanisms closely interconnected with each other.19
Our findings indicate the complexity of the pathophysiology of RLS, to which numerous biological mechanisms contribute, all equally important for the understanding of the disease.
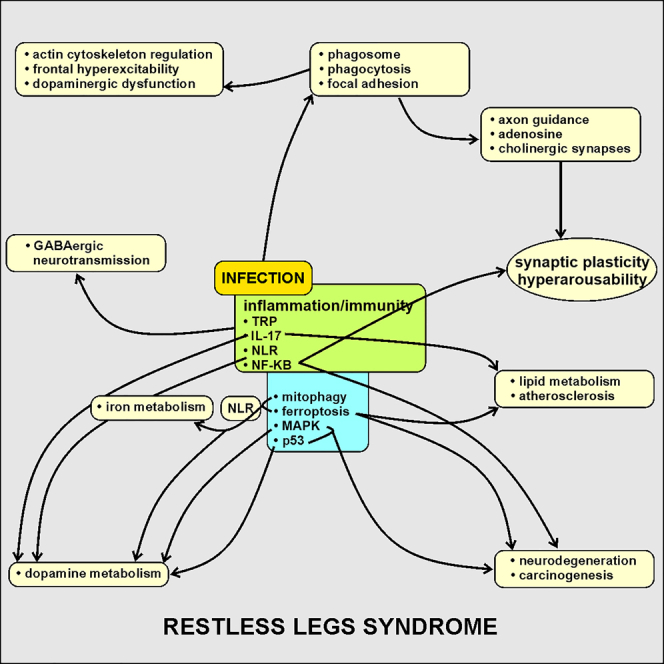
Our study confirms that genetic predisposition plays a key role in RLS and provides important biological evidence on its inflammatory nature; the high involvement of infectious agents, mainly viruses with neurotropism and the TORCH complex, could represent the trigger for inflammatory and immune reactions in genetically predisposed subjects and activate a series of biological pathways (especially IL-17, TRP, NF-κB, NLR, MAPK, p53, mitophagy, and ferroptosis) involved in neurotransmitter mechanisms, synaptic plasticity and axon guidance, as well as neurodegeneration, carcinogenesis, and metabolism (Figure 2). The different dysregulated biological pathways that we found also provide a biological explanation to the clinical and epidemiological data on the association between RLS and pregnancy, with alterations in iron metabolism and with immunological, neurodegenerative, and cardiovascular diseases.
Figure 2.
Proposed possible role of infectious agents, mainly viruses with neurotropism and the TORCH complex, as trigger for inflammatory and immune reactions in genetically predisposed subjects for RLS
A series of biological pathways (especially IL-17, TRP, NF-κB, NLR, MAPK, p53, mitophagy, and ferroptosis) involved in neurotransmitter mechanisms, synaptic plasticity, hyperarousability, and axon guidance, as well as neurodegeneration, carcinogenesis, and metabolism might be activated by infections.
We found a dysregulation of genes and mechanisms involved in neurogenesis and neuroplasticity, especially of crossroads structures in the limbic system and corticothalamic projections; it is therefore likely that, during the cerebral maturation of patients with RLS, there are genetically determined factors (such as those aforementioned) that modify the processes of neuroplasticity, leading to the creation of aberrant sensory-motor and associative areas, as well as to neurotransmitter alterations, hence the symptoms reported by patients.
Our study contributes, in addition, to expand the knowledge on why some pharmacological agents worsen the symptoms of RLS, unlike others and, above all, provides important insights on future therapeutic targets to be considered in this disorder, such as agonists of nicotinic acetylcholine receptors, ferroptosis inhibitors, drugs that reduce the inflammatory response mediated by IL-17 and therapies able to modulate the inflammasome (especially the STAT3/NLR pathway).
In light of the compelling results of our study, the rationale for future studies lies in the need to bridge the gap between the identified dysregulated mRNA transcriptome in RLS and the established genetic variants associated with the condition. By examining the intricacies of the transcriptomic landscape and aligning them with known genetic factors, future studies should aim to elucidate the molecular mechanisms linking genetic predisposition to the observed dysregulation in various pathways.
It is plausible to envision that future experiments may also target exploring the intricate regulatory networks governing gene expression through epigenomic approaches. These studies could provide crucial information allowing us to better understand the complexity of biological processes related to RLS. Finally, in light of our results, further genetic, proteomic and metabolomic studies are necessary in idiopathic RLS, to better characterize the possible role of neurodegeneration, carcinogenesis or metabolic mechanisms, as there are currently no data on this matter in the literature.
Limitations of this study
Finally, the main limitation of this study includes the relatively small sample size and the inclusion of only Caucasian individuals; however, the effect size of the differences found between RLS patients and controls support the validity of our findings which, anyway, need replication in order to be considered for a wider generalizability. Our stringent recruitment criteria, with participants being off medications in order to avoid potential confounding effects on the transcriptomic analysis, represent a strength of this study. Nonetheless, further analyses in larger cohorts applying complementary validation techniques and exploring the influence of factors that may contribute to transcriptome changes are necessary. Another limitation is that the number of patients recruited precludes correlation analysis with the disease severity, ferritin levels, or familiarity. It will be important to study these clinical characteristics which, correlated with the findings from the transcriptome analysis, might be of potential value in the objective assessment of the neurobiological profile and severity of RLS.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| Ficoll Plaque PLUS | GE Healthcare Life Sciences | Cat#GEHE17-1440-02 |
| TRIzol Reagent | Invitrogen Life Technologies | Cat#15596026 |
| Agilent RNA analysis kit | Agilent | Cat#5067-5576 |
| Illumina Stranded mRNA Library Prep Kit | Illumina | Cat#20040534 |
| NovaSeq 6000 SP Reagent Kit v1.5 | Illumina | Cat#20040719 |
| Deposited data | ||
| Raw and analyzed data | This paper | E-MTAB-13155 |
| A transcriptome analysis study on mRNA and long non-coding RNAs in patients with Parkinson’s disease | Salemi et al., 202225 | https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-11326 |
| Software and algorithms | ||
| Cutadapt | Readthedocs | https://cutadapt.readthedocs.io/en/v3.4/ |
| STAR version | GitHub | https://github.com/alexdobin/STAR |
| GenCode | GenCode | https://www.gencodegenes.org/human/release_37.html |
| Subread | Sourceforge | https://subread.sourceforge.net/ |
| DESeq2 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| ComplexHeatmap | Bioconductor | https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| ggplot2 | Tidyverse | https://ggplot2.tidyverse.org/ |
| R SVA | Bioconductor | https://bioconductor.org/packages/release/bioc/html/sva.html |
| pathfindR | CRAN.r-project.org | https://cran.r-project.org/web/packages/pathfindR/index.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Raffaele Ferri (rferri@oasi.en.it).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Raw sequencing data of RLS patients are publicly available at ArrayExpress with the accession number E-MTAB-13155, as listed in the key resources table. Raw sequencing data of controls are publicly available at ArrayExpress with the accession number E-MTAB-11326, as listed in the key resources table.
-
•
This study does not report original new code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Collection of samples
For this study, 17 caucasian drug-free patients (13 females and 4 males, mean age 55.8 years, range 24-76, mean IRLS severity scale 21.1, mean disease duration 6.6 years) were enrolled, along with 18 age- and sex-matched caucasian persons without RLS who had already served as controls in a previously published dataset by our group (E-MTAB-11326). RLS was diagnosed according to the International RLS Study Group diagnostic criteria,1 by means of a semi-structured clinical interview and a careful exclusion of RLS mimics. Exclusion criteria included: (a) a sleep disorder diagnosis other than RLS; (b) psychiatric, neurologic, cardiovascular, or neurodegenerative diagnosis; (c) neurodevelopmental delay; (d) use of CNS drugs within the year prior to the study or use of any drug or medication for 3 weeks before the polysomnographic recording. In addition, adult subjects with an apnea/hypopnea index >10/hour of sleep were excluded. A family history of RLS was reported by two patients. Controls included drug-free subjects without any sleep, physical, neurological, or psychiatric disorder. The subjects’ consent was obtained according to the Declaration of Helsinki and the study was approved by the ethical committee of the Oasi Research Institute.
Method details
Extraction of RNA
For all samples, (controls and patients), venous blood collection was carried out in the morning after a 12-hour fast. The samples, immediately after collection, were stratified using Ficoll-Paque (Ficoll Plaque PLUS–GE Healthcare Life Sciences, Piscataway, NJ, USA), and the PBMCs were stored at -80°C until RNA isolation. RNA was extracted using TRIzol reagent (TRIzol Reagent, Invitrogen Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s instructions. The samples were collected, stored, and extracted using identical procedures. RNA extraction occurred simultaneously for samples within the same group. RNA was stored at -80°C until further processing. We included a DNase treatment after the RNA isolation method to ensure sample purity and accurate quantification. Before starting the protocol, the yield and quality of RNA were evaluated using a NanoDrop One spectrophotometer (NanoDrop) and a TapeStation 4200 (Agilent Technologies, 5301 Stevens Creek Blvd, Santa Clara, CA, USA), respectively. Specifically, the 260/280 ratio of the samples was within the range of 1.8 to 2 as measured by the NanoDrop, while the RIN (RNA Integrity Number) was between 6 to 8 as determined using the TapeStation.
Transcriptome profiling
RNA sequencing and data analysis were performed at Genomix4Life Srl (Baronissi, Italy). Indexed libraries were prepared from 600 ng purified RNA with Illumina Stranded mRNA Library Prep Kit. After enrichment of mRNA by oligo dT magnetic beads, purified mRNA was fragmented and copied into first strand complimentary DNA using reverse transcriptase and random primers. In a second strand complimentary DNA synthesis step, dUTP replaced dTTP to achieve strand specificity, followed by adapter ligation and PCR amplification. The Illumina NovaSeq 6000 platform was used to sequence libraries pooled in equimolar quantity in a 2×75 paired-end format.
Quantification and statistical analysis
The raw sequence files generated (fastq files) underwent quality control analysis using FastQC. Low-quality reads, short reads (≤25 bp), and adaptor sequences were trimmed with cutadapt (v.3.4). Then, the fastq files were mapped on reference genome using the bioinformatics tool STAR (version 2.7.5c) with the standard parameters for paired reads. The genome reference was the assembly Human obtained from GenCode (HG38-Release 37 (GRCh38.p13). The quantification of genes regulated for each sequenced sample was performed using feature Counts algorithm (v.2.0). R was utilized to normalize the data, using negative binomial generalized linear models, considering all genes expressed in ≥25% of samples, by Bioconductor DESeq2 package which utilizes Benjamin-Hochberg correction for padj calculation, with default parameters. Transcripts showing fold change ≥1.50 or ≤−1.50 (|FC| ≥1.50), with adjusted p-values ≤0.05 (padj), were considered as differentially expressed. ComplexHeatmap and ggplot2 package in R were used to make HeatMaps and volcano plot, respectively. We investigated the possibility of correcting batch effects using the function svaseq (surrogate variable analysis) of the R SVA package (since they are two different sequences), but the results did not change. The R package pathfindR for comprehensive identification of enriched pathways in omics data, through active subnetworks, was used to perform the functional analysis. In particular, pathfindR analysis was based on the KEGG pathway database and selected genes set with padj≤0.001 and |fold-change|≥2.5. Only the enriched terms with adjusted p-value ≤0.05 were used for the downstream analysis such as hierarchical clustering of the terms. Furthermore, the pathfindR function score_terms was used to calculate the aggregated term scores per sample based on gene expression patterns. The raw data (fastq files) of mRNAs identified are available at ArrayExpresse E-MTAB-13155. The power calculation based on our data as total number of genes (23,331, see Results), proportion of non-differentially expressed genes (0.65, see Results), and false discovery rate level 0.05 showed that that to obtain a power >0.8 a sample size of 10 was required, as shown also in Figure S1.
Acknowledgments
This research was partially supported by a grant from the Italian Ministry of Health “Ricerca Corrente” (RC n. 2773776 to Dr. Ferri and RC 2779777 to Dr. Salemi).
Author contributions
Conceptualization, M.P.M. and R.F.; methodology, M.P.M., M.S., A.R., G.M., M.R., M.G.S., A.A., and R.F.; investigation, M.P.M., M.S., A.R., G.M., M.R., M.G.S., A.A., and R.F.; writing – original draft, M.P.M., M.S., A.R., G.M., and R.F.; writing – review and editing, M.P.M., M.S., G.L., A.R., G.M., M.R., M.G.S., A.A., L.M.D., O.B., L.F.S., and R.F.; funding acquisition, R.F.; resources, M.S. and G.M; supervision, M.P.M. and R.F.
Declaration of interests
The authors declare no competing interests.
Published: March 26, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109568.
Supplemental information
References
- 1.Allen R.P., Picchietti D.L., Garcia-Borreguero D., Ondo W.G., Walters A.S., Winkelman J.W., Zucconi M., Ferri R., Trenkwalder C., Lee H.B., International Restless Legs Syndrome Study Group Restless legs syndrome/Willis-Ekbom disease diagnostic criteria: updated International Restless Legs Syndrome Study Group (IRLSSG) consensus criteria--history, rationale, description, and significance. Sleep Med. 2014;15:860–873. doi: 10.1016/j.sleep.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 2.Mogavero M.P., Mezzapesa D.M., Savarese M., DelRosso L.M., Lanza G., Ferri R. Morphological analysis of the brain subcortical gray structures in restless legs syndrome. Sleep Med. 2021;88:74–80. doi: 10.1016/j.sleep.2021.10.025. [DOI] [PubMed] [Google Scholar]
- 3.Rizzo G., Plazzi G. Neuroimaging Applications in Restless Legs Syndrome. Int. Rev. Neurobiol. 2018;143:31–64. doi: 10.1016/bs.irn.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Lanza G., Ferri R. The neurophysiology of hyperarousal in restless legs syndrome: Hints for a role of glutamate/GABA. Adv. Pharmacol. 2019;84:101–119. doi: 10.1016/bs.apha.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Jiménez-Jiménez F.J., Alonso-Navarro H., García-Martín E., Agúndez J.A.G. Inflammatory factors and restless legs syndrome: A systematic review and meta-analysis. Sleep Med. Rev. 2023;68 doi: 10.1016/j.smrv.2022.101744. [DOI] [PubMed] [Google Scholar]
- 6.Silber M.H., Buchfuhrer M.J., Earley C.J., Koo B.B., Manconi M., Winkelman J.W., Scientific and Medical Advisory Board of the Restless Legs Syndrome Foundation The Management of Restless Legs Syndrome: An Updated Algorithm. Mayo Clin. Proc. 2021;96:1921–1937. doi: 10.1016/j.mayocp.2020.12.026. [DOI] [PubMed] [Google Scholar]
- 7.Allen R.P., Picchietti D.L., Auerbach M., Cho Y.W., Connor J.R., Earley C.J., Garcia-Borreguero D., Kotagal S., Manconi M., Ondo W., et al. Evidence-based and consensus clinical practice guidelines for the iron treatment of restless legs syndrome/Willis-Ekbom disease in adults and children: an IRLSSG task force report. Sleep Med. 2018;41:27–44. doi: 10.1016/j.sleep.2017.11.1126. [DOI] [PubMed] [Google Scholar]
- 8.Lanza G., Fisicaro F., Cantone M., Pennisi M., Cosentino F.I.I., Lanuzza B., Tripodi M., Bella R., Paulus W., Ferri R. Repetitive transcranial magnetic stimulation in primary sleep disorders. Sleep Med. Rev. 2023;67 doi: 10.1016/j.smrv.2022.101735. [DOI] [PubMed] [Google Scholar]
- 9.DelRosso L.M., Ferri R., Chen M.L., Kapoor V., Allen R.P., Mogavero M.P., Picchietti D.L. Clinical efficacy and safety of intravenous ferric carboxymaltose treatment of pediatric restless legs syndrome and periodic limb movement disorder. Sleep Med. 2021;87:114–118. doi: 10.1016/j.sleep.2021.08.030. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Borreguero D., Silber M.H., Winkelman J.W., Högl B., Bainbridge J., Buchfuhrer M., Hadjigeorgiou G., Inoue Y., Manconi M., Oertel W., et al. Guidelines for the first-line treatment of restless legs syndrome/Willis-Ekbom disease, prevention and treatment of dopaminergic augmentation: a combined task force of the IRLSSG, EURLSSG, and the RLS-foundation. Sleep Med. 2016;21:1–11. doi: 10.1016/j.sleep.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Chaturvedi P., Khan R., Sahu P., Ludhiadch A., Singh G., Munshi A. Role of Omics in Migraine Research and Management: A Narrative Review. Mol. Neurobiol. 2022;59:5809–5834. doi: 10.1007/s12035-022-02930-3. [DOI] [PubMed] [Google Scholar]
- 12.Mogavero M.P., DelRosso L.M., Bruni O., Salemi M., Salsone M., Novellino F., Zucconi M., Ferini Strambi L., Ferri R. Genetics and epigenetics of rare hypersomnia. Trends Genet. 2023;39:415–429. doi: 10.1016/j.tig.2023.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Allen R.P., La Buda M.C., Becker P., Earley C.J. Family history study of the restless legs syndrome. Sleep Med. 2002;3(Suppl):S3–S7. doi: 10.1016/s1389-9457(02)00140-5. [DOI] [PubMed] [Google Scholar]
- 14.Akçimen F., Dion P.A., Rouleau G.A. Progress in the genetics of restless legs syndrome: the path ahead in the era of whole-genome sequencing. Sleep. 2022;45 doi: 10.1093/sleep/zsac136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trenkwalder C., Allen R., Högl B., Clemens S., Patton S., Schormair B., Winkelmann J. Comorbidities, treatment, and pathophysiology in restless legs syndrome. Lancet Neurol. 2018;17:994–1005. doi: 10.1016/S1474-4422(18)30311-9. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z., Guan R., Pan L. Exploration of restless legs syndrome under the new concept: A review. Medicine. 2022;101 doi: 10.1097/MD.0000000000032324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earley C.J., Jones B.C., Ferré S. Brain-iron deficiency models of restless legs syndrome. Exp. Neurol. 2022;356 doi: 10.1016/j.expneurol.2022.114158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mondello S., Kobeissy F.H., Mechref Y., Zhao J., El Hayek S., Zibara K., Moresco M., Plazzi G., Cosentino F.I.I., Ferri R. Searching for Novel Candidate Biomarkers of RLS in Blood by Proteomic Analysis. Nat. Sci. Sleep. 2021;13:873–883. doi: 10.2147/NSS.S311801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellei E., Monari E., Ozben S., Koseoglu Bitnel M., Topaloglu Tuac S., Tomasi A., Bergamini S. Discovery of restless legs syndrome plasmatic biomarkers by proteomic analysis. Brain Behav. 2018;8 doi: 10.1002/brb3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin J.W., Lee J.H., Kim H., Lee D.H., Baek K.H., Sunwoo J.S., Byun J.I., Kim T.J., Jun J.S., Han D., Jung K.Y. Bioinformatic analysis of proteomic data for iron, inflammation, and hypoxic pathways in restless legs syndrome. Sleep Med. 2020;75:448–455. doi: 10.1016/j.sleep.2020.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Patton S.M., Cho Y.W., Clardy T.W., Allen R.P., Earley C.J., Connor J.R. Proteomic analysis of the cerebrospinal fluid of patients with restless legs syndrome/Willis-Ekbom disease. Fluids Barriers CNS. 2013;10:20. doi: 10.1186/2045-8118-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellei E., Bergamini S., Monari E., Tomasi A., Koseoglu M., Topaloglu Tuac S., Ozben S. Evaluation of potential cardiovascular risk protein biomarkers in high severity restless legs syndrome. J. Neural. Transm. 2019;126:1313–1320. doi: 10.1007/s00702-019-02051-7. [DOI] [PubMed] [Google Scholar]
- 23.Dong X., Mondello S., Kobeissy F., Ferri R., Mechref Y. Serum Glycomics Profiling of Patients with Primary Restless Legs Syndrome Using LC-MS/MS. J. Proteome Res. 2020;19:2933–2941. doi: 10.1021/acs.jproteome.9b00549. [DOI] [PubMed] [Google Scholar]
- 24.Roy A., Earley C.J., Allen R.P., Kaminsky Z.A. Developing a biomarker for restless leg syndrome using genome wide DNA methylation data. Sleep Med. 2021;78:120–127. doi: 10.1016/j.sleep.2020.12.017. [DOI] [PubMed] [Google Scholar]
- 25.Salemi M., Lanza G., Mogavero M.P., Cosentino F.I.I., Borgione E., Iorio R., Ventola G.M., Marchese G., Salluzzo M.G., Ravo M., Ferri R. A Transcriptome Analysis of mRNAs and Long Non-Coding RNAs in Patients with Parkinson's Disease. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23031535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinstock L.B., Brook J.B., Walters A.S., Goris A., Afrin L.B., Molderings G.J. Restless legs syndrome is associated with long-COVID in women. J. Clin. Sleep Med. 2022;18:1413–1418. doi: 10.5664/jcsm.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wipper B., Romero-Gutierrez C., Winkelman J.W. Restless legs syndrome severity in the National RLS Opioid Registry during the COVID-19 pandemic. Sleep Med. 2022;90:96–101. doi: 10.1016/j.sleep.2022.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Happe S., Kundmüller L., Reichelt D., Husstedt I.W., Evers S. Comorbidity of restless legs syndrome and HIV infection. J. Neurol. 2007;254:1401–1406. doi: 10.1007/s00415-007-0563-2. [DOI] [PubMed] [Google Scholar]
- 29.Kunisaki K.M. Do sleep disturbances contribute to comorbidities in HIV? Curr. Opin. HIV AIDS. 2023;18:81–86. doi: 10.1097/COH.0000000000000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hennessy M.D., Zak R.S., Gay C.L., Pullinger C.R., Lee K.A., Aouizerat B.E. Polymorphisms of interleukin-1 Beta and interleukin-17Alpha genes are associated with restless legs syndrome. Biol. Res. Nurs. 2014;16:143–151. doi: 10.1177/1099800413478827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardoso F. HIV-related movement disorders: epidemiology, pathogenesis and management. CNS Drugs. 2002;16:663–668. doi: 10.2165/00023210-200216100-00002. [DOI] [PubMed] [Google Scholar]
- 32.Wallace D.M., Alcaide M.L., Wohlgemuth W.K., Jones Weiss D.L., Uribe Starita C., Patel S.R., Stosor V., Levine A., Skvarca C., Long D.M., et al. Prevalence and correlates of restless legs syndrome in men living with HIV. PLoS One. 2021;16 doi: 10.1371/journal.pone.0258139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunisaki K.M., De Francesco D., Sabin C.A., Winston A., Mallon P.W.G., Anderson J., Bagkeris E., Boffito M., Doyle N., Haddow L., et al. Sleep Disorders in Human Immunodeficiency Virus: A Substudy of the Pharmacokinetics and Clinical Observations in People Over Fifty (POPPY) Study. Open Forum Infect. Dis. 2021;8 doi: 10.1093/ofid/ofaa561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sieminski M., Losy J., Partinen M. Restless legs syndrome in multiple sclerosis. Sleep Med. Rev. 2015;22:15–22. doi: 10.1016/j.smrv.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Khalesi Z., Tamrchi V., Razizadeh M.H., Letafati A., Moradi P., Habibi A., Habibi N., Heidari J., Noori M., Nahid Samiei M., et al. Association between human herpesviruses and multiple sclerosis: A systematic review and meta-analysis. Microb. Pathog. 2023;177 doi: 10.1016/j.micpath.2023.106031. [DOI] [PubMed] [Google Scholar]
- 36.Soldan S.S., Lieberman P.M. Epstein-Barr virus and multiple sclerosis. Nat. Rev. Microbiol. 2023;21:51–64. doi: 10.1038/s41579-022-00770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaan A., Rajnik M. StatPearls. StatPearls Publishing; 2023. TORCH Complex.https://www.ncbi.nlm.nih.gov/books/NBK560528/ [PubMed] [Google Scholar]
- 38.Baggiani M., Dell'Anno M.T., Pistello M., Conti L., Onorati M. Human Neural Stem Cell Systems to Explore Pathogen-Related Neurodevelopmental and Neurodegenerative Disorders. Cells. 2020;9:1893. doi: 10.3390/cells9081893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonough A., Lee R.V., Weinstein J.R. Microglial Interferon Signaling and White Matter. Neurochem. Res. 2017;42:2625–2638. doi: 10.1007/s11064-017-2307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinweg K., Nippita T., Cistulli P.A., Bin Y.S. Maternal and neonatal outcomes associated with restless legs syndrome in pregnancy: A systematic review. Sleep Med. Rev. 2020;54 doi: 10.1016/j.smrv.2020.101359. [DOI] [PubMed] [Google Scholar]
- 41.Malaquias M.J., Magrinelli F., Quattrone A., Neo R.J., Latorre A., Mulroy E., Bhatia K.P. Presynaptic Hemiparkinsonism Following Cerebral Toxoplasmosis: Case Report and Literature Review. Mov. Disord. Clin. Pract. 2023;10:285–299. doi: 10.1002/mdc3.13631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cece H., Tokay L., Yildiz S., Karakas O., Karakas E., Iscan A. Epidemiological findings and clinical and magnetic resonance presentations in subacute sclerosing panencephalitis. J. Int. Med. Res. 2011;39:594–602. doi: 10.1177/147323001103900228. [DOI] [PubMed] [Google Scholar]
- 43.Yang B., Yin H., Wang J., Gan J., Li J., Han R., Pei M., Song L., Yang H. A metabolic biomarker panel of restless legs syndrome in peritoneal dialysis patients. Metabolomics. 2022;18:79. doi: 10.1007/s11306-022-01938-z. [DOI] [PubMed] [Google Scholar]
- 44.Mills K.H.G. IL-17 and IL-17-producing cells in protection versus pathology. Nat. Rev. Immunol. 2023;23:38–54. doi: 10.1038/s41577-022-00746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manconi M., Garcia-Borreguero D., Schormair B., Videnovic A., Berger K., Ferri R., Dauvilliers Y. Restless legs syndrome. Nat. Rev. Dis. Primers. 2021;7:80. doi: 10.1038/s41572-021-00311-z. [DOI] [PubMed] [Google Scholar]
- 46.Jha M.K., Minhajuddin A., Gadad B.S., Greer T.L., Mayes T.L., Trivedi M.H. Interleukin 17 selectively predicts better outcomes with bupropion-SSRI combination: Novel T cell biomarker for antidepressant medication selection. Brain Behav. Immun. 2017;66:103–110. doi: 10.1016/j.bbi.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferri R., Mogavero M.P., Bruni O., Picchietti D.L., Kapoor V., DelRosso L.M. Leg Movements during Sleep in Children Treated with Serotonergic Antidepressants. Sleep. 2022;45 doi: 10.1093/sleep/zsab236. [DOI] [PubMed] [Google Scholar]
- 48.Ferri R., Mogavero M.P., Bruni O., Picchietti D.L., DelRosso L.M. Periodic leg movements during sleep associated with antidepressants: A meta-analysis. Neurosci. Biobehav. Rev. 2023;148 doi: 10.1016/j.neubiorev.2023.105126. [DOI] [PubMed] [Google Scholar]
- 49.DelRosso L.M., Mogavero M.P., Fickensher A., Bruni O., Schenck C.H., Ferri R. Effects of bupropion and SSRI antidepressants on leg movement activity and chin muscle tone during sleep in adolescents. J. Clin. Sleep Med. 2023;19:151–161. doi: 10.5664/jcsm.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walters A.S., Paueksakon P., Adler C.H., Moussouttas M., Weinstock L.B., Spruyt K., Bagai K. Restless Legs Syndrome Shows Increased Silent Postmortem Cerebral Microvascular Disease With Gliosis. J. Am. Heart Assoc. 2021;10 doi: 10.1161/JAHA.120.019627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Öz A., Çelik Ö. Downregulation of TRPM7, TRPM8, and TRPV1 channels modulate apoptotic parameters and neurodegenerative markers: Focus on neuronal differentiation and Parkinson's disease model. Cell Biol. Int. 2023;47:1502–1518. doi: 10.1002/cbin.12048. [DOI] [PubMed] [Google Scholar]
- 52.Salemi M., Mogavero M.P., Lanza G., Mongioì L.M., Calogero A.E., Ferri R. Examples of Inverse Comorbidity between Cancer and Neurodegenerative Diseases: A Possible Role for Noncoding RNA. Cells. 2022;11 doi: 10.3390/cells11121930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel A.A., Sakurai A., Himmel N.J., Cox D.N. Modality specific roles for metabotropic GABAergic signaling and calcium induced calcium release mechanisms in regulating cold nociception. Front. Mol. Neurosci. 2022;15 doi: 10.3389/fnmol.2022.942548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vlaming K.E., van Wijnbergen K., Kaptein T.M., Nijhuis M., Kootstra N.J., de Bree G.J., Geijtenbeek T.B. Crosstalk between TLR8 and RIG-I-like receptors enhances antiviral immune responses. Front. Med. 2023;10 doi: 10.3389/fmed.2023.1146457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta S., Guleria R.S. Involvement of Nuclear Factor-kappaB in Inflammation and Neuronal Plasticity Associated with Post-Traumatic Stress Disorder. Cells. 2022;11:2034. doi: 10.3390/cells11132034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suwała S., Rzeszuto J., Glonek R., Krintus M., Junik R. Is Restless Legs Syndrome De Facto Thyroid Disease? Biomedicines. 2022;10:2502. doi: 10.3390/biomedicines10102502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schell C., Schleich R., Walker F., Yazdi A.S., Lerche H., Röcken M., Axmann D., Ghoreschi K., Eberle F.C. Restless legs syndrome in psoriasis: an unexpected comorbidity. Eur. J. Dermatol. 2015;25:255–260. doi: 10.1684/ejd.2015.2525. [DOI] [PubMed] [Google Scholar]
- 58.Demir S., Kucuk A., Altas M., Cure E. Restless Leg Syndrome and Sleep Disorders in Patients with Rheumatoid Arthritis and Its Relation with Anemia Parameters. Acta Medica. 2021;64:137–144. doi: 10.14712/18059694.2021.24. [DOI] [PubMed] [Google Scholar]
- 59.Weinstock L.B., Bosworth B.P., Scherl E.J., Li E., Iroku U., Munsell M.A., Mullin G.E., Walters A.S. Crohn's disease is associated with restless legs syndrome. Inflamm. Bowel Dis. 2010;16:275–279. doi: 10.1002/ibd.20992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinstock L.B., Walters A.S. Restless legs syndrome is associated with irritable bowel syndrome and small intestinal bacterial overgrowth. Sleep Med. 2011;12:610–613. doi: 10.1016/j.sleep.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 61.Weinstock L.B., Walters A.S., Mullin G.E., Duntley S.P. Celiac disease is associated with restless legs syndrome. Dig. Dis. Sci. 2010;55:1667–1673. doi: 10.1007/s10620-009-0943-9. [DOI] [PubMed] [Google Scholar]
- 62.Ohto U. Activation and regulation mechanisms of NOD-like receptors based on structural biology. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.953530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y.H., Gao X., Tang Y.R., Chen F.Q., Yu Y., Sun M.J., Li Y. Resolvin D1 Alleviates Mechanical Allodynia via ALX/FPR2 Receptor Targeted Nod-like Receptor Protein 3/Extracellular Signal-Related Kinase Signaling in a Neuropathic Pain Model. Neuroscience. 2022;494:12–24. doi: 10.1016/j.neuroscience.2022.04.019. [DOI] [PubMed] [Google Scholar]
- 64.Possemato E., La Barbera L., Nobili A., Krashia P., D'Amelio M. The role of dopamine in NLRP3 inflammasome inhibition: Implications for neurodegenerative diseases. Ageing Res. Rev. 2023;87 doi: 10.1016/j.arr.2023.101907. [DOI] [PubMed] [Google Scholar]
- 65.Nolan R.A., Reeb K.L., Rong Y., Matt S.M., Johnson H.S., Runner K., Gaskill P.J. Dopamine activates NF-kappaB and primes the NLRP3 inflammasome in primary human macrophages. Brain Behav. Immun. Health. 2020;2 doi: 10.1016/j.bbih.2019.100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pike A.F., Longhena F., Faustini G., van Eik J.M., Gombert I., Herrebout M.A.C., Fayed M.M.H.E., Sandre M., Varanita T., Teunissen C.E., et al. Dopamine signaling modulates microglial NLRP3 inflammasome activation: implications for Parkinson's disease. J. Neuroinflammation. 2022;19:50. doi: 10.1186/s12974-022-02410-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang Z., Yin X., Wang M., Wang Y., Li F., Gao Y., Han G., Gao Z., Wang Z. beta-Hydroxybutyrate alleviates pyroptosis in MPP(+)/MPTP-induced Parkinson's disease models via inhibiting STAT3/NLRP3/GSDMD pathway. Int. Immunopharmacol. 2022;113 doi: 10.1016/j.intimp.2022.109451. [DOI] [PubMed] [Google Scholar]
- 68.Alonso-Navarro H., García-Martín E., Agúndez J.A.G., Jiménez-Jiménez F.J. Association between restless legs syndrome and other movement disorders. Neurology. 2019;92:948–964. doi: 10.1212/WNL.0000000000007500. [DOI] [PubMed] [Google Scholar]
- 69.Limousin N., Blasco H., Corcia P., Arnulf I., Praline J. The high frequency of restless legs syndrome in patients with amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2011;12:303–306. doi: 10.3109/17482968.2011.557736. [DOI] [PubMed] [Google Scholar]
- 70.Wang S., Zheng X., Huang J., Lin J., Yang T., Xiao Y., Jiang Q., Li C., Shang H. Restless legs syndrome and cognitive function among adults: a systematic review and meta-analysis. J. Neurol. 2023;270:1361–1370. doi: 10.1007/s00415-022-11484-2. [DOI] [PubMed] [Google Scholar]
- 71.Kim K.Y., Kim E.H., Lee M., Ha J., Jung I., Kim E. Restless leg syndrome and risk of all-cause dementia: a nationwide retrospective cohort study. Alzheimer's Res. Ther. 2023;15:46. doi: 10.1186/s13195-023-01191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo C., Chen L., Wang Y. Substance abuse and neurodegenerative diseases: focus on ferroptosis. Arch. Toxicol. 2023;97:1519–1528. doi: 10.1007/s00204-023-03505-4. [DOI] [PubMed] [Google Scholar]
- 73.Hao S., Liang B., Huang Q., Dong S., Wu Z., He W., Shi M. Metabolic networks in ferroptosis. Oncol. Lett. 2018;15:5405–5411. doi: 10.3892/ol.2018.8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khachatryan S.G., Ferri R., Fulda S., Garcia-Borreguero D., Manconi M., Muntean M.L., Stefani A. Restless legs syndrome: Over 50 years of European contribution. J. Sleep Res. 2022;31 doi: 10.1111/jsr.13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li K., Liu B., Wang F., Bao J., Wu C., Huang X., Hu F., Xu Z., Ren H., Yang X. Decreased serum ferritin may be associated with increased restless legs syndrome in Parkinson's disease (PD): a meta-analysis for the diagnosis of RLS in PD patients. Int. J. Neurosci. 2019;129:995–1003. doi: 10.1080/00207454.2019.1608200. [DOI] [PubMed] [Google Scholar]
- 76.Kenkhuis B., Bush A.I., Ayton S. How iron can drive neurodegeneration. Trends Neurosci. 2023;46:333–335. doi: 10.1016/j.tins.2023.02.003. [DOI] [PubMed] [Google Scholar]
- 77.Liu C., Li Z., Li B., Liu W., Zhang S., Qiu K., Zhu W. Relationship between ferroptosis and mitophagy in cardiac ischemia reperfusion injury: a mini-review. PeerJ. 2023;11 doi: 10.7717/peerj.14952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singleton A., Hardy J. Progress in the genetic analysis of Parkinson's disease. Hum. Mol. Genet. 2019;28:R215–R218. doi: 10.1093/hmg/ddz183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang W., Liu H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 80.Huang Y., Wang P., Morales R., Luo Q., Ma J. Map2k5-Deficient Mice Manifest Phenotypes and Pathological Changes of Dopamine Deficiency in the Central Nervous System. Front. Aging Neurosci. 2021;13 doi: 10.3389/fnagi.2021.651638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim T.W., Koo S.Y., Riessland M., Cho H., Chaudhry F., Kolisnyk B., Russo M.V., Saurat N., Mehta S., Garippa R., et al. TNF-NFkB-p53 axis restricts in vivo survival of hPSC-derived dopamine neuron. bioRxiv. 2023 doi: 10.1101/2023.03.29.534819. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li J., Cao F., Yin H.L., Huang Z.J., Lin Z.T., Mao N., Sun B., Wang G. Ferroptosis: past, present and future. Cell Death Dis. 2020;11:88. doi: 10.1038/s41419-020-2298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harrer P., Mirza-Schreiber N., Mandel V., Roeber S., Stefani A., Naher S., Wagner M., Gieger C., Waldenberger M., Peters A., et al. Epigenetic Association Analyses and Risk Prediction of RLS. Mov. Disord. 2023;38:1410–1418. doi: 10.1002/mds.29440. [DOI] [PubMed] [Google Scholar]
- 84.Ryan J., Wrigglesworth J., Loong J., Fransquet P.D., Woods R.L. A Systematic Review and Meta-analysis of Environmental, Lifestyle, and Health Factors Associated With DNA Methylation Age. J. Gerontol. A Biol. Sci. Med. Sci. 2020;75:481–494. doi: 10.1093/gerona/glz099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Esen R., Ediz L., Gulcu E., Demirdag F., Turktas U., Guner S., Ebinc S., Cifci A., Demir C. Restless legs syndrome in multiple myeloma patients. J. Clin. Med. Res. 2012;4:318–322. doi: 10.4021/jocmr1083w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aricò D., Raggi A., Siragusa M., Zucconi M., Ferri R. Restless legs syndrome as the presenting symptom of multiple myeloma. J. Clin. Sleep Med. 2013;9:383–385. doi: 10.5664/jcsm.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mogavero M.P., DelRosso L.M., Fanfulla F., Bruni O., Ferri R. Sleep disorders and cancer: State of the art and future perspectives. Sleep Med. Rev. 2021;56 doi: 10.1016/j.smrv.2020.101409. [DOI] [PubMed] [Google Scholar]
- 88.Silvani A., Ghorayeb I., Manconi M., Li Y., Clemens S. Putative Animal Models of Restless Legs Syndrome: A Systematic Review and Evaluation of Their Face and Construct Validity. Neurotherapeutics. 2023;20:154–178. doi: 10.1007/s13311-022-01334-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Romigi A., Vitrani G. Improvement of restless legs syndrome by varenicline as antismoking treatment. J. Clin. Sleep Med. 2013;9:1089–1090. doi: 10.5664/jcsm.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lyu S., Xing H., DeAndrade M.P., Liu Y., Perez P.D., Yokoi F., Febo M., Walters A.S., Li Y. The Role of BTBD9 in Striatum and Restless Legs Syndrome. eNeuro. 2019;6 doi: 10.1523/ENEURO.0277-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ramaswamy S., Colangelo C., Markram H. Data-Driven Modeling of Cholinergic Modulation of Neural Microcircuits: Bridging Neurons, Synapses and Network Activity. Front. Neural Circuits. 2018;12:77. doi: 10.3389/fncir.2018.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dasgupta R., Seibt F., Beierlein M. Synaptic Release of Acetylcholine Rapidly Suppresses Cortical Activity by Recruiting Muscarinic Receptors in Layer 4. J. Neurosci. 2018;38:5338–5350. doi: 10.1523/JNEUROSCI.0566-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lanza G., Cantone M., Lanuzza B., Pennisi M., Bella R., Pennisi G., Ferri R. Distinctive patterns of cortical excitability to transcranial magnetic stimulation in obstructive sleep apnea syndrome, restless legs syndrome, insomnia, and sleep deprivation. Sleep Med. Rev. 2015;19:39–50. doi: 10.1016/j.smrv.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 94.Ferri R., Cosentino F.I.I., Manconi M., Rundo F., Bruni O., Zucconi M. Increased electroencephalographic high frequencies during the sleep onset period in patients with restless legs syndrome. Sleep. 2014;37:1375–1381. doi: 10.5665/sleep.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ferri R., Manconi M., Aricò D., Sagrada C., Zucconi M., Bruni O., Oldani A., Ferini-Strambi L. Acute dopamine-agonist treatment in restless legs syndrome: effects on sleep architecture and NREM sleep instability. Sleep. 2010;33:793–800. doi: 10.1093/sleep/33.6.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lanza G., Bachmann C.G., Ghorayeb I., Wang Y., Ferri R., Paulus W. Central and peripheral nervous system excitability in restless legs syndrome. Sleep Med. 2017;31:49–60. doi: 10.1016/j.sleep.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 97.Montplaisir J., Boucher S., Gosselin A., Poirier G., Lavigne G. Persistence of repetitive EEG arousals (K-alpha complexes) in RLS patients treated with L-DOPA. Sleep. 1996;19:196–199. doi: 10.1093/sleep/19.3.196. [DOI] [PubMed] [Google Scholar]
- 98.Ferré S., Sarasola L.I., Quiroz C., Ciruela F. Presynaptic adenosine receptor heteromers as key modulators of glutamatergic and dopaminergic neurotransmission in the striatum. Neuropharmacology. 2023;223 doi: 10.1016/j.neuropharm.2022.109329. [DOI] [PubMed] [Google Scholar]
- 99.Ferré S., García-Borreguero D., Allen R.P., Earley C.J. New Insights into the Neurobiology of Restless Legs Syndrome. Neuroscientist. 2019;25:113–125. doi: 10.1177/1073858418791763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Falk J., Castellani V. In: Cellular Migration and Formation of Neuronal Connections. Rubenstein J.L.R., Rakic P., editors. Academic Press; 2013. Chapter 4 - Axon Guidance: Semaphorin/Neuropilin/Plexin Signaling; pp. 69–88. [DOI] [Google Scholar]
- 101.Schormair B., Zhao C., Bell S., Tilch E., Salminen A.V., Pütz B., Dauvilliers Y., Stefani A., Högl B., Poewe W., et al. Identification of novel risk loci for restless legs syndrome in genome-wide association studies in individuals of European ancestry: a meta-analysis. Lancet Neurol. 2017;16:898–907. doi: 10.1016/S1474-4422(17)30327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ono K., Clavairoly A., Nomura T., Gotoh H., Uno A., Armant O., Takebayashi H., Zhang Q., Shimamura K., Itohara S., et al. Development of the prethalamus is crucial for thalamocortical projection formation and is regulated by Olig2. Development. 2014;141:2075–2084. doi: 10.1242/dev.097790. [DOI] [PubMed] [Google Scholar]
- 103.Xie Q., Li Z., Wang Y., Zaidi S., Baranova A., Zhang F., Cao H. Preeclampsia Drives Molecular Networks to Shift Toward Greater Vulnerability to the Development of Autism Spectrum Disorder. Front. Neurol. 2020;11:590. doi: 10.3389/fneur.2020.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koyama Y. Signaling molecules regulating phenotypic conversions of astrocytes and glial scar formation in damaged nerve tissues. Neurochem. Int. 2014;78:35–42. doi: 10.1016/j.neuint.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 105.DelRosso L.M., Reuter-Yuill L.M., Cho Y., Ferri R., Mogavero M.P., Picchietti D.L. Clinical efficacy and safety of intravenous ferric carboxymaltose treatment for restless legs symptoms and low serum ferritin in children with autism spectrum disorder. Sleep Med. 2022;100:488–493. doi: 10.1016/j.sleep.2022.09.021. [DOI] [PubMed] [Google Scholar]
- 106.Wu K., Han W., Tian Q., Li Y., Lu W. Activity- and sleep-dependent regulation of tonic inhibition by Shisa7. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu K., Shepard R.D., Castellano D., Han W., Tian Q., Dong L., Lu W. Shisa7 phosphorylation regulates GABAergic transmission and neurodevelopmental behaviors. Neuropsychopharmacology. 2022;47:2160–2170. doi: 10.1038/s41386-022-01334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schmitz L.J.M., Klaassen R.V., Ruiperez-Alonso M., Zamri A.E., Stroeder J., Rao-Ruiz P., Lodder J.C., van der Loo R.J., Mansvelder H.D., Smit A.B., Spijker S. The AMPA receptor-associated protein Shisa7 regulates hippocampal synaptic function and contextual memory. Elife. 2017;6 doi: 10.7554/eLife.24192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.He H., An F., Huang Q., Kong Y., He D., Chen L., Song H. Metabolic effect of AOS-iron in rats with iron deficiency anemia using LC-MS/MS based metabolomics. Food Res. Int. 2020;130 doi: 10.1016/j.foodres.2019.108913. [DOI] [PubMed] [Google Scholar]
- 110.Forghani R., Chandrasekaran A., Papoian G., Giniger E. A new view of axon growth and guidance grounded in the stochastic dynamics of actin networks. Open Biol. 2023;13 doi: 10.1098/rsob.220359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim I.H., Rossi M.A., Aryal D.K., Racz B., Kim N., Uezu A., Wang F., Wetsel W.C., Weinberg R.J., Yin H., Soderling S.H. Spine pruning drives antipsychotic-sensitive locomotion via circuit control of striatal dopamine. Nat. Neurosci. 2015;18:883–891. doi: 10.1038/nn.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Trouillon R., Ewing A.G. Actin controls the vesicular fraction of dopamine released during extended kiss and run exocytosis. ACS Chem. Biol. 2014;9:812–820. doi: 10.1021/cb400665f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sydnor V.J., Larsen B., Bassett D.S., Alexander-Bloch A., Fair D.A., Liston C., Mackey A.P., Milham M.P., Pines A., Roalf D.R., et al. Neurodevelopment of the association cortices: Patterns, mechanisms, and implications for psychopathology. Neuron. 2021;109:2820–2846. doi: 10.1016/j.neuron.2021.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Paul K.C., Binder A.M., Horvath S., Kusters C., Yan Q., Rosario I.D., Yu Y., Bronstein J., Ritz B. Accelerated hematopoietic mitotic aging measured by DNA methylation, blood cell lineage, and Parkinson's disease. BMC Genom. 2021;22:696. doi: 10.1186/s12864-021-08009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cederberg K.L.J., Silvestri R., Walters A.S. Vitamin D and Restless Legs Syndrome: A Review of Current Literature. Tremor Other Hyperkinet. Mov. 2023;13:12. doi: 10.5334/tohm.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marano M., Pozzilli V., Magliozzi A., Tabacco G., Naciu A.M., Palermo A., Di Lazzaro V. Leg restlessness and hyperparathyroidism in Parkinson's disease, a further clue to RLS pathogenesis? Front. Neurol. 2023;14 doi: 10.3389/fneur.2023.1113913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Syed Abd Halim S.A., Abd Rashid N., Woon C.K., Abdul Jalil N.A. Natural Products Targeting PI3K/AKT in Myocardial Ischemic Reperfusion Injury: A Scoping Review. Pharmaceuticals. 2023;16:739. doi: 10.3390/ph16050739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zanigni S., Calandra-Buonaura G., Giannini G., Tonon C., Cortelli P., Provini F. The association between restless legs syndrome, cardiovascular and metabolic diseases: hypotheses and evidence from the literature. Arch. Ital. Biol. 2015;153:170–183. doi: 10.12871/0003982920152342. [DOI] [PubMed] [Google Scholar]
- 119.Ferini-Strambi L., Walters A.S., Sica D. The relationship among restless legs syndrome (Willis-Ekbom Disease), hypertension, cardiovascular disease, and cerebrovascular disease. J. Neurol. 2014;261:1051–1068. doi: 10.1007/s00415-013-7065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gottlieb D.J., Somers V.K., Punjabi N.M., Winkelman J.W. Restless legs syndrome and cardiovascular disease: a research roadmap. Sleep Med. 2017;31:10–17. doi: 10.1016/j.sleep.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guo J., Hu Z., Ren L., Zhao W., Zuo R., Guo S., Jia C., Gao W. Circulating tumor necrosis factor-alpha, interleukin-1beta, and interleukin-17A estimates increased major adverse cardiac event risk in acute myocardial infarction patients. J. Clin. Lab. Anal. 2023;37 doi: 10.1002/jcla.24853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Maiolino G., Bisogni V., Soranna D., Pengo M.F., Pucci G., Vettor R., Fava C., Colussi G.L., Bilo G., Lombardi C., et al. Effects of insomnia and restless legs syndrome on sleep arterial blood pressure: A systematic review and meta-analysis. Sleep Med. Rev. 2021;59 doi: 10.1016/j.smrv.2021.101497. [DOI] [PubMed] [Google Scholar]
- 123.Ferri R., Silvani A., Mogavero M.P., Rundo F., Bruni O., Picchietti D.L., DelRosso L.M. Heart rate changes associated with the different types of leg movements during sleep in children, adolescents and adults with restless legs syndrome. J. Sleep Res. 2021;30 doi: 10.1111/jsr.13379. [DOI] [PubMed] [Google Scholar]
- 124.DelRosso L.M., Mogavero M.P., Ferri R. Effect of Sleep Disorders on Blood Pressure and Hypertension in Children. Curr. Hypertens. Rep. 2020;22:88. doi: 10.1007/s11906-020-01100-x. [DOI] [PubMed] [Google Scholar]
- 125.Van Der Westhuizen E.T., Summers R.J., Halls M.L., Bathgate R.A.D., Sexton P.M. Relaxin receptors--new drug targets for multiple disease states. Curr. Drug Targets. 2007;8:91–104. doi: 10.2174/138945007779315650. [DOI] [PubMed] [Google Scholar]
- 126.Padhan P., Maikap D., Pathak M. Restless leg syndrome in rheumatic conditions: Its prevalence and risk factors, a meta-analysis. Int. J. Rheum. Dis. 2023;26:1111–1119. doi: 10.1111/1756-185X.14710. [DOI] [PubMed] [Google Scholar]
- 127.Bentley A.J., Rosman K.D., Mitchell D. Gender differences in the presentation of subjects with restless legs syndrome. Sleep Med. 2006;7:37–41. doi: 10.1016/j.sleep.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 128.Tempest N., Boyers M., Carter A., Lane S., Hapangama D.K. Premenopausal Women With a Diagnosis of Endometriosis Have a Significantly Higher Prevalence of a Diagnosis or Symptoms Suggestive of Restless Leg Syndrome: A Prospective Cross-Sectional Questionnaire Study. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.599306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Prosperetti C., Manconi M. Restless Legs Syndrome/Willis-Ekbom Disease and Pregnancy. Sleep Med. Clin. 2015;10:323–329. doi: 10.1016/j.jsmc.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 130.El Khoudary S.R., Nasr A. Cardiovascular Disease in Women: Does Menopause Matter? Curr. Opin. Endocr. Metab. Res. 2022;27 doi: 10.1016/j.coemr.2022.100419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ferri R., DelRosso L.M., Silvani A., Cosentino F.I.I., Picchietti D.L., Mogavero P., Manconi M., Bruni O. Peculiar lifespan changes of periodic leg movements during sleep in restless legs syndrome. J. Sleep Res. 2020;29 doi: 10.1111/jsr.12896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw sequencing data of RLS patients are publicly available at ArrayExpress with the accession number E-MTAB-13155, as listed in the key resources table. Raw sequencing data of controls are publicly available at ArrayExpress with the accession number E-MTAB-11326, as listed in the key resources table.
-
•
This study does not report original new code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.