Abstract
Mammalian type C retroviral envelope proteins contain a variable proline-rich region (PRR), located between the N-terminal receptor-binding domain and the more highly conserved C-terminal portion of the surface (SU) subunit. We have investigated the role of the PRR in the function of murine leukemia virus (MuLV) envelope protein. In the MuLVs, the PRR contains a highly conserved N-terminal sequence and a hypervariable C-terminal sequence. Despite this variability, the amphotropic PRR could functionally substitute for the ecotropic PRR. The hypervariable region of the PRR was not absolutely required for envelope protein function. However, truncations in this region resulted in decreased levels of both the SU and TM proteins in viral particles and increased amounts of the uncleaved precursor protein, Pr85. In contrast, the N-terminal conserved region was essential for viral infectivity. Deletion of this region prevented the stable incorporation of envelope proteins into viral particles in spite of normal envelope protein processing, wild-type levels of cell surface expression, and a wild-type ability to induce syncytia in an XC cell cocultivation assay. However, higher levels of the SU protein were shed into the supernatant, suggesting a defect in SU-TM interactions. Our data are most consistent with a role for the PRR in stabilizing the overall structure of the protein, thereby affecting the proper processing of Pr85, SU-TM interactions, and the stable incorporation of envelope proteins into viral particles. In addition, we have demonstrated that the PRR can tolerate the insertion of a peptide-binding domain, making this a potentially useful site for constructing targetable retroviral vectors.
Retroviruses enter their host cells through an interaction between the viral envelope protein and a specific cell surface receptor (1, 19, 42). Binding of the envelope protein to its receptor leads to the fusion of viral and host cell membranes, which allows internalization of the viral core. Retroviral envelope proteins are composed of two subunits, a surface (SU) protein which binds to the cellular receptor (42), and a transmembrane (TM) protein which is required for the membrane fusion events after receptor binding (43).
Within the type C mammalian retroviruses, the N-terminal portion of SU is responsible for receptor recognition (3, 23, 25). The more conserved C-terminal region of the protein is believed to associate with the TM protein and to be involved in the postbinding events that lead to fusion of the viral and cellular membranes (14, 30, 32, 36). The SU proteins of the type C mammalian retroviruses also contain a segment of proline-rich sequence between the N-terminal receptor binding domain and the more conserved C-terminal portion (21). This proline-rich region (PRR) is found in the SU proteins of the gibbon ape leukemia viruses (7), the feline leukemia viruses (FeLVs) (9, 26), and the different MuLV subtypes (5, 21, 28). The PRR contains a conserved N-terminal domain of 14 or 15 amino acids, but the remainder of the PRR sequence varies both in length and in sequence among different viruses (21, 28).
The role of the PRR in envelope protein function has not been determined. It has been speculated that the PRR might participate in postbinding events during viral entry, since point mutations and linker insertions in this region produce fusion-defective envelope proteins (2, 14) and can prevent the stable association of the SU and TM subunits (14). In addition, the PRR has been shown to be capable of influencing the receptor recognition properties of the N-terminal region of SU (3, 28), although whether this is a result of the PRR containing a direct receptor contact point or whether it is due to a more indirect effect on the structure of this region remains to be determined. Structural studies of a peptide corresponding to the FeLV PRR indicated that this region may exist as a polyproline β-turn helix with a propensity to oligomerize, that could fold into a separate domain in SU (11).
We wished to study in more detail the role of the PRR in MuLV envelope protein function. We looked initially at the interchangeability of the PRRs from the ecotropic and amphotropic MuLV envelope proteins and demonstrated that the amphotropic PRR could functionally substitute for the ecotropic region, despite differences in the length and sequence of the variable regions. In addition, analysis of a series of truncations of the variable C-terminal region revealed that over half of the sequence could be truncated without significant loss of envelope protein function, although further truncations had effects on envelope protein incorporation and the stability of SU-TM interaction. In contrast, we found that the N-terminal conserved region was absolutely essential for viral infectivity. Deletion of this sequence prevented the incorporation of envelope proteins into virions and completely abolished the viral titer.
One of the major goals of retroviral vector development is the generation of targetable vectors that could be used to achieve cell-specific gene delivery (4, 6). The apparent flexibility of the PRR variable sequence led us to investigate whether this region would be able to tolerate the insertion of a peptide that could redirect binding of the envelope protein to a new cell surface protein. As a proof of this principle, we inserted a 16-amino-acid collagen-binding peptide into different locations in the PRR. Retroviral vector particles containing such chimeric envelope proteins achieved a wild-type titer on NIH 3T3 cells and conferred specific binding to collagen, indicating that the PRR may be a promising site for ligand insertion when constructing targetable vectors.
MATERIALS AND METHODS
Cell lines.
NIH 3T3 and 293T cells were maintained in Dulbecco’s modified Eagle’s medium (GIBCO-BRL, Grand Island, N.Y.). XC cells were grown in Eagle’s minimal essential medium (GIBCO-BRL). Both media were supplemented with 10% fetal calf serum (HyClone, Logan, Utah) and 2 mM glutamine.
Envelope protein expression plasmids.
pCEE+ is an expression plasmid for the wild-type Moloney MuLV (MoMuLV) ecotropic envelope protein (23); pCAE contains the 4070A amphotropic envelope protein (25). Two restriction sites, AvrII and NgoMI, were generated as silent point mutations by recombinant PCR (18) at the 5′ and 3′ ends of the ecotropic PRR, respectively, creating plasmid pCEE*. The oligonucleotides used were 5′-GACTCAGATACCAAAACCTAGGACCCCGCGTC-3′ and 5′-CCCAACTTCCACCGGCCGGCACGGAAAATAGGCTGC-3′; the AvrII and NgoMI restrictions sites are underlined. The amphotropic PRR was amplified by PCR from plasmid pCAE with the addition of AvrII and NgoMI sites at the ends of the PCR fragment and used to replace the PRR in pCEE*, generating plasmid E/A-PRR. Similarly, C-terminal and N-terminal PRR truncation mutants of E/A-PRR were created by replacing the CEE* PRR with fragments of the amphotropic PRR of different lengths, generated as PCR products with terminal AvrII and NgoMI sites. All mutants were confirmed by sequencing the final constructs.
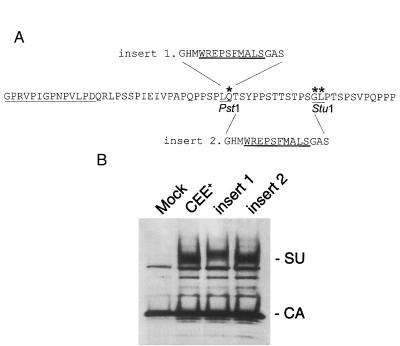
To facilitate the insertion of a 16-amino-acid peptide containing a collagen-binding domain from von Willebrand’s factor (39) into the PRR of E/A-PRR, the restrictions sites PstI and StuI were generated in the PRR as nonsilent mutations through recombinant PCR at the sites shown in Fig. 7. The oligonucleotides used were 5′-GCCACCTAGCCCCCTGCAGACCAGTTACCCCCCT-3′ for PstI and 5′-ACCAGTACACCCTCAGGCCTCCCTACAAGTCCAAGT-3′ for StuI. The collagen binding peptide sequence was created as two overlapping oligonucleotides, flanked by either PstI ends (insert 1) or PstI and StuI ends (insert 2). Insert 1 was encoded by the sequence 5′-GGGCCATATGTGGCGCGAACCGAGCTTCATGGCTCTGAGCGGTGCTAGCCTGCA-3′, and insert 2 was encoded by the sequence 5′-GGGCCATATGTGGCGCGAACCGAGCTTCATGGCTCTGAGCGGTGCTTCAGG-3′.
FIG. 7.
Insertion of a collagen-binding domain into the PRR of E/A-PRR. (A) A 16-amino-acid peptide containing a collagen-binding domain (underlined) from von Willebrand’s factor (39) was inserted into two sites in the PRR of E/A-PRR. PstI and StuI sites were created to facilitate these insertions. Insert 1 was at the PstI site, and insert 2 was a replacement of the PstI-StuI fragment. The amino acids marked with asterisks are nonsilent mutations created by the generation of the PstI and StuI sites. (B) Western blot analysis shows that both of the chimeric envelope proteins were incorporated efficiently into viral particles.
Retroviral vector production and titer determination.
Retroviral vectors were produced by transient transfection of 293T cells by calcium phosphate precipitation, essentially as described previously (16, 37). The plasmids used were the MoMuLV gag-pol expression plasmid pHIT60 (37); the retroviral vector pCnBg (16), which expresses lacZ and neo; and an env expression plasmid. At 16 h posttransfection, the cells were washed and fresh medium was added. At 48 h later, the culture supernatant was collected and filtered through a 0.45-μm-pore-size filter and either used immediately or stored at −70°C. The virus titer was determined as G418-resistant CFU per milliliter. For titer determination NIH 3T3 cells (3 × 104 cells per 30-mm well) were plated in 3 ml of medium; 18 h later, the medium was replaced with 1 ml of appropriately diluted viral supernatant containing Polybrene (8 μg/ml) and the mixture was incubated for 2 h at 37°C. An additional 2 ml of medium was then added, and the cultures were incubated for a further 18 h. The cells were then selected for 10 days in medium containing 0.6 mg of G418 per ml, and resistant colonies were scored by methylene blue staining.
Western blot analysis of envelope proteins in viral particles and cell lysates.
Retroviral particles generated by transient transfection of 293T cells were pelleted through 20% sucrose at 16,000 × g and 4°C for 30 min. Viral pellets were resuspended in 30 μl of 2× sodium dodecyl sulfate (SDS) gel-loading buffer (100 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 0.01% bromphenol blue, 1.4 M β-mercaptoethanol) and boiled for 5 min. Viral protein samples were resolved on a precast 8 to 16% polyacrylamide gel (Novex, San Diego, Calif.) and transferred onto an Immobilon membrane (Millipore, Bedford, Mass.). The blot was blocked overnight at 4°C in 5% powdered milk in TBS (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.25% Tween 20), and incubated with primary antibodies at room temperature for 2 h. After being washed in TBS buffer for 30 min, the blots were incubated with secondary antibodies for 1 h at room temperature and then given a further 30-min wash in TBS. Specific proteins were visualized with the enhanced chemiluminescence detection system (Amersham International plc., Arlington Heights, Ill.). The primary antibodies used were goat anti-Rauscher MuLV gp69/71, 1:3,000 dilution (Quality Biotech, Camden, N.J.; lot 79S656); goat anti-Raucher MuLV p30, 1:10,000 dilution (Quality Biotech; lot 78S221); and rat anti-AKR MuLV p15E (BABCO, Berkeley, Calif.; lot 42/114) (31). The secondary antibodies were horseradish peroxidase-conjugated rabbit anti-goat immunoglobulin G (IgG) (1:10,000) and horseradish peroxidase-conjugated goat anti-rabbit IgG (1:10,000) (Pierce, Rockford, Ill.).
To analyze the form of the envelope proteins present in cell lysates, 293T cells were transfected with envelope protein expression plasmids (30 μg per 10-cm plate) and lysates were prepared by incubating the cells in 500 μl of lysis buffer (20 mM Tris-HCl [pH 7.5], 1% Triton X-100, 0.05% SDS, 5 mg of sodium deoxycholate per ml, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride) for 10 min at 4°C and centrifuging them at 10,000 × g for 10 min to pellet the nuclei. Envelope proteins were denatured by treating the lysates with 0.5% SDS and 1% β-mercaptoethanol at 100°C for 5 min and then deglycosylated with 50 mM sodium phosphate (pH 7.5)–1% Nonidet P-40 (NP-40)–500 U of N-glycosidase F (New England Biolabs, Beverly, Mass.) at 37°C for 1 h. Envelope proteins were detected by Western blotting as described above.
Cocultivation fusion assay.
293T cells (6 × 105 cells) were plated in a 60-mm dish and transfected with 15 μg of envelope expression vector DNA by calcium phosphate precipitation. At 16 h posttransfection, the cells were washed and fresh medium was added. Nonirradiated XC cells (106 cells) were added to the plate 24 h posttransfection, and the culture was incubated for an additional 24 h at 37°C. Syncytia were observed and counted by light microscopy after staining of the cells with 1% methylene blue in methanol.
Cell surface expression of envelope proteins.
293T cells in a 60-mm plate were transiently transfected with 15 μg of an envelope expression vector. At 48 h posttransfection, the cells were harvested with enzyme-free cell dissociation solution (GIBCO-BRL) and washed with phosphate-buffered saline (PBS). The cells (106) were incubated at 4°C for 1 h with the rat monoclonal antibody 83A25 (10), which recognizes the C terminus of MoMuLV gp70 envelope protein. The cells were washed with 10% goat serum in PBS and incubated with fluorescein isothiocyanate-labeled goat anti-rat IgG (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.) at 4°C for 1 h. They were then washed again and fixed in 4% paraformaldehyde in PBS. The fluorescence intensity of the cell samples were measured with a FACStar Plus flow cytometer (Becton Dickinson, San Jose, Calif.).
Immunoprecipitation of SU from cell culture supernatants.
293T cells in a 60-mm plate were transiently transfected with 15 μg of an envelope expression vector, and SU was immunoprecipitated from culture supernatants as described previously (45). At 48 h posttransfection, the cells were washed with PBS and incubated in cell-labeling medium (Dulbecco’s modified Eagle’s medium without methionine and cysteine and containing 10% dialyzed fetal calf serum) for 1 h at 37°C. The cells were then incubated for 8 h in cell-labeling medium containing 100 μCi each of [35S]methionine and [35S]cysteine (Amersham), harvested, and lysed in immunoprecipitation buffer (10 mM Na2HPO4 [pH 7.4], 150 mM NaCl, 0.5% Triton X-100, 0.1% SDS, 0.05% sodium deoxycholate) for 20 min at 4°C. After centrifugation at 4°C for 10 min at 16,000 × g, the supernatants were incubated with 8 μl of goat anti-Rauscher MuLV gp69/71 antiserum (Quality Biotech; lot 79S656) and 20 μl of protein G-Sepharose (Sigma, St. Louis, Mo.) overnight at 4°C. The samples were then centrifuged at 80 × g for 2 min, and the pellets were resuspended and washed once each in washing buffer 1 (100 mM Tris-Cl [pH 7.5], 150 mM NaCl, 2 mM EDTA, 0.2% NP-40), washing buffer 2 (100 mM Tris-Cl [pH 7.5], 500 mM NaCl, 2 mM EDTA, 0.2% NP-40), and washing buffer 3 (10 mM Tris-Cl [pH 7.5]). The samples were centrifuged at 80 × g for 1 min and resuspended in 30 μl of 2× SDS gel-loading buffer and boiled for 5 min. They were resolved by SDS-polyacrylamide gel electrophoresis on 8 to 16% polyacrylamide gels. The gels were fixed in 5% acetic acid–5% isopropanol for 20 min, washed in water for 15 min, and soaked in Autofluor (National Diagnostics, Atlanta, Ga.) for 30 min to enhance the signal. The gel was dried and exposed to BioMax MR film (Eastman Kodak, Rochester, N.Y.) at −70°C overnight.
Collagen-binding assay.
Each well of a 24-well collagen-coated plate (Becton-Dickinson Labware, Bedford, Mass.) was incubated with 500 μl of retroviral vector supernatant for 1 h at room temperature, washed with PBS, and incubated for 1 h with 500 μl of hybridoma supernatant containing the rat monoclonal anti-SU antibody, 83A25 (10). Following a further PBS wash, the well was incubated with 200 μl of goat anti-rat antibody (Zymed, San Francisco, Calif.) at a 1:50 dilution for 30 min, washed with PBS, incubated with 200 μl of rat peroxidase anti-peroxidase antibody (Sternberger Monoclonals Inc., Lutherville, Mass.) at a 1:50 dilution for 30 min, and washed with PBS. Finally, 500 μl of diaminobenzidine/nickel chloride was added and the resulting color change was monitored at 600 nm with a Rainbow Spectra plate reader.
RESULTS
Generation of a chimeric ecotropic envelope protein containing the amphotropic PRR.
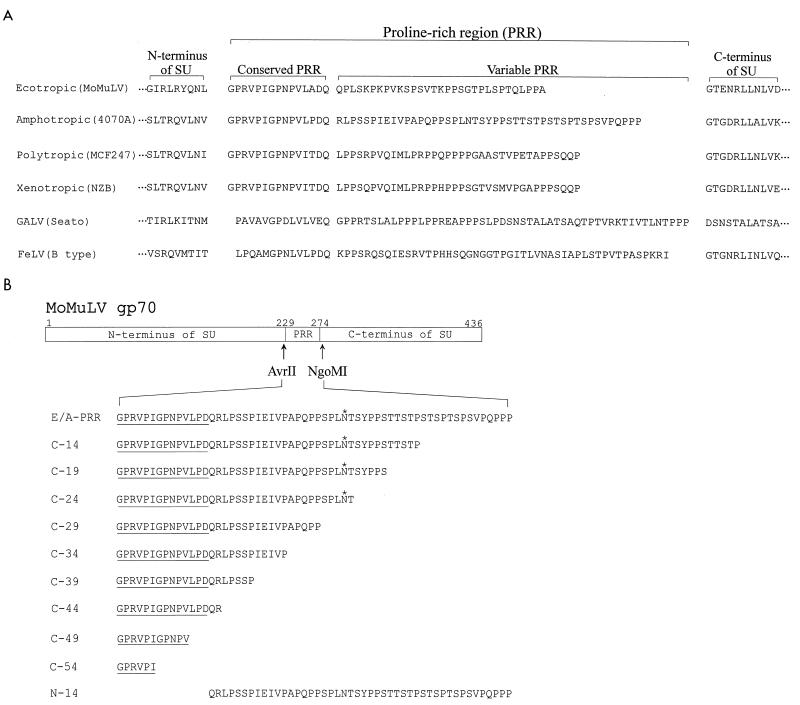
The MuLV envelope protein is composed of two subunits, the SU protein, gp70, and the TM protein, p15E, which are cleaved from a precursor protein, Pr85 (19). Amino acid sequence alignments of the SU proteins of the type C retroviruses have previously identified a variable PRR located between the N-terminal receptor-binding domains and the more highly conserved C-terminal regions (21). In the MuLVs, the PRR is composed of 30 to 60 amino acids, with a proline content of about 30%. The N-terminal 14 or 15 amino acids form a distinct motif, which is highly conserved in all the major subtypes of the MuLVs, while the remainder of the PRR varies in both length and sequence (Fig. 1A).
FIG. 1.
(A) Sequence alignment of the PRR from several mammalian type C retroviruses. The PRR links the N-terminal receptor-binding domain and the more highly conserved C-terminal part of the SU protein. The N-terminal region of the PRR is conserved among the different viruses and is highly conserved within the different MuLV subtypes shown. In contrast, the C-terminal portion of the PRR varies in both sequence and length. (B) Construction of envelope protein mutants. AvrII and NgoMI restriction enzyme sites were introduced into the MoMuLV SU to flank the PRR. These sites were used to replace the ecotropic PRR with either the full-length amphotropic PRR sequence (E/A-PRR) or C-terminal truncations of that sequence. The truncations are named according to the number of amino acids deleted from the C terminus, and construct N-14 has the N-terminal 14 conserved amino acids deleted. The asterisks indicate a potential N-linked glycosylation site present in the amphotropic PRR.
To investigate the requirements for the function of the PRR, we constructed a chimeric envelope protein, E/A-PRR, in which the amphotropic PRR was used to exactly replace the ecotropic MoMuLV PRR (Fig. 1B). The amphotropic sequence is 14 amino acids longer than the ecotropic PRR, contains an additional potential N-linked glycosylation site, and varies considerably in sequence from the ecotropic PRR in the C terminus. We produced retroviral vector particles containing the E/A-PRR protein and tested the function of the protein by analyzing the ability of these retroviral vectors to transduce NIH 3T3 cells. Vectors containing the E/A-PRR protein were able to transduce NIH 3T3 cells at titers similar to those for the wild-type ecotropic envelope protein (Table 1), indicating that the amphotropic sequence could functionally substitute for the ecotropic PRR and that the precise PRR sequence was not essential for the function of the MoMuLV envelope protein.
TABLE 1.
Summary of characteristics of envelope proteins
| Envelope protein | Viral titer (106 CFU/ml)a | Cell-cell fusionb | CSEc | Incorporationd of:
|
Cell lysatee | SU in culture supernatantf | ||
|---|---|---|---|---|---|---|---|---|
| SU | TM | Pr85 | ||||||
| Wild-type CEE+ | 3.9 ± 0.4 | +++ | +++ | ++++ | ++++ | − | Pr85 = SU | + |
| E/A-PRR | 1.8 ± 0.4 | +++ | +++ | ++++ | ++++ | − | Pr85 = SU | + |
| C-14 | 3.5 ± 0.6 | +++ | +++ | ++++ | ++++ | − | Pr85 = SU | + |
| C-19 | 2.8 ± 0.5 | +++ | +++ | +++ | +++ | − | Pr85 > SU | + |
| C-24 | 2.9 ± 0.5 | + | +++ | ++ | ++ | − | Pr85 > SU | ++ |
| C-29 | 2.4 ± 0.3 | ± | +++ | ++ | ++ | − | Pr85 > SU | ++ |
| C-34 | 0.8 ± 0.3 | − | ++ | + | + | + | Pr85 > SU | ++ |
| C-39 | 0.12 ± 0.03 | − | ++ | ± | ± | ++ | Pr85 > SU | + |
| C-44 | 0.19 ± 0.05 | − | +++ | ± | ± | +++ | Pr85 only | ± |
| C-49 | 0.23 ± 0.05 | − | +++ | ± | ± | +++ | Pr85 only | ± |
| C-54 | 0 | − | + | − | − | − | Pr85 only | ++++ |
| N-14 | 0 | +++ | +++ | − | − | − | Pr85 = SU | ++ |
Viral titer on NIH 3T3 cells of retroviral vectors containing envelope proteins. Each titer value is the average of three independent experiments ± standard error.
Extent of syncytium formation in 293T/XC cocultivation assays at 37°C. +++, 11 to 20 syncytia per field; +, 1 to 5 syncytia per field; +, only a few syncytia observed on the entire assay plate; −, no syncytia detected.
Cell surface expression (CSE) of envelope protein on transfected 293T cells, as determined by FACScan analysis. +++, wild-type level of cell surface expression; ++, 50% of the wild-type level; +, 20% of the wild-type level.
Envelope protein detected in viral particles by Western blot analysis, as determined by densitometry. ++++, wild-type level of protein; +++, 50% of the wild-type level; ++, 20% of the wild-type level; +, 5% of the wild-type level; ±, only trace amounts of envelope protein; −, no envelope protein.
Extent of Pr85-to-SU processing in 293T cell lysates.
SU immunoprecipitated from cell culture supernatants as determined by densitometry. +, wild-type level of SU; ++, 2×, wild-type level; ++++, 4× wild-type level; ±, trace amount of SU.
Minimal length of PRR required for envelope protein function.
Although the precise sequence of the ecotropic PRR was not required for the function of the MoMuLV envelope protein, the suggested role of the PRR as a linker between two different domains of SU may depend on both the high proline content of the region and the length, and therefore the flexibility, of this sequence. To investigate these requirements, we constructed a series of C-terminal truncations of the PRR sequence in E/A-PRR (Fig. 1B) and analyzed the ability of retroviral vectors carrying these proteins to transduce NIH 3T3 cells. This analysis revealed that extensive deletions of the C-terminal hypervariable region of the PRR could be made with little effect on viral infectivity (Table 1). The deletions of 29 and 34 of the 58 amino acids of the amphotropic PRR (constructs C-29 and C-34) resulted in viral titers that were 62 and 21% of those for the wild-type ecotropic envelope protein, respectively. Even the deletion of 49 residues in C-49 produced viral particles that gave titers that were 6% of the wild-type level, even though the deletion in C-49 removed all of the C-terminal variable region and 4 residues of the conserved N-terminal region. However, when the truncation was extended a further 5 amino acids into the N-terminal region (construct C-54), the envelope protein totally lost function and the viral titer became zero.
These data implicated the N-terminal motif in envelope protein function. To investigate more precisely the role of the conserved N terminus in envelope protein function, we deleted this region from E/A-PRR to produce mutant N-14 (Fig. 1B). Mutant N-14 was assessed for its ability to transduce NIH 3T3 cells and, similar to mutant C-54, was found to give no titer (Table 1).
PRR mutants differentially affect virus-cell and cell-cell fusion.
We examined the effects of the PRR deletions on the levels of cell surface expression of the envelope proteins and on the ability of the envelope proteins to direct cell-cell fusion. All of the mutant proteins were detected on the surface of transfected 293T cells, although mutants C-34 and C-39 were present at about 50% the level of the wild-type protein and mutant C-54 was present at only about 20% of the wild-type level (Table 1).
The ability of the proteins to direct cell-cell fusion was measured in a cocultivation assay between 293T cells expressing the envelope proteins and the indicator XC cell line which expresses the ecotropic receptor. XC cells fuse with cells expressing the ecotropic envelope protein on their surface, resulting in the formation of syncytia. This assay revealed that cell-cell fusion was more sensitive to C-terminal truncations of the PRR than was virus-cell fusion as measured by NIH 3T3 cell transduction. Defects in syncytium formation were first observed with the removal of only 24 residues, and truncations beyond 29 residues resulted in completely nonfusogenic proteins.
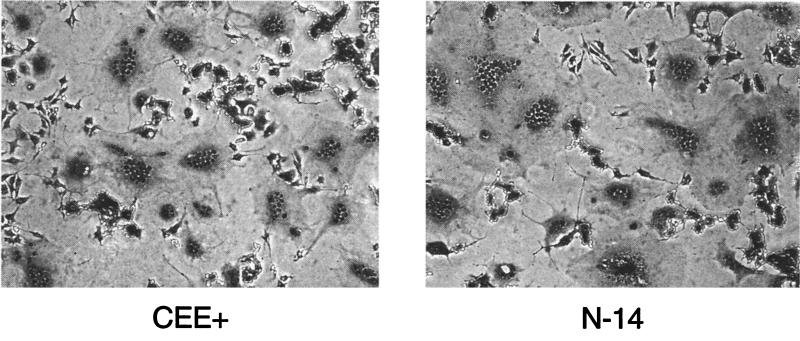
A different phenotype was observed for the N-terminal deletion mutant N-14. This protein was present at normal levels on the cell surface and resulted in wild-type levels of syncytia (Fig. 2). However, despite the fusogenicity of this protein in cell-cell assays, retroviral vector particles produced with this construct gave no titer on NIH 3T3 cells (Table 1).
FIG. 2.
Fusogenicity of the N-14 mutant. 293T cells expressing the wild-type CEE+ or N-14 mutant envelope proteins were cocultivated at 37°C with XC indicator cells, which express the ecotropic receptor. Both envelope proteins resulted in equivalent levels of syncytium formation.
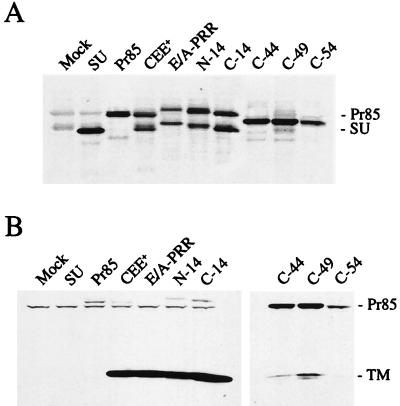
Effects of deletions of the PRR on envelope protein incorporation into viral particles.
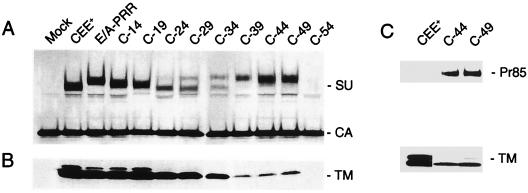
Western blot analysis was carried out to examine the levels of the C-terminally truncated envelope proteins within viral particles (Fig. 3). The SU subunit of the full-length protein, E/A-PRR, ran with a different mobility from the parental MoMuLV envelope protein, CEE+, presumably due to the presence of 14 additional amino acids, an extra N-linked glycosylation site in the amphotropic PRR, and several potential additional O-linked glycosylation sites (Fig. 1). Deletions C-14 and C-19 resulted in envelope proteins that were incorporated into the viral particles at levels comparable to that of the wild-type protein, but the level of envelope protein associated with viral particles gradually decreased for truncations C-24 to C-49, as evidenced by the lower levels of both the SU and TM subunits. For this group of proteins, an additional band was detected by the anti-SU antiserum, which migrated at a higher position than SU. The intensity of the band increased as more amino acids were deleted, and for constructs C-44 and C-49, the majority of the protein recognized by the anti-SU antiserum was the higher-mobility form.
FIG. 3.
Western blot analysis of virion-associated envelope proteins. (A and B) Viral particles were pelleted through 20% sucrose, and specific proteins were detected including the SU protein and the viral capsid protein, CA (A) and TM protein (B). The MuLV TM protein exists in two forms, full-length p15E and a truncated form p12E (15, 20), both of which can be seen. Mock transfection results in no envelope protein. (C) Viral proteins were probed with anti-TM antibody, revealing the presence of Pr85 in virions for constructs C-44 and C-49.
We investigated the nature of the higher-migrating band observed for proteins C-24 to C-49. Probing a Western blot of mutants C-44 and C-49 with an anti-TM antibody revealed that this protein was probably the uncleaved precursor envelope protein, Pr85 (Fig. 3C). It is possible that the presence of this precursor protein in particles reflected an underlying defect in the processing of Pr85 to the SU and TM subunits. Accordingly, we analyzed the form of the envelope proteins present in the lysates of transfected cells. To clearly distinguish between Pr85, SU, and various processing intermediates of Pr85, we treated the lysates with an N-glycosidase and also included marker protein constructs that expressed only the MoMuLV SU or Pr85 proteins (Fig. 4A). These results demonstrated that while approximately equal amounts of the Pr85 and SU proteins could be observed for the wild-type protein CEE+, the parental hybrid E/A-PRR, and the mutants N-14 and C-14, only a single band was observed for mutants C-44, C-49, and C-54. Reduced levels of Pr85 processing to SU were observed for the truncations C-19 to C-39 (Table 1 and data not shown). Probing the cell lysates with an anti-TM antibody revealed that the single species observed for mutants C-44, C-49, and C-54 was Pr85 (Fig. 4B).
FIG. 4.
Envelope proteins in cell lysates. (A) Cell lysates of 293T cells expressing envelope proteins were deglycosylated and probed with an anti-SU antibody. Construct SU is terminated at the natural SU-TM cleavage site and therefore produces only SU, while construct Pr85 contains a mutated cleavage site and produces only Pr85. Mock transfection results in no envelope protein. (B) Deglycosylated cell lysates were probed with anti-TM antibody, which detects Pr85 and the processed TM protein.
Mutant C-54, which contains a deletion extending into the N-terminal conserved region, did not result in any detectable SU, TM, or Pr85 proteins in viral particles (Fig. 3). Although Pr85 was seen in cell lysates (Fig. 4), C-54 differed from mutants C-44 and C-49 in that no Pr85 was subsequently incorporated into viral particles. In addition, this mutant was present at a much lower level on the surface of cells (Table 1). These defects alone could account for the lack of titer and the loss of the ability to promote cell-cell fusion observed with this mutant (Table 1).
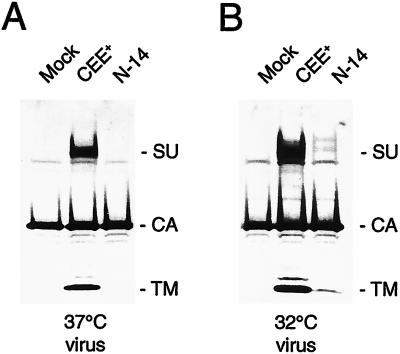
N-14 is a temperature-sensitive incorporation mutant.
A possible explanation for the lack of infectivity of the N-14 mutant, despite its ability to promote cell-cell fusion, is that this protein is not stably incorporated into viral particles. Accordingly, we used Western blot analysis to investigate the envelope protein content of the viral particles. The results revealed that there was no detectable envelope protein present in the viral particles (Fig. 5A), even though the N-14 mutant displayed normal processing of the precursor Pr85 to the mature SU protein in cell lysates (Fig. 4).
FIG. 5.
N-14 is a temperature-sensitive incorporation mutant. (A) N-14 is undetectable in viral particles produced at 37°C. (B) N-14 is incorporated into viral particles at a low level at 32°C.
We investigated the possibility that the defect in the N-14 envelope protein was a temperature-sensitive mutation by analyzing the envelope protein content of virions harvested at 32°C. Western blot analysis revealed that the N-14 mutant was present in viral particles at low levels at this temperature (Fig. 5B). In addition, N-14 virions harvested at 32°C gave a titer of 1.4 × 104 CFU/ml on NIH 3T3 cells, which was 10% of the value obtained for the wild-type envelope protein at this temperature. These results therefore demonstrated that N-14 is a temperature-sensitive incorporation mutant.
PRR affects SU-TM interactions.
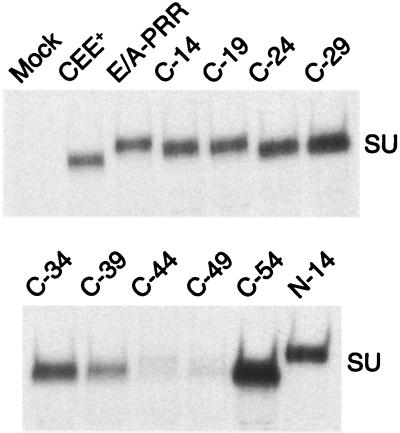
Previous linker insertion mutations in the PRR have been shown to result in a more labile SU-TM interaction, resulting in greater shedding of the SU subunit from viral particles (14). We therefore investigated the effects of the PRR mutants on SU-TM interactions by analyzing the amount of free SU protein that could be immunoprecipitated from the supernatants of cells expressing the envelope protein mutants (Fig. 6).
FIG. 6.
Immunoprecipitation of SU protein from cell culture supernatants. SU proteins were immunoprecipitated from the supernatants of 293T cells transiently transfected with various envelope protein expression plasmids.
This analysis revealed that the progressive shortening of the C-terminal region of the PRR up to mutant C-29 resulted in an increased tendency to shed SU into the culture supernatant. Mutants C-34 and C-39 did not appear to continue this trend, since they produced smaller amounts of free SU in the supernatant than did mutant C-29, but both of these proteins were present at notably lower levels on the cell surface. Mutants C-44 and C-49 produced very little free SU in the supernatants of transfected cells. However, both of these proteins demonstrated very poor processing of Pr85 to SU, as evidenced by the lack of SU detected in cell lysates (Fig. 4) and the high levels of incorporation of Pr85 into viral particles (Fig. 3).
Higher levels of SU were seen in the supernatants of cells transfected with mutant N-14 and in particular with mutant C-54. While neither protein is present in viral particles, the two proteins have distinct properties (Table 1; Fig. 4). For C-54, no processed SU could be detected in cell lysates and very little envelope protein was present on the cell surface. In contrast, mutant N-14 produced normal levels of both Pr85 and SU in cell lysates and was readily detectable on the cell surface.
Insertion of a collagen-binding peptide into the PRR.
Since the PRR could tolerate changes in both the actual sequence of its C-terminal region and its overall length, we tested whether this region would allow the insertion of a peptide-binding domain, in order to generate a targetable retroviral vector. As a proof of the principle, a 16-amino-acid peptide containing the collagen-binding domain from von Willebrand’s factor (39) was inserted into two different sites in the PRR of protein E/A-PRR (Fig. 7A). The chimeric envelope proteins so generated were efficiently incorporated into retroviral vectors (Fig. 7B) and gave titers on NIH 3T3 cells that were similar to those obtained with the wild-type protein (Table 2), demonstrating that insertions in the PRR did not disrupt envelope protein function.
TABLE 2.
Characterization of the chimeric envelope proteins containing a collagen-binding domain
| Envelope proteina | Viral titerb (CFU/ml) | Binding to collagenc |
|---|---|---|
| CEE+ | 4.0 × 106 | 0 |
| Insert 1 | 1.6 × 106 | 0.512 ± 0.007 |
| Insert 2 | 1.5 × 106 | 0.530 ± 0.054 |
For the structures of insert 1 and insert 2, see Fig. 7A.
Viral titer on NIH 3T3 cells of retroviral vectors containing the chimeric envelope proteins.
Capacity of the chimeric envelope proteins to bind to collagen as measured by the collagen-binding assay (see Materials and Methods). The numbers shown are the mean of three independent determinations of optical density at 600 nm ± standard error.
To assess whether the inserted peptide could confer binding to collagen, we analyzed the ability of retroviral particles pseudotyped with the chimeric envelope proteins to bind to collagen-coated plates. Both of the chimeric envelope proteins bound virions to the plates, while the wild-type MoMuLV envelope protein did not (Table 2).
DISCUSSION
Three distinct domains are present in the SU proteins of the type C mammalian retroviruses. The N-terminal region, which displays the most variability between different viruses, interacts with the specific viral receptor and thereby determines the tropism of the virus (42), while the more highly conserved C-terminal region associates with the TM subunit (30, 32). These two regions are separated by a PRR of variable length and sequence. The location and high proline content of the PRR have led to speculation that this region could serve as a flexible linker or hinge region, important for transmitting the conformational changes that are thought to occur in the retroviral envelope protein complex after binding to a receptor (13, 35). As such, it may function in a similar way to the hinge regions of immunoglobulins, which introduce flexibility into the antigen-binding arms to allow optimal interactions with antigens (40).
Our analysis of the role of the PRR in the MuLVs initially investigated the ability of the amphotropic PRR to functionally substitute for the shorter ecotropic PRR in the MoMuLV envelope protein. We constructed a chimeric protein, E/A-PRR, which proved to be fully functional and to produce retroviral vectors with titers that were 46% of those for the wild-type ecotropic parent. This result indicated that it is not the actual sequence of the PRR that is important for function but, rather, some overall feature of the region, such as the high proline content. We also demonstrated that a 16-amino-acid peptide could be inserted into E/A-PRR, making the PRR 31 amino acids longer than the original ecotropic region without affecting envelope protein function. The ability of the PRR to tolerate this insertion suggests that this region could be used for inserting novel peptide-binding domains to generate targetable retroviral vectors.
We observed that E/A-PRR migrated at a higher position on an SDS-polyacrylamide gel than did the wild-type MoMuLV envelope protein. This result is most probably due to the presence of the additional N-linked glycosylation site that is predicted to be in the amphotropic PRR. This assumption is consistent with the observation that the truncated amphotropic PRR present in protein C-24, which no longer contains the consensus N-linked glycosylation site, produced a protein that ran at the same size as the wild-type protein. Alternatively, it has been reported that point mutations in the ecotropic PRR can affect the mobility of both Pr85 and SU and that this was the result of differential glycosylation (2).
The PRR contains two distinct sequence motifs. The N-terminal 14 or 15 amino acids are conserved in all of the type C mammalian retroviruses, while the C-terminal region of between 31 and 61 residues varies considerably in its primary sequence. The PRR has been proposed to form a polyproline β-turn helix (11), with 10 strong turns predicted in both the amphotropic sequence (41) and the FeLV sequence (11). Of these, three are present in the N-terminal region. It is likely that the PRR forms an exposed loop between the N- and C-terminal domains of SU (11). The presence of N-linked glycosylation sites in several PRR sequences and the high tendency of this region to induce neutralizing antibodies in FeLV (26, 27) are consistent with the PRR being surface exposed. Furthermore, we have demonstrated that the insertion of a collagen-binding domain from von Willebrand’s factor into this region allowed the resulting envelope protein to bind to collagen, which is also indicative of an accessible region.
The C-terminal deletion mutants we produced in E/A-PRR had several effects on envelope protein function. Initially, the deletion of just 14 amino acids, which shortened the amphotropic PRR to the same length as the original ecotropic PRR, restored the titer to near-wild-type levels. Subsequent truncations of up to 29 amino acids had very little effect on the titer. However, the sequential truncation of the C-terminal region did result in decreased levels of the processed SU and TM subunits of the envelope protein being incorporated into virions. In addition, we observed an increasing amount of the uncleaved precursor protein, Pr85, in virions. The decreases in both cell-cell fusion and transduction efficiency that we observed for mutants C-24 to C-49 could be secondary to these incorporation defects. We could always detect some form of envelope protein present on the cell surface for all of the mutants, although C-54 was detected at much lower levels. However, it is possible that the major species detected for mutants C-39 to C-49 was the uncleaved precursor Pr85.
Cleavage of the precursor envelope protein into the mature SU and TM subunits is essential for retroviral infectivity. While mutation of the cleavage site can force the incorporation of a low level of uncleaved precursor protein into virions, the resulting particles are noninfectious (references 8, 12, and 24 and data not shown). Truncation of the PRR C-terminal region in mutants C-24 to C-49 resulted in a decreased efficiency of Pr85- to-SU processing in cell lysates and the appearance of increasing amounts of Pr85 in viral particles. Despite the high levels of Pr85 and low levels of SU and TM proteins in virions, retroviral vectors containing the C-terminal truncation mutants up to C-49 remained infectious. It seems likely that the low level of properly cleaved protein detected for mutants C-39 to C-49 was sufficient to allow viral entry. These results indicate that virus-cell fusion can occur efficiently even when only a small amount of processed envelope protein is present. This observation contrasts with the results from the cell-cell fusion assay, in which we detected hardly any syncytia for mutant C-29 and none for mutant C-34, despite their abilities to produce almost wild-type titers in retroviral vectors. These results indicate that cell-cell fusion has different requirements from virus-cell fusion, as we (46) and others (14, 29, 44) have previously noted.
Removal of the N-terminal domain of the PRR in mutant N-14 resulted in an envelope protein that was not incorporated into virions at all at 37°C, although low levels could be detected at 32°C. This result occurred despite the presence of normal levels of Pr85 processing in cell lysates and wild-type levels of cell surface protein. Furthermore, the cell surface N-14 protein was fully able to induce cell-cell fusion in a cocultivation assay. Higher levels of SU were shed into the supernatant of transfected cells than occurred with the wild-type protein, suggesting that one defect in this protein may be a decreased stability in the interaction between the SU and TM subunits. Previous linker insertions in the N-terminal region of the MoMuLV PRR have also been reported to produce viruses with a temperature-sensitive phenotype (14). These mutant virions incorporated only SU and were infectious at 32°C. Similar to the N-14 mutant, they also had a higher tendency to shed SU into the supernatant than did the wild-type virus. However, these mutants induced syncytia only at 32°C, in contrast to N-14, which was also fusogenic at 37°C.
Decreased affinity between the SU and TM subunits can result in the loss of SU, leaving the remaining TM protein unstable and rapidly degraded, as has previously been suggested for mutants of MoMuLV (14) and human immunodeficiency virus type 1 (17). However, this explanation is at odds with the apparently normal cell surface levels of the N-14 mutant and its ability to induce cell-cell fusion. It is conceivable that the N-14 deletion somehow prevented the incorporation of the envelope protein into budding viral cores, although defects in envelope protein incorporation have been observed only with certain mutations in the cytoplasmic tail of the MoMuLV protein (20). An alternative explanation could be that the association of the N-14 protein with virions decreased the stability of this protein. Envelope protein incorporated into MuLV cores has the C-terminal 16 amino acids of the cytoplasmic tail (the R peptide) removed by the viral protease. This processing enhances the fusogenicity of the wild-type envelope protein (33, 34) and could possibly strain an already weakened SU-TM interaction in mutant N-14, leading to the loss of SU and the rapid degradation of TM.
A defect in SU-TM association could also account for the phenotype observed with protein C-54. This truncation retained only 6 amino acids of the N-terminal conserved domain of the PRR with none of the C-terminal region and only 1 of the 10 predicted β-turns in this region (11). Mutant C-54 resulted in the largest amounts of SU shed into the culture supernatant and had only a very low level of envelope protein that could be detected on the cell surface. In addition, only Pr85 and no processed SU could be detected in cell lysates. Since the protein was detected at only very low levels on the cell surface and in virions, it was not surprising that C-54 was unable to cause either cell-cell fusion or virus-cell fusion.
The underlying cause of the defect in mutant C-54 is unclear. It is possible that this large truncation in the PRR destabilized SU-TM interactions so that processed envelope protein was not stably expressed on the cell surface. This could result in both a low level of detection of cell surface envelope protein, a lack of SU protein detected in the cell lysates, and a large amount of free SU shed into the culture supernatant. It is also possible that different pathways in the cell can be used to transport the envelope protein and that the envelope protein shed into the supernatant is transported differently from the protein that ultimately ends up on the cell surface. Notably, two distinct populations have previously been detected for membrane-associated and secreted simian immunodeficiency virus SU (38).
Previous mutational analyses of this region have demonstrated a role for the PRR in SU-TM association, the stable incorporation of envelope protein into virions, envelope protein-mediated cell-cell fusion, and viral infectivity (2, 14). Our analysis of the E/A-PRR deletion mutants indicates that the primary defects in these mutants are the ability of processed envelope protein to be incorporated into virions and stable SU-TM associations. Similar effects on both Pr85 cleavage and SU-TM association have been reported for mutations in the C-terminal region of the Friend MuLV SU (22). These effects are consistent with the PRR being important for the overall folding of the envelope protein, which in turn could influence the rate of transport of the protein through the export pathway, the efficiency of SU-TM cleavage, and the consequent stability of the noncovalently linked SU and TM proteins in the final envelope protein complex. The partially temperature-sensitive phenotype of the N-14 mutant also supports the notion that this region is important for protein conformation.
Our data do not support a primary defect in fusion or binding for the PRR mutants. Even the truncation of 49 of the 61 amino acids from the C-terminus of the PRR gave a titer that was still 10% of the E/A-PRR titer in a retroviral vector, and the N-14 mutant produced wild-type levels of syncytia in a fusion assay. In addition, although the PRR has been shown to be able to influence the receptor-binding properties of certain MuLV subtypes (3, 16), it is likely that this effect is also secondary to an influence on the overall protein structure. However, we cannot rule out the possibility that the PRR plays a different role in different MuLV subtypes. Our analysis is consistent with a role for the PRR in the proper processing of Pr85 and the stable association of the SU and TM subunits, with downstream effects on fusion and infectivity.
ACKNOWLEDGMENTS
We thank Ling Lu and Michael J. Skotzko for technical assistance and Lingtao Wu for helpful discussions.
This work was supported by Genetic Therapy, Inc. (GTI)/Novartis (Gaithersburg, Md.) and by NIH grant CA59318-04.
REFERENCES
- 1.Albritton L M, Kim J W, Tseng L, Scadden D, Cunningham J M. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen K B. A domain of murine retrovirus surface protein gp70 mediates cell fusion, as shown in a novel SC-1 cell fusion system. J Virol. 1994;68:3175–3182. doi: 10.1128/jvi.68.5.3175-3182.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battini J L, Heard J M, Danos O. Receptor choice determinants in the envelope proteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benedict, C. A., Y. Zhao, N. Kasahara, P. M. Cannon, and W. F. Anderson. Development of retroviral vectors that target hematopoietic stem cells. Hematopoietic cell therapy. In A. D. Ho and R. E. Champlin (ed.), Hematopoietic stem cell therapy, in press. Cambridge University Press, Cambridge, United Kingdom.
- 5.Bosselman R A, van Straten F, van Beveren C, Verma I M, Vogt M. Analysis of the env gene of a molecularly cloned and biologically active Moloney mink cell focus-forming proviral DNA. J Virol. 1982;44:19–31. doi: 10.1128/jvi.44.1.19-31.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosset F L, Russell S J. Targeting retrovirus entry. Gene Ther. 1996;3:946–956. [PubMed] [Google Scholar]
- 7.Delassus S, Sonigo P, Wain-Hobson S. Genetic organization of gibbon ape leukemia virus. Virology. 1989;53:205–213. doi: 10.1016/0042-6822(89)90236-5. [DOI] [PubMed] [Google Scholar]
- 8.Dong J, Dubay J W, Perez L G, Hunter E. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein define a requirement for dibasic residues for intracellular cleavage. J Virol. 1992;66:865–874. doi: 10.1128/jvi.66.2.865-874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elder J H, McGee J S, Munson M, Bittle J L, Grant C. Localization of neutralization regions of the envelope gene of feline leukemia virus by using anti-synthetic peptide antibodies. J Virol. 1987;61:8–15. doi: 10.1128/jvi.61.1.8-15.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans L H, Morrison R P, Malik F G, Portis J, Britt W J. A neutralizeable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J Virol. 1990;64:6176–6183. doi: 10.1128/jvi.64.12.6176-6183.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fontenot J D, Tjandra N, Ho C, Andrews P C, Montelaro R C. Structure and self assembly of a retrovirus (FeLV) proline rich neutralization domain. J Biomol Struct Dyn. 1994;11:821–837. doi: 10.1080/07391102.1994.10508035. [DOI] [PubMed] [Google Scholar]
- 12.Freed E O, Risser R. The role of envelope glycoprotein processing in murine leukemia virus infection. J Virol. 1987;61:2852–2856. doi: 10.1128/jvi.61.9.2852-2856.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert J M, Hernandez D L, Balliet K W, Bates P, White J M. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J Virol. 1995;69:7410–7415. doi: 10.1128/jvi.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray K D, Roth M J. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J Virol. 1993;67:3489–3496. doi: 10.1128/jvi.67.6.3489-3496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green N, Shinnick T M, Witte O, Ponticelli A, Sutcliffe J G, Lerner R A. Sequence-specific antibodies show that maturation of Moloney murine leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han J, Cannon P M, Lai K, Zhao Y, Eiden M V, Anderson W F. Identification of envelope protein residues required for the expanded range of 10A1 murine leukemia virus. J Virol. 1997;71:8103–8108. doi: 10.1128/jvi.71.11.8103-8108.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho S N, Hunt H D, Horton R M, Pullen J K, Please L R. Site-directed mutagenesis by overlap extension using polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 19.Hunter E, Swanstrom R. Retrovirus envelope glycoproteins. Curr Top Microbiol Immunol. 1992;157:187–253. doi: 10.1007/978-3-642-75218-6_7. [DOI] [PubMed] [Google Scholar]
- 20.Januszeski M M, Cannon P M, Chen D, Rozenberg Y, Anderson W F. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J Virol. 1997;71:3613–3619. doi: 10.1128/jvi.71.5.3613-3619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch W, Hunsmann G, Friedrich R. Nucleotide sequence of the envelope gene of Friend murine leukemia virus. J Virol. 1983;45:1–9. doi: 10.1128/jvi.45.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li I, Pinter A, Kayman S C. The critical N-linked glycan of murine leukemia virus envelope protein promotes both folding of the C-terminal domains of the precursor polyprotein and stability of the postcleavage envelope complex. J Virol. 1997;71:7012–7019. doi: 10.1128/jvi.71.9.7012-7019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacKrell A J, Soong N W, Curtis C M, Anderson W F. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J Virol. 1996;70:1768–1774. doi: 10.1128/jvi.70.3.1768-1774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCune J M, Rabin L B, Feinberg M B, Lieberman M, Kosek J C, Reyes G R, Weissman I L. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53:55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- 25.Morgan R A, Nussbaum O, Muenchau D D, Shu L, Couture L, Anderson W F. Analysis of the functional and host range-determining regions of the murine ecotropic and amphotropic retrovirus envelope proteins. J Virol. 1993;67:4712–4721. doi: 10.1128/jvi.67.8.4712-4721.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nick S, Klaws J, Freibel K, Birr C, Hunsmann G, Bayer H. Virus neutralizing and enhancing epitopes characterized by synthetic oligopeptides derived from the feline leukemia virus glycoprotein sequence gp70. J Gen Virol. 1990;71:77–83. doi: 10.1099/0022-1317-71-1-77. [DOI] [PubMed] [Google Scholar]
- 27.Nunberg J H, Rogers G, Gilbert J H, Snead R M. Method to map antigenic determinants recognized by monoclonal antibodies: localization of a determinant virus neutralization on the feline leukemia virus envelope protein. Proc Natl Acad Sci USA. 1984;81:3675–3679. doi: 10.1073/pnas.81.12.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ott D, Friedrich R, Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990;64:757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park B H, Matuschke B, Lavi E, Gaulton G N. A point mutation in the env gene of a murine leukemia virus induces syncytium formation and neurologic disease. J Virol. 1994;68:7516–7524. doi: 10.1128/jvi.68.11.7516-7524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinter A, Fleissner E. The presence of disulfide-linked gp70-p15(E) complexes in AKR murine leukemia virus. Virology. 1977;83:417–422. doi: 10.1016/0042-6822(77)90187-8. [DOI] [PubMed] [Google Scholar]
- 31.Pinter A, Honnen W J, Tung J S, O’Donnell P V, Hammerling U. Structural domains of endogenous murine leukemia virus gp70s containing specific antigenic determinants defined by monoclonal antibodies. Virology. 1982;116:499–516. doi: 10.1016/0042-6822(82)90143-x. [DOI] [PubMed] [Google Scholar]
- 32.Pinter A, Kopelman R, Li Z, Kayman S C, Sanders D A. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzyme. J Virol. 1997;71:8073–8077. doi: 10.1128/jvi.71.10.8073-8077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragheb J A, Anderson W F. Uncoupled expression of Moloney murine leukemia virus envelope polypeptides SU and TM: a functional analysis of the role of TM domains in viral entry. J Virol. 1994;68:3207–3219. doi: 10.1128/jvi.68.5.3207-3219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sattentau Q J, Moore J P. Conformational change induced in the human immunodeficiency virus envelope protein by soluble CD4. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schilz T F, Jameson B A, Lopalco L, Siccardi A G, Weissman R A, Moore J P. Conserved structural features in the interaction between retroviral surface and transmembrane glycoproteins. AIDS Res Hum Retroviruses. 1992;18:1571–1580. doi: 10.1089/aid.1992.8.1571. [DOI] [PubMed] [Google Scholar]
- 37.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three plasmid expression system for the production of high titre retroviral vectors. Nucleic Acids Res. 1995;23:626–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spies P C, Compans R W. Alternate pathways of secretion of simian immunodeficiency virus envelope glycoproteins. J Virol. 1993;67:6536–6541. doi: 10.1128/jvi.67.11.6535-6541.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takagi J, Asai H, Saito Y. A collagen/gelatin-binding decapeptide derived from bovine propolypeptide of von Willebrand factor. Biochemistry. 1992;32:8530–8534. doi: 10.1021/bi00151a021. [DOI] [PubMed] [Google Scholar]
- 40.Tucker P W, Sligntom J L, Blattner F R. Mouse IgA heavy chain gene sequence: implications for evolution of immunoglobulin hinge exons. Proc Natl Acad Sci USA. 1981;78:7684–7688. doi: 10.1073/pnas.78.12.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valsesia-Wittmann S, Morling F J, Hatziioannous T, Russell S J, Cosset F. Receptor co-operation in retrovirus entry: recruitment of an auxiliary entry mechanism after retargeted binding. EMBO J. 1997;160:1214–1223. doi: 10.1093/emboj/16.6.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss R A. Cellular receptors and viral glycoproteins involved in retrovirus entry. In: Levy J A, editor. The Retroviridae. New York, N.Y: Plenum Press; 1993. pp. 1–108. [Google Scholar]
- 43.White J M. Membrane fusion. Science. 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 44.Wilson C A, Marsh J W, Eiden M V. The requirements for viral entry differ from those for virally induced syncytium formation in NIH 3T3/DTras cells exposed to Moloney murine leukemia virus. J Virol. 1992;66:7262–7269. doi: 10.1128/jvi.66.12.7262-7269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y, Lee S, Anderson W F. Functional interactions between monomers of the retroviral envelope protein complex. J Virol. 1997;71:6967–6972. doi: 10.1128/jvi.71.9.6967-6972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu N L, Cannon P M, Chen D, Anderson W F. Mutational analysis of the fusion peptide of Moloney murine leukemia virus transmembrane protein p15E. J Virol. 1998;72:1632–1639. doi: 10.1128/jvi.72.2.1632-1639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]