Abstract
The human cytomegalovirus (HCMV) gCIII envelope complex is composed of glycoprotein H (gH; gpUL75), glycoprotein L (gL; gpUL115), and a third, 125-kDa protein not related to gH or gL (M. T. Huber and T. Compton, J. Virol. 71:5391–5398, 1997; L. Li, J. A. Nelson, and W. J. Britt, J. Virol. 71:3090–3097, 1997). Glycosidase digestion analysis demonstrated that the 125-kDa protein was a glycoprotein containing ca. 60 kDa of N-linked oligosaccharides on a peptide backbone of 65 kDa or less. Based on these biochemical characteristics, two HCMV open reading frames, UL74 and TRL/IRL12, were identified as candidate genes for the 125-kDa glycoprotein. To identify the gene encoding the 125-kDa glycoprotein, we purified the gCIII complex, separated the components by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and subjected gH and the 125-kDa glycoprotein to amino acid microsequence analysis. Microsequencing of an internal peptide derived from purified 125-kDa glycoprotein yielded the amino acid sequence LYVGPTK. A FASTA search revealed an exact match of this sequence to amino acids 188 to 195 of the predicted product of the candidate gene UL74, which we have designated glycoprotein O (gO). Anti-gO antibodies reacted in immunoblots with a protein species migrating at ca. 100 to 125 kDa in lysates of HCMV-infected cells and with 100- and 125-kDa protein species in purified virions. Anti-gO antibodies also immunoprecipitated the gCIII complex and recognized the 125-kDa glycoprotein component of the gCIII complex. Positional homologs of the UL74 gene were found in other betaherpesviruses, and comparisons of the predicted products of the UL74 homolog genes demonstrated a number of conserved biochemical features.
The envelope glycoproteins of the herpesviruses play multiple critical roles in the viral life cycle, including attachment, penetration, cell-to-cell spread, and envelopment and maturation of nascent viral particles. To understand the life cycles of these complex viruses, it is necessary to have a thorough knowledge of the structures and functions of the envelope glycoproteins. Biochemical studies of human cytomegalovirus (HCMV) virions revealed that the viral envelope contains at least 10 glycoproteins, many of which are organized into three predominant high-molecular-weight, disulfide-bonded complexes designated gCI, gCII, and gCIII (23). Given the resolution of earlier analyses and the large coding capacity of HCMV (9), it is very likely that other proteins are also present in the viral envelope. To date, however, only six virion envelope glycoproteins have been mapped to the viral genome: gp48 (UL4) (8), GCR33 (UL33) (37), glycoprotein B (gB; UL55) (4, 12, 36), glycoprotein H (gH; UL75) (13, 41, 44), glycoprotein M (gM; UL100) (1, 27, 32), and glycoprotein L (gL; UL115) (24, 28, 33, 50).
The gH-gL complexes of herpesviruses, including HCMV, have been implicated in viral fusion events, including entry and cell-to-cell spread (18–20, 29, 30, 38, 41, 42, 46, 47). To understand the specific molecular function(s) of HCMV gH-gL, it is necessary to define the structural organization of these glycoproteins as they exist in the virus. Earlier studies demonstrated that the 240-kDa gCIII envelope complex contained the gH and gL homologs of HCMV (13, 23, 24, 33, 41, 44). However, coexpression of gH and gL in recombinant systems did not reconstitute gCIII, suggesting that the gCIII complex contained other gene products which were distinct from gH and gL (24, 33). Detailed characterization of gCIII clearly demonstrated that a 125- to 145-kDa protein was present in gCIII from HCMV-infected cells and purified virions (23, 24, 33). The 125-kDa protein was shown to be antigenically and structurally unrelated to either gH or gL, explaining the lack of gCIII reconstitution by coexpression of gH and gL and suggesting that a third HCMV gene product was contained in the complex (24, 33).
In this study, we report that the 125-kDa protein is encoded by the HCMV UL74 gene. The biochemical features of the 125-kDa protein revealed it was a glycoprotein and suggested the UL74 open reading frame (ORF) as a possible candidate gene. Amino acid microsequencing of the purified 125-kDa glycoprotein confirmed that this glycoprotein was the product of the UL74 gene. Additionally, an anti-UL74 antibody immunoprecipitated the 240-kDa gCIII complex and specifically recognized the 125-kDa glycoprotein component of gCIII. Together, these data corroborate that the HCMV UL74 gene product, which we have designated glycoprotein O (gO), is the third glycoprotein component of the gCIII complex. Interestingly, the HCMV gO gene has positional homologs in the betaherpesvirus subfamily, and a comparison of the predicted products of the gO homolog genes revealed a number of shared biochemical features.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Immortalized fibroblast (IF) cells were cultured as previously described (10). The AD169 strain of HCMV was grown and titered as previously described (10). Gradient-purified virions were isolated as previously described (11). Monoclonal antibodies 14-4b (5) and AP865 (51), generously supplied by W. Britt, and polyclonal antibody 26388, kindly provided by A. Minson, were described previously (24).
Immunoblotting and immunoprecipitations.
Immunoblotting and immunoprecipitations were performed essentially as previously described (24). In brief, proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (with or without reducing agents) and electrotransferred to nitrocellulose (Millipore) for immunoblotting. Primary antibodies were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit antibodies (Pierce). Renaissance Western blot chemiluminesence reagent (NEN) was used to detect the peroxidase conjugates. For immunoprecipitations, IF cells were radiolabeled with 50 μCi of [35S]methionine-cysteine (NEN)/ml for 16 h. Radioimmunoprecipitation assay (RIPA) (24) cell lysates were precleared with protein G (formalin-killed cell suspension; Sigma) and supplemented with 0.5% bovine serum albumin. The precipitating antibody was added to the lysates, and antibody-antigen complex was recovered with immobilized protein A (Pierce). The protein A pellets were washed extensively with RIPA lysis buffer and resuspended in SDS-PAGE sample buffer (with or without reducing agents) in preparation for SDS-PAGE. Resultant gels were dried and imaged via a GS-525 Molecular Imager (Bio-Rad).
N-Glycosidase F digestion of biotinylated proteins.
IF cells were infected with HCMV at a multiplicity of infection (MOI) of 3. At 4 days postinfection, infected cell monolayers were washed with PBS-MC (phosphate-buffered saline [PBS] supplemented with 1 mM MgCl2 and 0.1 mM CaCl2). EZ-link sulfo-NHS-LC biotin (Pierce) in PBS-MC (2 mg/ml) was added to the cells, which were then incubated at 37°C for 1 h. The biotin solution was removed, and the cells were washed extensively with PBS-MC. To quench the biotinylation reaction, 10 mM glycine in PBS-MC was added to the cells for 10 min at 37°C. The glycine solution was removed, and the cells were washed extensively with PBS-MC. N-Glycosidase F (Boehringer Mannheim) digestions were performed as instructed by the manufacturer. Briefly, immunoprecipitated proteins recovered by immobilized protein A were heated at 95°C for 3 min in a PBS solution containing 0.45% SDS and 90 mM β-mercaptoethanol. After centrifugation to remove the immobilized protein A, the denatured protein solution was diluted twofold with PBS and made 50 mM in EDTA and 1% in Nonidet P-40. Samples were digested with 10 U of N-glycosidase F for 16 h at 37°C. An additional 3 U of N-glycosidase F was added, and the digestions were incubated for an additional 2 h.
Isolation of 125-kDa protein from immunoaffinity-purified gCIII complex.
Seven milligrams of antibody 14-4b (5) was coupled to 1 ml of CNBr-activated Sepharose 4B (Pharmacia Biotech) as instructed by the manufacturer. One hundred T150 tissue culture flasks containing near-confluent cultures of IF cells were infected with HCMV AD169 at an MOI of approximately 3. At 5 days postinfection, the infected cells were recovered, pelleted, and resuspended in RIPA buffer supplemented with a protease inhibitor cocktail (PIC) (24) and 10 mM iodoacetamide for 30 min on ice; insoluble material was removed by centrifugation at 43,000 × g. The clarified lysate was applied to the 14-4b column and circulated continuously for 16 h by a peristaltic pump. The column was washed with 30 column volumes of RIPA-PIC-iodoacetamide. Bound protein was eluted by 100 mM glycine (pH 2.5) in RIPA-PIC-iodoacetamide containing 1/10 the amount of detergents and immediately neutralized by adding a 1/10 volume of 1 M Tris (pH 8.5). Fractions containing the gCIII complex (as judged by analytical nonreducing SDS-PAGE) were pooled, concentrated with Centricon filters (Amicon), and subjected to nonreducing SDS-PAGE (6% gel). To minimize the incidence of oxidants and free radicals, which may block the N termini of proteins, all preparative acrylamide gels were allowed to polymerize for 24 h before use, and 0.1 mM thioglycolate (Sigma) was added to the cathode running buffer of these preparative gels. The region of the gel containing the gCIII complex was excised and inserted into the well of a 10% acrylamide gel. This gel slice was subjected to a secondary reducing SDS-PAGE as described previously (24). The resultant gel was electrotransferred to Immobilon-Psq (Millipore) in 25 mM N-ethylmorpholine (pH to 8.3 with formic acid)–10% methanol. Transferred proteins were visualized on the membrane by staining with 0.1% amido black (Sigma) in 40% methanol–10% acetic acid followed by extensive washes in double-distilled H2O. The membrane was then dried, and appropriate sections were excised for N-terminal and internal microsequence analysis at the Protein/DNA Technology Center of Rockefeller University (16, 17).
Production of a UL74-specific polyclonal serum.
A portion of the HCMV UL74 gene (corresponding to the codons for amino acids 32 to 466) was amplified by PCR with Pfu polymerase (Stratagene) from HCMV AD169 DNA, prepared as described previously (24). The UL74 PCR product was cloned into plasmid pET28a (Novagen), which placed the UL74 gene downstream of the coding sequence for a six-histidine tag. pET28a-UL74 was transformed into Escherichia coli BL21(DE3), and production of recombinant UL74-His protein was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside for 2 h. The insoluble inclusion bodies, containing the recombinant UL74-His protein, were solubilized in 6 M urea and purified by Ni chelate chromatography as instructed by the manufacturer (Novagen). Purified UL74-His protein was injected into a New Zealand White rabbit for production of an anti-UL74 polyclonal serum.
Computer analyses of protein sequences.
Peptidesort and GAP analyses of protein sequences were performed with the Genetics Computer Group (GCG) programs (GCG, Oxford Molecular Group, Inc., Madison, Wis.) (15).
RESULTS
Identification of candidate genes encoding the 125-kDa component of gCIII.
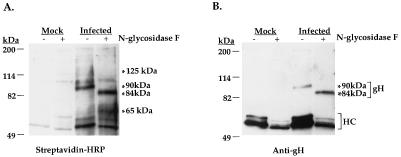
Previously we determined that the 125-kDa protein was antigenically distinct from gH and gL and was likely the product of an HCMV gene (24). The diffuse banding pattern of the 125-kDa protein in SDS-PAGE suggested that it was a glycoprotein. To confirm this hypothesis and facilitate genetic identification of the 125-kDa protein, we performed glycosidase digestion of the 125-kDa protein. To this end, biotinylated gCIII complex was immunoprecipitated and digested with N-glycosidase F to remove both high-mannose and complex asparagine (N)-linked oligosaccharides. When the blots were probed with streptavidin-HRP to detect all biotinylated proteins, we detected in undigested immunoprecipitates from infected cells two protein species of 125 and 90 kDa, which represent the 125-kDa protein and gH, respectively (Fig. 1A). N-Glycosidase F-digested proteins revealed two protein species of 65 and 84 kDa which likely correspond to deglycosylated forms of the 125-kDa protein and gH. To confirm that the 90- and 84-kDa protein species were gH, the blot in Fig. 1A was stripped and probed with a gH-specific antibody (Fig. 1B). Thus, the 65-kDa protein species represents 125-kDa protein lacking N-linked chains (the presence of other posttranslational modifications, such as O-linked oligosaccharides, was not assessed), confirming, as suggested by its banding pattern, that it is a glycoprotein containing approximately 60 kDa of N-linked glycosylation on a peptide backbone of 65 kDa or less. This biochemical profile is similar to that found by Li et al., who showed that the 125-kDa protein contained both simple and complex N-linked sugars with a core protein size of approximately 64 kDa (33). Estimating that a single N-linked oligosaccharide chain adds between 2 and 4 kDa to the mass of a protein, this analysis suggested that the gene encoding the 125-kDa glycoprotein would contain 15 to 30 N-linked glycosylation sites, with a primary sequence of ca. 590 amino acids or less. Examination of the HCMV genome revealed only two ORFs matching these characteristics. The UL74 gene predicts a protein of 466 amino acids and 18 potential N-linked glycosylation sites, and the TRL/IRL12 gene predicts a protein of 416 amino acids and 23 potential N-linked glycosylation sites. Both genes were cloned and expressed as recombinant proteins for use as antigens for the production of UL74 (this report) and TRL/IRL12-specific antibodies (data not shown).
FIG. 1.
N-Glycosidase digestion of the 125-kDa protein. Mock-infected and HCMV-infected IF cells were surface biotinylated and immunoprecipitated with antibody 14-4b. Immunoprecipitated proteins were incubated overnight in the presence or absence of N-glycosidase, subjected to reducing SDS-PAGE, and electroblotted to nitrocellulose. (A) The immunoblot was probed with streptavidin-HRP for detection of biotinylated proteins. (B) The blot in panel A was stripped and reprobed with an anti-gH antibody followed by an HRP-conjugated goat anti-mouse antibody. HC, immunoglobulin heavy chain of the immunoprecipitating antibody.
Purification and microsequencing of the 125-kDa glycoprotein.
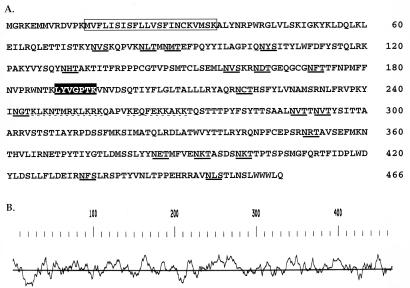
To obtain isolated 125-kDa glycoprotein, the gCIII complex was purified from HCMV-infected fibroblasts by monoclonal antibody affinity chromatography. The gCIII complex eluted from the column was excised from the gel after nonreducing SDS-PAGE, subjected to a second reducing SDS-PAGE, and then transferred to an Immobilon membrane in preparation for microsequence analysis. This procedure resulted in purification and separation of the three complex components (data not shown). To ensure that we had isolated the gCIII complex and that our purification methodology was not inherently detrimental to subsequent microsequence analysis, the portion of the membrane containing the protein corresponding to gH was initially subjected to N-terminal microsequencing (17). Microsequencing of this sample yielded the amino acid sequence XXEALDPHAFHLLLN, which corresponds to amino acids 32 (E) to 44 (N) of gH. This sequence is ca. 14 amino acids downstream of the NH3 terminus predicted after cleavage of the proposed signal peptide (amino acids 1 to 18) (13). This result suggests that the actual signal peptide is longer than predicted or that the N terminus of gH is proteolytically processed posttranslationally. Similar analysis of the 125-kDa glycoprotein indicated that it was N-terminally blocked. To microsequence an internally derived peptide, the 125-kDa glycoprotein was digested with endoproteinase Lys-C and the resultant peptides were purified by high-pressure liquid chromatography (16). Sequence from a single purified peptide yielded the unambiguous amino acid sequence LYVGPTK. A FASTA database search with this peptide sequence revealed an exact match to amino acids 189 to 195 of the predicted product of the HCMV UL74 gene (9) (Fig. 2B), one of the candidate genes identified by glycosidase analysis of the 125-kDa glycoprotein.
FIG. 2.
(A) Amino acid sequence of the predicted product of the HCMV UL74 gene. The sequence matching the internal peptide sequence derived from the 125-kDa glycoprotein is boxed in black. The hydrophobic domain is boxed, and potential N-linked glycosylation sites are underlined. The two potential heparin-binding sequences are underlined with a dashed line. Numbers on the right denote amino acid residues. (B) Kyte-Doolittle hydropathy analysis of the UL74 amino acid sequence. Areas above and below the line denote hydrophilic and hydrophobic domains, respectively. The scale above the line graph indicates the amino acid residue.
Predicted features of the HCMV UL74 gene product.
The UL74 gene encodes a 466-amino-acid protein with a calculated polypeptide backbone of 54.2 kDa (Fig. 2a). Kyte-Doolittle hydropathy analysis (31) (Fig. 2B) revealed a hydrophobic sequence near the N terminus (amino acids 14 to 30) that may serve as a signal sequence. No other significant hydrophobic sequences that could potentially act as membrane-spanning domains were detected. Thus, the UL74 glycoprotein either is soluble and membrane associated via its interaction with gH or is a type II transmembrane protein since the hydrophobic domain is inset 14 amino acids from the initiator methionine. These possibilities are under investigation. The primary amino acid sequence contains 18 potential N-linked glycosylation sites. There are two additional potential N-linked glycosylation sites within the UL74 primary sequence, but these are followed by proline residues and thus cannot serve as sites of oligosaccharide addition (3). There is also a clustering of serine and threonine residues between amino acids 271 and 337, suggesting potential O-linked glycosylation. The UL74 gene product is predicted to be a basic protein, based on an isoelectric point of 10.3, calculated by Peptidesort in the GCG package (15). Interestingly, a number of the basic lysine and arginine residues present in the UL74 sequence are concentrated into two stretches of amino acids (between residues 245 and 270) which are similar to consensus heparin-binding sequences (6) (Fig. 2B). The UL74 sequence also contains six cysteine residues (one is in the predicted hydrophobic sequence) which are likely important for disulfide bonding. Five of the six cysteine residues reside in the N-terminal half of the sequence.
A nomenclature for HCMV gene products was established in 1993 (49). Genes encoding glycoproteins are given a “gp” designation along with the corresponding ORF; thus, the UL74 gene product would be designated gpUL74. To maintain consistency with the naming of other herpesvirus envelope glycoproteins, we chose to also designate the protein with a “g” preceding a capital letter. Since the last herpesvirus glycoprotein to be named was gN (2, 26), we have designated the UL74 glycoprotein as glycoprotein O (gO).
Characterization of gO in HCMV-infected IF cells.
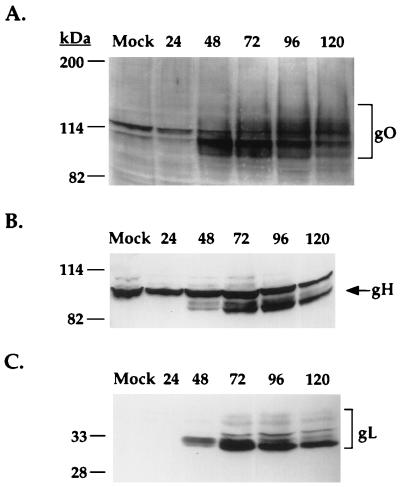
Antibodies produced to a recombinant form of UL74 (gO) were analyzed in immunoblot analysis of HCMV-infected cell lysates (Fig. 3A). The gO antibody specifically reacted with a protein species migrating broadly between 100 and 125 kDa; this species apparently comprises several constituents that may be attributable to processing intermediates or alternates. Pulse-chase experiments are required to determine the relationships between the various forms (25). The gO-immunoreactive species first became apparent at 48 h postinfection and reached maximum levels at between 72 and 96 h postinfection (Fig. 3A). Additionally, the other gCIII glycoprotein components, gH and gL, were expressed with similar kinetics as gO (Fig. 3B and C). This time course of expression of gO, gH, and gL is consistent with the expression of viral structural proteins, which are usually expressed at maximum levels at late times during infection.
FIG. 3.
Time course of expression of gO, gH, and gL. IF cells were infected with HCMV (MOI of 3) or mock infected and then lysed at 24-h intervals postinfection as indicated above the lanes. Lysates were resolved by reducing SDS-PAGE and electrotransferred, and the blots were reacted with anti-gO antibody (A), anti-gH antibody AP865 (B) or anti-gL antibody 26388 (C).
gO is present in purified HCMV virions.
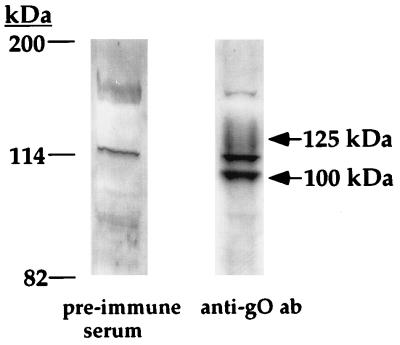
Since the 125-kDa glycoprotein is part of the mature gCIII complex found in virions, gO is predicted to be present in purified, banded viral particles. The presence of gO in the mature viral particle was verified by immunoblotting purified HCMV virions with the gO antibody. Figure 4 shows that the gO antibody was specifically reactive with a 125-kDa species which is consistent in size and migration pattern with the 125-kDa glycoprotein component of gCIII. The gO antibody also reacted with an approximately 100-kDa species, which is similar in size to the 100-kDa form of gO present in infected cells (25). This result suggests that alternatively processed forms of gO may be present in mature virions; experiments to examine this possibility are under way.
FIG. 4.
Immunoblots of purified HCMV virions. Gradient-purified virions were lysed in reducing SDS-PAGE sample buffer, resolved by SDS-PAGE, and electrotransferred to nitrocellulose. The blots was probed with either preimmune serum (as a control) or anti-gO antibody (ab).
The gO antibody immunoprecipitates the 240-kDa gCIII complex and recognizes the 125-kDa glycoprotein component of gCIII.
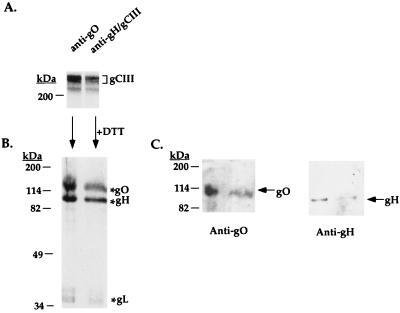
To confirm that gO was the 125-kDa third component of the gCIII complex, the gO antibody was used in immunoprecipitations of lysates of radiolabeled HCMV-infected IF cells. As shown in Fig. 5A, the gO antibody immunoprecipitated a 240-kDa species which comigrates with the authentic gCIII complex (24). To verify that the 240-kDa species immunoprecipitated by the gO antibody is the gCIII complex, the section of the dried gel containing the 240-kDa species was excised, reduced, and subjected to a subsequent SDS-PAGE; as a control, the 240-kDa gCIII complex immunoprecipitated by antibody 14-4b was also excised and reduced. Figure 5B shows that reduction of the 240-kDa species immunoprecipitated by the gO antibody yielded three proteins, one of approximately 125 kDa, one of 90 kDa, and a doublet at approximately 36 kDa, which represent gO, gH, and gL, respectively. An identical profile was seen upon reduction of the gCIII complex immunoprecipitated by antibody 14-4b (Fig. 5B). As further confirmation that the 125-kDa glycoprotein represented gO, duplicates of the excised and reduced samples from Fig. 5B were immunoblotted with either the gO antibody or a gH antibody. Figure 5C shows that the 125-kDa glycoprotein derived from either immunoprecipitate was detected with the gO antibody. Similarly, the 90-kDa gH protein was detected with the anti-gH antibodies from both immunoprecipitates. These data prove that the 125-kDa glycoprotein present in the gCIII envelope complex, which we have designated gO, is encoded by the UL74 gene.
FIG. 5.
Immunoprecipitation of the gCIII complex by the gO antibody. HCMV-infected cells were metabolically labeled with [35S]Met-Cys, lysed, and immunoprecipitated with either the anti-gO antibody or anti-gH/gCIII antibody 14-4b. (A) The proteins immunoprecipitated with the indicated antibodies were resolved by nonreducing SDS-PAGE. (B) The ∼240-kDa species precipitated by either the anti-gO or anti-gH immunoprecipitation was excised from the dried gel, reduced, and resolved by a second reducing SDS-PAGE. (C) Duplicates of the gel in panel B were electrotransferred to nitrocellulose and probed with the anti-gO antibody or anti-gH antibody AP865.
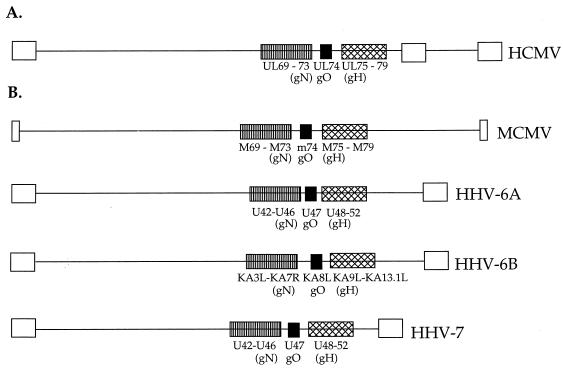
The HCMV gO gene has positional homologs in other betaherpesviruses.
We examined the genomes of other herpesviruses for homologs of the HCMV UL74 gene. Interestingly, the UL74 gene is positioned within the HCMV genome between two blocks of genes which are conserved in all herpesviruses (9, 39) (Fig. 6A). The conserved gene block upstream of the UL74 gene includes the UL69 to UL73 (gN homolog) ORFs, and the conserved gene block downstream of the UL74 gene includes the UL75 (gH) to UL79 ORFs (9, 39) (Fig. 6A). Examination of the genomes of closely related betaherpesviruses, including murine cytomegalovirus (MCMV) (45), human herpesvirus 6A (HHV-6A) (21, 22), HHV-6B (14, 35), and HHV-7 (40), revealed the same organization of genes as in HCMV, with a gO positional homolog gene flanked by the gN gene block and the gH gene block (Fig. 6B). In the alphaherpesviruses herpes simplex virus type 1 and varicella-zoster virus, and in the gammaherpesviruses Epstein-Barr virus (EBV) and HHV-8, the genomic organization of the two conserved blocks of genes is divergent from that in HCMV (9, 39, 48). If gO positional homolog genes exist in these four herpesviruses, the gO homolog gene should be found contiguous with either the gH gene or the gN homolog gene. However, none of the genes contiguous to the gH and gN genes in these four herpesviruses encodes for a heavily glycosylated protein with an N-terminal signal sequence and predicted basic isoelectric point (data not shown). Thus, there do not appear to be gO positional homolog genes within the alpha- or gammaherpesviruses, although these herpesviruses may encode glycoproteins which are analogous to gO in either structure or function. Indeed, it has been well established that the EBV BZLF2 gene (not a positional homolog of the HCMV UL74 gene) encodes a glycoprotein, gp42, which is the third component of the EBV gH-gL complex (34).
FIG. 6.
Schematic representation of betaherpesvirus genomes showing positional homology of the gO genes. Open boxes represent terminal and internal repeat regions of the genomes; striped boxes represent the conserved block of genes including the gN homolog; black boxes represent the gO gene; hatched boxes represent the conserved block of genes including the gH gene. (A) HCMV genome; (B) MCMV, HHV-6A, HHV-6B, and HHV-7 genomes.
Analyses of the amino acid sequences of the HHV-6A, HHV-6B, HHV-7, and MCMV gO positional homolog genes indicated, on average, 40% similarity and 20% identity at the amino acid level to HCMV gO, analyzed by GAP in the GCG package (15) (Table 1). This relatively low level of identity is not surprising, since low amino acid identity is often seen between herpesvirus glycoprotein homologs. For instance, HCMV gH and HCMV gL have approximately 27 and 28% amino acid identity, respectively, with the other betaherpesvirus gH and gL homologs, as determined by GAP analysis (15) (data not shown). Alignment analyses of the gO homolog amino acid sequences to determine regions of similarity did not reveal any obvious conserved domains (data not shown). However, the predicted products of the gO positional homolog genes do share a number of biochemical features (Table 1). The gO homologs are predicted to be heavily glycosylated proteins, containing between 7 and 23 potential N-linked glycosylation sites (Table 1). The gO homolog amino acid sequences also contain clusters of serine and threonine residues, suggesting potential O-linked glycosylation, and five to six cysteine residues that reside almost exclusively in the N-terminal half of the predicted proteins (data not shown). Also, the gO homologs appear to be basic proteins, having calculated isoelectric points of approximately 10 as estimated by Peptidesort in the GCG package (15) (Table 1).
TABLE 1.
Characteristics of predicted products of gO positional homolog genes
| Virus | Gene designation | No. of amino acids | No. of NXT/S sites | pIa | No. of cysteine residues | % amino acid similarityb/% amino acid identityb to HCMV UL74 |
|---|---|---|---|---|---|---|
| HCMV | UL74 | 466 | 18 | 10.3 | 6 | NAc |
| HHV-6A | U47 | 651 | 18 | 10.3 | 5 | 39.1/19.4 |
| HHV-6B | KA8L | 738 | 23 | 9.4 | 5 | 36.1/19.4 |
| HHV-7 | U47 | 313 | 11 | 10.3 | 6 | 42.7/22.5 |
| MCMV | m74 | 438 | 7 | 9.8 | 6 | 43.4/22.0 |
Determined by PeptideStructure (GCG).
Determined by GAP (GCG).
NA, not applicable.
DISCUSSION
In this communication, we have reported the genetic identity of a 125-kDa HCMV envelope glycoprotein which associates with gH and gL to form the viral gCIII envelope complex. Through multiple lines of evidence, we have demonstrated that this 125-kDa glycoprotein component is the product of the HCMV UL74 gene. First, biochemical characterization of the 125-kDa glycoprotein suggested the HCMV UL74 gene as a potential gene candidate. Second, amino acid microsequence of a peptide derived from the purified 125-kDa glycoprotein matched a sequence within the predicted product of the UL74 gene. Third, antibodies specific for UL74 immunoprecipitated the gCIII complex and recognized the 125-kDa glycoprotein component of gCIII. We have designated the HCMV UL74 glycoprotein as gO, consistent with the nomenclature established for herpesvirus glycoproteins.
The finding that the HCMV gO gene has positional homologs that share predicted biochemical features suggests that other herpesviruses will be found to contain a third glycoprotein component in their respective gH-gL complexes. Interestingly, these gO positional homologs appear to present only in other betaherpesviruses, suggesting that the gO homologs represent the first-described betaherpesvirus-specific envelope glycoprotein homologs. It remains to be determined if the alphaherpesviruses encode a protein that is analogous to gO. However, it has been well established that the EBV glycoprotein gp42 is a third component of the EBV gH-gL complex (34). Comparisons of gp42 with gO do not reveal any obvious similarities. gp42 is much smaller (42 versus 125 kDa) and contains considerably less carbohydrate (10 kDa of N-linked glycosylation versus at least 60), and it does not require disulfide bonding for association with EBV gH and gL (34). Examination of the gp42 amino acid sequence does not reveal it to be a basic protein (calculated isoelectric point of 8.6 [data not shown]). The only apparent biochemical similarity between gp42 and gO is that both are predicted to be type II transmembrane proteins (34). Most likely, comparisons of the functions of gp42 and gO will indicate the true relatedness of these two glycoproteins.
What role(s) does gO play in HCMV infection? Since no molecular functions have been assigned to gO, we can only speculate on the potential functionality(s) of gO. Due to the association of gO with the gH-gL complex, a crucial participant in the viral fusion machinery (29, 30, 41), one obvious function of gO could be to facilitate entry and/or cell-to-cell spread of infection. In particular, gO may be required strictly for entry into certain specialized cell types. HCMV has been found in association with a number of diverse cell types in vivo, and it is likely that the virus can employ different modes of entry to gain access into these divergent cell types. Such a role for gO would mirror the functionality of EBV gp42, which has been shown to be required for entry of virus into one permissive cell type (B cells) but not the other permissive cell type (epithelial cells) (34, 52). Alternatively, demonstration that gO has heparin-binding capabilities, as predicted by primary sequence analysis, could imply a role in attachment or stabilization of virus prior to fusion. Efforts are under way to assay the specific molecular functions of gO and of the gH-gO-gL complex in HCMV infection.
This study highlights an important limitation in HCMV research. There are 57 potential glycoproteins in the commonly used laboratory strain AD169 (9) and as many as 70 glycoprotein ORFs in clinical isolates (7). This unparalleled glycoprotein-coding capacity supports the hypothesis that HCMV has the potential for functional compensation and functional redundancy, a phenomenon documented for herpesvirus envelope proteins, but implies that HCMV also has the capability for specialized functional roles tailored to replication and pathogenic features in the biology of HCMV infection. It is noteworthy, therefore, that few gene products are characterized with respect to biosynthesis within infected cells and incorporation into the virion. In conjunction with an unknown number of individual glycoproteins, there are three major disulfide-linked envelope complexes, each with implicated functional roles in entry and spread of infection. Prior to this report, only the gCI complex, which contains a dimer of gB in association with cellular annexin II (4, 23, 43), was structurally and genetically defined. Here we report a completed genetic composition of gCIII. The gCII complex, which has a suggested role in heparin binding, has at least three protein components, only one of which is mapped to the viral genome (1, 27, 32). Thus, before the mechanism of viral entry and spread can be fully elucidated, whether in model, permissive fibroblasts or in biologically targeted cell types such as monocytes/macrophages, endothelial cells, and epithelial cells, the composition of the envelope must be defined and the genes encoding the proteins must be identified.
ACKNOWLEDGMENTS
This study was supported in part by Public Health Service grant A1-34998. Protein sequence determination was performed by the Protein/DNA Technology Center of Rockefeller University.
We gratefully acknowledge the members of the Compton lab for their assistance and expertise in cell scraping.
REFERENCES
- 1.Baines J D, Roizman B. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J Virol. 1993;67:1441–1452. doi: 10.1128/jvi.67.3.1441-1452.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett B C, Dolan A, Telford E A R, Davison A J, McGeoch D J. A novel herpes simplex virus gene (UL49A) encodes a putative membrane protein with counterparts in other herpesviruses. J Gen Virol. 1992;73:2167–2171. doi: 10.1099/0022-1317-73-8-2167. [DOI] [PubMed] [Google Scholar]
- 3.Bause E. Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem J. 1983;209:331–336. doi: 10.1042/bj2090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britt W J. Neutralizing antibodies detect a disulfide-linked glycoprotein complex within the envelope of human cytomegalovirus. Virology. 1984;135:369–378. doi: 10.1016/0042-6822(84)90193-4. [DOI] [PubMed] [Google Scholar]
- 5.Britt W J, Vulger L, Butfiloski E J, Stephens E B. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J Virol. 1990;64:1079–1085. doi: 10.1128/jvi.64.3.1079-1085.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardin A D, Weintraub H J R. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 7.Cha T-A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang C P, Vesole D H, Nelson J, Oldstone M B, Stinski M F. Identification and expression of a human cytomegalovirus early glycoprotein. J Virol. 1989;63:3330–3337. doi: 10.1128/jvi.63.8.3330-3337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 10.Compton T. An immortalized human fibroblast cell line is permissive for human cytomegalovirus infection. J Virol. 1993;67:3644–3648. doi: 10.1128/jvi.67.6.3644-3648.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Compton T, Nepomuceno R R, Nowlin D M. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology. 1992;191:387–395. doi: 10.1016/0042-6822(92)90200-9. [DOI] [PubMed] [Google Scholar]
- 12.Cranage M P, Kouzarides T, Bankier A T, Satchwell S, Weston K, Tomlinson P, Barrell B, Hart H, Bell S E, Minson A C, et al. Identification of the human cytomegalovirus glycoprotein B gene and induction of neutralizing antibodies via its expression in recombinant vaccinia virus. EMBO J. 1986;5:3057–3063. doi: 10.1002/j.1460-2075.1986.tb04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cranage M P, Smith G L, Bell S E, Hart H, Brown C, Bankier A T, Tomlinson P, Barrell B G, Minson T C. Identification and expression of a human cytomegalovirus glycoprotein with homology to the Epstein-Barr virus BXLF2 product, varicella-zoster virus gpIII, and herpes simplex virus type 1 glycoprotein H. J Virol. 1988;62:1416–1422. doi: 10.1128/jvi.62.4.1416-1422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dambaugh T R, O’Brian J J, Anton E D, Greenamoyer C A, Lindquester G J, Pellett P E. Genetic content of a 20.9 kb segment of human herpesvirus 6B strain Z29 spanning the homologs of human herpesvirus 6A genes U40-57 and containing the origin of replication. Arch Virol. 1996;142:103–123. doi: 10.1007/s007050050062. [DOI] [PubMed] [Google Scholar]
- 15.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez J, Andrews L, Mische S M. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal Biochem. 1994;214:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez J, Gharahdaghi F, Mische S M. Routine identification of proteins from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels or polyvinyl difluoride membranes using matrix assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF-MS) Electrophoresis. 1998;19:1036–1045. doi: 10.1002/elps.1150190619. [DOI] [PubMed] [Google Scholar]
- 18.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuller A O, Lee W. Herpes simplex virus type 1 entry through a cascade of virus-cell interactions requires different roles of gD and gH in penetration. J Virol. 1992;66:5002–5012. doi: 10.1128/jvi.66.8.5002-5012.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller A O, Santos R E, Spear P G. Neutralizing antibodies specific for glycoprotein H of herpes simplex virus permit viral attachment to cell but prevent penetration. J Virol. 1989;63:3435–3443. doi: 10.1128/jvi.63.8.3435-3443.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gompels U A, Carss A L, Sun N, Arrand J R. Infectivity determinants encoded in a conserved gene block of human herpesvirus-6. DNA Sequence. 1992;3:25–39. doi: 10.3109/10425179209039693. [DOI] [PubMed] [Google Scholar]
- 22.Gompels U A, Nicholas J, Lawrence G, Jones M, Jones M, Thomson B J, Martin M E D, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 23.Gretch D R, Kari B, Rasmussen L, Gehrz R C, Stinski M F. Identification and characterization of three distinct families of glycoprotein complexes in the envelopes of human cytomegalovirus. J Virol. 1988;62:875–881. doi: 10.1128/jvi.62.3.875-881.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber M T, Compton T. Characterization of a novel third member of the human cytomegalovirus glycoprotein H-glycoprotein L complex. J Virol. 1997;71:5391–5398. doi: 10.1128/jvi.71.7.5391-5398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huber, M. T., and T. Compton. 1998. Unpublished data.
- 26.Jons A, Granzow H, Kuchling R, Mettenleiter T C. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J Virol. 1996;70:1237–1241. doi: 10.1128/jvi.70.2.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kari B, Li W, Cooper J, Goertz R, Radeke B. The human cytomegalovirus UL100 gene encodes the gC-II glycoproteins recognized by group 2 monoclonal antibodies. J Gen Virol. 1994;75:3081–3086. doi: 10.1099/0022-1317-75-11-3081. [DOI] [PubMed] [Google Scholar]
- 28.Kaye J F, A. G U, Minson A C. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with the HCMV UL115 gene product. J Gen Virol. 1992;73:2693–2698. doi: 10.1099/0022-1317-73-10-2693. [DOI] [PubMed] [Google Scholar]
- 29.Keay S, Baldwin B. Anti-idiotype antibodies that mimic gp86 of human cytomegalovirus inhibit viral fusion but not attachment. J Virol. 1991;65:5124–5128. doi: 10.1128/jvi.65.9.5124-5128.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keay S, Merigan T C, Rasmussen L. Identification of cell surface receptors for the 86-kilodalton glycoprotein of human cytomegalovirus. Proc Natl Acad Sci USA. 1989;86:10100–10103. doi: 10.1073/pnas.86.24.10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 32.Lehner R, Meyer H, Mach M. Identification and characterization of a human cytomegalovirus gene encoding for a membrane protein that is conserved among human herpesviruses. J Virol. 1989;63:3792–3800. doi: 10.1128/jvi.63.9.3792-3800.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Nelson J A, Britt W J. Glycoprotein H-related complexes of human cytomegalovirus: identification of a third protein in the gCIII complex. J Virol. 1997;71:3090–3097. doi: 10.1128/jvi.71.4.3090-3097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q, Turk S M, Hutt-Fletcher L M. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol. 1995;69:3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindquester G J, Inoue N, Allen R D, Castelli J W, Stamey F R, Dambaugh T R, O’Brian J J, Danovich R M, Frenkel N, Pellett P E. Restriction endonuclease mapping and molecular cloning of the human herpesvirus 6 variant B strain Z29 genome. Arch Virol. 1996;141:367–379. doi: 10.1007/BF01718406. [DOI] [PubMed] [Google Scholar]
- 36.Mach M, Utz U, Fleckenstein B. Mapping of the major glycoprotein gene of human cytomegalovirus. J Gen Virol. 1986;67:1461–1467. doi: 10.1099/0022-1317-67-7-1461. [DOI] [PubMed] [Google Scholar]
- 37.Margulies B J, Browne H, Gibson W. Identification of the human cytomegalvirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology. 1996;225:111–125. doi: 10.1006/viro.1996.0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller N, Hutt-Fletcher L M. A monoclonal antibody to glycoprotein gp85 inhibits fusion but not attachment to Epstein-Barr virus. J Virol. 1988;62:2366–2372. doi: 10.1128/jvi.62.7.2366-2372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mocarski E S., Jr . Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 40.Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus 7. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pachl C, Probert W S, Hermsen K M, Masiarz F R, Rasmussen L, Merigan T C, Spaete R R. The human cytomegalovirus strain Towne glycoprotein H gene encodes glycoprotein p86. Virology. 1989;169:418–426. doi: 10.1016/0042-6822(89)90167-0. [DOI] [PubMed] [Google Scholar]
- 42.Peeters B, de Wind N, Broer R, Gielkens A, Moormann R. Glycoprotein H of pseudorabies virus is essential for entry and cell-to-cell spread of the virus. J Virol. 1992;66:3888–3892. doi: 10.1128/jvi.66.6.3888-3892.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pietropaolo R L, Compton T. Direct interaction between human cytomegalovirus glycoprotein B and cellular annexin II. J Virol. 1997;71:9803–9807. doi: 10.1128/jvi.71.12.9803-9807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rasmussen L, Nelson R, Kelsall D, Merigan T. Murine monoclonal antibody to a single protein neutralizes the infectivity of human cytomegalovirus. Proc Natl Acad Sci USA. 1984;81:876–880. doi: 10.1073/pnas.81.3.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez J E, Moninger T, Grose C. Entry and egress of varicella virus blocked by same anti-gH monoclonal antibody. Virology. 1993;196:840–844. doi: 10.1006/viro.1993.1543. [DOI] [PubMed] [Google Scholar]
- 47.Roop C, Hutchinson L, Johnson D C. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J Virol. 1993;67:2285–2297. doi: 10.1128/jvi.67.4.2285-2297.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russo J J, Bohenzky R A, Chien M-C, Chien J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spaete R R, Gehrz R C, Landini M P. Human cytomegalovirus structural proteins. J Gen Virol. 1994;75:3287–3308. doi: 10.1099/0022-1317-75-12-3287. [DOI] [PubMed] [Google Scholar]
- 50.Spaete R R, Perot K, Scott P I, Nelson J A, Stinski M F, Pachl C. Coexpression of truncated human cytomegalovirus gH with the UL115 gene product or the truncated human fibroblast growth factor receptor results in the transport of gH to the cell surface. Virology. 1993;193:853–861. doi: 10.1006/viro.1993.1194. [DOI] [PubMed] [Google Scholar]
- 51.Urban M, Britt W, Mach M. The dominant linear neutralizing antibody-binding site of glycoprotein gp86 of human cytomegalovirus is strain specific. J Virol. 1992;66:1303–1311. doi: 10.1128/jvi.66.3.1303-1311.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Hutt-Fletcher L M. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J Virol. 1988;72:158–163. doi: 10.1128/jvi.72.1.158-163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]