Abstract
[3H]palmitic acid was metabolically incorporated into the viral fusion protein (F) of Edmonston or freshly isolated measles virus (MV) during infection of human lymphoid or Vero cells. The uncleaved precursor F0 and the F1 subunit from infected cells and extracellular virus were both labeled, indicating that palmitoylation can take place prior to F0 cleavage and that palmitoylated F protein was incorporated into virus particles. [3H]palmitic acid was released from F protein upon hydroxylamine or dithiothreitol treatment, indicating a thioester linkage. In cells transfected with the cloned MV F gene, in which the cysteines located in the intracytoplasmic and transmembrane domains (Cys 506, 518, 519, 520, and 524) were replaced by serine, a major reduction of [3H]palmitic acid incorporation was observed for F mutated at Cys 506 and, to a lesser extent, at Cys 518 and Cys 524. We also observed incorporation of [3H]palmitic acid in the F1 subunit of canine distemper virus F protein. Cell fusion induced by cotransfection of cells with MV F and H (hemagglutinin) genes was significantly reduced after replacement of Cys 506 or Cys 519 with serine in the MV F gene. Transfection with the F gene with a mutation for Cys 518 abolished cell fusion, although less mutant protein was detected on the cell surface. These results suggest that the F protein transmembrane domain cysteines 506 and 518 participate in structures involved in cell fusion, possibly mediated by palmitoylation.
A number of viral and cellular proteins have been found to be modified by palmitoylation and myristoylation. This list includes viral membrane proteins, cell receptors, and a number of proteins involved in cell signaling or metabolic regulation (12, 21, 22). Myristoylation is a cotranslational modification occurring at the amino terminus, while palmitoylation takes place posttranslationally and prior to transport of the protein through the Golgi system. Palmitoylation involves the addition of a 16-carbon saturated fatty acyl moiety via thioester or ester linkage to cysteine, serine, or threonine residues and, in contrast to myristoylation, it may be reversible (39). Although the possible roles of fatty acid acylations are not currently known, there are suggestions that protein palmitoylation may modify protein conformation and functions such as protein targeting (5, 11, 16, 18), lateral diffusion on the membrane (36), protein oligomerization (1, 26), or cytoplasmic protein association with membranes for transmembrane proteins (38).
A function for palmitoylation of viral proteins has not yet been demonstrated. There are suggestions that this posttranslational modification may influence the infectivity of influenza virus particles (46) or tissue invasiveness (14). For Sindbis virus, it was shown that replacing acylated cysteine residues of both the 6K protein and the E2 glycoprotein influences virus assembly and budding (10, 13). A possible function for palmitoylation of viral proteins in cell fusion activity has been reported by some researchers for influenza virus (23) and was found negative for vesicular stomatitis virus (VSV) (41), influenza virus (24, 27), and murine leukemia virus (45).
Some paramyxoviruses, such as Newcastle disease virus (7, 37) and simian virus 5, but not Sendai virus (40), have palmitoylated membrane glycoproteins. Data on fatty acylation of morbillivirus proteins have not been reported. We show here that palmitic acid, but not myristic acid, is incorporated in measles virus (MV) proteins. Palmitoylation was found only in the F0 precursor and the F1 subunit of MV and canine distemper virus (CDV). The fusion protein allows virus entry and cell-to-cell transmission of the virus by inducing membrane fusion. The F protein of MV is a tri- or tetrameric type 1 membrane glycoprotein whose monomers are bound by hydrophobic forces and which is synthesized in the endoplasmic reticulum as an inactive precursor, F0. The fully glycosylated F0 is cleaved into F1 and F2 subunits, which are joined by disulfide bonds to form the functional F protein (30, 43). We report here that palmitic acid is incorporated through thioester bonds into F0 and F1, and transfection of the F gene, subjected to site-directed mutagenesis to replace cysteines by serines, indicated that the majority binds to Cys 506 and, to a lesser degree, to Cys 518 and 524 of the MV F protein. To elucidate the possible functions of this modification, we cotransfected cells with MV H and mutated F genes and measured the induction of cell fusion. We have found that F protein Cys 506 and 518 are important for cell fusion activity, possibly through their palmitoylation.
MATERIALS AND METHODS
Cells, virus, and plasmid vectors.
MOLT4 and Dakiki cells were maintained in RPMI medium supplemented with glutamine, penicillin-streptomycin, and 10% fetal calf serum. Growth of the Edmonston strain and the recent MV isolate Ma93F (32) was done as described previously (8). Growth of the Onderstepoort strain of CDV was done on Vero cell monolayers in Dulbecco’s modified Eagle medium supplemented with 2% fetal calf serum, glutamine, and gentamycin. The recombinant vaccinia virus vTF7-3 (which expresses T7 RNA polymerase) was grown in Vero cells, as described by Fuerst et al. (9). Plasmids containing the complete coding region of the F protein gene (nucleotides [nt] 537 to 2369) (3F21) and the H protein gene (nt 1 to 1948) (3H18) of the MV Edmonston strain were constructed. cDNAs obtained by reverse transcription-PCR with primers having restriction enzyme sites were cloned downstream of the T7 polymerase promotor of vector pGEM4Z (Promega).
Metabolic labeling of cells and immunoprecipitation of proteins.
Cells were [35S]methionine-cysteine labeled for 5 h, and proteins were immunoprecipitated (radioimmunoprecipitation assay) as described previously (6) with 20 μCi of TRAN35S-label (1,200 Ci/mmol; ICN)/ml in a medium containing one-fifth the normal concentration of Met and Cys. [3H]palmitic acid labeling of 2 × 106 MOLT4 and Dakiki cells was done, unless otherwise specified, for 5 h with 1 mCi of [9,10-3H]palmitic acid (50 Ci/mmol; Amersham) in medium containing 1 mM sodium pyruvate and 1% nonessential amino acids. CDV-infected Vero cells showing about 70% cytopathic effects were labeled for 5 h with 1 mCi [9,10-3H]palmitic acid/5 × 106 cells in medium containing 1 mM pyruvate and 1% nonessential amino acids.
Guinea pig anti-MV antibodies used for immunoprecipitation of MV proteins were from M. A. Bioproducts. Anti-F protein monoclonal antibodies (MAbs) (kindly provided by R. Buckland, Institute Pasteur, Lyon, France) Ost-2 (from A. D. M. E. Osterhaus, Erasmus University, Rotterdam, The Netherlands) and 263-5 (19) were used for immunoprecipitation and fluorescence-activated cell sorter analysis, respectively.
Site-specific mutagenesis.
Mutant F genes were derived from plasmid pGEM-3F21 (wild type) with the Transformer mutagenesis kit (Clontec) and the selection primer 5′-ATTTCACACCGCCCATGGTGCACTC-3′, following the supplier’s instructions. The 27-nt oligomer (nt 2078 to 2104) 5′-CTGATTGCAGTGTCTCTTGGAGGGTTG-3′ was used to generate the Cys 506 mutant (the specific mutated codon is underlined) (F gene residues are from the consensus sequence of Radecke and Billeter [28]). The 37-nt oligomer (nt 2109 to 2145) 5′-GGATCCCCGCTTTAATATCTTGCTGCAGGGGGCGTTG-3′ was used to generate the Cys 518 mutant. The oligomer (nt 2109 to 2145) 5′-GGATCCCCGCTTTAATATGTTCCTGCAGGGGGCGTTG-3′ was used to generate the Cys 519 mutant. The oligomer (nt 2109 to 2145) 5′-GGATCCCCGCTTTAATATGTTGCTCCAGGGGGCGTTG-3′ was used to generate the Cys 520 mutant, and the 23-mer (nt 2135 to 2157) 5′-AGGGGGCGTTCTAACAAAAAGGG was used to generate the Cys 524 mutant. To verify that only a single change existed, mutant genes were sequenced in their entirety by dideoxynucleotide chain-terminating sequencing (34) and manual analysis or by using an automated DNA sequencer (model ABI377-18; Perkin-Elmer). To verify the identity of the mutated proteins and to assay their reactivity with the anti-F MAb Ost-2, wild-type and mutant fusion proteins were synthesized in vitro with a transcription-translation coupled reticulocyte lysate system (TNT; Promega).
Transient expression of F protein mutants.
Fusion genes were expressed by the recombinant vaccinia virus-encoding T7 polymerase system (9). MOLT4 cells were transfected with plasmid DNA by using Lipofectin (GIBCO-BRL) essentially as recommended by the manufacturer. For every 2.5 × 105 cells infected with vaccinia virus vTF7-3 at a multiplicity of infection of 10 PFU/cell, a mixture of 1 to 2 μg of DNA and 5 μg of Lipofectin was used. Twenty hours after transfection, the cells were labeled for 3 h with 60 μCi of [3H]palmitic acid or 30 μCi of TRAN35S-label and processed as indicated.
Fusion assays.
Fusion assays were performed with MOLT4 cells. Vaccinia virus vTF7-3 infection of MOLT4 cells produced very limited cytopathic effects 20 h postinfection, and syncytium formation by cotransfection of both fusion and hemagglutinin genes is reproducible. Several concentrations of F and H plasmid DNAs were tested for their fusion effects, and a 1:1 ratio was chosen as optimum for fusion assays. Exponentially growing cells (2.5 × 105) were cotransfected with 0.5 μg of plasmid DNA expressing F protein and 0.5 μg of plasmid DNA expressing H protein. For flow cytometry analysis of surface F protein expression, transfected cells were incubated with anti-F MAb 263-5 and subsequently with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G for indirect staining.
RESULTS
Fusion proteins of measles virus and CDV are metabolically labeled with [3H]palmitic acid.
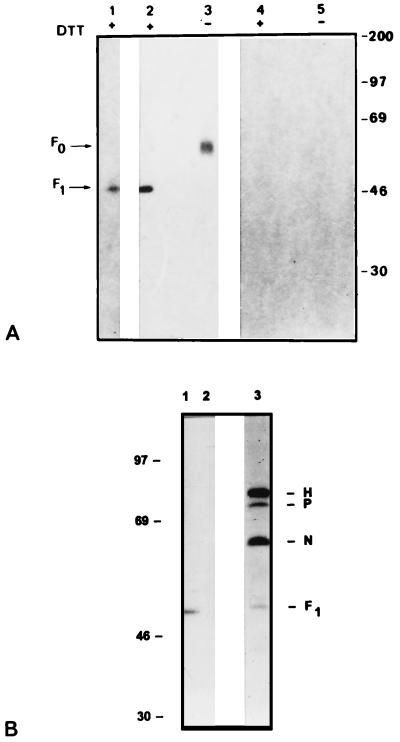
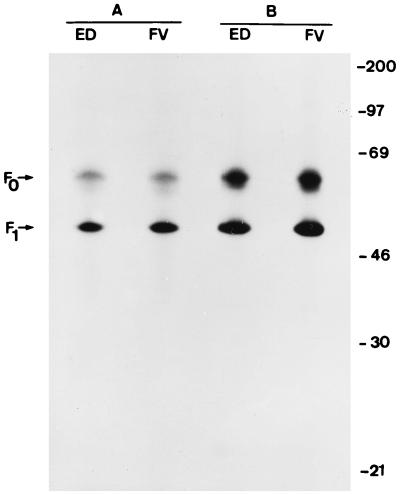
We labeled MOLT4 cells infected with the MV Edmonston strain or Vero cells infected with the CDV Onderstepoort strain with [3H]palmitic acid or [3H]myristic acid under conditions in which little metabolism of exogenous fatty acid occurs. Cell extracts and pelleted extracellular virus particles were immunoprecipitated with anti-MV guinea pig serum or anti-MV F MAb. The immunoprecipitates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing or nonreducing conditions. In MV-infected MOLT4 cells labeled with [3H]palmitic acid, radioactivity was incorporated only into F protein (Fig. 1A) (also see Fig. 4). This result agrees with the unpublished observation of Fabian Wild (cited in reference 3). We detected [3H]palmitic acid in F1 and F0 from both cell-associated and extracellular virus. No radioactivity was detected in any MV proteins when infected cells were labeled with [3H]myristic acid. When other cell lines were labeled, such as the human B Dakiki cells or monkey Vero cells (not shown) infected with Edmonston or primary MV isolates, the radioactivity was incorporated into the cleaved and uncleaved forms of F protein, the F1 and F0 proteins (Fig. 2). The stoichiometry of F protein palmitoylation cannot be determined by metabolic radiolabeling because the specific activity of the intracellular pool is not known. For comparison, the G protein of the Indiana strain of VSV, which is known to be modified by a single palmitate moiety (33, 35), was labeled in parallel in infected Vero cells. The relative ratios of [3H]palmitate to [35S]methionine in both MV F1 and VSV G were compared. Correcting for the number of methionines in each protein, our preliminary estimate is that F1 protein carries an average of 1.6 molecules of palmitate (data not shown). In Vero cells infected with CDV, the [3H]palmitic acid was also incorporated into F1 protein (Fig. 1B).
FIG. 1.
(A) PAGE of [3H]palmitic acid (lanes 1 to 3)- and [3H]myristic acid (lanes 4 and 5)-labeled polypeptides in Edmonston MV-infected MOLT4 cells. Extracellular (lane 1) and intracellular (lanes 2 to 5) viral proteins were labeled for 15 h, immunoprecipitated with anti-MV guinea pig serum, and analyzed under reducing (+) or nonreducing (−) conditions. A 25-day exposure of the fluorogram is shown. Molecular masses (in kilodaltons) are on the right. (B) Incorporation of [3H]palmitic acid into F protein of CDV. [3H]palmitic acid-labeled intracellular polypeptides in uninfected (lane 2) and CDV-infected (lane 1) Vero cells and TRAN35S-labeled proteins from CDV-infected Vero cells (lane 3) are shown. CDV proteins were immunoprecipitated with anti-MV guinea pig serum and analyzed in a SDS–10% polyacrylamide slab gel. A 30-day exposure of the fluorogram is shown. Molecular masses (in kilodaltons) are on the left.
FIG. 4.
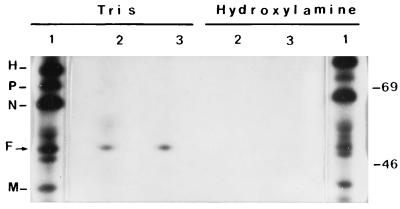
Sensitivity of MV acylated proteins to hydroxylamine. Extracts from MV Edmonston-infected MOLT4 cells labeled with TRAN35S-label (1) or [3H]palmitate (2 and 3) were immunoprecipitated with anti-MV antibodies; the immunocomplexes were treated with 1 M Tris-HCl (pH 8) or 1 M hydroxylamine (pH 8) at room temperature for 3 h and analyzed by SDS-PAGE and fluorography. Molecular masses (in kilodaltons) are on the right.
FIG. 2.
Incorporation of [3H]palmitic acid into F protein of Edmonston strain and wild-type isolate FV. [3H]palmitic acid-labeled intracellular polypeptides in Edmonston (ED) and primary isolate Ma93F (FV) virus-infected Dakiki cells (2 × 106) were immunoprecipitated with the anti-F MAb Ost-2 (A), and the supernatant was immunoprecipitated with the anti-MV guinea-pig serum (B). A 20-day exposure of the fluorogram is shown. Molecular masses (in kilodaltons) are on the right.
Identification of the lipid moiety of [3H]palmitic acid-labeled F protein.
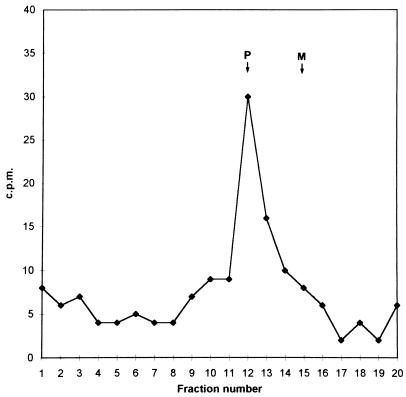
Since fatty acids can be metabolically altered and interconvert to longer-chain variants, it was necessary to chemically characterize the protein-associated radioactivity. Gel-purified [3H]palmitic acid-labeled F protein was subjected to complete acid hydrolysis, and the fatty acid released was identified by organic solvent extraction and reverse-phase thin-layer chromatography. The majority of incorporated radioactivity migrated as the palmitic acid standard, with a peak at 6 cm from the origin, while the myristic acid standard migrated 7.5 cm (Fig. 3). Under the labeling conditions employed, most of the radioactivity incorporated into the F protein thus remained as palmitic acid.
FIG. 3.
Identification of fatty acid removed from F protein as palmitate. Thin-layer chromatography of fatty acids from hydrolyzed [3H]palmitic acid-labeled F protein from MV Edmonston-infected MOLT4 cells. Markers of palmitic (P) and myristic (M) acids were detected under UV light after rhodamine G impregnation, and radioactivity was estimated in a beta radiation counter.
Acylation of MV F protein occurs through thioester linkage on cysteine residues.
To investigate the nature of the fatty acid linkage to F protein, the sensitivity of the palmitate-labeled proteins to the action of hydroxylamine or reducing agents such as dithiothreitol (DTT) was examined. If acylation of F protein had occurred on the cysteine residues via a thioester bond, as in the cases of other viral and cellular proteins, the linkage would be susceptible to cleavage with hydroxylamine at neutral pH. In contrast, acylation of serine residues occurs through a hydroxyester bond, which is unaffected by this treatment. Myristic acid, which is commonly attached through amide linkages, is also not susceptible to cleavage by this treatment. To examine the stability of the linkage, MV proteins labeled with either [3H]palmitic acid or [35S]methionine-cysteine were immunoprecipitated and the immunocomplexes were treated with hydroxylamine and analyzed by SDS-PAGE and fluorography. Alternatively, the material was run in parallel gels, and after fixation, the gels were treated for 16 h at room temperature with 1 M hydroxylamine (pH 8) or 1 M Tris-HCl (pH 8). The [3H]palmitic acid label was cleaved by hydroxylamine, whereas the [35S] label was unaffected by this treatment (Fig. 4). The susceptibility to hydroxylamine at neutral pH indicated that palmitic acid was attached to F protein through thioester linkage. To confirm this result, we investigated the sensitivity of [3H]palmitic acid label to reducing agents. Densitometric tracing of autoradiography (Fig. 5) revealed that over 85% of [3H]palmitic acid is released from F protein after treatment with 0.2 M DTT. Coomassie blue staining of F protein bands showed that there was no significant loss of protein following DTT treatment (not shown). Since oxygen ester linkages are known to resist such treatment, these results strongly support the idea that cysteine residues are the palmitic acid linkage sites in MV F protein.
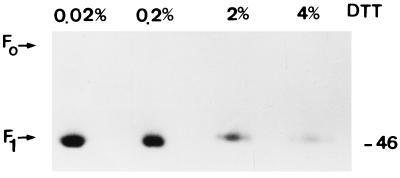
FIG. 5.
Sensitivity of acylated F protein to DTT. Fusion protein immunocomplexes from [3H]palmitic acid-labeled MV Edmonston-infected MOLT4 cells were treated with increasing concentrations of DTT. Samples were heated to 95°C for 5 min and subjected to SDS-PAGE and fluorography. The molecular mass (in kilodaltons) is on the right.
Replacement by serine of cysteine 506 or 518, located at MV F protein transmembrane-cytoplasmic domains, inhibited palmitoylation.
Despite the increasing number of palmitoylation sites identified, there appears to be no consensus motif for predicting candidate cysteine residues, which may be acylated. It has been reported for several viral glycoproteins that palmitic acid is attached to the region between the transmembrane anchor and the cytoplasmic tail of the molecule. To identify the palmitoylated cysteine residues of MV F protein, we have investigated palmitate incorporation into wild-type and mutant F proteins in which the cysteine residues located in the transmembrane-intracytoplasmic domain (Cys 506, 518, 519, 520, and 524) have been mutated to serine (Fig. 6). After site-directed mutagenesis, each mutation was confirmed by sequencing the entire mutated F gene; Fig. 7 shows the sequences of mutated codons and the reactivity with specific F MAbs of the in vitro-transcribed and -translated F mutants.
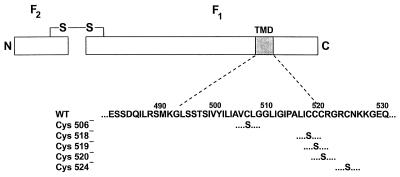
FIG. 6.
Schematic diagram of mutations introduced at the transmembrane domain of MV Edmonston fusion protein. The F protein is shown as two rectangles, denoting the F2 and F1 subunits at the amino and carboxyl domains, bound by a disulfide bridge. The amino acid sequence surrounding the predicted transmembrane domain (TMD) is listed; the numbers represent the positions of residues from the N-terminal methionine. For the mutants, only mutated residues are depicted. The nomenclature of the mutants specifies the original residue at the position indicated.
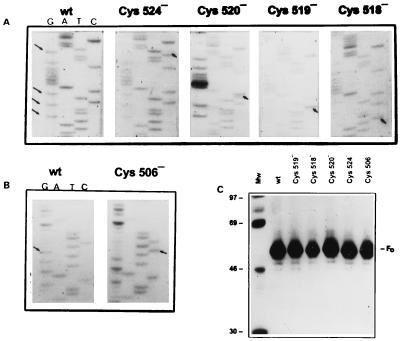
FIG. 7.
Analysis of F gene plasmid mutants. (A) Sequence ladders of wild-type (wt) plasmid 5′-AATAT(G)TT(G)CT(G)CAGGGGGCGTT(G)AAC-3′ and mutant plasmids Cys 524, Cys 520, Cys 519, and Cys 518. (B) Sequence ladders of wild-type plasmid 5′-TTGCAGTGT(G)TCTT-3′ and mutant plasmid Cys 506. Arrows indicate mutated bases. (C) Protein synthesized in vitro by plasmid DNA in a TNT system (Promega) after immunoprecipitation with anti-F MAb. Mw, molecular weight markers (shown in thousands on the left).
The F protein expressed by transfection of MOLT4 cells with cloned F protein cDNA can also be labeled with [3H]palmitic acid (not shown), indicating that no other viral proteins are required for acylation. Cells cotransfected with H genes and control or mutated F genes were labeled in parallel with [3H]palmitic acid and [35S]methionine plus cysteine to monitor the level of F protein palmitoylation and expression. After immunoprecipitation, samples were fractionated by SDS–10% PAGE under reducing conditions; 3H label incorporated into the F protein band was measured by scintillation counting of excised bands after transfer to polyvinylidene difluoride membranes, and 35S radioactivity was measured by autoradiography and densitometric analysis. Because the expression levels differed significantly, to normalize the [3H]palmitate incorporation, the ratio of palmitate incorporation to methionine incorporation was determined for the mutants and the wild-type F (Table 1). Figure 8 shows the fluorographic analysis of [3H]palmitic acid and TRAN35S-label incorporation into mutants 506, 518, and 524 from experiments run in parallel. Mutation at Cys 506 and, to a lesser extent, at Cys 518 inhibited the incorporation of [3H]palmitic acid into F protein. In addition, a minor reduction of palmitate incorporation was observed for the mutation at Cys 524. Individual substitution of the conserved cysteines 519 and 520 does not affect palmitoylation of F protein mutants.
TABLE 1.
Palmitoylation of fusion protein mutants
| F protein | [3H]palmitic acida,b (cpm) | [35S]Met and Cysb,c (relative optical density) | 3H/35S (%) |
|---|---|---|---|
| Wild type | 312 ± 78 | 1.65 ± 0.02 | 189 (100) |
| Cys 506− | 52 ± 13 | 1.30 ± 0.03 | 40 (21) |
| Cys 518− | 122 ± 30 | 0.88 ± 0.08 | 139 (73) |
| Cys 519− | 237 ± 59 | 1.31 ± 0.04 | 182 (96) |
| Cys 520− | 244 ± 61 | 1.14 ± 0.03 | 214 (113) |
| Cys 524− | 263 ± 66 | 1.72 ± 0.02 | 153 (81) |
Values represent the specific radioactivity incorporated into F0 and F1 by 4 × 106 transfected MOLT4 cells labeled for 3 h with 1 mCi of [3H]palmitic acid. The proteins recognized by anti-F MAb Ost-2 were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and measured on a β-radiation scintillation counter.
Values are presented as means ± standard deviations from three separate experiments.
Densitometric values of F0 and F1 from SDS-PAGE autoradiography of 2 × 106 transfected MOLT4 cells labeled with TRAN35S-label after radioimmunoprecipitation assay with anti-F MAb Ost-2.
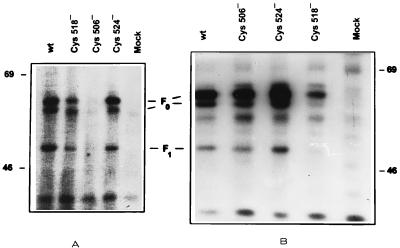
FIG. 8.
Palmitoylation of F mutants Cys 506, Cys 518, and Cys 524. MOLT4 cells cotransfected with the H gene and wild-type (wt) or mutant F plasmid DNA were labeled with [3H]palmitic acid (A) or TRAN35S-label (B). Cell lysates were immunoprecipitated with MAb against F protein and subjected to SDS-PAGE, and the fluorograms were exposed for 30 and 5 days, respectively. Mock transfection was performed in the absence of plasmid DNA. Molecular masses (in kilodaltons) are on the right and left.
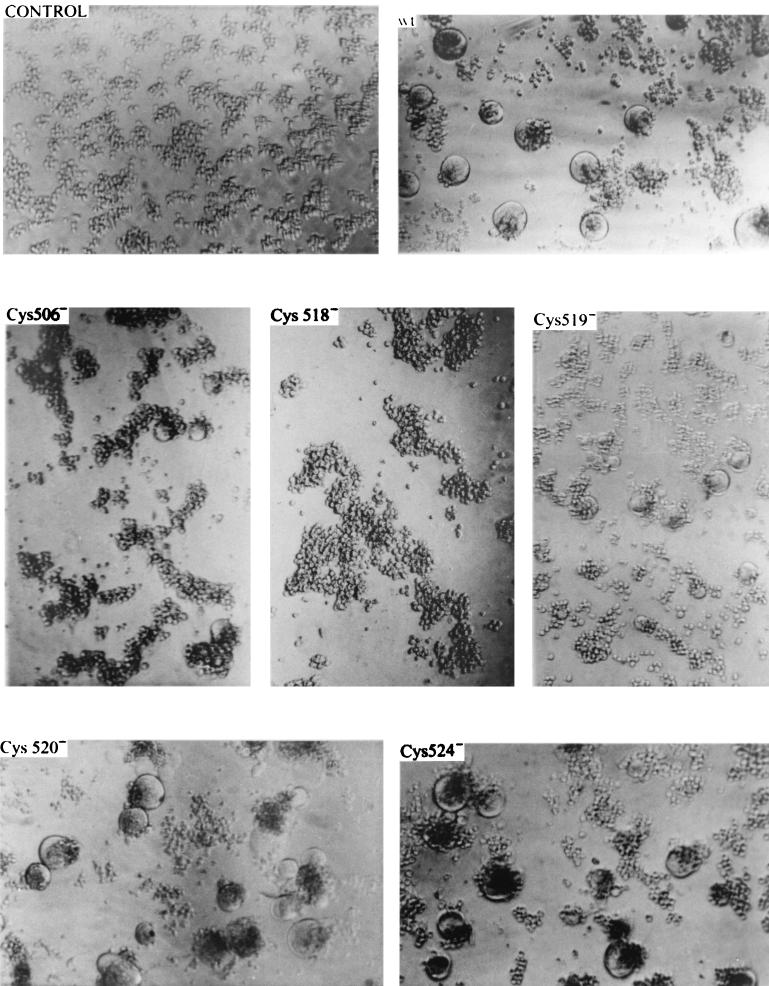
Mutation of Cys 506 or Cys 518 at the MV F protein transmembrane-cytoplasmic domains inhibited the induction of syncytium formation.
The expression of F protein mutants at the cell surface of transfected MOLT4 cells is similar to that in wild-type F, with the exception of that of the Cys 518− mutant, which is somewhat reduced as observed in flow cytometry experiments (Table 2). The induction of syncytia is markedly reduced, however, for the Cys 518− mutant and, to a lesser extent, for Cys 506 and 519 mutants, as shown in Table 2 and Fig. 9. The inability of the Cys 518− mutant to cause fusion does not appear to be an effect of the concentration of F, since increasing the amount of Cys 518 plasmid DNA from 0.5 to 1.5 μg does not augment syncytium induction (data not shown). These results suggest that these cysteines at the transmembrane-intracytoplasmic domains participate in structures important for MV cell fusion activity.
TABLE 2.
Quantitation of expression and activities of fusion protein mutants
| DNA | F cell-surface fluorescence (%)a | Syncytia (%)b |
|---|---|---|
| Wild type | 49.8 (100) | 810 ± 54 (100) |
| Cys 506− | 51.8 (104) | 366 ± 7 (45) |
| Cys 518− | 37.4 (75) | 12 ± 4 (1.5) |
| Cys 519− | 43.3 (86.8) | 513 ± 32 (63) |
| Cys 520− | 48.0 (96.2) | 755 ± 15 (93) |
| Cys 524− | 41.3 (83) | 785 ± 5 (97) |
Values are presented as the percentage of positively stained cells within the gated area of live cells.
Number of syncytia (>4 nuclei) per 2.5 × 105 transfected MOLT4 cells. The values are means ± standard deviations from three separate experiments.
FIG. 9.
Fusion of MOLT4 cells induced by wild-type (wt) and mutant F proteins. Typical areas of the culture were photographed at 20 h postinfection with a Fluorovert-FS (Leitz) inverted microscope with contrast-phase optics and a magnification of ×200.
DISCUSSION
We have shown that the fusion protein of primary isolates and a vaccine strain of MV is palmitoylated during infection of B and T cells and fibroblasts; CDV fusion protein is also palmitoylated. These results suggest that this acylation may have a function in the infection cycle of morbillivirus. MV F protein incorporates palmitic acid through thioester linkage. The C terminus of the F protein contains cysteine residues that are highly conserved among the morbilliviruses (2, 20), one in the cytoplasmic tail (Cys 524 for MV) and four in the putative transmembrane domain (Cys 506, 518, 519, and 520 for MV), with Cys 519 and 520 present in all members of the genus. Employing site-specific mutagenesis, we have identified three cysteine residues in the transmembrane-cytoplasmic domain as acylation sites of MV fusion protein. Palmitoylation of MV fusion protein appears to take place mainly at Cys 506 and, to a lesser extent, at Cys 518 and Cys 524. The residues Cys 519 and 520 appear not to be palmitoylated. Although the assignment of disulfide bridges in the fusion protein of MV is currently unknown, our findings suggest that these invariant residues may participate in this type of structure. In any case, the assignment of palmitoylation sites and the overall level of acylation determined by specific mutagenesis are relative, since the acylation efficiency may be altered by changes in one of the cysteine residues (14, 15, 24).
It is known that palmitic acid modifies some cellular and viral membrane proteins, but the possible functions of this fatty acid acylation have not been determined. We have studied the induction of cell fusion by F mutants that lack cysteine at the palmitoylation sites. As F mutant protein lacking a cysteine at position 524 could induce fusion, it is unlikely that palmitoylation at this site would be of importance for fusion. The position 518 F mutant does not induce fusion, suggesting that palmitoylation at this site may be required for cell fusion. If we assume that Cys 506 is palmitoylated on a majority of the molecules, then Cys 518 would be palmitoylated on only a minority. However, mutation of Cys 518 appears to have a major effect on membrane fusion. It therefore seems unlikely that the palmitoylation at Cys 518 is responsible for the effect. Alternatively, cysteine 518 itself may be required for this function. Such a result might be expected if the mutation altered or destabilized the overall conformation of the protein. In fact, the cellular level of the mutant Cys 518 protein detected by immunoprecipitation and surface immunofluorescence was reduced by about 50%, possibly due to altered stability or a modification of antigenic sites. The failure to overcome the lack of fusion following an increase in the concentration of mutant genes favors the latter alternative. Finally, the cysteine residue at position 506 shows the highest palmitoylation level. When Cys 506 was replaced by serine, the mutant was found to induce fusion at a lower level, suggesting that palmitoylation at position 506 is not absolutely required for fusion, but that it has an accessory role. Our observation that F protein from extracellular virus is also palmitoylated suggests that this acylation may affect both cell-cell and virus-cell membrane fusion.
In addition to the fusion sequence at the amino terminus of the F1 polypeptide (25, 31), other sequences, such as the cysteine-rich region that appears to be the site of interaction with the MV H protein (42) and the leucine zipper structure N-terminal to the transmembrane region (4, 44), seem to play critical roles in maintaining a biologically active structure (17, 43). Our results suggest that the cysteines at the transmembrane domain, Cys 506, 518, and 519, are also functionally involved in promotion of cell fusion. In contrast, transmembrane cysteine 520 and intracytoplasmic Cys 524 do not appear to be specifically required for syncytium formation.
Since palmitoylation of F protein may participate in virus-promoted cell fusion, it will be of interest to evaluate the stoichiometry of F protein palmitoylation during lytic and persistent infections by parental and nonfusogenic MV variants (8). To elucidate the biological significance of palmitoylation, it would be interesting to introduce these mutations in the F protein transmembrane and cytoplasmic domains into the MV-infective cloned DNA (29).
ACKNOWLEDGMENTS
We thank B. Moss for providing vaccinia virus vTF7-3 and E. Díaz de Espada (Intervet-AKZO, The Netherlands) for a seed of Onderstepoort CDV.
J.O. was supported by a predoctoral training grant from the Fondo de Investigaciones Sanitarias, and J.C. was a recipient of a fellowship from the Conserjería de Sanidad-Comunidad de Madrid. This work was supported by grants from the Fondo de Investigaciones Sanitarias, FIS-96/1592 to R.F.-M. and FIS-94/555 to M.L.C.
REFERENCES
- 1.Bach R, Koningsberg W H, Nemerson Y. Human tissue contains thioester linked palmitate and stearate on the cytoplasmic half-cysteine. Biochemistry. 1988;27:4227–4231. doi: 10.1021/bi00412a004. [DOI] [PubMed] [Google Scholar]
- 2.Bolt G, Blixenkrone-Moller M, Gottschalck E, Wishaupt R G A, Welsh M J, Earle J A P, Rima B K. Nucleotide and deduced amino acid sequences of the matrix (M) and fusion (F) protein genes of cetacean morbillivirus isolated from a porpoise and a dolphin. Virus Res. 1994;34:291–304. doi: 10.1016/0168-1702(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 3.Buckland R, Geralt C, Barker R, Wild T F. Fusion glycoprotein of measles virus: nucleotide sequence of the gene and comparison with other paramyxoviruses. J Gen Virol. 1987;68:1695–1703. doi: 10.1099/0022-1317-68-6-1695. [DOI] [PubMed] [Google Scholar]
- 4.Buckland R, Malvoisin E, Beauverger P, Wild T F. A leucine zipper structure present in the measles virus fusion protein is not required for its tetramerization but is essential for fusion. J Gen Virol. 1992;73:1703–1707. doi: 10.1099/0022-1317-73-7-1703. [DOI] [PubMed] [Google Scholar]
- 5.Cadwallader K A, Paterson H, Macdonald S G, Hancock J F. N-terminally myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol Cell Biol. 1994;14:4722–4730. doi: 10.1128/mcb.14.7.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celma M L, Fernández-Muñoz R. Measles virus gene expression in lytic and persistent infections of a human lymphoblastoid cell line. J Gen Virol. 1992;73:2203–2209. doi: 10.1099/0022-1317-73-9-2203. [DOI] [PubMed] [Google Scholar]
- 7.Chatis P A, Morrison T C. Fatty acid modification of Newcastle disease virus glycoproteins. J Virol. 1982;43:342–347. doi: 10.1128/jvi.43.1.342-347.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernández-Muñoz R, Celma M L. Measles virus from a long-term persistently infected human T lymphoblastoid cell line, in contrast to the cytocidal parental virus, establishes an immediate persistence in the original cell line. J Gen Virol. 1992;73:2195–2202. doi: 10.1099/0022-1317-73-9-2195. [DOI] [PubMed] [Google Scholar]
- 9.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaedigk-Nitschko K, Ding M, Levy M A, Schlesinger M J. Site-directed mutations in the Sindbis virus 6K protein reveal sites for acylation and the underacylated protein affect virus release and virion structure. Virology. 1990;175:282–291. doi: 10.1016/0042-6822(90)90210-i. [DOI] [PubMed] [Google Scholar]
- 11.Grosenbach D W, Ulaeto D, Hruby D E. Palmitylation of the vaccinia virus 37-KDa major envelope antigen. Identification of a conserved acceptor motif and biological relevance. J Biol Chem. 1997;272:1956–1964. doi: 10.1074/jbc.272.3.1956. [DOI] [PubMed] [Google Scholar]
- 12.Hruby D E, Franke C A. Viral acylproteins: greasing the wheels of assembly. Trends Microbiol. 1993;1:20–25. doi: 10.1016/0966-842x(93)90020-r. [DOI] [PubMed] [Google Scholar]
- 13.Ivanova L, Schlesinger M J. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J Virol. 1993;67:2546–2551. doi: 10.1128/jvi.67.5.2546-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin H, Subbarao K, Bagal S, Leser G P, Murphy B R, Lamb R A. Palmitylation of the influenza virus hemagglutinin (H3) is not essential for virus assembly or infectivity. J Virol. 1996;70:1406–1414. doi: 10.1128/jvi.70.3.1406-1414.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karnik S S, Ridge K D, Bhattacharya S, Khorana H G. Palmitoylation of bovine opsin and its cysteine mutants in COS cells. Proc Natl Acad Sci USA. 1993;90:40–44. doi: 10.1073/pnas.90.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laakkonen P, Ahola T, Kääriäinen L. The effects of palmitoylation on membrane association of Semliki forest virus RNA capping enzyme. J Biol Chem. 1996;271:28567–28571. doi: 10.1074/jbc.271.45.28567. [DOI] [PubMed] [Google Scholar]
- 17.Lamb R A. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Hughes T E, Sessa W C. The first 35 amino acids and fatty acylation sites determine the molecular targeting of endothelial nitric oxide synthetase into the Golgi region of cells: a green fluorescent protein study. J Cell Biol. 1997;137:1525–1535. doi: 10.1083/jcb.137.7.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malvoisin E, Wild F. Contribution of measles virus fusion protein in protective immunity: anti-F monoclonal antibodies neutralize virus infectivity and protect mice against challenge. J Virol. 1990;64:5160–5162. doi: 10.1128/jvi.64.10.5160-5162.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer G, Diallo A. The nucleotide sequence of the fusion protein gene of the peste des petits ruminants virus: the long untranslated region in the 5′-end of the F protein gene of morbillivirus seems to be specific to each virus. Virus Res. 1995;37:23–38. doi: 10.1016/0168-1702(95)00013-g. [DOI] [PubMed] [Google Scholar]
- 21.Milligan G, Parenti M, Magee A I. The dynamic role of palmitoylation in signal transduction. Trends Biochem Sci. 1995;20:181–186. doi: 10.1016/s0968-0004(00)89004-0. [DOI] [PubMed] [Google Scholar]
- 22.Mumby S M. Reversible palmitoylation of signaling proteins. Curr Opin Cell Biol. 1997;9:148–154. doi: 10.1016/s0955-0674(97)80056-7. [DOI] [PubMed] [Google Scholar]
- 23.Naeve C W, Williams D. Fatty acids on the A/Japan/305/57 influenza virus hemagglutinin have a role in membrane fusion. EMBO J. 1990;9:3857–3866. doi: 10.1002/j.1460-2075.1990.tb07604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naim H Y, Amarneh B, Ktistakis N T, Roth M G. Effects of altering palmitoylation sites on biosynthesis and function of the influenza virus hemagglutinin. J Virol. 1992;65:2491–2500. doi: 10.1128/jvi.66.12.7585-7588.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norrby E. The effect of a carbobenzoxy tripeptide on the biological activities of measles virus. Virology. 1971;44:599–608. doi: 10.1016/0042-6822(71)90374-6. [DOI] [PubMed] [Google Scholar]
- 26.Olson E N, Glaser L, Merlie J P. Alfa and beta subunits of the nicotic acetylcholine receptor contain covalently bound lipid. J Biol Chem. 1984;259:5364–5367. [PubMed] [Google Scholar]
- 27.Philipp H C, Schroth B, Veit M, Krumbiegel M, Herrmann A, Schmidt M F G. Assessment of fusogenic properties of influenza virus hemagglutinin deacylated by site-directed mutagenesis and hydroxylamine treatment. Virology. 1995;210:20–28. doi: 10.1006/viro.1995.1313. [DOI] [PubMed] [Google Scholar]
- 28.Radecke F, Billeter M A. Appendix: measles virus antigenome and protein consensus sequences. Curr Top Microbiol Immunol. 1995;191:181. doi: 10.1007/978-3-642-78621-1_12. [DOI] [PubMed] [Google Scholar]
- 29.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dötsch C, Christiansen G, Billeter M A. Rescue of measles virus from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson C, Hull D, Greer P, Hasel K, Berkovich A, Englund G, Bellini W, Rima B, Lazzarini R. The nucleotide sequence of the mRNA encoding the fusion protein of measles virus (Edmonston strain): a comparison of fusion proteins from several different paramyxoviruses. Virology. 1986;155:508–523. doi: 10.1016/0042-6822(86)90212-6. [DOI] [PubMed] [Google Scholar]
- 31.Richardson C D, Scheid A, Choppin P W. Specific inhibition of paramyxovirus and myxovirus replication by oligopeptides with amino acid sequences similar to those at the N-termini of the F1 or HA2 viral polypeptides. Virology. 1980;105:205–222. doi: 10.1016/0042-6822(80)90168-3. [DOI] [PubMed] [Google Scholar]
- 32.Rima B K, Earle J A P, Yeo R P, Herlihy L, Baczko K, ter Meulen V, Carabaña J, Caballero M, Celma M L, Fernández-Muñoz R. Temporal and geographical distribution of measles virus genotypes. J Gen Virol. 1995;76:1173–1180. doi: 10.1099/0022-1317-76-5-1173. [DOI] [PubMed] [Google Scholar]
- 33.Rose J K, Adams G A, Gallione C J. The presence of cysteine in the cytoplasmic domain of vesicular stomatitis virus glycoprotein is required for palmitate addition. Proc Natl Acad Sci USA. 1984;81:2050–2054. doi: 10.1073/pnas.81.7.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt M F G, Schlesinger M J. Fatty acid binding of vesicular stomatitis virus glycoprotein: a new type of post-translational modification of the viral glycoprotein. Cell. 1979;17:813–819. doi: 10.1016/0092-8674(79)90321-0. [DOI] [PubMed] [Google Scholar]
- 36.Scullion B F, Hou Y, Paddington J K, Rose J K, Jacobson K. Effects of mutations in three domains of vesicular stomatitis glycoprotein on its lateral diffusion in the plasma membrane. J Cell Biol. 1987;105:69–75. doi: 10.1083/jcb.105.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sergel T, Morrison T G. Mutations in the cytoplasmic domain of the fusion glycoprotein of Newcastle disease virus depress syncytia formation. Virology. 1995;210:264–272. doi: 10.1006/viro.1995.1343. [DOI] [PubMed] [Google Scholar]
- 38.Shum L, Turck C W, Derynck R. Cysteine 153 and 154 of transmembrane transforming growth factor alfa are palmitoylated and mediate cytoplasmic protein association. J Biol Chem. 1996;271:28502–28508. doi: 10.1074/jbc.271.45.28502. [DOI] [PubMed] [Google Scholar]
- 39.Towler D A, Gordon J L, Adams S P, Glaser L. The biology and enzymology of eukaryotic protein acylation. Annu Rev Biochem. 1988;57:69–99. doi: 10.1146/annurev.bi.57.070188.000441. [DOI] [PubMed] [Google Scholar]
- 40.Veit M, Schmidt M F, Rott R. Different palmitoylation of paramyxovirus glycoproteins. Virology. 1989;168:173–176. doi: 10.1016/0042-6822(89)90417-0. [DOI] [PubMed] [Google Scholar]
- 41.Whitt M A, Rose J K. Fatty acylation is not required for membrane fusion activity or glycoprotein assembly into VSV virions. Virology. 1991;185:875–878. doi: 10.1016/0042-6822(91)90563-q. [DOI] [PubMed] [Google Scholar]
- 42.Wild T F, Fayolle J, Beauverger P, Buckland R. Measles virus fusion: role of the cysteine-rich region of the fusion glycoprotein. J Virol. 1994;68:7546–7548. doi: 10.1128/jvi.68.11.7546-7548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wild T F, Buckland R. Functional aspects of envelope-associated measles virus proteins. Curr Top Microbiol Immunol. 1995;191:51–64. doi: 10.1007/978-3-642-78621-1_4. [DOI] [PubMed] [Google Scholar]
- 44.Wild T F, Buckland R. Inhibition of measles virus infection and fusion with peptides corresponding to the leucine zipper region of the fusion protein. J Gen Virol. 1997;78:107–111. doi: 10.1099/0022-1317-78-1-107. [DOI] [PubMed] [Google Scholar]
- 45.Yang C, Compans R W. Palmitoylation of the murine leukemia virus envelope glycoprotein transmembrane subunits. Virology. 1996;221:87–97. doi: 10.1006/viro.1996.0355. [DOI] [PubMed] [Google Scholar]
- 46.Zurcher T, Luo G, Palese P. Mutations at palmitoylation sites of the influenza virus hemagglutinin affect virus formation. J Virol. 1994;68:5748–5754. doi: 10.1128/jvi.68.9.5748-5754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]