Abstract
Cinnamaldehyde is a natural compound extracted from cinnamon bark essential oil, acclaimed for its versatile properties in both pharmaceutical and agricultural fields, including antimicrobial, antioxidant, and anticancer activities. Although potential of cinnamaldehyde against plant pathogenic bacteria like Agrobacterium tumefaciens and Pseudomonas syringae pv. actinidiae causative agents of crown gall and bacterial canker diseases, respectively has been documented, indepth studies into cinnamaldehyde’s broader influence on plant pathogenic bacteria are relatively unexplored. Particularly, Pectobacterium spp., gram-negative soil-borne pathogens, notoriously cause soft rot damage across a spectrum of plant families, emphasizing the urgency for effective treatments. Our investigation established that the Minimum Inhibitory Concentrations (MICs) of cinnamaldehyde against strains P. odoriferum JK2, P. carotovorum BP201601, and P. versatile MYP201603 were 250 μg/ml, 125 μg/ml, and 125 μg/ml, respectively. Concurrently, their Minimum Bactericidal Concentrations (MBCs) were found to be 500 μg/ml, 250 μg/ml, and 500 μg/ml, respectively. Using RNA-sequencing analysis, we identified 1,907 differentially expressed genes in P. carotovorum BP201601 treated with 500 μg/ml cinnamaldehyde. Notably, our results indicate that cinnamaldehyde upregulated nitrate reductase pathways while downregulating the citrate cycle, suggesting a potential disruption in the aerobic respiration system of P. carotovorum during cinnamaldehyde exposure. This study serves as a pioneering exploration of the transcriptional response of P. carotovorum to cinnamaldehyde, providing insights into the bactericidal mechanisms employed by cinnamaldehyde against this bacterium.
Keywords: Cinnamaldehyde, Pectobacterium spp., Minimum Inhibitory Concentration (MIC), Minimum Bactericidal Concentration (MBC), nitrate reductase pathways
Introduction
Cinnamaldehyde, a natural compound extracted from cinnamon bark essential oil, is receiving significant attention across various areas, most notably within the pharmaceutical and food industries, due to its multifaceted properties. These include antimicrobial, anti-inflammatory, antioxidant, anticancer, and anti-obesity activities [1-3]. Cinnamaldehyde has demonstrated potent antimicrobial effects against bacterial strains such as Streptococcus mutans [1], which is associated with the human oral cavity, and Enterococcus faecalis, known to colonize the urinary tract, cause endocarditis, and persist in root canal infections [4]. Its anti-inflammatory action, notably the effect of trans-cinnamaldehyde on lipopolysaccharide-stimulated macrophages, is attributed to the suppression of the phosphorylation of mitogen-activated protein kinases (MAPKs) [2]. Furthermore, cinnamaldehyde 's therapeutic potential extends to vascular smooth muscle cells (VSMCs), which are implicated in the development of atherosclerosis. In this context, cinnamaldehyde activates nuclear factor erythroid 2-related factor 2 (Nrf2) antioxidant enzymes and diminishes proliferation and intimal hyperplasia in Zucker Diabetic Fatty (ZDF) rats following balloon injury [5]. In the area of metabolic health, cinnamaldehyde 's preventive effects against diabetes and obesity are gaining attention, but understanding the underlying mechanisms remains an ongoing effort [3]. From a regulatory perspective, cinnamaldehyde has secured approval as a safe natural active compound from authorities such as the Food and Drug Administration (FDA) in the United States and Europe with a recommended daily intake of 1.25 mg/kg [3], cinnamaldehyde has found its place as a flavor additive in everyday chemicals and dietary supplement products.
In the field of agriculture, studies are ongoing regarding the antimicrobial properties of cinnamaldehyde against plant pathogens. It exerts antagonistic effects against fungi like Aspergillus flavus and Phytophthora capsici, making it a potent shield against post-harvest spoilage [6, 7]. Moreover, cinnamaldehyde strengthens the innate defenses of specific crops. For instance, post-harvest citrus fruits treated with cinnamaldehyde exhibit enhanced resistance to pathogens like Pecicillium digitatum and Geotrichum citri-aurantii which cause green mold and sour rot, respectively [8] This resilience is further correlated with increased enzymatic activities, namely phenylalanine ammonia-lyase (PAL), peroxidase (POD), polyphenol oxidase (PPO), β-1, 3-glucanase (GLU) and chitinase (CHT), which strengthening their defensive stance [8]. Recent discoveries have further highlighted cinnamaldehydés inhibitory effects on Botrytis cineria in cherry tomatoes and Colletotrichum fructicola in Chinese sorghum [9, 10]. Additionally, it exhibits distinct action on Fusarium sambucinum in potatoes, where it disrupts ergosterol biosynthesis, essential for cell membrane stability [11]. Expanding its range, cinnamaldehydés antimicrobial effect extends beyond fungi, exhibiting activity against both gram-positive and gram-negative bacteria. Within this spectrum, its actions against plant pathogens such as Agrobacterium tumefaciens, implicated in crown gall disease, and Pseudomonas syringae pv. actinidiae, responsible for bacterial canker disease, are particularly noteworthy [12, 13]. Preliminary studies offer promising insights into cinnamaldehydés antimicrobial potency; however, the exact mechanisms remain elusive [12, 13].
With this foundation, we examine cinnamaldehydés antimicrobial capacities against Pectobacterium spp. This bacterium, being facultative anaerobic, triggers soft rot disease in a wide range of hosts such as cabbage, potato, radish, carrot, tomato, cauliflower, bell pepper, and cucumber, leading to significant crop damage [14-16]. Soft rot disease can be indirectly controlled through methods such as water management and plant nutrition; however, as of today, complete control is difficult to achieve [17]. Since cinnamaldehyde is a natural compound, it would be suitable for use in several crops if it demonstrated antibacterial activity against Pectobacterium spp. By employing RNA-sequencing, we aim to unveil the antimicrobial mechanisms that cinnamaldehyde uses against Pectobacterium. Through this pursuit, we hope to enrich and potentially revolutionize sustainable crop protection paradigms with the use of the natural compound cinnamaldehyde.
Materials and Methods
Bacterial Strains and Growth Conditions
Strains of Pectobacterium odoriferum JK2, P. carotovorum BP201601, and P. versatile MYP201603 were obtained from Highland Agriculture Research Institute, Rural Development Administration. All Pectobacterium spp. were cultivated in Luria-Bertani (LB) medium (Difco Laboratories, USA) and incubated at 30°C for 24 h.
Minimum Inhibitory Concentration and Minimum Bactericidal Concentration Test
We evaluated the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values of cinnamaldehyde against P. carotovorum spp. according to recommended procedure from the Clinical and Laboratory Standards Institute [18]. Cinnamaldehyde (Sigma-Aldrich) was prepared in 2% Ethyl alcohol (EtOH) and tested across a range of concentrations from 31.25 to 1000 μg/ml. We then introduced bacterial solutions (at a density of 1 × 106 to 1 × 108 CFU/ml) and allowed them to incubate at 30°C for a duration of 24 h. To ensure the validity of our experimental design, an EtOH control (consisting of the sterile culture medium and EtOH) was included. The MIC was identified as the minimum concentration at which no noticeable bacterial growth occurred. The MBC was subsequently established as the smallest concentration that prevented any bacterial colony formation upon subsequent cultivation on agar surfaces. All tests were conducted in triplicate during each session, and the findings presented are an aggregate of three independent experiments.
RNA Preparation for RNA Sequencing
P. carotovorum BP201601 strain was treated with 500 μg/ml cinnamaldehyde or 2% EtOH(control) and subsequently incubated at 30°C in a shaking incubator. After an 8 h of treatment, cells were harvested by centrifugation at 8,000 ×g for 10 min, and bacterial pellets were stored in a -80°C deep freezer until RNA extraction. RNA was extracted using the Trizol reagent, the QIAzol Lysis Reagent, and the RNeasy Mini Kit (Qiagen, USA) following the manufacturer’s instructions. RNA samples were prepared with three biological replicates in each group. The total RNA concentration was determined using Quant-IT RiboGreen (Invitrogen, USA). The integrity of the total RNA was assessed using the TapeStation RNA screentape (Agilent Technologies, USA). For each sample, a library was prepared from 1 μg of total RNA using the Illumina TruSeq Stranded mRNA Sample Prep Kit (Illumina Inc., USA). Initially, bacterial rRNA-depleted samples were prepared with the NEBNext rRNA Depletion kit (NEB, USA). Post rRNA depletion, the residual RNA was fragmented using divalent cations at high temperatures.
cDNA Synthesis and Library Construction and Annotation
The fragmented RNA was reverse transcribed into first-strand cDNA using SuperScript II reverse transcriptase (Invitrogen) and random primers. This step was followed by second-strand cDNA synthesis employing DNA Polymerase I, RNase H, and dUTP. The cDNA fragments underwent end repair, the addition of an 'A' base, and adapter ligation. The resultant products were purified and enriched with PCR, yielding the final cDNA library. Library concentrations were quantified using the KAPA Library Quantification kits (KAPA Biosystems, USA) for Illumina Sequencing platforms, as per the qPCR Quantification Protocol Guide (KAPA Biosystems), and validated with the TapeStation D1000 ScreenTape (Agilent Technologies, USA). Indexed libraries were sequenced on an Illumina NovaSeq (Illumina, Inc.) with paired-end (2×100 bp) sequencing by Macrogen Incorporated. The raw reads obtained from the sequencer were preprocessed to discard low-quality and adapter sequences. The refined reads were then aligned to the P. carotovorum subsp. carotovorum PCC21 genome using Bowtie 1.1.2, an ultrafast, memory-efficient short read aligner leveraging the Burrows-Wheeler index and a quality-aware backtracking algorithm. Reference genome sequences and gene annotations were sourced from the NCBI Genome assembly and the NCBI RefSeq database, respectively. Post-alignment, HTSeq v0.10.0 was utilized to assemble the aligned reads into transcripts and quantify their abundance. Both gene-level and transcript-level quantifications were expressed in terms of raw read count, FPKM (Fragments Per Kilobase of transcript per Million mapped reads), and TPM (Transcripts Per Million).
Analysis of Differentially Expression Genes (DEGs) and Functional Annotation
The relative abundances of genes were quantified in Read Count using StringTie. We conducted statistical analyses to identify differentially expressed genes (DEGs), utilizing estimates of abundance for each gene across samples with Read Count values of zero in more than one sample were excluded from analysis. To facilitate log2 transformation, 1 was added to the Read Count value of each filtered gene. The filtered data underwent log2 transformation and were then subjected to Relative Log Expression (RLE) normalization. The statistical significance of differential gene expression was determined using the nbinomWaldTest within the DESeq2 package, examining fold changes under the null hypothesis that there were no differences among the groups. The False Discovery Rate (FDR) was controlled by adjusting the p-values using the Benjamini-Hochberg algorithm. For the set of DEGs, hierarchical clustering analysis was carried out using complete linkage with Euclidean distance as a measure of similarity. Gene enrichment, functional annotation analysis, and pathway analysis for the significant gene list were conducted based on the blastGO platform (http://geneontology.org/) and the KEGG pathway database (https://www.genome.jp/kegg/).
Hierarchical Clustering
Hierarchical clustering analysis was conducted using complete linkage with Euclidean distance as a measure of similarity. This was utilized to showcase the expression patterns of differentially expressed transcripts that met criteria of |fold change| ≥2 and raw p < 0.05.
Volcano Plot
We utilized a volcano plot to visualize the differences between groups, plotting the log2 fold change (X-axis) against the -log10 p-value (Y-axis) obtained from the average comparisons of each group. All data analysis and visualization of differentially expressed genes were conducted using R version 4.2.2 (www.r-project.org).
Expression Analysis Using qRT-PCR
The synthesis of first-strand cDNA for qRT-PCR was carried out using 1 μg of total RNA and SuperScript II reverse transcriptase (Invitrogen), following the protocol provided by the manufacturer. The expression of genes was analyzed using the CFX96 Real-time PCR detection system and SYBR green fluorescent dye (Bio-Rad). The PCR program included an initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 sec, annealing at 57°C for 30 sec, and extension at 72°C for 30 sec. We used the 16S rRNA gene from P. carotovorum BP201601 as an internal control to normalize gene expression. Gene expression levels were calculated using the ΔΔCt method, and to facilitate comparison, we expressed them as relative values against the expression levels on Control or cinnamaldehyde. The specific sequences of the primers used are provided in Table S3.
Data Availability Statement
The whole transcriptome sequence raw data were deposited in the NCBI database (https://www.ncbi.nlm.nih.gov/sra) with accession numbers SAMN38089555 and SAMN38089557 for Control and cinnamaldehyde treated samples, respectively.
Results
Antibacterial activity of Cinnamaldehyde against Pectobacterium spp.
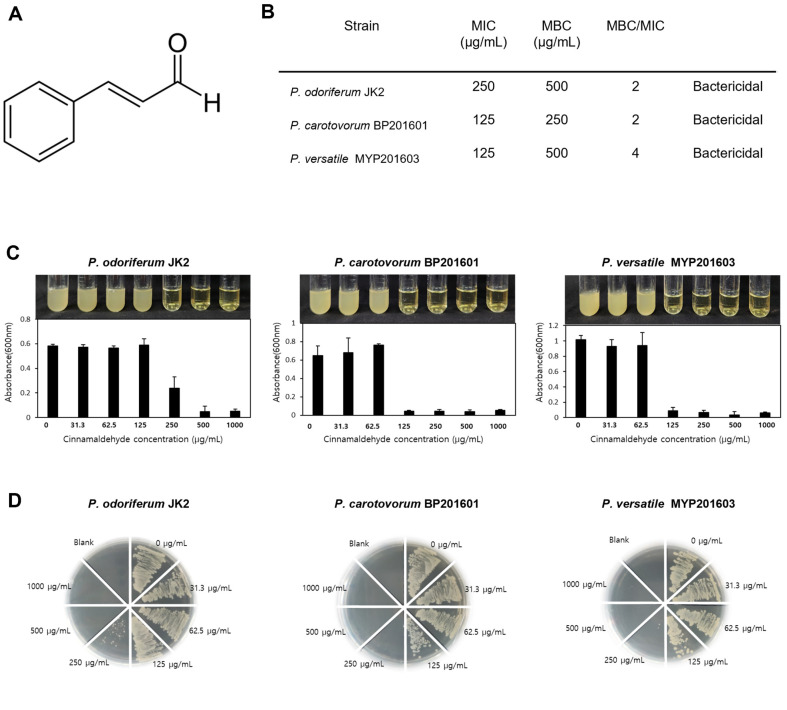
Pectobacterium odoriferum JK2, P. carotovorum BP201601, and P. versatile MYP201603 were isolated from various regions in Korea (Gangneung, Boseong, Miryang) and from different hosts (potato or cabbage). The genome sequences of these three Pectobacterium strains have also been deposited in the NCBI database (P. carotovorum JK2 - CP034938.1, P. carotovorum BP201601 - CP034236.1, P. odoriferum MYP201603 - CP051628.1). We tested the antimicrobial activity of cinnamaldehyde (Fig. 1A) against these three Pectobacterium strains as part of our approach for further transcriptome analysis.
Fig. 1. Bactericidal activity of Cinnamaldehyde against Pectobacterium spp.
(A) Chemical structure of cinnamaldehyde. (B) MIC and MBC value of P. odoriferum JK2, P. carotovorum BP201601, P. versatile MYP201603 against cinnamaldehyde. (C) inhibition of growth of P. odoriferum JK2, P. carotovorum BP201601, P. versatile MYP201603 by cinnamaldehyde at the given concentrations was measured in LB broth after 24 h of incubation at 30°C. Bacterial turbidity was measured at OD = 600 nm, and present as the mean ± SE of three independent experiments. (D) Bactericidal concentrations of cinnamaldehyde for Pectobacterium spp. were determined by inoculating on agar plate from each replicates in broth dilution plate that shows a complete absence of growth and was incubated at 30°C for 24 h.
The MIC values were determined to be 250 μg/ml for P. odoriferum JK2 and 125 μg/ml for both P. carotovorum BP201601 and P. versatile MYP201603 (Fig. 1B and 1C). In contrast, MBC values were found to be 500 μg/ml, 250 μg/ml, and 500 μg/ml for P. odoriferum JK2, P. carotovorum BP201601, and P. versatile MYP201603, respectively (Fig. 1B and 1D). The resulting MBC:MIC ratios suggest a bactericidal effect of cinnamaldehyde on all three Pectobacterium spp. strains. This conclusion is supported by the general consensus that an MBC to MIC ratio ≤ 4 typically indicates bactericidal activity [19].
To assess the bactericidal potency of cinnamaldehyde against densely cultured Pectobacterium spp., bacterial concentrations between 1 × 107 CFU/ml and 1 × 108 CFU/ml were exposed to either 125 μg/ml or 500 μg/ml of cinnamaldehyde. Within 8 h, the growth of P. carotovorum BP201601 was significantly inhibited by a concentration of 500 μg/ml cinnamaldehyde, whereas the 125 μg/ml concentration proved ineffective for cultures at 1 × 108 CFU/ml (Fig. S1A and S1B). Additionally, 24 h after treatment with 500 μg/ml of cinnamaldehyde, bactericidal effects on 1 × 108 CFU/ml cultures were evident in both P. odoriferum JK2 and P. versatile MYP201603 (Fig. S1A). Furthermore, at both 125 μg/ml and 500 μg/ml concentrations of cinnamaldehyde, all three bacterial strains demonstrated susceptibility when cultured at 1 × 107 CFU/ml, showing effects as early as 4 h post-exposure (Fig. S1A and S1B).
RNA Sequencing and Mapping Analysis
To unravel the mechanism underlying cinnamaldehyde’s bactericidal activity, we chose P. carotovorum BP201601, as it showed the lowest MBC value among the three tested Pectobacterium spp. (Fig. 1B). Cinnamaldehyde at 500 μg/ml has been observed to have bactericidal activity at densely cultured concentrations (1 × 108 CFU/ml)(Fig. S1A). Consequently, P. carotovorum BP201601 was exposed to this concentration of cinnamaldehyde, known to exert bactericidal effects, and samples were collected at the 8-h mark for RNA sequencing.
Each sample library yielded between 21,826,458 to 36,637,298 raw reads. After trimming, the read counts ranged from 7,247,982 to 36,287,172, with Q20 > 98.74% and Q30 > 95.75% for each library (Table S1). These trimmed reads were aligned to the reference genome P. carotovorum BP201601.1 (GCF_009931195.1) using the Bowtie aligner. The alignment revealed that 63.34-95.48% of reads matched the reference genome (Table S2). Reads suppressed due to multiple mappings accounted for 0.17-0.53%, and 4.26-36.44% of the reads failed to align (Table S2).
Analysis of Differentially Expressed Genes (DEGs)
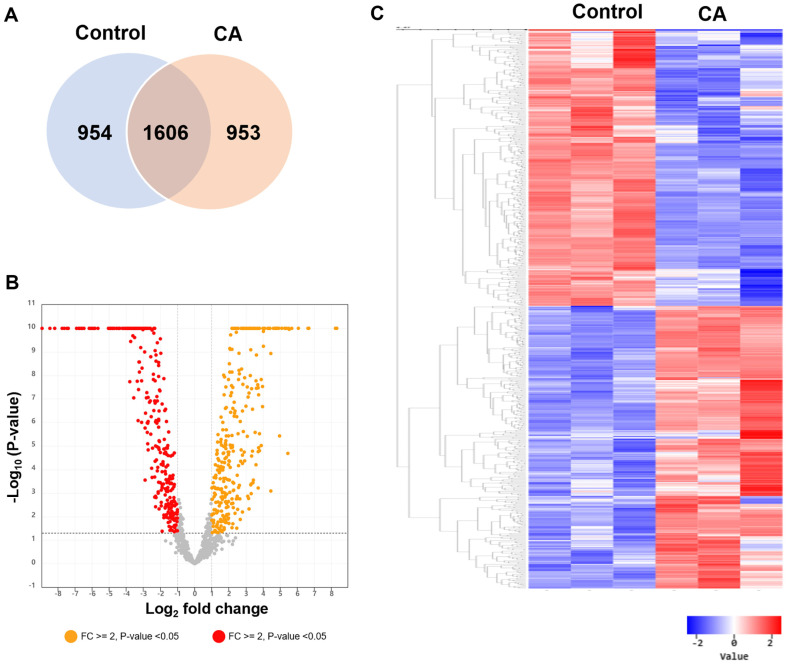
Upon comparing the gene expressions of cinnamaldehyde treated and untreated P. carotovorum BP201601, we identified a total of 1,907 DEGs. Of these, 953 genes were upregulated, while 954 were downregulated (Fig. 2A). The distribution and significance of these DEGs were visualized through a volcano plot and a hierarchical clustering heatmap (Fig. 2B and 2C). Annotation of the 1,907 DEGs using the NCBI-nr database highlighted that genes associated with nitrate reductase were distinctly upregulated in the cinnamaldehyde treated group (Supplementary S1). This suggests potential metabolic shifts in P. carotovorum in response to cinnamaldehyde exposure. A detailed list of all DEGs can be found in Supplementary S1. Overall, the data underscores the pronounced transcriptional adjustments of P. carotovorum BP201601 upon cinnamaldehyde treatment.
Fig. 2. Differentially expressed genes (DEGs) in P. carotovorum BP201601 following cinnamaldehyde treatment.
(A) Venn diagram showing the overlap of DEGs between the cinnamaldehyde (CA)-treated and untreated (Control) groups. (B) Volcano plot displaying DEGs in P. carotovorum BP201601 after cinnamaldehyde treatment. Genes with a log2 fold change of ≥2 or ≤−2 and a p-value of <0.05 are classified as DEGs. Up- and down-regulated DEGs are represented by red and yellow dots, respectively. (C) Heat map illustrating hierarchical clustering based on 1,907 DEGs with a log2 fold change of ≥2 or ≤−2 and a p-value of <0.05. Red and blue colors indicate up- and down-regulated DEGs in the cinnamaldehyde-treated group compared to the untreated group, respectively.
Gene Ontology (GO) Annotation and KEGG Analysis of DEGs
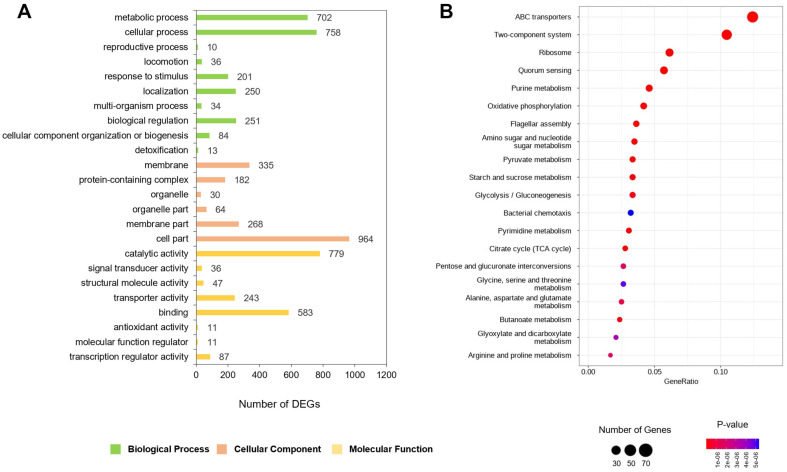
GO labeling represents a globally accepted gene function classification system, comprised of three primary sectors: biological process (BP), cellular component (CC), and molecular function (MF). To explore the functional implications of the most pronounced up- or downregulated genes, we conducted a GO enrichment assessment based on the Fisher’s exact test, using a p value threshold of ≤ 0.05. Summarily, the DEGs were distributed across 47 GO terms: 21 under BP, 14 under CC, and 12 under MF. As depicted in Fig. 3A and Supplementary File 2, the predominant terms within BP include “cellular process (GO:0009987)”, “metabolic process (GO:0008152)”, and “biological regulation (GO:0065007)”, with respective DEG counts of 758, 702, and 251. Within the CC part, “cell part (GO:0044464)” “membrane (GO:0016020)”, “membrane part (GO:0044425)” with DEG counts of 964, 335, and 268, respectively (Fig. 3A and Supplementary S2). Within MF, the terms “catalytic activity (GO:0003824)”, “binding (GO:0005488)”, and “transporter activity (GO:0005215)” stood out, with corresponding DEG counts of 779, 583, and 243 (Fig. 3A and Supplementary S2).
Fig. 3. Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of 1,907 DEGs.
(A) GO annotation analysis of the 1,907 DEGs was carried out using the blastGO platform. The figure presents 24 identified GO terms from the three primary categories: “cellular components,” “molecular function,” and “biological process.” The numbers of up- and down-regulated DEGs in each identified GO term are also shown. (B) KEGG pathway enrichment analysis of the 1,907 DEGs was conducted using KEGG pathway database. The top 20 enriched pathways were determined based on p-value, rich factor, and the count of enriched genes.
To further understand the pathways potentially altered by cinnamaldehydés impact on P. carotovorum BP201601 growth, we mapped DEGs to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, performing a pathway enrichment analysis. This resulted in the identification of thirty significantly enriched pathways (bonferroni value ≤ 0.05) (Supplementary S3). These included pathways such as “Two-component system”, which plays a crucial role in bacterial signal transduction [20]; the “Ribosome”, essential for protein synthesis [21]; the “ABC transporters”, vital for nutrient uptake and drug resistance [22]; and the “Oxidative phosphorylation”, a key component of energy metabolism [23]. In our analysis of the top 20 KEGG pathways (Fig. 3B), we found that fourteen pathways – "purine metabolism," "oxidative phosphorylation," "amino sugar and nucleotide sugar metabolism," "pyruvate metabolism," "starch and sucrose metabolism," "glycolysis/gluconeogenesis," "pyrimidine metabolism," "citrate cycle," "pentose and glucoronate interconversions," "glycine, serine and threonine metabolism," "alanine, aspartate and glutamate metabolism," "butanoate metabolism," "glyoxylate and dicarboxylate metabolism," and "arginine and proline metabolism" are involved in metabolic pathways. These results indicate that cinnamaldehyde induces significant metabolic changes in P. carotovorum BP201601. Detailed representation and specifics of these enriched pathways can be referenced in Supplementary File S3.
Exploring the Impact of Cinnamaldehyde on Metabolic Pathways in P. carotovorum
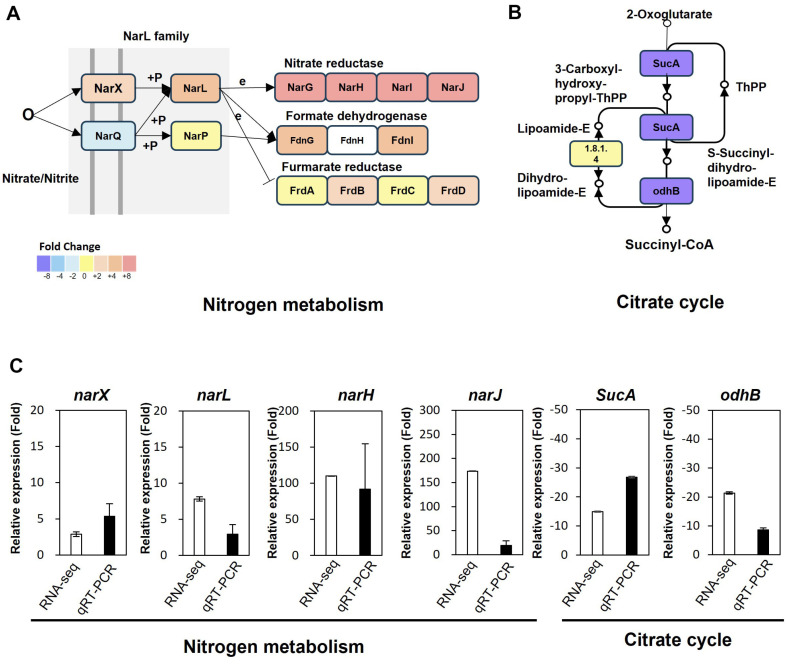
In the top 20 up-regulated DEGs, genes associated with nitrogen-reductase, specifically gene-EH203_10035, gene-EH203_10045 (narJ), gene-EH203_10040 (narH), gene-EH203_09350 (napA), and gene-EH203_09335, exhibited notable increases in expression levels in cinnamaldehyde-treated samples, with respective fold changes of 297.4, 173.45, 109.93, 79.45, and 60.13, when compared to the control (Supplementary S1). Given the recognized upregulation of the narKGHJI gene cluster under oxygen deprivation, our attention gravitated towards metabolic pathways activated in hypoxic conditions [24].
Within the "nitrogen metabolism" pathway, genes such as gene-EH203_10025 (narX), gene-EH203_10020 (narL), gene-EH203_10035 (narG), gene-EH203_10040 (narH), gene-EH203_10050 (narI), and gene-EH203_10045 (narJ) involved in nitrate reductase functions, demonstrated a marked upregulation in cinnamaldehyde-treated samples (Supplementary S1 and Fig. 4A). Additionally, other pathways activated under oxygen-deficient conditions, including formate dehydrogenase N (FDH-N, subunit FdnGHI) with genes gene-EH203_14760 (fdnG), gene-EH203_14770 (fdnI), and fumarate reductase genes gene-EH203_02410 (frdB), gene-EH203_02400 (frdD), were significantly activated post cinnamaldehyde treatment [25].
Fig. 4. Pathways highlighting the differentially expressed genes identified in this study.
(A) Up-regulated genes in the nitrogen metabolism pathway and. (B) Down-regulated genes in the citrate cycle pathway in RNA-seq. The scale represents the average fold change (FC) values between samples treated with cinnamaldehyde and the controls. The gene depicted in this figure are detailed in Supplementary File 4. (C) The gene expression data from RNA-seq and qRT-PCR validation of genes involved in the nitrate metabolism and citrate cycle pathways are shown, with relative values compared against the expression levels in the control. Error bars represent the standard error of the mean from three biological replicates.
The “citrate cycle" is pivotal for cellular energy generation in aerobic conditions. Intriguingly, cinnamaldehyde exposure led to a pronounced downregulation of the genes gene-EH203_15330 (sucA) and gene-EH203_15325 (odhB) (Supplementary S1 and Fig. 4B). Both sucA and odhB have integral roles within the citrate cycle, mediating the conversion of succinyl-CoA to succinate and the decarboxylation of 2-oxoglutarate to succinyl-CoA and CO2, respectively [26]. This evident downregulation may signal a cinnamaldehyde induced disturbance in aerobic cellular respiration. The expression of genes involved in nitrate reductase pathway and citrate cycle were also validated using qRT-PCR (Fig. 4C).
Discussion
P. carotovorum spp., a gram-negative facultative anaerobic bacterium, presents a significant challenge in agriculture due to its pathogenicity. The unearthing of cinnamaldehydés antibacterial efficacy against this bacterium, as underscored by our study, offers a promising alternative for combating bacterial-induced plant diseases. Notably, cinnamaldehydés influence extends beyond simple bactericidal action, especially evident at higher bacterial concentrations.
In environments infested with bacteria, as seen in our experiments with densities reaching 1 × 108 CFU/ml, cinnamaldehyde demonstrated bactericidal action against multiple strains of Pectobacterium spp. (Fig. S1). Our RNA sequencing revealed a pronounced metabolic shift, notably the increase in nitrate reductase genes, highlighting the bacterium's adaptability. While cinnamaldehyde seems to inhibit aerobic respiration by targeting citrate cycle genes such as sucA and odhB, the bacteria appear to adapt to this condition by upregulating anaerobic pathways, specifically nitrate reduction [28, 29]. Indeed, Nitrate metabolism is an adaptation mechanism used by many bacteria for survival in anaerobic environments [28]. Indeed, many other bacteria such as Bacillus subtilis, Actinobacillus pleuropneumoniae, utilize nitrate metabolism under anaerobic conditions showing increased nitrate reductase (Nar) gene expression [27, 28]. Furthermore, under hypoxic conditions where lipids might act as an alternate energy substrate replacing oxygen, the "Lipoic acid metabolism" pathway genes, gene-EH203_15675 (LipB) and gene-EH203_15680 (LipA), also showed enhanced expression in cinnamaldehyde treated samples (Supplementary File 1 and Fig. S2). The heightened emphasis on lipoic acid metabolism pathways seems to underscore a shift towards anaerobic metabolism (Fig. S2) [30], especially in the face of cinnamaldehyde-induced oxygen deprivation condition.
In other mechanisms studies, bactericidal actions of cinnamaldehyde against Listeria monocytogenes suggested inhibition on energy generation, specifically inhibition of glucose uptake or utilization, and effect membrane permeabilities [31]. Similarly, top 20 KEGG pathway analysis in our study, cinnamaldehyde treated P. carotovorum BP201601 showed 89, 75 DEGs on “ABC transporters” and “Two-component system”, respectively. Which are related in glucose uptake and membrane permeability [32, 33].
Quorum sensing is defined as cell-to-cell communications that occurs through the secretion of autoinducers when bacterial populations reach a threshold level. In previous studies, cinnamaldehyde has been shown to have anti-quorum sensing activities in both human and plant pathogenic bacteria, such as Aeromonas hydrophila, Pseudomonas aeruginosa, P. fluorescence, A.tumefaciens [34-37]. Quorum sensing plays a vital role in bacterial virulence, such as biofilm formation. Anti-quorum sensing strategies, therefore, could serve as effective anti-infective approaches. In our studies, we observed significant changes in 41 DEGs related to quorum sensing pathways upon cinnamaldehyde exposure (Supplementary S3).
Cinnamaldehyde is also suggested promising molecules in pharmaceutical area. As their bactericidal effect on pathogenic E.coli strain 042 which cause diarrhea, cinnamaldehyde-treated mice showed lower-levels of colonization by than the untreated groups [38]. In pathway studies, cinnamaldehyde inhibits the synthesis of RNA, DNA and protein leading to bacteria to total collapse [38]. Similarly, In metabolomics studies using LC-MS, cinnamaldehyde changed E.coli metabolism through interactions with proteins, nucleic acids, lipids, and carbohydrates [39]. In our studies, KEGG pathway on DEGs showed “Purine metabolism”, “Pyrimidine metabolism”, it is a crucial component for RNA and DNA, and “Ribosome”, “Alanine, aspartate and glutamate metabolism”, and “Arginine and proline metabolism” which are related in protein synthesis. Similar to previous studies [38, 39], cinnamaldehyde may also effect on RNA, DNA, and protein synthesis in P.carotovorum BP201601, however, the studies of exact mechanisms are needed for further studies.
The antibacterial activity through metabolic change presents a paradigm shift from traditional antibiotics. Unlike many conventional antimicrobial agents that target specific bacterial functions or structures [40, 41], cinnamaldehyde seems to impose a metabolic stress, pushing bacteria towards potentially less efficient or unsustainable metabolic pathways. While the bacteria attempt to adapt, the consequent over-reliance on these pathways might create an unsustainable intracellular environment, leading to their eventual downfall. This mode of action offers a compelling advantage in the contemporary battle against antibiotic resistance. Many bacterial strains, having been exposed to conventional antibiotics, have developed multi-drug resistance (MDR) [42]. For example, streptomycin has been considered to control soft rot disease in agriculture [43], however, streptomycin-resistant Pectobacterium strains have also been identified [44]. Therefore, the use of antibiotics, especially in open fields, is very dangerous because it accelerates the development of bacteria resistance. Cinnamaldehyde, with its unique mode of action that redirects bacterial metabolism rather than directly targeting growth, might be less prone to engender resistance, positioning it as a potential alternative or adjunct to traditional antibiotics [45]. Moreover, if cinnamaldehyde proves to be environmentally stable, its application could extend beyond targeted treatments to broad-spectrum agricultural pest management, possibly with minimal environmental impacts. Its potential impacts on bacterial virulence factor production, and efficacy against a broader spectrum of phytopathogens, if explored further, could establish cinnamaldehyde as a cornerstone in sustainable agricultural strategies. Future endeavors should delve deeper into cinnamaldehydés precise intracellular targets, further elucidating its unique mode of action. In an era where MDR is increasingly challenging the effectiveness of established antimicrobials, cinnamaldehyde emerges not just as an alternative but as an exemplar of how we might rethink our approach to bacterial pathogens.
Supplemental Materials
Supplementary data for this paper are available on-line only at http://jmb.or.kr.
Acknowledgments
This research was supported by grants from the Agenda Project (Agenda Project Nos. PJ01679901) of the Rural Development Administration (RDA), South Korea.
Footnotes
Author Contributions
J.J. performed most experiments, interpreted results, designed the study and wrote the manuscript; D.J. performed MIC test; S.K. revised manuscripts.
Conflict of Interest
The authors have no financial conflicts of interest to declare.
References
- 1.He Z, Huang Z, Jiang W, Zhou W. Antimicrobial activity of cinnamaldehyde on Streptococcus mutans biofilms. Front. Microbiol. 2019;10:2241. doi: 10.3389/fmicb.2019.02241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim ME, Na JY, Lee JS. Anti-inflammatory effects of trans-cinnamaldehyde on lipopolysaccharide-stimulated macrophage activation via MAPKs pathway regulation. Immunopharmacol. Immunotoxicol. 2018;40:219–224. doi: 10.1080/08923973.2018.1424902. [DOI] [PubMed] [Google Scholar]
- 3.Zhu R, Liu H, Liu C, Wang L, Ma R, Chen B, et al. Cinnamaldehyde in diabetes: A review of pharmacology, pharmacokinetics and safety. Pharmacol. Res. 2017;122:78–89. doi: 10.1016/j.phrs.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 4.Akshaya BS, Premraj K, Iswarya C, Muthusamy S, Ibrahim H-IM, Khalil HE, et al. Cinnamaldehyde inhibits Enterococcus faecalis biofilm formation and promotes clearance of its colonization by modulation of phagocytes in vitro. Microb. Pathog. 2023;181:106157. doi: 10.1016/j.micpath.2023.106157. [DOI] [PubMed] [Google Scholar]
- 5.Buglak NE, Jiang W, Bahnson ESM. Cinnamic aldehyde inhibits vascular smooth muscle cell proliferation and neointimal hyperplasia in Zucker Diabetic Fatty rats. Redox Biol. 2018;19:166–178. doi: 10.1016/j.redox.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Wang M, Li M, Zhao T, Zhou L. Cinnamaldehyde inhibits the growth of Phytophthora capsici through disturbing metabolic homoeostasis. PeerJ. 2021;9:e11339. doi: 10.7717/peerj.11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu S, Yang K, Chen L, Liu M, Geng Q, He X, et al. Cinnamaldehyde, a promising natural preservative against Aspergillus flavus. Front. Microbiol. 2019;10:2895. doi: 10.3389/fmicb.2019.02895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan B, Gao Z, Reymick OO, Ouyang Q, Chen Y, Long C, et al. Cinnamaldehyde promotes the defense response in postharvest citrus fruit inoculated with Penicillium digitatum and Geotrichum citri-aurantii. Pestic. Biochem. Physiol. 2021;179:104976. doi: 10.1016/j.pestbp.2021.104976. [DOI] [PubMed] [Google Scholar]
- 9.Zhao W, Hu A, Ren M, Wei G, Xu H. First report on Colletotrichum fructicola causing anthracnose in Chinese sorghum and its management using phytochemicals. J. Fungi. 2023;9:279. doi: 10.3390/jof9020279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo H, Qin X, Wu Y, Yu W, Liu J, Xi Y, et al. Biocontrol of gray mold of cherry tomatoes with the volatile organic monomer from Hanseniaspora uvarum, Trans-Cinnamaldehyde. Food Bioproc. Tech. 2019;12 doi: 10.1007/s11947-019-02319-6. [DOI] [Google Scholar]
- 11.Wei J, Bi Y, Xue H, Wang Y, Zong Y, Prusky D. Antifungal activity of cinnamaldehyde against Fusarium sambucinum involves inhibition of ergosterol biosynthesis. J. Appl. Microbiol. 2020;129:256–265. doi: 10.1111/jam.14601. [DOI] [PubMed] [Google Scholar]
- 12.Ahmed B, Jailani A, Lee JH, Lee J. Inhibition of growth, biofilm formation, virulence, and surface attachment of Agrobacterium tumefaciens by cinnamaldehyde derivatives. Front. Microbiol. 2022;13:1001865. doi: 10.3389/fmicb.2022.1001865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song YR, Choi MS, Choi GW, Park IK, Oh CS. Antibacterial activity of cinnamaldehyde and estragole extracted from plant essential oils against Pseudomonas syringae pv. actinidiae causing bacterial canker disease in kiwifruit. Plant Pathol J. 2016;32:363–370. doi: 10.5423/PPJ.NT.01.2016.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaczek-Moczydłowska MA, Fleming CC, Young GK, Campbell K, O'Hanlon R. Pectobacterium and Dickeya species detected in vegetables in Northern Ireland. Eur. J. Plant Pathol. 2019;154:635–647. doi: 10.1007/s10658-019-01687-1. [DOI] [Google Scholar]
- 15.Kang M, Kim SJ, Lee JY, Yoon S-R, Kim SH, Ha J-H. Inactivation of Pectobacterium carotovorum subsp. carotovorum on Chinese cabbage (Brassica rapa L. subsp. pekinensis) by wash treatments with phenolic compounds. Food Sci. Technol. 2018;93:229–236. doi: 10.1016/j.lwt.2018.03.045. [DOI] [Google Scholar]
- 16.Chandrashekar BS, PrasannaKumar MK, Parivallal PB, Pramesh D, Banakar SN, Patil SS, et al. Host range and virulence diversity of Pectobacterium carotovorum subsp. brasiliense strain RDKLR infecting radish in India, and development of a LAMPbased diagnostics. J. Appl. Microbiol. 2022;132:4400–4412. doi: 10.1111/jam.15553. [DOI] [PubMed] [Google Scholar]
- 17.Youdkes D, Helman Y, Burdman S, Matan O, Jurkevitch E. Potential control of potato soft rot disease by the obligate predators bdellovibrio and like organisms. Appl. Environ. Microbiol. 2020;86:e02543–19. doi: 10.1128/AEM.02543-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wayne P. Clinical and laboratory standards institute: Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document M100-S20 2010
- 19.Sader HS, Fritsche TR, Jones RN. Potency and bactericidal activity of iclaprim against recent clinical gram-positive isolates. Antimicrob Agents Chemother. 2009;53:2171–2175. doi: 10.1128/AAC.00129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanzi F, Vladimirov S, Knudsen CR, Goldman YE, Cooperman BS. Protein synthesis by single ribosomes. RNA. 2003;9:1174–1179. doi: 10.1261/rna.5800303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garmory HS, Titball RW. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect. Immun. 2004;72:6757–6763. doi: 10.1128/IAI.72.12.6757-6763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson DF. Oxidative phosphorylation: regulation and role in cellular and tissue metabolism. J. Physiol. 2017;595:7023–7038. doi: 10.1113/JP273839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood NJ, Alizadeh T, Bennett S, Pearce J, Ferguson SJ, Richardson DJ, et al. Maximal expression of membrane-bound nitrate reductase in Paracoccus is induced by nitrate via a third FNR-like regulator named NarR. J. Bacteriol. 2001;183:3606–3613. doi: 10.1128/JB.183.12.3606-3613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maklashina E, Berthold DA, Cecchini G. Anaerobic expression of Escherichia coli succinate dehydrogenase: functional replacement of fumarate reductase in the respiratory chain during anaerobic growth. J. Bacteriol. 1998;180:5989–5996. doi: 10.1128/JB.180.22.5989-5996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buck D, Spencer ME, Guest JR. Cloning and expression of the succinyl-coA synthetase genes of Escherichia coli K12. Microbiol. 1986;132:1753–1762. doi: 10.1099/00221287-132-6-1753. [DOI] [PubMed] [Google Scholar]
- 27.Marino M, Ramos HC, Hoffmann T, Glaser P, Jahn D. Modulation of anaerobic energy metabolism of Bacillus subtilis by arfM (ywiD) J. Bacteriol. 2001;183:6815–6821. doi: 10.1128/JB.183.23.6815-6821.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Tang H, Yan C, Han W, Peng L, Xu J, et al. The metabolic adaptation in response to nitrate is critical for Actinobacillus pleuropneumoniae growth and pathogenicity under the regulation of NarQ/P. Infect. Immun. 2022;90:e00239–00222. doi: 10.1128/iai.00239-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez CA, Miller BM, Rivera-Chávez F, Velazquez EM, Byndloss MX, Chávez-Arroyo A, et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science. 2016;353:1249–1253. doi: 10.1126/science.aag3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snoep JL, van Bommel M, Lubbers F, Teixeira de Mattos MJ, Neijssel OM. The role of lipoic acid in product formation by Enterococcus faecalis NCTC 775 and reconstitution in vivo and in vitro of the pyruvate dehydrogenase complex. J. Gen. Microbiol. 1993;139 Pt 6:1325–1329. doi: 10.1099/00221287-139-6-1325. [DOI] [PubMed] [Google Scholar]
- 31.Gill AO, Holley RA. Mechanisms of bactericidal action of cinnamaldehyde against Listeria monocytogenes and of eugenol against L. monocytogenes and Lactobacillus sakei. Appl. Environ. Microbiol. 2004;70:5750–5755. doi: 10.1128/AEM.70.10.5750-5755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verdon G, Albers SV, Dijkstra BW, Driessen AJM, Thunnissen A-MWH. Crystal Structures of the ATPase subunit of the glucose ABC transporter from sulfolobus solfataricus: Nucleotide-free and nucleotide-bound conformations. J. Mol. Biol. 2003;330:343–358. doi: 10.1016/S0022-2836(03)00575-8. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Ha U, Zeng L, Jin S. Regulation of membrane permeability by a two-component regulatory system in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2003;47:95–101. doi: 10.1128/AAC.47.1.95-101.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Topa SH, Palombo EA, Kingshott P, Blackall LL. Activity of cinnamaldehyde on quorum sensing and biofilm susceptibility to antibiotics in Pseudomonas aeruginosa. Microorganisms. 2020;8:455. doi: 10.3390/microorganisms8030455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T, Wang D, Liu N, Ma Y, Ding T, Mei Y, et al. Inhibition of quorum sensing-controlled virulence factors and biofilm formation in Pseudomonas fluorescens by cinnamaldehyde. Int. J. Food Microbiol. 2018;269:98–106. doi: 10.1016/j.ijfoodmicro.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Zhou S, Yang Q, Liu Y, Yang Y, Xu N, et al. Cinnamaldehyde decreases the pathogenesis of Aeromonas hydrophila by inhibiting quorum sensing and biofilm formation. Fishes. 2023;8:122. doi: 10.3390/fishes8030122. [DOI] [Google Scholar]
- 37.Ahmed B, Jailani A, Lee JH, Lee J. Inhibition of growth, biofilm formation, virulence, and surface attachment of Agrobacterium tumefaciens by cinnamaldehyde derivatives. Front. Microbiol. 2022;13:1001865. doi: 10.3389/fmicb.2022.1001865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira WA, Pereira CDS, Assunção RG, da Silva ISC, Rego FS, Alves LSR, et al. New Insights into the antimicrobial action of cinnamaldehyde towards Escherichia coli and its effects on intestinal colonization of mice. Biomolecules. 2021;11:302. doi: 10.3390/biom11020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mousavi F, Bojko B, Bessonneau V, Pawliszyn J. Cinnamaldehyde characterization as an antibacterial agent toward E. coli metabolic profile using 96-blade solid-phase microextraction coupled to liquid chromatography-mass spectrometry. J. Proteome Res. 2016;15:963–975. doi: 10.1021/acs.jproteome.5b00992. [DOI] [PubMed] [Google Scholar]
- 40.Kapoor G, Saigal S, Elongavan A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin0 Pharmacol. 2017;33:300–305. doi: 10.4103/joacp.JOACP_349_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Epand RM, Walker C, Epand RF, Magarvey NA. Molecular mechanisms of membrane targeting antibiotics. Biochim. Biophys. Acta - Biomembr. 2016;1858:980–987. doi: 10.1016/j.bbamem.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 42.Nikaido H. Multidrug resistance in bacteria. Annu. Rev. Biochem. 2009;78:119–146. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Czajkowski R, Pérombelon MCM, van Veen JA, van der Wolf JM. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and dickeya species: a review. Plant Pathol. 2011;60:999–1013. doi: 10.1111/j.1365-3059.2011.02470.x. [DOI] [Google Scholar]
- 44.Vu NT, Roh E, Thi TN, Oh CS. Antibiotic resistance of Pectobacterium Korean strains susceptible to the bacteriophage phiPccP-1. Res. Plant Dis. 2022;28:166–171. doi: 10.5423/RPD.2022.28.3.166. [DOI] [Google Scholar]
- 45.Usai F, Di Sotto A. trans-cinnamaldehyde as a novel candidate to overcome bacterial resistance: An overview of in vitro studies. Antibiotics (Basel) 2023;12:254. doi: 10.3390/antibiotics12020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole transcriptome sequence raw data were deposited in the NCBI database (https://www.ncbi.nlm.nih.gov/sra) with accession numbers SAMN38089555 and SAMN38089557 for Control and cinnamaldehyde treated samples, respectively.