Abstract
During late- and post-ripening stages, grape berry undergoes profound biochemical and physiological changes whose molecular control is poorly understood. Here, we report the role of NAC61, a grapevine NAC transcription factor, in regulating different processes involved in berry ripening progression. NAC61 is highly expressed during post-harvest berry dehydration and its expression pattern is closely related to sugar concentration. The ectopic expression of NAC61 in Nicotiana benthamiana leaves resulted in low stomatal conductance, high leaf temperature, tissue collapse and a higher relative water content. Transcriptome analysis of grapevine leaves transiently overexpressing NAC61 and DNA affinity purification and sequencing analyses allowed us to narrow down a list of NAC61-regulated genes. Direct regulation of the stilbene synthase regulator MYB14, the osmotic stress-related gene DHN1b, the Botrytis cinerea susceptibility gene WRKY52, and NAC61 itself was validated. We also demonstrate that NAC61 interacts with NAC60, a proposed master regulator of grapevine organ maturation, in the activation of MYB14 and NAC61 expression. Overall, our findings establish NAC61 as a key player in a regulatory network that governs stilbenoid metabolism and osmotic, oxidative, and biotic stress responses that are the hallmark of late- and post-ripening grape stages.
Keywords: Abiotic stress, biotic stress, Botrytis cinerea, grapevine, late ripening, NAC61, post-harvest dehydration, stilbenoid metabolism
NAC61 regulates stilbene biosynthesis and abiotic/biotic stress responses that hallmark late- and post-ripening developmental stages in grapes. NAC61 self-activates and synergistically interacts with the master ripening regulator NAC60.
Introduction
Fruit ripening is an irreversible, highly regulated process involving physiological and biochemical changes maximizing fruit organoleptic traits to attract herbivores and facilitate seed dispersal (Giovannoni, 2004). The main changes that take place during ripening include fruit degreening, colored pigment accumulation, textural changes (leading to softening), and composition changes, such as the depletion of organic acids and the accumulation of sugars and aroma compounds. This complex program peaks when mature seeds are ready to be dispersed. Nonetheless, at advanced ripening, tissue softening and eventual decay make fruits susceptible to attack by opportunistic pathogens and, consequently, an enhancement of the constitutive defense against pathogens is inherent in the ripening program. Several processes that take place during ripening, such as chloroplast and cell wall disassembly, reactive oxygen species (ROS) increase, protein degradation, and the activation of the secondary metabolism, resemble senescence-like processes (Gómez et al., 2014). However, some specific metabolic activities and the fact that only subsets of senescence-related genes are activated during ripening suggest that, although partially recruiting processes and metabolisms typically associated with senescing tissues, ripening is a distinct process that precedes, and may predispose the fruit to, subsequent senescence (Gapper et al., 2013; Forlani et al., 2019).
To reach its final composition, grape berry, which is a typical non-climacteric fruit, undergoes a developmental process comprising a vegetative and a ripening growth phase (Zenoni et al., 2021). The vegetative phase involves pericarp growth due to rapid cell division and the accumulation of organic acids, tannins, and other phenolic compounds. The ripening phase features several physical, physiological, and compositional changes such as pericarp tissue softening, cell expansion, loss of organic acids, anthocyanin accumulation in the skin, and the progressive accumulation of sugars, reaching levels normally in excess of 20% in the juice (Conde et al., 2007). Several studies have revealed that the onset of ripening, known as veraison, coincides with a profound transcriptomic rearrangement featuring the rapid down-regulation of genes strongly expressed during the vegetative phase of berry development and the up-regulation of genes participating in the ripening program (Fasoli et al., 2012, 2018; Massonnet et al., 2017). Moreover, additional extensive transcriptomic and metabolic changes have been shown to occur in berries of clusters left on the vine beyond ripening, or harvested and placed in dehydrating rooms (Zamboni et al., 2008; Zenoni et al., 2016), thus revealing that the developmental program of the grape berry is not terminated at the fruit commercial ripening stage. These studies identified certain transcription factors (TFs) as putative master regulators of the grape berry developmental progression and the metabolisms featured at each developmental stage (Palumbo et al., 2014; Zenoni et al., 2016; Massonnet et al., 2017; Fasoli et al., 2018). Among these TFs, several members of the NAC (NAM/ATAF1/CUC2) TF gene family are included. NACs are plant-specific TFs with a wide range of activities during plant and fruit development (Olsen et al., 2005; Forlani et al., 2021). The tomato Non-ripening (NOR) was the first NAC TF to be described as a master regulator of fruit ripening (White, 2002; Kumar et al., 2018; Gao et al., 2020), and was recently shown to play a role in leaf senescence as well (Ma et al., 2019). SlNAC1 (also known as SlNAC033) has been shown to have a role in heat and chilling tolerance (Ma et al., 2013; Liang et al., 2015), in defense against bacterial pathogens (Huang et al., 2013), and in fruit softening and pigmentation (Ma et al., 2014; Meng et al., 2016). The Arabidopsis AtNAP (NAC-like, Activated by AP3/PI, ANAC029) has been shown to promote both silique maturation and leaf senescence (Guo and Gan, 2006; Kou et al., 2012). The strawberry FaNAC035 was demonstrated to regulate ripening by controlling fruit softening as well as pigment and sugar accumulation, through the regulation of abscisic acid (ABA) biosynthesis and signalling, and cell-wall degradation and modification (Martín-Pizarro et al., 2021). The NAC TFs are also involved in drought and oxidative stress responses (Tran et al., 2004; Balazadeh et al., 2011; Mao et al., 2012; Puranik et al., 2012) and in the regulation of ROS metabolism (Fang et al., 2015).
In grapevine, the genes NAC33 and NAC60 have been functionally investigated as putative master regulators of the vegetative-to-mature transition in several plant organs (D’Incà et al., 2021, 2023). NAC33 plays a major role in the leaf and fruit, terminating photosynthetic activity and organ growth, whereas for NAC60, which is able to complement the nor mutant phenotype in tomato, a dual role as an orchestrator of both ripening- and senescence-related processes has been proposed. Moreover, it has been shown that the NAC60 homodimer is the prevalent form in berries during ripening, although the ability of NAC60 to form heterodimers with NAC03 and NAC33 has also been demonstrated, suggesting the existence of a NAC60–NAC regulatory network (D’Incà et al., 2023). Interestingly, NAC60 and the as yet uncharacterized NAC61 were also identified as markers of post-harvest dehydration (Zenoni et al., 2016).
Here, we report the functional characterization of the grapevine NAC TF NAC61, providing evidence of its role in the regulation of berry late- and post-ripening processes. We studied the function of NAC61 through investigating the expression and co-expression pattern of NAC61, its ectopic overexpression in Nicotiana benthamiana, its transient overexpression in grapevine plants, and by performing DNA affinity purification and sequencing (DAP-seq). We identified direct targets of NAC61 and demonstrated its ability to activate genes acting in stilbene biosynthesis and osmotic, heat, and oxidative/biotic stress responses. We also investigated the upstream regulation of NAC61 and demonstrated its activation by NAC61 itself and by NAC60, showing that abiotic and biotic factors may influence its expression.
Materials and methods
Plant material
Nicotiana benthamiana plants were grown as previously described (Amato et al., 2019). All the assays and technical measurements were performed on leaves of 5-week-old healthy N. benthamiana plants. Different sets of plants were used for each experiment. Vitis vinifera cv. ‘Thompson Seedless’ plantlets were micropropagated in vitro and cultivated in HB medium (Hoos and Blaich, 1988) in a growth chamber at 25 °C with a 16 h photoperiod. Vitis vinifera cv. ‘Thompson Seedless’ embryogenic calli were grown as previously described (Amato et al., 2019). Vitis vinifera cv. ‘Syrah’ fruiting cuttings were propagated as previously described (Mullins and Rajasekaran, 1981). Vitis vinifera cv. ‘Corvina’ berries (mature and low-/high-temperature dried) were collected for cDNA preparation and transcriptomic analysis (Shmuleviz et al., 2023). Vitis vinifera cv. ‘Müller-Thurgau’ vines were grown in Monzambano (Mantova province, north-east Italy) and grapes were harvested in the 2017 season at full maturity.
Isolation and cloning
The NAC61 (VIT_08s0007g07640) coding sequence (CDS) was isolated from grapevine cv. ‘Syrah’ ripening berry cDNA, and the regulatory regions of NAC61, DHN1b (VIT_04s0023g02480), MYB14 (VIT_07s0005g03340), and WRKY52 (VIT_17s0000g01280) were isolated from cv. ‘Syrah’ genomic DNA. cDNA and genomic DNA were extracted from cv. ‘Syrah’ fruiting cuttings prepared as previously described (D’Incà et al., 2021). Amplification was performed by using the KAPA HiFi DNA polymerase (KAPA Biosystems, Wilmington, MA, USA) and primer sets listed in Supplementary Table S1. The isolated sequences were directionally cloned into the pENTR/D-TOPO Gateway entry vector (Invitrogen, Waltham, MA, USA) and transferred by site-specific LR recombination into a specific binary vector (ThermoFisher Scientific). For agroinfiltration of N. benthamiana and grapevine cv. ‘Thompson Seedless’ plantlets, the NAC61 CDS was transferred into the pK7GW2.0 binary overexpression vector. For the dual-luciferase reporter assay (DLRA), the NAC61, DHN1b, MYB14, and WRKY52 target gene regulatory regions were transferred into the pPGWL7.0 reporter vector to control the expression of the firefly luciferase gene (LUC). For the bimolecular fluorescence complementation (BiFC) assay, the NAC61 sequence was transferred into the pnYGW vector.
The NAC60 CDS isolation, cloning into the pENTR/D-TOPO entry vector, and site-specific LR recombination into the pK7GW2.0 binary overexpression vector was previously performed (D’Incà et al. 2023).
Transient overexpression
The pK7WG2.0 vectors containing 35S:NAC61 or a non-coding sequence (control) were transferred to Agrobacterium tumefaciens strain C58C1 by electroporation. For transient expression in grapevine cv. ‘Thompson Seedless’, 5-week-old in vitro-grown plantlets were vacuum infiltrated as previously described (Amato et al., 2016), and molecular analyses were carried out on leaf samples collected 7 d after agroinfiltration. For transient expression in N. benthamiana, three fully expanded young leaves were syringe infiltrated as previously described (Amato et al., 2016), and phenotypic analysis was carried out over 3 d after agroinfiltration. To validate the expression of NAC61, N. benthamiana leaf tissues were collected 2 d after infiltration and immediately pulverized under liquid nitrogen. RNA was isolated from 100 mg of ground leaf material by using TRI Reagent® (Merck) as recommended by the manufacturer. cDNA synthesis was conducted according to Amato et al. (2016), and gene expression in comparison to the ACTIN internal control was determined by reverse transcription–PCR using the primer sets listed in Supplementary Table S1.
Stomatal conductance and thermal camera measurements
The stomatal conductance measurements were carried out on control and 35S:NAC61-expressing N. benthamiana leaves for 6 d post-infiltration by using a portable leaf porometer (SC-1, METER Group, Inc., Pullman, WA, USA). Three biological replicates (different plants), each with three technical replicates (different leaves), were performed for each sample (nine replicates for each sample). Thermal images were taken of control and 35S:NAC61-expressing N. benthamiana leaves for 6 d post-infiltration with a thermal camera (FLIR E6 Wifi, FLIR Systems, Sweden). Six biological replicates (different plants), each with three technical replicates (different leaves), were performed for each sample (18 replicates for each sample).
Relative water content measurement
Relative water content (RWC) measurement was performed on control and 35S:NAC61-expressing N. benthamiana leaves as previously described (Xu et al., 2022). Three biological replicates (different plants), each with three technical replicates (different leaves), were performed for each sample (nine replicates for each sample). Leaves were sampled 2 d after agroinfiltration, immediately weighed (fresh weight; ‘fw’) and then immersed in distilled water for 2 h at room temperature. The weight of hydrated leaves (‘w’) was measured. The leaves were then dried for 24 h at 60 °C and dry weight (‘dw’) was measured. The RWC was calculated as [(fw–dw)/(w–dw)]×100. The evaluation of local water accumulation was performed by the agroinfiltration of 35S:NAC61 and control vectors into delimited portions of the same leaf. Three biological replicates (different plants), each with three technical replicates (different leaves), were performed, resulting in nine replicates for each sample. Infiltrated leaves were inspected daily and photographs of the abaxial face were taken to observe the progression of the phenotype in agroinfiltrated tissues.
Ion leakage assay
Ion leakage assays were performed on control and 35S:NAC61-expressing N. benthamiana leaf discs according to Imanifard et al. (2018). Six biological replicates (different plants), each with three technical replicates (different leaves), were performed for each sample (18 replicates for each sample). Leaf discs (~5 mm in diameter) were collected 24 h after leaf agroinfiltration (T0) and immersed in 50 ml of non-ionic double-distilled water for 30 min at 25 °C with shaking at 90 rpm to eliminate ions released because of physical damage. The 18 leaf discs from each sample were distributed into three wells of a multi-well plate containing 2 ml of distilled water per well (one replicate per well; the six discs were each from an independent plant to avoid plant-specific effects). The conductivity of the solution was measured using a conductivity meter (Horiba Scientific, Edison, NJ, USA) immediately after plate preparation and during the subsequent 24 h under constant light (50 μmol m–2 s–1) at 25 °C with shaking at 90 rpm.
3,3ʹ-Diaminobenzidine assay
3,3ʹ-Diaminobenzidine (DAB) staining was performed on control and 35S:NAC61-expressing N. benthamiana leaves as previously described (Daudi and O’Brien, 2012). Three biological replicates (different plants), each with three technical replicates (different leaves), were performed for each sample, (nine replicates per sample). Leaf discs (~2 cm in diameter) were collected and used for the assay. A DAB solution was used to evaluate the production of H2O2 2 d after agroinfiltration. The assay was performed in 12-well plates and the DAB staining solution was vacuum infiltrated. The plate was covered with aluminum foil and incubated for 5 h at room temperature with shaking at 100 rpm. At the end of the staining, the discs were placed in falcon tubes containing 25 ml of 80% ethanol to degrade all the chlorophylls. Finally, the staining was quantified by using ImageJ software (https://imagej.net/ij/index.html).
Real-time quantitative polymerase chain reaction
Leaf and berry tissues were harvested and pulverized under liquid nitrogen. For gene expression analysis, RNA was isolated from 100 mg of ground leaf material (for the cv. ‘Thompson Seedless’ transcriptomic analysis) and 200 mg of ground berry material (for the expression analysis on cv. ‘Müller-Thurgau’ drying berries), using the Spectrum Plant Total RNA kit (Merck KGaA, Darmstadt, Germany). Gene expression was determined by real-time quantitative polymerase chain reaction (RT–qPCR) as previously described (Zenoni et al., 2011) using the primer sets listed in Supplementary Table S1. Data are presented as the mean ±SD of three biological replicates. Figures show the normalized expression pattern using UBIQUITIN1 (UBI1; VIT_16s0098g01190) as internal control. UBIQUITIN1 has been previously demonstrated to be a good housekeeping gene, while very similar results were obtained with ELONGATION FACTOR1 (EF1; VIT_12s0035g01130) in all the experimental conditions. High correlation coefficients (R2) were obtained by performing linear regression between the UBI1- and EF1-normalized NAC61 expression data.
Transcriptomic analysis on NAC61 transiently overexpressing grapevine plants
Microarray analysis was performed with the RNA used for RT–qPCR. For transient expression, the three most highly overexpressing plants and three control lines were selected and used as biological replicates. The cDNA synthesis, labelling, hybridization, and washing steps were performed according to the Agilent Microarray-Based Gene Expression Analysis Guide (https://www.agilent.com/cs/library/usermanuals/Public/G4140-90040_GeneExpression_OneColor_6.9.pdf). Each sample was hybridized to an Agilent custom microarray four-pack 44K format (Agilent Sure Print HD 4X44K 60-mer; cat. no. G2514F-048771) (Dal Santo et al., 2016) and scanned using an Agilent Scanner (G2565CA; Agilent Technologies, Santa Clara, CA, USA).
DNA affinity purification and sequencing
Young leaves of cv. ‘Syrah’ were harvested and pulverized using liquid nitrogen. Genomic DNA was extracted from 1 g of powdered material as described by D’Incà et al., (2021) and Illumina libraries were prepared as previously described (Galli et al., 2018). The NAC61 sequence was transferred from the pENTR/D-TOPO vector to the Gateway-compatible destination vector pIX-HALO (Bartlett et al., 2017). The HALO-NAC61 and GST-HALO (used as negative control) fusion proteins were translated in vitro using the TNTR SP6 coupled reticulocyte lysate system (Promega). Two replicates were used for the TF and input libraries (generated with the GST-HALO empty vector). The DAP-seq was performed according to a previously described procedure (Galli et al., 2018), and a total of 7.0 (control replicate 1), 5.6 (control replicate 2), 24 (NAC61 replicate 1), and 22 (NAC61 replicate 2) million reads were obtained. DAP-seq bioinformatic analysis was performed as previously described (Orduña et al., 2022, 2023; D’Incà et al., 2023). Briefly, DAP-seq libraries were aligned to the PN40024 12X.v2 reference genome using bowtie2 (Langmead and Salzberg, 2012), with post-processing to remove reads with a MAPQ score <30. Peak detection was performed using GEM peak caller (Guo et al., 2012) version 3.4 with the 12X.v2 genome assembly using the following parameters: ‘–q 1 –t 1 –k_min 6 –kmax 20 –k seqs 600 –k_neg_dinu_shuffle’, limited to nuclear chromosomes. The replicates were analyzed as multi-replicates with the GEM replicate mode. Detected peaks were associated to the closest gene model from PN40024 v1 on the 12X.0 assembly transposed to the 12X.2 assembly annotation file using the BioConductor package ChIPpeakAnno (L.J. Zhu et al., 2010) with default parameters.
Dual-luciferase reporter assay
The pK7WG2.0 vectors containing the NAC61 and NAC60 (VIT_08s0007g07670) CDS and the pPGWL7.0 vectors harboring the DHN1b, MYB14 and WRKY52 regulative regions were transferred to A. tumefaciens strain C58C1 by electroporation. The DLRA was performed on three fully expanded infiltrated N. benthamiana leaves from three different plants, as previously described (Cavallini et al., 2015). The assay was performed on fresh leaf discs collected 72 h after Agrobacterium-mediated infection and following the manufacturer’s instructions (Promega). A reference vector overexpressing the Renilla reniformis luciferase gene (REN) was used to normalize LUC luminescence. REN and LUC luminescence were detected using a Tecan Infinite ® M200 PLEX instrument. Each test was performed in biological triplicate and each value was measured in triplicate.
Grapevine protoplast transfection and bimolecular fluorescence complementation assay
For the BiFC assay, the NAC61 and NAC60 CDSs were cloned into the pnYGW and pGWcY Gateway vectors, respectively. Vitis vinifera cv. ‘Thompson Seedless’ protoplasts were isolated from embryogenic calli and transfected (Bertini et al., 2019), cultured in multi-well plates in the dark at 25 °C, and analyzed 1 d after transfection. The yellow fluorescent protein (YFP) signal was detected using a Leica TCS SP5 AOBS confocal microscope (argon laser, 514 nm excitation source, 550–570 nm collection bandwidth, auto gain).
Vitis vinifera cv. ‘Müller-Thurgau’ berry post-harvest dehydration and noble rot induction
Approximately 405 kg of berry bunches of V. vinifera cv. ‘Müller-Thurgau’ were harvested in August 2017 in Custoza (Italy) when the total soluble solids content, measured using a DBR35 digital refractometer (Giorgio Bormac, Carpi, Italy), was 18.25 ± 0.05 °Brix. The bunches were arranged in plastic boxes and transferred to a ventilated dehydration facility at the farm ‘La Prendina’ in Monzambano (Mantova, Italy) for dehydration under controlled conditions [14–15 °C, 53–60% relative humidity (RH)] provided by a DEUM 5 HP machine (Sordato, Verona, Italy). Three randomly selected replicates of ~100 berries each were sampled weekly from the start of the trial until 30 d after the induction of noble rot to determine the total soluble solids and the total acidity. In addition, three dedicated boxes were weighed weekly using a CH50K50 electronic balance (Kern, Balingen, Germany). After 29 d of dehydration, half of the boxes were covered with plastic film and water-filled trays were placed inside to increase the RH and induce noble rot (Negri et al., 2017), while the remaining plastic boxes (control berries) were left under normal dehydrating conditions. The two different environmental conditions were imposed for a further 28 d. The RH in both conditions was monitored using Hobo Pro v2 sensors connected to data loggers (Onset Computer Corporation, Bourne, MA, USA). Control and noble-rot-induced berries were sampled in three biological triplicates for transcriptomic analysis at 7, 21, and 28 d (t1, t2, and t3, respectively) from noble rot induction. Each replicate consisted of 50 randomly collected berries that were immediately deseeded and frozen and then pulverized under liquid nitrogen. For measurement of the ratio of glycerol to d-gluconic acid, a glycerol assay kit (Merck) and a d-gluconic acid/d-glucono-δ-lactone assay kit (Megazyme International) were used according to the manufacturers’ instructions. A 1 g sample of powdered berry pericarp material was diluted in 10 ml of buffer containing 500 µl Carrez 1 solution, 500 µl Carrez 2 solution, and 1 ml 0.1 M NaOH topped up to 10 ml with water, and filtered with standard filter paper. After treatment with polyvinylpolypyrrolidone to remove colored solutes, samples were filtered again.
Gene co-expression networks, binding motif comparison, and promoter analyses
NAC61 gene co-expression networks (GCNs) were extracted from the AggGCNs app (Orduña et al., 2023). Gene set enrichment analysis conducted in this study was conducted with the gprofiler2 R package (Kolberg et al., 2020), using the MapMan manually curated annotation described by Orduña et al. (2023) with default settings. A significance threshold of 0.05 was chosen for P-values adjusted with the Benjamini–Hochberg correction procedure (Benjamini and Hochberg, 1995). Enriched sequences found in NAC61 binding sites were compared with those found in Arabidopsis thaliana using the RSAT Plants NGS- ChIP-seq Peak-Discovery software (https://rsat.eead.csic.es/plants/) (Santana-Garcia et al., 2022). The top 600 best-scored peak sequences (–50 bp<peak center<+50 bp) were retrieved from the DAP-seq analysis and used with default parameters. Most significant, frequent, and middle-centered motifs were selected. An untargeted binding discovery analysis and a targeted binding comparison analysis were also performed on the NAC61 promoter using the RSAT Plants Motif discovery oligo-analysis software (https://rsat.eead.csic.es/plants/oligo-analysis_form.cgi), using the default parameters (selecting ‘Vitis vinifera PN40024.v4.55’ as organism), and the RSAT Plants Pattern Matching Matrix-scan (https://rsat.eead.csic.es/plants/matrix-scan_form.cgi), for surveying the ‘Arabidopsis PBM’, ‘Athamap’ and ‘Cistrome’ dataset.
Statistical analysis
We performed pairwise t-tests to compare differences in leaf stomatal conductance, thermal imaging, RWC, ion leakage and DAB measurements. To identify differences in gene expression in RT–qPCR and DLRA data we performed one-sample t-tests. Feature extraction and statistical analysis of the microarray data were conducted by using the Limma package in R (Ritchie et al., 2015). P-values were normalized using the Benjamini–Hochberg correction (Benjamini and Hochberg, 1995) and the differentially expressed genes (DEGs) were identified by adjusted P-value <0.1 and selected by fold change (FC) >|1.5|.
Results
NAC61 is up-regulated in post-veraison stages and correlates with osmotic stress in grape berries
The NAC61 expression pattern, according to the global gene expression atlas of V. vinifera (Fasoli et al., 2012), shows an increase during berry development and also in other organs, such as seeds, rachis, stems and roots, whereas weak expression is found in flower organs (Fig. 1A). In berry tissues of different grape cultivars, the NAC61 expression level sharply increases at veraison (Fig. 1A; Supplementary Fig. S1A, B) (Massonnet et al., 2017; Fasoli et al., 2018) and shows a second step of up-regulation after harvest, reaching the highest expression level at the end of the post-harvest dehydration process [post-harvest withering (PHW) stages; Fig. 1A]. By examining a transcriptomic dataset from a genotype × environment study (Dal Santo et al., 2018), we observed that NAC61 belongs to a stage-specific cluster of genes, whose expression increases after veraison and is thus poorly affected by environmental conditions (Supplementary Fig. S1C). We then investigated the relationship between the NAC61 expression level and the sugar content in berries, based on transcriptomic and technological data retrieved from previous studies. We found a high positive correlation during ripening when the berry exceeds a sugar content of 15–18 °Brix (Fig. 1B) (Fasoli et al., 2018). This close relationship between sugar concentration and NAC61 expression is maintained until the end of ripening and is also observed in berries during PHW, in which the highest expression level coincides with the highest sugar concentration, independent of the genotype considered (Fig. 1C) (Zenoni et al., 2016). Inspection of the transcriptomic dataset from a study aiming specifically at dissecting the effect of time and dehydration level in post-harvest dehydrating grape berries (Zenoni et al., 2020) revealed that NAC61 expression is more strongly correlated with berry weight loss level (an indirect measure of sugar concentration) than with time, further strengthening the above-reported observations (Fig. 1D; Supplementary Fig. S1D).
Fig. 1.
NAC61 expression analysis. (A) NAC61 expression behavior in grapevine organs throughout development (bar plot) and compared in the heatmap (logarithmic value) with that of NAC60 and NAC33. The data were retrieved from the atlas transcriptomic dataset of cv. ‘Corvina’ (Fasoli et al., 2012). Each value represents the mean ±SD of three biological replicates. (B) Correlation between NAC61 expression level and sugar content in grape berries sampled from fruit set to maturity in cv. ‘Cabernet Sauvignon’ and cv. ‘Pinot noir’ (Fasoli et al., 2018). The black line represents the trend of the averaged values of the two varieties. The R2 values shown correspond to the fitting of different polynomial regressions to each corresponding group of samples (orange for cv. ‘Cabernet Sauvignon’ samples, blue for cv. ‘Pinot noir’ samples, and black for the entire set of samples). (C) Correlation between NAC61 expression level and sugar content in grape berries sampled during post-harvest dehydration in six different varieties (Zenoni et al., 2016). (D) Correlation between NAC61 expression level and berry weight loss in cv. ‘Corvina’ berries sampled during traditional long and forced short post-harvest dehydration processes (Zenoni et al., 2020). Expression values were determined by microarray analysis and each value represents the mean ±SD from three biological replicates. (E) NAC61 GCNs based on berry, leaf, and tissue-independent (TI) datasets. Left, Venn diagram showing exclusive and shared genes based on the three datasets; right, three-dimensional plot of co-expressed genes in which NAC, WRKY, and ZIP family members already described as having involvement in berry ripening and/or stress responses are indicated.
Extracting the NAC61 gene-centered networks from berry (67 experiments), leaf (42 experiments), and tissue-independent (131 experiments) datasets through the AggGCN app within the VitViz platform (http://www.vitviz.tomsbiolab.com/), we identified a total of 810 NAC61 co-expressed genes mainly belonging to the ‘Transcription regulation’ and ‘Transcription factors’ functional categories (Supplementary Dataset S1; Supplementary Fig. S2). Most of the NAC61 co-expressed TFs belong to the NAC, zinc finger, WRKY and MYB families (Fig. 1E; Supplementary Dataset S1). Moreover, we found 98 genes previously defined as key regulators of berry ripening and 575 genes (71% of the total) that are differentially modulated during the post-harvest dehydration process (Palumbo et al., 2014; Zenoni et al., 2016; Massonnet et al., 2017; Fasoli et al., 2018) (Supplementary Dataset S1). Accordingly, the stilbene synthases (STSs) regulators MYB15 (VIT_05s0049g01020), WRKY03 (VIT_01s0010g03930), and WRKY43 (VIT_14s0068g01770) are also co-expressed with NAC61.
The NAC family multispecies phylogenetic tree (https://tomsbiolab.com/wp-content/uploads/2021/10/Fig.-S4.png) (Supplementary Fig. S3) revealed that NAC61 is located close to ANAC046 and NAC33 (VIT_19s0027g00230), both of which are involved in the senescence process (Oda-Yamamizo et al., 2016; D’Incà et al., 2021), to ORS1, which codes for an H2O2-responsive NAC TF also controlling senescence in Arabidopsis (Balazadeh et al., 2011), and to OsNAC2, a positive regulator of drought and salt tolerance through ABA-mediated pathways in rice (Jiang et al., 2019).
NAC61 heterologous expression induces a water-soaking-like phenotype and programmed cell death in N. benthamiana leaves
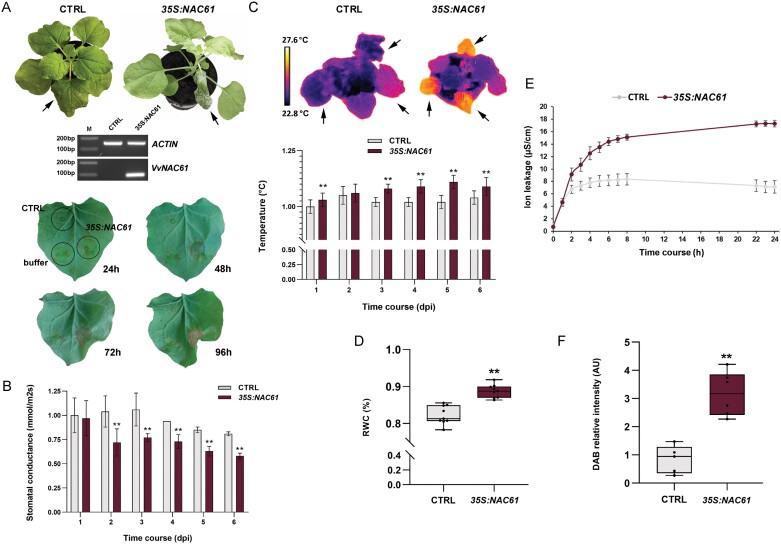
The role of NAC61 was studied through transient heterologous expression in N. benthamiana plants. At 3 d after agroinfiltration, transgenic leaves showed leaf tissue collapse with loss of turgor (Fig. 2A). To highlight the effect of NAC61, leaves were simultaneously agroinfiltrated with NAC61, the control vector, and the agroinfiltration buffer. At 2 d post-infiltration (48 h), part of the leaves near the site of NAC61 infiltration showed a darker color, suggesting a local accumulation of water that preceded tissue collapse, observed at 3 d (72 h) and 4 d (96 h) after infiltration (Fig. 2A).
Fig. 2.
NAC61 ectopic expression in N. benthamiana plants. (A) Control and NAC61-expressing N. benthamiana plants 3 d after infection (top panel), RT–PCR validating the NAC61 ectopic expression in comparison to the control (middle panel), and spot-infiltrated leaves after 24, 48, 72, and 96 h (bottom panel). (B) Stomatal conductance measurements in NAC61-expressing leaves compared with control leaves. (C) Thermal camera visualization (top panel) and leaf temperature measurements (bottom panel) in NAC61-expressing leaves compared with controls. (D) RWC measurements in NAC61-expressing leaves compared with controls at 2 d after agroinfiltration. (E) Ion leakage measurements in NAC61-expressing leaves compared with controls from T0 (24 h after leaf agroinfiltration) to 24 h. (F) DAB staining determining H2O2 accumulation in NAC61-expressing leaves compared with control leaves at 2 d after agroinfiltration. Each value represents the mean ±SD of three biological replicates tested in technical replicate (n=3). Asterisks indicate statistically significant differences (**P<0.01; t-test). Data shown in B, C, and E have been normalized to the control value at the starting time point.
Interestingly, starting from 2 d after NAC61 agroinfiltration, transgenic leaves displayed a lower stomatal conductance compared with the control leaves (Fig. 2B). In addition, a significantly higher leaf temperature was registered in transgenic leaves (Fig. 2C). To investigate whether NAC61 expression favors water retention within plant tissue, we measured the RWC in transgenic and control leaves at 2 d after infiltration. The analysis revealed a significantly higher water content in NAC61-overexpressing leaves compared with controls (Fig. 2D), resembling a water-soaking phenotype.
Moreover, in line with the cell death observed at the whole-leaf level at 4 d post-infection (Fig. 2A), NAC61 expression significantly increased ion leakage as early as 2 d post-infection, suggesting a possible loss of membrane integrity (Fig. 2E). Finally, a higher level of H2O2 (detected by DAB staining) was observed in NAC61-expressing plants in comparison to the controls (Fig. 2F), in line with the above-described phenotype, as H2O2 is a key ROS involved in both stomatal closure and programmed cell death (Liu and Zhang, 2021).
Transient NAC61 overexpression in V. vinifera affects stilbenoid-related gene expression
Grapevine cv. ‘Thompson Seedless’ plants transiently overexpressing NAC61 (Supplementary Fig. S4A, B) showed 1157 DEGs compared with control plants (Supplementary Dataset S2). Among the DEGs, 530 genes were up-regulated and 627 were down-regulated. As previously reported for NAC33 and NAC60 (D’Incà et al., 2021, 2023), no clear phenotypic alterations were observed in NAC61-overexpressing ‘Thompson Seedless’ leaves. Gene category MapMan distribution and DEGs enrichment analysis reveal that up-regulated genes are mainly involved in ‘Secondary metabolism’, in particular ‘Stilbenoid biosynthesis’, and in ‘Oxidoreductases activity’, in particular ‘Laccases’, whereas down-regulated genes are mainly represented by ‘Photosynthesis-related mechanisms’ and other primary metabolism-related processes, such as ‘Lipid and carbohydrate metabolisms’ (Fig. 3A; Supplementary Dataset S3).
Fig. 3.
Transcriptomic responses to NAC61 overexpression in leaves of grapevine cv. ‘Thompson seedless’. (A) Functional enrichment analysis of up-regulated and down-regulated DEGs. (B) Heatmap of up-regulated DEGs involved in phenylpropanoid synthesis, regulation, and modification (Supplementary Dataset S2). Markers of the PHW process (Zenoni et al., 2016) and genes belonging to the STS GRN (Pilati et al., 2021) are highlighted according to the color code in the Venn diagram.
We highlight that NAC61 highly affects the expression of five phenylalanine ammonia lyases (PALs), corresponding to the first and committed step in the phenylpropanoid pathway, and 12 STSs, genes encoding the key enzymes leading to stilbenoid biosynthesis (Fig. 3B; Supplementary Dataset S2). The five PALs and eight of the 12 STSs were previously described as markers of post-harvest dehydration (Zenoni et al., 2016). Up-regulated DEGs also included 33 laccases (LACs), which are proposed to be involved in the oxidative polymerization of phenolic compounds (Keylor et al., 2015), six of which were also described as markers of post-harvest dehydration (Fig. 3B; Supplementary Dataset S2). Overall, 34 out of the 75 molecular markers of post-harvest berry dehydration were found to be up-regulated by NAC61 (Supplementary Dataset S2).
By inspecting the recently proposed STS gene regulatory network (GRN), built by the OneGenE tool (Pilati et al., 2021), we found 117 of the 530 up-regulated genes (Fig. 3B; Supplementary Dataset S2), including the stilbenoid regulators MYB14, WRKY03, and WRKY43. Moreover, 13 out of all of the up-regulated LACs are also found in the STS GRN (Supplementary Dataset S2), indicating their potential role in stilbene polymerization (i.e. the production of viniferins), as previously suggested (Zenoni et al., 2016; Pilati et al., 2021; Orduña et al., 2022). The up-regulation of MYB14 and a laccase gene (LAC25; VIT_18s0001g01280) in the NAC61-overexpressing plants was validated by RT–qPCR (Supplementary Fig. S4C). Of note, the ectopic expression of NAC61 led to the over-expression of NAC61 itself.
Examination of the NAC61 cistrome for identifying NAC61 high-confidence targets
To identify putative direct targets of NAC61, we inspected its genome-wide binding landscape (cistrome) by carrying out DAP-seq. We identified 8558 binding events assigned to 6734 genes (Fig. 4A). The distribution of NAC61 DNA-binding events, with respect to their position from the transcription start site (TSS) of the identified genes, showed a preferential localization in proximal upstream regions and inside genes (Fig. 4A; Supplementary Dataset S4). Within the ‘inside gene’ category, most binding events were found at the very start of the gene feature, that is, close to 100 bp. By inspecting the 600 top-scoring peaks, we identified the major binding motif [CA(C/A)G(C/T)(A/C)A] (Fig. 4B), correlated with A. thaliana ANAC46, which controls cell death during leaf senescence (Huysmans et al., 2018), ANAC55, which is involved in ABA and jasmonic acid responses (Jiang et al., 2009), and ANAC047, the closest homologue of NAC60 (D’Incà et al., 2023) (Supplementary Fig. S5) according to RSAT Plants phylogenetic footprints. To identify the putative targets of NAC61, we focused on genes for which the TF bound to their promoter region (from –3 kb to +100 bp relative to the TSS), and identified 2471 peaks and 2263 unique genes (Supplementary Dataset S4). The use of MapMan ontology of these genes highlighted functional enrichment in the descriptors ‘Transcriptional regulation (zinc fingers, NACs, MYBs, ERFs)’, ‘Solute transport’, ‘Protein homeostasis’, and ‘External stimuli and pathogen response’ (Fig. 4C).
Fig. 4.
NAC61 DAP-seq analyses. (A) Distribution of NAC61 DNA-binding events with respect to their position from the TSS of their assigned genes. The distribution of peak positions is represented in the pie chart. (B) De novo forward binding motif obtained from the inspection of the top 600 scoring peaks of the NAC61 library using the RSAT tool. (C) Functional enrichment analysis of genes to which NAC61 bound (from –3 kb to +100 bp relative to the TSS).
To define NAC61 high-confidence targets (HCTs), we then overlapped the 1157 cv. ‘Thompson Seedless’ DEGs and the 2263 NAC61-bound unique genes (Fig. 5A). A total of 129 HCTs were thus identified (29 of which were in common with at least one of the GCNs; Supplementary Dataset S5). Interestingly, NAC61 itself and other 15 annotated TF genes were identified among the 78 HCTs, up-regulated in NAC61-overexpressing plants, and further assigned to six clusters according to their expression profile in the global gene expression atlas (Fasoli et al., 2012; Fig. 5B). This allowed us to focus on the genes most closely correlated with NAC61 throughout the development of different grapevine organs.
Fig. 5.
Identification and validation of HCTs. (A) Venn diagram showing the number of common genes between the DAP-seq bound genes (peaks in the region from –3 kb to +100 bp relative to the TSS), DEGs (FC ≥1.5 and adjusted P-value <0.1), and GCNs (berry, leaf, and tissue-independent datasets) (Supplementary Dataset S5). The NAC61 HCTs are in the grey-shaded sections. (B) Heatmap representing the atlas expression (Fasoli et al., 2012) of the HCTs up-regulated by the overexpression of NAC61 in cv. ‘Thompson seedless’ leaves. The clusterization of HCTs was performed by using the Expression Atlases App (Corvina) within the VitViz platform (http://www.vitviz.tomsbiolab.com/), using the z-score data transformation and clustering by row. The 29 genes shared by the three datasets are highlighted in bold. Markers of the PHW process (Zenoni et al., 2016) and genes belonging to the STS GRN (Pilati et al., 2021) are indicated with violet and green circles, respectively. (C) NAC61 DNA-binding events shown as density plots and delimited between –3 kb and +100 bp from the TSS of NAC61, WRKY52, DHN1b, and MYB14. The peaks were identified by GEM and were pointed out with their corresponding signal score in the proximal promoter regions. Asterisks indicate the most significant peaks obtained by the DAP-seq analysis. The negative control corresponds to an input library generated with an empty GST-HALO vector. (D) NAC61, WRKY52, DHN1b, and MYB14 promoter activation by NAC61 tested by DLRA in infiltrated N. benthamiana leaves. LUC values are reported relative to the REN value. Each value represents the mean ±SD of three biological replicates tested in technical replicate (n=3). Asterisks indicate statistically significant differences (*P<0.05, **P<0.01; t-test).
Among the HCTs belonging to the NAC61 cluster (Cluster V), we found several candidate genes putatively involved in abiotic and biotic stress responses, such as two zinc fingers (VIT_13s0019g00480; VIT_06s0004g04180), an aldo/keto reductase (AKR; VIT_05s0062g00980), an alternative oxidase (AOX1A; VIT_02s0033g01400), pathogenesis-related protein 4 (PR4;VIT_14s0081g00030), encoding a chitinase, and WRKY52, recently reported as a Botrytis cinerea susceptibility gene (Wang et al., 2018). Albeit less closely correlated with NAC61 expression, other clusters included transcripts functionally associated with stress responses. In Cluster VI, we found a harpin inducing protein 1-like 9 (VIT_08s0007g02360), a lysine histidine transporter 1 (LHT1; VIT_06s0061g01210), and the previously described DHN1b, all highly expressed in berry during post-harvest dehydration and in seed after veraison. Moreover, the aquaporins NIP5 (VIT_02s0025g03260) and DELTA-TIP (VIT_09s0002g04020), the nodulin MtN21 (VIT_01s0026g00520), and a lipoxygenase (LOXA; VIT_06s0004g01510) were identified in Cluster I; a myo-inositol oxygenase (MIOX; VIT_11s0016g02800), a glutathione S-transferase (GST2; VIT_07s0005g00030), a glutaredoxin (VIT_10s0003g00390), a calmodulin (CML101; VIT_01s0010g02930), a salt tolerance zinc finger (VIT_18s0001g09230), and the AVR9/CF-9 rapidly elicited protein (VIT_01s0011g06140) were identified in Cluster II; and an aldehyde dehydrogenase (ALDH2B8; VIT_01s0026g00220), NADH glutamate dehydrogenase (VIT_16s0039g02750), Monopteros (VIT_04s0043g00940), a triacylglycerol lipase (VIT_07s0005g01240), and a kelch repeat-containing F-box family protein (VIT_14s0068g02150) were identified in Cluster IV. The above-described stilbenoid-related genes STS36 (VIT_16s0100g01100), MYB14, WRKY43, and WRKY03 are also found among the NAC61 HCTs. These genes, together with a flavin-containing monooxygenase (VIT_07s0104g01260), MAP kinase substrate 1 (MKS1; VIT_01s0011g03650), peroxidase 12 (Prx12; VIT_18s0072g00160), nitrilase 4 (NIT4; VIT_02s0109g00430), and the MYB164 (VIT_17s0000g03560), were characterized by an increase of expression in berry skin during post-harvest dehydration, in senescing leaf, and in root (Cluster III). The NAC61-binding signal found in the promoters of WRKY52, DHN1b, MYB14, and NAC61 (Fig. 5C; Supplementary Fig. S6) was confirmed by DLRA, showing a significant activation by NAC61 (Fig. 5D).
NAC61 self-activates and synergistically interacts with the grape berry ripening master regulator NAC60
NAC61 was recently identified as a putative target of NAC60 (Fig. 6A) (D’Incà et al., 2023) and shows a delayed expression pattern in comparison to that of NAC60 in developing berries (Fig. 1A). Here, we demonstrated that NAC60 directly controls the activation of NAC61 (i) by inspecting the expression of NAC61 in previously produced transgenic grapevines with altered NAC60 activity and (ii) by DLRA. In comparison to the wild type, a significantly higher NAC61 expression level was observed in leaves stably overexpressing NAC60, whereas a significantly lower NAC61 expression level was observed in leaves expressing the NAC60 dominant repressor to overcome endogenous NAC60 activity (Fig. 6B; Supplementary Fig. S7). A dominant repressor (also known as negative dominant) was created by fusing the NAC60 C-terminal to the plant-specific EAR repressor domain and placing this chimeric repressor under the control of the endogenous NAC60 promoter (D’Incà et al., 2023). Moreover, by performing DLRA, we demonstrated the ability of NAC60 to significantly transactivate (1.98-fold) the NAC61 regulatory region (Fig. 6C).
Fig. 6.
The NAC61–NAC60 regulatory complex regulates NAC61 and MYB14 activation. (A) NAC60 DNA-binding events shown as density plots and delimited between –3 kb and +100 bp from the TSS of NAC61. The NAC60 binding motifs were searched for in three different genomic libraries (a, berry gDNA; b and c are biological replicates of leaf gDNA; D’Incà et al., 2023) (B) NAC61 expression level in grapevine leaves stably overexpressing NAC60 (OX:NAC60) and expressing the NAC60 dominant repressor (NAC60.EAR), determined by RT–qPCR. Each value is relative to the expression of UBIQUITIN1 (VIT_16s0098g01190) and represents the mean ±SD of three biological replicates (D’Incà et al., 2023). Asterisks indicate statistically significant differences (*P<0.05; t-test) in comparison to the control. (C) NAC61 promoter transactivation by NAC60 tested by DLRA in infiltrated N. benthamiana leaves. LUC values are reported relative to the REN value. Each value represents the mean ±SD of three biological replicates tested in technical replicate (n=3). Asterisks indicate statistically significant differences (**P<0.01; t-test). (D) BiFC analysis in grapevine protoplasts showing NAC61–NAC60 protein interaction. Corresponding controls are also shown. Images show a representative case of YFP signal being detected in the cell nucleus by confocal laser scanning. (E) NAC61 and MYB14 promoter activation tested by DLRA in infiltrated N. benthamiana leaves. The activity of NAC61 alone (also reported in Fig. 5D) and combined NAC61–NAC60 activity were tested. LUC values are reported relative to the REN value. Each value represents the mean ±SD of three biological replicates tested in technical replicate (n=3). Asterisks indicate statistically significant differences (**P<0.01; t-test). The data reported in C and E were derived from the same experiment and control values are therefore the same.
Both NAC61 and NAC60 were found to be markers of the berry post-ripening phase (Zenoni et al., 2016), and NAC60 was also among the 92 common genes of the three NAC61 GCN datasets. Using a BiFC assay, we demonstrated the physical interaction between the two NACs and found that a NAC60–NAC61 heterocomplex is localized into the nucleus (Fig. 6D). Accordingly, we also showed that NAC60 significantly increases the ability of NAC61 to activate itself (Figs 5C, 6E). Indeed, we found that NAC61 is induced 18.57-fold by NAC61 alone, whereas the heterodimeric complex formed with NAC60 promotes a synergistic action which resulted in a 293.43-fold induction (Fig. 6E). Moreover, NAC60 was previously reported to activate MYB14 alone (D’Incà et al., 2023). Here, we found that NAC60 significantly increased the ability of NAC61 to activate the MYB14 promoter (4.29-fold) in comparison to the action of NAC61 alone (2-fold) (Fig. 6E). By investigating the NAC61 regulatory region (–3.0 kb to the TSS), we found two binding sites for A. thaliana ANAC047 (Supplementary Fig. S8), coinciding with the already identified NAC60-binding site and perfectly matching the NAC60-binding locations. We also found binding sites for RAP2.6, ABI3VP1/VRN1, and DEAR4, which are involved in ABA, osmotic, drought and salt stress responses (Q. Zhu et al., 2010; J. Wang et al., 2020; S. Wang et al., 2020; Zhang et al., 2020), for RRTF1 and RAP2.3, which mediate plant defense responses against B. cinerea and other pathogens (León et al., 2020; J. Wang et al., 2020), for ORA47, a regulator of general stress responses induced by methyl jasmonate (Zeng et al., 2022), and for DREB2C, AP2EREBP, and G2like_tnt.At3g13040, which are involved in response to drought and dehydration (Riechmann and Meyerowitz, 1998; Kizis et al., 2001; Dietz et al., 2010; Lee et al., 2010; Wang et al., 2022), corroborating the role of NAC61 in abiotic and biotic stress responses (Supplementary Fig. S8).
NAC61 expression is enhanced by high temperature and B. cinerea infection during berry post-harvest dehydration
Taking advantage of the experimental plan from a recent study (Shmuleviz et al., 2023), we analyzed the expression of NAC61 in grape berries subjected to post-harvest dehydration in two different temperature regimens. The results showed that NAC61 expression is significantly induced by high temperature, similar to its HCT MYB14 (Fig. 7A; Supplementary Fig. S9A).
Fig. 7.
Trends in the expression of NAC61 and target genes during post-harvest dehydration conducted in different conditions. (A) NAC61 and MYB14 expression levels during post-harvest dehydration performed under high- and low-temperature conditions (Shmuleviz et al., 2023). Each value is relative to the expression of UBIQUITIN1 (VIT_16s0098g01190) and data presented are the mean ±SD of three biological replicates. Asterisks indicate significant differences (*P<0.05; t-test). (B) Experimental plan for noble rot induction. Berries of cv. ‘Müller Thurgau’ were collected at full maturity and put in a dehydrating room for 29 d to reach 30% weight loss. Then, half of the berries were covered to induce noble rot. The three stages (t0, t1, and t2) of infected and control berries collected for further analyses are shown with representative images. (C) Glycerol to d-gluconic acid ratio assessed as an indicator of noble rot development. Each value corresponds to the mean ±SD of three replicates. Asterisks indicate significant differences (*P<0.05; t-test). (D) NAC61, MYB14, and WRKY52 expression levels in noble-rot-induced berries tested at different phases of B. cinerea infection in cv. ‘Müller-Thurgau’ berries (noble-rot-induced samples) compared with control berries. Each value is relative to the expression of UBIQUITIN1 (VIT_16s0098g01190) and represents the mean ±SD of three biological replicates. Asterisks indicate statistically significant differences (*P<0.05, **P<0.01; t-test) of the noble-rot-induced samples compared with the controls.
Because previous reports suggested that NAC61 is induced during grape infection by B. cinerea, in conditions of noble rot development but not of grey mold (Blanco-Ulate et al., 2015; Kelloniemi et al., 2015), we set up specific experimental conditions to induce noble rot in harvested berries (Negri et al., 2017) and analyzed the expression of NAC61 and some of its putative targets. Bunches of cv. ‘Müller-Thurgau’ were harvested at full maturity and placed in a ventilated dehydrating facility under controlled conditions (Fig. 7B). After 29 d of dehydration, when the soluble solid content reached ~24 °Brix, half of the bunches were covered with plastic film to naturally increase the RH, which is required for noble rot induction (Fig. 7B). In these conditions, dehydration and juice solute concentration were limited (Supplementary Fig. S7B, C), and the first signs of noble rot appeared on berries 7 d after coverage (t1). After a further 14 d, infected berries reached the characteristic pourri plein stage (t2), and 1 week later, the clearly shriveled berries reached the pourri rôtì stage (t3) (Fig. 7B) (Negri et al., 2017). The glycerol to d-gluconic acid ratio was assessed as an indicator of noble rot development (Fig. 7C) (Ribéreau-Gayon et al., 2006). The analysis of transcript levels by RT–qPCR in control and noble-rot-induced berries showed a strong up-regulation of NAC61 and the NAC61 targets MYB14 and WRKY52 during noble rot development (Fig. 7D; Supplementary Fig. S9A).
Discussion
NAC61 controls the stilbenoid biosynthetic pathway as a conserved feature of late and post-ripening
Stilbenoids are a group of polyphenols synthesized by STSs in response to biotic, abiotic, and developmental cues. STSs are also expressed in the absence of external stimuli in a tissue- and cultivar-dependent manner (Versari et al., 2001; Pezet et al., 2003; Jean-Denis et al., 2006; Gatto et al., 2008; Eisenmann et al., 2019). In grapevine, STSs represent a large gene family encompassing 41 isoforms (Vannozzi et al., 2012), most of which are regulated by subgroup 2 R2R3-MYB TFs (i.e. MYB13/MYB14/MYB15; Orduña et al., 2022). Among the 12 transiently activated STSs in NAC61-overexpressing leaves (Fig. 3B), STS36 (Huang et al., 2018) has been identified as a candidate NAC61 HCT. Additionally, our results demonstrate that NAC61 binds to the regulatory regions of MYB14 (which is also directly regulated; Fig. 5), WRKY03 and WRKY43, all of which cooperate in enhancing STSs expression (Vannozzi et al., 2018). These results allow us to place NAC61 in a high hierarchical position for the regulation of stilbene synthesis. Also corroborating this NAC61-dependent transcriptional cascade, 117 out of 530 genes up-regulated by NAC61 overexpression belong to the recently described STSs GRN including more than 1000 structural and regulatory genes potentially involved in stilbenoid metabolism (Fig. 3B; Supplementary Dataset S2) (Pilati et al., 2021). As well as the above-described HCTs (STS36, MYB14, WRKY03, and WRKY43), we identified two other R2R3-MYB members, MYB163 and MYB164, previously associated with the phenylpropanoid pathway (Wong et al., 2016).
In grapevine, LACs have been proposed to control the oxidative polymerization of both monomeric stilbenes and monolignols, producing viniferins and lignin, respectively (Keylor et al., 2015). Vitis vinifera LAC family members were recently assigned to the stilbenoid- and lignin-related subgroups based on sequence similarity and co-expression (Pilati et al., 2021). Interestingly, among the 33 LACs up-regulated in NAC61-overexpressing grapevine plants, 23 belong to the stilbenoid subgroup, whereas none of these belong to the lignin-related subgroup. Nevertheless, the absence of LAC genes among the HCTs indicates that NAC61-mediated LAC regulation may occur indirectly, likely through MYB14 as shown by Orduña et al. (2022).
Among the NAC61 HCTs, we also found AKR, AOX1A, AT-hook protein 1 (AHP1), a flavin-containing monooxygenase, harpin inducing protein 1-like 9, kelch repeat-containing f-box family protein, and LHT1 to be present in the STSs GRN (Supplementary Dataset S5). Although the role of these genes in stilbenoid metabolism, is not clear, and further investigations are needed, their belonging to the NAC61 HCTs strongly supports the master regulatory role of NAC61 in the synthesis and modification of stilbenes. The accumulation of stilbenoids is a hallmark of the late- and post-ripening stages; thus, the genes related to their synthesis can be considered true markers of these developmental transitions. Consistently, 34 out of the 75 PHW molecular markers defined by Zenoni et al. (2016) are up-regulated by NAC61 overexpression, including JAZ4, eight STSs, six LACs, the dirigent protein DIR16, four nitrilases, an osmotin, Prx12, and a pathogenesis-related protein, in addition to the previously mentioned MYB14, MYB164, WRKY03, and WRKY43 (Fig. 3B; Supplementary Dataset S2).
NAC61 is responsive to osmotic stress
During the late- and post-ripening stages, grape berries are subjected to a progressive increase of solute concentration, resulting in a severe osmotic stress. Several pieces of evidence arising from our study strongly indicate the involvement of NAC61 in osmotic stress responses: (i) the close correlation between sugar concentration and NAC61 expression; (ii) the earlier activation of NAC61 in berry flesh, where sugars and other metabolites accumulate (in comparison to the skin); (iii) the higher RWC in N. benthamiana NAC61-expressing leaves in comparison to control leaves; and (iv) the identification of NAC61 HCTs potentially involved in the osmotic stress response through different strategies/mechanisms.
Four zinc finger protein-coding genes were found among the HCTs: two salt tolerance zinc fingers, a zinc finger C2H2 type, and the C2H2-type zinc finger ZAT11, belonging to the branch of the C2H2 family containing the ZAT domain, whose role in response to abiotic stresses in several plant species has been widely described (Liu et al., 2022). Interestingly, cis-regulatory motifs of ZAT TFs were found in dehydrin (DHN) genes recently characterized in Brachypodium grasses (Decena et al., 2021). DHNs are ubiquitously expressed in periods of low intracellular water content (Yang et al., 2012; Liu et al., 2017; Smith and Graether, 2022) and their role in coping with osmotic stress has been demonstrated in several species (Tiwari and Chakrabarty, 2021). In our study, we identified and validated DHN1b as a target of NAC61. DHN1b is up-regulated in withering grape berries (Zamboni et al., 2008) and in leaves and berries subjected to water stress (Xiao and Nassuth, 2006; Savoi et al., 2017). Further investigation to elucidate a putative cooperation of NAC61 and ZAT11 in regulating DHN1b expression in osmotic stress conditions would be worthwhile in future studies.
The NAC61 HCTs include also LHT1, a major candidate for root acquisition of aspartate and a transporter of aspartate, asparagine, and glutamate in rice (Guo et al., 2020). In grapevine, the induction of a lysine histidine transporter and other amino acid transporters in salt-stressed plants has been observed (Aydemir et al., 2020). This finding, together with the increased RWC in the NAC61-overexpressing N. benthamiana leaves, suggests that NAC61 could exert its role in the water/osmotic stress response by regulating amino acid accumulation. Accordingly, an asparagine synthase is the HCT most strongly up-regulated by NAC61 overexpression. This enzyme catalyzes the synthesis of asparagine from aspartate, and asparagine is one of the most represented amino acids in grape berries (Bouloumpasi et al., 2015). Together, these two amino acids are widely described as drought-responsive metabolites (Han et al., 2021). In addition, a glutamate dehydrogenase (GDH), which catalyzes the oxidative deamination of glutamate to generate α-ketoglutarate (Sullivan et al., 2015), thus providing the carbon for de novo synthesis of aspartate, was also identified among the NAC61 HCTs.
Osmotic stress responses could also involve auxin metabolism, which in turn may contribute to drought tolerance through regulation of stomatal closure. Interestingly, an auxin efflux carrier and the ARF TF Monopteros, which inhibits stomatal development (Zhang et al., 2014), were found among the NAC61 HCTs. Similarly, the down-regulation of the HCTs histidine phosphotransfer AHP4, whose knockout narrows stomatal apertures, heightens leaf temperatures during water stress, and increases leaf RWC (Ha et al., 2022), and the phosphatase PP2CA/AHG3, whose repression activates ABA-mediated signaling pathway leading to stomatal closure and water retention (Jung et al., 2020), may contribute to NAC61 function in osmotic stress responses. Interestingly, hypoxia-related genes are found among NAC61-induced DEGs, such as a Hypoxia-responsive gene and three dehydration-responsive proteins (RD22), indicating a decrease in oxygen supply, likely due to water saturation of the apoplast (van den Dries et al., 2013).
The remodeling of lipid composition, to maintain the fluidity and stability of cell membranes, is another change adopted by plants to respond to osmotic stress. In this regard, among the HCTs, we found a triacylglycerol lipase, whose induction in grape berry under water deficit was previously reported (Savoi et al., 2017). During prolonged drought, the membranes could be subjected to degradative processes due not only to lipolytic activity but also peroxidative activity. In this context, we could hypothesize an involvement of two NAC61 HCTs, Prx12 and LOXA, both of which are also known to be involved in pathogen responses. The activity of these two enzymes could be related to the cell death observed in NAC61-overexpressing leaves 3 d post-agroinfiltration.
NAC61 regulates redox state and defense genes during noble rot development
The significant increase of H2O2 (indicated by DAB staining) observed in NAC61-overexpressing N. benthamiana leaves and the NAC61-mediated up-regulation of five Peroxidases (Prxs), including the HCT Prx12, may account for apoplastic ROS production (Survila et al., 2016). This strongly suggests a direct involvement of NAC61 in ROS accumulation. Interestingly, Prxs are a well-known class of pathogen-related (PR) proteins induced in host plant tissues by pathogen infection (Lüthje and Martinez-Cortes, 2018), suggesting a direct involvement of NAC61 in biotic stress responses as well. Accordingly, besides genes related to stilbenoid synthesis and osmotic stress that may account for grapevine defense against pathogens (Yang et al., 2012), we also found several other HCTs related to biotic stress responses, such as PR4, the biotic-stress-responsive calmodulin-like CML101 (Vandelle et al., 2018), the harpin inducing protein 1-like 9 and AKR, also identified as NAC60 targets (D’Incà et al., 2023), an Avr9/Cf-9 rapidly elicited protein, LOXA, and MKS1.
The up-regulation of genes encoding PR proteins has been previously evidenced in healthy berries during PHW as part of a general response to biotic stresses (Zenoni et al., 2016). We could then hypothesize that NAC61 is involved in ROS metabolism/homeostasis, on the one hand, by regulating the expression of genes involved in defense, while also activating mechanisms for ROS detoxification. Indeed, several genes involved in ROS scavenging/detoxification were found to be HCTs, such MIOX, a flavin-containing monooxygenase, AOX1A, an aldehyde dehydrogenase (ALDH288), a glutaredoxin, a GDH, and GST2 (Zhang et al., 2012; Munir et al., 2020; Vanlerberghe et al., 2020; Wang et al., 2023).
ROS are also crucial signals for the induction of the hypersensitive response, a programmed cell death process that facilitates plant infection by necrotrophic pathogens (Soosaar et al., 2005), including B. cinerea (Govrin and Levine, 2000), which is responsible for grey mold in grapes. However, in particular conditions, infection with the fungus leads to the development of noble rot, which promotes biochemical and metabolic changes in grape berries associated with interesting organoleptic features conferred upon sweet white wines (e.g. Amarone and Sauternes wines) thanks to a weaker (or even controlled) infection. The strong induction of NAC61 in botrytized berries, together with the HCT WRKY52, which has been characterized as a grapevine susceptibility gene of B. cinerea (Wang et al., 2018), indicates a possible direct involvement of NAC61 in providing favorable conditions for noble rot development. This hypothesis is further supported by the identification among the HCTs of MKS1, whose overexpression in A. thaliana was shown to increase susceptibility to B. cinerea (Petersen et al., 2010) and to promote the up-regulation of Prx12 specifically during noble rot (Blanco-Ulate et al., 2015; Lovato et al., 2019). Moreover, VqSTS36 was shown to enhance susceptibility to B. cinerea in A. thaliana and tomato (Huang et al., 2018). On the other hand, NAC61 could also contribute to a weaker infection by simultaneously mitigating the favorable conditions for B. cinerea growth through the regulation of PR4, which encodes a chitinase, which could inhibit the growth of fungal hyphae (Grover, 2012), and LOXA, which is involved in the biosynthesis of jasmonate, known to mediate defense against necrotrophic pathogens (Antico et al., 2012). Furthermore, in A. thaliana the overexpression of JAZ8 represses defense responses against B. cinerea through its interaction with AtWRKY75 (Chen et al., 2020). Considering that WRKY52 is one of the closest homologues of AtWRKY75 (Vannozzi et al., 2018), and that two JAZs, namely JAZ2 and JAZ4, are up-regulated by NAC61, a similar mechanism that allows noble rot development could be suggested in grape berries.
Interestingly, Botrytis elliptica infection of Lilium regale down-regulates miR164 (Gao et al., 2017), and the transient overexpression of miR164f in apple leaves enhances their susceptibility to Alternaria alternata AP, possibly due to the down-regulation of a NAC TF (Zhou et al., 2023). Moreover, several NAC members belonging to the same clade as NAC61 are post-transcriptionally regulated by the miR164 family (Kim et al., 2009; Sun et al., 2012; Lira et al., 2017). We could therefore assume a scenario in which B. cinerea may affect miR164 expression in the late berry ripening stage to allow an increase in the NAC61 transcript level. Similarly, low-temperature storage conditions, which ultimately lead to a delay in fruit senescence, repress the strawberry genes FaNAC087 and FaNAC038 due to an increase of their negative regulator miR164 (Xu et al., 2013; Li et al., 2017). Since NAC61 also shows a lower expression in berries experiencing post-harvest dehydration under low-temperature conditions, control by miR164 might be conserved in grape.
NAC-dependent transcriptional network behind berry aging and stress responses
The GCNs obtained from different grapevine organs revealed a strong correlation between NAC61 and many genes previously associated with the late- and post-ripening developmental stages (Supplementary Dataset S1). Consistently, NAC61 HCTs include several genes involved in stilbenoid metabolism as well as osmotic and biotic stress responses, which characterize these late processes (Fig. 8), thus suggesting NAC61 as a key regulator triggering the molecular mechanisms controlling ripening progression.
Fig. 8.
Proposed model of NAC61 mechanism of action. NAC61 high-confidence targets (HCTs) related to stilbenoid metabolism and stress responses that are inherent in late- and post-ripening phases are highlighted. The regulatory mechanisms controlling NAC61 expression are also depicted. Validated mechanisms of transcriptional regulation are shown with black solid lines and hypothetical mechanisms are shown with dotted lines. Asterisks indicate genes putatively also involved in biotic stress responses (*) and abiotic/osmotic stress responses (**). The orange dotted line represents hypothetical physical interactions.
Interestingly, NAC60, which belongs to the NAC61 GCN, not only activates the expression of NAC61 but also forms heterodimers with it, providing a mechanism for the transactivation of their common target MYB14. Moreover, the NAC61 self-activation is greatly increased by a proposed NAC60–NAC61 heterodimer (Fig. 8). These pieces of evidence, together with previously described NAC60–NAC03, NAC60–NAC33, and NAC33–NAC03 interactions, suggest that NAC61 participates in a NAC–NAC regulatory network, whose mechanism of action and additional players have just begun to be elucidated.
In addition, other NACs are co-expressed with NAC61, and thus represent putative partners. These include NAC11, previously described as a berry ‘switch gene’ by Massonnet et al. (2017), NAC17, which is involved in salinity and drought stress responses (Ju et al., 2020), and NAC26, which has been proposed as a determinant of berry size variation (Tello et al., 2015; Muñoz-Espinoza et al., 2020) and a regulator of seed and fruit development through the interaction with MADS9 (Zhang et al., 2021). Therefore, further lines of research should focus on the characterization of NAC61 downstream target genes and interacting proteins to elucidate the molecular mechanisms underlying berry aging and stress responses.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. NAC61 expression pattern during berry development.
Fig. S2. Gene Ontology (GO) enrichment of the NAC61 co-expressed genes.
Fig. S3. NAC61-containing cluster in the NACs phylogenetic tree.
Fig. S4. NAC61 overexpression in cv. ‘Thompson Seedless’ grapevine leaves.
Fig. S5. NAC61 binding motif discovery analysis and motif comparison with published A. thaliana datasets.
Fig. S6. Scheme of the promoter regions amplified for the NAC61, DHN1b, MYB14 and WRKY52 transient activation experiment (Fig. 5D).
Fig. S7. Linear regression between the UBIQUITIN1 and EF1 expression level in the cv. ‘Syrah’ grape samples.
Fig. S8. NAC61 regulative region analysis for A. thaliana ANAC047 and stress-related proteins cis-elements performed with the RSAT software.
Fig. S9. Transcriptomic and technological details on post-harvest withering grape sample.
Table S1. List of used primers.
Dataset S1. NAC61 co-expressed genes. The GCNs were obtained separately by
Dataset S2. Transcriptomic analysis of NAC61-overexpressing and control cv. ‘Thompson Seedless’ leaves.
Dataset S3. Gene category MapMan distribution and enrichment analysis of DEGs.
Dataset S4. NAC61 DAP-seq bound genes.
Dataset S5. List of defined HCTs and detail of genes grouped in Fig. 5A.
Acknowledgements
The authors thank Mario Pezzotti (University of Verona, Italy) for supporting this study and for critical discussions, and Ron Shmuleviz for providing the cv. ‘Corvina’ mature and low/high-temperature dried berries cDNA. The authors would like to thank the Genomics and Transcriptomics platform of Centro Piattaforme Tecnologiche (CPT), University of Verona. All the bioinformatic analyses were performed on the HPC cluster Garnatxa at the Institute for Integrative Systems Biology (I2SysBio).
Contributor Information
Chiara Foresti, Department of Biotechnology, University of Verona, Verona, Italy.
Luis Orduña, Institute for Integrative Systems Biology (I2SysBio), Universitat de València-CSIC, Valencia, Spain.
José Tomás Matus, Institute for Integrative Systems Biology (I2SysBio), Universitat de València-CSIC, Valencia, Spain.
Elodie Vandelle, Department of Biotechnology, University of Verona, Verona, Italy.
Davide Danzi, Department of Biotechnology, University of Verona, Verona, Italy.
Oscar Bellon, Department of Biotechnology, University of Verona, Verona, Italy.
Giovanni Battista Tornielli, Department of Biotechnology, University of Verona, Verona, Italy.
Alessandra Amato, Department of Biotechnology, University of Verona, Verona, Italy.
Sara Zenoni, Department of Biotechnology, University of Verona, Verona, Italy.
John Lunn, MPI of Molecular Plant Physiology, Germany.
Author contributions
CF, GBT, AA, and SZ designed the research; CF, DD, OB, and AA performed the research; CF, LO, JTM, AA, and SZ analyzed data; CF, LO, and JTM contributed with analytic and computational tools and analyzed data; CF, JTM, EV, GBT, AA, and SZ wrote the paper. All authors read and agreed to the published version of the manuscript.
Conflict of interest
The authors have no conflict of interest to declare.
Funding
This work was supported by Grant Ricerca di Base ‘Definition of master regulator genes of fruit ripening in grapevine’, University of Verona, awarded to SZ; by PRIN 2017 ‘Regulation of gene expression in grapevine: analysis of genetic and epigenetic determinants’; by grants PID2021-128865NB-I00 and RYC-2017-23645 awarded to JTM; and PRE2019-088044 fellowship awarded to LO from the Ministerio de Ciencia, Innovación y Universidades (MCIU, Spain), Agencia Estatal de Investigación (AEI, Spain), and Fondo Europeo de Desarrollo Regional (FEDER, European Union). This article is based on work from Innovators Grant IG17111 GRAPEDIA, supported by COST (European Cooperation in Science and Technology).
Data availability
Microarray data for the transient expression experiments on V. vinifera cv. ‘Thompson Seedless’ are available at GEO under accession no. GSE232165. DAP-seq raw data have been submitted to GEO, including metadata of samples and conducted analysis (bioinformatic parameters) according to the FAIR principles, under accession no. GSE230185. DAP-seq results on NAC61 can be visualized in the DAPBrowse tool available at the Vitis Visualization Platform (http://www.vitviz.tomsbiolab.com/). The role for NAC61 has been deposited in the Gene Reference Catalogue found at the Grape Genomics Encyclopedia portal (http://grapedia.org/). All other data supporting the findings of this study are available within the paper and within its supplementary data published online.
References
- Amato A, Cavallini E, Walker AR, et al. 2019. The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. The Plant Journal 99, 1220–1241. [DOI] [PubMed] [Google Scholar]
- Amato A, Cavallini E, Zenoni S, Finezzo L, Begheldo M, Ruperti B, Tornielli GB.. 2016. A grapevine TTG2-like WRKY transcription factor is involved in regulating vacuolar transport and flavonoid biosynthesis. Frontiers in Plant Science 7, 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antico CJ, Colon C, Banks T, Ramonell KM.. 2012. Insights into the role of jasmonic acid-mediated defenses against necrotrophic and biotrophic fungal pathogens. Frontiers in Biology 7, 48–56. [Google Scholar]
- Aydemir BÇ, Özmen CY, Kibar U, Mutaf F, Büyük PB, Bakır M, Ergül A.. 2020. Salt stress induces endoplasmic reticulum stress-responsive genes in a grapevine rootstock. PLoS One 15, e0236424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B.. 2011. ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Molecular Plant 4, 346–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett A, O’Malley RC, Huang SC, Galli M, Nery JR, Gallavotti A, Ecker JR.. 2017. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nature Protocols 12, 1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Methodological) 57, 289–300. [Google Scholar]
- Bertini E, Tornielli GB, Pezzotti M, Zenoni S.. 2019. Regeneration of plants from embryogenic callus-derived protoplasts of Garganega and Sangiovese grapevine (Vitis vinifera L) cultivars. Plant Cell, Tissue and Organ Culture 138, 239–246. [Google Scholar]
- Blanco-Ulate B, Amrine KC, Collins TS, et al. 2015. Developmental and metabolic plasticity of white-skinned grape berries in response to Botrytis cinerea during noble rot. Plant Physiology 169, 2422–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouloumpasi E, Soufleros EH, Tsarchopoulos C, Biliaderis CG.. 2015. Primary amino acid composition and its use in discrimination of Greek red wines with regard to variety and cultivation region. VITIS: Journal of Grapevine Research 41, 195–202. [Google Scholar]
- Cavallini E, Matus JT, Finezzo L, Zenoni S, Loyola R, Guzzo F, Schlechter R, Ageorges A, Arce-Johnson P, Tornielli GB.. 2015. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiology 167, 1448–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang L, Xiang S, Chen Y, Zhang H, Yu D.. 2020. The transcription factor WRKY75 positively regulates jasmonate-mediated plant defense to necrotrophic fungal pathogens. Journal of Experimental Botany 72, 1473–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C, Silva P, Fontes N, Dias ACP, Tavares RM, Sousa MJ, Agasse A, Delrot S, Gerós H.. 2007. Biochemical changes throughout grape berry development and fruit and wine quality. Food 1, 1–22. [Google Scholar]
- Dal Santo S, Fasoli M, Negri S, D’Incà E, Vicenzi N, Guzzo F, Tornielli GB, Pezzotti M, Zenoni S.. 2016. Plasticity of the berry ripening program in a white grape variety. Frontiers in Plant Science 7, 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo S, Zenoni S, Sandri M, et al. 2018. Grapevine field experiments reveal the contribution of genotype, the influence of environment and the effect of their interaction (G×E) on the berry transcriptome. Plant Journal 93, 1143–1159. [DOI] [PubMed] [Google Scholar]
- Daudi A, O’Brien JA.. 2012. Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio-Protocol 2, e263. [PMC free article] [PubMed] [Google Scholar]
- Decena MA, Gálvez-Rojas S, Agostini F, Sancho R, Contreras-Moreira B, Des Marais DL, Hernandez P, Catalán P.. 2021. Comparative genomics, evolution, and drought-induced expression of Dehydrin genes in model Brachypodium grasses. Plants 10, 2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Vogel MO, Viehhauser A.. 2010. AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245, 3–14. [DOI] [PubMed] [Google Scholar]
- D’Incà E, Cazzaniga S, Foresti C, Vitulo N, Bertini E, Galli M, Gallavotti A, Pezzotti M, Battista Tornielli G, Zenoni S.. 2021. VviNAC33 promotes organ de-greening and represses vegetative growth during the vegetative-to-mature phase transition in grapevine. New Phytologist 231, 726–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Incà E, Foresti C, Orduña L, et al. 2023. The transcription factor VviNAC60 regulates senescence- and ripening-related processes in grapevine. Plant Physiology 192, 1928–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann B, Czemmel S, Ziegler T, Buchholz G, Kortekamp A, Trapp O, Rausch T, Dry I, Bogs J.. 2019. Rpv3-1 mediated resistance to grapevine downy mildew is associated with specific host transcriptional responses and the accumulation of stilbenes. BMC Plant Biology 19, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Liao K, Du H, Xu Y, Song H, Li X, Xiong L.. 2015. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. Journal of Experimental Botany 66, 6803–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasoli M, Dal Santo S, Zenoni S, et al. 2012. The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24, 3489–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasoli M, Richter CL, Zenoni S, Bertini E, Vitulo N, Dal Santo S, Dokoozlian N, Pezzotti M, Tornielli GB.. 2018. Timing and order of the molecular events marking the onset of berry ripening in grapevine. Plant Physiology 178, 1187–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlani S, Masiero S, Mizzotti C.. 2019. Fruit ripening: the role of hormones, cell wall modifications, and their relationship with pathogens. Journal of Experimental Botany 70, 2993–3006. [DOI] [PubMed] [Google Scholar]
- Forlani S, Mizzotti C, Masiero S.. 2021. The NAC side of the fruit: tuning of fruit development and maturation. BMC Plant Biology 21, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M, Khakhar A, Lu Z, Chen Z, Sen S, Joshi T, Nemhauser JL, Schmitz RJ, Gallavotti A.. 2018. The DNA binding landscape of the maize AUXIN RESPONSE FACTOR family. Nature Communications 9, 4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Cui Q, Cao Q-Z, Liu Q, He H-B, Zhang D-M, Jia G-X.. 2017. Transcriptome-wide analysis of Botrytis elliptica responsive microRNAs and their targets in Lilium regale Wilson by high-throughput sequencing and degradome analysis. Frontiers in Plant Science 8, 753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wei W, Fan Z, et al. 2020. Re-evaluation of the nor mutation and the role of the NAC-NOR transcription factor in tomato fruit ripening. Journal of Experimental Botany 71, 3560–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper NE, Mc Quinn RP, Giovannoni JJ.. 2013. Molecular and genetic regulation of fruit ripening. Plant Molecular Biology 82, 575–591. [DOI] [PubMed] [Google Scholar]
- Gatto P, Vrhovsek U, Muth J, et al. 2008. Ripening and genotype control stilbene accumulation in healthy grapes. Journal of Agricultural and Food Chemistry 56, 11773–11785. [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. 2004. Genetic regulation of fruit development and ripening. Plant Cell 16, S170–S180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez MD, Vera-Sirera F, Pérez-Amador MA.. 2014. Molecular programme of senescence in dry and fleshy fruits. Journal of Experimental Botany 65, 4515–4526. [DOI] [PubMed] [Google Scholar]
- Govrin EM, Levine A.. 2000. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Current Biology 10, 751–757. [DOI] [PubMed] [Google Scholar]
- Grover A. 2012. Plant chitinases: genetic diversity and physiological roles. Critical Reviews in Plant Sciences 31, 57–73. [Google Scholar]
- Guo N, Hu J, Yan M, Qu H, Luo L, Tegeder M, Xu G.. 2020. Oryza sativa Lysine-Histidine-type Transporter 1 functions in root uptake and root-to-shoot allocation of amino acids in rice. The Plant Journal 103, 395–411. [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S.. 2006. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal 46, 601–612. [DOI] [PubMed] [Google Scholar]
- Guo Y, Mahony S, Gifford DK.. 2012. High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Computational Biology 8, e1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CV, Mostofa MG, Nguyen KH, et al. 2022. The histidine phosphotransfer AHP4 plays a negative role in Arabidopsis plant response to drought. The Plant Journal 111, 1732–1752. [DOI] [PubMed] [Google Scholar]
- Han M, Zhang C, Suglo P, Sun S, Wang M, Su T.. 2021. l-Aspartate: an essential metabolite for plant growth and stress acclimation. Molecules 26, 1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoos G, Blaich R.. 1988. Metabolism of stilbene phytoalexins in grapevines: oxidation of resveratrol in single-cell cultures. Vitis 27, 1–12 [Google Scholar]
- Huang L, Yin X, Sun X, Yang J, Rahman MZ, Chen Z, Wang X.. 2018. Expression of a grape VqSTS36-increased resistance to powdery mildew and osmotic stress in Arabidopsis but enhanced susceptibility to Botrytis cinerea in Arabidopsis and tomato. International Journal of Molecular Sciences 19, 2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Miao M, Kud J, Niu X, Ouyang B, Zhang J, Ye Z, Kuhl JC, Liu Y, Xiao F.. 2013. SlNAC1, a stress-related transcription factor, is fine-tuned on both the transcriptional and the post-translational level. New Phytologist 197, 1214–1224. [DOI] [PubMed] [Google Scholar]
- Huysmans M, Buono RA, Skorzinski N, Radio MC, De Winter F, Parizot B, Mertens J, Karimi M, Fendrych M, Nowack MK.. 2018. NAC transcription factors ANAC087 and ANAC046 control distinct aspects of programmed cell death in the Arabidopsis columella and lateral root cap. The Plant Cell 30, 2197–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanifard Z, Vandelle E, Bellin D.. 2018. Measurement of hypersensitive cell death triggered by avirulent bacterial pathogens in Arabidopsis. Methods in Molecular Biology 1743, 39–50. [DOI] [PubMed] [Google Scholar]
- Jean-Denis JB, Pezet R, Tabacchi R.. 2006. Rapid analysis of stilbenes and derivatives from downy mildew-infected grapevine leaves by liquid chromatography–atmospheric pressure photoionisation mass spectrometry. Journal of Chromatography A 1112, 263–268. [DOI] [PubMed] [Google Scholar]
- Jiang D, Zhou L, Chen W, Ye N, Xia J, Zhuang C.. 2019. Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in rice via ABA-mediated pathways. Rice 12, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Li H, Bu Q, Li C.. 2009. The RHA2a-interacting proteins ANAC019 and ANAC055 may play a dual role in regulating ABA response and jasmonate response. Plant Signaling & Behavior 4, 464–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YL, Yue XF, Min Z, Wang XH, Fang YL, Zhang JX.. 2020. VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiology and Biochemistry 146, 98–111. [DOI] [PubMed] [Google Scholar]
- Jung C, Nguyen NH, Cheong JJ.. 2020. Transcriptional regulation of protein phosphatase 2C genes to modulate abscisic acid signaling. International Journal of Molecular Sciences 21, 9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelloniemi J, Trouvelot S, Héloir MC, et al. 2015. Analysis of the molecular dialogue between gray mold (Botrytis cinerea) and grapevine (Vitis vinifera) reveals a clear shift in defense mechanisms during berry ripening. Molecular Plant-Microbe Interactions 28, 1167–1180. [DOI] [PubMed] [Google Scholar]
- Keylor MH, Matsuura BS, Stephenson CR.. 2015. Chemistry and biology of resveratrol-derived natural products. Chemical Reviews 115, 8976–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG.. 2009. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057. [DOI] [PubMed] [Google Scholar]
- Kizis D, Lumbreras V, Pagès M.. 2001. Role of AP2/EREBP transcription factors in gene regulation during abiotic stress. FEBS Letters 498, 187–189. [DOI] [PubMed] [Google Scholar]
- Kolberg L, Raudvere U, Kuzmin I, Vilo J, Peterson H.. 2020. gprofiler2 -- an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler [version 2; peer review: 2 approved]. F1000Research 9, 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou X, Watkins CB, Gan S-S.. 2012. Arabidopsis AtNAP regulates fruit senescence. Journal of Experimental Botany 63, 6139–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Tamboli V, Sharma R, Sreelakshmi Y.. 2018. NAC-NOR mutations in tomato Penjar accessions attenuate multiple metabolic processes and prolong the fruit shelf life. Food Chemistry 259, 234–244. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-j, Kang J-y, Park H-J, Kim MD, Bae MS, Choi H-i, Kim SY.. 2010. DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiology 153, 716–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León J, Costa-Broseta A, Castillo MC.. 2020. RAP23 negatively regulates nitric oxide biosynthesis and related responses through a rheostat-like mechanism in Arabidopsis. Journal of Experimental Botany 71, 3157–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lai T, Song H, Xu X.. 2017. MiR164 is involved in delaying senescence of strawberry (Fragaria ananassa) fruit by negatively regulating NAC transcription factor genes under low temperature. Russian Journal of Plant Physiology 64, 251–259. [Google Scholar]
- Liang XQ, Ma NN, Wang GD, Meng X, Ai XZ, Meng QW.. 2015. Suppression of SlNAC1 reduces heat resistance in tomato plants. Biologia Plantarum 59, 92–98. [Google Scholar]
- Lira BS, Gramegna G, Trench BA, et al. 2017. Manipulation of a senescence-associated gene improves fleshy fruit yield. Plant Physiology 175, 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Khan AR, Gan Y.. 2022. C2H2 zinc finger proteins response to abiotic stress in plants. International Journal of Molecular Sciences 23, 2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang L, Zhang T, Yang X, Li D.. 2017. Functional characterization of KS-type dehydrin ZmDHN13 and its related conserved domains under oxidative stress. Scientific Reports 7, 7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang H.. 2021. Reactive oxygen species and nitric oxide as mediators in plant hypersensitive response and stomatal closure. Plant Signaling & Behavior 16, 1985860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovato A, Zenoni S, Tornielli GB, Colombo T, Vandelle E, Polverari A.. 2019. Specific molecular interactions between Vitis vinifera and Botrytis cinerea are required for noble rot development in grape berries. Postharvest Biology and Technology 156, 110924. [Google Scholar]
- Lüthje S, Martinez-Cortes T.. 2018. Membrane-bound class III peroxidases: unexpected enzymes with exciting functions. International Journal of Molecular Sciences 19, 2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Feng H, Meng X, Li D, Yang D, Wu C, Meng Q.. 2014. Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biology 14, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma NN, Zuo YQ, Liang XQ, Yin B, Wang GD, Meng QW.. 2013. The multiple stress-responsive transcription factor SlNAC1 improves the chilling tolerance of tomato. Physiologia Plantarum 149, 474–486. [DOI] [PubMed] [Google Scholar]
- Ma X, Balazadeh S, Mueller-Roeber B.. 2019. Tomato fruit ripening factor NOR controls leaf senescence. Journal of Experimental Botany 70, 2727–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R.. 2012. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. Journal of Experimental Botany 63, 2933–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Pizarro C, Vallarino JG, Osorio S, et al. 2021. The NAC transcription factor FaRIF controls fruit ripening in strawberry. The Plant Cell 33, 1574–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet M, Fasoli M, Tornielli GB, Altieri M, Sandri M, Zuccolotto P, Paci P, Gardiman M, Zenoni S, Pezzotti M.. 2017. Ripening transcriptomic program in red and white grapevine varieties correlates with berry skin anthocyanin accumulation. Plant Physiology 174, 2376–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C, Yang D, Ma X, Zhao W, Liang X, Ma N, Meng Q.. 2016. Suppression of tomato SlNAC1 transcription factor delays fruit ripening. Journal of Plant Physiology 193, 88–96. [DOI] [PubMed] [Google Scholar]
- Mullins MG, Rajasekaran K.. 1981. Fruiting cuttings: revised method for producing test plants of grapevine cultivars. American Journal of Enology and Viticulture 32, 35–40. [Google Scholar]
- Munir S, Mumtaz MA, Ahiakpa JK, Liu G, Chen W, Zhou G, Zheng W, Ye Z, Zhang Y.. 2020. Genome-wide analysis of Myo-inositol oxygenase gene family in tomato reveals their involvement in ascorbic acid accumulation. BMC Genomics 21, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Espinoza C, Di Genova A, Sánchez A, Correa J, Espinoza A, Meneses C, Maass A, Orellana A, Hinrichsen P.. 2020. Identification of SNPs and InDels associated with berry size in table grapes integrating genetic and transcriptomic approaches. BMC Plant Biology 20, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri S, Lovato A, Boscaini F, et al. 2017. The induction of noble rot (Botrytis cinerea) infection during postharvest withering changes the metabolome of grapevine berries (Vitis vinifera L., cv. Garganega). Frontiers in Plant Science 8, 1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda-Yamamizo C, Mitsuda N, Sakamoto S, Ogawa D, Ohme-Takagi M, Ohmiya A.. 2016. The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Scientific Reports 6, 23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K.. 2005. NAC transcription factors: structurally distinct, functionally diverse. Trends in Plant Science 10, 79–87. [DOI] [PubMed] [Google Scholar]
- Orduña L, Li M, Navarro-Paya D, et al. 2022. Direct regulation of shikimate, early phenylpropanoid, and stilbenoid pathways by Subgroup 2 R2R3-MYBs in grapevine. The Plant Journal 110, 529–547. [DOI] [PubMed] [Google Scholar]
- Orduña L, Santiago A, Navarro-Payá D, Zhang C, Wong DCJ, Matus JT.. 2023. Aggregated gene co-expression networks predict transcription factor regulatory landscapes in a non-model plant species. Journal of Experimental Botany 74, 6522–6540. [DOI] [PubMed] [Google Scholar]
- Palumbo MC, Zenoni S, Fasoli M, Massonnet M, Farina L, Castiglione F, Pezzotti M, Paci P.. 2014. Integrated network analysis identifies fight-club nodes as a class of hubs encompassing key putative switch genes that induce major transcriptome reprogramming during grapevine development. The Plant Cell 26, 4617–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K, Qiu JL, Lütje J, Fiil BK, Hansen S, Mundy J, Petersen M.. 2010. Arabidopsis MKS1 is involved in basal immunity and requires an intact N-terminal domain for proper function. PLoS One 5, e14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezet R, Perret C, Jean-Denis JB, Tabacchi R, Gindro K, Viret O.. 2003. δ-Viniferin, a resveratrol dehydrodimer: one of the major stilbenes synthesized by stressed grapevine leaves. Journal of Agricultural and Food Chemistry 51, 5488–5492. [DOI] [PubMed] [Google Scholar]
- Pilati S, Malacarne G, Navarro-Payá D, Tomè G, Riscica L, Cavecchia V, Matus JT, Moser C, Blanzieri E.. 2021. Vitis OneGenE: a causality-based approach to generate gene networks in Vitis vinifera sheds light on the laccase and dirigent gene families. Biomolecules 11, 1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M.. 2012. NAC proteins: regulation and role in stress tolerance. Trends in Plant Science 17, 369–381. [DOI] [PubMed] [Google Scholar]
- Ribéreau-Gayon P, Dubourdieu D, Doneèche BB, Lonvaud AA.. 2006. The grape and its maturation. In: Handbook of enology: the microbiology of wine and vinifications, Vol. 1. Chichester: John Wiley & Sons, 241–297. [Google Scholar]
- Riechmann JL, Meyerowitz EM.. 1998. The AP2/EREBP family of plant transcription factors. Biological Chemistry 379, 633–646. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santana-Garcia W, Castro-Mondragon JA, Padilla-Gálvez M, et al. 2022. RSAT 2022: regulatory sequence analysis tools. Nucleic Acids Research 50, W670–W676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoi S, Wong DCJ, Degu A, Herrera JC, Bucchetti B, Peterlunger E, Fait A, Mattivi F, Castellarin SD.. 2017. Multi-omics and integrated network analyses reveal new insights into the systems relationships between metabolites, structural genes, and transcriptional regulators in developing grape berries (Vitis vinifera L.) exposed to water deficit. Frontiers in Plant Science 8, 1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmuleviz R, Amato A, Commisso M, D’Incà E, Luzzini G, Ugliano M, Fasoli M, Zenoni S, Tornielli GB.. 2023. Temperature affects organic acid, terpene and stilbene metabolisms in wine grapes during postharvest dehydration. Frontiers in Plant Science 14, 1107954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Graether SP.. 2022. The disordered dehydrin and its role in plant protection: a biochemical perspective. Biomolecules 12, 1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soosaar JL, Burch-Smith TM, Dinesh-Kumar SP.. 2005. Mechanisms of plant resistance to viruses. Nature Reviews Microbiology 3, 789–798. [DOI] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG.. 2015. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162, 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Korir NK, Han J, Shangguan LF, Kayesh E, Leng XP, Fang JG.. 2012. Characterization of grapevine microR164 and its target genes. Molecular Biology Reports 39, 9463–9472. [DOI] [PubMed] [Google Scholar]
- Survila M, Davidsson PR, Pennanen V, Kariola T, Broberg M, Sipari N, Heino P, Palva ET.. 2016. Peroxidase-generated apoplastic ROS impair cuticle integrity and contribute to DAMP-elicited defenses. Frontiers in Plant Science 7, 1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tello J, Torres-Pérez R, Grimplet J, Carbonell-Bejerano P, Martínez-Zapater JM, Ibáñez J.. 2015. Polymorphisms and minihaplotypes in the VvNAC26 gene associate with berry size variation in grapevine. BMC Plant Biology 15, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari P, Chakrabarty D.. 2021. Dehydrin in the past four decades: from chaperones to transcription co-regulators in regulating abiotic stress response. Current Research in Biotechnology 3, 249–259. [Google Scholar]
- Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K.. 2004. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. The Plant Cell 16, 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandelle E, Vannozzi A, Wong D, Danzi D, Digby AM, Dal Santo S, Astegno A.. 2018. Identification, characterization, and expression analysis of calmodulin and calmodulin-like genes in grapevine (Vitis vinifera) reveal likely roles in stress responses. Plant Physiology and Biochemistry 129, 221–237. [DOI] [PubMed] [Google Scholar]
- van den Dries N, Giannì S, Czerednik A, Krens FA, de Klerk GJ.. 2013. Flooding of the apoplast is a key factor in the development of hyperhydricity. Journal of Experimental Botany 64, 5221–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Dahal K, Alber NA, Chadee A.. 2020. Photosynthesis, respiration and growth: a carbon and energy balancing act for alternative oxidase. Mitochondrion 52, 197–211. [DOI] [PubMed] [Google Scholar]
- Vannozzi A, Dry IB, Fasoli M, Zenoni S, Lucchin M.. 2012. Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biology 12, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannozzi A, Wong DCJ, Höll J, Hmmam I, Matus JT, Bogs J, Ziegler T, Dry I, Barcaccia G, Lucchin M.. 2018. Combinatorial regulation of stilbene synthase genes by WRKY and MYB transcription factors in grapevine (Vitis vinifera L). Plant & Cell Physiology 59, 1043–1059. [DOI] [PubMed] [Google Scholar]
- Versari A, Parpinello GP, Tornielli GB, Ferrarini R, Giulivo C.. 2001. Stilbene compounds and stilbene synthase expression during ripening, wilting, and UV treatment in grape cv. Corvina. Journal of Agricultural and Food Chemistry 49, 5531–5536. [DOI] [PubMed] [Google Scholar]
- Wang J, Nan N, Shi L, Li N, Huang S, Zhang A, Liu Y, Guo P, Liu B, Xu Z-Y.. 2020. Arabidopsis BRCA1 represses RRTF1-mediated ROS production and ROS-responsive gene expression under dehydration stress. New Phytologist 228, 1591–1610. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhou Y, Ding Y, Chen C, Chen X, Su N, Zhang X, Pan Y, Li J.. 2023. Novel flavin-containing monooxygenase protein FMO1 interacts with CAT2 to negatively regulate drought tolerance through ROS homeostasis and ABA signaling pathway in tomato. Horticulture Research 10, uhad037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Guo T, Wang Z, Kang J, Yang Q, Shen Y, Long R.. 2020. Expression of three related to ABI3/VP1 genes in Medicago truncatula caused increased stress resistance and branch increase in Arabidopsis thaliana. Frontiers in Plant Science 11, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tu M, Wang D, Liu J, Li Y, Li Z, Wang Y, Wang X.. 2018. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnology Journal 16, 844–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Zhao S, Liu JF, et al. 2022. Genome-wide identification of Tomato Golden 2-Like transcription factors and abiotic stress related members screening. BMC Plant Biology 22, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ. 2002. Recent advances in fruit development and ripening: an overview. Journal of Experimental Botany 53, 1995–2000. [DOI] [PubMed] [Google Scholar]
- Wong DCJ, Schlechter R, Vannozzi A, Höll J, Hmmam I, Bogs J, Tornielli GB, Castellarin SD, Matus JT.. 2016. A systems-oriented analysis of the grapevine R2R3-MYB transcription factor family uncovers new insights into the regulation of stilbene accumulation. DNA Research 23, 451–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Nassuth A.. 2006. Stress- and development-induced expression of spliced and unspliced transcripts from two highly similar dehydrin 1 genes in V. riparia and V. vinifera. Plant Cell Reports 25, 968–977. [DOI] [PubMed] [Google Scholar]
- Xu J, Ji Z, Wang C, Xu F, Wang F, Zheng Y, Tang Y, Wei Z, Zhao T, Zhao K.. 2022. WATER-SOAKED SPOT1 controls chloroplast development and leaf senescence via regulating reactive oxygen species homeostasis in rice. Frontiers in Plant Science 13, 918673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yin L, Ying Q, Song H, Xue D, Lai T, Xu M, Shen B, Wang H, Shi X.. 2013. High-throughput sequencing and degradome analysis identify miRNAs and their targets involved in fruit senescence of Fragaria ananassa. PLoS One 8, e70959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, He M, Zhu Z, Li S, Xu Y, Zhang C, Singer SD, Wang Y.. 2012. Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness to various forms of abiotic and biotic stress. BMC Plant Biology 12, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni A, Minoia L, Ferrarini A, Tornielli GB, Zago E, Delledonne M, Pezzotti M.. 2008. Molecular analysis of post-harvest withering in grape by AFLP transcriptional profiling. Journal of Experimental Botany 59, 4145–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Chen H, Wang Y, Hicks D, Ke H, Pruneda-Paz J, Dehesh K.. 2022. ORA47 is a transcriptional regulator of a general stress response hub. The Plant Journal 110, 562–571. [DOI] [PubMed] [Google Scholar]
- Zenoni S, Amato A, D’Incà E, Guzzo F, Tornielli GB.. 2020. Rapid dehydration of grape berries dampens the post-ripening transcriptomic program and the metabolite profile evolution. Horticulture Research 7, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenoni S, Amato A, Tornielli GB.. 2021. 1.39 - Grape berry transcriptome. In: Cifuentes A, ed. Comprehensive foodomics. Oxford: Elsevier, 558–571. [Google Scholar]
- Zenoni S, D’Agostino N, Tornielli GB, et al. 2011. Revealing impaired pathways in the an11 mutant by high-throughput characterization of Petunia axillaris and Petunia inflata transcriptomes. The Plant Journal 68, 11–27. [DOI] [PubMed] [Google Scholar]
- Zenoni S, Fasoli M, Guzzo F, et al. 2016. Disclosing the molecular basis of the postharvest life of berry in different grapevine genotypes. Plant Physiology 172, 1821–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, He SB, Li L, Yang HQ.. 2014. Auxin inhibits stomatal development through MONOPTEROS repression of a mobile peptide gene STOMAGEN in mesophyll. Proceedings of the National Academy of Sciences, USA 111, E3015–E3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Dong R, Wang Y, Li X, Ji M, Wang X.. 2021. NAC domain gene VvNAC26 interacts with VvMADS9 and influences seed and fruit development. Plant Physiology and Biochemistry 164, 63–72. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mao L, Wang H, Brocker C, Yin X, Vasiliou V, Fei Z, Wang X.. 2012. Genome-wide identification and analysis of grape aldehyde dehydrogenase (ALDH) gene superfamily. PLoS One 7, e32153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li W, Gao X, Xu M, Guo Y.. 2020. DEAR4, a member of DREB/CBF family, positively regulates leaf senescence and response to multiple stressors in Arabidopsis thaliana. Frontiers in Plant Science 11, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Cao L, Hu K, Yu X, Qu S.. 2023. miR164–NAC21/22 module regulates the resistance of Malus hupehensis against Alternaria alternata by controlling jasmonic acid signaling. Plant Science 330, 111635. [DOI] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR.. 2010. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Zhang J, Gao X, Tong J, Xiao L, Li W, Zhang H.. 2010. The Arabidopsis AP2/ERF transcription factor RAP26 participates in ABA, salt and osmotic stress responses. Gene 457, 1–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data for the transient expression experiments on V. vinifera cv. ‘Thompson Seedless’ are available at GEO under accession no. GSE232165. DAP-seq raw data have been submitted to GEO, including metadata of samples and conducted analysis (bioinformatic parameters) according to the FAIR principles, under accession no. GSE230185. DAP-seq results on NAC61 can be visualized in the DAPBrowse tool available at the Vitis Visualization Platform (http://www.vitviz.tomsbiolab.com/). The role for NAC61 has been deposited in the Gene Reference Catalogue found at the Grape Genomics Encyclopedia portal (http://grapedia.org/). All other data supporting the findings of this study are available within the paper and within its supplementary data published online.