Abstract
A requisite step in reovirus infection of the murine intestine is proteolysis of outer-capsid proteins to yield infectious subvirion particles (ISVPs). When converted to ISVPs by intestinal proteases, virions of reovirus strain type 3 Dearing (T3D) lose 90% of their original infectivity due to cleavage of viral attachment protein ς1. In an analysis of eight field isolate strains of type 3 reovirus, we identified one additional strain, type 3 clone 31 (T3C31), that loses infectivity and undergoes ς1 cleavage upon conversion of virions to ISVPs. We examined the ς1 deduced amino acid sequences of T3D and the eight field isolate strains for a correlation between sequence variability and ς1 cleavage. The ς1 proteins of T3D and T3C31 contain a threonine at amino acid position 249, whereas an isoleucine occurs at this position in the ς1 proteins of the remaining strains. Thr249 occupies the d position of a heptad repeat motif predicted to stabilize ς1 oligomers through α-helical coiled-coil interactions. This region of sequence comprises a portion of the fibrous tail domain of ς1 known as the neck. Substitution of Thr249 with isoleucine or leucine resulted in resistance to cleavage by trypsin, whereas replacement with asparagine did not affect cleavage susceptibility. These results demonstrate that amino acid position 249 is an independent determinant of T3D ς1 cleavage susceptibility and that an intact heptad repeat is required to confer cleavage resistance. We performed amino-terminal sequence analysis on the ς1 cleavage product released during trypsin treatment of T3D virions to generate ISVPs and found that trypsin cleaves ς1 after Arg245. Thus, the sequence polymorphism at position 249 controls cleavage at a nearby site in the neck region. The relevance of these results to reovirus infection in vivo was assessed by treating virions with the contents of a murine intestinal wash under conditions that result in generation of ISVPs. The pattern of ς1 cleavage susceptibility generated by using purified protease was reproduced in assays using the intestinal wash. These results provide a mechanistic explanation for ς1 cleavage during exposure of virions to intestinal proteases and may account for certain strain-dependent patterns of reovirus pathogenesis.
Following oral inoculation into newborn mice, mammalian reoviruses undergo primary replication in intestinal tissue and spread to the central nervous system (64). However, not all reovirus strains are capable of productive infection in intestinal tissue. Prototype type 3 reovirus strain Dearing (T3D) grows poorly in the murine intestine after oral inoculation. In contrast, prototype type 1 reovirus strain Lang (T1L) grows well in intestinal tissue and is capable of systemic spread (9, 32, 35, 54). Proteolytic processing of reovirus virions in the intestinal lumen (6, 10) or in the endocytic compartment (3, 11, 15, 56, 59) results in generation of infectious subvirion particles (ISVPs) (48) and is required for reovirus to establish productive infection of either animal hosts (1, 6) or cultured cells (3, 19, 59). Under conditions that result in generation of ISVPs, treatment of T3D virions in vitro with either chymotrypsin or trypsin is associated with cleavage of viral attachment protein ς1 and a 10-fold decrease in viral infectivity (46). Identical treatment of T1L is not associated with ς1 cleavage or reduced infectivity. Strain-dependent differences in ISVP infectivity loss and ς1 cleavage cosegregate in genetic analyses with the ς1-encoding S1 gene, which indicates that cleavage susceptibility of T3D ς1 protein is an intrinsic property of this molecule and that infectivity loss experienced by T3D ISVPs is causally linked to ς1 cleavage (46). Furthermore, studies of T1L × T3D reassortant viruses show that the S1 gene is the primary determinant of strain-specific differences in growth of reovirus in the murine intestine (9). These findings suggest that the infectivity loss experienced by T3D following oral inoculation results from susceptibility of its ς1 protein to cleavage by intestinal proteases.
The ς1 protein is a fibrous protein with a head-and-tail morphology (4, 14, 24, 25). In virions, ς1 exists as an oligomer (7, 39, 58) located at the vertices of the virion icosahedron (20, 25). Results from genetic and biochemical studies of ς1 protein suggest the presence of two discrete receptor-binding domains in ς1 of type 3 reovirus. A domain in the tail is important for binding sialic acid (16, 17, 46, 55), and a domain in the head is important for binding an unidentified receptor on L cells (21, 45, 60, 67, 70) and determining viral tropism within the murine central nervous system (8, 33). Binding of sialic acid by ς1 is the basis for hemagglutination (HA) by type 3 reovirus (2, 17, 26, 27, 49, 50) and growth of type 3 reovirus in murine erythroleukemia cells (16, 55).
Following treatment of T3D virions with chymotrypsin to generate ISVPs, monoclonal antibody (MAb) G5, which binds the T3D ς1 head (8), is markedly diminished in its capacity to bind viral particles and neutralize infectivity (46). This result suggests that the ς1 head is released from the viral particle following protease treatment of T3D virions. However, T3D ISVPs retain the ability to bind cell surface sialic acid (46), which suggests that the ς1 tail domain remains associated with ISVPs. Studies using protease treatment of ς1 purified from virions (70) and expressed ς1 protein (22) indicate the presence of a highly protease-sensitive region near the middle of ς1 primary sequence. Based on predictions of ς1 secondary structure (47) and image reconstructions of ς1 protein visualized by electron microscopy (24), the protease-sensitive sequences are proposed to represent a head-proximal flexible portion of the tail termed the neck. These results have been reconciled in a model of T3D ISVP formation in which ς1 is cleaved within the neck region between receptor-binding domains in the head and tail (46). However, the site of ς1 cleavage on T3D ISVPs has not been identified, and the mechanism of cleavage sensitivity is unknown.
In this study, we performed experiments to determine the mechanism of ς1 susceptibility to cleavage by intestinal proteases during generation of ISVPs. Our results suggest that ς1 cleavage sensitivity is influenced by subunit interactions in the ς1 oligomer. Furthermore, results from these studies strongly support the existence of discontiguous receptor-binding domains in the ς1 head and tail (16, 46).
MATERIALS AND METHODS
Cells and viruses.
Spinner culture-adapted L cells were grown in either suspension or monolayer cultures, using Joklik’s modified Eagle’s minimal essential medium (Irvine Scientific, Santa Ana, Calif.) supplemented to contain 5% fetal bovine serum (Summit Biotechnology, Fort Collins, Colo.), 2 mM l-glutamine, and 100 U of penicillin, 100 μg of streptomycin, and 0.25 μg of amphotericin per ml (Irvine). Spodoptera frugiperda (Sf21) insect cells were grown in either suspension or monolayer cultures, using Grace’s medium (Gibco, Grand Island, N.Y.) supplemented to contain 10% fetal bovine serum plus 100 U of penicillin, 100 μg of streptomycin, and 0.25 μg of amphotericin per ml. Reovirus strain T3D is a laboratory stock. Field isolate strains type 3 clone 9 (T3C9), type 3 clone 18 (T3C18), type 3 clone 31 (T3C31), type 3 clone 43 (T3C43), type 3 clone 44 (T3C44), type 3 clone 45 (T3C45), type 3 clone 84 (T3C84), and type 3 clone 93 (T3C93) were obtained originally from the collection of Leon Rosen (18, 51–53). Purified virion preparations were made from second- and third-passage L-cell lysate stocks of twice-plaque-purified reovirus as previously described (25). To obtain purified virions containing 35S-labeled proteins, Easy Tag Express-[35S] protein labeling mix (NEN, Boston, Mass.) was added to cell suspensions (∼12.5 μCi per ml) at the initiation of infection.
Baculovirus vector strains were derived from Autographa californica nuclear polyhedrosis virus (AcMNPV; Clontech Laboratories, Palo Alto, Calif.). Recombinant baculoviruses containing wild-type and mutant S1 gene cDNAs were generated by cloning into pBacPAK8 and pBacPAK9 baculovirus transfer vectors (Clontech), followed by lipofection-mediated cotransfer of plasmid recombinants and linearized BacPAK6 AcMNPV DNA (Clontech) into Sf21 cells according to the supplier’s instructions. After 5 days of incubation, recombinant virus clones were isolated by plaque purification on monolayers of Sf21 cells and amplified by two passages in Sf21 cells.
Digestion of reovirus virions with intestinal proteases.
Purified reovirus virions at a concentration of 2 × 1012 particles per ml in virion storage buffer (150 mM NaCl, 10 mM MgCl2, 10 mM Tris [pH 7.5]) were digested at 37°C with 200 μg of Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK)-treated bovine α-chymotrypsin (Sigma Chemical Co., St. Louis, Mo.) per ml for various intervals. Digestion reactions were stopped by adding 5 mM phenylmethylsulfonyl fluoride (Sigma) to the treatment mixtures and cooling at 0°C.
Determination of virus titer after protease treatment of reovirus virions.
Virus titer after protease treatment of virions was determined by plaque assay as previously described (62).
SDS-PAGE of reovirus structural proteins.
Discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as previously described (37). In preparation for electrophoresis, 35S-labeled viral particles in virion storage buffer were mixed 1:1 with 2× sample buffer (250 mM Tris [pH 6.8], 4% 2-mercaptoethanol, 2% SDS, 0.02% bromophenol blue) and incubated at 65°C for 5 min. Samples then were loaded into wells of a 10% polyacrylamide gel and electrophoresed at 25-mA constant current until the dye front reached the bottom of the gel. Gels were dried onto filter paper (Bio-Rad Laboratories, Hercules, Calif.) under vacuum and exposed to Biomax MR film (Eastman Kodak Co., Rochester, N.Y.).
HA assay.
HA assays using virions and ISVPs were performed as previously described (46).
Cloning and mutagenesis of S1 gene cDNAs.
The ς1-encoding S1 gene cDNAs of strains T3D, T3C9, and T3C84 were generated by using reverse transcription-PCR (36) and cloned into the pCR2.1 vector (Invitrogen, San Diego, Calif.). S1 genes were amplified with primers specific for the noncoding regions of the T3D S1 gene.
Site-directed mutants of the T3D S1 gene were produced by using the splice-overlap-extension PCR technique (30). Primers bearing desired mutations in the S1 gene were used in independent primary reactions to generate S1 gene fragments having sequence complementarity over the terminal 20 nucleotides. The complementary primer sets used for mutagenesis were as follows (nucleotides differing from wild-type T3D S1 sequence are underlined): 5′AGGCGCAATTGAGCAAAGTT3′/5′AACTTTGCTCAATTGCGCCT3′ (Thr249→Ile), 5′AGGCGCACTTGAGCAAAGTT3′/5′AACTTTGCTCAAGTGCGCCT3′ (Thr249→Leu), and 5′AGGCGCAAATGAGCAAAGTT3′/5′AACTTTGCTCATTTGCGCCT3′ (Thr249→Asn). Reaction mixes included 0.5 μg of recombinant pCR2.1 plasmid template, 0.2 μg of S1-specific primers, 200 μM each deoxynucleoside triphosphate, and 2.5 U of Pfu DNA polymerase (Stratagene, La Jolla, Calif.) in a total volume of 50 μl of Pfu reaction buffer (Stratagene). Reactions were subjected to 40 iterations of a thermal cycle consisting of 94°C for 1 min, 55°C for 2 min, and 72°C for 3 min. The final cycle was followed by incubation at 72°C for 20 min. Primary PCR products were resolved in a 1% Tris-borate-EDTA agarose gel containing ethidium bromide, and the desired amplification product was allowed to migrate onto DE-81 chromatography paper (Whatman, Maidstone, England) inserted into the gel, followed by elution in a solution of 1 M LiCl, 10 mM Tris (pH 7.6), and 1 mM EDTA in 20% ethanol. Eluted DNA was concentrated by ethanol precipitation and reconstituted in water.
Secondary PCR products were amplified in reaction mixtures containing 1 pmol each of the two purified primary PCR products, 0.2 μg of S1-specific primers complementary to T3D S1 gene segment termini, 200 μM each deoxynucleoside triphosphate, and 5 U of Taq DNA polymerase (Promega, Madison, Wis.) in a total volume of 50 μl of PCR Optimizer buffer (Invitrogen) adjusted to pH 9.0 and 2 mM MgCl2. Thermal cycling parameters were identical to those listed for the primary reactions. Products from the secondary PCR were gel purified and cloned into the pCR2.1 vector. Sequence fidelity was confirmed for ς1-encoding regions of all S1 gene cDNAs in recombinant pCR2.1 constructs; nucleotide sequences were determined by automated analysis using an ABI model 377 (PE-Applied Biosystems, Norwalk, Conn.) or using phage T7 DNA polymerase (U.S. Biochemical, Cleveland, Ohio) and [35S]ATP. Error-free S1 gene cDNAs then were cloned into baculovirus transfer vectors.
Expression and purification of recombinant ς1 proteins.
First- or second-passage recombinant baculovirus stocks were used to infect Sf21 cell monolayers (107 cells) at a multiplicity of infection of ≥1 PFU per cell. After 20 h of incubation, culture medium was replaced with methionine-free Grace’s medium (Gibco) supplemented to contain 10% fetal bovine serum, 100 U of penicillin, 100 μg of streptomycin, and 0.25 μg of amphotericin per ml, and 15 μCi of [35S]methionine protein labeling mix per ml. After an additional 48 h of incubation, cells were harvested and resuspended in 1 ml of phosphate-buffered saline containing 5 mM phenylmethylsulfonyl fluoride and Complete, Mini, EDTA-free protease inhibitor cocktail (Boehringer Mannheim, Indianapolis, Ind.). Cells were lysed by sonication, and supernatants were cleared of debris by centrifugation. T1L ς1-specific MAb 5C6 (63) conjugated to cyanogen bromide-activated Sepharose (Pharmacia, Uppsala, Sweden) was used to deplete supernatants of contaminant proteins that adsorb nonspecifically to antibody. Supernatants then were incubated with type 3 ς1-specific MAb G5 (13) conjugated to Sepharose. Beads containing adsorbed ς1 protein were washed five times with buffer consisting of 50 mM Tris (pH 8), 1.2 M NaCl, 0.4% SDS, 0.2% Triton X-100, and 5 mM EGTA, followed by three washes with a solution of 50 mM triethanolamine (pH 11.6), 0.5 M NaCl, and 0.1% Triton X-100. Beads then were washed three times with virion storage buffer.
Treatment of expressed ς1 proteins with proteases.
Aliquots of ς1-containing Sepharose beads in virion storage buffer were incubated at 10°C with 0, 7.5, 22.5, or 67.5 μg of TLCK-treated bovine α-chymotrypsin per ml for 180 min or at 4°C with 0, 2, 6, or 18 μg Nα-p-tosyl-l-sulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated bovine trypsin (Sigma) per ml for 60 min. Reactions were mixed 1:1 with modified 2× protein sample buffer (125 mM Tris [pH 6.8], 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 80 mM MgCl2, 0.02% bromophenol blue) and incubated at 100°C for 5 min. Reaction products were resolved in an SDS–10% polyacrylamide gel and visualized by autoradiography.
Identification of protease cleavage sites in ς1 protein of T3D ISVPs.
Purified virions of T3D at a concentration of 2 × 1013 particles per ml were digested at 15°C with 100 μg of TPCK-treated bovine trypsin (Sigma) per ml for 30 min in virion storage buffer supplemented to contain 0.05 mM TPCK. Reactions were terminated by the addition of TLCK to a final concentration of 0.5 mM. ISVPs were centrifuged at 125,000 × g for 1 h to form a pellet. Supernatants containing the ς1 cleavage product were incubated with MAb G5 (1.0 mg of antibody per ml) conjugated to Sepharose at 4°C overnight. Beads were washed three times in radioimmunoprecipitation assay buffer (150 mM NaCl, 10 mM Tris [pH 7.4], 0.1% [wt/vol] SDS, 1% [wt/vol] sodium deoxycholate, 1% [vol/vol] Nonidet P-40) and then incubated at 70°C for 10 min in sample buffer without 2-mercaptoethanol. Supernatants were subjected to electrophoresis in an SDS–20% polyacrylamide gel at 25-mA constant current for 24 h, followed by equilibration of the gel in transfer buffer (25 mM Tris [pH 8.3], 192 mM glycine) at 4°C for 30 min. Proteins were electroblotted onto a polyvinylidene difluoride membrane by using a Mini Transblot apparatus (Bio-Rad) operated at 30-V constant voltage overnight. Protein bands were visualized by Coomassie blue staining, and a band migrating at approximately 25 kDa was excised and subjected to sequence analysis by Edman degradation using a Procise 492 protein sequencer (PE-Applied Biosystems, Foster City, Calif.).
The procedure for isolating a ς1 cleavage product also was performed with purified 35S-labeled T3D virions. In this case, ISVPs contained in trypsin digests of virions were not removed prior to addition of MAb G5. Polypeptides recovered with MAb G5 were subjected to electrophoresis under reducing conditions in an SDS–14% polyacrylamide gel and subjected to autoradiography. The Coomassie blue-stained ς1 cleavage product on the polyvinylidene difluoride membrane was verified by comparison with the autoradiogram.
Assessment of ς1 cleavage upon treatment of virions with a murine intestinal wash.
Three-day-old NIH Swiss mice (Harlan Sprague Dawley) were euthanized, and the entire small and large intestine was resected. Contents of 20 intestines were harvested in a total volume of 5 ml of virion storage buffer by repeated flushing using a 1-ml syringe and 25-gauge needle. Suspended material was removed by centrifugation at 23,000 × g for 30 min, and clarified supernatants were used in reactions containing 4 × 1010 purified 35S-labeled reovirus virions. Virions were digested at 20°C for 3.5 h in a total volume of 12 μl of virion storage buffer containing 0.5, 1.5, or 10 μl of intestinal wash. Digestion reactions were mixed 1:1 with modified 2× protein sample buffer, incubated at 100°C for 8 min, and subjected to electrophoresis in SDS–10% polyacrylamide gels, followed by autoradiography to visualize viral proteins.
RESULTS
Type 3 reovirus field isolate strains vary in infectivity and ς1 cleavage during treatment with intestinal proteases to generate ISVPs.
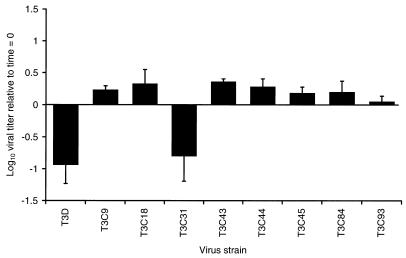
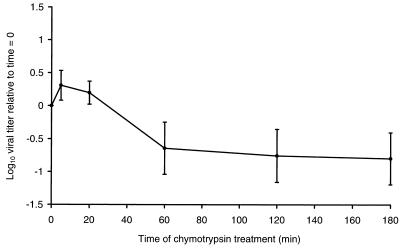
Treatment of virions of reovirus strain T3D with either chymotrypsin or trypsin under conditions to generate ISVPs results in approximately a 10-fold loss in infectivity and cleavage of ς1 protein (46). To determine whether other type 3 reovirus strains also lose infectivity when converted to ISVPs, purified virions of eight type 3 field isolate strains were treated with chymotrypsin under conditions to generate ISVPs, and aliquots of the treatment mixtures were titrated on L-cell monolayers (Fig. 1). Strain T3C31 was the only strain other than T3D to exhibit a decrease in infectivity. Similar to ISVPs of T3D, which lost about 90% of pretreatment infectivity, ISVPs of T3C31 lost approximately 84% of the original infectivity of virions. Changes in T3C31 infectivity recapitulated the kinetic profile observed when chymotrypsin was used to generate ISVPs of T3D (46); a slight increase in viral titer occurred at early time points of protease treatment, followed by a rapid decline in titer that approached its lowest point by 60 min of treatment (Fig. 2). Infectivity of the remaining field isolate strains was slightly (as much as twofold in the cases of T3C18 and T3C43) increased after treatment with chymotrypsin to generate ISVPs. Consistent with the effect of chymotrypsin on viral infectivity, strains T3D and T3C31 lost ≥90% of the original infectivity following treatment with trypsin (data not shown). Thus, infectivity loss associated with T3D ISVPs also is common to T3C31 ISVPs but is not a universal property of type 3 reoviruses.
FIG. 1.
Changes in viral infectivity during generation of ISVPs by using chymotrypsin. Purified virions of T3D, T3C9, T3C18, T3C31, T3C43, T3C44, T3C45, T3C84, and T3C93 at a concentration of 2 × 1012 particles per ml were treated with chymotrypsin at 37°C for 180 min. Infectious titers of virion preparations before and after treatment were determined by plaque assay using L cells. Changes in viral infectivity are expressed as the ratio of log10 viral titer at 180 and 0 min of chymotrypsin treatment. Shown are the means and standard deviations of three independent experiments.
FIG. 2.
Time course of change in T3C31 infectivity during generation of ISVPs by using chymotrypsin. Purified virions of strain T3C31 at a concentration of 2 × 1012 particles per ml were treated with chymotrypsin at 37°C. At the times indicated, reactions were terminated, and infectious titers of virion preparations were determined by plaque assay using L cells. Changes in viral infectivity are expressed as the ratio of log10 viral titer relative to 0 min of chymotrypsin treatment. Shown are the means and standard deviations of three independent experiments.
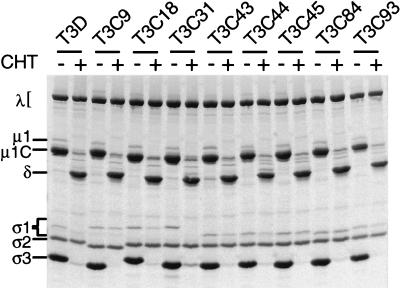
To determine whether differences in viral infectivity of type 3 reovirus strains correlate with differences in susceptibility of their ς1 proteins to proteolytic cleavage, viral proteins in chymotrypsin treatment mixtures were analyzed by SDS-PAGE (Fig. 3). Findings consistent with generation of ISVPs, loss of outer-capsid protein ς3, and appearance of the stable cleavage product, δ, of outer-capsid protein μ1C were observed in these experiments. Following treatment with chymotrypsin, bands corresponding to ς1 proteins of T3D and T3C31 were lost, whereas bands corresponding to ς1 proteins of the remaining seven strains were not. Thus, changes in viral infectivity after chymotrypsin treatment correlate with the status of ς1 protein observed by SDS-PAGE: strains that exhibit decreased infectivity have cleaved ς1 proteins. These findings are in agreement with previous studies of virions and ISVPs of T1L and T3D (46) and demonstrate a consistent correlation between changes in viral infectivity and ς1 cleavage susceptibility during generation of ISVPs.
FIG. 3.
Electrophoretic analysis of viral structural proteins of type 3 reovirus strains following treatment with chymotrypsin to generate ISVPs. Purified 35S-labeled virions of T3D, T3C9, T3C18, T3C31, T3C43, T3C44, T3C45, T3C84, and T3C93 at a concentration of 2 × 1012 particles per ml were treated with chymotrypsin (CHT) at 37°C for 60 min. Equal numbers of treated and untreated viral particles (2 × 1011) were dissociated in sample buffer and loaded into wells of an SDS–10% polyacrylamide gel. After electrophoresis, gels were prepared for fluorography and exposed to film. Viral proteins are labeled.
Protease treatment of type 3 field isolate strains increases their capacity to produce HA.
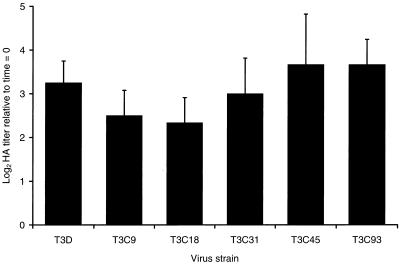
T3D ISVPs demonstrate an increase in HA titer relative to T3D virions (46, 63), which suggests that the HA domain in ς1 protein is altered by a conformational change in ς1 or further exposed by proteolysis of ς3 and μ1/μ1C proteins during generation of ISVPs. However, it is also possible that cleavage of T3D ς1 protein facilitates the enhanced capacity of T3D ISVPs to produce HA. To determine whether chymotrypsin-mediated cleavage of ς1 protein plays a role in increased HA titer, we treated purified virions of the five HA-positive field isolate strains (T3C9, T3C18, T3C31, T3C45, and T3C93) with chymotrypsin to generate ISVPs and then tested the treatment mixtures for their capacity to agglutinate human type O erythrocytes (Fig. 4). Commensurate with an increase in T3D HA titer of approximately 10-fold, the HA titer of the type 3 field isolate strains increased 4- to 16-fold following chymotrypsin treatment. Therefore, in contrast to changes in viral infectivity, the increased capacity of type 3 ISVPs to produce HA relative to virions is independent of ς1 cleavage status. These data suggest that a domain of ς1 protein important for HA is altered with respect to conformation or environment during conversion of virions to ISVPs.
FIG. 4.
Changes in viral HA capacity during generation of ISVPs by using chymotrypsin. Purified virions of T3D, T3C9, T3C18, T3C31, T3C45, and T3C93 at a concentration of 2 × 1012 particles per ml were treated with chymotrypsin at 37°C for 180 min. HA activity of virion preparations was determined by endpoint titration using human type O erythrocytes and serial dilutions of virus. Changes in HA activity are expressed as the ratio of log2 HA titer at 180 and 0 min of chymotrypsin treatment. Shown are the means and standard deviations of three independent experiments.
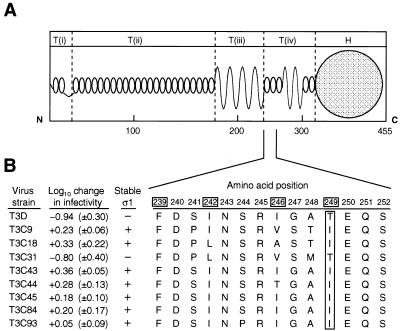
Comparison of the deduced amino acid sequences of ς1 proteins link infectivity loss and ς1 cleavage to amino acid 249.
The correlation of sequence variability with biologic polymorphisms has provided important information about structure-function relationships in reovirus ς1 protein (16, 17, 68). We previously determined the deduced ς1 amino acid sequences of the eight field isolate strains used in this study and found a high degree of conservation among these strains and T3D (85 to 99% sequence identity in pairwise sequence comparisons [18]). Therefore, to identify sequences in ς1 associated with infectivity loss and ς1 cleavage, we examined the ς1 amino acid sequences of T3D and the field isolate strains for residues unique to the chymotrypsin-sensitive strains. The ς1 sequences of six of these strains, T3D, T3C43, T3C44, T3C45, T3C84, and T3C93, are very similar, showing variation at only 19 of 455 total amino acid positions in ς1 (18). Given that only T3D among these strains exhibits chymotrypsin-mediated infectivity loss and ς1 cleavage, we examined the six ς1 sequences for residues unique to T3D. Val22, Ile88, Thr249, and Thr408 were found to be unique to T3D, compared to Ile22, Thr88, Ile249, and Ala408 in the other strains; thus, one or more of these residues were considered likely to be associated with chymotrypsin sensitivity of T3D ς1. When ς1 sequences of the more distantly related group of strains, T3C9, T3C18, and T3C31, were examined, we noted that the ς1 protein of chymotrypsin-sensitive strain T3C31 has a threonine at amino acid position 249; strains T3C9 and T3C18 possess an isoleucine at that position, as do strains T3C43, T3C44, T3C45, T3C84, and T3C93 (Fig. 5). Thus, Thr249 is unique to T3D and T3C31, which suggests that the amino acid residue at position 249 determines susceptibility of ς1 protein to proteolytic cleavage.
FIG. 5.
Identification of residues important for infectivity loss and ς1 cleavage of reovirus strains T3D and T3C31. (A) Model of ς1 structure. Predicted ς1 secondary structure (47) and morphologic domains of ς1 [T(i), T(ii), T(iii), T(iv), and H] described previously (24) are shown and scaled proportionally to the domains identified in electron microscopic images of ς1 isolated from virions (24). Amino acid positions are scaled according to their predicted relationships to individual ς1 morphologic domains (47). (B) Alignment of ς1 amino acid sequences. Deduced ς1 amino acid sequences of strains T3D and T3C31 were aligned with those of T3C9, T3C18, T3C43, T3C44, T3C45, T3C84, and T3C93 (18) and examined for correlation of sequence variability with viral infectivity changes and ς1 cleavage susceptibility during the generation of ISVPs by using chymotrypsin. The ς1 proteins of T3D and T3C31 contain a threonine residue at position 249, whereas all other ς1 proteins contain an isoleucine at that position. Shown is an alignment of amino acid residues 239 through 252, which are proposed to form α-helical coiled coil comprising a portion of the ς1 neck (47). In the alignment of ς1 sequences, residues in boxes are found in the a or d position of a heptad repeat motif characteristic of α-helical coiled coils (42). The mean (±standard deviation) ratio of log10 viral titer at 180 and 0 min of chymotrypsin treatment is shown for each strain.
Analysis of cleavage susceptibility of expressed ς1 proteins altered at amino acid position 249.
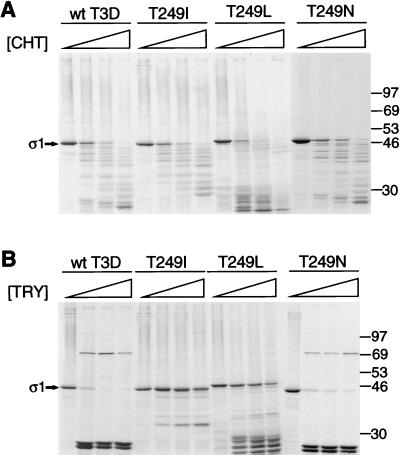
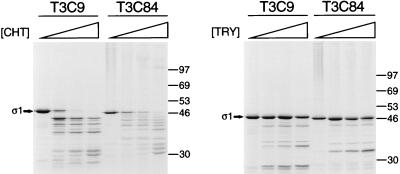
The importance of amino acid residue 249 in cleavage of ς1 by intestinal proteases was assessed using baculovirus-expressed, purified T3D ς1 proteins modified by site-directed mutagenesis. Consistent with the chymotrypsin sensitivity of virion-associated T3D ς1 protein (Fig. 3), expressed wild-type T3D ς1 protein was cleaved by chymotrypsin, resulting in the accumulation of cleavage products of ≤30 kDa as assessed by SDS-PAGE (Fig. 6A). Mutant ς1 proteins in which Thr249 was replaced with isoleucine, leucine, or asparagine also were cleaved by chymotrypsin. To determine whether chymotrypsin sensitivity is a feature of type 3 ς1 proteins expressed and tested under conditions used in this study, expressed ς1 proteins of strains T3C9 and T3C84 were tested in protease assays using chymotrypsin and trypsin (Fig. 7). Neither T3C9 nor T3C84 virions lose infectivity upon conversion to ISVPs by using chymotrypsin (Fig. 1), and their ς1 proteins are resistant to cleavage (Fig. 3). However, expressed T3C9 and T3C84 ς1 proteins were cleaved following treatment with chymotrypsin. These results indicate that cleavage of the Thr249→Ile and Thr249→Leu mutant T3D ς1 proteins by chymotrypsin is an inherent property of expressed ς1 protein and that the role of amino acid position 249 in cleavage of ς1 by chymotrypsin cannot be addressed using this experimental system.
FIG. 6.
Cleavage susceptibility of expressed T3D ς1 protein altered by site-directed mutagenesis. Wild-type (wt) and mutant 35S-labeled ς1 proteins of T3D were expressed in Sf21 insect cells by using baculovirus vectors and purified by using anti-ς1 MAb G5. The threonine residue at amino acid position 249 of T3D ς1 was substituted with isoleucine (T249I), leucine (T249L), or asparagine (T249N). (A) MAb G5-conjugated Sepharose containing expressed ς1 protein was treated with various concentrations of chymotrypsin (CHT) at 10°C for 180 min. Treatment mixtures were heated at 100°C in sample buffer and subjected to electrophoresis in an SDS–10% polyacrylamide gel. Digestion products were visualized by autoradiography. Positions of molecular weight standards (in kilodaltons) are shown. Bands corresponding to full-length ς1 are indicated.  , 0 to 67.5 μg of chymotrypsin per ml. (B) MAb G5-conjugated Sepharose containing expressed ς1 protein was treated with various concentrations of trypsin (TRY) at 4°C for 60 min. Treatment mixtures were processed as described for panel A.
, 0 to 67.5 μg of chymotrypsin per ml. (B) MAb G5-conjugated Sepharose containing expressed ς1 protein was treated with various concentrations of trypsin (TRY) at 4°C for 60 min. Treatment mixtures were processed as described for panel A.  , 0 to 18 μg of trypsin per ml.
, 0 to 18 μg of trypsin per ml.
FIG. 7.
Cleavage susceptibility of expressed T3C9 and T3C84 ς1 proteins. 35S-labeled ς1 proteins of T3C9 and T3C84 were expressed in Sf21 insect cells by using baculovirus vectors and purified by using anti-ς1 MAb G5. MAb G5-conjugated Sepharose containing expressed ς1 protein was treated with various concentrations of either chymotrypsin (CHT) at 10°C for 180 min or trypsin (TRY) at 4°C for 60 min. Treatment mixtures were heated at 100°C in sample buffer and subjected to electrophoresis in an SDS–10% polyacrylamide gel. Digestion products were visualized by autoradiography. Positions of molecular weight standards (in kilodaltons) are shown. Bands corresponding to full-length ς1 are indicated.  , 0 to 67.5 μg of chymotrypsin per ml or 0 to 18 μg of trypsin per ml.
, 0 to 67.5 μg of chymotrypsin per ml or 0 to 18 μg of trypsin per ml.
ISVPs of T3D generated by using trypsin exhibit the properties of viral infectivity loss and ς1 cleavage characteristic of ISVPs generated by using chymotrypsin (46). Therefore, we tested the effect of trypsin on expressed ς1 proteins, examining first the control proteins, T3C9 ς1 and T3C84 ς1 (Fig. 7). These ς1 proteins exhibited resistance to cleavage by trypsin, which indicated that this enzyme would be suitable to test the role of position 249 in ς1 protease susceptibility. Wild-type T3D ς1 protein was cleaved by trypsin into two major fragments in the range of 25 kDa (Fig. 6B). These stable cleavage products most likely represent the amino-terminal 25/26-kDa and carboxy-terminal 23/24-kDa fragments observed previously following trypsin treatment of T3D ς1 proteins purified from virions (70) or expressed by recombinant baculovirus (22). Upon replacement of Thr249 with Ile, which is found at position 249 in the seven chymotrypsin-resistant ς1 proteins, T3D ς1 was resistant to cleavage by trypsin, showing very little loss in band intensity at the highest concentration of enzyme used (18 μg per ml). In contrast, only a small fraction of intact wild-type ς1 remained at the lowest enzyme concentration (2 μg per ml), and a full-length ς1 band was barely detectable at higher concentrations. Thus, an isoleucine at amino acid position 249 confers resistance to ς1 cleavage by the intestinal protease, trypsin.
We next ascertained whether there exists an absolute requirement for isoleucine to confer protease resistance. Thr249 was substituted with the isoleucine isomer, leucine. Like the Thr249→Ile mutant ς1 protein, the Thr249→Leu mutant was resistant to cleavage by trypsin and underwent only moderate proteolysis at the highest concentration of enzyme used (Fig. 6B). These results suggest that the critical determinant of protease resistance is the presence of an apolar residue at position 249. To test this hypothesis, we used an additional ς1 mutant substituted at position 249 with the polar amino acid, asparagine. When treated with trypsin, the Thr249→Asn mutant exhibited a cleavage profile virtually indistinguishable from that of wild-type ς1 (Fig. 6B). Thus, our findings suggest that susceptibility of T3D ς1 to proteolytic cleavage depends on the type of amino acid at position 249, where apolar and polar residues respectively confer cleavage resistance and sensitivity.
Identification of a cleavage site in the ς1 protein of T3D ISVPs generated by protease treatment of virions.
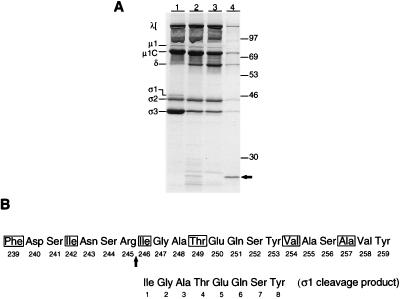
The site at which trypsin cleaves T3D ς1 during the generation of ISVPs was determined by amino-terminal sequence analysis of the ς1 cleavage product liberated by protease treatment of purified virions. Virions were treated with trypsin, and the carboxy-terminal fragment of ς1 was captured by using MAb G5, which binds the virion-distal head domain (8) (Fig. 8A). Trypsin treatment of T3D virions resulted in the loss of ς3 protein and appearance of the μ1C cleavage product, δ, consistent with the formation of ISVPs. A single ς1 cleavage product of approximately 25 kDa was purified from the digestion reaction using MAb G5 and subjected to eight cycles of Edman microsequencing. This analysis revealed the unambiguous amino acid sequence Ile-Gly-Ala-Thr-Glu-Gln-Ser-Tyr, which corresponds exactly to T3D ς1 amino acid residues 246 to 253 (Fig. 8B). An arginine residue occupies amino acid position 245 of T3D ς1, and cleavage at this site is congruous with the action of trypsin at the carboxy-terminal side of basic residues (12). The sequence contained in residues 246 to 253 is not repeated elsewhere in T3D ς1. Therefore, results from amino-terminal sequence analysis indicate that virion-associated ς1 protein is cleaved by trypsin between Arg245 and Ile246, which are proposed to form a portion of the ς1 neck (47).
FIG. 8.
Amino-terminal sequence analysis of ς1 cleavage products liberated during the generation of T3D ISVPs by using trypsin. (A) Isolation of a ς1 cleavage product by using anti-ς1 MAb G5. Purified 35S-labeled virions of T3D at a concentration of 2 × 1013 particles per ml were treated with 100 μg of trypsin per ml at 15°C for 30 min. Cleavage products were purified by using MAb G5-conjugated Sepharose, resolved in an SDS–14% polyacrylamide gel, and visualized by autoradiography. Lane 1, 4 × 1011 untreated viral particles; lane 2, 4 × 1011 viral particles treated with trypsin; lane 3, supernatant from trypsin digest (shown in lane 2) after incubation with MAb G5-conjugated Sepharose; lane 4, trypsin-generated virion cleavage products (from a total of 2 × 1012 viral particles) bound to MAb G5-conjugated sepharose. Viral proteins are labeled. Positions of molecular weight standards (in kilodaltons) are indicated. An arrow indicates the ς1 cleavage product (lane 4) isolated by using MAb G5-conjugated Sepharose. This band was used as a reference to identify the Coomassie blue-stained ς1 cleavage product (see Materials and Methods) subjected to amino-terminal sequence analysis. (B) Identification of the trypsin cleavage site in virion-associated T3D ς1 protein. Amino-terminal residues 1 through 8 of the trypsin-generated ς1 cleavage product are aligned with a region of sequence proposed to form the ς1 neck, amino acids 239 to 259 (47). Residues in boxes occur in the a or d position of a heptad repeat motif characteristic of α-helical coiled coils (42). This alignment indicates that trypsin cleaves ς1 between Arg245 and Ile246 during the generation of ISVPs. The cleavage site in ς1 primary sequence is indicated by an arrow.
Use of a murine intestinal wash to test the stability of type 3 ς1 protein during proteolytic conversion of virions to ISVPs.
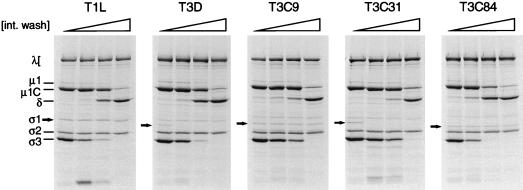
To determine whether a natural milieu of intestinal enzymes would reproduce the pattern of ς1 cleavage susceptibility observed after treatment of type 3 reovirus virions with purified chymotrypsin, virions were treated with the contents of an intestinal lavage from newborn mice. As a control for resistance of ς1 protein to proteolytic cleavage, we first treated virions of strain T1L with the intestinal wash (Fig. 9). The ς1 protein of this strain was shown previously to remain intact when ISVPs are generated in vitro by using chymotrypsin or trypsin (46) and when ISVPs are generated in the intestinal lumen of newborn mice (10). SDS-PAGE analysis of viral proteins showed that purified virions of T1L were converted to ISVPs upon treatment with the intestinal wash. The ς1 protein of these particles appeared fully intact, which indicates that ISVPs generated by this method are suitable for study of ς1 susceptibility to proteolysis. Purified virions of T3D, T3C9, T3C31, and T3C84 also were converted to ISVPs upon treatment with the intestinal wash (Fig. 9). Bands corresponding to full-length T3C9 and T3C84 ς1 proteins were not significantly different from those of untreated virions, even at the highest concentration of intestinal wash (∼83% [vol/vol]). However, the ς1 proteins of T3D and T3C31 were very susceptible to cleavage under these conditions; ς1 protein bands of these strains were not detectable at the lowest concentration of intestinal wash tested (∼4% [vol/vol]). Other than loss of ς1 protein, there were no remarkable differences between the ISVP protein profiles of the ς1-unstable and ς1-stable type 3 strains. The same cleavage profiles were observed when ISVPs were recovered in intestinal washes following intragastric inoculation of newborn mice with 35S-labeled virions (data not shown). These results mirror those obtained in assays using purified intestinal enzymes and strongly suggest that strain-specific differences in ς1 stability observed in assays using in vitro-generated ISVPs are also exhibited in the intestinal lumen.
FIG. 9.
Analysis of viral structural proteins following generation of ISVPs by using a murine intestinal wash. Purified 35S-labeled virions of T1L, T3D, T3C9, T3C31, or T3C84 at a concentration of 3.3 × 1012 particles per ml were treated with various concentrations of a murine intestinal wash (int. wash) at 20°C for 3.5 h. Aliquots of 12 μl were heated at 100°C in sample buffer and subjected to electrophoresis in an SDS–10% polyacrylamide gel, followed by autoradiography to visualize viral proteins. Viral proteins are labeled. The ς1 protein is indicated by an arrow.  , 0 to 83% (vol/vol) intestinal wash.
, 0 to 83% (vol/vol) intestinal wash.
DISCUSSION
We report here the identification of an amino acid residue in the neck region of T3D ς1 protein, Thr249, that determines susceptibility of ς1 to proteolytic cleavage during ISVP formation. Thr249 is not the site of protease action; rather, this residue mediates cleavage at a nearby site, Arg245. Algorithms to predict secondary structure suggest that the ς1 neck region is composed of sequences that form a four-stranded cross β sheet flanked by two short segments of α-helical coiled coil (47), with Thr249 occupying a position expected to stabilize interhelical contacts through hydrophobic interactions (Fig. 5). The pattern of ς1 protease sensitivity exhibited by type 3 reovirus ISVPs generated by using purified chymotrypsin was recapitulated when ISVPs were generated by using the contents of a murine intestinal wash. Our findings show that cleavage susceptibility of T3D ς1 protein is controlled indirectly by amino acid 249, perhaps through an effect on subunit interactions. Furthermore, these results establish a molecular model to explain viral infectivity loss in vitro (46) and in vivo (9, 32).
Identification of an amino acid residue in T3D ς1 protein that determines susceptibility to cleavage by protease.
The identification of a sequence polymorphism that correlates with ς1 cleavage susceptibility was facilitated by the characterization of type 3 reovirus strains that vary in infectivity loss and cleavage of ς1 following protease treatment of virions to generate ISVPs. By comparing deduced ς1 amino acid sequences of these strains, we found that both infectivity loss and ς1 cleavage exhibited by strains T3D and T3C31 are correlated with sequence polymorphism at a single amino acid position in ς1, residue 249 (Fig. 5). This correlation strongly suggests that amino acid 249 is the sole determinant of type 3 ς1 cleavage by protease but does not exclude the potential contribution of other amino acid positions where sequence similarity (as opposed to sequence identity) is correlated with cleavage sensitivity. In concordance with the sequence correlation, expressed ς1 protein substituted at Thr249 with an isoleucine was not cleaved by trypsin, despite nearly complete cleavage of wild-type protein (Fig. 6B). This result confirms that the amino acid residue at position 249 is an independent determinant of ς1 cleavage by protease.
Though conferring resistance to cleavage by trypsin, the Thr249→Ile replacement did not prevent cleavage by chymotrypsin (Fig. 6A). This result was unexpected since field isolate reovirus strains containing an isoleucine residue at ς1 amino acid position 249 have chymotrypsin-resistant ς1 proteins. However, our results do not necessarily suggest different mechanisms of ς1 cleavage sensitivity with respect to trypsin and chymotrypsin. It is possible that baculovirus-expressed type 3 ς1 protein adopts a conformation slightly different from that of virion-associated ς1 and that this conformation confers sensitivity to chymotrypsin cleavage by a mechanism unrelated to sequence polymorphism at position 249. Alternatively, resistance of type 3 ς1 protein to cleavage by chymotrypsin may require association of ς1 with other proteins of the reovirus virion, such as outer-capsid protein ς3 or core-spike protein λ2, both of which likely interact with ς1 (20, 29, 34, 38, 63).
Sites of ς1 cleavage during protease treatment of T3D virions to generate ISVPs.
We determined the site of ς1 cleavage on T3D ISVPs to better understand properties of receptor binding by virion and subvirion particles and to ascertain how amino acid position 249 influences ς1 cleavage susceptibility. An approximately 25-kDa trypsin-generated ς1 fragment, purified with a monoclonal antibody that binds the ς1 head (8), was found to contain an isoleucine residue at its amino terminus corresponding to Ile246 (Fig. 8B). Arg245 is most likely the singular site of trypsin cleavage since trypsin was found to cleave at this exact position in expressed T3D ς1 protein (22) and additional processing of the trypsin-generated carboxy-terminal fragment was not observed for expressed T3D ς1 protein (21, 22, 40; Fig. 6B) or ς1 purified from virions (70). Therefore, results of this analysis argue that trypsin cleaves virion-associated ς1 between Arg245 and Ile246 and that the particle-associated ς1 cleavage product consists of amino acids 1 to 245.
We attempted to determine the site of ς1 cleavage on ISVPs by chymotrypsin. However, it was not possible to isolate a stable chymotrypsin cleavage product of ς1 (5). This result is in agreement with the outcome of experiments using expressed ς1 protein in which we observed an array of cleavage products after chymotrypsin treatment, unlike the two stable fragments generated by trypsin (Fig. 6 and 7). A different set of experimental conditions will be necessary to identify the site of chymotrypsin cleavage in virion-associated ς1.
Relationship of the neck region to receptor-binding domains in type 3 ς1 protein.
In studies of attachment by T3D ISVPs, we found that following ς1 cleavage, sequences in ς1 that bind sialic acid remain particle associated despite loss of a receptor-binding domain in the ς1 head (46). Residues in ς1 shown to determine the capacity of type 3 reovirus to bind sialylated receptors, Asn198, Arg202, and Pro204, are contained in a region of predicted cross β sheet in the ς1 tail immediately amino terminal to the neck (16, 17). Thus, we have proposed a model of type 3 ς1 structure in which the neck region bridges discrete attachment domains, one in the tail that binds sialic acid and another in the head that binds an unidentified receptor (16, 46). This model is supported by results of amino-terminal sequence analysis of T3D ς1 proteolytic cleavage products liberated during the generation of ISVPs (Fig. 8B). The cleavage site mapped by using trypsin is carboxy terminal to sequences in the tail that determine sialic acid binding and amino terminal to sequences in the head that bind a receptor on L cells (41, 45, 67, 70). Results of the sequence determination are also consistent with the finding that HA titers of T3D and T3C31 ISVPs were not lower than those of intact virions (Fig. 4). In fact, ς1 sequences that bind sialic acid appear to be more accessible to sialylated ligands on the erythrocyte surface since the HA titer increased when type 3 reovirus virions were converted to ISVPs by using chymotrypsin. Based on these results, we conclude that type 3 ς1 protein possesses a modular arrangement of receptor-binding domains and that binding of sialic acid is a function of the tail, perhaps directly involving sequences predicted to form a β-sheet motif.
Mechanism of type 3 ς1 protein susceptibility to cleavage by proteases.
The amino terminal of the two proposed coiled-coil units in the ς1 neck contains three classical heptad repeats in which apolar residues occupy the first and fourth amino acid positions (47). Residue 249 is located in the d position of the second heptad repeat (Fig. 5). Since trypsin and chymotrypsin preferentially cleave adjacent to basic and bulky aromatic residues, respectively (12), a threonine at position 249 in T3D ς1 is not predicted to be the site of protease action. Concordantly, no cleavage at this site was indicated by amino-terminal sequence analysis of T3D ς1 cleavage products (Fig. 8B). One plausible mechanism of cleavage sensitivity is that the presence of threonine disrupts hydrophobic contacts between apposed α helices, which then allows protease to attack a neighboring target sequence. Conversely, the presence of an isoleucine is predicted to favor a more stable interhelical association and thereby shield the protease target site. This model is supported by the finding that protease resistance could be engineered by replacement of Thr249 with isoleucine as well as another hydrophobic amino acid, leucine, but not by replacement with a polar amino acid, asparagine (Fig. 6B). Additionally, a trypsin cleavage site (Arg245) in virion-associated T3D ς1 protein was identified within four residues of Thr249, which is consistent with the prediction that interfacial contacts of an α-helical coiled coil would be locally destabilized by the occurrence of a polar residue in the d position of a heptad repeat. The ς1 oligomer has been modeled as either a coiled-coil homotrimer (40, 58) or a pair of parallel coiled-coil homodimers (24). It is possible that a polar residue at position 249 makes protease cleavage sites accessible by causing a localized decrease in the compactness of either a dimeric or trimeric coiled coil or by indirectly perturbing the stable association between pairs of coiled-coil dimers.
Our experiments do not directly address putative conformational differences in the ς1 neck in its cleavage-susceptible and cleavage-resistant states, and it is possible that the effect of Thr249 is to disrupt intramolecular ς1 structure. Confirmation of the mechanism of ς1 cleavage susceptibility will require further characterization of the neck region by using biophysical techniques.
Cleavage of ς1 protein and reovirus pathogenesis.
The cleavage status of type 3 ς1 protein in vivo is unknown. Virions of T1L orally inoculated into newborn mice are rapidly converted to ISVPs in the intestinal lumen (10). Furthermore, proteolytic processing of virions to ISVPs is an obligate step in reovirus infectivity in the intestine (1, 6). A reasonable inference from these findings is that the ς1 proteins of T3D and T3C31 are cleaved in the murine intestine. We tested this possibility by treating virions with the contents of a murine intestinal lavage under conditions that lead to the generation of ISVPs. Examination of viral structural proteins by SDS-PAGE showed that the ς1 proteins of both strains were cleaved under these conditions; however, the ς1 proteins of strains T3C9 and T3C84 remained intact (Fig. 9). This pattern of ς1 cleavage sensitivity replicates results obtained for both virions and expressed ς1 protein in in vitro cleavage assays with purified protease (Fig. 3, 6B, and 7). Therefore, these results indicate that ς1 proteins of type 3 reovirus strains are differentially susceptible to cleavage during the course of natural infection.
T3D is avirulent when infection is initiated in the intestine (31, 54, 68), even though this virus is neurotropic (23, 61, 65) and highly neurovirulent (23, 31, 44, 57, 62) following intramuscular or intracranial inoculation. Therefore, cleavage of T3D ς1 protein by intraluminal proteases, as shown here, may contribute to the avirulence of this strain in newborn mice inoculated orally. This model is supported by linkage of the S1 gene to differences in T1L and T3D growth and spread following peroral inoculation (9, 32). Verification of this hypothesis will require genetic analysis of the in vivo cleavage susceptibility of T3D ς1. However, if ς1 stability in the intestinal lumen is a virulence determinant of reovirus, it is not necessarily the predominant influence on virulence after oral inoculation; 50% lethal doses reported for type 3 strains (including T3D, T3C9, T3C31, and T3C84) (68) do not correlate with patterns of ς1 cleavage susceptibility observed in this study. We are conducting studies on a variant of T3D adapted to growth in murine intestinal tissue (28) to better understand the precise relationship between ς1 stability and reovirus pathogenesis.
Function of the ς1 neck domain.
The neck region likely exhibits considerable flexibility since this portion of the protein is highly sensitive to proteolysis (22; Fig. 8B) and shows enhanced curvature in analyses of ς1 visualized by electron microscopy (14, 24, 25). Flexibility in the ς1 neck may optimally orient sequences in the head and tail for receptor engagement and viral entry. This situation would be similar to attachment by bacteriophage T4, where the latent receptor-binding domain in the short tail fiber is repositioned to an active orientation by extension of the fiber at a proposed hinge (43). Another function of the ς1 neck may be to promote intraendosomal virion disassembly by undergoing conformational adjustments that alter interactions between ς1 and neighboring outer-capsid components, such as the ς3 protein (34, 38, 63). Indeed, mutations in both ς1 and ς3 proteins confer the ability of virions to bypass blocks to disassembly within the endosome (66, 69), suggesting a cooperative interaction of ς1 with ς3 in the dissociation of outer-capsid proteins. Additional sites in the ς1 tail, near the middle and near the virion-proximal amino terminus, also show evidence of flexibility (24), and concerted adjustments at these positions and the neck region may facilitate the dramatic conformational change in ς1 observed when the outer capsid is degraded by protease to produce an ISVP (25). Functions of the ς1 neck in reovirus infection will become clearer as the structure and conformational dynamics in this region of ς1 are elucidated.
ACKNOWLEDGMENTS
We thank Mehmet Goral, Patrick Green, and Gerald Stubbs for critical review of the manuscript. We also thank Joy Duong of the Elizabeth B. Lamb Center for Pediatric Research and Eric Howard and Masaaki Tamura of the Protein Chemistry Laboratory, Department of Biochemistry, Vanderbilt University School of Medicine, for expert technical assistance.
This research was supported by PHS awards AI39533 (M.L.N.) and AI38296 (J.D.C. and T.S.D.) from the National Institute of Allergy and Infectious Diseases, PHS award T32 GM07347 from the National Institute of General Medical Studies for the Vanderbilt Medical-Scientist Training Program (G.S.B. and D.T.D.), and Vanderbilt Diabetes Research and Training Center grant P60 DK20593. This work also was supported by the National Science Foundation and University Research Council, Vanderbilt University (E.S.B.), an Amos Christie fellowship from the Department of Pediatrics at Vanderbilt University School of Medicine (T.H.S.), and the Elizabeth B. Lamb Center for Pediatric Research.
REFERENCES
- 1.Amerongen H M, Wilson G A R, Fields B N, Neutra M R. Proteolytic processing of reovirus is required for adherence to intestinal M cells. J Virol. 1994;68:8428–8432. doi: 10.1128/jvi.68.12.8428-8432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong G D, Paul R W, Lee P W. Studies on reovirus receptors of L cells: virus binding characteristics and comparison with reovirus receptors of erythrocytes. Virology. 1984;138:37–48. doi: 10.1016/0042-6822(84)90145-4. [DOI] [PubMed] [Google Scholar]
- 3.Baer G S, Dermody T S. Mutations in reovirus outer-capsid protein ς3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J Virol. 1997;71:4921–4928. doi: 10.1128/jvi.71.7.4921-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjea A C, Brechling K A, Ray C A, Erikson H, Pickup D J, Joklik W K. High-level synthesis of biologically active reovirus protein ς1 in a mammalian expression vector system. Virology. 1988;167:601–612. [PubMed] [Google Scholar]
- 5.Barton, E. S., and T. S. Dermody. Unpublished observation.
- 6.Bass D M, Bodkin D, Dambrauskas R, Trier J S, Fields B N, Wolf J L. Intraluminal proteolytic activation plays an important role in replication of type 1 reovirus in the intestines of neonatal mice. J Virol. 1990;64:1830–1833. doi: 10.1128/jvi.64.4.1830-1833.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassel-Duby R, Nibert M, Homcy C, Fields B, Sawutz D. Evidence that the sigma 1 protein of reovirus serotype 3 is a multimer. J Virol. 1987;61:1834–1841. doi: 10.1128/jvi.61.6.1834-1841.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassel-Duby R, Spriggs D R, Tyler K L, Fields B N. Identification of attenuating mutations on the reovirus type 3 S1 double-stranded RNA segment with a rapid sequencing technique. J Virol. 1986;60:64–67. doi: 10.1128/jvi.60.1.64-67.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodkin D K, Fields B N. Growth and survival of reovirus in intestinal tissue: role of the L2 and S1 genes. J Virol. 1989;63:1188–1193. doi: 10.1128/jvi.63.3.1188-1193.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodkin D K, Nibert M L, Fields B N. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J Virol. 1989;63:4676–4681. doi: 10.1128/jvi.63.11.4676-4681.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borsa J, Sargent M D, Lievaart P A, Copps T P. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology. 1981;111:191–200. doi: 10.1016/0042-6822(81)90664-4. [DOI] [PubMed] [Google Scholar]
- 12.Branden C, Tooze J. Introduction to protein structure. N.Y: Garland; 1991. pp. 231–246. [Google Scholar]
- 13.Burstin S J, Spriggs D R, Fields B N. Evidence for functional domains on the reovirus type 3 hemagglutinin. Virology. 1982;117:146–155. doi: 10.1016/0042-6822(82)90514-1. [DOI] [PubMed] [Google Scholar]
- 14.Centonze V E, Chen Y, Severson T F, Borisy G G, Nibert M L. Visualization of individual reovirus particles by low-temperature, high-resolution scanning electron microscopy. J Struct Biol. 1995;115:215–225. doi: 10.1006/jsbi.1995.1046. [DOI] [PubMed] [Google Scholar]
- 15.Chang C T, Zweerink H J. Fate of parental reovirus in infected cell. Virology. 1971;46:544–555. doi: 10.1016/0042-6822(71)90058-4. [DOI] [PubMed] [Google Scholar]
- 16.Chappell J D, Gunn V L, Wetzel J D, Baer G S, Dermody T S. Mutations in type 3 reovirus that determine binding to sialic acid are contained in the fibrous tail domain of viral attachment protein ς1. J Virol. 1997;71:1834–1841. doi: 10.1128/jvi.71.3.1834-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dermody T S, Nibert M L, Bassel-Duby R, Fields B N. A ς1 region important for hemagglutination by type 3 reovirus strains. J Virol. 1990;64:5173–5176. doi: 10.1128/jvi.64.10.5173-5176.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dermody T S, Nibert M L, Bassel-Duby R, Fields B N. Sequence diversity in S1 genes and S1 translation products of eleven type 3 reovirus strains. J Virol. 1990;64:4842–4850. doi: 10.1128/jvi.64.10.4842-4850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dermody T S, Nibert M L, Wetzel J D, Tong X, Fields B N. Cells and viruses with mutations affecting viral entry are selected during persistent infections of L cells with mammalian reoviruses. J Virol. 1993;67:2055–2063. doi: 10.1128/jvi.67.4.2055-2063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dryden K A, Wang G, Yeager M, Nibert M L, Coombs K M, Furlong D B, Fields B N, Baker T S. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duncan R, Horne D, Strong J E, Leone G, Pon R T, Yeung M C, Lee P W K. Conformational and functional analysis of the C-terminal globular head of the reovirus cell attachment protein. Virology. 1991;182:810–819. doi: 10.1016/0042-6822(91)90622-i. [DOI] [PubMed] [Google Scholar]
- 22.Duncan R, Lee P W K. Localization of two protease-sensitive regions separating distinct domains in the reovirus cell-attachment protein ς1. Virology. 1994;203:149–152. doi: 10.1006/viro.1994.1465. [DOI] [PubMed] [Google Scholar]
- 23.Flamand A, Gagner J P, Morrison L A, Fields B N. Penetration of the nervous systems of suckling mice by mammalian reoviruses. J Virol. 1991;65:123–131. doi: 10.1128/jvi.65.1.123-131.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser R D, Furlong D B, Trus B L, Nibert M L, Fields B N, Steven A C. Molecular structure of the cell-attachment protein of reovirus: correlation of computer-processed electron micrographs with sequence-based predictions. J Virol. 1990;64:2990–3000. doi: 10.1128/jvi.64.6.2990-3000.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furlong D B, Nibert M L, Fields B N. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988;62:246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentsch J R, Pacitti A F. Effect of neuraminidase treatment of cells and effect of soluble glycoproteins on type 3 reovirus attachment to murine L cells. J Virol. 1985;56:356–364. doi: 10.1128/jvi.56.2.356-364.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentsch J R, Pacitti A F. Differential interaction of reovirus type 3 with sialylated receptor components on animal cells. Virology. 1987;161:245–248. doi: 10.1016/0042-6822(87)90192-9. [DOI] [PubMed] [Google Scholar]
- 28.Haller B L, Barkon M L, Li X-Y, Hu W M, Wetzel J D, Dermody T S, Virgin H W., IV Brain- and intestine-specific variants of reovirus serotype 3 strain Dearing are selected during chronic infection of severe combined immunodeficient mice. J Virol. 1995;69:3933–3937. doi: 10.1128/jvi.69.6.3933-3937.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes E C, Lee P W K, Miller S E, Joklik W K. The interaction of a series of hybridoma IgGs with reovirus particles: demonstration that the core protein λ2 is exposed on the particle surface. Virology. 1981;108:147–155. doi: 10.1016/0042-6822(81)90534-1. [DOI] [PubMed] [Google Scholar]
- 30.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 31.Hrdy D B, Rubin D H, Fields B N. Molecular basis of reovirus neurovirulence: role of the M2 gene in avirulence. Proc Natl Acad Sci USA. 1982;79:1298–1302. doi: 10.1073/pnas.79.4.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kauffman R S, Wolf J L, Finberg R, Trier J S, Fields B N. The ς1 protein determines the extent of spread of reovirus from the gastrointestinal tract of mice. Virology. 1983;124:403–410. doi: 10.1016/0042-6822(83)90356-2. [DOI] [PubMed] [Google Scholar]
- 33.Kaye K M, Spriggs D R, Bassel-Duby R, Fields B N, Tyler K L. Genetic basis for altered pathogenesis of an immune-selected antigenic variant of reovirus type 3 Dearing. J Virol. 1986;59:90–97. doi: 10.1128/jvi.59.1.90-97.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kedl R, Schmechel S, Schiff L. Comparative sequence analysis of the reovirus S4 genes from 13 serotype 1 and serotype 3 field isolates. J Virol. 1995;69:552–559. doi: 10.1128/jvi.69.1.552-559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keroack M, Fields B N. Viral shedding and transmission between hosts determined by reovirus L2 gene. Science. 1986;232:1635–1638. doi: 10.1126/science.3012780. [DOI] [PubMed] [Google Scholar]
- 36.Kowalik T F, Yang Y-Y, Li J K-K. Molecular cloning and comparative sequence analyses of bluetongue virus S1 segments by selective synthesis of specific full-length DNA copies of dsRNA genes. Virology. 1990;177:820–823. doi: 10.1016/0042-6822(90)90557-8. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Lee P W K, Hayes E C, Joklik W K. Protein ς1 is the reovirus cell attachment protein. Virology. 1981;108:156–163. doi: 10.1016/0042-6822(81)90535-3. [DOI] [PubMed] [Google Scholar]
- 39.Leone G, Duncan R, Lee P W K. Trimerization of the reovirus cell attachment protein (ς1) induces conformational changes in ς1 necessary for its cell-binding function. Virology. 1991;184:758–761. doi: 10.1016/0042-6822(91)90447-j. [DOI] [PubMed] [Google Scholar]
- 40.Leone G, Duncan R, Mah D C, Price A, Cashdollar L W, Lee P W K. The amino-terminal heptad repeat region of reovirus cell attachment protein ς1 is responsible for ς1 oligomer stability and possesses intrinsic oligomerization function. Virology. 1991;182:336–345. doi: 10.1016/0042-6822(91)90677-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leone G, Maybaum L, Lee P W K. The reovirus cell attachment protein possesses two independently active trimerization domains: basis of dominant negative effects. Cell. 1992;71:479–488. doi: 10.1016/0092-8674(92)90516-f. [DOI] [PubMed] [Google Scholar]
- 42.Lupus A. Coiled coils: new structures and new functions. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 43.Makhov A M, Trus B L, Conway J F, Simon M N, Zurabishvili T G, Mesyanzhinov V V, Steven A C. The short tail-fiber of bacteriophage T4: molecular structure and a mechanism for its conformational transition. Virology. 1993;194:117–127. doi: 10.1006/viro.1993.1241. [DOI] [PubMed] [Google Scholar]
- 44.Morrison L A, Fields B N, Dermody T S. Prolonged replication in the mouse central nervous system of type 3 reoviruses isolated from persistently infected L-cell cultures. J Virol. 1993;67:3019–3026. doi: 10.1128/jvi.67.6.3019-3026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagata L, Masri S A, Pon R T, Lee P W K. Analysis of functional domains on reovirus cell attachment protein ς1 using cloned S1 gene deletion mutants. Virology. 1987;160:162–168. doi: 10.1016/0042-6822(87)90056-0. [DOI] [PubMed] [Google Scholar]
- 46.Nibert M L, Chappell J D, Dermody T S. Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved ς1 protein. J Virol. 1995;69:5057–5067. doi: 10.1128/jvi.69.8.5057-5067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nibert M L, Dermody T S, Fields B N. Structure of the reovirus cell-attachment protein: a model for the domain organization of ς1. J Virol. 1990;64:2976–2989. doi: 10.1128/jvi.64.6.2976-2989.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nibert M L, Fields B N. Early steps in reovirus infection of cells. In: Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 341–364. [Google Scholar]
- 49.Paul R W, Choi A H, Lee P W K. The α-anomeric form of sialic acid is the minimal receptor determinant recognized by reovirus. Virology. 1989;172:382–385. doi: 10.1016/0042-6822(89)90146-3. [DOI] [PubMed] [Google Scholar]
- 50.Paul R W, Lee P W K. Glycophorin is the reovirus receptor on human erythrocytes. Virology. 1987;159:94–101. doi: 10.1016/0042-6822(87)90351-5. [DOI] [PubMed] [Google Scholar]
- 51.Rosen L, Abinanti F R. Natural and experimental infection of cattle with human types of reovirus. Am J Hyg. 1960;71:424–429. doi: 10.1093/oxfordjournals.aje.a120108. [DOI] [PubMed] [Google Scholar]
- 52.Rosen L, Abinanti F R, Hovis J F. Further observations on the natural infection of cattle with reoviruses. Am J Hyg. 1963;77:38–48. doi: 10.1093/oxfordjournals.aje.a120294. [DOI] [PubMed] [Google Scholar]
- 53.Rosen L, Hovis J F, Mastrota F M, Bell J A, Huebner R J. Observations on a newly recognized virus (Abney) of the reovirus family. Am J Hyg. 1960;71:258–265. doi: 10.1093/oxfordjournals.aje.a120109. [DOI] [PubMed] [Google Scholar]
- 54.Rubin D H, Fields B N. Molecular basis of reovirus virulence: role of the M2 gene. J Exp Med. 1980;152:853–868. doi: 10.1084/jem.152.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubin D H, Wetzel J D, Dworkin C, Williams W V, Cohen J A, Dermody T S. Binding of type 3 reovirus by a domain of the ς1 protein important for hemagglutination leads to infection of murine erythroleukemia cells. J Clin Invest. 1992;90:2536–2542. doi: 10.1172/JCI116147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silverstein S C, Astell C, Levin D H, Schonberg M, Acs G. The mechanism of reovirus uncoating and gene activation in vivo. Virology. 1972;47:797–806. doi: 10.1016/0042-6822(72)90571-5. [DOI] [PubMed] [Google Scholar]
- 57.Spriggs D R, Fields B N. Attenuated reovirus type 3 strains generated by selection of haemagglutinin antigenic variants. Nature. 1982;297:68–70. doi: 10.1038/297068a0. [DOI] [PubMed] [Google Scholar]
- 58.Strong J E, Leone G, Duncan R, Sharma R K, Lee P W K. Biochemical and biophysical characterization of the reovirus cell attachment protein ς1: evidence that it is a homotrimer. Virology. 1991;184:23–32. doi: 10.1016/0042-6822(91)90818-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sturzenbecker L J, Nibert M, Furlong D, Fields B N. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J Virol. 1987;61:2351–2361. doi: 10.1128/jvi.61.8.2351-2361.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turner D L, Duncan R, Lee P W K. Site-directed mutagenesis of the C-terminal portion of reovirus protein ς1: evidence for a conformation-dependent receptor binding domain. Virology. 1992;186:219–227. doi: 10.1016/0042-6822(92)90076-2. [DOI] [PubMed] [Google Scholar]
- 61.Tyler K L, Bronson R T, Byers K B, Fields B N. Molecular basis of viral neurotropism: experimental reovirus infection. Neurology. 1985;35:88–92. doi: 10.1212/wnl.35.1.88. [DOI] [PubMed] [Google Scholar]
- 62.Virgin H W, IV, Bassel-Duby R, Fields B N, Tyler K L. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing) J Virol. 1988;62:4594–4604. doi: 10.1128/jvi.62.12.4594-4604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Virgin H W, IV, Mann M A, Fields B N, Tyler K L. Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J Virol. 1991;65:6772–6781. doi: 10.1128/jvi.65.12.6772-6781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Virgin H W, IV, Tyler K L, Dermody T S. Reovirus. In: Nathanson N, editor. Viral pathogenesis. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 669–699. [Google Scholar]
- 65.Weiner H L, Powers M L, Fields B N. Absolute linkage of virulence and central nervous system tropism of reoviruses to viral hemagglutinin. J Infect Dis. 1980;141:609–616. doi: 10.1093/infdis/141.5.609. [DOI] [PubMed] [Google Scholar]
- 66.Wetzel J D, Wilson G J, Baer G S, Dunnigan L R, Wright J P, Tang D S H, Dermody T S. Reovirus variants selected during persistent infections of L cells contain mutations in the viral S1 and S4 genes and are altered in viral disassembly. J Virol. 1997;71:1362–1369. doi: 10.1128/jvi.71.2.1362-1369.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams W V, Guy H R, Rubin D H, Robey F, Myers J N, Kieber-Emmons T, Weiner D B, Greene M I. Sequences of the cell-attachment sites of reovirus type 3 and its antiidiotypic/antireceptor antibody: modeling of their three-dimensional structures. Proc Natl Acad Sci USA. 1988;85:6488–6492. doi: 10.1073/pnas.85.17.6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson G A R, Morrison L A, Fields B N. Association of the reovirus S1 gene with serotype 3-induced biliary atresia in mice. J Virol. 1994;68:6458–6465. doi: 10.1128/jvi.68.10.6458-6465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson G J, Wetzel J D, Puryear W, Bassel-Duby R, Dermody T S. Persistent reovirus infections of L cells select mutations in viral attachment protein ς1 that alter oligomer stability. J Virol. 1996;70:6598–6606. doi: 10.1128/jvi.70.10.6598-6606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yeung M C, Lim D, Duncan R, Shahrabadi M S, Cashdollar L W, Lee P W K. The cell attachment proteins of type 1 and type 3 reovirus are differentially susceptible to trypsin and chymotrypsin. Virology. 1989;170:62–70. doi: 10.1016/0042-6822(89)90352-8. [DOI] [PubMed] [Google Scholar]