Abstract
Dormancy represents a fascinating adaptive strategy for organisms to survive in unforgiving environments. After a period of dormancy, organisms often exhibit exceptional resilience. This period is typically divided into hibernation and aestivation based on seasonal patterns. However, the mechanisms by which organisms adapt to their environments during dormancy, as well as the potential relationships between different states of dormancy, deserve further exploration. Here, we selected Perccottus glenii and Protopterus annectens as the primary subjects to study hibernation and aestivation, respectively. Based on histological and transcriptomic analysis of multiple organs, we discovered that dormancy involved a coordinated functional response across organs. Enrichment analyses revealed noteworthy disparities between the two dormant species in their responses to extreme temperatures. Notably, similarities in gene expression patterns pertaining to energy metabolism, neural activity, and biosynthesis were noted during hibernation, suggesting a potential correlation between hibernation and aestivation. To further explore the relationship between these two phenomena, we analyzed other dormancy-capable species using data from publicly available databases. This comparative analysis revealed that most orthologous genes involved in metabolism, cell proliferation, and neural function exhibited consistent expression patterns during dormancy, indicating that the observed similarity between hibernation and aestivation may be attributable to convergent evolution. In conclusion, this study enhances our comprehension of the dormancy phenomenon and offers new insights into the molecular mechanisms underpinning vertebrate dormancy.
Keywords: Hibernation, Aestivation, Multi-organs, Convergent evolution
INTRODUCTION
Hibernation is a common phenomenon in nature and an important means for organisms to survive winter conditions (Possmayer et al., 2010). Hibernation has garnered extensive research interest across multiple disciplines, including medicine, biology, and ecology, due to its potential applications in wide-ranging endeavors, from interstellar travel to the treatment of terminal illnesses. Thus, scholars have undertaken comprehensive research on hibernation to elucidate its underlying mechanism and potential functions. At the macro level, organisms entering hibernation typically display reduced activity and metabolic rates until the end of winter (Geiser, 2004). At the micro level, hibernation involves complex physiological processes that require the coordination of various organs and biological pathways. Jansen et al. (2019) observed notable variations in gene expression related to urea production and actin synthesis in three organs of grizzly bears during hibernation, implying complex multi-organ coordination. Furthermore, although hibernation is associated with decreased bone density due to inactivity, studies have not shown a significant decline in locomotor capabilities post-revival. Research on Daurian ground squirrels has shown that inhibition of the Wnt signaling pathway during hibernation leads to elevated levels of the GSK-3β protein, mitigating calcium loss (Gao et al., 2023). In addition, water maintenance during hibernation plays a crucial role in successful overwintering. While extremely cold temperatures can exacerbate the transpiration of liquid water, hibernating animals counteract this by accumulating substantial urea in their bodies, increasing body fluid concentration and reducing the risk of freezing (Zhong & Wang, 2022). Interestingly, hibernation may influence longevity, with hibernating species often exhibiting longer than expected lifespans. Population studies on yellow-bellied marmots have shown significant age stagnation during hibernation in females (Pinho et al., 2022), while studies on hibernating bats have revealed a slowing down of senescence (Sullivan et al., 2022). Recent discoveries have also noted the hibernation capabilities of certain fish species, such as Perccottus glenii (Dybowski, 1877), which can enter a state of hibernation in frozen waters, primarily in northern China (Li et al., 2018). This fish demonstrates a similar hibernation process as that of amphibians and reptiles, capable of surviving in a completely frozen state (Chai et al., 2020). Further research on the resilience of Perccottus glenii to endure freezing temperatures has been conducted using multi-omics analysis (Jiang et al., 2023).
Research into the natural world has expanded our understanding of biological dormancy beyond just hibernation. Studies have revealed that certain organisms residing in hot regions voluntarily enter a state of high temperature-induced dormancy, known as aestivation, prior to the onset of the dry season to mitigate the detrimental effects of extreme heat, standing in contrast to cold temperature-initiated hibernation (Delaney et al., 1974). Currently, there are fewer animals capable of hibernation compared to those possessing the ability to aestivate. Among vertebrates, only one species, Protopterus annectens (Owen, 1839), exhibits a distinct aestivation phenomenon (Chew et al., 2004). Protopterus annectens is an ancient fish native to south-central Africa, distinguished not only by its aestivation capability but also by its primitive lungs. These lungs enable Protopterus annectens to meet its oxygen requirements in situations of low water oxygen levels or when gill respiration is impractical. It is these aforementioned characteristics that render this species exceptionally adaptable to inhospitable environments. Despite limited research on aestivation, studies on Protopterus annectens have provided key insights into this form of dormancy. The major challenge during summer dormancy is coping with high temperatures and water scarcity. Urea has been identified as playing a crucial role in maintaining osmotic pressure and retaining water in Protopterus annectens, with increases in urea synthesis enzyme levels observed across various phases of aestivation (Loong et al., 2012). The expression levels of aqp1 and aqp3 exhibit changes in Protopterus annectens in response to dehydration and rehydration during aestivation (Chng et al., 2016). Additionally, the nitric oxide synthase/nitric oxide (NOS/NO) pathway undergoes tissue-specific modifications in Protopterus annectens during this period, in sync with organ repositioning (Garofalo et al., 2015). Moreover, myostatin levels are up-regulated in Protopterus annectens during aestivation periods to counteract muscle atrophy in vivo and preserve muscle structure (Ong et al., 2017). Research has also indicated that blood flow rate decreases during long-term hibernation, with a potential risk of injury from the sudden increase in flow upon reawakening. Notably, studies have suggested that a reduction in nkaα expression may be induced to mitigate the risks of ischemia and perfusion injury during the dormancy and revival processes (Hiong et al., 2014).
Macroscopic physiological phenomena that share similar characteristics often suggest comparable underlying molecular mechanisms (Berens et al., 2015; Pankey et al., 2014). Despite the distinct triggers for hibernation and aestivation, both forms of dormancy are characterized by motor stagnation and reduced metabolism, hinting at a possible link between the two. In this study, Perccottus glenii was investigated to explore the mechanisms of hibernation, while Protopterus annectens was studied in the context of aestivation. Comparative transcriptomics was used to identify and analyze general physiological changes that occur during dormancy, as well as to delineate the molecular differences between hibernation and aestivation. This study aims to provide a scientific foundation for a deeper understanding of dormancy across different organisms.
MATERIALS AND METHODS
Ethics statement
All procedures were performed following relevant guidelines and regulations and under the approval of the Ethics Committee of the Institute of Hydrobiology, Chinese Academy of Sciences (Approval No.: Y21304506).
Tissue acquisition of hibernation samples
Active and hibernating Perccottus glenii fish samples were collected from Keshan County, Qiqihar City, Heilongjiang Province, China, in September 2020 and February 2021, respectively. For each collection session, six wild adult fish samples were obtained without distinguishing sex. Immediately upon collection, tissue samples from six different organs, including the gill, gut, heart, kidney, skin, and spleen, were excised and preserved in liquid nitrogen at the collection site.
Tissue acquisition of aestivation samples
Adult specimens of Protopterus annectens (body length 50–100 cm) were collected from South Central Africa and imported through a local fish farm in Guangzhou, China. The samples were maintained in the laboratory in glass aquaria filled with dechlorinated water (2.3×10-3 mol/L Na+, 5.4×10-4 mol/L K+, 9.5×10-4 mol/L Ca2+, 8×10-5 mol/L Mg2+, 3.4×10-3 mol/L Cl-, and 6×10-4 mol/L HCO3-, pH 7.0, 25°C), which was changed daily, with no separation by sex. The fish underwent a month-long acclimatization period to the laboratory conditions, during which they were fed frozen fish daily and maintained under artificial lighting that simulated the duration and intensity of daylight corresponding to their native habitat. Food was withdrawn 96 h before any experimental procedures for both control and aestivating Protopterus annectens to ensure sufficient time for the gut to be completely cleared (Chng et al., 2017b). Control fish (n=6) raised in freshwater were used as controls and euthanized with an excess of neutralized 0.05% MS222, with tissue samples then collected and stored at −80°C. To induce aestivation, some Protopterus annectens were individually placed in plastic tanks (Length 29 cm×Width 19 cm×Height 17.5 cm) containing 15 mL of dechlorinated tap water, adjusted to 0.3‰ salinity with seawater, at 27–29°C and 85%–90% humidity. Within approximately 6 days, the fish formed dried brown mucus cocoons, marking the commencement of the aestivation period, which lasted for 6 months. To ensure high humidity (>90%) levels within the tank, 1–2 mL of water was sprayed onto the side of the tank daily. After 6 months, the mud cocoons were manually opened, and the fish were euthanized using 0.05% MS222 neutralizer (n=6 for each group). Eleven different tissue samples (brain, eye, gill, gut, heart, kidney, liver, lung, muscle, skin, and spinal cord) were collected and stored at −80°C.
Transcriptome data acquisition
Individual frozen tissue samples of Protopterus annectens and Perccottus glenii were homogenized in 1 mL of TRIzol reagent (Takara, China). RNA was extracted according to the manufacturer’s instructions and treated with RNase-free DNase I (NEB, USA) for 30 min at 37°C to remove residual DNA. RNA quality was determined using 1.2% EtBr-agarose gels, and RNA quantity (A260/A280) was measured with a Nanodrop 2000 spectrophotometer (Thermo Scientific, USA) and 2100 Bioanalyzer (Agilent Technologies, USA). Poly(A) mRNA was purified from total RNA using a NEBNext poly(A) mRNA Magnetic Isolation Module (NEB, USA). Furthermore, cDNA libraries were prepared using the NEBNext mRNA Library Prep Master Mix Set for Illumina (NEB, USA) and NEBNext Multiplex Oligos for Illumina (NEB, USA). Fragment lengths in the cDNA libraries were measured using 1.8% EtBr-agarose gels. Quantitative real-time polymerase chain reaction (qPCR) was performed using the Library Quantification Kit-Illumina GA Universal (Kapa, USA). Qualified cDNA libraries were then immobilized in an Illumina cBot to generate clusters and sequenced using the Illumina HiSeqTM 2500 platform (Illumina, USA) at Biomarker Technologies. Raw sequencing data were uploaded to the NCBI database. In addition, the scope of the study was expanded to include liver transcriptomic data of Ursus arctos (PRJNA413091), Bufo gargarizans (PRJNA378697), Nanorana parkeri (PRJCA010139), Alligator sinensis (PRJNA556093), and Perccottus glenii (PRJNA818152) during hibernation and non-hibernation from the NCBI database (https://www.ncbi.nlm.nih.gov/) for further exploration.
Transcriptome assembly and expression estimation
FastQC (v.0.12.0) was used to check the quality of the raw RNA-seq reads. Only paired-end reads longer than 50 bp at either end after trimming were retained for subsequent analysis. High-quality paired reads from samples were separately mapped to the reference genome using HISAT2 (v.2.2.1). StringTie (v.2.28.0) was then used to generate gene expression level counts in fragments per kilobase of transcript per million fragments mapped (FPKM). Principal component analysis (PCA) was performed based on the expression patterns of all genes. Differentially expressed genes (DEGs) in the samples were screened using the R package DESeq2 (v.1.36.0). A false discovery rate (FDR)-adjusted P-value of less than 0.05 and |Log2FoldChange| of less than 1 were set as the level of significance.
Functional enrichment and orthogroup identification
EggNog (https://eggnog-mapper.embl.de/) was used for online annotation of the coding sequences of each species sample. The R package AnnotationForge (v.1.38.1) was then used for further analysis of functional enrichment required by the database. Enrichment of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) in DEGs was estimated using the annotations of all identified transcripts as a background. TransDecoder (v.5.7.0) was used to predict open reading frames (ORFs), with these predicted ORFs then aligned against the UniProt protein database using BLASTP to provide homology evidence. OrthoFinder was used to categorize genes into “groups” (orthogroups) to facilitate analysis of expression levels across various species. This approach involves classifying genes that evolved from a single ancestral gene sequence into orthogroups based on the analysis of the ancestral species.
Real-time quantitative PCR assay
cDNA samples were generated from mRNA using reverse transcriptase (Promega, USA) in the presence of an RNase inhibitor (Invitrogen, USA) and oligo(dT) and dNTP cocktail (Takara, China). To validate the expression patterns discovered using RNA sequencing (RNA-seq) analyses, qPCR was performed using SYBR Green (Roche, Switzerland) chemistry on a Light Cycle® 480 II (Roche, Switzerland). Based on the sequence of the assembled transcriptome, we used Premier 5 to design the required primers for subsequent experiments(Van-Beers et al., 1998). Three replicates for each gene were analyzed, with beta-actin serving as the internal control. The PCR cycling program consisted of 45 three-step cycles of 95°C for 10 s, 55°C for 10 s, and 72°C for 20 s. To confirm signal specificity, a melting program was run after completion of the PCR cycles. Student t-test was used to detect differences in gene expression between two different samples.
RESULTS
Sample data characteristics
Following transcriptome sequencing of all tissue samples using the Illumina Hiseq platform, a total of 126 RNA-seq datasets were generated, including 90 Protopterus annectens datasets and 36 Perccottus glenii datasets. Approximately 558 Gb of total data were obtained, including 450 Gb of Protopterus annectens data and 108 Gb of Perccottus glenii data. Transcriptome data mapping (including NCBI transcriptome data) to the genome of each species using HISAT2 showed a high mapping rate, suggesting that the transcriptome data were of good quality with high confidence.
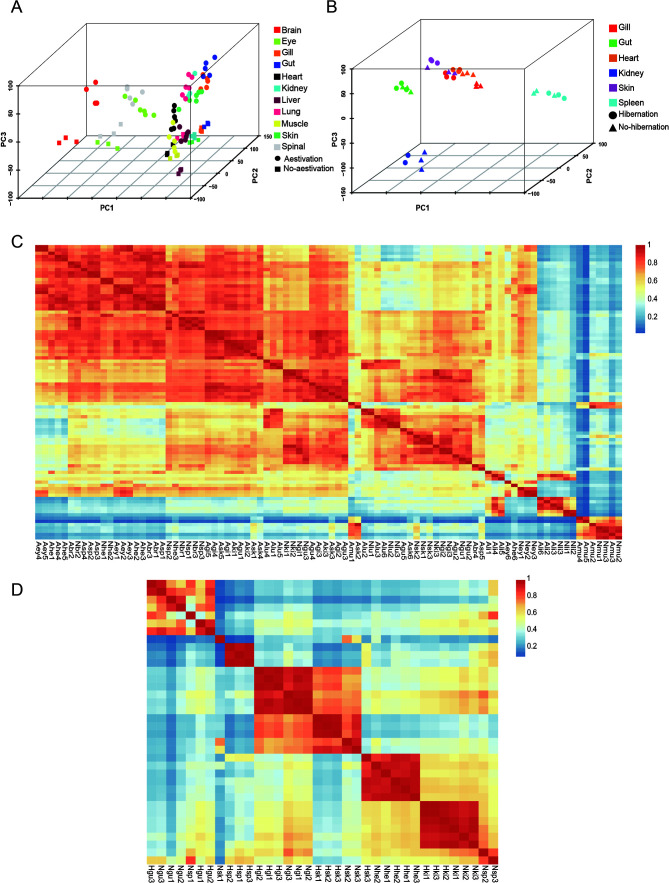
After filtering and refining the raw data, we ended up with a total of 17 different organ clean reads from the two studied species. Three-dimensional principal component analysis (PCA) was used to assess the distribution of organ samples, revealing satisfactory clustering for each sample group of Protopterus annectens and Perccottus glenii . Moreover, clear distinctions were observed between organs in different physiological states (Figure 1A, B). Additional analysis was conducted to assess the extent of correlation among biological replicates from different organs in varying physiological states. Results from inter-group correlation clustering analysis revealed satisfactory correlation within the transcriptome data, with good correlation among biological replicates (Figure 1C, D). Pearson correlation coefficients exceeded 0.9, suggesting minimal differences within sample groups and suitability for subsequent differential analysis.
Figure 1.
Schematic representation of data quality
A, B: Multi-organ PCA plots of Protopterus annectens and Perccottus glenii , respectively. C: Sample correlation thermograms of Protopterus annectens. Abr: Aestivating brain; Aey: Aestivating eye; Agi: Aestivating gill; Agu: Aestivating gut; Ahe: Aestivating heart; Aki: Aestivating kidney; Ali: Aestivating liver; Alu: Aestivating lung; Amu: Aestivating muscle; Ask: Aestivating skin; Asp: Aestivating spinal cord. Nbr: Non-aestivating brain; Ney: Non-aestivating eye; Ngi: Non-aestivating gill; Ngu: Non-aestivating gut; Nhe: Non-aestivating heart; Nki: Non-aestivating kidney; Nli: Non-aestivating liver; Nlu: Non-aestivating lung; Nmu: Non-aestivating muscle; Nsk: Non-aestivating skin; Nsp: Non-aestivating spinal cord. D: Sample correlation thermograms of Perccottus glenii . Hgi: Hibernating gill; Hgu: Hibernating gut; Hhe: Hibernating heart; Hki: Hibernating kidney; Hsk: Hibernating skin; Hsp: Hibernating spleen. Ngi: Non-hibernating gill; Ngu: Non-hibernating gut; Nhe: Non-hibernating heart; Nki: Non-hibernating kidney; Nsk: Non-hibernating skin; Nsp: Non-hibernating spleen.
Changes in gene expression during hibernation and aestivation
Transcriptomic analysis of Perccottus glenii was conducted during the hibernation state. Results showed that 1 030 genes were differentially expressed in the gill tissue (352 up-regulated and 678 down-regulated), 622 genes were differentially expressed in the gut tissue (240 up-regulated and 382 down-regulated), 1 223 genes were differentially expressed in the heart tissue (283 up-regulated and 940 down-regulated), 958 genes were differentially expressed in the kidney (318 up-regulated and 614 down-regulated), 2 343 genes were differentially expressed in the skin (843 up-regulated and 1 500 down-regulated), and 1 709 genes were differentially expressed in the spleen (614 up-regulated and 1 095 down-regulated).
Transcriptomic analysis was also conducted for Protopterus annectens during aestivation, with a total of 41 627 genes found to be differentially expressed during aestivation. Notably, 2 736 genes were differentially expressed in the brain (1 328 up-regulated and 1 408 down-regulated), 4 475 genes were differentially expressed in the eye (2 039 up-regulated and 2 436 down-regulated), 4 099 genes were differentially expressed in the gill (1 999 up-regulated and 2 100 down-regulated), 3 812 genes were differentially expressed in the gut (1 849 up-regulated and 1 963 down-regulated), 2 430 genes were differentially expressed in the heart (1 292 up-regulated and 1 138 down-regulated), 4 156 genes were differentially expressed in the kidney (1 871 up-regulated and 2 285 down-regulated), 3 179 genes were differentially expressed in the liver (1 768 up-regulated and 1 411 down-regulated), 3 772 genes were differentially expressed in the lung (1 879 up-regulated and 1 893 down-regulated), 3 152 genes were differentially expressed in the muscle (1 583 up-regulated and 1 569 down-regulated), 4 652 genes were differentially expressed in the skin (2 294 up-regulated and 2 358 down-regulated), and 2 428 genes were differentially expressed in the spinal cord (1 339 up-regulated and 1 089 down-regulated).
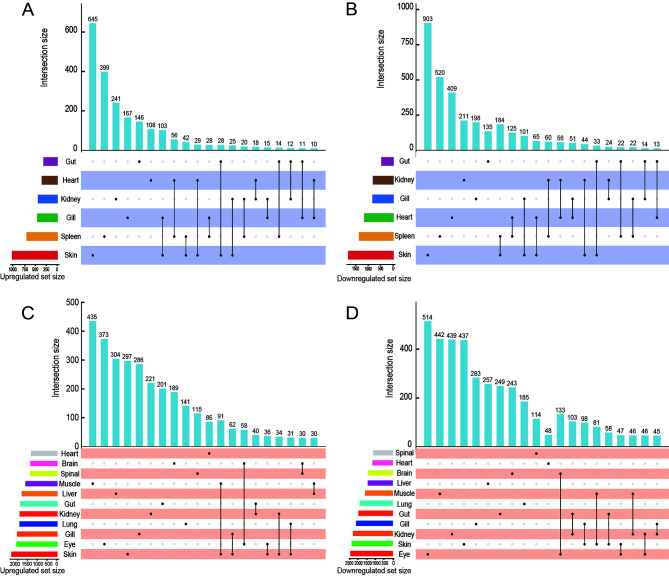
Subsequent analysis focused on identifying potential key genes within the DEGs associated with each dormancy type, with an additional goal of uncovering DEGs common to both hibernation and aestivation. However, the findings did not align with our initial expectations. As shown in the UpSet diagram (Figure 2), there was a preponderance of specific DEGs over shared ones across the various organs in both dormancy states. In Perccottus glenii , only 39 DEGs were identified (33 down-regulated and six up-regulated). Analysis revealed that more than 50% of the co-up-regulated DEGs were implicated in regulating cell number, whereas the majority of co-down-regulated DEGs were involved in the maintenance of cell structure stability and stress response. In Protopterus annectens, 37 co-up-regulated DEGs were identified, the majority of which were involved in apoptosis inhibition and stress resilience, while over half of the 34 co-down-regulated DEGs were involved in biosynthesis reduction and transport processes. These findings suggest that control of energy consumption is a feature of various organs during aestivation. A comparison of the common DEGs in each organ between the two dormancy states revealed no shared genes between the two species. Nonetheless, the co-DEGs identified in both were classified into five functional categories highly relevant to dormancy (Table 1). These results highlight the significant variability in DEGs across different dormancy states, with some overarching functional similarities.
Figure 2.
Upset plots of genes showing expression change during dormancy in each organ
A: Up-regulated genes in Perccottus glenii during hibernation. B: Down-regulated genes in Perccottus glenii during hibernation. C: Up-regulated genes in Protopterus annectens during aestivation. D: Down-regulated genes in Protopterus annectens during aestivation.
Table 1. Main types and numbers of co-DEGs during dormancy.
| Species | Metabolism | Cell cycle | Cellular immunity | Cytoskeleton | Signaling pathways |
| Protopterus annectens | 7 | 15 | 8 | 7 | 11 |
| Perccottus glenii | 6 | 4 | 9 | 5 | 7 |
Differences in physiological functions during dormancy
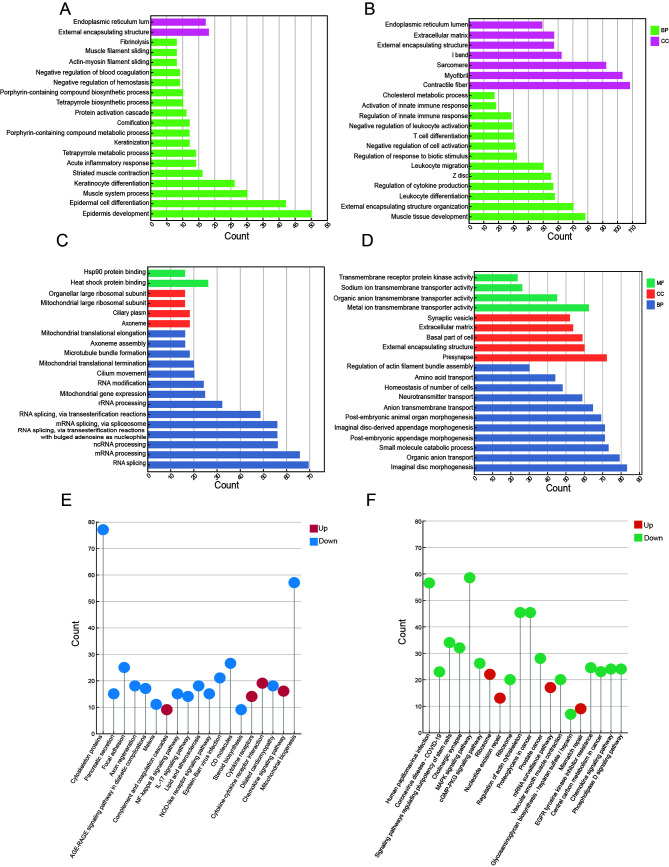
Enrichment analysis was performed to elucidate the functional significance of DEGs in the two dormancy phases. We identified 449 significantly up-regulated GO terms and 1 856 significantly down-regulated GO terms in Perccottus glenii (Figure 3A, B). Specifically, in the gill tissue, up-regulated genes were enriched in activation of the inflammatory response, suppression of trauma response, and regulation of complement activation, while down-regulated genes were significantly enriched in differentiation, activation, and migration of immune cells. In the gut, up-regulated genes were associated with the preservation of extracellular matrix structure and activity of diverse protease inhibitors, while down-regulated genes were significantly enriched in lipid metabolism and immune responses. In the heart, down-regulated genes contributed to cardiac function suppression during hibernation, particularly the immune activities of granulocytes and lymphocytes, as well as inhibition of immune cell apoptosis, thus facilitating the preservation of immunity while conserving energy. In the kidney, up-regulated genes were enriched in GO terms related to the maintenance of organ structure and regulation of muscle contraction-relaxation, while down-regulated genes were enriched in GO terms related to biological stress and immune pathways. In the spleen, up-regulated genes were enriched in hemoglobin synthesis and metabolism, whereas down-regulated genes were enriched in cell response to growth factor stimulation and connective tissue development. In the skin, down-regulated genes were associated with muscle cell activity and development, while up-regulated genes were enriched in functions involved in stress resistance of surface cells, including epidermal cell differentiation and keratinization, as well as immune and inflammatory responses.
Figure 3.
Functional enrichment analysis plots
A: Up-regulated GO enrichment results for Perccottus glenii . B: Down-regulated GO enrichment results for Perccottus glenii . C: Up-regulated GO enrichment results for Protopterus annectens. D: Down-regulated GO enrichment results for Protopterus annectens. E: KEGG enrichment results for Perccottus glenii . F: KEGG enrichment results for Protopterus annectens.
Subsequent KEGG pathway analysis for Perccottus glenii revealed significant down-regulation and enrichment in lipid biosynthetic pathways, particularly steroid biosynthesis, in the gill, gut, kidney, and skin (Figure 3E). The heart and spleen exhibited down-regulation and enrichment in multiple signaling pathways, including the NF-κB, TNF, and IL-17 pathways, with the heart additionally exhibiting down-regulation and enrichment in blood lipid metabolism and apoptosis pathways. In the skin, up-regulated genes were enriched in the chemokine signaling pathway and cytokine signal-receptor action processes, while down-regulated genes were enriched in pathways related to cytoskeleton maintenance and steroid biosynthesis processes. These findings strongly suggest that skin plays a crucial role in Perccottus glenii hibernation under conditions of complete freezing.
The above results indicate that, during hibernation, Perccottus glenii undergoes pronounced physiological changes, with a focus on enhancing tolerance to cold stress and hypoxia in specific tissues through processes such as keratinization and acute inflammatory response. Additionally, various high-energy physiological functions, such as immune response, proliferation and differentiation, metabolism, and structural maintenance, are inhibited or reduced to conserve energy.
For Protopterus annectens, GO enrichment analysis identified adaptations to high temperature and drought (Figure 3C, D). Tissues responsible for detecting and transmitting physiological changes were highly responsive, with minor changes leading to significant functional differences. Notably, RNA modification processes (i.e., RNA splicing, methylation, and capping) were enriched in Protopterus annectens neuron-rich organs (brain, eye, and spinal cord), while neurotransmitter transmission (inclusive of neurotransmitter synthesis, synaptic vesicle formation, and synaptic signal release) was associated with up-regulated RNA modification processes. Genes associated with learning and memory were significantly and uniformly down-regulated across all three tissues, potentially accounting for the altered neurotic behavior in animals during rest periods. The main organs responsible for gas exchange, namely the gills and lungs, were enriched in RNA modifications, binding of heat shock protein, and transcriptional promotion processes, while down-regulated DEGs were enriched in organ structure development, transmembrane transport of organic matter, and metabolism. The gut, liver, and kidney, as metabolic organs, mainly showed down-regulation in the synthesis and degradation of organic matter, such as polyols, organic acids, and lipids, as well as a reduction in immune responses, with an up-regulation in RNA modification and ribosomal biogenesis. The heart, muscle, and skin, key connective tissue organs, exhibited significant enrichment in heat shock protein binding, ribosome precursor generation, and RNA polymerase synthesis, with down-regulated genes enriched in organogenesis, cell response to stimulation, and energy metabolism.
KEGG enrichment analysis provided broader insights into the physiological alterations in various organs of Protopterus annectens during aestivation (Figure 3F). Results showed that up-regulated pathways were predominantly involved in spliceosome and mRNA surveillance pathways, whereas the down-regulated pathways were more organ-specific. Notably, genes implicated in various signaling pathways, such as the cGMP-PKG, chemokine, and MAPK signaling pathways, were significantly down-regulated and enriched in both the brain and eyes. Additionally, the gills, kidney, lung, and skin exhibited down-regulation in pathways related to epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI) resistance and organic metabolism. In the heart, down-regulated genes were enriched in circadian pathways.
Thus, both the GO and KEGG enrichment results revealed that up-regulated genes in Protopterus annectens were primarily enriched in RNA-related processes, while down-regulated genes were enriched in various signaling pathways.
Potential relationship between hibernation and aestivation
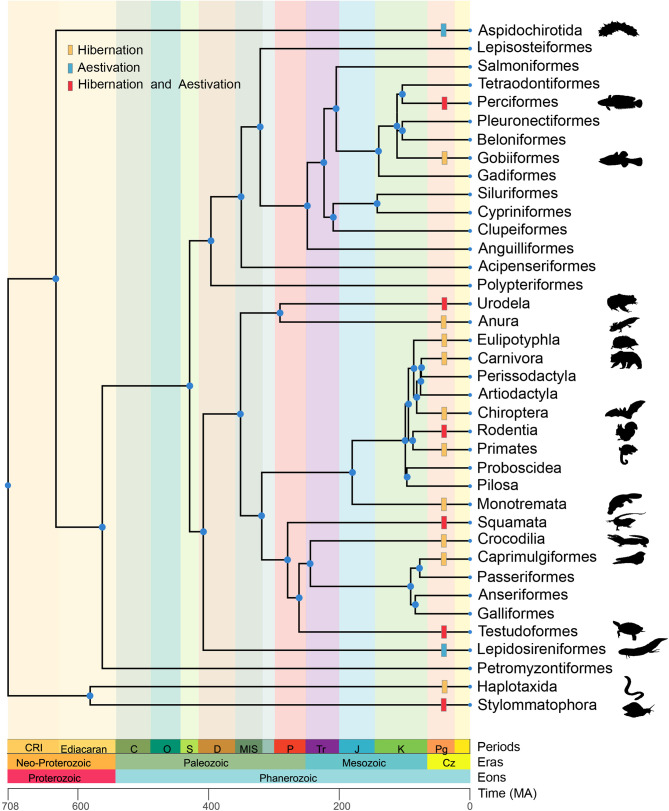
The above findings indicate functional similarities in the dormancy states of the two species, with both exhibiting symptoms of reduced activity, prolonged lethargy, and other comparable traits, providing evidence of a shared nature hibernation and aestivation. Therefore, we next investigated the relationship between hibernation and aestivation through transcriptomic analysis, building upon existing dormancy research. We hypothesized that the similarity between hibernation and aestivation may be attributed to convergent evolution. To test this, an inventory of species known to undergo dormancy was compiled, and TimeTree (http://timetree.org/) was used to construct a phylogenetic tree for these species (Figure 4), illustrating the widespread occurrence of dormancy. Notably, the divergence times of nodes on the phylogenetic tree suggested that the development of dormancy may have evolved as an adaptive response to environmental conditions rather than strict genetic inheritance. Our results also indicated that species exhibiting dormancy tend to have constrained mobility and are incapable of migrating to more appropriate habitats before the onset of dry or winter seasons. Furthermore, dormancy tended to be more prevalent among terrestrial animals than aquatic ones, likely reflecting differences in environmental challenges faced by organisms in gaseous or aqueous habitats.
Figure 4.
Phylogenetic relationships of species possessing dormancy ability versus species without dormancy ability
Phylogenetic topological relationships were established by selecting typical species to represent the whole order. Yellow, blue, and red markers on branches indicate respective dormancy types in species within that taxon. MA: Million years ago.
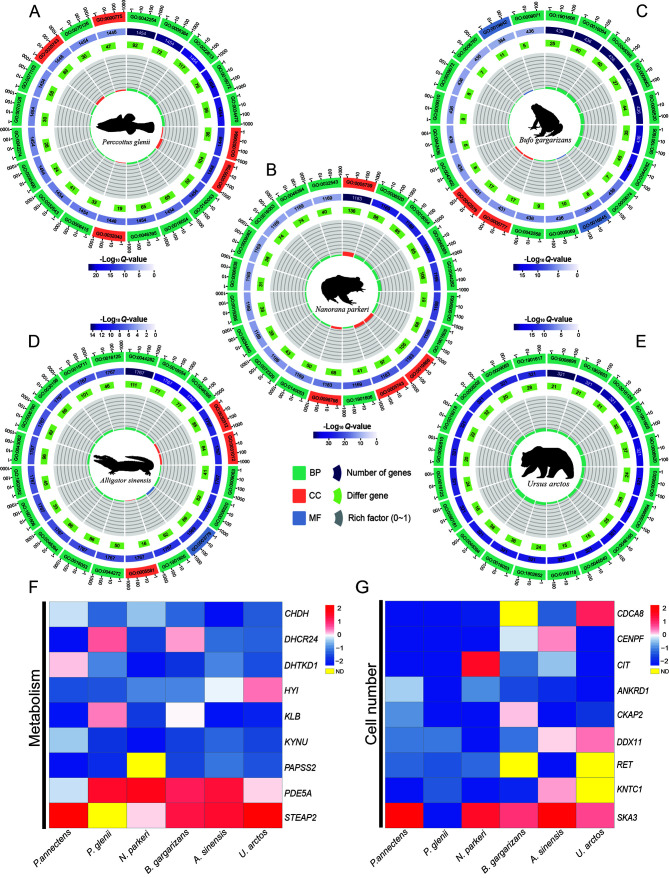
The liver, as the organ with the most extensive metabolic function and highest capacity for self-regeneration, plays a crucial role in the body. A hallmark of dormancy in organisms is the marked reduction in metabolic activities, leading to significant changes in liver function. Given the systemic impact of dormancy, we specifically focused on the liver for further examination. Transcriptomic data from the livers of five species (U. arctos, A. sinensis, N. parkeri, B. gargarizans, and Perccottus glenii ) were obtained from NCBI for analysis. GO functional enrichment analysis demonstrated distinct liver function changes across all species, despite their similar physiological state (Figure 5A–E), primarily evident in the up-regulated terms. Notably, Perccottus glenii showed enrichment in protein modification and protease complexes, whereas N. parkeri and B. gargarizans showed enrichment in immune function and muscle fibers, respectively, A. sinensis showed enrichment in both RNA and protein modifications, and U. arctos was enriched in ion channels and organic matter transport processes. Conversely, the patterns of down-regulation were remarkably consistent across species, showing universal reduction in biosynthetic and catabolic processes. KEGG pathway enrichment analysis further indicated that each species could be broadly split into four primary functions: anabolism, catabolism, signaling pathways, and cell structure (Table 2).
Figure 5.
Multi-species analysis plots
A–E: GO enrichment results from the NCBI database. First circle from the outside indicates top 20 GO terms enriched; numbers outside the circle are a sitting scale for the number of DEGs. Different colors represent different ontologies. Second circle indicates the total number of genes and Q-values in each GO term. Third circle represents number of DEGs Fourth circle represents enrichment factor value for each GO term. For background grid lines, each grid represents 0.1. F, G: Figure shows log-transformed ratios of dormant-phase expressed genes (up-regulated in red, down-regulated in blue) in two functional classes: metabolism (F) and cell number (G) .
Table 2. Main types and numbers of KEGG pathways in liver of dormant species.
| Species | Anabolism | Catabolism | Signaling pathways | Cell structure |
| N/A: Not available. | ||||
| Perccottus glenii | 4 | N/A | 5 | 4 |
| Nanorana parkeri | 6 | 13 | 4 | 3 |
| Bufo gargarizans | N/A | 1 | 1 | 1 |
| Alligator sinensis | 2 | 4 | 5 | 6 |
| Ursus arctos | 6 | 12 | 2 | 1 |
To further investigate the association between hibernation and aestivation, a comprehensive review was conducted of liver transcriptome data across all species studied. Following OrthoFinder analysis (Gallant et al., 2014), 1 133 orthologous genes were identified across the six species. Combined with the DEG results, 66 orthologous genes demonstrated significant differential expression. Considering the differences in expression levels, 24 orthologous genes with consistent expression were eventually identified. According to the gene function interpretation provided by the online database, the selected orthologous genes were primarily involved in metabolic processes and cellular regulation (Figure 5F, G). Additionally, the expression of genes responsible for nervous system development (CIT) was suppressed, explaining the lethargy observed in most animals after dormancy upon awakening. Thus, these findings demonstrate that hibernation and aestivation exhibit convergent evolution in function, predominantly pertaining to metabolism and cell count regulation.
Real-time quantitative PCR
Gene expression analysis showed substantial differences in gene expression between dormant and non-dormant periods. To verify the accuracy of the screened DEGs, and considering the actual situation of the samples, a random subset of DEGs was selected from seven tissues for qPCR detection. Notably, gene expression was highly consistent between the RNA-seq and qPCR results (Supplementary Figure S1), confirming the reliability of the RNA-seq dataset.
DISCUSSION
In this study, transcriptomic analysis was utilized to examine the physiological modifications in Protopterus annectens and Perccottus glenii during dormancy from a holistic standpoint, as well as to investigate the correlation and disparities between hibernation and aestivation. Analysis of DEGs demonstrated significant differences between the two dormancy states, highlighting the influence of both species-specific and environmental factors. Despite these variances, notable similarities in gene expression patterns and internal physiological alterations were observed between hibernation and aestivation, further emphasizing the interconnected yet distinct nature of these dormancy phenomena.
Global changes during dormancy
Dormancy has evolved as an adaptive response, with traits exhibited during this state representing adaptations to environmental conditions (Geiser, 2013). Thus, changes in the external environment directly influence how organisms respond to the habitat in which they live. Seasonal temperature extremes, such as heatwaves and droughts in summer and low temperatures and freezing conditions in winter, can intensify survival stress. Regarding organ-specific changes, the physiological adaptations observed in this study varied across different organs, with each exhibiting unique changes. While it was difficult to establish a clear correlation between changes in individual organs based solely on analysis of DEGs and functional enrichment, by considering the organism as a whole and accounting for environmental factors, we identified connections between the physiological changes occurring in each organ during the dormancy period.
Understanding the response of Perccottus glenii to freezing injury is critical, as it is one of the few animals, aside from certain amphibians and reptiles, that hibernates by freezing. Our results showed that during hibernation, the physiological changes in various organs were well-coordinated, with external organs (i.e., gill and skin) demonstrating greater tolerance to freezing injury compared to the internal organs. The skin and gills, being in direct contact with ice, exhibited a pronounced ability to respond to external stimuli, indicating an adaptive increase in resistance. Conversely, internal organs such as the heart, kidneys, spleen, and gut primarily focused on preserving homeostasis, ensuring basal physiological activities while minimizing energy consumption. Thus, during hibernation, Perccottus glenii prioritizes the maintenance of basic functions whilst increasing the resistance and immunity of external organs against external factors.
In contrast to Perccottus glenii, Protopterus annectens demonstrated no significant improvement in immune response within its specific organs. The physiological changes observed in Protopterus annectens during aestivation were more complementary, likely due to the distinct environmental demands of aestivation. Previous research has noted that Protopterus annectens secretes a considerable amount of mucus to create a protective mucus cocoon during aestivation, a behavior observed in natural and laboratory environments (Chng et al., 2017a), serving as a barrier against external stressors. Additionally, Protopterus annectens experiences a notable alteration in its respiration pattern during aestivation. Under water-scarce conditions, gill respiration becomes highly challenging for Protopterus annectens, prompting a switch to lung respiration as the dominant mode. Consequently, the gills cease their respiratory function and are solely involved in maintaining osmotic pressure during aestivation (Chng et al., 2017b). In addition, numerous processes related to transcription and RNA modification were observed in various Protopterus annectens organs. Given that the biosynthesis pathway is energetically costly, its continuation during aestivation contrasts with the need for organisms to minimize energy consumption. This suggests that the observed alterations in Protopterus annectens play a crucial role in maintaining internal homeostasis throughout aestivation, warranting further investigation to uncover the reasons for these findings.
Differences in different dormant states
The differences between hibernation and aestivation largely stem from the diverse environmental stresses experienced by these different states of dormancy, characterized by temperature extremes that elicit diverse physiological responses. Notably, hibernating organisms must withstand the cold, while aestivating organisms must adapt to heat. Extreme temperatures not only disrupt nutrient intake but also impair physiological functions. During winter, Perccottus glenii exhibits exceptional resilience to frigid temperatures, completely freezing within ice during hibernation, resulting in visible skin wrinkling (Storey & Storey, 1986). The formation of intracellular ice crystals represents a critical survival challenge, potentially causing irreversible cellular damage (Storey & Storey, 1996). To minimize freezing-induced damage, Perccottus glenii typically accumulates cryoprotectants, such as glucose, polyols, and amino acids, within the cells (Jiang et al., 2023), increasing body fluid concentration to hinder ice crystal growth and simultaneously enhance water retention. Although the accumulation of organic matter can effectively decrease cellular injury due to freezing, hibernation significantly slows metabolism, affecting overall immune system function (Li et al., 2023) and increasing cellular vulnerability to microbial assaults (Psychrophiles). The barrier function of skin is crucial, with Perccottus glenii skin displaying increased keratinization, epidermalization, and immune response, offering protection against pathogens, harmful substances, and mechanical damage.
In contrast to the cold challenges of winter, summer heat poses risks of dehydration, skin tissue damage, potential carcinogenesis, and protein structure destabilizations, interrupting essential physiological activities (Horton et al., 2023). While shelter can offer protection from UV rays, escaping extreme heat is far more difficult. Animals that undergo aestivation have evolved various adaptations to cope with high-temperature stress. A recent study on Protopterus annectens did not find any thermal activation of the TRPV1 gene (thermoreceptor) in either high-temperature (44°C) or acidic environments (pH=6), indicating a lower thermal sensitivity of the TRPV1 gene compared to that in non-aestivating animals, contributing to its high-temperature tolerance (Hori & Saitoh, 2023). Moreover, while elevated temperatures can expedite oxidative damage in skin tissues, studies have not detected significant skin damage in Protopterus annectens under natural aestivation conditions. Fluorescence analysis of skin tissues of Protopterus annectens after varying periods of aestivation detected significant differences; short-term aestivation led to skin epidermis thinning and mucous cell reduction, whereas long-term aestivation resulted in skin epidermis regeneration. The research also noted an increase in aldehyde oxidase gene expression and heat shock protein expression, enhancing the skin’s resistance to oxidative stress (Amelio & Garofalo, 2023).
Similarities between different dormant states
Biological evolution is influenced by various factors, including natural selection, gene mutations, gene flow, and genetic drift (Stern & Crandall, 2018). Among these, natural selection is the primary driving force for biological evolution, profoundly impacting the evolutionary trajectory of populations across each region and era. In essence, organisms in favorable external environments are subjected to more selective evolutionary processes, while those in harsh environments fact limited evolutionary options (Balart-García et al., 2023). As a result, extreme conditions may result in similar evolutionary trajectories among diverse organisms adapting to the environment. This observation provides an indirect rationale for why adaptive evolution in adverse environments typically results in convergent evolution. Hibernation and aestivation represent two distinct forms of dormancy triggered by seasonal challenges, such as food scarcity and the inability to sustain normal physiological functions in extreme temperatures (Storey, 2003; Sturla et al., 2002). The consistent responses of diverse organisms to environmental shifts under similar external conditions indicate parallel physiological adaptations. Based on a review of existing studies, we propose that similarities between hibernation and aestivation may be classified according to the following characteristics.
Metabolic inhibition constitutes a highly prevalent and important survival strategy for animals facing a shift in environmental conditions from favorable to unfavorable (Navas & Carvalho, 2010). Reducing metabolic activity can help significantly lower internal energy consumption in adverse environments, potentially extending an animal’s life span (Storey & Storey, 2012). Analysis revealed a notable enrichment in DEGs associated with metabolic processes in both hibernating and aestivating animals. Interestingly, each dormant animal exhibited a remarkably consistent approach, uniformly reducing metabolism-related activities across all organs, with pronounced suppression of three significant nutrient assimilation and heterotrophy processes. Similarly, we found that water metabolism during dormancy was very slow. Previous studies on Protopterus annectens explored the roles of aquaporins 1 and 3 and urea transporters in regulating water and urea during dormancy, finding that their transcript and protein levels varied with dormancy duration to minimize water loss and enhance waste processing and hydration upon awakening (Chng et al., 2016, 2017a). Research on grizzly bears and alligators has shown that while urea synthesis is blocked during hibernation, urinary excretion almost completely ceases (Jansen et al., 2019; Lin et al., 2020), leading to high levels of urea in the body, increased osmotic pressure, and reduced water loss. Our results also demonstrated the suppression of other energy-consumptive processes, including the cell cycle, signaling, and gene expression-related pathways, with down-regulation of positive regulators of the mTOR, adipocytokine, and glucagon metabolism signaling pathways, crucial in energy metabolism and cell proliferation. These findings suggest a substantial reduction in adenosine triphosphate (ATP) synthesis in the body when animals enter dormancy, with non-critical energy-consuming pathways notably curtailed to conserve resources for essential life maintenance.
The complex physiological changes that occur in multicellular organisms are built upon the cooperation of individual cells (Ahmed et al., 2021). However, a significant proportion of cells perish more rapidly than the organism’s natural aging process, requiring continuous cell proliferation to maintain stable cell numbers throughout life (Myster & Duronio, 2010). As such, there is a clear conflict between the high-energy demands of cell proliferation and the low-energy demands of organisms during dormancy, with previous studies and our findings demonstrating notable cell cycle inhibition during dormancy (Charles & Don, 1954). Nevertheless, merely inhibiting cell proliferation alone does not guarantee cell count stability during the dormancy maintenance period, as apoptosis and cell necrosis still pose challenges. Interestingly, nature seems to have devised effective solutions. Our observations showed significant down-regulation of genes related to the apoptotic pathway in dormant animals, suggesting a crucial strategy for maintaining somatic cell stability during dormancy. Inhibition of the apoptotic pathway is primarily divided into somatic and immune cells. Inhibiting somatic cell apoptosis can to some extent prolong the natural cell survival cycle and ensure the necessary number of basal cells in the organism. Moreover, the inhibition of apoptosis of immune cells can maintain a certain degree of immunity with minimal energy consumption, preventing the emergence of massive cell death caused by microbial infection during dormancy (Patterson et al., 1957).
Organisms entering hibernation or aestivation experience a prolonged period of activity reduction, attributed to the suppression of neural activity at the onset of dormancy that regulates the onset, maintenance, and cessation of their dormant states (Ma et al., 2005). Our analysis revealed a significant down-regulation in DEGs involved in neurotransmitter transport (PRKCE, RAB3GAP2, and STXBP5), synaptogenesis (SYNGR1, HCN1, and EPN1), and biorhythm (CLOCK, CSNK1E, and ARNTL) in Protopterus annectens, leading to reduced neural signaling. The downregulation of ion transmembrane transport pathways was evident in organs rich in nerve cells, suggesting that the blockade of ion transport further inhibited neural activity. In addition, previous studies have shown that central nervous system inhibition also plays an important role during hibernation in animals. For example, activation of the adenosine A1 receptor, which inhibits excitatory neurotransmission, induces entry into hibernation in hamsters (Tamura et al., 2005). In Perccottus glenii , this receptor gene family has undergone significant expansion, strengthening the inhibitory capacity of neural signaling for inducing and maintaining hibernation (Jiang et al., 2023).
From a cellular stress response perspective, extreme temperatures can induce significant physiological stress in organisms, leading to prolonged periods of ischemia-reperfusion, hypoxia-reoxygenation, and mechanical damage (Zhang et al., 2021). To mitigate stress-induced cellular damage, organisms activate a series of protective measures. Heat shock proteins, functioning as molecular chaperones across diverse organisms, play a vital role in this defense. We found that genes encoding different heat shock proteins and stress response-related proteins, including UNC45B, HSPH1, BAG3, DNAJB1, and TRAP1, were notably up-regulated during dormancy, facilitating correct protein structure formation and preventing polypeptide chain denaturation. Prolonged dormancy can also trigger ischemia in the body, exacerbating DNA damage (Karanova, 2009) and leading to biological cell death if not repaired. Functional enrichment analysis revealed a significant up-regulation in the DNA damage repair pathway, addressing the possible harm caused by DNA damage. Furthermore, higher levels of antioxidant gene expression and antioxidant accumulation were observed in hibernating Perccottus glenii , improving resilience to oxidative processes during dormancy (Jiang et al., 2023).
CONCLUSIONS
Hibernation and aestivation represent evolutionary adaptations by organisms to seasonal temperature extremes. While various studies have explored the physiological, cellular, and molecular adaptive mechanisms of specific tissues in organisms entering dormancy (hibernation or aestivation), few studies have explored the post-dormancy physiological changes or the similarities and differences between the two dormancy states from a holistic point of view. Here, using transcriptomics, we analyzed the environmental adaptation mechanisms of Protopterus annectens and Perccottus glenii in their respective dormant states, and further analyzed the similarities and differences between hibernation and aestivation using publicly available data from the NCBI database. Our results revealed several similarities in gene expression changes related to metabolism, stress response, neural activity, and cell number control that were particularly pronounced in the two dormant species. In conclusion, this work provides a theoretical basis for understanding how organisms adapt to extreme environments and contributes to further research into dormancy and the development of artificial dormancy techniques.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
Acknowledgments
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
S.P.H. and L.D.Y. were the leaders and designers of the research; C.W., L.H.G., and H.F.J. collected the samples; Y.H.N. analyzed the data; Y.H.N., G.G.L., and L.D.Y. wrote the paper. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the National Natural Science Foundation of China (32170480, 31972866), Youth Innovation Promotion Association, Chinese Academy of Sciences (http://www.yicas.cn), Young Top-notch Talent Cultivation Program of Hubei Province, and Wuhan Branch, Supercomputing Center, Chinese Academy of Sciences, China
Contributor Information
Guo-Gang Li, Email: ligg@qhnu.edu.cn.
Lian-Dong Yang, Email: yangld@ihb.ac.cn.
Shun-Ping He, Email: clad@ihb.ac.cn.
DATA AVAILABILITY
All sequencing data were deposited in the NCBI BioProject database with accession numbers PRJNA1012339 (Perccottus glenii), PRJNA1012340 (Protopterus annectens), as well as at GSA (CRA014545) and ScienceDB (DOI:10.57760/sciencedb.15544).
References
- Ahmed R, Reza HM, Shinohara K, et al Cellular senescence and its impact on the circadian clock. Journal of Biochemistry. 2021;171(5):493–500. doi: 10.1093/jb/mvab115. [DOI] [PubMed] [Google Scholar]
- Amelio D, Garofalo F Morpho-functional changes of lungfish Protopterus dolloi skin in the shift from freshwater to aestivating conditions. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2023;266:110846. doi: 10.1016/j.cbpb.2023.110846. [DOI] [PubMed] [Google Scholar]
- Balart-García P, Aristide L, Bradford TM, et al Parallel and convergent genomic changes underlie independent subterranean colonization across beetles. Nature Communications. 2023;14(1):3842. doi: 10.1038/s41467-023-39603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens AJ, Hunt JH, Toth AL Comparative transcriptomics of convergent evolution: different genes but conserved pathways underlie caste phenotypes across lineages of eusocial insects. Molecular Biology and Evolution. 2015;32(3):690–703. doi: 10.1093/molbev/msu330. [DOI] [PubMed] [Google Scholar]
- Chai LH, Huang PY, Bao X Tolerant ability and physiological and biochemical responses of Chinese sleeper Perccottus glenii to icing up and hypoxia environment. Journal of Dalian Ocean University. 2020;35(2):218–222. [Google Scholar]
- Charles PL, Don WF The effect of hibernation on the growth of sarcoma in the hamster. Cancer Research. 1954;14(1):25–28. [PubMed] [Google Scholar]
- Chew SF, Chan NKY, Loong AM, et al Nitrogen metabolism in the African lungfish (Protopterus dolloi) aestivating in a mucus cocoon on land. Journal of Experimental Biology. 2004;207(5):777–786. doi: 10.1242/jeb.00813. [DOI] [PubMed] [Google Scholar]
- Chng YR, Ong JLY, Ching B, et al Molecular characterization of aquaporin 1 and aquaporin 3 from the gills of the African lungfish, Protopterus annectens, and changes in their branchial mRNA expression levels and protein abundance during three phases of aestivation. Frontiers in Physiology. 2016;7:532. doi: 10.3389/fphys.2016.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng YR, Ong JLY, Ching B, et al Aestivation induces changes in the mRNA expression levels and protein abundance of two isoforms of urea transporters in the gills of the African lungfish. Protopterus annectens. Frontiers in Physiology. 2017a;8:71. doi: 10.3389/fphys.2017.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng YR, Ong JLY, Ching B, et al Molecular characterization of three Rhesus glycoproteins from the gills of the African lungfish, Protopterus annectens, and effects of aestivation on their mRNA expression levels and protein abundance. PLoS One. 2017b;12(10):e0185814. doi: 10.1371/journal.pone.0185814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney RG, Lahiri S, Fishman AP Aestivation of the African lungfish Protopterus Aethiopicus: cardiovascular and respiratory functions. Journal of Experimental Biology. 1974;61(1):111–128. doi: 10.1242/jeb.61.1.111. [DOI] [PubMed] [Google Scholar]
- Gallant JR, Traeger LL, Volkening JD, et al Genomic basis for the convergent evolution of electric organs. Science. 2014;344(6191):1522–1525. doi: 10.1126/science.1254432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XL, Wang SQ, Shen SQ, et al Differential bone remodeling mechanism in hindlimb unloaded and hibernating Daurian ground squirrels: a comparison between artificial and natural disuse within the same species. Journal of Comparative Physiology B. 2023;193(3):329–350. doi: 10.1007/s00360-023-01482-9. [DOI] [PubMed] [Google Scholar]
- Garofalo F, Amelio D, Icardo JM, et al Signal molecule changes in the gills and lungs of the African lungfish Protopterus annectens, during the maintenance and arousal phases of aestivation. Nitric Oxide. 2015;44:71–80. doi: 10.1016/j.niox.2014.11.017. [DOI] [PubMed] [Google Scholar]
- Geiser F Metabolic rate and body temperature reduction during hibernation and daily torpor. Annual Review of Physiology. 2004;66:239–274. doi: 10.1146/annurev.physiol.66.032102.115105. [DOI] [PubMed] [Google Scholar]
- Geiser F Hibernation. Current Biology. 2013;23(5):R188–R193. doi: 10.1016/j.cub.2013.01.062. [DOI] [PubMed] [Google Scholar]
- Hiong KC, Ip YK, Wong WP, et al Brain Na+/K+-ATPase α-subunit isoforms and aestivation in the African lungfish. Protopterus annectens. Journal of Comparative Physiology B. 2014;184(5):571–587. doi: 10.1007/s00360-014-0809-0. [DOI] [PubMed] [Google Scholar]
- Hori S, Saitoh O Decreased heat sensitivity of lungfish TRPV1 revealed by the heterologous expression system. Biochemical and Biophysical Research Communications. 2023;647:16–22. doi: 10.1016/j.bbrc.2023.01.060. [DOI] [PubMed] [Google Scholar]
- Horton L, Brady J, Kincaid CM, et al The effects of infrared radiation on the human skin. Photodermatology, Photoimmunology & Photomedicine. 2023;39(6):549–555. doi: 10.1111/phpp.12899. [DOI] [PubMed] [Google Scholar]
- Jansen HT, Trojahn S, Saxton MW, et al Hibernation induces widespread transcriptional remodeling in metabolic tissues of the grizzly bear. Communications Biology. 2019;2(1):336. doi: 10.1038/s42003-019-0574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HF, Lv WQ, Wang Y, et al Multi-omics investigation of freeze tolerance in the amur sleeper, an aquatic ectothermic vertebrate. Molecular Biology and Evolution. 2023;40(3):msad040. doi: 10.1093/molbev/msad040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanova MV Free amino acid composition in blood and muscle of the gobi Precottus glehni at the period of preparation and completion of hibernation. Journal of Evolutionary Biochemistry and Physiology. 2009;45(1):67–77. doi: 10.1134/S0022093009010062. [DOI] [PubMed] [Google Scholar]
- Li AQ, Leng HX, Li ZL, et al Temporal dynamics of the bat wing transcriptome: Insight into gene-expression changes that enable protection against pathogen. Virulence. 2023;14(1):2156185. doi: 10.1080/21505594.2022.2156185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HJ, He Y, Jiang JM, et al Molecular systematics and phylogenetic analysis of the Asian endemic freshwater sleepers (Gobiiformes: Odontobutidae) Molecular Phylogenetics and Evolution. 2018;121:1–11. doi: 10.1016/j.ympev.2017.12.026. [DOI] [PubMed] [Google Scholar]
- Lin JQ, Huang YY, Bian MY, et al A unique energy-saving strategy during hibernation revealed by Multi-Omics analysis in the Chinese alligator. iScience. 2020;23(6):101202. doi: 10.1016/j.isci.2020.101202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loong AM, Hiong KC, Wong WP, et al Differential gene expression in the liver of the African lungfish, Protopterus annectens, after 6 days of estivation in air. Journal of Comparative Physiology B. 2012;182(2):231–245. doi: 10.1007/s00360-011-0613-z. [DOI] [PubMed] [Google Scholar]
- Ma YL, Zhu XW, Rivera PM, et al Absence of cellular stress in brain after hypoxia induced by arousal from hibernation in Arctic ground squirrels. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology. 2005;289(5):R1297–R1306. doi: 10.1152/ajpregu.00260.2005. [DOI] [PubMed] [Google Scholar]
- Myster D L, Duronio R J Cell cycle: To differentiate or not to differentiate? Current Biology. 2010;10(8):R302–R304. doi: 10.1016/s0960-9822(00)00435-8. [DOI] [PubMed] [Google Scholar]
- Ong JLY, Chng YR, Ching B, et al Molecular characterization of myostatin from the skeletal muscle of the African lungfish, Protopterus annectens, and changes in its mRNA and protein expression levels during three phases of aestivation. Journal of Comparative Physiology B. 2017;187(4):575–589. doi: 10.1007/s00360-017-1057-x. [DOI] [PubMed] [Google Scholar]
- Pankey MS, Minin VN, Imholte GC, et al Predictable transcriptome evolution in the convergent and complex bioluminescent organs of squid. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(44):E4736–E4742. doi: 10.1073/pnas.1416574111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson WB, Lyman CP, Patterson HR Growth of human tumors in hibernating hamsters. Proceedings of the Society for Experimental Biology and Medicine. 1957;96(1):94–97. doi: 10.3181/00379727-96-23402. [DOI] [PubMed] [Google Scholar]
- Pinho GM, Martin JGA, Farrell C, et al Hibernation slows epigenetic ageing in yellow-bellied marmots. Nature Ecology & Evolution. 2022;6(4):418–426. doi: 10.1038/s41559-022-01679-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possmayer F, McCaig L, Yao LJ, et al. 2010. Coping with the cold: effect of hibernation on pulmonary surfactant in the thirteen-lined ground squirrel. Biophysical Journal, 98(3 Suppl 1): 76a.
- Stern DB, Crandall KA The evolution of gene expression underlying vision loss in cave animals. Molecular Biology and Evolution. 2018;35(8):2005–2014. doi: 10.1093/molbev/msy106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey KB. 2003. Mammalian hibernation: Transcriptional and translational controls. In: Roach RC, Wagner PD, Hackett PH. Hypoxia: Through the Lifecycle. New York: Springer, 21–38.
- Storey KB, Storey JM Freeze tolerant frogs: cryoprotectants and tissue metabolism during freeze–thaw cycles. Canadian Journal of Zoology. 1986;64(1):49–56. doi: 10.1139/z86-008. [DOI] [Google Scholar]
- Storey KB, Storey JM Natural freezing survival in animals. Annual Review of Ecology and Systematics. 1996;27:365–386. doi: 10.1146/annurev.ecolsys.27.1.365. [DOI] [Google Scholar]
- Storey KB, Storey JM Aestivation: signaling and hypometabolism. Journal of Experimental Biology. 2012;215(9):1425–1433. doi: 10.1242/jeb.054403. [DOI] [PubMed] [Google Scholar]
- Sturla M, Paola P, Carlo G, et al Effects of induced aestivation in Protopterus annectens: A histomorphological study. Journal of Experimental Zoology. 2002;292(1):26–31. doi: 10.1002/jez.1139. [DOI] [PubMed] [Google Scholar]
- Sullivan IR, Adams DM, Greville LJS, et al Big brown bats experience slower epigenetic ageing during hibernation. Proceedings of the Royal Society B:Biological Sciences. 2022;289(1980):20220635. doi: 10.1098/rspb.2022.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Shintani M, Nakamura A, et al Phase-specific central regulatory systems of hibernation in Syrian hamsters. Brain Research. 2005;1045(1-2):88–96. doi: 10.1016/j.brainres.2005.03.029. [DOI] [PubMed] [Google Scholar]
- Van-Beers EH, Rings EHHM, Posthuma G, et al Intestinal carbamoyl phosphate synthase I in human and rat: Expression during development shows species differences and mosaic expression in duodenum of both species. Journal of Histochemistry & Cytochemistry. 1998;46(2):231–240. doi: 10.1177/002215549804600212. [DOI] [PubMed] [Google Scholar]
- Zhang J, Gupta A, Storey KB Freezing stress adaptations: critical elements to activate Nrf2 related antioxidant defense in liver and skeletal muscle of the freeze tolerant wood frogs. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2021;254:110573. doi: 10.1016/j.cbpb.2021.110573. [DOI] [PubMed] [Google Scholar]
- Zhong QM, Wang JL. 2022. Seasonal flexibility of kidney structure and factors regulating water and salt in Eremias multiocellata. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 274: 111301.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.
Data Availability Statement
All sequencing data were deposited in the NCBI BioProject database with accession numbers PRJNA1012339 (Perccottus glenii), PRJNA1012340 (Protopterus annectens), as well as at GSA (CRA014545) and ScienceDB (DOI:10.57760/sciencedb.15544).