Abstract
Cyclooxygenase (COX) inhibitors are ubiquitous in aquatic systems and have been detected in fish tissues. The exposure of fish to these pharmaceuticals is concerning because COX inhibitors disrupt the synthesis of prostaglandins (PGs), which modulate a variety of essential biological functions, including reproduction. In the present study, we investigated the effects of well-characterized mammalian COX inhibitors on female fathead minnow reproductive health. Fish (n=8) were exposed for 96 h to water containing indomethacin (IN; 100 μg/l), ibuprofen (IB; 200 μg/l) or celecoxib (CX; 20 μg/l), and evaluated for effects on liver metabolome and ovarian gene expression. Metabolomic profiles of IN, IB and CX were not significantly different from control or one another. Exposure to IB and CX resulted in differential expression of comparable numbers of genes (IB = 433, CX= 545). In contrast, 2558 genes were differentially expressed in IN-treated fish. Functional analyses (canonical pathway and gene set enrichment) indicated extensive effects of IN on PG synthesis pathway, oocyte meiosis and several other processes consistent with physiological roles of PGs. Transcriptomic data were congruent with PG data; IN reduced plasma prostaglandin F2α concentration, whereas IB and CX did not. Five putative AOPs were developed linking the assumed molecular initiating event of COX inhibition, with PG reduction and the adverse outcome of reproductive failure via reduction of: 1) ovulation, 2) reproductive behaviors mediated by exogenous or endogenous prostaglandins, and 3) oocyte maturation in fish. These pathways were developed using, in part, empirical data from the present study and other publicly-available data.
Keywords: cyclooxygenase inhibition, prostaglandins, reproduction, fish, adverse outcome pathway, nonsteroidal anti-inflammatory drugs
Introduction
Cyclooxygenase (COX; also known as prostaglandin-endoperoxide synthase; PTGS) catalyzes conversion of arachidonic acid into endoperoxide prostaglandin G2, a precursor for synthesis of bioactive PGs (Simmons et al., 2004). Prostaglandins modulate variety of essential biological functions (including reproduction, immune function and vasodilation; Simmons et al., 2004, Takahashi et al., 2010, Gomez-Abellán et al., 2016). There are multiple COX isoforms in vertebrates (Havird et al., 2008); the COX-1 isoform is typically expressed constitutively and is involved in maintenance of homeostatic functions, while COX-2 is largely inducible (e.g., during discrete stages of gamete maturation, by inflammation; Simmons et al., 2004).
Inhibition of COX activity and PG synthesis is of special concern in fish because COX inhibitors are ubiquitous in fish tissues (e.g., Brozinski et al., 2012; Corcoran et al., 2010) and aquatic systems (typically nonsteroidal anti-inflammatory drugs present in the low μg/L range, with maximal concentrations nearing 100 μg/L in treated wastewater effluents; Corcoran et al., 2010; Koné et al., 2013), and because PGs play an important role in fish reproduction. In teleosts, PGs have been shown to mediate ovulation (e.g., Takahashi et al., 2013), and there is some evidence indicating their involvement in oocyte maturation (Lister and Van Der Kraak, 2008; Sorbera et al., 2001). Importance of PGs for fish ovulation has been conclusively demonstrated by studies that inhibited ovulation by impairing either PG synthesis (using the mammalian COX inhibitor IN; Lister and Van Der Kraak, 2008; Patino et al., 2003; Stacey and Pandey, 1975), or both PG synthesis and signaling (Fujimori et al., 2011). Prostaglandins also play an instrumental role in modulating fish reproductive behavior and synchronization of reproduction between sexes. For instance, endogenous PGs elicit spawning behavior in female fish (e.g., Kobayashi et al., 2002), while exogenous PGs (excreted by ovulating females) act as priming pheromones that facilitate physiological and behavioral reproductive readiness in males (Moore et al., 2002; Stacey and Sorensen, 2009). More specifically, waterborne PGF2α and its metabolites released by ovulating females facilitate synchronization of reproductive readiness by increasing luteinizing hormone, 17,20β-dihydroxy-4-pregnen-3-one, testosterone, and 11-ketotestosterone, expressible sperm and reproductive behaviors in receiving male fish (Kobayashi et al., 2002; Lado et al., 2013; Moore et al., 2002; Stacey and Sorensen, 2009).
Several studies have indicated that exposure to mammalian COX-inhibitors can adversely impact fish reproduction (e.g., Flippin et al., 2007; Yokota et al., 2015). Medaka ovulation assays (in vitro) indicated that diclofenac sodium, ketoprofen, salicylic acid, mefenamic acid, and acetylsalicylic acid exert anti-ovulatory activity, which correlated with data published on the inhibitory activity on human COX (Yokota et al., 2015). Flippin et al. (2007) observed that a 6-week exposure of adult medaka to waterborne ibuprofen (IB; 100 μg/l) led to a decreased number of spawning events. Lister and Van Der Kraak (2008) noted that exposure to indomethacin (IN; 100 μg/l) reduced cumulative egg output in zebrafish upon 16-d waterborne exposure. While the effects on egg output have been noted, molecular, cellular and organismal events underlying the observed reproductive impairments have not been fully elucidated. For example, it remains unclear whether mammalian COX-inhibitors can alter fish COX-activity (Knight and Van Der Kraak, 2015). Consequently, it has been postulated that their adverse reproductive effects on fish may be mediated via modes of action independent of COX-inhibition (e.g., Knight and Van Der Kraak, 2015).
To characterize mechanisms by which mammalian COX-inhibitors may impact reproduction of female fish, we evaluated effects of short-term, waterborne, exposure to IN and IB (non-selective mammalian COX-inhibitors), and celecoxib (CX; predominantly inhibits the COX-2 isoform) in adult teleost fish (Pimephales promelas - fathead minnow). We assessed effects on ovarian transcriptomic and hepatic metabolomic profiles, ovarian COX activity, and circulating PGF2α, 17β-estradiol (E2) and vitellogenin (VTG; egg yolk protein precursor). We hypothesized that exposure to mammalian COX-inhibitors would impact COX-activity, circulating prostaglandins and gene sets associated with PG synthesis and ovulation. Conversely, because the most sensitive targets of COX-inhibitors are COX activity and PGs, we did not anticipate significant effects on vitellogenesis and associated measures (plasma E2 and VTG). In vitro steroidogenesis assays indicate that IN has no impacts on E2 production (Aggregated Computational Toxicology Resource (ACToR; Judson et al., 2012) accessed May 14 2016), and that it inhibits COX activity and PGE2 synthesis at concentrations that are at least 10–30 times lower than concentrations that initiate next most sensitive molecular targets (i.e., interleukins, peroxisome proliferator-activated receptors).
In addition to generating empirical data specific to the action of COX-inhibitors in fish, we utilized publically available, high-throughput bioactivity data (ACToR), curated depositories of chemical-gene interactions (Comparative Toxicogenomics Database (CTD); Davis et al., 2013), and sequence similarity evaluation tool (Sequence Alignment to Predict Across Species Susceptibility; SeqAPASS; LaLone et al., 2016) to compare our empirical, fish-related findings to mammalian data, and to develop adverse outcome pathway (AOP) for COX inhibition. Development of AOPs has potential to support and enhance the use of mechanistic data in regulatory decision-making (Becker et al., 2015; Villeneuve et al. 2014a, b). It involves organizing knowledge about mechanistic relationships between initial chemical-biological interactions (molecular initiating events; MIEs), intermediary key events (KEs), and adverse outcomes (AOs) relevant to risk assessment (Ankley et al., 2010).
Methods
Chemical Selection and Identification of Exposure Concentrations
Because interactions of mammalian COX inhibitors (many of which are non-steroidal anti-inflammatory pharmaceuticals; NSAIDs) with fish COX are not well characterized, we utilized mammalian mechanistic data to guide chemical selection. We selected NSAIDs with varying COX inhibition mechanisms with regard to selectivity and time-dependence of binding to COX. Ibuprofen is a non-selective, reversible competitive inhibitor; it inhibits COX-1 and COX-2 activity immediately after addition, and is eliminated from the COX active site rapidly (i.e., is not time-dependent; Selinsky et al., 2001). Indomethacin is also a non-selective, reversible competitive inhibitor, but unlike IB it typically requires several minutes to bind, and takes hours to dissociate from the active site. Celecoxib, demonstrates distinct, three-step inhibitory mechanisms typical of diarylheterocyclic inhibitors; CX’s selectivity for COX-2 is achieved by a slow irreversible binding step which has been attributed to strong interactions with the sulfonamide (or sulfone) group that fits into the side-pocket of COX-2 (Selinsky et al., 2001; Walker et al., 2001).
Water exposure concentrations were selected based on a combination of factors. The main objective was to initiate a biological response based upon COX inhibition, without causing acute or overt toxicity due, for example, to narcosis or other reactive pathways. This could be particularly challenging with IB and IN, where margins of safety (for mammals) are among the smallest observed among all pharmaceuticals (Berninger and Brooks, 2010). Because fish toxicity data for these COX inhibitors were either not available (i.e., CX), or limited to a handful of studies and/or endpoints, we selected exposure concentrations for the present study based on both in vivo results (where possible) and human therapeutic concentrations (“read-across” approach; e.g., Hugget et al., 2003). While the “read-across” approach makes several assumptions that may affect accuracy of its toxicity estimate (e.g., the assumption is that there is a target conservation and that the interaction between drug and target will result in pharmacological response before a toxicological one; Rand-Weaver et al., 2013), it can provide an informative starting point for exposure concentration selection in absence of in vivo data for species of interest. To predict an effective dose for fish, the human peak plasma concentration (Cmax) and the log octanol-water partition coefficient (logKow) of the test chemicals were used to determine an aqueous effect threshold (AqET) - the concentration likely to result in an effect based on the pharmaceutical’s therapeutic mechanism of action. Approaches developed by Huggett et al. (2003) and modified by Berninger et al. (2011) were used for these calculations and predicted AqET concentrations of 2.8, 257, and 1.4 μg/l for CX, IB, and IN, respectively. Because all three compounds are weak bases, the equations were modified following published approaches (e.g., Berninger et al., 2011; Supplementary Information) resulting in logD adjusted AqET values of 2.8, 5902, and 142.6 μg/l for CX, IB, and IN, respectively. For IB and IN, concentrations where in vivo reproductive effects had been observed (100 μg/l; Flippin et al., 2007; Lister and Van Der Kraak, 2008) were also considered in exposure concentration selection. For IB, the predicted AqET and in vivo study responses matched fairly well (Flippin et al. (2007) observed decreased frequency of egg production at 100 μg/l IB), but given the short time course (4 d) of our experiment, and much higher logD adjusted AqET, a higher concentration (200 μg/l IB) was selected. For IN, the in vivo effect concentrations fell between the AqET and the adjusted AqET, thus a concentration of 100 μg/l was identified as likely to elicit COX inhibition during the 4 d exposure period; this is in agreement with in vivo studies that observed reduced egg output by IN at 100 μg/l (e.g., Lister and Van Der Kraak, 2008). For CX, a final value of 20 μg/l was ultimately selected, given its relatively larger margin of safety and our desire to be above a COX inhibition threshold. No in vivo fish reproduction data were available to further evaluate selected CX concentration.
Animals
All laboratory procedures involving animals were reviewed and approved by the local Animal Care and Use Committee in accordance with Animal Welfare Act regulations and Interagency Research Animal Committee guidelines. Adult, sexually mature fathead minnows (age 7 months, females) were obtained from an on-site culture facility at the U.S. Environmental Protection Agency (EPA) lab in Duluth, MN.
Chemical Exposures
Each chemical super-stock was prepared by dissolving “neat” chemical in UV-treated Lake Superior water (IB 10 mg/l; IN 1 mg/l and CX 1 mg/l) without use of carrier solvents. Stocks were sonicated for 2 h at 35°C to assure that chemicals were dissolved. Just prior to onset of chemical exposure, these stocks were further diluted with UV-treated Lake Superior water to achieve the target concentrations of 200 μg /l (IB), 100 μg /l (IN), and 20 μg /l (CX).
One day prior to the onset of exposure, eight adult female fish were placed in each of eight exposure aquaria (20-l aquaria). Chemical treatments (Control (C), IB, IN or CX) were randomly assigned, and each chemical exposure was conducted in duplicate. Throughout the exposure the fish were kept at 25°C under 16:8 h light:dark photoperiod, and fed adult brine shrimp (San Francisco Bay Brand, Newark, CA) twice a day. Control (UV-treated Lake Superior) or chemical enriched water was delivered to exposure tanks at a flow rate of 45 ml/min (complete tank renewal was achieved every 3.5 h). Chemical stocks, a water blank, chemical-spiked water samples, and a duplicate sample from each one of the exposure tanks (C, IB, IN and CX) were collected daily for confirmatory measurement of chemical concentrations.
After the 96 h of exposure, fish were removed from the test tanks, and anesthetized with tricaine methane sulfonate (100 mg/l buffered with 200 mg NaHCO3/l). Ovulation status was determined by presence/absence of expressible eggs upon applying gentle pressure on abdomen. Blood was collected from the caudal vein of the fish with a heparinized microcapillary tube and centrifuged to obtain plasma. Plasma was stored in microcentrifuge tubes at −80°C for VTG, E2 and PG analyses. Ovaries and livers were removed, divided equally amongst three microcentrifuge tubes, cryopreserved in liquid nitrogen, and stored at −80°C for metabolomics, transcriptomics and COX-activity analyses.
Chemical Analyses
Analysis of IN, IB and CX in water was performed by liquid chromatography. Sample aliquots of 50 μl were directly injected onto an Agilent 1100 Series HPLC with a Zorbax SB-C18 (2.1 × 75 mm, 3.5 μm) column. Separations were achieved at 200 μl/min using isocratic elution of 56% acetonitrile in 0.1% formic acid. Retention times (min) were 3.95, 4.40, and 5.0 for IN, IB, and CX, respectively. Concentrations of CX were determined by liquid chromatography-mass spectrometry (LC-MS) using negative polarity atmospheric pressure photoionization (APPI). Toluene was supplied as a dopant at 50 μl/min for the APPI source. The MS was operated in SIM (selective ion monitoring) mode at 380 and 381 m/z. Source parameters were optimized by flow injection analysis and were as follows: fragmentor voltage 100, gas temperature 350° C, vaporizer temperature 325° C, drying gas 10 l/min, nebulizer pressure 60 PSIG, capillary voltage 4000 v, gain 14. Ibuprofen and IN were both determined using a diode array detector (DAD) at 220 nm. Quantification of all compounds was performed by an external standard method. Concentrations of matrix spikes averaged 94.0 ± 0.05% (mean ± SD, n = 10), 101 ± 0.04% (n = 10), and 101 ± 0.04% (n = 10) for CX, IB, and IN, respectively. Duplicate analysis agreement was 96 ± 0.05% for CX (n = 5) and > 99% for both IB (n = 5) and IN (n = 5).
Ovarian COX Activity and Plasma E2, VTG and PG Analyses
For COX activity measurement, ovarian tissue collected from C, IB, IN and CX fish, was extracted following the procedure described by Lister and Van der Kraak (2008). Ovary pieces were homogenized with 0.1 M Tris-HCl buffer containing 1 mM EDTA (pH 7.8) at a ratio of 10 μl/mg tissue. The homogenates were centrifuged at 10,000 × g for 15 min at 4°C. The supernatant was removed, stored on wet ice and analyzed immediately. Ovarian total COX enzyme activity was measured and quantified using fluorescent activity assay kit designed to detect combined COX-1 and COX −2 activities (Cayman Chemicals, Ann Arbor, MI, USA).
Plasma prostaglandin F2α (PGF2α) concentrations were measured using an enzyme-linked immunosorbent assay (ELISA) per manufacturer’s instructions (Cayman Chemicals, Ann Arbor, MI, USA). Plasma VTG concentrations were determined using an ELISA with a polyconal antibody to fathead minnow VTG, and purified fathead minnow VTG as a standard (Ankley et al., 2001). Plasma E2 concentrations were measured by radioimmunoassay (Jensen et al., 2001). Statistical significance of effects of chemical treatment on COX activity, E2, PGF2α and VTG was evaluated using two-way ANOVA (with ovulation status, and chemical treatment as factors) followed by the Dunnett’s post-hoc test. The differences were considered significant if p < 0.05. All data were evaluated for normality (normal probability of residuals was examined) and homogeneity of variance (Lavene’s test). Because these requirements were violated in some cases, the data was also analyzed using non-parametric Kruskal-Wallis ANOVA (with chemical treatment as a factor). Because the outcomes of the non-parametric test were congruent with those of two-way ANOVA, which is robust and can tolerate some departure from the requirements of normality and homoscedasticity, we report the results of two-way ANOVA for COX, E2, VTG and PG analyses.
Metabolomics
Individual liver samples were extracted in a 96-well plate format using a dual-phase extraction process to profile changes in relative abundances of hepatic endogenous metabolites (Ekman et al., 2012). Polar extracts were vacuum dried and then reconstituted in 200 μl of 0.1 M sodium phosphate buffered deuterium oxide (pH 7.4) that contained 50 μM sodium 3-(trimethylsilyl) propionate-2,2,3,3-d4 (TSP). Samples were analyzed with 1H-NMR spectroscopy based on an automated push-through direct injection technique (Teng et al., 2012), using acquisition parameters reported in Supporting Information titled Metabolomics Methods.
NMR data analysis - The spectra were zero-filled, line broadened at 0.3Hz, and Fourier transformed (ACD/1D NMR Manager, Advanced Chemistry Development). Using an automated routine, the spectra were phase- and baseline-corrected, referenced to TSP, and binned at a width of 0.005 ppm. We excluded several regions of binned data to eliminate a residual water peak (4.82–4.88 ppm), residual methanol peak (3.33–3.37 ppm), and a large resonance from betaine that exhibited high leverage and variability (3.26–3.29 ppm). Remaining bins were then normalized to unit total integrated intensity. We used principal component analysis (PCA) to compare metabolite profiles across the four exposure classes (SIMCA-13.0, Umetrics Inc.), as this statistical framework has detected biological and metabolomic responses to a wide variety of contaminant exposures in other studies (e.g., Davis et al., 2013, Ekman et al., 2011, Viant et al., 2006). Cross-validation was used to determine the number of PCA components and cumulative Q2 (Q2cum) was used to assess model over-fitting. Outliers were identified with a Hotelling’s T2 test at the 95% confidence interval. A one-way analysis of variance (ANOVA) with a post-hoc Tukey’s test (R v.2.15.1, http://www.R-project.org) assessed whether average PCA score values differed among the exposure classes; a separate analysis was run for first and second component scores. To identify individual endogenous metabolites that were significantly affected by an exposure, we also generated t-test-filtered difference spectra based on the binned and normalized spectra. On a pairwise basis, we subtracted the average spectrum of control females from the spectrum of each female within a given exposure class. We then applied a t-test to each spectral bin to assess whether those average differences (i.e., exposed vs. control) were significantly different. For significantly different spectral bins, we identified metabolite peaks using Chenomx NMR Suite 7.0 (Chenomx Inc.). The suitability and interpretation of these statistical approaches have been justified and discussed elsewhere (Collette et al., 2010, Eriksson et al., 2006).
Transcriptomics
For microarray analyses, total RNA was isolated from female ovary samples (n= 7–8 per treatment) using an RNeasy Mini RNA purification kit (Qiagen Inc., Valencia, CA, USA). RNA quality was assessed with an Agilent 2100 Bioanalyzer (Agilent, Wilmington, DE, USA) and quantity was determined using a Nano-Drop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE). Total RNA was stored at −80°C until analyzed with oligonucleotide microarrays. Microarray analysis was conducted at the University of Florida (Gainesville, FL, USA) using a 15K feature fathead minnow microarray (Gene Expression Omnibus, Accession platform number GPL9248) designed in Dr. Nancy Denslow’s laboratory (University of Florida, Gainesville, FL, USA) and manufactured by Agilent Technologies (Palo Alto, CA, USA). RNA samples of 500 ng were used for cDNA synthesis; cRNA labeling, amplification, and hybridization were performed following the manufacturer’s kits and protocols (Quick Amp Labeling Kit; One-Color Microarray-Based Gene Expression Analysis Protocol v 5.7; Agilent Technologies). An Agilent Technologies G2505B scanner was used to scan microarray images at 5-μm resolution and data were extracted using Agilent Feature Extraction software version 9.5 (Agilent Technologies). Consistent with the “Minimum Information About a Microarray Experiment Standards” (Brazma et al., 2001), text versions of the Agilent raw data from this study have been deposited in the Gene Expression Omnibus (GSE72976).
Microarray data were imported into JMP Genomics version 4.1 (SAS Institute Inc., Cary, NC). Raw intensity data were subjected to a logarithm base-2 transformation followed by median normalization. Genes differentially transcribed (DEGs) between controls and fish exposed to either IB, IN, or CX were identified by one-way ANOVA (p < 0.05). The Database for Annotation, Visualization and Integrated Discovery (DAVID version 6.7; Huang et al., 2009) was used to identify statistical enrichment of groups of genes that were populating pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Tanabe and Kanehisa, 2012). Human homologs and human-specific pathways were used for these analyses. Human homologs were identified using the NCBI Human Reference Sequence database; e-value < 1e-4, and identity match > 75% were used as a criteria for homolog identification. Only genes represented on the microarray platform were used as background genes for KEGG analyses. Enrichment was considered significant if Expression Analysis Systematic Explorer (EASE) score < 0.1 (a recommended default cutoff); EASE score is a more conservative equivalent of one-tail Fisher Exact probability value (Hosack et al., 2003).
Because the primary purpose of the above microarray-generated data analyses was to generate mechanistic hypotheses we did not apply stringent multiple adjustment statistics/criteria to identify DEGs and associated enriched KEGG Pathways, as we wanted to limit number of false negatives for these exploratory analyses. Conversely, when conducting gene set enrichment analyses (GSEA; see below for details), where we tested pre-selected hypotheses (i.e., that exposure to COX-inhibitors would lead to enrichment of predefined sets of genes associated with fish reproductive function) we used a more conservative approach - false discovery rate (FDR) cutoff of 25% was used for GSEA (Subramanian et al., 2005).
To determine whether and how COX inhibitors interfere with sets of gene that modulate fish reproduction (Ingenuity Pathway Analyses (IPA; Ingenuity Systems, Inc., USA) and KEGG are poorly populated with fish-specific pathways) we conducted GSEA using gene sets corresponding to features represented within a conceptual model of the teleost fish brain-pituitary-gonadal-hepatic (BPGH) axis (Villeneuve et al., 2012). Briefly, these gene sets contain genes involved in normal BPGH functioning, as well as those indicative of exposure to endocrine-active chemicals. The gene sets are organized based on tissue type (ovary, liver, and brain compartments), and key functional categories (e.g., steroid biosynthesis, oocyte growth, vitellogenesis, PG-related; Villeneuve et al., 2012).
For GSEA analyses, the microarray raw data were normalized using Fastlo11 implemented in R (http://www.r-project.org/). GSEA was conducted using GSEA version 2.0 software (http://www.broadinstitute.org/gsea/) following published procedures (Subramanian et al., 2005). Normalized gene expression matrix files, and files defining the phenotypes (experimental cases) were constructed using custom R code (Villeneuve et al., 2012). These files, along with a .gmt file that included 23 gene sets (Villeneuve et al. 2012), were imported into the GSEA software. All GSEA analyses were performed using 1000 permutations, with the permutation type set to phenotype. Gene sets with size less than 10 or more than 500 were excluded from GSEA analyses. To determine significantly enriched gene sets binary combinations of phenotypes were considered in each analysis (i.e., C vs. IB, C vs. CX, C vs. IN); a FDR cutoff of 25% was used.
Finally, to broaden the scope of the analyses, IPA software was used to identify additional enriched canonical pathways, as well as putative chemicals and transcription factors potentially responsible for the observed gene expression changes.
Derivation and Evaluation of Putative AOPs
Putative AOPs (Villeneuve et al., 2014b) were developed following OECD guidelines (OECD 2013, 2014) as a part of OECD Project 1.29. “Catalog of putative AOPs that will enhance the utility of US EPA ToxCast high throughput screening data for hazard identification”. Each of these were deposited in the AOP Wiki (https://aopwiki.org) – a central repository for AOPs developed as a part of a joint effort between European Commission – DG Joint Research Centre (JRC) and U.S. EPA. Unique numbers have automatically been assigned to the AOPs by AOP-Wiki (63, 100, 101, 102 and 103); these numbers are subsequently used throughout when referring to these AOPs.
Preliminary assessment of the relative level of confidence in the newly developed putative AOPs was conducted following the weight of evidence considerations outlined by Becker et al. (2015). We evaluated the biological plausibility of the putative AOPs; this involved determining whether relationships between upstream and downstream KEs are consistent with established understanding of unperturbed biological pathways. We also addressed the essentiality of the KEs in a context of each AOP; essentiality is typically evaluated by determining whether downstream KEs/AOs can be prevented if upstream KEs are blocked. Typically, weight of evidence analysis for well-characterized AOPs also involves assessment of empirical support (e.g., toxicity studies with contaminants that disrupt MIE of interest) for temporal, dose-response, and incidence concordance. Given the putative nature of our AOPs, and the nature of this manuscript (research paper) we place emphasis on presentation/interpretation of the empirical data generated, and only incorporate the evaluation of empirical evidence in the discussion in a cursory fashion. Based on the collaborative ethos of the wiki platform for AOP development, it is expected that other investigators will contribute additional data and analyses that can help support or refute the relationships depicted. Nevertheless, major uncertainties, inconsistencies and data gaps related to the putative AOPs were identified and are presented.
Evaluation of Sequence/Structural Conservation of MIE
With knowledge that the chemicals of interest (IN, IB, CX) act on mammalian COX enzymes to deliver their therapeutic benefits, protein sequence and structural conservation was evaluated by comparing the human PG G/H synthase 1 (COX-1; National Center for Biotechnology Information (NCBI) accession NP_000953) and the human PG G/H synthase 2 (COX-2; NCBI accession NP_000954) enzymes across species using SeqAPASS tool (LaLone et al., 2016). Comparisons between the human proteins were conducted, initially, by assessing the percent similarity between primary amino acid sequences and then at the level of the functional domains (NCBI Conserved Domain Database, cd09816 for both COX-1 and COX-2), which contain the heme binding site, substrate binding site, and homodimer interface for each sequence. Further, individual amino acid residues deemed to be critical for inhibition of COX-2 were evaluated for conservation across species using NCBI Constraint-Based Local Alignment Tool, COBALT (Rowlinson et al., 2003). Specifically, the human COX-2 sequence (NP_000954) was used as the template, identifying residues that aligned with Tyr-348, Try-355, Tyr-385, and Ser-530 across taxa (Supplemental Data, COX-2 Individual AA).
Preliminary Evaluation of Functional Conservation
To evaluate concordance of the empirical, fish-specific data generated by our study with the published literature, we also queried the Comparative Toxicogenomics Database (CTD; http://ctdbase.org/), which largely curates chemical-gene/protein interaction data from empirical mammalian studies (Davis, 2013). We used CTD to extract genes and pathways associated with the chemical (IN, CX, or IB) exposure. Lastly, we used the ACToR’s Interactive Chemical Safety for Sustainability Dashboard (iCSS; http://actor.epa.gov/dashboard/) to investigate whether there were commonalities between IN fish gene expression and in vitro mammalian HTS data for IN provided via the iCSS Dashboard. This dashboard provides access to results of high throughput chemical screening data generated by Toxicity Forecaster (ToxCast) and Toxicity Testing in the 21st century (Tox21) programs (Kavlock et al., 2012; Schmidt, 2009).
Because the present manuscript is focused on the impacts of COX-inhibitors on reproduction-related genes and endpoints, data and assessment of non-reproduction related processes are not discussed in detail. However, these results, and statistical analyses relevant to other processes are presented in the supplementary information as they may be of interest to the community and may inform the development of other AOPs (Supplementary Tables 1–3).
RESULTS
Mean concentrations (μg/L ± SD) of chemicals in exposure tanks were 213 (19.1) for IB, 91.9 (6.6) for IN, and 29.3 (4.9) for CX. There was one mortality in the IB treatment; it occurred within 1.5 h of introduction of the fish to the exposure tank. The number of ovulating females (defined as females that had expressible eggs) on the morning of tissue collection (Day 4 of exposure) was similar across treatments; 56% for control and for 50% for IB, IN, and CX treatments.
Ovarian COX Activity
Chemical treatment had a significant effect on ovarian COX activity in IN and CX vs. control fish (Two-Way ANOVA, Figure 1A). There was also a significant effect of ovulation status on COX activity, and an interaction between ovulation status and COX inhibition. However, post-hoc tests did not detect significant differences between nonovulated controls vs. nonovulated IB, IN or CX-treated fish and ovulated controls vs. ovulated IB, IN or CX- treated fish. Thus, throughout the manuscript we refer to COX as not affected by IN, CX or IB. Both, log transformed and raw data yielded same results.
Figure 1.
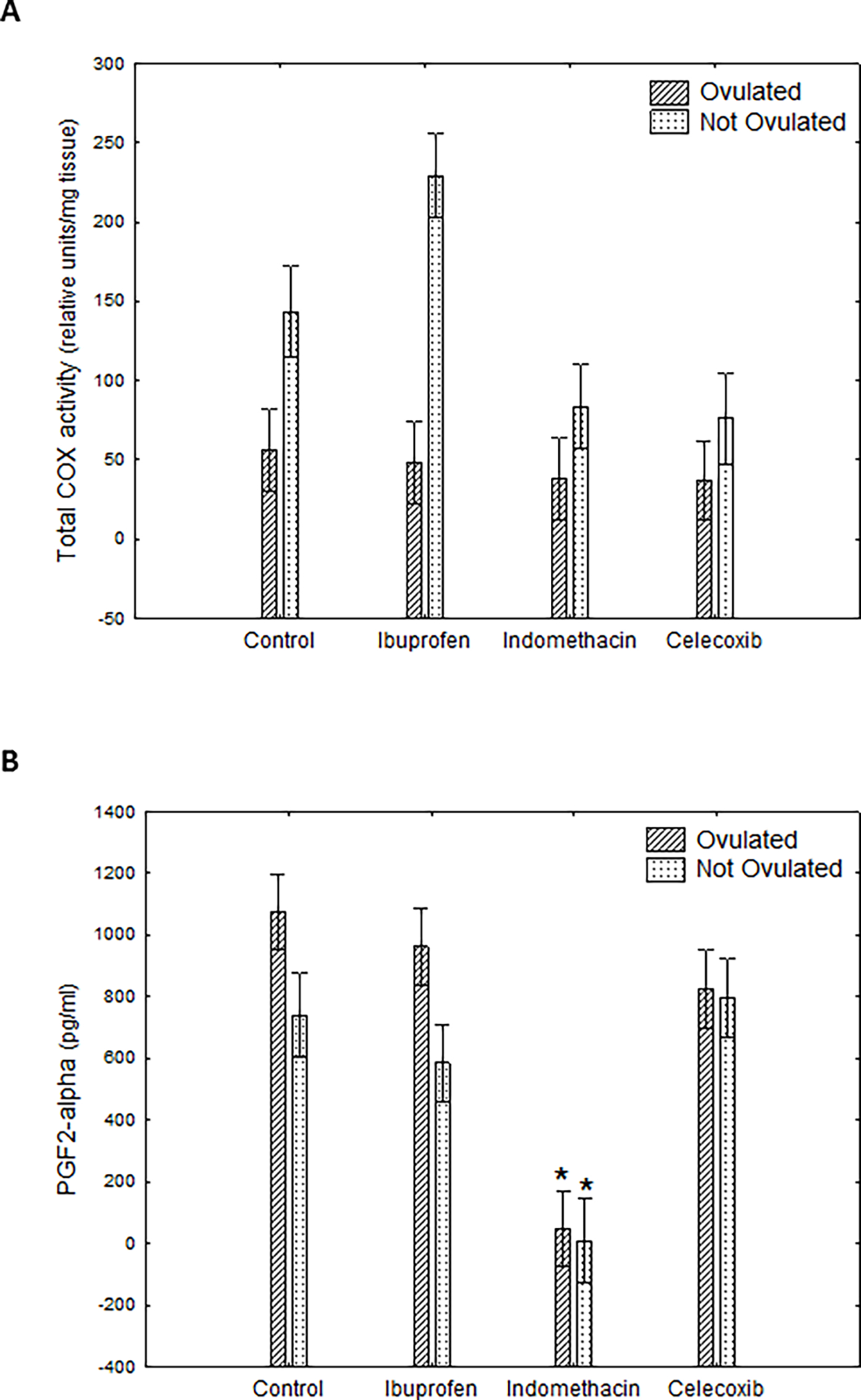
Effects of mammalian cyclooxygenase inhibitors on (A) ovarian COX activity (N=47), and, (B) circulating prostaglandin-F2α (N=64) in female fathead minnows (two-way ANOVA followed by the Dunnett’s post-hoc test). Specific treatments significantly different (Dunnett’s post-hoc test, p < 0.05) from their respective controls (i.e., ovulated treated versus ovulated controls, and non-ovulated treated versus non-ovulated controls) are labeled with an asterisk (*).
Plasma E2, VTG, PG Analyses
Both ovulation status and chemical treatment had an effect on plasma PGF2α concentrations, but there was no significant interaction between the two factors. Plasma PGF2α concentrations were higher in ovulated females. Only IN exposure significantly reduced concentrations of PGF2α; this effect was observed in both ovulated and non-ovulated fish (Figure 1B). There was no significant effect of ovulation status, chemical treatment, or their interaction on plasma E2 or VTG concentrations (Supplementary Figures 1A and 1B).
Metabolomics
Principal component analysis indicated that liver metabolomic profiles of females exposed to IN, IB or CX were similar to control fish; exposure classes were not significantly different from one another along either PC1 or PC2 (ANOVA PC1: F2, 43 = 0.571, p = 0.569; ANOVA PC2: F2, 43 = 0.871, p = 0.426). IN fish exhibited the most separation from other treatments (along PC2) suggesting that IN had the most extensive, albeit small, effects on the liver metabolome (Supplementary Figure 2A). Average difference spectra (exposed minus lab-control) of hepatic metabolites for IN, IB and CX females indicated that only a few metabolites changed relative to the controls (Supplementary Figure 2B). Due to the paucity of compelling liver metabolome responses, ovarian transcriptomic endpoints and other apical measurements ultimately served as the primary basis for AOP development. Nevertheless, while NMR methodologies may not be able to detect subtle effects, especially not the effects on the less abundant metabolites, the lack of effects observed via NMR has some utility for data interpretation - it indicates that the concentrations of COX-inhibitors were likely below those that would cause overt toxicity. This suggests that concentration we used in this study likely minimized initiation of less sensitive, secondary targets, and predominantly interacted with primary targets (COX activity/PG synthesis in this case), at least in case of IN.
Transcriptomics
When compared to controls, exposure to IB or CX resulted in differential expression of a comparable numbers of genes (IB = 433, CX= 545). In contrast, there were 2558 differentially expressed genes (DEGs) in IN-treated fish compared to controls, indicating more extensive effects. About 28% of IB DEGs and 26% of CX DEGs were also differentially expressed in IN fish (Supplementary Table 1); 24 DEGs were common to all three chemical treatments.
Gene set enrichment analyses (GSEA) –
Focused investigation of impacts of IN, IB and CX on gene sets involved in fish BPGH functioning (using gene lists from Villeneuve et al., 2012) indicated that multiple reproduction-related gene sets were altered (Table 1). The ‘Gonad prostaglandin-related’ gene set was significantly enriched in IN fish when compared to C (Table 1, Supplementary Table 2). The genes driving enrichment of the PG gene set were mildly (max 1.8 fold vs. control), but consistently up-regulated in IN-treated fish (e.g., PG-endoperoxide synthases 2 and 2a, carbonyl reductase 1, and PG D2 and E synthases). Interestingly, only one gene contributing to the PG gene set enrichment was down-regulated (0.85 fold vs. control) in IN fish - phospholipase A2 group IVa. GSEA for IB and CX did not reveal any effects on PG related gene set.
Table 1.
Overview of enrichment of predefined sets of genes associated with fish reproductive function - gene set enrichment analysis (GSEA) for microarray data.
| Gene Set (total number of genes in each gene set) | IN | IB | CX |
|---|---|---|---|
| Gonad All (78) | C | ||
| Gonad All With Neighbors (227) | C | ||
| Gonad Steroid Biosynthesis (11) | C | ||
| Gonad Prostaglandin-Related (10) | IN | ||
| Gonad Oocyte Maturation (21) | |||
| Gonad Oocyte Growth (10) | |||
| Gonad Cholesterol Synthesis (12) | CX | ||
| Gonad Minus Cholesterol Synthesis (67) | C | ||
| Gonad Ccnb1 Plus Neighbors (106) | |||
| Gonad Acvrl1 Plus Neighbors (12) | C | C | |
| Gonad Oocyte Maturation Plus Neighbors (143) | |||
| Gonad Oocyte Growth Plus Neighbors (113) | |||
| Gonad Cholesterol Uptake Plus Neighbors (15) | C | ||
| Gonad Steroid Biosynthesis Plus Neighbors (12) | C | ||
| Gonad Ccnb1 Zp3 Plus Non-Redundant Neighbors (118) | CX | ||
| Liver All (38) | C | ||
| Liver Cholesterol Biosynthesis (13) | |||
| Liver Steroid Metabolism (17) | |||
| Pituitary All (26) | |||
| Brain All (19) | |||
| Hypothalamic-Pituitary-Gonadal-Hepatic All (138) | C |
Gray shading indicates significant enrichment in the control group compared to the treated; black shading indicates significant enrichment in the treated group relative to control; lack of shading indicates no significant enrichment (false discovery rate < 25% was considered statistically significant). C - Control, IN - Indomethacin, IB - Ibuprofen, CX- Celecoxib.
When comparing IB and control fish, enrichment was observed only in controls (Table 1). The genes that contributed to core enrichment of gene sets ‘gonad steroid biosynthesis’ and ‘gonad steroid biosynthesis with neighbors’ included cytochrome P450, Family 19, Subfamily A, Polypeptide 1a (cyp19a1a) and cytochrome P450 17a1 (cyp17a1), hydroxy-delta-5-steroid dehydrogenase, 3 beta steroid delta-isomerase (3bhsd), and 11-beta-hydroxysteroid dehydrogenase-like (11bhsd-like) – all were downregulated in IB fish when compared to control.
CX had a somewhat different profile than IB; two gene sets were enriched in CX fish (Table 1) and four were enriched in controls (including ‘gonad activin receptor1 plus neighbors’, set, which was also enriched in controls when compared to IB; Table 1). Genes driving enrichment of the ‘cholesterol uptake’ gene set (e.g., apolipoprotein Eb (apoeb), steroidogenic acute regulatory protein (star)) were down-regulated in CX-treated fish. Two gene sets were enriched/upregulated in CX-exposed fish: 1) ‘gonad cholesterol synthesis’, and 2) ‘gonad_ccnb1_zp3_plus_non-redundant_neighbors’ - genes in this category are functionally associated with: 1) ribosomes, ribosomal proteins and translation, 2) zona pellucida and 3) rRNA binding.
In conclusion, GSEA pointed to a variety of responses indicative of effects on the BPGH functioning, but only IN–exposed fish exhibited enrichment of gene sets specifically associated with PG synthesis (additional GSEA outputs are provided for IN-exposed fish in Supplementary Table 2).
Canonical pathway enrichment –
Exposure to IN, IB and CX largely resulted in enrichment of: 1) canonical pathways associated with processes normally modulated by PGs (e.g., KEGG pathways for renin-angiotensin, cardiac contraction, oocyte meiosis, cell adhesion molecules, nucleotide excision repair; Iyer et al., 2000; Wennmalm, 2013; Zou et al., 2002), or 2) pathways indicative of known therapeutic effects/side effects of COX-inhibitors (e.g., beneficial uses for treatment of symptoms of neurodegenerative diseases and oxidative stress; Simmons et al., 2004; Supplementary Table 2). More specifically, three KEGG pathways were enriched in IB-treated fish: ERbB signaling, cell adhesion molecules and long term depression (Supplementary Table 2). Despite the low number of DEGs (545) in CX fish, seven KEGG pathways were significantly enriched, including neuroactive ligand-receptor interaction, renin-angiotensin systems, cell-adhesion molecules and the gonadotropin releasing hormone (GnRH) signaling pathway (Supplementary Table 2). Indomethacin impacted eight pathways, including Huntington’s, Alzheimer’s and Parkinson’s disease, ribosome, oxidative phosphorylation, cardiac muscle contraction and oocyte meiosis (Supplementary Table 2). Closer examination of the oocyte meiosis pathway enrichment indicates dysregulation by IN was not limited to a discrete portion of the pathway; several regulatory (androgen receptor; ar) and signaling molecules (adenylate cyclase [ac], protein kinase A [pka], ribosomal protein S6 kinase, 90kDa, polypeptide 6 [rsk 1/2], as well as the downstream genes (anaphase-promoting complex-cyclosome [apc/c], budding uninhibited by benzimidazoles [bub3], mitotic arrest deficient [mad1/2], and separase) involved in oocyte meiosis were impacted by IN-exposure (Figure 2).
Figure 2.
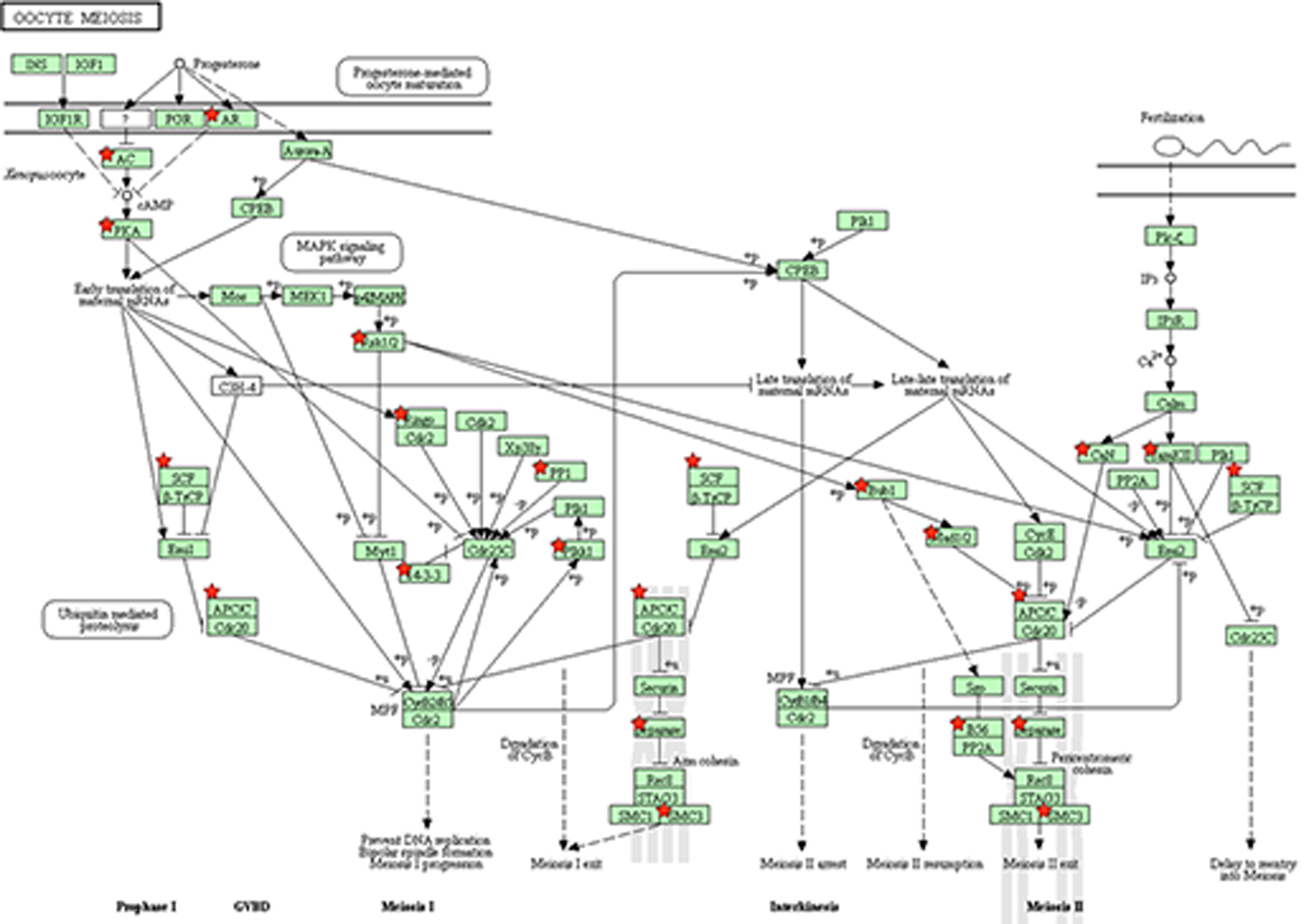
Enrichment of oocyte meiosis pathway in indomethacin-exposed fish. The functional annotation analyses (Database for Annotation, Visualization and Integrated Discovery; DAVID) indicated statistically significant enrichment of oocyte meiosis pathway (Kyoto Encyclopedia of Genes and Genomes). EASE Score (a more conservative equivalent of one-tail Fisher Exact probability value used by DAVID) was considered significant at p < 0.1. Differentially expressed genes contributing to oocyte meiosis pathway enrichment are labeled with a red star adjacent to the gene symbol.
Additional canonical pathways associated with reproductive function were enriched in IN fish (IPA analyses). These included androgen signaling, estrogen receptor signaling, GnRH signaling, dopamine receptor signaling, peroxisome proliferator-activated receptors/ retinoid X receptor (PPAR/RXR) activation, and glucocorticoid receptor and corticotropin releasing hormone signaling (Supplementary Table 3).
Derivation of Putative AOPs for COX Inhibition Leading to Reproductive Impairment
Because IN was the only chemical treatment that had an effect on circulating PG, and because those responses were congruent with the transcriptomic data (altered PG synthesis pathway; Table 1), we used IN as a model COX inhibitor for AOP development. Putative AOPs for COX inhibition leading to reproductive impairment via disruption of: 1) ovulation (Fig 3A; https://aopkb.org/aopwiki/index.php/Aop:63), 2) pheromonally-mediated synchronization of behavior and reproductive readiness (Figure 3B; https://aopkb.org/aopwiki/index.php/Aop:101), 3) performance of female spawning behavior (Figure 3C; https://aopkb.org/aopwiki/index.php/Aop:100) and 4) oocyte maturation (Figure 4, https://aopkb.org/aopwiki/index.php/Aop:102; https://aopkb.org/aopwiki/index.php/Aop:103) were developed.
Figure 3.

Putative adverse outcome pathways for cyclooxygenase inhibition leading to reproductive dysfunction via inhibition of (A) ovulation, (B) female spawning behavior, and (C) pheromone release in reproductively mature female fish. MIE - molecular initiating event, KE - key event, AO – adverse outcome.
Figure 4.
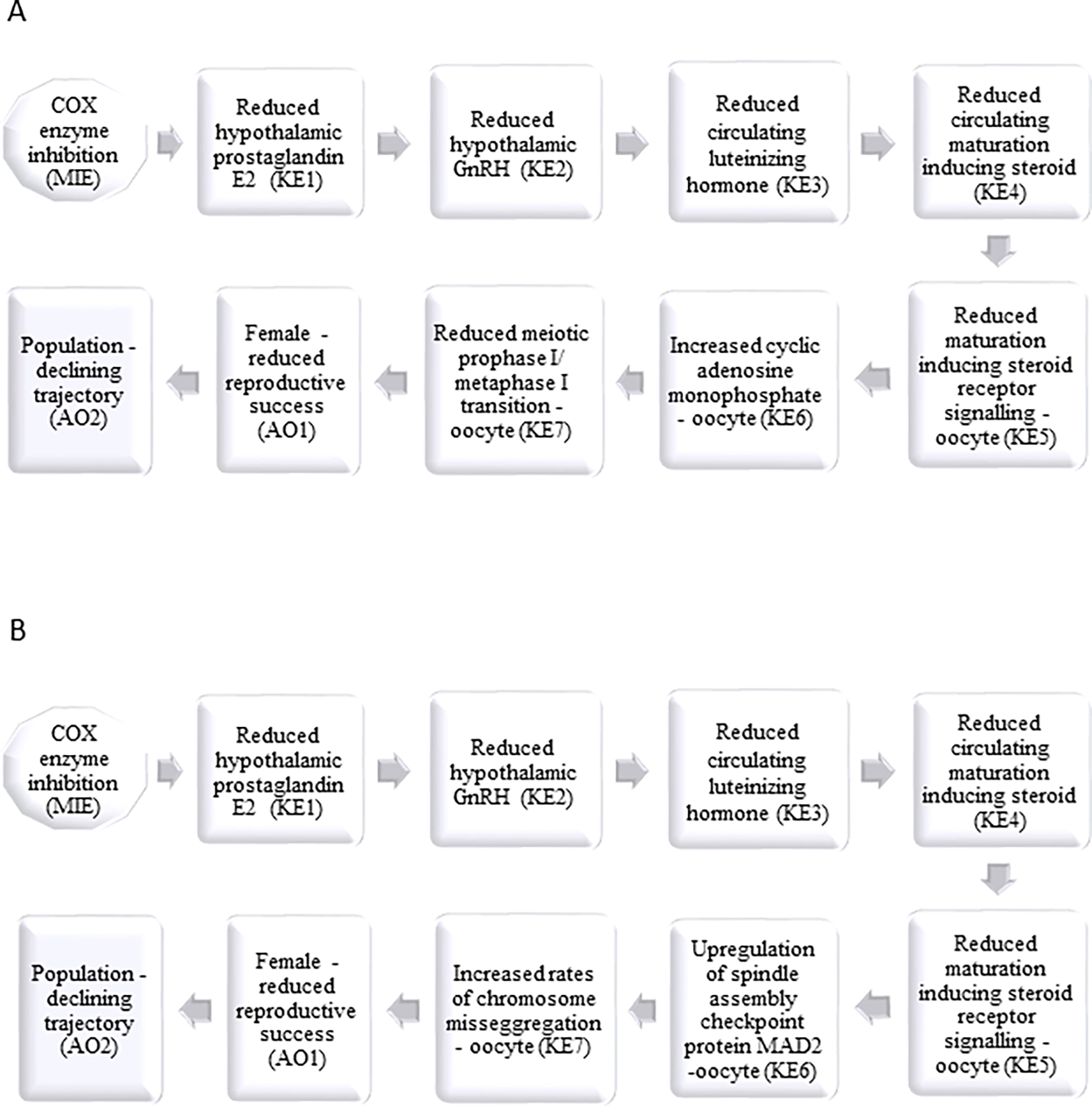
Putative adverse outcome pathways for cyclooxygenase inhibition leading to reproductive dysfunction via interference with of oocyte meiosis via (A) reduced meiotic prophase I/ metaphase I transition (reduced germinal vesicle breakdown) due to cyclic adenosine monophosphate increase, and (B) upregulation of spindle assembly checkpoint proteins and aneuploidy in reproductively mature female fish. MIE - molecular initiating event; KE - key event; AO – adverse outcome; E2– 17β-estradiol; GnRH – gonadotropin releasing hormone; MAD2 - mitotic arrest deficient-like 2.
At this early stage of AOP development, we elected to use total COX inhibition as the MIE. Currently there is only a weak empirical evidence that fish COX activity can be impacted by human COX-inhibitors (as determined by the indirect assessment of the peroxidase activity of the COX). However, based on a handful of empirical studies in which impacts on fish COX activity were observed (e.g., Flippin et al., 2007; Galus, 2014), the similarity of fish and mammalian COX functional domains, and the need to definitively address this gap in knowledge we designated decreased COX activity, and not PG reduction, as the MIE (for further discussion of this decision please see “Evaluation of Sequence/Structural Conservation of MIE” and Discussion sections). Moreover, we retained COX inhibition as the assumed MIE because of the high strength of empirical evidence for its inhibition by NSAIDs in humans, and because of the nature of the fish studies that reported lack of inhibition. Specifically, most of the fish studies have used COX-activity assays that were not optimized for fish (Patel, 2014), were conducted, and did not employ temporally or concentration-intense designs in well-characterized tissues that would allow one to confidently reject this hypothesis. In addition, conceptually one would not necessarily expect to observe effects on MIE and KEs at the same point in time. Indeed, the expected, temporally concordant pattern may be that effects such as that on COX activity is transient and by day 4 offset by compensatory responses, but nonetheless still influencing downstream events (i.e., circulating PGs). Furthermore, in many cases highly dynamic molecular scale responses, such as COX activity, will be much better studied/demonstrated in vitro, and thus lack of evidence for inhibition of COX with in vivo fish studies is not necessarily unexpected.
We did not attempt to resolve which COX isoform was impacted as there are typically 3–4 isoforms of COX in fish (Ishikawa et al., 2007), and their identities and specific roles in fish reproduction have not been fully characterized. A handful of studies that attempted to elucidate differential role of COX-1 vs. COX-2 in fish reproduction have indicated that both isoforms have roles in reproduction (zebrafish; Lister and Van der Kraak, 2008), and that their roles may be species-specific (Fujimori et al., 2011). Overall, COX-2 has been more strongly implicated as a driver of ovulation in some teleost species (i.e., medaka; Fujimori et al., 2011; Takahashi et al., 2013).
For the first three AOPs (Fig 3A–3C) COX inhibition is identified as the MIE which leads to the first KE - reduction of PG synthesis. Prostaglandins PGE2 and PGF2α are suggested for the first KE because they have been either reduced upon exposure to IN in our study, or because literature has indicated that they serve as the mediators of ovulation (e.g., PGE2- Fujimori et al., 2011; PGF2α - Crespo et al., 2015), female-specific spawning behavior (e.g., PGF2α - Juntti et al., 2016) and/or have a pheromonal function in fish (e.g., PGF2α - Stacey and Sorensen, 2009). After KE1 the AOPs diverge: 1) in ovarian theca cells KE1 leads to reduced stimulation of PG receptors in granulosa cells and thus impairment of ovulation (AOP 63; Fig. 3A), 2) in plasma and the preoptic area of the brain it leads to reduced stimulation of PG receptors, which results in reduced performance of female-specific spawning behaviors and subsequent decrease in female’s ability to attract spawning mates (AOP 100; Fig 3B), and 3) in urine it leads to reduced output of PG-based pheromones and subsequent decreases in female’s ability to attract/secure spawning mates (AOP 101; Fig. 3C). The AOPs defined above share the two AOs - reduced reproductive success and subsequent declining trajectory of the populations.
Two putative AOPs that involve dysregulation of GnRH and subsequent impairment of oocyte maturation (Figure 4) were developed in response to extensive transcriptomic effects along oocyte meiosis (Figure 2) and GnRH signaling pathways (Supplementary Table 3) in IN-exposed fish, as well as the evidence from mammalian literature indicating involvement of PGs in GnRH release and GnRH neuronal function (Clasadonte et al., 2011a, b). These two AOPs form an AOP network that shares the MIE and the first five KEs describing reduced PG synthesis, GnRH and LH secretion, and subsequent reduction of maturation inducing steroids (MIS). Because IN-exposed females exhibited significant alterations in GnRH signaling and downregulation of nuclear AR expression, as well as dysregulation of the androgen signaling pathways associated with both genomic and non-genomic androgen signaling we propose that impacts on oocyte maturation may be associated with these changes. Both, androgens (in mammals, amphibians and some fish; Hammes 2004; Tokumoto et al., 2011) and/or 17α, 20β-dihydroxy-4-pregnen-3-one (17,20βP and its derivatives in fish; Suwa and Yamashita 2007; Tokumoto et al., 2012) have been shown to serve as MIS. Several frog and fish studies indicate that androgens may play an important role in promoting oocyte maturation (most likely via classical nuclear AR agonism; Lutz et al., 2001; Hammes et al., 2004). For above reasons, COX inhibition and subsequent reduction in nuclear AR and/or membrane PR signaling (Figure 4) is hypothesized to lead to 1) increased intracellular cAMP (supported by the upregulation of adenylate cyclase and protein kinase A mRNA in IN-exposed fish) and reduced meiotic prophase I/ metaphase I transition (measurable by decreased germinal vesicle breakdown [GVBD] rates; Fig 4A, AOP 102), or 2) upregulation of spindle assembly checkpoint proteins (supported by the upregulation of Mad-2 in IN-exposed fish), and thus inhibition of anaphase onset of oocyte meiosis and interference with chromosome segregation (Fig 4B, AOP 103). Both of these AOPs culminate in reduced reproductive success and subsequent declining trajectory of the populations.
Evaluation of Sequence/Structural Conservation of MIE
SeqAPASS analyses of cross-species primary amino acid sequence similarity compared to the human COX-1 and COX-2 indicated a substantial degree of conservation across vertebrate species for both enzymes (Supplementary Figure 3). Sequence similarity ranging between 53.6 and 15.5% compared to human COX-2 was also identified with invertebrates (Bivalvia, Branchiostomidae, Anthozoa, Malacostraca, Ascidiacea, and Gastropoda). Ten invertebrates were identified as ortholog candidates (Supplementary Figure 3A and B). Of the vertebrate species sharing sequence similarity to the human COX-1 and COX-2, 90 and 54 of these protein sequences were identified as ortholog candidates, respectively (Supplemental Data Set 1, Primary Amino Acid Sequence Comparisons). Because ortholog sequences are more likely to maintain similar function, these results provide a line of evidence to indicate that vertebrates and some invertebrates preserve a relevant MIE for interaction with COX inhibitors.
Evaluation of sequence similarity within regions identified as the key functional domains for COX-1 indicated 69.7 to 96.6% similarity among ortholog candidates for species within the taxonomic groups Mammalia, Aves, Crocodylia, Testudines, Amphibia, Actinopteri, Lepidosauria, Chondrichthyes, and Chondrichthyes (Supplementary Figure 3C). Consistent with the primary amino acid sequence evaluation, COX-2 maintains conservation of the functional domain among both vertebrates and invertebrates (ortholog candidates between 47.4 and 97.7% similarity; Supplementary Figure 3D). These data provide an additional line of evidence that COX enzymes are conserved among vertebrates and some invertebrates, predicting they may similarly interact with known mammalian COX inhibitors.
SeqAPASS was also used to align four key residues linked to functional conformations relative to binding of COX-inhibitors. Specifically, site-directed mutagenesis and evaluation of a crystal structure of murine COX-2, identified amino acid residues important for chelation of ligands and direct chemical interaction. Using the human COX-2 as a template protein sequence, we aligned all species identified as sharing sequence similarity at the primary amino acid sequence level, specifically identifying residues aligning with tyrosine (Tyr) at position 348, Tyr at position 355, Tyr at position 385, and serine (Ser) at position 530. This analysis showed that all four structurally and functionally important residues were conserved across the majority of vertebrate species (including fish) and invertebrates from Bivalvia, Branchiostomidae, Gastropoda, Anthozoa (Table 2; Supplemental Data Set 2 - Individual residue comparison). The four human COX-2 amino acid residues aligned with 26 fish species (Organism class, Actinopteri). Overall, these data identified high conservation of key COX-2 residues comparing other vertebrates and certain invertebrates to humans, further supporting that chemical perturbation of COX-2 as a MIE could be well conserved across a variety of species.
Table 2.
Comparison of key amino acid residues across select vertebrate species from different taxonomic groups using human prostaglandin G/H synthase 2 as the template sequence.
| NCBI Accession | Protein Name | Species Name | aClass | Y348 | Y355 | Y385 | S530 |
|---|---|---|---|---|---|---|---|
|
| |||||||
| NP_000954.1 | prostaglandin G/H synthase 2 precursor | Homo sapiens | Mammalia | Y | Y | Y | S |
| XP_004028113.1 | PREDICTED: prostaglandin G/H synthase 2 | Gorilla gorilla gorilla | Mammalia | Y | Y | Y | S |
| XP_524999.3 | PREDICTED: prostaglandin G/H synthase 2 | Pan troglodytes | Mammalia | Y | Y | Y | S |
| XP_006023936.1 | PREDICTED: prostaglandin G/H synthase 2 isoform X2 | Alligator sinensis | Crocodylidae | Y | Y | Y | S |
| XP_006267636.1 | PREDICTED: prostaglandin G/H synthase 2 | Alligator mississippiensis | Crocodylidae | Y | Y | Y | S |
| XP_009504608.1 | PREDICTED: prostaglandin G/H synthase 2 isoform X1 | Phalacrocorax carbo | Aves | Y | Y | Y | S |
| EOB04442.1 | Prostaglandin G/H synthase 2, partial | Anas platyrhynchos | Aves | Y | Y | Y | S |
| XP_003208549.1 | PREDICTED: prostaglandin G/H synthase 2-like | Meleagris gallopavo | Aves | Y | Y | Y | S |
| NP_001161191.1 | prostaglandin G/H synthase 2 isoform 1 precursor | Gallus gallus | Aves | Y | Y | Y | S |
| XP_009086668.1 | PREDICTED: prostaglandin G/H synthase 2 | Serinus canaria | Aves | Y | Y | Y | S |
| XP_002190785.1 | PREDICTED: prostaglandin G/H synthase 2 | Taeniopygia guttata | Aves | Y | Y | Y | S |
| XP_007056755.1 | PREDICTED: prostaglandin G/H synthase 2 | Chelonia mydas | Testudines | Y | Y | Y | S |
| XP_005313923.1 | PREDICTED: prostaglandin G/H synthase 2 | Chrysemys picta bellii | Testudines | Y | Y | Y | S |
| XP_006110924.1 | PREDICTED: prostaglandin G/H synthase 2 | Pelodiscus sinensis | Testudines | Y | Y | Y | S |
| XP_003223472.1 | PREDICTED: prostaglandin G/H synthase 2 | Anolis carolinensis | Lepidosauria | Y | Y | Y | S |
| ETE58294.1 | Prostaglandin G/H synthase 2, partial | Ophiophagus hannah | Lepidosauria | Y | Y | Y | S |
| XP_007435833.1 | PREDICTED: prostaglandin G/H synthase 2 | Python bivittatus | Lepidosauria | Y | Y | Y | S |
| XP_006009311.1 | PREDICTED: prostaglandin G/H synthase 2 isoform X1 | Latimeria chalumnae | Lepidosauria | Y | Y | Y | S |
| NP_001020675.1 | prostaglandin G/H synthase 2 precursor | Danio rerio | Actinopteri | Y | Y | Y | S |
| XP_003445100.2 | PREDICTED: prostaglandin G/H synthase 2-like | Oreochromis niloticus | Actinopteri | Y | Y | Y | S |
| AAS21313.2 | cyclooxygenase 2 | Fundulus heteroclitus | Actinopteri | Y | Y | Y | S |
| NP_001118139.1 | prostaglandin G/H synthase 2 precursor | Oncorhynchus mykiss | Actinopteri | Y | Y | Y | S |
| NP_001165867.1 | prostaglandin G/H synthase 2 precursor | Oryzias latipes | Actinopteri | Y | Y | Y | S |
| NP_001025697.1 | prostaglandin G/H synthase 2 precursor | Xenopus (Silurana) tropicalis | Amphibia | Y | Y | Y | S |
| NP_001086946.1 | prostaglandin G/H synthase 2 precursor | Xenopus laevis | Amphibia | Y | Y | Y | S |
| YP_004935976.1 | prostaglandin G/H synthase 2 | Cercopithecine herpesvirus 5 | Herpesvirales | Y | Y | Y | S |
| AFL03426.1 | Rh10 | Macacine herpesvirus 3 | Herpesvirales | Y | Y | Y | S |
| XP_007893749.1 | PREDICTED: prostaglandin G/H synthase 2 | Callorhinchus milii | Chondrichthyes | Y | Y | Y | S |
| AAL37727.1 | AF420317_1 cyclooxygenase | Squalus acanthias | Chondrichthyes | Y | Y | Y | S |
| ACH73268.1 | cyclooxygenase-2 | Myxine glutinosa | Myxinformes | Y | Y | Y | S |
| ACP28169.2 | cyclooxygenase | Crassostrea gigas | Bivalvia | Y | Y | Y | S |
| XP_002586987.1 | hypothetical protein BRAFLDRAFT_129952 | Branchiostoma floridae | Branchiostomidae | Y | Y | Y | S |
| AHW81484.1 | cyclooxygenase-c | Branchiostoma belcheri tsingtauense | Branchiostomidae | Y | Y | Y | S |
| XP_009047579.1 | hypothetical protein LOTGIDRAFT_139178, partial | Lottia gigantea | Gastropoda | Y | Y | Y | S |
| XP_005090976.1 | PREDICTED: prostaglandin G/H synthase 1-like | Aplysia californica | Gastropoda | Y | Y | Y | S |
| AAF93168.1 | cyclooxygenase | Gersemia fruticosa | Anthozoa | Y | Y | Y | S |
| AAF93169.1 | cyclooxygenase | Plexaura homomalla | Anthozoa | Y | Y | Y | S |
| AHA44500.1 | cyclooxygenase | Penaeus monodon | Malacostraca | Y | Y | Y | S |
| ADB65785.1 | cyclooxygenase | Caprella sp. KV-2010a | Malacostraca | Y | Y | Y | S |
| ADB65786.1 | cyclooxygenase | Gammarus sp. KV-2010a | Malacostraca | Y | Y | Y | S |
| XP_002123273.1 | PREDICTED: prostaglandin G/H synthase 2-like | Ciona intestinalis | Ascidiacea | Y | F | Y | S |
| EFX85708.1 | putative cyclooxygenase | Daphnia pulex | Branchiopoda | Y | Y | Y | A |
|
| |||||||
| EHJ75729.1 | putative oxidase/peroxidase | Danaus plexippus | Insecta | W | E | H | F |
| XP_006558125.1 | PREDICTED: myeloperoxidase-like isoform X2 | Apis mellifera | Insecta | F | W | H | F |
| XP_003487986.1 | PREDICTED: hypothetical protein LOC100740410 | Bombus impatiens | Insecta | F | W | H | F |
| XP_004933948.2 | PREDICTED: peroxidase-like | Bombyx mori | Insecta | W | E | H | F |
| XP_005179733.1 | PREDICTED: uncharacterized protein LOC101892480 | Musca domestica | Insecta | F | W | H | F |
| EAT44728.2 | AAEL003933-PA | Aedes aegypti | Insecta | F | W | H | F |
| XP_003741036.1 | PREDICTED: uncharacterized protein LOC100899356 | Metaseiulus occidentalis | Arachnida | L | P | W | F |
| XP_002400188.1 | peroxidase, putative | Ixodes scapularis | Arachnida | F | P | W | F |
| AAO33164.1 | major ampullate gland peroxidase | Nephila senegalensis | Arachnida | Y | S | F | F |
| CBI34960.3 | unnamed protein product | Vitis vinifera | Rosids | W | T | Y | A |
| CDX93419.1 | BnaA06g06290D | Brassica napus | Rosids | W | T | Y | A |
| XP_008360922.1 | PREDICTED: alpha-dioxygenase 2-like | Malus domestica | Rosids | W | T | Y | A |
| XP_003553937.1 | PREDICTED: alpha-dioxygenase 1-like | Glycine max | Rosids | W | T | Y | A |
| XP_007210768.1 | hypothetical protein PRUPE_ppa020149mg | Prunus persica | Rosids | W | T | Y | A |
| BAJ90503.1 | predicted protein | Hordeum vulgare subsp. vulgare | Liliopsida | W | T | Y | A |
| CDM84254.1 | unnamed protein product | Triticum aestivum | Liliopsida | W | T | Y | A |
| EAY82977.1 | hypothetical protein OsI_38200 | Oryza sativa Indica Group | Liliopsida | W | T | Y | A |
| AFW75180.1 | hypothetical protein ZEAMMB73_220081 | Zea mays | Liliopsida | W | T | Y | A |
Note - Table abbreviations: Y = Tyrosine; W = Tryptophan; F = Phenylalanine; L = Leucine; E = Glutamic Acid; P = Proline; S = Serine; T = Threonine; H = Histidine; A = Alanine; Black shading indicates an amino acid residue that shares a similar side chain with the selected residue from the human template sequence. Green shading indicates amino acid residue that does not match the selected residue from the human template sequence.
Functional Conservation
To further evaluate the degree to which our proposed AOPs (early KEs in particular) might be applicable not only to fish, but higher vertebrates, we compared outcomes of our empirical IN fish data with human high throughput screening (HTS) data and CTD-curated chemical-gene interaction data (largely mammalian). Comparison of the IN-related empirical fish data with HTS outputs (http://actor.epa.gov/dashboard/) indicated many similarities between the human HTS (Supplementary Table 4) and fish datasets. The most sensitive targets of IN in HTS assays were PGs (i.e., PGE2 measured indirectly by assessing PG E2 receptor stimulation; EC50= 0.02 μM) and COX-1 activity (EC50 = 0.07 μM; COX −2 was not evaluated). Other sensitive targets of IN included cytokines (IL6 and CSF1; EC50 = 0.6–0.7 μM). At higher concentrations (EC50 = 2.8 – 18 μM), IN resulted in PPAR activation, matrix metalloproteinase upregulation, and alteration of endpoints associated with cell adhesion, dopamine transport and cell cycle. Majority of the above targets and/or pathways associated with those targets were also altered in or experimental fish exposed to IN (based on the transcriptomic data; Supplementary Tables 1–3).
Evaluation of CTD chemical-gene interaction outputs for IN indicated that the following reproduction-related pathways that were enriched in IN-exposed fish were also curated in CTD as being impacted by IN: steroid hormone biosynthesis, PPAR signaling, GnRH signaling, arachidonic acid metabolism, progesterone-mediated oocyte maturation and oocyte meiosis (Supplementary Table 5).
DISCUSSION
Lack of the effects on COX activity, plasma PGF2α and PG-related gene sets (via GSEA) in CX- and IB-exposed fish indicates that concentrations of CX and IB may have been below those needed to initiate the MIE of interest (COX) and subsequent PG reductions. However, it is notable that in both, IB and CX fish, canonical pathway enrichment analyses detected changes in gene expression indicative of exposure to COX-inhibitors (but GSEA analyses did not). This finding is not unexpected, as canonical analyses and GSEA approaches evaluated different sets of genes. The PG-related set for GSEA was largely comprised of a handful of genes involved in PG synthesis, whereas enriched canonical pathways captured a broad variety of processes that are associated with PG function or therapeutic targets of COX-inhibitors. Furthermore, as we have noted before, GSEA was statistically more conservative than canonical pathway analyses in terms of limiting false discovery rates. Unfortunately, our COX activity data did not provide definitive clarification as to whether the MIE of interest was influenced by IB and CX. There are several possible explanations for this. First, basal levels of COX activity in fish tissues are close to the limit of detection for the COX-activity assay, and thus unlikely to provide accurate and/or precise interpolation of COX activity (Patel, 2014). Second, the differences in the COX activity may be difficult to detect because COX activity is spatially and temporally dynamic, especially in the ovary of asynchronous spawning fish (Lister and Van der Kraak, 2008) such as the fathead minnow. Typically, COX and PGs are elevated only shortly before ovulation (e.g., Fujimori et al., 2011). Third, the homogenization and dilution of the samples may have impacted the evaluation of COX-activity inhibition – the inhibition is not based on covalent modification so it is plausible that tissue handling altered binding of COX-inhibitors.
Below, we examine extant literature to evaluate the proposition that the exposure concentrations of CX and IB were below those needed to initiate targets of interest (COX and PG) in fish. There are no published fish CX studies that could help with evaluation of this proposition. Existing IB studies (Bhandari and Venables, 2011; Patel, 2014) indicate that the “read-across” predicted IB concentration used in the present study was within the order of magnitude of the effective dose for fish, but did not offer a definitive answer. The exposure concentration used in our study (i.e. 200 μg/l IB) fell between a concentration that was too low (100 μg/L IB) to reach plasma IB level within the human Cmax range, and the concentration that was sufficient to do so (270 μg/l IB) (fathead minnow, 48h waterborne exposure; Patel, 2014). It is important to note that a large individual variability in IB plasma concentrations was observed for the 270 μg/L IB treatment - only one individual had IB plasma levels within Cmax range, whereas others were outside of the range (two were below and one was above Cmax; Patel, 2014). Patel (2014) could not reliably evaluate impacts on COX activity due to low basal levels of COX, but reported decreased gill PG metabolite levels upon exposure to 370 μg/l IB, but not 9 μg/l IB. Prostaglandin metabolite levels of fish exposed to IB concentrations similar to those used in the present study (200 μg/l) were not evaluated by Patel (2014). Bhandari and Venables (2011) found that exposure to 50 μg/L and 100 μg/L IB reduced PGE2 in the gills of a bluntnose minnow, Pimephales notatus (a species closely related to P. promelas), and based on the gill IB and PGE2 measurements concluded that “read-across” prediction based on human data underestimated sensitivity of the bluntnose minnow COX to IB. Because of lack of empirical evidence and poor understanding of intra- and inter-specific variation in absorption, distribution, metabolism, and sensitivity of fish COX to IB and CX, it remains uncertain whether the concentrations of CX and IB used in the present study were sufficient to impact COX and related processes. Therefore, only IN data (where significant impacts on plasma PGF2α and PG-related gene sets, and reproduction-related canonical pathways were observed) were used to inform AOP development.
Putative AOPs - Evidence
Reduction of PG synthesis, following exposure to cyclooxygenase inhibitors has been consistently demonstrated in mammals and is a basis for therapeutic use of these compounds (reviewed in Simmons et al., 2004). Fish studies that support this KE relationship are largely lacking. Although fish studies have consistently reported dose-dependent decreases in PGs upon exposure to mammalian COX inhibitors (e.g., Bhandari and Venables 2011; Grosser et al., 2002; Lister and Van der Kraak, 2008; Morthorst et al., 2013; Patel et al., 2014), empirical evidence that reduced PG synthesis is driven by COX inhibition in fish is very weak (including the present study). Some of the best evidence for the COX inhibition by mammalian COX-inhibitors in fish comes from studies on embryonic zebrafish where COX activity was measureable throughout gastrulation and segmentation, and was inhibited by both COX-1 (SC-560) and COX-2 specific (DuP-697) inhibitors (Galus, 2014). The lack of empirical data that support COX inhibition in fish (by mammalian COX inhibitors) is unexpected given the strong and consistent evidence for reduction of PGs in fish, as well as a high level of similarity of functional domains in teleost and mammalian COX (as indicated by the SeqAPASS analyses). Due to weak evidence for COX inhibition, additional time- and dose-intensive studies (including in vitro studies) with a variety of mammalian COX-inhibitors and variety of tissues are warranted. Furthermore, optimization of COX-activity assays that can be used for evaluation of basal COX-activity and/or assays that incorporate induction of inflammatory response may be needed to settle whether mammalian COX inhibitors can impact fish COX, and subsequent reduction in PGs (that has been widely and consistently documented in fish).
Transcriptomic data for IN-exposed females support the proposition that mammalian COX inhibitors can impact PG pathways in fish. Upregulation of genes involved in PG synthesis (e.g., ptges2/2a and PG synthase E) observed in the present study, likely reflects compensatory mechanisms to the IN-induced reduction of PG synthesis. For example, increased transcription of ptges2 and/or PG synthases has been reported as a compensatory response to COX inhibition by aspirin and CX in mammalian models (Johnson et al., 1997; Reese et al., 2001). Such compensatory effects combined with the ability of NSAIDs to retard degradation of COX protein (Kang et al., 2007) could have also interfered with the ability to detect effects on COX activity in fish.
It is biologically plausible that a reduction in PGs could lead to reduced stimulation of PG receptors. While specific and distinct roles of PGs and their receptors in mammalian reproduction are well elucidated (Funk et al., 2001; Richards et al., 2002; Sugimoto et al., 2015), comparable fish data are lacking. Collectively, fish studies indicate that there are species-specific differences in PGs and PG receptor types that mediate ovulation in fish. Both, PGFs (e.g., in trout; Cetta and Goetz, 1982) and PGEs (e.g., in medaka; Fujimori et al., 2011) and their respective receptors have been shown to be essential for ovulation in teleosts. In medaka, essentiality of PGE2 and its receptor (E type receptor 4; EP4) for ovulation has been demonstrated (Hagiwara et al., 2014). In vitro exposure of large-sized ovarian follicles to IN reduced PGE2 and ovulation, co-administration of PGE2 with IN rescued ovulation, and exposure to EP4 receptor antagonists diminished ovulation (Fujimori et al., 2011). Overall, there is a strong body of evidence that PGs and their receptors are indispensable for teleost follicle rupture (Takahashi et al., 2013) and that several pharmaceuticals designed to inhibit human COX enzymes (e.g., IN, NS-398, SC-560, diclofenac sodium, mefenamic acid) can block/decrease ovulation in fish likely via decreased PG production (Crespo et al., 2015; Joy and Singh, 2013; Lister and Van Der Kraak, 2008; Patino et al., 2003; Yokota et al., 2015) and reduced stimulation of PG receptors (Fujimori et al., 2011). Dose-dependence and temporal concordance of these KEs have been extensively characterized in mammalian models, but represent a data gap in fish studies.
In addition to impacting reproductive outcomes via inhibiting ovulation, the reduction of endogenous PGs could lead to reduced performance of spawning behavior in female fish (AOP 100; Fig. 3B). The importance of the ovulatory PGF2α surge for performance of female-specific spawning behaviors (and thus synchronization of female sexual activity with egg presence) is thoroughly and consistently documented in externally-fertilizing teleosts (e.g., Juntti et al., 2016; Kobayashi et al., 2002; Sorensen and Goetz, 1993). Essentiality of PGF2α for female spawning behavior has been demonstrated in goldfish where administration of IN blocked performance of female spawning behavior, which could be rescued by injection of PGF2α (Stacey, 1976). Furthermore, the studies of cichlid fish with mutant putative receptor for PGF2α (ptgfr) had shown that PGF2α signaling is necessary and sufficient for initiation of the final stages of female reproductive behavior (Juntti et al., 2016).
It is also biologically plausible that reduced synthesis and release of PGF2α (and its metabolites) by females could lead to reduced olfactory stimulation of males (AOP 101, Fig 3C), and thus impact female’s ability to attract/secure spawning mates, and to induce appropriate behavioral and physiological effects in males (i.e., both, increases in socio-sexual interaction, and circulating LH and milt volume are normally induced by pheromonal PGs) that ensure interspecific synchronization of spawning readiness (reviewed in Stacey and Sorensen, 2009). Essentiality of PG mediated pheromonal stimulation for reproduction in fish, and its disruption by IN has been demonstrated extensively (reviewed in Stacey and Sorensen, 2009).
In the present study, IN induced many gene expression changes indicative of interference with oocyte maturation. We propose that these changes are linked to PG reductions we observed, and could lead to: 1) reduced ability of oocyte to overcome cAMP mediated meiotic arrest (AOP 102), and 2) interference with spindle assembly checkpoint (AOP 103). In the present study IN-exposed fish exhibited upregulation of spindle assembly checkpoint proteins mad2 (15-fold) and bub3. Upregulation of mad2 mRNA in mammals has been shown to lead to cell cycle arrest in meiosis I (Wassman et al., 2003) and chromosome missegregation events in meiosis I (which can result in aneuploid metaphase II oocytes; Niault et al., 2007). Further evidence for interference with meiosis progression (in IN-exposed fish) was indicated by upregulated apc/c and dysregulation of rsk1 (Sun and Kim, 2012; Tunquist and Maller et al., 2003). These findings of cell cycle disruption are in concordance with high throughput mammalian data for IN (e.g., effects on cell cycle), and mammalian studies which documented effects of NSAIDs on cancer cell cycle via alteration of proteins involved in kinetochore/centromere assembly (Bienek et al., 2014) and cell cycle arrest (Narayanan et al., 2003). Nevertheless, it remains to be determined whether these are COX-inhibition/PG related effects. Unlike their ovulatory/behavioral function, the role of PGs in fish oocyte maturation (AOPs 102 and 103), while previously suggested (e.g., Lister and Van Der Kraak, 2008; Sorbera et al., 2001), is not well elucidated. Recent in vitro studies of role of PGs in amphibians demonstrated that PGF2a can induce GVBD and meiosis resumption up to metaphase II, and that IN administration can lead to dose-dependent decrease in arachidonic acid induced GVBD (Ortiz et al., 2014). Inhibition of in vitro spontaneous oocyte maturation of full-grown follicles by IN has also been observed in fish (Lister and Van der Kraak, 2008). These in vitro observations, indicate that effects of IN on the genes associated with oocyte maturation observed in this study, may not have to be controlled centrally (via HPG axis) as proposed in AOPs 102 and 103.
Because of evidence for a role of PGs in amphibian and mammalian oocyte maturation, and empirical transcriptomics fish data indicating impacts of IN on oocyte maturation (present study) we developed two putative AOPs linking COX-inhibition to impairment of oocyte maturation (102 and 103; Figure 3D, 3E). Prostaglandins have been shown to stimulate hypothalamic GnRH release (Ojeda et al., 1975) and GnRH neuron activity (Clasadonte et al., 2011b), and to modulate autoregulation of GnRHR and differential release of LH and FSH in mammalian models (Naidich et al., 2010; Naor et al., 2007). While these mechanisms have not been elucidated in fish, Pati and Habibi (2002) suggested involvement of arachidonic metabolism in fish GnRH signaling. Overall, evidence that supports KE relationships (especially those relating to interplay between GnRH and PGs) outlined in oocyte-maturation AOPs is extremely limited in fish, but given the extensive effects of IN on expression of genes associated with GnRH signaling and oocyte meiosis, and clearly defined role of PGs in oocyte maturation in mammals and amphibians, these AOPs deserve further attention.
Evaluation of Alternative MIE and/or Pleiotropic Potential of NSAIDs in Fish
Another important part of AOP development involves critical evaluation of potential alternative mechanisms that may explain observed toxicological effects (OECD, 2013). Such alternatives may either suggest other types of pathway-based assays needed to adequately detect impacts of this class of compounds and/or may identify other branches in the overall AOP network that must be considered to accurately predict related toxicological outcomes. The majority of the transcriptomic, pathway-level effects for all three chemicals (IN, CX and IB) were associated with processes like muscle contraction, renin-angiotensin system, reproduction, inflammation and neurodegenerative diseases, which are normally modulated by PGs (Funk et al., 2001), and/or constitute known therapeutic uses of COX-inhibitors (Cudaback et al., 2014; Simmons et al., 2004). This observation may seem surprising, as we did not always detect effects of CX and IB on COX inhibition and/or PGs - the main therapeutic targets for NSAIDs. There are several potential explanations. First, it is possible that observed transcriptomic effects in IB and CX fish were driven by COX inhibition/PG decreases, but that our experimental design, in terms of duration and dose, was not optimal for detection of COX inhibition/PGs. Temporally intensive studies with endocrine-active compounds have shown how factors like compensatory response can strongly influence dose-response time course behaviors, particularly for early KEs at the molecular and biochemical level that are subject to highly dynamic control (Ankley and Villeneuve, 2015).
Alternatively, the effects of IB and CX on PG-related processes could have been a result of COX-inhibition independent mechanisms. Such mechanisms have been described for NSAIDs in mammalian systems and the best understood ones include: 1) Inhibition of enzymes involved in arachidonic acid metabolism (i.e., lipooxygenases; Sagi et al., 2003), 2) Activation of PPARα/ɣ (Lehman et al., 1997; Romeiro et al., 2008), and 3) Inhibition of nuclear factor κB complex (NF-κB; Kopp and Ghosh, 1994). It is well established that, aside from COX inhibition, anti-inflammatory and reproductive effects of NSAIDs may be mediated by the activation of PPARs (Jaradat et al., 2001; Komar et al., 2005; Lehman et al., 1997) or NF-κB (Kopp et al. 1994; Sagi et al., 2003). For example, NSAIDs can modulate expression of ptgs2 via PPARy (Pang et al., 2003) or NF-κB (Tanabe et al., 2002). Decreases in the expression and activity of aromatase via activation of PPARα (Toda et al., 2003) and/or by disrupting the interaction of NF-κB with the aromatase promoter II (Fan et al., 2005) have also been noted. We observed both downregulation of aromatase expression (IB fish) and increased expression of ptges2 (IN fish). Overall, transcriptomics data from CX or IB-exposed fish, supported the proposition that COX-inhibition independent mechanisms (PPAR and NFκB) may have been involved. Transcriptomics data also indicated significant impacts on androgen signaling pathways in IN-exposed fish; anti-androgenic effects of NSAIDs via suppression of nuclear AR expression and signaling are well documented in mammalian cancer models (e.g., Kashiwagi et al., 2013; Pan et al., 2003).
Conclusion
We have outlined five putative AOPs linking COX-inhibition to reproduction-related AOs. While literature and analyses conducted in the present study indicate high level of sequence/structural/functional conservation of COX across vertebrates, direct empirical evidence for COX inhibition in fish remains weak, and majority of fish studies have not demonstrated the well-established phenomena of COX inhibition by NSAIDs observed in mammalian models. Further, time- and dose- intensive studies, with adequately optimized assays, are warranted to conclusively determine whether mammalian COX inhibitors can induce COX-inhibition in fish and whether this is the mechanism by which widely documented PG reduction is initiated. The assessment of confidence in the biological plausibility of AOPs 63, 100, and 101 indicated strong support. The evidence for essentiality of the KEs portrayed in these three pathways is ranked as moderate to high – there is a particularly large number of studies that established that the downstream KEs portrayed in these pathways (e.g., ovulation, spawning behavior) do not occur if PG synthesis is blocked/attenuated. In terms of biological domain of applicability of these three AOPs, we have found strong evidence that AOP 63 is applicable to female vertebrates, with biological plausibility for humans, rats, mice and select amphibians (Xenopus laevis and Rhinella arenarum) and teleost fish (e.g., Oryzas latipes, Danio rerio, Salmo sp.) being particularly strong (Crespo et al., 2015; Duffy et al., 2015; Hester et al., 2010; Lister and Van Der Kraak, 2008; Pall et al., 2001; Yokota et al., 2015). AOPs 101 and 102 are applicable to female Teleosts, cyprinids and cichlids in particular, where essentiality of PGs for performance of reproductive behavior, and pheromonal signaling of reproductive readiness has been extensively documented (e.g., Juntti et al., 2016). There is a strong evidence for essentiality and conservation of function of GnRH >> LH >>MIS >>oocyte maturation cascade captured in AOPs 102 and 103 across female vertebrates (Jalabert et al., 1991; Bobe et al., 2008). Conversely, evidence for biological plausibility of KER “reduced hypothalamic PGE2 leading to reduced hypothalamic GnRH” outlined in the oocyte maturation-related AOPs (102–103) is lacking for most vertebrates with the exception of mammals (Clasadonte et al., 2011b; Naor et al., 2007, Naidich et al., 2010). The role of PGs in modulation of hypothalamic GnRH, and its involvement in regulation of female reproduction and behavior warrants further study. It is important to note that there is strong in vitro evidence that PGs play a role in oocyte maturation and activation and pronuclei formation in female mammals and amphibians indicating that there may be another pathway, separate from the HPG-mediated one proposed in AOPs 102 and 103, that could explain extensive effects of IN on oocyte meiosis observed in the present study (Ortiz et al., 2015). There are no studies that systematically and/or concurrently evaluated effects of mammalian COX inhibitors on KE portrayed in these pathways (102, 103) in mammals or fish. Overall, evidence for temporal, dose-response, and incidence concordance for oocyte maturation AOPs (102 and 103) is lacking in both fish and mammals. Our data also allude to the possibility of alternative/pleiotropic mechanisms through which NSAIDs may elicit biological responses, which may ultimately prove adverse. Specifically, our data suggest that potential interactions with PPAR, AR, and steroidogenic enzymes (e.g., aromatase) might also need to be considered when trying to predict the toxicity of these compounds. Relative sensitivity of these molecular targets to NSAIDs (vs. COX inhibition) in fish and other non-human models, and their potential to lead to apical AOs relevant to ecological risk assessment should be elucidated further.
Supplementary Material
Acknowledgements
We thank Dr. Kellie Fay and three anonymous reviewers for a thoughtful peer review. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government. The contents neither constitute, nor necessarily reflect, US EPA policy.
Funding
This work was supported by the University of St. Thomas; US Environmental Protection Agency (EPA) (Office of Research and Development’s Chemical Safety for Sustainability Research Program) and Oak Ridge Institute Science and Education.
Footnotes
Supplementary Data Description
Supplementary Information contains detailed description of an aqueous effect threshold (AqET) and metabolomics methods.
Supplementary Tables contain the following: List of differentially expressed genes (Supplementary Table 1); KEGG Pathway and Gene Set Enrichment Analyses (Supplementary Table 2); Ingenuity Pathway Analysis outputs (Supplementary Table 3); ActoR iCSS Dashboard Exports for IN and CX (Supplementary Table 4); and results of CTD pathway enrichment analyses for IN, IB and CX (Supplementary Table 5).
Supplementary Figures contain data for circulating vitellogenin and estradiol (Supplementary Figure 1); metabolomics results (Supplementary Figure 2); and primary amino acid sequence alignments with human prostaglandin G/H synthase 1 (COX-1) precursor, prostaglandin G/H synthase 2 (COX-2) precursor; and alignments for COX-1 functional domain (prostaglandin endoperoxide synthase) and COX-2 functional domain (prostaglandin endoperoxide synthase) across select taxa (Supplementary Figure 3).
Supplemental Data Set 1 contains sequence alignments between primary amino acid sequences (Prostaglandin G_H synthase 1 precursor_Homo sapiens.csv and Prostaglandin G_H synthase 1 precursor_Homo sapiens.csv) and functional domains (cd09816_prostaglandin_endoperoxide_synthase for COX-1.csv and cd09816_prostaglandin_endoperoxide_synthase for COX-2.csv) comparing the human sequence to all species with sequence similarity.
Supplemental Data Set 2 includes Out_Test taxa.csv files containing alignments between key individual amino acid residues of all species from the named taxonomic group compared to the human COX-2.
References
- Ankley GT, Jensen KM, Kahl MD, Korte JJ, and Makynen EA (2001). Description and evaluation of a short-term reproduction test with the fathead minnow (Pimephales promelas). Environ. Toxicol. Chem. 20, 1276–1290. [PubMed] [Google Scholar]
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder PK, Serrrano JA, Tietge JE, and Villeneuve DL (2010). Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem. 29, 730–741. [DOI] [PubMed] [Google Scholar]
- Ankley GT and Villeneuve DL (2015). Temporal changes in biological responses and uncertainty in assessing risks of endocrine-disrupting chemicals: insights from intensive time-course studies with fish. Toxicol. Sci. 144, 259–275. [DOI] [PubMed] [Google Scholar]
- Becker RA, Ankley GT, Edwards SW, Kennedy S, Linkov I, Meek B, Sachana M, Segner H, Van Der Burg B, Villeneuve DL, Watanabe H, and Barton-Maclaren TS (2015). Increasing Scientific Confidence in Adverse Outcome Pathways: Application of Tailored Bradford-Hill Considerations for Evaluating Weight of Evidence. Regul. Toxicol. Pharmacol. 72, 514–537. [DOI] [PubMed] [Google Scholar]
- Berninger JP and Brooks BW (2010). Leveraging mammalian pharmaceutical toxicology and pharmacology data to predict chronic fish responses to pharmaceuticals. Toxicol. Lett. 193, 69–78. [DOI] [PubMed] [Google Scholar]
- Berninger JP, Du B, Connors KA, Eytcheson SA, Kolkmeier MA, Prosser KN, Valenti TW, Chambliss CK, and Brooks BW (2011). Effects of the antihistamine diphenhydramine on selected aquatic organisms. Environ. Toxicol. Chem. 30, 2065–2072. [DOI] [PubMed] [Google Scholar]
- Bhandari K and Venables B (2011). Ibuprofen bioconcentration and prostaglandin E2 levels in the bluntnose minnow Pimephales notatus. Comp. Biochem. Physiol. C: Pharmacol. Toxicol. 153, 251–257. [DOI] [PubMed] [Google Scholar]
- Bieniek J, Childress C, Swatski MD, and Yang W (2014). COX-2 inhibitors arrest prostate cancer cell cycle progression by down-regulation of kinetochore/centromere proteins. Prostate. 74, 999–1011. [DOI] [PubMed] [Google Scholar]
- Bobe J, Jalabert B and Fostier A (2008). Oogenesis: post-vitellogenic events leading to a fertilizable oocyte. Fish Reproduction. Science Publishers, Enfield. 1–36. [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, and Causton HC (2001). Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29, 365–371. [DOI] [PubMed] [Google Scholar]
- Brozinski J, Lahti M, Meierjohann A, Oikari A, and Kronberg L (2012). The anti-inflammatory drugs diclofenac, naproxen and ibuprofen are found in the bile of wild fish caught downstream of a wastewater treatment plant. Environ. Sci. Technol. 47, 342–348. [DOI] [PubMed] [Google Scholar]
- Cetta F and Goetz FW (1982). Ovarian and plasma prostaglandin E and F levels in brook trout (Salvelinus fontinalis) during pituitary-induced ovulation. Biol. Reprod. 27, 1216–1221. [DOI] [PubMed] [Google Scholar]
- Clasadonte J, Sharif A, Baroncini M, and Prevot V (2011a). Gliotransmission by prostaglandin E2: a prerequisite for GnRH neuronal function? Front. Endocrinol. 2, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasadonte J, Poulain P, Hanchate NK, Corfas G, Ojeda SR, and Prevot V (2011b). Prostaglandin E2 release from astrocytes triggers gonadotropin-releasing hormone (GnRH) neuron firing via EP2 receptor activation. Proc. Natl. Acad. Sci. U. S. A. 108, 16104–16109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran J, Winter MJ, and Tyler CR (2010). Pharmaceuticals in the aquatic environment: a critical review of the evidence for health effects in fish. Crit. Rev. Toxicol. 40, 287–304. [DOI] [PubMed] [Google Scholar]
- Collette TW, Teng Q, Jensen KM, Kahl MD, Makynen EA, Durhan EJ, Villeneuve DL, Martinović-Weigelt D, Ankley GT. and Ekman DR. (2010). Impacts of an anti-androgen and an androgen/anti-androgen mixture on the metabolite profile of male fathead minnow urine. Environ. Sci. Technol. 44, 6881–6886. [DOI] [PubMed] [Google Scholar]
- Crespo D, Goetz FW, Planas JV (2015). Luteinizing hormone induces ovulation via tumor necrosis factor α-dependent increases in prostaglandin F2α in a nonmammalian vertebrate. Sci. Rep. 5, 1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudaback E, Jorstad NL, Yang Y, Montine TJ, and Keene CD (2014). Therapeutic implications of the prostaglandin pathway in Alzheimer’s disease. Biochem. Pharmacol. 88, 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Murphy CG, Johnson R, Lay JM, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, Rosenstein MC, and Wiegers TC (2012). The comparative toxicogenomics database: update 2013. Nucleic Acids Res. gks994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Collette TW, Villeneuve DL, Cavallin JE, Teng Q, Jensen KM, Kahl MD, Mayasich JM, Ankley GT, Ekman DR (2013). Field-based approach for assessing the impact of treated pulp and paper mill effluent on endogenous metabolites of fathead minnows (Pimephales promelas). Environ. Sci. Technol. 47, 10628–10636. [DOI] [PubMed] [Google Scholar]
- Duffy DM (2015). Novel contraceptive targets to inhibit ovulation: the prostaglandin E2 pathway. Hum. Reprod. Update, p.dmv026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman DR, Villeneuve DL, Teng Q, Ralston-Hooper KJ, Martinović-Weigelt D, Kahl MD, Jensen KM, Durhan EJ, Makynen EA, Ankley GT, Collette TW (2011). Use of gene expression, biochemical, and metabolite profiles to enhance exposure and effects assessment of the model androgen 17 beta-trenbolone in fish.” Environ. Toxicol. Chem. 30, 319–329. [DOI] [PubMed] [Google Scholar]
- Ekman D, Hartig P, Cardon M, Skelton D, Teng Q, Durhan E, Jensen K, Kahl M, Villeneuve D, and Gray L Jr (2012). Metabolite profiling and a transcriptional activation assay provide direct evidence of androgen receptor antagonism by bisphenol A in fish. Environ. Sci. Technol. 46, 9673–9680. [DOI] [PubMed] [Google Scholar]
- Eriksson L, Kettaneh-Wold N, Trygg J, Wikström C, and Wold S (2006). Multi-and megavariate data analysis: Part I: basic principles and applications. 2nd ed. Umetrics, Umeå, Sweden [Google Scholar]
- Fan W, Yanase T, Morinaga H, Mu Y, Nomura M, Okabe T, Goto K, Harada N, and Nawata H (2005). Activation of peroxisome proliferator-activated receptor-γ and retinoid X receptor inhibits aromatase transcription via nuclear factor-κB. Endocrinology. 146, 85–92. [DOI] [PubMed] [Google Scholar]
- Flippin JL, Huggett D, and Foran CM (2007). Changes in the timing of reproduction following chronic exposure to ibuprofen in Japanese medaka, Oryzias latipes. Aquat. Toxicol. 81, 73–78. [DOI] [PubMed] [Google Scholar]
- Fujimori C, Ogiwara K, Hagiwara A, Rajapakse S, Kimura A, and Takahashi T (2011). Expression of cyclooxygenase-2 and prostaglandin receptor EP4b mRNA in the ovary of the medaka fish, Oryzias latipes: possible involvement in ovulation. Mol. Cell. Endocrinol. 332, 67–77. [DOI] [PubMed] [Google Scholar]
- Funk CD (2001). Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 294, 1871–1875. [DOI] [PubMed] [Google Scholar]
- Galus M (2014). Chronic Exposure to Environmentally Relevant Pharmaceutical Concentrations Effects Reproductive and Developmental Physiology in Zebrafsih (Danio rerio). (Doctoral dissertation, McMaster University Hamilton) http://hdl.handle.net/11375/16282 [Google Scholar]
- Gómez-Abellán V, and Sepulcre MP (2016). The role of prostaglandins in the regulation of fish immunity.” Mol. Immunol. 69, 139–145. [DOI] [PubMed] [Google Scholar]
- Grosser T, Yusuff S, Cheskis E, Pack MA, and FitzGerald GA (2002). Developmental expression of functional cyclooxygenases in zebrafish. Proc. Natl. Acad. Sci. U. S. A. 99, 8418–8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara A, Ogiwara K, Katsu Y and Takahashi T (2014). Luteinizing hormone-induced expression of Ptger4b, a prostaglandin E2 receptor indispensable for ovulation of the medaka Oryzias latipes, is regulated by a genomic mechanism involving nuclear progestin receptor. Biol. Reprod. 90, 126. [DOI] [PubMed] [Google Scholar]
- Hammes SR (2004). Steroids and oocyte maturation—a new look at an old story. Mol. Endocrinol. 18, 769–775. [DOI] [PubMed] [Google Scholar]
- Havird JC, Miyamoto MM, Choe KP, and Evans DH (2008). Gene duplications and losses within the cyclooxygenase family of teleosts and other chordates. Mol. Biol. Evol. 25, 2349–2359. [DOI] [PubMed] [Google Scholar]
- Hester KE, Harper MJ and Duffy DM (2010). Oral administration of the cyclooxygenase-2 (COX-2) inhibitor meloxicam blocks ovulation in non-human primates when administered to simulate emergency contraception. Hum. Reprod. 25, 360–367. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Sherman BT, Lane HC, Lempicki RA (2003). Identifying biological themes within lists of genes with EASE. Genome Biol. 4, (10)1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, and Lempicki RA (2009). Extracting biological meaning from large gene lists with DAVID. Current. Protoc. Bioinformatics. 13.11. 1–13.11. 13. [DOI] [PubMed] [Google Scholar]
- Huggett D, Cook J, Ericson J, and Williams R (2003). A theoretical model for utilizing mammalian pharmacology and safety data to prioritize potential impacts of human pharmaceuticals to fish. Hum. Ecol. Risk Assess. 9, 1789–1799. [Google Scholar]
- Ishikawa TO, Griffin KJ, Banerjee U and Herschman HR (2007). The zebrafish genome contains two inducible, functional cyclooxygenase-2 genes. Biochem. Bioph. Res. Co. 352, 181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SN, Yamada K, Diz DI, Ferrario CM, and Chappell MC (2000). Evidence that prostaglandins mediate the antihypertensive actions of angiotensin-(1–7) during chronic blockade of the renin-angiotensin system. J. Cardiovasc. Pharmacol. 36, 109–117. [DOI] [PubMed] [Google Scholar]
- Jalabert BE, Fostier AL, Breton BE, Weil CL (1991). Oocyte maturation in vertebrates. Vertebrate endocrinology: fundamentals and biomedical implications. 4 (part A), 23–90. [Google Scholar]
- Jaradat MS, Wongsud B, Phornchirasilp S, Rangwala SM, Shams G, Sutton M, Romstedt KJ, Noonan DJ, and Feller DR (2001). Activation of peroxisome proliferator-activated receptor isoforms and inhibition of prostaglandin H 2 synthases by ibuprofen, naproxen, and indomethacin. Biochem. Pharmacol. 62, 1587–1595. [DOI] [PubMed] [Google Scholar]
- Jensen KM, Korte JJ, Kahl MD, Pasha MS, and Ankley GT (2001). Aspects of basic reproductive biology and endocrinology in the fathead minnow (Pimephales promelas). Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 128, 127–141. [DOI] [PubMed] [Google Scholar]
- Johnson RD, Polakoski K, Everson WV, and Nelson DM (1997). Aspirin induces increased expression of both prostaglandin H synthase-1 and prostaglandin H synthase-2 in cultured human placental trophoblast. Obstet. Gynecol. 177, 78–85. [DOI] [PubMed] [Google Scholar]
- Judson RS, Martin MT, Egeghy P, Gangwal S, Reif DM, Kothiya P, Wolf M, Cathey T, Transue T, and Smith D (2012). Aggregating data for computational toxicology applications: The US Environmental Protection Agency (EPA) aggregated computational toxicology resource (ACToR) system. Int. J. Mol. Sci. 13, 1805–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy KP and Singh V (2013). Functional interactions between vasotocin and prostaglandins during final oocyte maturation and ovulation in the catfish Heteropneustes fossilis. Gen. Comp. Endocrinol. 186, 126–135. [DOI] [PubMed] [Google Scholar]
- Juntti SA, Hilliard AT, Kent KR, Kumar A, Nguyen A, Jimenez MA, Loveland JL, Mourrain P, and Fernald RD (2016). A Neural Basis for Control of Cichlid Female Reproductive Behavior by Prostaglandin F2α. Curr. Biol. 26, 943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Mbonye UR, DeLong CJ, Wada M, and Smith WL (2007). Regulation of intracellular cyclooxygenase levels by gene transcription and protein degradation. Prog. Lipid Res. 46, 108–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi E, Shiota M, Yokomizo A, Itsumi M, Inokuchi J, Uchiumi T, and Naito S (2013). Prostaglandin receptor EP3 mediates growth inhibitory effect of aspirin through androgen receptor and contributes to castration resistance in prostate cancer cells. Endocr. Relat. Cancer. 20, 431–441. [DOI] [PubMed] [Google Scholar]
- Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, Knudsen T, Martin M, Padilla S, and Reif D (2012). Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol. 25, 1287–1302. [DOI] [PubMed] [Google Scholar]
- Knight OM and Van Der Kraak G (2015). The role of eicosanoids in 17α, 20β-dihydroxy-4-pregnen-3-one-induced ovulation and spawning in Danio rerio. Gen. Comp. Endocrinol. 213, 50–58. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Sorensen PW, and Stacey NE (2002). Hormonal and pheromonal control of spawning behavior in the goldfish. Fish Physiol. Biochem. 26, 71–84. [Google Scholar]
- Komar CM (2005). Peroxisome proliferator-activated receptors (PPARs) and ovarian function--implications for regulating steroidogenesis, differentiation, and tissue remodeling. Reprod. Biol. Endocrinol. 3, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koné M, Cologgi DL, Lu W, Smith DW, Ulrich AC (2013). Pharmaceuticals in Canadian sewage treatment plant effluents and surface waters: occurrence and environmental risk assessment. Environ. Technol. Rev. 2, 17–27. [Google Scholar]
- Kopp E and Ghosh S (1994). Inhibition of NF-kappa B by sodium salicylate and aspirin. Science. 265, 956–959. [DOI] [PubMed] [Google Scholar]
- Lado WE, Zhang D, Mennigen JA, Zamora JM, Popesku JT, and Trudeau VL (2013). Rapid modulation of gene expression profiles in the telencephalon of male goldfish following exposure to waterborne sex pheromones. Gen. Comp. Endocrinol. 192, 204–213. [DOI] [PubMed] [Google Scholar]
- LaLone CA, Villeneuve DL, Lyons D, Helgen HW, Robinson SL, Swintek JA, Saari TW and Ankley GT (2016). Sequence Alignment to Predict Across Species Susceptibility (SeqAPASS): A web-based tool for addressing the challenges of cross-species extrapolation of chemical toxicity. Toxicol. Sci. p.kfw 119. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Lenhard JM, Oliver BB, Ringold GM, and Kliewer SA (1997). Peroxisome proliferator-activated receptors α and γ are activated by indomethacin and other non-steroidal anti-inflammatory drugs. J. Biol. Chem. 272, 3406–3410. [DOI] [PubMed] [Google Scholar]
- Lister A and Van Der Kraak G (2008). An investigation into the role of prostaglandins in zebrafish oocyte maturation and ovulation. Gen. Comp. Endocrinol. 159, 46–57. [DOI] [PubMed] [Google Scholar]
- Lutz LB, Cole LM, Gupta MK, Kwist KW, Auchus RJ, and Hammes SR (2001). Evidence that androgens are the primary steroids produced by Xenopus laevis ovaries and may signal through the classical androgen receptor to promote oocyte maturation. Proc. Natl. Acad. Sci. U. S. A. 98, 13728–13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A, Olsén KH, Lower N, and Kindahl H (2002). The role of F-series prostaglandins as reproductive priming pheromones in the brown trout. J. Fish Biol. 60, 613–624. [Google Scholar]
- Morthorst JE, Lister A, Bjerregaard P, and Van Der Kraak G (2013). Ibuprofen reduces zebrafish PGE 2 levels but steroid hormone levels and reproductive parameters are not affected. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 157, 251–257. [DOI] [PubMed] [Google Scholar]
- Naidich M, Shterntal B, Furman R, Pawson AJ, Jabbour HN, Morgan K, Millar RP, Jia J, Tomic M, and Stojilkovic S (2010). Elucidation of mechanisms of the reciprocal cross talk between gonadotropin-releasing hormone and prostaglandin receptors. Endocrinology. 151, 2700–2712. [DOI] [PubMed] [Google Scholar]
- Naor Z, Jabbour HN, Naidich M, Pawson AJ, Morgan K, Battersby S, Millar MR, Brown P, and Millar RP (2007). Reciprocal cross talk between gonadotropin-releasing hormone (GnRH) and prostaglandin receptors regulates GnRH receptor expression and differential gonadotropin secretion. Mol. Endocrinol.. 21, 524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan BA, Condon MS, Bosland MC, Narayanan NK, and Reddy BS (2003). Suppression of N-methyl-N-nitrosourea/testosterone-induced rat prostate cancer growth by celecoxib: effects on cyclooxygenase-2, cell cycle regulation, and apoptosis mechanism(s). Clin. Cancer Res. 9, 3503–3513. [PubMed] [Google Scholar]
- Niault T, Hached K, Sotillo R, Sorger PK, Maro B, Benezra R, and Wassmann K (2007). Changing Mad2 levels affects chromosome segregation and spindle assembly checkpoint control in female mouse meiosis I. PLoS One. 2, e1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD, 2013. Guidance Document on Developing and Assessing Adverse Outcome Pathways. Series on Testing and Assessment No. 184. Available at: http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?cote=env/jm/mono(2013)6&doclanguage=en
- OECD, 2014. Users’ Handbook Supplement to the Guidance Document for Developing and Assessing AOPs. Available at: https://aopkb.org/common/AOP_Handbook.pdf [Google Scholar]
- Ojeda S, Harms P, and McCann S (1975). Effect of Inhibitors of Prostaglandin Synthesis on Gonadotropin Release in the Rat 1. Endocrinology. 97, 843–854. [DOI] [PubMed] [Google Scholar]
- Ortiz ME, Arias-Torres AJ and Zelarayán LI (2015). Role of arachidonic acid cascade in Rhinella arenarum oocyte maturation. Zygote, 23, 603–614. [DOI] [PubMed] [Google Scholar]
- Pall M, Fridén BE, Brännström M (2001). Induction of delayed follicular rupture in the human by the selective COX-2 inhibitor rofecoxib: a randomized double-blind study. Hum. Reprod. 16, 1323–1328. [DOI] [PubMed] [Google Scholar]
- Pan Y, Zhang JS, Gazi MH, and Young CY (2003). The cyclooxygenase 2-specific nonsteroidal anti-inflammatory drugs celecoxib and nimesulide inhibit androgen receptor activity via induction of c-Jun in prostate cancer cells. Cancer Epidemiol. Biomarkers Prev. 12, 769–774. [PubMed] [Google Scholar]
- Pang L, Nie M, Corbett L, and Knox AJ (2003). Cyclooxygenase-2 expression by nonsteroidal anti-inflammatory drugs in human airway smooth muscle cells: role of peroxisome proliferator-activated receptors. J. Immunol. 170, 1043–1051. [DOI] [PubMed] [Google Scholar]
- Patel A (2014). The applicability of the” read-across hypothesis” for assessing the effects of human pharmaceuticals on fish (Doctoral dissertation, Brunel University London). [Google Scholar]
- Pati D and Habibi HR (2002). Involvement of protein kinase C and arachidonic acid pathways in the gonadotropin-releasing hormone regulation of oocyte meiosis and follicular steroidogenesis in the goldfish ovary. Biol. Reprod. 66, 813–822. [DOI] [PubMed] [Google Scholar]
- Patino R, Yoshizaki G, Bolamba D, and Thomas P (2003). Role of arachidonic acid and protein kinase C during maturation-inducing hormone-dependent meiotic resumption and ovulation in ovarian follicles of Atlantic croaker. Biol. Reprod. 68, 516–523. [DOI] [PubMed] [Google Scholar]
- Rand-Weaver M, Margiotta-Casaluci L, Patel A, Panter GH, Owen SF and Sumpter JP (2013). The read-across hypothesis and environmental risk assessment of pharmaceuticals. Environ. Sci. Technol. 47, 11384–11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese J, Zhao X, Ma WG, Brown N, Maziasz TJ, and Dey SK (2001). Comparative Analysis of Pharmacologic and/or Genetic Disruption of Cyclooxygenase-1 and Cyclooxygenase-2 Function in Female Reproduction in Mice 1. Endocrinology. 142, 3198–3206. [DOI] [PubMed] [Google Scholar]
- Richards JS, Russell DL, Ochsner S, and Espey LL (2002). Ovulation: new dimensions and new regulators of the inflammatory-like response. Annu. Rev. Physiol. 64, 69–92. [DOI] [PubMed] [Google Scholar]
- Romeiro NC, Sant’Anna CM, Lima LM, Fraga CA, and Barreiro EJ (2008). NSAIDs revisited: Putative molecular basis of their interactions with peroxisome proliferator-activated gamma receptor (PPARγ). Eur. J. Med. Chem. 43, 1918–1925. [DOI] [PubMed] [Google Scholar]
- Rowlinson SW, Kiefer JR, Prusakiewicz JJ, Pawlitz JL, Kozak KR, Kalgutkar AS, Stallings WC, Kurumbail RG, and Marnett LJ (2003). A novel mechanism of cyclooxygenase-2 inhibition involving interactions with Ser-530 and Tyr-385. J. Biol. Chem. 278, 45763–45769. [DOI] [PubMed] [Google Scholar]
- Sagi SA, Weggen S, Eriksen J, Golde TE, and Koo EH (2003). The non-cyclooxygenase targets of non-steroidal anti-inflammatory drugs, lipoxygenases, peroxisome proliferator-activated receptor, inhibitor of kappa B kinase, and NF kappa B, do not reduce amyloid beta 42 production. J. Biol. Chem. 278, 31825–31830. [DOI] [PubMed] [Google Scholar]
- Schmidt CW (2009). TOX 21: new dimensions of toxicity testing. Environ. Health Perspect. 117, A348–A353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinsky BS, Gupta K, Sharkey CT, and Loll PJ (2001). Structural analysis of NSAID binding by prostaglandin H2 synthase: time-dependent and time-independent inhibitors elicit identical enzyme conformations. Biochemistry. 40, 5172–5180. [DOI] [PubMed] [Google Scholar]
- Simmons DL, Botting RM, and Hla T (2004). Cyclooxygenase isozymes: the biology of prostaglandin synthesis and inhibition. Pharmacol. Rev. 56, 387–437. [DOI] [PubMed] [Google Scholar]
- Sorbera LA, Asturiano JF, Carrillo M, and Zanuy S (2001). Effects of polyunsaturated fatty acids and prostaglandins on oocyte maturation in a marine teleost, the European sea bass (Dicentrarchus labrax). Biol. Reprod. 64, 382–389. [DOI] [PubMed] [Google Scholar]
- Sorensen PW and Goetz FW (1993). Pheromonal and reproductive function of F prostaglandins and their metabolites in teleost fish. J. Lipid Mediat. 6, 385–393. [PubMed] [Google Scholar]
- Stacey NE, and Pandey S. (1975). Effects of indomethacin and prostaglandins on ovulation of goldfish. Prostaglandins. 9, 597–607. [DOI] [PubMed] [Google Scholar]
- Stacey N and Sorensen P (2009). Hormonal Pheromones in Fish. In Hormones, Brain and Behavior (Pfaff DW, Arnold AP, Etgen AM, Fahrbach SE and Rubin RT, Eds), 2nd ed., pp. 639–681. Academic Press, San Diego, CA. [Google Scholar]
- Stacey N (1976). Effects of indomethacin and prostaglandins on the spawning behaviour of female goldfish. Prostaglandins. 12, 113–126. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, and Lander ES (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Inazumi T, and Tsuchiya S (2015). Roles of prostaglandin receptors in female reproduction. J. Biochem. 157, 73–80. [DOI] [PubMed] [Google Scholar]
- Sun SC and Kim NH (2012). Spindle assembly checkpoint and its regulators in meiosis. Hum. Reprod. Update. 18, 60–72. [DOI] [PubMed] [Google Scholar]
- Suwa K and Yamashita M (2007). Regulatory mechanisms of oocyte maturation and ovulation. In The Fish Oocyte (Babin PJ, Cerda J, and Lubzens E Eds.), pp. 323–347. Springer. [Google Scholar]
- Takahashi T, Fujimori C, Hagiwara A, and Ogiwara K (2013). Recent advances in the understanding of teleost medaka ovulation: the roles of proteases and prostaglandins. Zool. Sci. 30, 239–247. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Igarashi H, Amita M, Hara S, and Kurachi H (2010). Roles of prostaglandins during oocyte maturation: lessons from knockout mice. Journal of Mammalian Ova Research. 27, 11–20. [Google Scholar]
- Tamba S, Yodoi R, Morimoto K, Inazumi T, Sukeno M, Segi-Nishida E, Okuno Y, Tsujimoto G, Narumiya S, and Sugimoto Y (2010). Expression profiling of cumulus cells reveals functional changes during ovulation and central roles of prostaglandin EP2 receptor in cAMP signaling. Biochimie. 92, 665–675. [DOI] [PubMed] [Google Scholar]
- Tanabe M and Kanehisa M (2012). Using the KEGG database resource. Current Protocols in Bioinformatics. 1.12. 1–1.12. 43. [DOI] [PubMed] [Google Scholar]
- Tanabe T and Tohnai N (2002). Cyclooxygenase isozymes and their gene structures and expression. Prostaglandins Other Lipid Mediat. 68, 95–114. [DOI] [PubMed] [Google Scholar]
- Teng Q, Ekman DR, Huang W, and Collette TW (2012). Push-through direct injection NMR: an optimized automation method applied to metabolomics. Analyst. 137, 2226–2232. [DOI] [PubMed] [Google Scholar]
- Toda K, Okada T, Miyaura C, & Saibara T (2003). Fenofibrate, a ligand for PPARα, inhibits aromatase cytochrome P450 expression in the ovary of mouse. J. Lipid Res. 44, 265–270. [DOI] [PubMed] [Google Scholar]
- Tokumoto T, Tokumoto M, Oshima T, Shimizuguchi K, Fukuda T, Sugita E, Suzuki M, Sakae Y, Akiyama Y, and Nakayama R (2012). Characterization of multiple membrane progestin receptor (mPR) subtypes from the goldfish ovary and their roles in the induction of oocyte maturation. Gen. Comp. Endocrinol. 177, 168–176. [DOI] [PubMed] [Google Scholar]
- Tokumoto T, Yamaguchi T, Ii S, and Tokumoto M. (2011). In vivo induction of oocyte maturation and ovulation in zebrafish. PLoS One. 6, e25206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunquist BJ and Maller JL (2003). Under arrest: cytostatic factor (CSF)-mediated metaphase arrest in vertebrate eggs. Genes Dev. 17, 683–710. [DOI] [PubMed] [Google Scholar]
- Viant MR, Pincetich CA, Tjeerdema RS (2006). Metabolic effects of dinoseb, diazinon and esfenvalerate in eyed eggs and alevins of Chinook salmon (Oncorhynchus tshawytscha) determined by H-1 NMR metabolomics. Aquat. Toxicol. 77, 359–371. [DOI] [PubMed] [Google Scholar]
- Villeneuve DL, Garcia-Reyero N, Martinović-Weigelt D, Li Z, Watanabe KH, Orlando EF, LaLone CA, Edwards SW, Burgoon LD, and Denslow ND (2012). A graphical systems model and tissue-specific functional gene sets to aid transcriptomic analysis of chemical impacts on the female teleost reproductive axis. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 746, 151–162. [DOI] [PubMed] [Google Scholar]
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, Landesmann B, Lettieri T, Munn S, and Nepelska M (2014a). Adverse outcome pathway (AOP) development I: strategies and principles. Toxicol. Sci. 142, 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, Landesmann B, Lettieri T, Munn S, Nepelska M, Ottinger MA, Vergauwen L, and Whelan M (2014b). Adverse outcome pathway development II: best practices. Toxicol. Sci. 142, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MC, Kurumbail RG, et al. (2001). “A three-step kinetic mechanism for selective inhibition of cyclo-oxygenase-2 by diarylheterocyclic inhibitors.” Biochem. J. 357, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann K, Niault T, and Maro B (2003). Metaphase I arrest upon activation of the Mad2-dependent spindle checkpoint in mouse oocytes. Curr. Biol. 13, 1596–1608. [DOI] [PubMed] [Google Scholar]
- Wennmalm Å (2013). Prostaglandins and the heart. Hearts Heart-like Organs. 2, 41–92. [Google Scholar]
- Yokota H, Eguchi S, Hasegawa S, Okada K, Yamamoto F, Sunagawa A, Tanaka M, Yamamoto R and Nakano E (2015). Assessment of in vitro antiovulatory activities of nonsteroidal anti-inflammatory drugs and comparison with in vivo reproductive toxicities of medaka (Oryzias latipes). Environ. Toxicol. doi: 10.1002/tox.22173. [DOI] [PubMed] [Google Scholar]
- Zou M, Shi C, and Cohen RA (2002). High glucose via peroxynitrite causes tyrosine nitration and inactivation of prostacyclin synthase that is associated with thromboxane/prostaglandin H2 receptor–mediated apoptosis and adhesion molecule expression in cultured human aortic endothelial cells. Diabetes. 51, 198–203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


