Abstract
Salmonella enterica serovar Typhimurium is one of the top Salmonella serovars annually linked to poultry production and corresponding human illnesses. Because of this, vaccination of commercial poultry against Salmonella Typhimurium has been a focal point in recent years. There are several commercially available Salmonella Typhimurium vaccines available for use in poultry production. Among these are modified live vaccines, including Poulvac ST (Zoetis), Megan Egg (AviPro), and Megan Vac 1 (AviPro). In this study, analyses of 27 field isolates of Salmonella Typhimurium from poultry sources indicated evidence for the persistence of some vaccine-origin strains through the commercial production cycle. Further analyses of 26,812 database isolates indicated vaccine-origin isolates are persisting frequently through processing, are present on retail meat products, and are even occasionally found in human patients. A novel polymerase chain reaction (PCR) was created and validated which enables simultaneous identification of Salmonella enterica sp., the Salmonella Typhimurium serovar, and differentiation of wild type Salmonella Typhimurium from live attenuated vaccines involving mutations in the cya/crp or aroA genes. The PCR was developed considering whole genome differences between the vaccines and wild type field isolates and was validated using different field isolates and recovered vaccine strains. This method enables poultry producers to rapidly determine if recovered field isolates have a vaccine origin.
Key words: Salmonella, vaccine, poultry, persistence, Typhimurium
INTRODUCTION
Salmonella enterica subsp. enterica is a major causative agent of foodborne illnesses worldwide. Salmonella infections in humans are obtained from a wide variety of sources, but among the primary sources of disease are poultry products (Collaboration, 2021). Vaccination is a critically important tool in poultry production for mitigating against Salmonella serovars presenting a human health risk. Many commercially available options exist for vaccination against Salmonella in poultry. These options include killed, subunit, and live attenuated vaccines. Live attenuated vaccines are preferred over killed vaccines for their cross protective capabilities, ease of oral application, longer lasting immunity, and reduced production costs (Desin, et al., 2013). It has been well established that live attenuated vaccines not only exert protection against their targeted serovar, but also exert some level of cross-protection against other Salmonella serovars (Hassan and Curtiss, 1994; Hassan and Curtiss, 1997).
Megan Vac 1 and Megan Egg (AviPro) are commercially available, live attenuated S. Typhimurium vaccines developed through genetic modification of 2 key genes important for metabolism, crp and cyaA (Curtiss and Kelly, 1987). These vaccines have been shown to elicit protection against S. Typhimurium and/or Enteritidis in laying hens (Holt et al., 2003), breeders, and their progeny broilers (Dorea et al., 2010). PoulVac ST is a commercially available, live attenuated S. Typhimurium vaccine developed through genetic modification of the aroA-serC operon, also critical for metabolism (Alderton et al., 1991). Similar to Megan Vac 1 and Megan Egg, Poulvac ST has been shown to provide both homologous and heterologous protection in broilers (Methner et al., 2001; Muniz et al., 2017). All of the above-mentioned vaccines, therefore, are valuable tools in the poultry industry for reducing Salmonella loads and subsequent contamination of retail products.
Commercialized live attenuated vaccines have been vetted thoroughly to ensure that they are genetically stable and highly unlikely to revert to a virulent state. However, an intrinsic risk to any live attenuated poultry vaccine is its potential to persist within flocks in which it is applied (Desin et al., 2013). There is little to no published evidence that live attenuated vaccine strains persist in flocks, although previous work suggested that persistence of Megan Vac 1 in vaccinated molting hens minimally occurred up to 2 wk post-vaccination (Holt et al., 2003). Based upon previous work studying crp/cyaA (Megan Egg/Megan Vac 1) versus aroA-attenuated (Poulvac ST) S. Typhimurium strains in poultry, it would be expected that crp-cyaA-based strains persist longer and induce a stronger immune response (Curtiss, 2024). However, no real-life data have been published to confirm their extended persistence. Furthermore, the respective companies have commercial approaches to differentiate their vaccine strains from wild type S. Typhimurium isolates, but there are no published protocols for this in the scientific literature. Therefore, the objective of this study was to examine the occurrence of commercial S. Typhimurium vaccine persistence using collected field isolates and U.S. national surveillance data, and to develop a rapid PCR for differentiating vaccine strains from true field isolates.
MATERIALS AND METHODS
DNA Extraction and Sequencing
This work was reviewed by the University of Minnesota Institutional Animal Care and Use Committee and deemed to be exempt from a need for protocol approval. All field isolates (n = 27) and isolates of vaccine origin (n = 3) were sequenced in this study using Illumina short-read technology. Field isolates were obtained from environmental sampling programs of commercial turkey companies in the USA, collected from live commercial production barns. Age of birds at the time of sampling ranged from 3-18 weeks. All flocks sampled were currently vaccinating against Salmonella using Megan Vac 1, Megan Egg, or Poulvac ST. Vaccine-origin strains were recovered from newly opened commercial vials by culturing overnight on XLT-4 agar and choosing a single isolated colony from plates. DNA was extracted from overnight TS broth (BD Difco) cultures of a single colony using the DNeasy Blood & Tissue kit (Qiagen, Valencia, CA) following the manufacturer's instructions. Genomic DNA libraries were created using the Nextera XT DNA library preparation kit and Nextera XT index kit v3 (Illumina, San Diego, CA), and sequencing was performed using 2 × 300-bp dual-index runs on an Illumina MiSeq.
Retrieval of Database Genomes
The NCBI Pathogen Detection tool was used to recover genomes of Salmonella Typhimurium deposited by the USDA Food Safety Inspection Services (FSIS), US Food and Drug Administration (FDA), and the Centers for Disease Control and Prevention (CDC). Isolates were first filtered for only those predicted to belong to the Typhimurium serovar. For FSIS and FDA isolates, the database was filtered for a collection date between 2015 and 2023, and further filtered for those having a poultry origin. For CDC isolates, the database was filtered for a collection date between 2017 and 2023 for those having a human origin. This search resulted in the retrieval of 2,006 S. Typhimurium genome assemblies from USDA FSIS, 348 genomes from US FDA, and 24,458 genomes from CDC.
Genome Assembly and Quality Assessment
Raw FASTQ files for each genome sequenced in this study were trimmed and quality filtered using Trimmomatic (v0.33) (Bolger et al., 2014), including removal of Illumina adapters, with a sliding window of 4 and average Phred quality score of 20, and 36 as the minimum read length. Assemblies of each genome were performed using Shovill (v1.0.4), specifying the SPAdes assembler (Bankevich et al., 2012), with default parameters (https://github.com/tseemann/shovill). Assembly quality was assessed with QUAST (v5.0.0) (Gurevich et al., 2013).
Genetic Feature Identification
Whole genome comparisons were performed between vaccine-derived genomes and the genomes of field isolates of S. Typhimurium. MAUVE alignments were first used to compare gross differences in wild type versus vaccine-origin genomes sequenced in this study (n = 30), with a focus on the cya, crp, and aroA genes. These same genomes were also compared at the gene level for any insertions, deletions, or re-arrangements in vaccine versus field isolates. Based upon the results of the MAUVE comparisons, ABRicate (v.0.8.13) (https://github.com/tseemann/abricate) was used to search against 26,812 database genomes with a minimum identity of 90% and minimum coverage of 90% to screen isolate genome assemblies for indications of disruptions in aroA, cyaA, or crp. If signals of disruption at expected gene positions were detected, isolate genomes were further investigated with MAUVE to confirm disruptions indicative of vaccine origin.
Phylogenetic Analyses
Single nucleotide polymorphisms (SNP) were identified in each sample identified as containing vaccine-origin crp/cyaA gene insertions using Snippy (v4.4.0), with a minimum sequencing depth of 10x (https://github.com/tseemann/snippy) and the assembled Megan Vac 1 vaccine strain used as the reference. A core SNP alignment was then created for all suspected vaccine-origin isolates (n = 175 US FSIS database isolates, n = 29 US FDA database isolates, and n = 25 CDC isolates, plus the Megan Egg and Poulvac ST genomes). SNP differences were compared between isolates using snp-dists (Supplementary Data).
Development of a PCR Typing Scheme
Using MAUVE, the precise vaccine strain gene mutation sites were identified for cyaA/crp (Megan Vac 1 and Megan Egg) and aroA (Poulvac ST). PCR primers were designed spanning the insertion sites for these genes. The PCR panel also included a universal marker for S. enterica subsp. enterica (invA) and a gene marker specific for S. Typhimurium (STM4497) (Kim, et al., 2006). PCR primers were designed using Primer3 (Koressaar and Remm, 2007). The panel was validated by screening a subset of strains from this study (n = 18). Amplification of targets was accomplished in a 25 uL reaction volume. Each reaction included 10.675 uL of nuclease-free water, 5.0 uL of 5X Green GoTaq Flexi Buffer, 4.0 uL of 25 mM MgCl2, 2.5 uL of the primer pool which contained 3 uM of each primer, 0.625 uL of 10 mM dNTPs, 0.2 uL of 5 U/ul GoTaq G2 Hot Start DNA Polymerase, and 2.0 uL of template DNA. The reactions were performed using a T100 thermal cycler (Bio-Rad, San Francisco, CA) using the following cycling parameters: 95°C for 5 min; 30 cycles of 95°C for 35 s, 57°C for 30 s, 72°C for 40 s; and a final cycle of 72°C for 10 min. Samples were subjected to agarose gel electrophoresis in 2% TBE agarose, and amplicons were compared to a 100-bp ladder (New England Biolabs Inc, Ipswich, MA). An isolate was considered to contain a gene of interest if it produced an amplicon of the expected size (Table 1).
Table 1.
Primers used in the development of a vaccine scheme targeting vaccine-origin Salmonella Typhimurium.
| Gene | Forward primer sequence | Reverse primer sequence | Product size |
|---|---|---|---|
| aroA | AATCCCTGACGTTACAACCC | TCCGGCATTACCGAGAAAC | 284 |
| crp | CCACATTCATAAGTACCCGTCA | TTGTAAGCGACGAGCCATC | 322 |
| invA | CCGCATTGTTGATTGCGATTAG | GGCTCTTCGGCACAAGTAATA | 378 |
| STM4497 | GCGTGAACACCTGAAGTATCT | GCCTGAAGGCGCGATAATA | 419 |
| cyaA | CTGGAACGGGTCACTGAATAC | GTTGTAGAGATACCAGCCTGAAC | 489 |
Data Availability
Raw reads from isolates sequenced in this study are available at the NCBI Short Read Archive (SRA) under BioProject accession no. PRJNA1081114.
RESULTS AND DISUSSION
Identification of Distinguishing Mutations in Vaccine Strains
Draft genomic sequences generated from the recovery of the live attenuated vaccine strains in this study were first compared to the completed genome of a known wild type S. Typhimurium type strain LT2 (NCBI GenBank AE006468.2) (McClelland, et al., 2001). Although many small-scale differences were identified between the strains, we focused on the integrity of the aroA, crp, and cyaA genes, as these genes were previously reported as the targets for attenuation in the commercial vaccine strains (Curtiss, 2024). In the Poulvac ST strain, the aroA gene was disrupted at position 1,062,727 bp in the LT2 genome (STM0978). In the Megan Vac 1 and Megan Egg strains, the crp gene was disrupted at position 3,615,908 bp in the LT2 genome (STM3466), and the cyaA gene was disrupted at position 4,147,559 bp in the LT2 genome (STM3939). Other known wild type S. Typhimurium in the NCBI database were examined and none possessed these gene disruptions. All observed gene disruptions involved insertions of IS10-like insertion sequences, likely reflective of the use of transposon-based Tn10 gene inactivation used to create the attenuated strains (Alderton et al., 1991; Hassan and Curtiss, 1990; Porter et al., 1993). These strains were presumably further modified with chemical curing for removal of the tetracycline resistance cassette commonly contained in laboratory Tn10 insertion methods (Maloy and Nunn, 1981), since these genes were absent from the genomes. While the use of Illumina sequencing prevented us from determining the exact lengths and annotations of the inserted transposon sequences in each gene disruption, they were at least 1,329 bp in length, but likely larger than this because the original Tn10 used for gene inactivation has duplicate inverted copies of IS10 surrounding a tetracycline resistance cassette. This length enables the use of PCR to identify isolates in which these genes have been disrupted versus those which remain intact.
Twenty-seven field isolates of S. Typhimurium were also analyzed using Illumina whole genome sequencing and then compared to the genome sequences of the vaccine strains and wild type LT2. These isolates were provided by 2 major turkey-producing companies spanning 2019 to 2023. Of the 27 field isolates examined, 17 isolates contained insertions in the crp and cyaA genes identical to the Megan Egg/Megan Vac 1 vaccine strains. At the whole genome level, isolates differed from 0 to 7 single nucleotide polymorphisms (SNPs) from the vaccine strain. In contrast, the true wild type isolates which did not possess the crp and cyaA insertions possessed 265-500 SNP differences compared to the Megan Egg vaccine strain. Additionally, the 2 AviPro vaccine strains (Megan Egg and Megan Vac 1) differed by zero SNPs, whereas the AviPro vaccine strains and Poulvac ST vaccine strain differed by 2,026 SNPs (Supplementary Data).
Presence of Vaccine-Origin Strains in USDA-FSIS Samples
We next analyzed 2,006 genomes of S. Typhimurium from isolates deposited by the USDA FSIS. These isolates were identified by filtering the NCBI Pathogen Detection Database for all S. Typhimurium isolates collected by FSIS, between 2015 and 2023, from retail poultry sources and live production. Of these, 1,877 isolates were labeled as chicken source and 129 were labeled as turkey source. In total, 123/1,877 (6.6%) of chicken-source isolates possessed the cyaA/crp gene insertions indicating a vaccine origin similar to the Megan Egg or Megan Vac 1 vaccine (Table 2). In contrast, 52/129 (40.3%) of turkey-source isolates possessed these same gene insertions. Vaccine origin isolates included those from samples labeled as chicken carcass (n = 35), comminuted chicken (n = 35), raw intact chicken (n = 49), and comminuted turkey (n = 49). No isolates were found that had the aroA gene insertion indicating a vaccine origin similar to the Poulvac ST vaccine. Isolates matching the Megan Egg or Megan Vac 1 vaccine strain profile via crp/cyaA gene insertions were then subjected to whole genome SNP analysis. Isolates differed from the Megan Vac 1 vaccine strain by only 0 to 3 SNPs (Supplementary Data).
Table 2.
Summary of vaccine origin isolates identified among S. Typhimurium from FSIS sampling efforts categorized by year of isolation, 2015–2023.
| Year | Number of chicken-source isolates | Number (%) of chicken-source isolates with vaccine origin | Number of turkey-source isolates | Number of turkey-source isolates with vaccine origin |
|---|---|---|---|---|
| 2015 | 53 | 8 (15.1) | 3 | 1 (33.3) |
| 2016 | 101 | 0 (0.0) | 0 | 0 (0.0) |
| 2017 | 220 | 7 (3.2) | 4 | 0 (0.0) |
| 2018 | 261 | 12 (4.6) | 11 | 0 (0.0) |
| 2019 | 198 | 16 (8.1) | 27 | 10 (37.0) |
| 2020 | 200 | 17 (8.5) | 25 | 14 (56.0) |
| 2021 | 212 | 16 (7.5) | 29 | 11 (37.9) |
| 2022 | 545 | 35 (6.4) | 27 | 15 (55.6) |
| 2023 | 87 | 12 (13.8) | 3 | 1 (33.3) |
| Total | 1,877 | 123 (6.6) | 129 | 52 (40.3) |
Presence of Vaccine-Origin Strains in USDA-FDA Samples
A similar search was performed for isolates of poultry origin deposited by the USA FDA. In total, 348 isolates were identified spanning 2015 to 2021. Of those, 217 isolates were derived from retail chicken products, 54 were from retail turkey products, and the remaining isolates were either undefined or were from live production. From these isolates, 29/348 (8.3%) were identified as possessing gene insertions indicating Megan Vac 1 or Megan Egg vaccine strain origin (Table 3). From retail chicken products, 4/217 isolates (1.8%) were of vaccine origin, and from retail turkey products, 1/54 isolates (1.9%) were of vaccine origin. Remaining isolates matching vaccine origin profiles were labeled as undefined chicken (n = 7), chicken organ or swab (n=6), chicken environment (n = 4), poultry environment (n = 2), and turkey organ or swab (n = 5). Isolates matching the Megan Egg or Megan Vac 1 vaccine strain profile via crp/cyaA gene insertions were then subjected to whole genome SNP analysis. Isolates differed from the vaccine strain by 0 to 4 SNPs.
Table 3.
Summary of vaccine origin isolates identified among S. Typhimurium from FDA sampling efforts categorized by year of isolation, 2015–2021.
| Year | Number of isolates | Number (%) of isolates with vaccine origin |
|---|---|---|
| 2015 | 78 | 8 (1.0) |
| 2016 | 51 | 4 (7.8) |
| 2017 | 69 | 8 (11.6) |
| 2018 | 57 | 3 (5.3) |
| 2019 | 62 | 4 (6.5) |
| 2020 | 7 | 0 (0.0) |
| 2021 | 24 | 2 (8.3) |
| Total | 348 | 29 (8.3) |
Presence of Vaccine-Origin Strains in CDC Samples
A similar search was performed for isolates of human origin deposited by the CDC. In total, 24,458 isolates were identified spanning 2017-2023. Of those, all isolates were derived from human infection. From these isolates, 25/24,458 (0.1%) were identified as possessing gene insertions indicating vaccine strain origin similar to Megan Vac 1 or Megan Egg. Of the 25 isolates, 12 were from blood infection, 6 were from stool samples, 5 were from urine samples, and 1 was from a wound infection. Isolates matching the Megan Egg or Megan Vac 1 vaccine strain profile via crp/cyaA gene insertions were then subjected to whole genome SNP analysis. Isolates differed from the vaccine strain by 0-2 SNPs.
Development of a Multiplex PCR to Differentiate Vaccine-Origin From Wild Type Field Salmonella Typhimurium
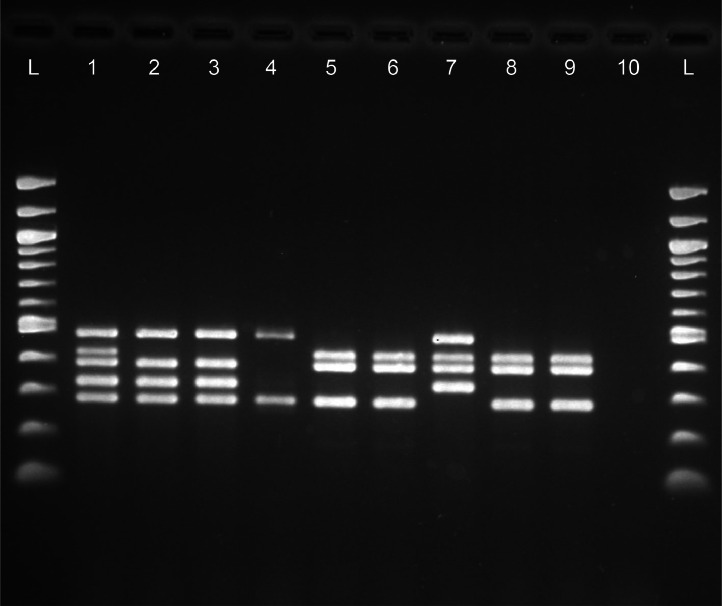
We then designed a multiplex PCR assay with targets identifying S. enterica (invA), S. Typhimurium (STM4497), and the 3 gene targets for inactivation in vaccine strains (aroA, cyaA, and crp). Primers were designed spanning the known sites of gene inactivation for aroA, cyaA, and crp. In this assay, a wild type Salmonella Typhimurium will be positive for all 5 gene targets. Isolates which amplify invA, but do not amplify STM4497, are Salmonella but not Typhimurium. Salmonella Typhimurium isolates amplifying aroA, but not crp or cyaA, are of vaccine origin (Megan Vac 1 or Megan Egg). Salmonella Typhimurium isolates amplifying crp or cyaA, but not aroA, are also of vaccine origin (Poulvac ST). This PCR panel was validated using isolates derived from the 3 vaccine strains (Megan Egg, Megan Vac 1, and Poulvac ST), a known wild type field isolate of S. Typhimurium, 2 known field strains with vaccine origin, a Salmonella Reading isolate, a Salmonella Braenderup isolate, and an E. coli laboratory strain (K-12 MG1655). All isolates displayed the expected gene products based upon their genomic profiles using WGS data (Figure 1).
Figure 1.
Endpoint multiplex PCR subjected to agarose gel electrophoresis to distinguish field versus vaccine origin S. Typhimurium strains. L = 100-bp ladder; 1 = true S. Typhimurium field isolate (positive control); 2 = S. Reading field isolate (S. Typhimurium negative control); 3 = S. Braenderup H9812 (S. Typhimurium negative control); 4 = E. coli K-12 MG1655 (Salmonella negative control); 5 = Megan Egg vaccine; 6 = Megan Vac 1 vaccine; 7 = Poulvac ST vaccine; 8 and 9 = vaccine-origin field isolates; 10 = no template negative control.
Discussion
This study was prompted by recent concerns that commercial S. Typhimurium vaccine persistence was resulting in field-positive Salmonella results in commercial turkey production. This is not a novel concept, as it was already well known that these types of vaccines have the capacity to persist during the flock cycle. However, the extent of persistence of vaccine-origin strains have not been widely reported. Here, we found that vaccine strain persistence appears to have occurred in both broiler and commercial turkey production, with evidence of strains persisting until slaughter and even through processing. Interestingly, the prevalence of vaccine-origin isolates found in turkey-source samples was substantially higher than those from chicken-source samples. This could reflect an enhanced ability for these vaccine-origin strains to persist in commercial turkey production. However, because this study was retrospective and utilized database isolates from which the sampling strategies are unclear, the results could also be heavily biased. It does suggest further investigation into this finding is warranted.
A limitation of this study was that it did not incorporate long-read sequencing to identify the precise length of the mutations used to generate the attenuated vaccine strains. This prevented us from fully describing the transposon-based insertion sequence; however, it did not impact the results of this study or the utility of the PCR tool, since the insertions are clearly longer than what would be detected using this PCR protocol. It is also possible that such mutations occur in wild type S. Typhimurium isolates. While possible, this is exceedingly unlikely for several reasons, including the attenuation it creates on the bacteria (which would not be a beneficial mutation likely to exceed in nature) and the likelihood of such mutations occurring within the same 2 precise genomic sites used for generation of the vaccine strains. We also confirmed that no known wild type S. Typhimurium in the NCBI database contain such mutations.
All of the vaccine origin S. Typhimurium identified in this study matched the genomic profiles of Megan Vac 1 or Megan Egg vaccines, and no evidence of Poulvac vaccine persistence was found. This could be reflective of inherent differences between cya/crp and aroA attenuated Salmonella vaccines, as it has been suggested that cya/crp vaccines exert stronger immune response but persist longer following administration (Curtiss, 2024). However, the market share of these vaccines in broiler and turkey production are unknown but likely different, and this could also impact how frequently vaccine strains persist and are identified in national surveillance databases. Therefore, these results should be taken with caution and not used to infer that aroA attenuated vaccines cannot persist in poultry production. It should also be noted that other live attenuated vaccines containing mutations in these genes, falling between the primer sets used here, would also be detected as vaccine origin strains. Thus, this assay should not be interpreted as necessarily specific for the respective vaccines studied here. However, all of the isolates identified as vaccine origin possessed mutations within identical and precise regions of cya and crp, providing a high level of confidence towards their origin.
Based on US surveillance programs, the rate of vaccine persistence was higher in samples at or prior to slaughter than in retail meat samples. This is not unexpected, as these vaccines are designed to be deficient and unlikely to persist in their hosts. Based upon the national surveillance data examined here, persistence of certain vaccine-origin strains within poultry production is fairly frequent, yet few isolates persist through processing. If this is the case, the underlying reasons for this are unknown, but are likely related to the various processing interventions used to reduce pathogen loads on retail meat products. It is quite likely that a metabolically deficient Salmonella, such as those derived from vaccines, would be less likely to survive the stressors encountered during processing. We also identified 25 human-source isolates deposited from CDC in the NCBI database with high genetic similarity (0–2 SNPs) and corresponding cya/crp mutations, indicating they were likely of vaccine origin. Epidemiological and patient data linked to these isolates were unavailable, so the sources of these infections are unknown. However, it is suggestive that such vaccines may have the capacity to cause human disease, either via foodborne acquisition or fecal-oral contact. Fortunately, the frequency of apparent vaccine origin isolates found in human patients appears to be very low. However, this finding warrants further investigation regarding the sources of infection and host immune status prior to infection.
The implications of the frequent occurrence of vaccine-origin Salmonella Typhimurium at slaughter are important. Current USDA-FSIS performance standards occur at this stage, and the presence of a positive sample contaminated with a vaccine-origin Salmonella Typhimurium would negatively impact that processing establishment. Vaccines are critically important tools for Salmonella control in poultry production, and future regulatory efforts should account for differences between vaccine-origin and wild type Salmonella Typhimurium, as well as other commercial live attenuated vaccines.
Acknowledgments
ACKNOWLEDGMENTS
This work is supported by Agricultural and Food Research Initiative (AFRI) grant 2020-67017-33235 from the USDA National Institute of Food and Agriculture, and contract NU50CK2022005931 from the Centers for Disease Control and Prevention. Bioinformatics was supported using tools available from the University of Minnesota's Minnesota Supercomputing Institute. Additional resources were provided for this study from the Mid-Central Research and Outreach Center in Willmar, Minnesota.
DISCLOSURES
The authors of this manuscript declare no competing interests.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2024.103707.
Appendix. Supplementary materials
REFERENCES
- Alderton M.R., Fahey K.J., Coloe P.J. Humoral responses and salmonellosis protection in chickens given a vitamin-dependent Salmonella typhimurium mutant. Avian Dis. 1991;35:435–442. [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaboration, T. I. F. S. A. 2021. Foodborne illness source attribution estimates for 2019 for Salmonella, Escherichia coli O157, Listeria monocytogenes, and Campylobacter using multi-year outbreak surveillance data, United States. U.S. Department of Health and Human Services’ Centers for Disease Control and Prevention and U.S. Food and Drug Administration, U.S. Department of Agriculture's Food Safety and Inspection Service., United States. Accessed March 2024. https://www.cdc.gov/foodsafety/ifsac/pdf/P19-2019-report-TriAgency-508.pdf.
- Curtiss R. Vaccines to control Salmonella in poultry. Avian Dis. 2024;67:427–440. doi: 10.1637/aviandiseases-D-23-99988. [DOI] [PubMed] [Google Scholar]
- Curtiss R., 3rd, Kelly S.M. Salmonella typhimurium deletion mutants lacking adenylate cyclase and cyclic AMP receptor protein are avirulent and immunogenic. Infect. Immun. 1987;55:3035–3043. doi: 10.1128/iai.55.12.3035-3043.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desin T.S., Koster W., Potter A.A. Salmonella vaccines in poultry: past, present and future. Expert Rev. Vaccines. 2013;12:87–96. doi: 10.1586/erv.12.138. [DOI] [PubMed] [Google Scholar]
- Dorea F.C., Cole D.J., Hofacre C., Zamperini K., Mathis D., Doyle M.P., Lee M.D., Maurer J.J. Effect of Salmonella vaccination of breeder chickens on contamination of broiler chicken carcasses in integrated poultry operations. Appl. Environ. Microbiol. 2010;76:7820–7825. doi: 10.1128/AEM.01320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan J.O., Curtiss R., 3rd. Control of colonization by virulent Salmonella typhimurium by oral immunization of chickens with avirulent delta cya delta crp S. typhimurium. Res Microbiol. 1990;141:839–850. doi: 10.1016/0923-2508(90)90119-b. [DOI] [PubMed] [Google Scholar]
- Hassan J.O., Curtiss R., 3rd. Development and evaluation of an experimental vaccination program using a live avirulent Salmonella typhimurium strain to protect immunized chickens against challenge with homologous and heterologous Salmonella serotypes. Infect. Immun. 1994;62:5519–5527. doi: 10.1128/iai.62.12.5519-5527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan J.O., Curtiss R., 3rd Efficacy of a live avirulent Salmonella typhimurium vaccine in preventing colonization and invasion of laying hens by Salmonella typhimurium and Salmonella enteritidis. Avian Dis. 1997;41:783–791. [PubMed] [Google Scholar]
- Holt P.S., Gast R.K., Kelly-Aehle S. Use of a live attenuated Salmonella typhimurium vaccine to protect hens against Salmonella enteritidis infection while undergoing molt. Avian Dis. 2003;47:656–661. doi: 10.1637/7002. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Park S.H., Lee T.H., Nahm B.H., Chung Y.H., Seo K.H., Kim H.Y. Identification of Salmonella enterica serovar Typhimurium using specific PCR primers obtained by comparative genomics in Salmonella serovars. J. Food Prot. 2006;69:1653–1661. doi: 10.4315/0362-028x-69.7.1653. [DOI] [PubMed] [Google Scholar]
- Koressaar T., Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Maloy S.R., Nunn W.D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Sanderson K.E., Spieth J., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Hou S., Layman D., Leonard S., Nguyen C., Scott K., Holmes A., Grewal N., Mulvaney E., Ryan E., Sun H., Florea L., Miller W., Stoneking T., Nhan M., Waterston R., Wilson R.K. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- Methner U., Berndt A., Steinbach G. Combination of competitive exclusion and immunization with an attenuated live Salmonella vaccine strain in chickens. Avian Dis. 2001;45:631–638. [PubMed] [Google Scholar]
- Muniz E.C., Verdi R., Leao J.A., Back A., Nascimento V.P.D. Evaluation of the effectiveness and safety of a genetically modified live vaccine in broilers challenged with Salmonella Heidelberg. Avian Pathol. 2017;46:676–682. doi: 10.1080/03079457.2017.1348598. [DOI] [PubMed] [Google Scholar]
- Porter S.B., Tinge S.A., Curtiss R., 3rd Virulence of Salmonella typhimurium mutants for White Leghorn chicks. Avian Dis. 1993;37:265–273. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads from isolates sequenced in this study are available at the NCBI Short Read Archive (SRA) under BioProject accession no. PRJNA1081114.