Abstract
Hepatitis B virus (HBV) is a hepatotropic DNA virus that replicates by reverse transcription. It chronically infects >296 million people worldwide including ~850,000 in the USA, and kills 820,000 annually world-wide. Current nucleos(t)ide analog (NA) or pegylated interferon α therapies do not eradicate the virus and would benefit from a complimentary antiviral drug. We performed a preliminary screen of 28 dispirotripiperazines against HBV, identifying 9 hits with EC50 of 0.7 - 25 μM. Compound 11826096 displays the most potent activity and represents a promising lead for future optimization. While the mechanism of action is unknown, preliminary assays limit possible targets to activities involved in RNA accumulation, translation, capsid assembly, and/or capsid stability. In addition, we built machine learning models to determine if they were able to predict the activity of this series of compounds. The novelty of these molecules indicated they were outside of the applicability domain of these models.
Graphical Abstract
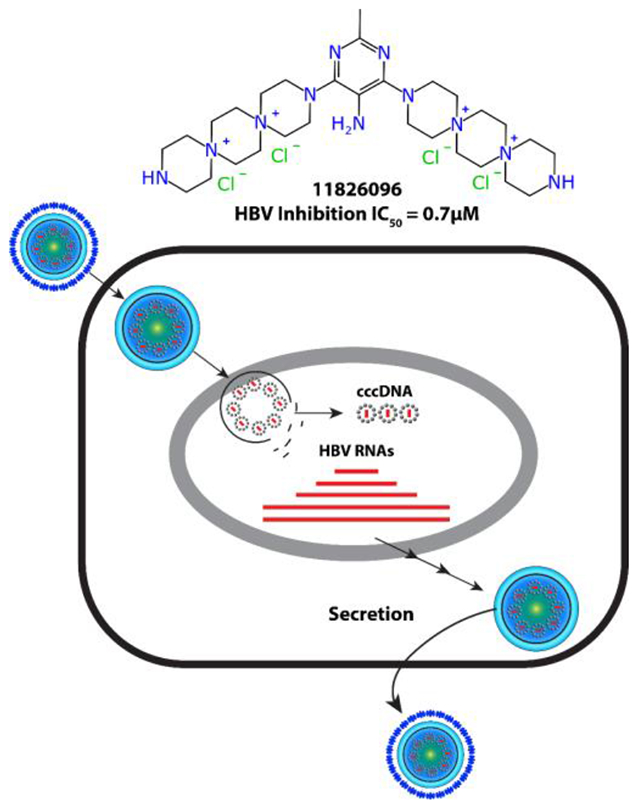
Introduction
Hepatitis B is a contagious liver disease caused by the hepatitis B virus (HBV) which affects approximately 296 million people worldwide including ~850,000 in the USA, and it also kills about 852,000 globally annually 1, 2. HBV replication 3, 4 requires viral entry, formation of the viral covalently closed circular DNA (cccDNA), production of the 4 viral RNAs by transcription from the cccDNA, translation and processing of the 7 viral proteins, nucleocapsid assembly, reverse transcription within capsids, recycling of nucleocapsids into the nucleus to help maintain the cccDNA pool, envelopment of nucleocapsids after reverse transcription is complete, and secretion of virions. Blocking any one of these steps will prevent production of infectious virus, and hence they are all potential targets for novel anti-HBV agents. Current standard of care therapy employs either pegylated interferon α or nucleos(t)ide analog (NA) drugs that target the viral DNA polymerase, with the NAs dominating therapy due to their better safety profile. The NAs usually drive HBV to near or below the clinical detection limit. However, replication is not completely suppressed, and HBV titers rebound if the drugs are withdrawn, so NA therapy is usually life-long. Nevertheless, treatment does cure ~6% of patients, so many more patients could be helped or cured by suppressing HBV further through the use of new drugs in combination5.It is therefore a high priority to develop next-generation therapies which would combine NAs with novel molecules that act on one or more targets and are under a different set of pharmacological constraints 6. These efforts include screening to identify novel replication inhibitors 7–14.
Dispirotripiperazines (DSTPs) are spirocyclic quaternary ammonium salts composed of piperazine or homopiperazine rings linked by shared nitrogen atoms. The DSTP ring system is commonly flanked by terminal N-acyl or N-heteroaryl groups, which can be varied to affect their physicochemical properties. They can be divided into three classes based on the relative number of 6- and 7- membered rings, with “6-6-6” DSTPs containing three piperazine rings, “7-6-6” containing two piperazine rings and one homopiperazine, and “7-6-7” containing one piperazine ring and two homopiperazine rings (Figure 1). The “7-7-7” DSTPs are highly unstable and therefore have not been investigated significantly.
Figure 1.
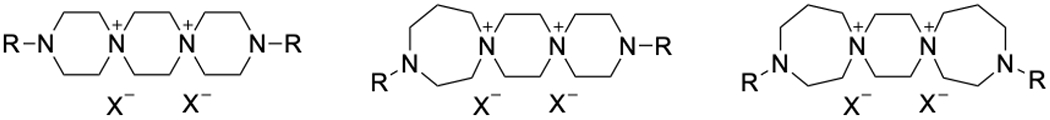
General structures for 6-6-6 (left), 7-6-6 (center), and 7-6-7 (right) DSTPs.
The first DSTP, spirazidine, was discovered in 1963 during an antitumor study based on the 1,4-bis(2-chloroethyl)piperazine scaffold 15. In subsequent studies, spirazidine displayed a wide spectrum of anti-cancer activity and relatively low toxicity, garnering interest from a number of groups as a potential drug lead 16. To that end, DSTP derivatives developed by Mikhalev et al.14 such as prospidine and spirobromine are still used as antitumor drugs (Figure 2).
Figure 2.
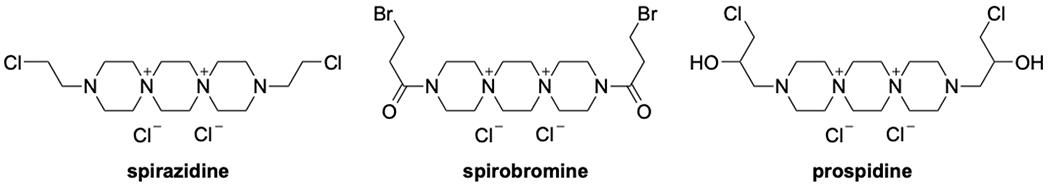
Structures of first generation 6-6-6 DSTPs.
In recent years, DSTPs have been investigated as potential antiviral agents. This scaffold displays promising activity against a number of viruses, including herpesviruses (HSV),cytomegalovirus, papillomavirus (HPV), and certain immunodeficiency viruses such as HIV 17, 18. This antiviral activity is attributed to the ability of dispirotripiperazines to bind heparan sulfate proteoglycans (HSPGs)16. HSPGs are primary binding sites for many viral pathogens, meaning compounds which bind HSPGs could prevent infection by blocking viral entry in the cell 19. However, DSTPs failed to inhibit certain viruses including varicela zoster virus, Epstein-Barr virus, and some HIV strains 20, indicating their mechanism(s) of action may not result exclusively from viral binding inhibition. DSTPs represent attractive targets for further development of antiviral compounds including as anti-HBV therapies.
The synthesis and SAR of spirobromine and similar DSTP analogs have been well studied for activity against select viruses such as HSV and HPV. We have previously disclosed a detailed review of these studies 21, which is summarized in Figure 3. In general, the DSTP ring system is the central pharmacophore which can bear alkyl, acyl, and heteroaryl groups on the terminal nitrogen atoms. Schmidtke et al. demonstrated in 2002 that nitropyrimidine derivatives had stronger antiviral activity, with 2-methyl-5-nitropyrimidine derivatives being optimal 22. Furthermore, attaching two DSTP ring systems to a central 2-methyl-5-nitropyrimidine resulted in a metabolically stable derivative with potent activity for subsequent biological evaluation 23. We have now also used the public data for HBV inhibition to generate machine learning models and predict the activity of the DSTP analogs.
Figure 3.
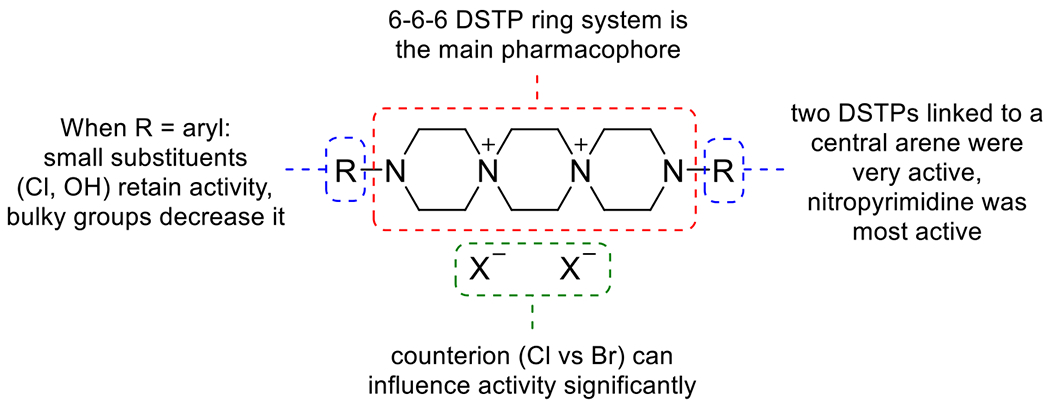
SAR of N-aryl DSTPs.
Results
DSTP synthesis
While the existing SAR for 6-6-6 DSTPs has been well described for HSV, activity against HBV has been largely unexplored despite a recognized need for HBV antiviral drugs. Moreover, while DSTPs containing 6-7-6 and 7-6-7 have been studied for anticancer properties, their antiviral activity has not been reported. With these considerations in mind, a panel of 6-6-6, 6-7-6, and 7-6-7 DSTP analogs with different heteroarenes were synthesized and tested for anti-HBV activity as well as cytotoxicity. Given that 5-nitropyrimidine displayed antiviral activity in previous studies, we wanted to explore other electron-deficient heterocycles such as triazines and pyridines. These analogs can be divided into two groups which contain either 1) a DSTP ring system between two heteroarenes or 2) two DSTP ring systems attached to a single heteroarene. The DSTP rings were synthesized according to a literature procedure24 starting from the respective cyclic diamine (Scheme 1).
Scheme 1.
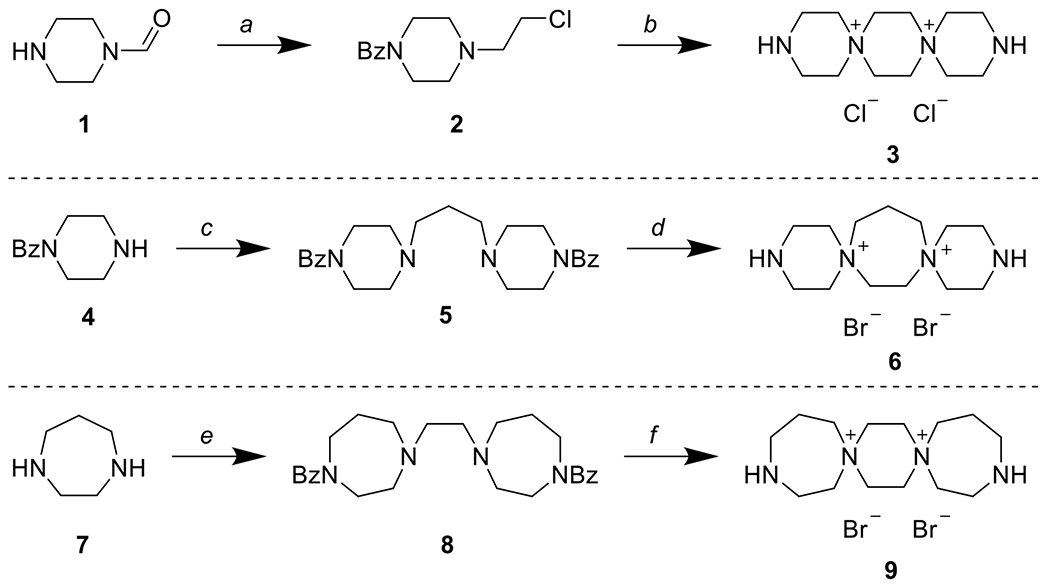
Synthesis of 6-6-6 (3), 6-7-6 (6), and 7-6-7 (9) DSTPs. a) 1. BzCl, NaHCO3, CHCl3 2. HCl, MeOH 3. ClCH2CH2OH, KOH, EtOH 4. SOCl2, CHCl3 b) 1. NaOH, EtOH 2. aq. HCl 3. LiOH, H2O c) 1,3-dibromopropane, NaHCO3, EtOH d) 1. 1,2-dibromoethane 2. 10% aq. HBr 3. LiOH, H2O e) 1. BzCl, AcOH, then NaOH 2. 1,2-dibromoethane, NaHCO3 f) 1. 1,2-dibromoethane 2. 10% aq. HBr 3. LiOH, H2O
Synthesis of the 6-6-6 DSTPs commenced with the benzoyl protection of 1-formylpiperazine (1), followed by removal of the formyl group with aqueous HCl. Alkylation of the free nitrogen with 2-chloroethanol followed by treatment with thionyl chloride afforded primary chloride 2. Finally, base-promoted cyclization followed by acidic cleavage of the benzoyl group afforded 6-6-6 DSTP 3. Synthesis of the 6-7-6 DSTPs started by reacting 1-benzoylpiperazine (4) with 1,3-dibromopropane to give diamine 5. Alkylation with 1,2-dibromoethane followed by acidic cleavage of the benzoyl groups afforded 6-7-6- DSTP 6. Synthesis of the 7-6-7 DSTPs started with the benzoyl protection of homopiperazine (7) followed by alkylation with 1,2-dibromoethane to give diamine 8. Subsequent alkylation in neat 1,2-dibromoethane followed by benzoyl deprotection afforded 7-6-7 DSTP 9.
The desired 6-6-6, 6-7-6, or 7-6-7 DSTPs were then reacted with mono- or dichloroarenes in the presence of triethylamine to yield a panel of mono- or bis-DSTP derivatives, respectively (Scheme 2).
Scheme 2.
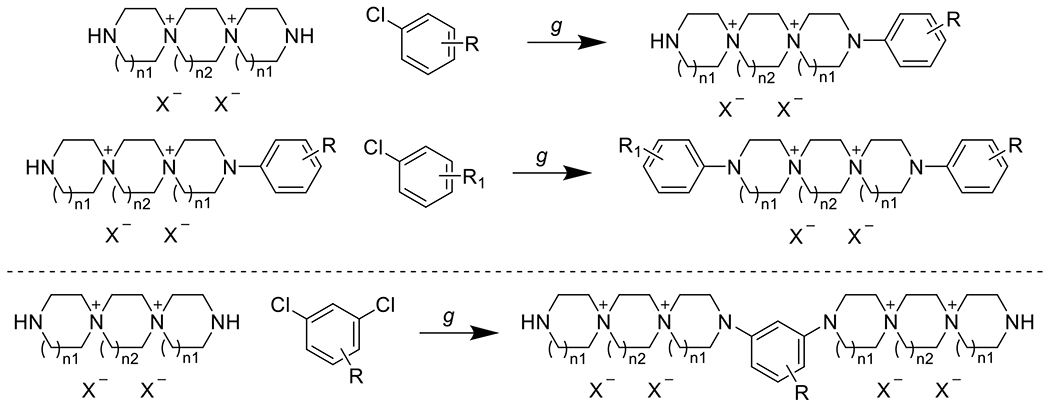
Coupling of chloroarenes to yield mono- (top) and bis- (bottom) DSTP derivatives. g) Et3N, EtOH/H2O or dioxane/H2O
DSTP HBV testing
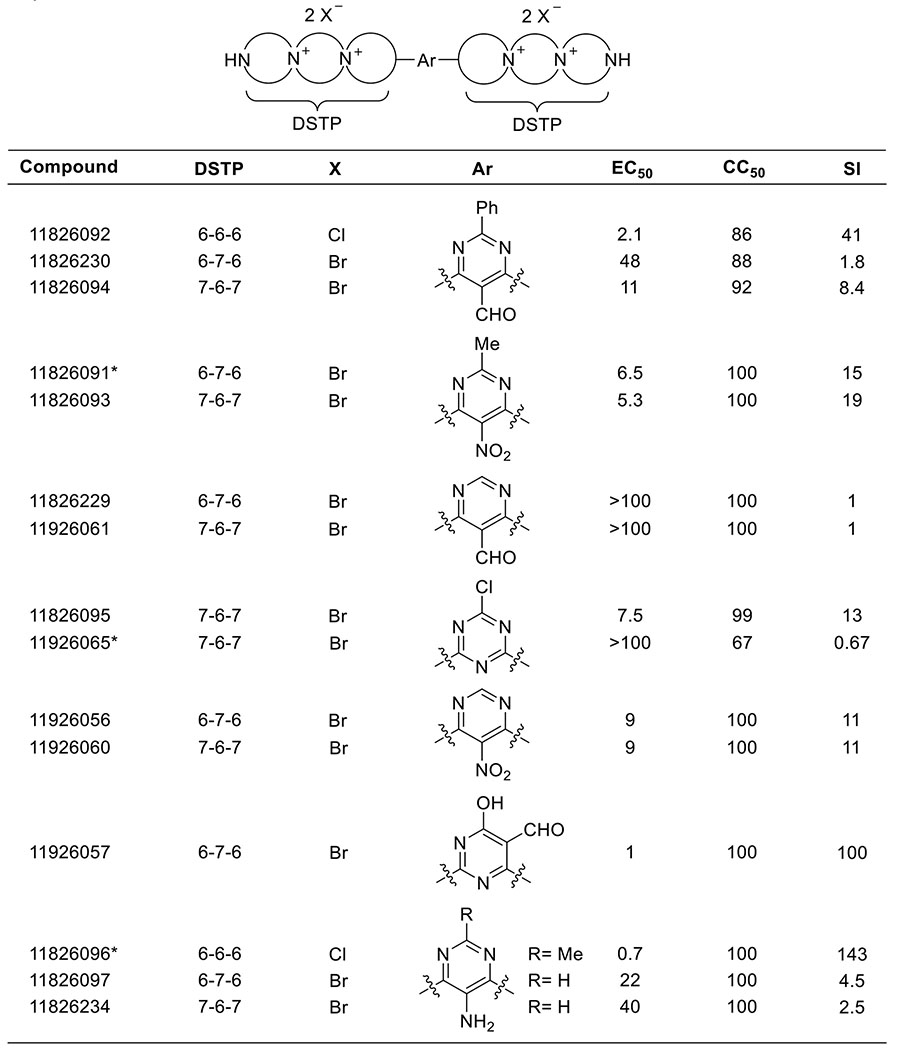
With the desired DSTP compounds in hand, the panel was tested for anti-HBV activity in a tissue culture HBV replication inhibition assay using a stably transfected, tetracycline-repressible cell line (HepDES19) in which HBV replication is launched by removing tetracycline from the medium 25. Unfortunately, none of the synthesized mono-DSTP derivatives displayed activity against HBV (Table S1). However, a number of bis-DSTP compounds displayed promising antiviral activity (Table 1), several of which we independently tested in HepG2 2.2.15 cells (Table 2). 5-nitropyrimidine derivatives displayed activity against HBV with both 7-6-7 and 6-7-6 ring systems, which is consistent with their activity against HSV and other HSPG-binding viruses. Excitingly, compound 11826096 displayed the most potent antiviral activity while exhibiting low cytotoxicity, suggesting that analogs of this heterocycle could be a potential antiviral drug lead. Overall, these studies suggest that antiviral activity is heavily dependent upon the identity of the heterocycle and the DSTP ring system (e.g. 7-6-7 vs 6-7-6).
Table 1.
HBV activity (EC50, μM) and cytotoxicity (CC50, μM) for bis-DSTP derivatives in HepDES19 cells.
|
Tested as dihydrochloride salt
Table 2.
Activity against Hepatitis B Virus ayw1 in HepG2 2.2.15 cells (data from NIAID).
| Compound | EC50 (μM) | EC90 (μM) | CC50 (μM) | SI50 | SI90 |
|---|---|---|---|---|---|
| 11826096 | 1.86 | 15.67 | >100.00 | >54 | >6 |
| 11826091 | 6.53 | 58.15 | >100.00 | >15 | >2 |
| 11826097 | 22.04 | 78.12 | >100.00 | >5 | >1 |
| 11826092 | 2.13 | 26.5 | >100.00 | >47 | >4 |
| 3TC | 0.01771 | 0.541 | >2.00 | >113 | >4 |
Mechanism of action studies
The mechanism of action for DSTPs against HBV is currently unknown. The dispirotripiperazines suppress both “+” and “−“ DNA strands similarly, eliminating the HBV ribonuclease H (RNaseH) activity as the target because RNaseH inhibition preferentially suppresses the viral plus-polarity DNA 14. DNA chain elongation inhibitors such as the NAs also preferentially suppress plus-polarity DNA because synthesis of the 2 strands is sequential, with minus-polarity DNA templating plus-DNA synthesis. Therefore, equal suppression of both strands eliminates DNA chain elongation as a target. Capsid accumulation was measured for 11826093 and 11826095 (promising hits) by measuring accumulation of intracellular HBV capsids by capsid blotting 26, which resolves assembled capsid particles from free capsid monomers in cells. Capsid blotting entails lysing cells replicating HBV and resolving the cytoplasmic extracts by agarose gel electrophoresis. The resolved proteins are transferred to a membrane and detected with an anti-capsid protein primary antibody, horseradish peroxidase-labeled secondary antibody, and chemiluminescence. 11826095 suppressed capsid accumulation (Figure 4), as did 11826091 in a separate assay, but to a lesser extent than anticipated based on the compound concentration used. Steps involved in virion secretion are eliminated in our studies as possible targets because our viral replication inhibition assays employ a viral genome in which the viral surface glycoproteins have been genetically ablated for biosafety reasons, eliminating envelopment and virion secretion. Our preliminary data limits possible targets for the DSTPs against HBV to activities involved in accumulation of viral RNA, protein, capsids, and/or encapsidation of the viral RNA.
Figure 4. Capsid blot revealing modest suppression of HBV capsid accumulation by 11826095 (CP193) in HepDES19 cells.
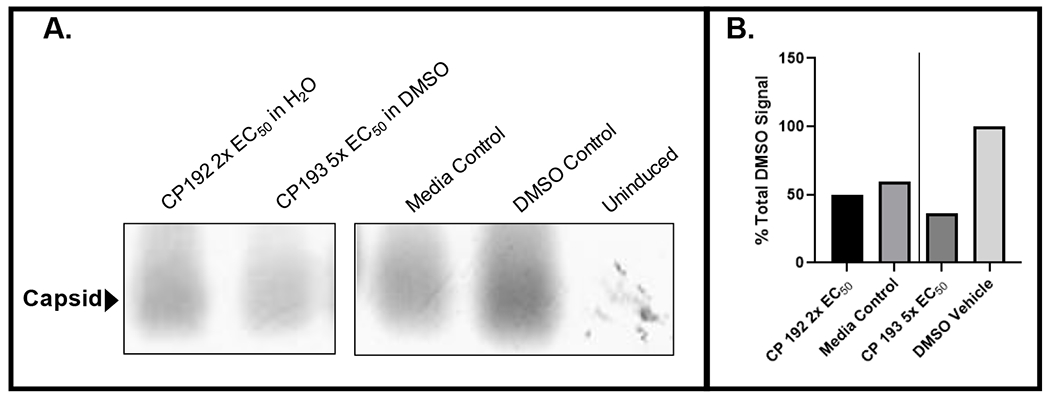
A. Capsid blot. B. Quantification of data in A. 11826095 (CP193) was dissolved in DMSO and 11826093 (CP192) was dissolved in water.
Machine learning
Training sets for HBV inhibition from ChEMBL varied between 1830 – 2567 unique compounds. Cross-validation statistics were excellent for both 1 and 10μM thresholds (Figure S1, S2), with Support vector classification (SVC) performing among the best. Regression models also showed good cross-validation statistics (Figure S3), with support vector regression (SVR) outperforming the other tested algorithms (Figure 5). Using the DSTP series as an external validation set for these models showed poor predictivity (Figure 5A). This is likely attributed to poor structural similarity overlap with the training data. This overlap can be visualized using a t-SNE plot, which compresses molecular descriptors (ECFP6) to a lower dimensional space giving a representation of the chemical property space covered (Figure 5B). The DSTP series is distinctively different from the compounds used for model training, suggesting that these molecules are indeed too chemically different to be predicted correctly by these models and are likely outside the applicability domain of the models. Based on this we built a model using the DSTP compounds alone to assess if this would have the potential to predict novel, more potent DSTP compounds. While small, this model does have reasonable cross-validation statistics (Figure S4) suggesting it may be more helpful in the selection process for future compounds in this class.
Figure 5. Machine learning models for HBV inhibition.
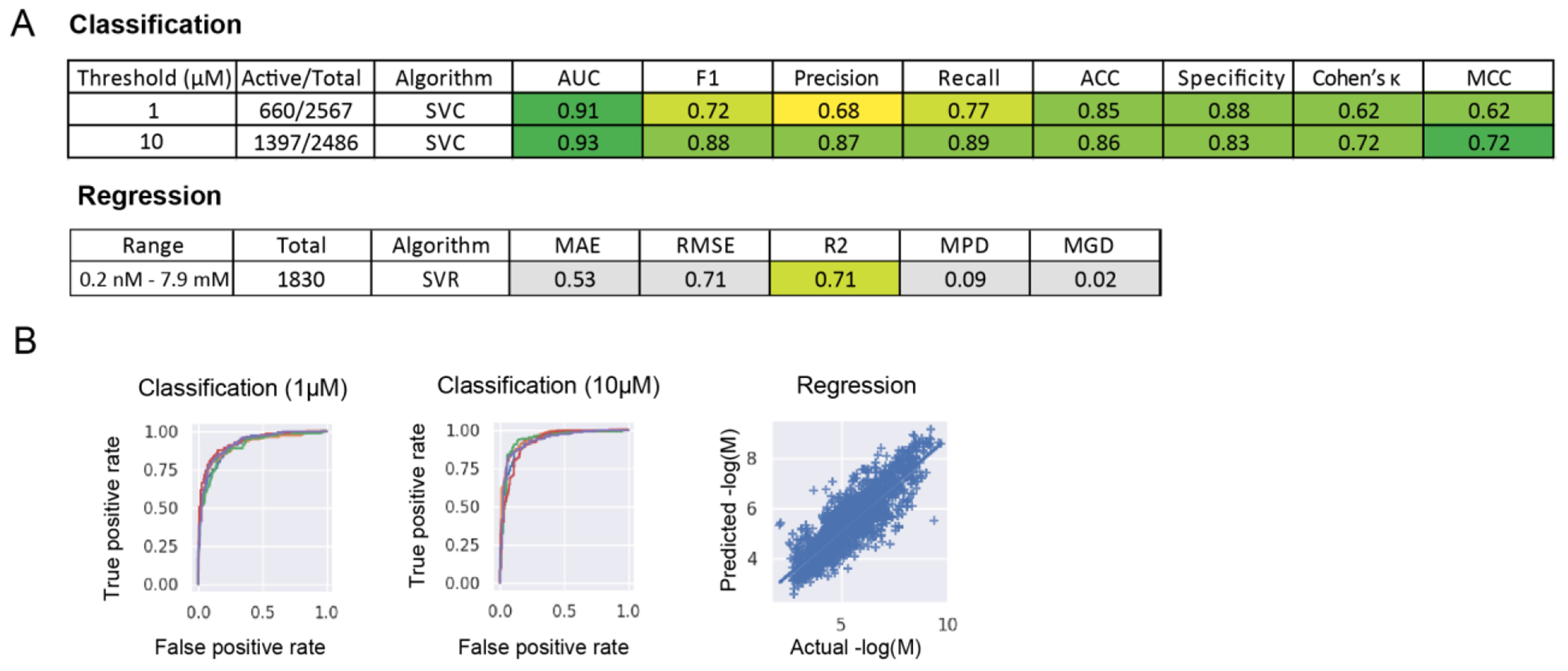
(A) Threshold or range, number of compounds, algorithm, and nested, 5-fold cross validation statistics for HBV inhibition modelsbuilt with training data from ChEMBL. (B) ROC plots for classification models (SVC) built with a 1μM or 10μM threshold (each line represents a single fold) and the predicted versus actual AC50 values (−logM) for a SVR regression model.(SVC=Support vector machine classification, SVR=Support vector machine regression, AUC=ROC “area under the curve”, F1= F1 score, ACC=Accuracy, MCC=Matthews Correlation Coefficient, RMSE= Root of the Mean of the Square of Errors, MAE=Mean of Absolute value of Errors, MGD=Mean Gamma deviance regression loss and MPD=Mean Poisson deviance regression loss)
Discussion and Conclusions
HBV treatment is dominated by monotherapy with NAs (lamivudine, adefovir, telbivudine, entecavir, and two prodrug forms of tenofovir), with entecavir and tenofovir dominating the market. They suppress HBV replication by 4-5 log10 in 70-90% of patients, usually to below the clinical detection limit 27–31. Therapy also reduces levels of HBV’s transcriptional template, the cccDNA by ~1 log10 after 1-2 years 32–34. However, resistance readily develops to the cheaper, older drugs such as lamivudine 35, and HBV is cleared in only 3-6% of patients 29–31, 36. Furthermore, NAs reduce chances of death from liver failure or hepatocellular carcinoma by just ~2–4-fold after 10 years 37–40. The costs of this partial suppression of disease progression are indefinite drug administration 28 and potential side effects from decades of drug exposure. Additionally, the nucleo(s)tide analogs are not fully benign. For example, Baraclude® (Entecavir, the dominant anti-HBV drug in the USA), can cause lactic acidosis and hepatomegaly with steatosis [Boxed Warning; Baraclude® packet insert (2015)].
HBV replication persists even when NAs have suppressed viral titers to undetectable levels. This is revealed by sequential accumulation of resistance mutations to NAs in the absence of detectable viremia 41–43, and is confirmed by analyses of viral DNA in the liver showing replenishment of the cccDNA via reverse transcription in the absence of detectable viremia 44. Therefore, failure to clear HBV in the absence of viremia is in part due to incomplete inhibition of viral replication. This means that further inhibiting replication through combination therapies will improve success rates for therapy, possibly leading to a clinical cure of HBV infections when drugs with different mechanisms of action are used in combination 45.
Machine learning models built using HBV inhibition data from ChEMBL showed excellent cross-validation statistics, suggesting that these may be potentially useful to assist in finding new inhibitors (Figure 5). Unfortunately, likely due to poor representation in the training set, the activity of the DSTP series was unable to be predicted accurately by these models. Following this, we therefore built models on the DSTP data alone and these had reasonable cross-validation statistics (Figure 6). These models can be externally tested in future with additional compounds.
Figure 6. Dispirotripiperazine (DSTP) series used as an external test set for machine learning models for HBV inhibition.
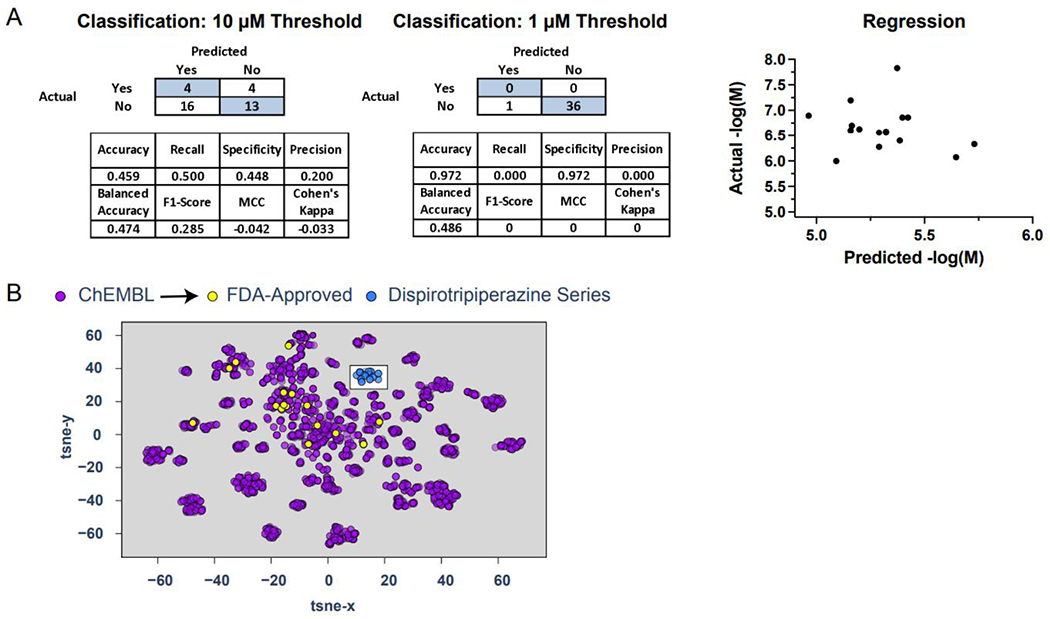
(A) Truth tables, statistical analysis (classification) and correlational visualization (regression) for the tested DSTPs. (B) A t-SNE visualization showing the ChEMBL HBV inhibition training set, FDA approved drugs and the tested DSTPs.
We have described the synthesis and anti-HBV evaluation of a small panel of mono- and bis- 6-6-6, 6-7-6, and 7-6-7 DSTPs containing heteroaryl groups with various substituents. While none of the synthesized mono-DSTP derivatives had activity against HBV, some bis-DSTP derivatives displayed low micromolar activity against HBV, with compound 11826096 displaying the most potent activity. Additionally, the low cytotoxicity associated with the compounds in cell culture supports the further investigation of DSTPs as antiviral drug leads. While this report provides largely preliminary in vitro activity, we hope that small molecules such as DSTPs could eventually be used in combination with NAs or other new anti-HBV agents to treat HBV.
The mechanism of action for these DSTPs against HBV replication is unknown at this early stage of development. However, the screening system employed (HepDES19 cells) limits the parts of the viral replication cycle that may be targeted. Screening with HepDES19 cells involves induction of HBV replication from a tetracycline-repressible transgene that carries mutations blocking production of the viral surface glycoproteins. Therefore, this system eliminates stages of the viral replication cycle upstream of transcription of the viral pregenomic RNA, viral envelopment, and virion secretion as potential targets of the DSTPs. The simultaneous suppression of the HBV plus- and minus-polarity DNA strands eliminates inhibition of viral DNA elongation and ribonuclease H activity as targets because inhibiting these steps preferentially suppresses the plus polarity DNA strand. Finally, capsid blotting (Figure. 4) reveals a modest suppression of capsid accumulation that is insufficient to account for the magnitude of reduction in viral DNA synthesis. Consequently, candidate targets for the DSTPs include encapsidation of the pregenomic RNA, protein-priming of DNA replication, and preferential destabilization of DNA-containing intracellular capsids (many capsids are empty in this cell system).
Future work on DSTPs could focus on the further elucidation of the mechanism of action and combination studies to assess for synergy with other HBV inhibitors. In the short term, we hope to expand upon our SAR with more DSTP analogs and define the stage of HBV replication targeted by DSTPs. The chemistry and purification we describe for the DSTPs is relatively complicated and may need to be simplified for future commercial utility. Alternatively, this complex chemistry could be commercially desirable as it represents a barrier for competitors for this class of compounds.
Experimental Section
Chemical Synthesis
General Procedure 1.
Mono-DSTPs were synthesized utilizing a modified procedure from Schmidtke et al.21 A solution of DSTP (1.0 equiv) in H2O was added to a suspension of chloroarene (2.15 equiv) in EtOH. Then, Et3N was added (2.05 equiv) and the reaction was allowed to proceed at room temperature for 4 hours. Then acetone was added, and the resulting precipitate was filtered and recrystallized from H2O/EtOH to yield the desired mono-DSTP. All compounds are >95% pure by HPLC.
General Procedure 2.
Bis-DSTPs were synthesized utilizing a modified procedure from Makarov et al.23 A solution of chloroarene (1.0 equiv) in EtOH was added to a solution of DSTP (2.0 equiv) in H2O and the reaction was heated to reflux for 2 hours. Then Et3N (4.0 equiv) was added and allowed to reflux for an additional 20 minutes. After cooling to room temperature, the solvent was evaporated under reduced pressure and MeOH was added. The resulting precipitate was filtered off and washed with MeOH and acetone to yield the desired bis-DSTP. All compounds are >95% pure by HPLC.
Activity against Hepatitis B Virus ayw1 in HepG2 2.2.15 cells
The anti-HBV assay was performed by NIAID as previously described 46, 47 with modifications to use real-time qPCR (TaqMan) to measure extracellular HBV DNA copy number associated with virions released from HepG2 2.2.15 cells. The HepG2 2.2.15 cell line is a stable human hepatoblastoma cell line that contains two copies of the HBV wild-type strain ayw1 genome and constitutively produces high levels of HBV. Antiviral compounds blocking any late step of viral replication such as transcription, translation, pregenome encapsidation, reverse transcription, particle assembly and release can be identified and characterized using this cell line.
Briefly, HepG2 2.2.15 cells are plated in 96-well microtiter plates at 1.5×104 cells/well in Dulbecco’s Modified Eagle’s Medium supplemented with 2% FBS, 380 μg/mL G418, 2.0 mM L-Glutamine, 100 units/mL Penicillin, 100 μg/mL Streptomycin, and 0.1 mM non-essential amino acids. Only the interior wells are utilized to reduce “edge effects” observed during cell culture; the exterior wells are filled with complete medium to help minimize sample evaporation. After 16-24 hours the confluent monolayer of HepG2 2.2.15 cells is washed, and the medium is replaced with complete medium containing various concentrations of a test compound in triplicate. lamivudine (3TC) is used as the positive control, while media alone is added to cells as a negative control (virus control, VC). Three days later the culture medium is replaced with fresh medium containing the appropriately diluted test compounds. Six days following the initial administration of the test compound, the cell culture supernatant is collected, treated with pronase and then used in a real-time quantitative TaqMan qPCR assay. The PCR-amplified HBV DNA is detected in real-time by monitoring increases in fluorescent signal that result from the exonucleolytic degradation of a quenched fluorescent probe molecule that hybridizes to the amplified HBV DNA. For each PCR amplification, a standard curve is simultaneously generated using dilutions of purified HBVDNA. Antiviral activity is calculated from the reduction in HBV DNA levels (EC50& EC90 values determined). A tetrazolium dye (MTS; 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium; CellTiter®96 Reagent, Promega) uptake assay is then employed to measure cell viability using the same assay plate, and the viability data is used to calculate compound cytotoxicity (CC50). The Selectivity Index (SI50) is calculated as CC50/EC50.
Activity against Hepatitis B Virus in HepDES19 cells.
HBV inhibition potential was measured in HepDES19 cells, a HepG2 derivative carrying a tetracycline-repressible HBV genomic expression cassette 25. Replication was detected after 3 days of compound exposure in cells induced to replicate HBV by tetracycline withdrawal using a strand-preferential quantitative PCR assays described 48 to help define the steps during HBV replication that were possible targets of the DSTPs. Cytotoxicity was measured using an MTS assay as described previously 48.
Capsid quantification.
HBV nucleocapsid levels were measured in HepDES19 cells replicating HBV after five days of compound treatment. Cells were lysed with core prep lysis buffer (10mM Tris pH 7.4, 1% Tween 20, 150mM NaCl) and 0.7 μL Protease Inhibitor Cocktail (Sigma-Aldrich, P8340). Lysates were centrifuged and a final concentration of 10mM CaCl2 and 35 units of micrococcal nuclease (New England BioLabs M0247S) was added to destroy non-encapsidated nucleic acids. Protein levels were quantified via a Bradford assay (Bio-Rad) using the manufactures protocol. Protein normalized cytoplasmic lysates were resolved by native agarose gel electrophoresis, then transferred onto a nitrocellulose membrane (GE Amersham) by capillary transfer using 10x SSC buffer (1.5M NaCl and 150mM sodium citrate). Nucleocapsid levels were measured by incubation with Mouse anti-HBcAg primary antibody (Tokyo Future Style, T2221) and Anti-Mouse IgG, AP conjugated secondary antibody (Promega, S3721) and detected by using BCIP and NBT Color Development Substrate (Promega, S3771). Bands were quantified by densitometry.
Data curation
Datasets for HBV inhibition (EC50/IC50 for target CHEMBL613497) were downloaded from the ChEMBL (version 32) database 49. Entries that defined either a numerical value for “EC50” or “IC50” value column were retained. Each dataset was sanitized using our proprietary software “E-Clean” which uses open-source RD-Kit tools in order to remove duplicate compounds and salts, as well as neutralize charges. For classification model training data, post compound standardization, a binary value was assigned based on defined threshold, AC50 value (converted to -logM) and qualifier (“=”,”>”,”<”). For duplicate compounds, as defined by InChIKey, a required 80% agreement was required to retain the compound. This was done to remove ambiguous compounds. As the % agreement was contingent on threshold, the total number of compounds per final dataset varied slightly. For regression model building, the compounds with a “=” qualifier and a value (−logM) were averaged together (geometric mean) and the rest of the compounds were discarded. Prior to model building, datasets were further standardized within the latest version of the Assay Central software which uses the Indigo Toolkit 50.
Machine learning
Our proprietary software Assay Central was used to generate multiple machine learning algorithms that are integrated to build classification and regression models that have been described in detail previously 51. The algorithms used included Bernoulli naïve Bayes, Linear Logistic Regression, AdaBoost Decision Tree, Random Forest, Support Vector Machine, Deep Neural Networks and XGBoost. Machine learning model validation was performed using a nested 5-fold cross validation. Nested 5-fold cross validation initially selects a random, stratified 20% hold out set that is removed from the training set prior to model building. The model is then built with the other 80% of the training data and the hyperparameters (if applicable) are optimized using a grid search using 5-fold dataset splits (20% validation sets). This optimized model is then used to predict the initial 20% hold out set and then repeated until all compounds have been in a hold-out set (total 20 models trained). The final nested 5-fold cross validation scores are an average of each of the hold-out set metrics. Due to its high computational requirement deep learning (DL) uses a 20% leave out set instead. Models were built using ECFP6 descriptors only and metrics generated as described previously 51. The applicability domain was calculated based on the reliability-density neighborhood (RDN) method which considers the model overlap and the individual bias and precision of the overlapping fingerprints (Figure 5) 52.
t-SNE visualization
t-SNE 53 embeds data into a lower-dimensional space (Figure 6). 1024 ECFP6 fingerprints were generated for all compounds. The ECFP6 fingerprints were then embedded into a 2-dimensional vector using t-SNE. All t-SNE values were generated using the scikit-learn library in python with default hyperparameters (n_components = 2, perplexity = 30, early exaggeration = 12.0, learning rate = 200, n_iter = 1000).
Supplementary Material
Acknowledgments
We kindly acknowledge Dr. Mindy Davis and Amanda Ullloa as well as their colleagues for assistance with antiviral testing services through NIAID. We also kindly acknowledge NIH funding: R44GM122196 from NIGMS, 1R01NS102164 from NINDS, and R01AI122669. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Collaborations Pharmaceuticals, Inc. has utilized the non-clinical and pre-clinical services program offered by the National Institute of Allergy and Infectious Diseases. We kindly acknowledge our colleagues for developing the software used in this work.
Abbreviations Used
- ACC
accuracy
- AUC
area under the curve
- BCIP
5-bromo-4-chloro-3-indolyl phosphate
- cccDNA
covalently closed circular DNA
- DSTP
dispirotripiperazine
- FBS
fetal bovine serum
- HSPG
heparan sulfate proteoglycan
- MAE
mean of absolute value of errors
- MCC
Matthews correlation coefficient
- MGD
mean gamma deviance regression loss
- MPD
mean Poisson deviance regression loss
- NBT
nitro-blue tetrazolium
- NIAID
National Institute of Allergy and Infectious Diseases
- NA
nucleoside/nucleotide analog
- qPCR
quantitative polymerase chain reaction
- RDN
reliability-density neighborhood
- RMSE
root of the mean of the square of errors
- RNaseH
ribonuclease H
- ROC
receiver operating characteristic
- SI
selectivity index
- SVC
support vector machine classification
- t-SNE
t-distributed stochastic neighbor embedding
Footnotes
Supplemental structural and analytical data for compounds synthesized, data for mono DSTP molecules are available in docx and csv file format.
Competing interests
S.E. is owner, and T.J. and T.R.L. employees of Collaborations Pharmaceuticals, Inc. All others have no competing interests.
References
- 1.CDC. Hepatitis b. https://www.cdc.gov/nchs/fastats/hepatitis.htm (accessed 25 May 2023).
- 2.WHO. Hepatitis b. https://www.who.int/news-room/fact-sheets/detail/hepatitis-b (accessed 25 May 2023).
- 3.Summers J; Mason WS Replication of the genome of a hepatitis b-like virus by reverse transcription of an rna intermediate. Cell 1982, 29, 403–415. [DOI] [PubMed] [Google Scholar]
- 4.Seeger C; Zoulim F; Mason WS Hepadnaviruses. In Fields virology, 6 ed.; Knipe DM; Howley PM, Eds. Lippincott Williams & Wilkins: Philadelphia PA, 2013; pp 2185–2221. [Google Scholar]
- 5.Kwon H; Lok AS Hepatitis b therapy. Nat Rev Gastroenterol Hepatol 2011, 8, 275–284. [DOI] [PubMed] [Google Scholar]
- 6.Levrero M; Subic M; Villeret F; Zoulim F Perspectives and limitations for nucleo(t)side analogs in future hbv therapies. Curr Opin Virol 2018, 30, 80–89. [DOI] [PubMed] [Google Scholar]
- 7.Tavis JE; Cheng X; Hu Y; Totten M; Cao F; Michailidis E; Aurora R; Meyers MJ; Jacobsen EJ; Parniak MA; Sarafianos SG The hepatitis b virus ribonuclease h is sensitive to inhibitors of the human immunodeficiency virus ribonuclease h and integrase enzymes. PLoS Pathog 2013, 9, e1003125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai CW; Lomonosova E; Moran EA; Cheng X; Patel KB; Bailly F; Cotelle P; Meyers MJ; Tavis JE Hepatitis b virus replication is blocked by a 2-hydroxyisoquinoline-1,3(2h,4h)-dione (hid) inhibitor of the viral ribonuclease h activity. Antiviral Res 2014, 108, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y; Cheng X; Cao F; Huang A; Tavis JE Beta-thujaplicinol inhibits hepatitis b virus replication by blocking the viral ribonuclease h activity. Antiviral Res 2013, 99, 221–229. [DOI] [PubMed] [Google Scholar]
- 10.Lu G; Lomonosova E; Cheng X; Moran EA; Meyers MJ; Le Grice SF; Thomas CJ; Jiang JK; Meck C; Hirsch DR; D’Erasmo MP; Suyabatmaz DM; Murelli RP; Tavis JE Hydroxylated tropolones inhibit hepatitis b virus replication by blocking the viral ribonuclease h activity. Antimicrob Agents Chemother 2015, 59, 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tavis JE; Lomonosova E The hepatitis b virus ribonuclease h as a drug target. Antiviral Res. 2015, 118, 132–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomonosova E; Daw J; Garimallaprabhakaran AK; Agyemang NB; Ashani Y; Murelli RP; Tavis JE Efficacy and cytotoxicity in cell culture of novel alpha-hydroxytropolone inhibitors of hepatitis b virus ribonuclease h. Antiviral Res 2017, 144, 164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards TC; Lomonosova E; Patel JA; Li Q; Villa JA; Gupta AK; Morrison LA; Bailly F; Cotelle P; Giannakopoulou E; Zoidis G; Tavis JE Inhibition of hepatitis b virus replication by n-hydroxyisoquinolinediones and related polyoxygenated heterocycles. Antiviral Res 2017, 143, 205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards TC; Mani N; Dorsey B; Kakarla R; Rijnbrand R; Sofia MJ; Tavis JE Inhibition of hbv replication by n-hydroxyisoquinolinedione and n-hydroxypyridinedione ribonuclease h inhibitors. Antiviral Res 2019, 164, 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikhalev VA, D MI, Smolina NE. Synthesis of dispiro-tripiperazine derivatives with cancerolytic properties. (preliminary report). Med Prom SSSR 1963, 1, 17–20. [PubMed] [Google Scholar]
- 16.Mikhalev VA, C. VA, Dorokhova MI, Smolina NE & Ya O. Tikhonova Haloalkylamines and their transformation products. Pharmaceutical Chemistry Journal 1972, 6, 355–359. [Google Scholar]
- 17.Schmidtke M, K. A, Meerbach A, Egerer R, Stelzner A, and Makarov V. Binding of a n,n’-bisheteryl derivative of dispirotripiperazine to heparan sulfate residues on the cell surface specifically prevents infection of viruses from different familie. Virology 2003, 311, 132–143. [DOI] [PubMed] [Google Scholar]
- 18.Levkovskaya LG, M. IE, Pushkina TV, Nikolaeva IS, Yu L. Krylova, Fomina AN & S. T Safonova Synthesis and antiviral activity of bisquaternary salts of 3,12-bis(3’-haloacyl)-3,12-diaza-6,9-diazoniadispiro[5.2.5.2]hexadecane. Pharmaceutical Chemistry Journal 1989, 23, 241–244. [Google Scholar]
- 19.Park PW Glycosaminoglycans and infection. Frontiers in Bioscience 2016, 21, 1260–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selinka HC; Florin L; Patel HD; Freitag K; Schmidtke M; Makarov VA; Sapp M Inhibition of transfer to secondary receptors by heparan sulfate-binding drug or antibody induces noninfectious uptake of human papillomavirus. J Virol 2007, 81, 10970–10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egorova A; Bogner E; Novoselova E; Zorn KM; Ekins S; Makarov V Dispirotripiperazine-core compounds, their biological activity with a focus on broad antiviral property, and perspectives in drug design (mini-review). European Journal of Medicinal Chemistry 2021, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidtke M; Riabova O; Dahse HM; Stelzner A; Makarov V Synthesis, cytotoxicity and antiviral activity of n,n’-bis-5-nitropyrimidyl derivatives of dispirotripiperazine. Antiviral Res 2002, 55, 117–127. [DOI] [PubMed] [Google Scholar]
- 23.Schmidtke M, R. O, Dahse H-M, Stelzner A, Makarov V. Synthesis, cytotoxicity and antiviral activity of n,n’-bis-5-nitropyrimidyl derivatives of dispirotripiperazine. Antiviral Research 2002, 55, 117–127. [DOI] [PubMed] [Google Scholar]
- 24.Vadim Makarov VN, Bolgarin Roman, Novoselova Elena. Pyrimidyl-di(diazaspiro-alkanes) with antiviral activity. 2017. [Google Scholar]
- 25.Guo H; Jiang D; Zhou T; Cuconati A; Block TM; Guo JT Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis b virus: An intermediate of covalently closed circular DNA formation. J Virol 2007, 81, 12472–12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo J; Xi J; Gao L; Hu J Role of hepatitis b virus capsid phosphorylation in nucleocapsid disassembly and covalently closed circular DNA formation. PLoS Pathog 2020, 16, e1008459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox N; Tillmann H Emerging pipeline drugs for hepatitis b infection. Expert.Opin.Emerg.Drugs 2011, 16, 713–729. [DOI] [PubMed] [Google Scholar]
- 28.Kwon H; Lok AS Hepatitis b therapy. Nat.Rev.Gastroenterol.Hepatol 2011, 8, 275–284. [DOI] [PubMed] [Google Scholar]
- 29.van Bommel F; De Man RA; Wedemeyer H; Deterding K; Petersen J; Buggisch P; Erhardt A; Huppe D; Stein K; Trojan J; Sarrazin C; Bocher WO; Spengler U; Wasmuth HE; Reinders JG; Moller B; Rhode P; Feucht HH; Wiedenmann B; Berg T Long-term efficacy of tenofovir monotherapy for hepatitis b virus-monoinfected patients after failure of nucleoside/nucleotide analogues. Hepatology 2010, 51, 73–80. [DOI] [PubMed] [Google Scholar]
- 30.Woo G; Tomlinson G; Nishikawa Y; Kowgier M; Sherman M; Wong DK; Pham B; Ungar WJ; Einarson TR; Heathcote EJ; Krahn M Tenofovir and entecavir are the most effective antiviral agents for chronic hepatitis b: A systematic review and bayesian meta-analyses. Gastroenterology 2010, 139, 1218–1229. [DOI] [PubMed] [Google Scholar]
- 31.Marcellin P; Heathcote EJ; Buti M; Gane E; De Man RA; Krastev Z; Germanidis G; Lee SS; Flisiak R; Kaita K; Manns M; Kotzev I; Tchernev K; Buggisch P; Weilert F; Kurdas OO; Shiffman ML; Trinh H; Washington MK; Sorbel J; Anderson J; Snow-Lampart A; Mondou E; Quinn J; Rousseau F Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis b. N.Engl.J.Med 2008, 359, 2442–2455. [DOI] [PubMed] [Google Scholar]
- 32.Werle-Lapostolle B; Bowden S; Locarnini S; Wursthorn K; Petersen J; Lau G; Trepo C; Marcellin P; Goodman Z; Delaney WE; Xiong S; Brosgart CL; Chen SS; Gibbs CS; Zoulim F Persistence of cccdna during the natural history of chronic hepatitis b and decline during adefovir dipivoxil therapy. Gastroenterology 2004, 126, 1750–1758. [DOI] [PubMed] [Google Scholar]
- 33.Cheng PN; Liu WC; Tsai HW; Wu IC; Chang TT; Young KC Association of intrahepatic cccdna reduction with the improvement of liver histology in chronic hepatitis b patients receiving oral antiviral agents. J.Med.Virol. 2011, 83, 602–607. [DOI] [PubMed] [Google Scholar]
- 34.Wong DK; Yuen MF; Ngai VW; Fung J; Lai CL One-year entecavir or lamivudine therapy results in reduction of hepatitis b virus intrahepatic covalently closed circular DNA levels. Antivir.Ther. 2006, 11, 909–916. [PubMed] [Google Scholar]
- 35.Devi U; Locarnini S Hepatitis b antivirals and resistance. Curr Opin Virol 2013, 3, 495–500. [DOI] [PubMed] [Google Scholar]
- 36.Wursthorn K; Jung M; Riva A; Goodman ZD; Lopez P; Bao W; Manns MP; Wedemeyer H; Naoumov NV Kinetics of hepatitis b surface antigen decline during 3 years of telbivudine treatment in hepatitis b e antigen-positive patients. Hepatology 2010, 52, 1611–1620. [DOI] [PubMed] [Google Scholar]
- 37.Evans AA; London WT; Gish RG; Cohen C; Block TM Chronic hbv infection outside treatment guidelines: Is treatment needed? Antivir Ther 2013, 18, 229–235. [DOI] [PubMed] [Google Scholar]
- 38.Liaw YF; Sung JJ; Chow WC; Farrell G; Lee CZ; Yuen H; Tanwandee T; Tao QM; Shue K; Keene ON; Dixon JS; Gray DF; Sabbat J; Cirrhosis Asian Lamivudine Multicentre Study, G. Lamivudine for patients with chronic hepatitis b and advanced liver disease. N Engl J Med 2004, 351, 1521–1531. [DOI] [PubMed] [Google Scholar]
- 39.Lok AS Does antiviral therapy for hepatitis b and c prevent hepatocellular carcinoma? J Gastroenterol Hepatol 2011, 26, 221–227. [DOI] [PubMed] [Google Scholar]
- 40.Lok AS; McMahon BJ Chronic hepatitis b: Update 2009. Hepatology 2009, 50, 661–662. [DOI] [PubMed] [Google Scholar]
- 41.Ghany M; Liang TJ Drug targets and molecular mechanisms of drug resistance in chronic hepatitis b. Gastroenterology 2007, 132, 1574–1585. [DOI] [PubMed] [Google Scholar]
- 42.Monto A; Schooley RT; Lai JC; Sulkowski MS; Chung RT; Pawlotsky JM; McHutchison JG; Jacobson IM Lessons from hiv therapy applied to viral hepatitis therapy: Summary of a workshop. Am.J.Gastroenterol. 2010, 105, 989–1004. [DOI] [PubMed] [Google Scholar]
- 43.Zoulim F; Locarnini S Hepatitis b virus resistance to nucleos(t)ide analogues. Gastroenterology 2009, 137, 1593–1608. [DOI] [PubMed] [Google Scholar]
- 44.Coffin CS; Mulrooney-Cousins PM; Peters MG; van MG; Roberts JP; Michalak TI; Terrault NA Molecular characterization of intrahepatic and extrahepatic hepatitis b virus (hbv) reservoirs in patients on suppressive antiviral therapy. J.Viral Hepat. 2011, 18, 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Block TM; Gish R; Guo H; Mehta A; Cuconati A; Thomas London W; Guo JT Chronic hepatitis b: What should be the goal for new therapies? Antiviral Res 2013, 98, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korba BE; Milman G A cell culture assay for compounds which inhibit hepatitis b virus replication. Antiviral Res 1991, 15, 217–228. [DOI] [PubMed] [Google Scholar]
- 47.Korba BE; Gerin JL Use of a standardized cell culture assay to assess activities of nucleoside analogs against hepatitis b virus replication. Antiviral Res 1992, 19, 55–70. [DOI] [PubMed] [Google Scholar]
- 48.Li Q; Edwards TC; Ponzar NL; Tavis JE A mid-throughput hbv replication inhibition assay capable of detecting ribonuclease h inhibitors. J Virol Methods 2021, 292, 114127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mendez D; Gaulton A; Bento AP; Chambers J; De Veij M; Felix E; Magarinos MP; Mosquera JF; Mutowo P; Nowotka M; Gordillo-Maranon M; Hunter F; Junco L; Mugumbate G; Rodriguez-Lopez M; Atkinson F; Bosc N; Radoux CJ; Segura-Cabrera A; Hersey A; Leach AR Chembl: Towards direct deposition of bioassay data. Nucleic Acids Res 2019, 47, D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lane TR; Urbina F; Rank L; Gerlach J; Riabova O; Lepioshkin A; Kazakova E; Vocat A; Tkachenko V; Cole S; Makarov V; Ekins S Machine learning models for mycobacterium tuberculosis in vitro activity: Prediction and target visualization. Mol Pharm 2022, 19, 674–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lane T; Russo DP; Zorn KM; Clark AM; Korotcov A; Tkachenko V; Reynolds RC; Perryman AL; Freundlich JS; Ekins S Comparing and validating machine learning models for mycobacterium tuberculosis drug discovery. Mol Pharm 2018, 15, 4346–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aniceto N; Freitas AA; Bender A; Ghafourian T A novel applicability domain technique for mapping predictive reliability across the chemical space of a qsar: Reliability-density neighbourhood. Journal of Cheminformatics 2016, 8, 69. [Google Scholar]
- 53.van der Maaten L; Hinton G Visualizing data using t-sne. J Machine Learning Research 2008, 9, 2579–2605. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


