Abstract
Interferons (IFNs) are a family of multifunctional cytokines with antiviral activities. The K9 open reading frame of Kaposi’s sarcoma-associated herpesvirus (KSHV) exhibits significant homology with cellular IFN regulatory factors (IRFs). We have investigated the functional consequence of K9 expression in IFN-mediated signal transduction. Expression of K9 dramatically repressed transcriptional activation induced by IFN-α, -β, and -γ. Further, it induced transformation of NIH 3T3 cells, resulting in morphologic changes, focus formation, and growth in reduced-serum conditions. The expression of antisense K9 in KSHV-infected BCBL-1 cells consistently increased IFN-mediated transcriptional activation but drastically decreased the expression of certain KSHV genes. Thus, the K9 gene of KSHV encodes the first virus-encoded IRF (v-IRF) which functions as a repressor for cellular IFN-mediated signal transduction. In addition, v-IRF likely plays an important role in regulating KSHV gene expression. These results suggest that KSHV employs an unique mechanism to antagonize IFN-mediated antiviral activity by harboring a functional v-IRF.
Interferons (IFNs) are a family of cytokines that exhibit such diverse biological effects as the inhibition of cell growth and protection against viral infection. Newly synthesized IFN interacts with cellular surface receptors, resulting in the synthesis of a group of cellular proteins (7). An important regulatory step unique to the response to IFN treatment is the activation of a transcriptional factor that recognizes a conserved cis-acting DNA element located within the regulatory sequences of target genes (7). Alpha and beta IFNs (IFN-α and IFN-β) and gamma IFN (IFN-γ) regulate the expression of overlapping sets of genes and elicit similar yet distinct biological activities (7). The conserved cis-acting IFN-stimulated response element (ISRE) binds to members of the IFN regulatory factor (IRF) family, which include IRF-1, IRF-2, the IFN consensus sequence binding protein (ICSBP), and the IFN-α-stimulated gene factor 3γ (ISGF3γ) (8, 10, 31). IRF-1, a transcriptional activator, and IRF-2, its antagonistic repressor, have been identified as regulators of IFN-α and IFN-inducible genes (12). ICSBP has been shown to repress IFN- and IRF-1-mediated activation of a number of reporter genes that are driven by the conserved cis-acting DNA element, suggesting that ICSBP acts by interfering with the function of these activators (8). IRF-1, IRF-2, and ICSBP are constitutively expressed in the nucleus, while ISGF3γ is expressed as a latent cytoplasmic protein that is translocated into the nucleus as part of a multisubunit complex formed upon IFN treatment (6).
The IFN-γ activation site (GAS) overlaps with the ISRE but binds to distinct trans-acting factors called STATs (signal transducers and activators of transcription) (6). STATs transduce a signals from cytokine receptors to DNA transcription regulatory elements. STAT proteins are cytoplasmic proteins that are activated by the phosphorylation of a specific tyrosine residue by a Jak family kinase (14). Phosphorylated STATs dimerize and translocate to the nucleus, where they bind to GAS or the ISRE and direct transcription (14).
Many lines of epidemiological evidence suggest an infectious etiology for Kaposi’s sarcoma (KS). DNA sequences of a novel member of the herpesvirus group, called KS-associated herpesvirus (KSHV) or human herpesvirus 8, have been widely identified in KS tumors from human immunodeficiency virus-positive and -negative patients (4, 5, 39). KSHV has also been consistently identified in body cavity-based lymphomas and some forms of Castleman’s disease (4, 5). Analyses of KSHV genomic sequences indicate that KSHV is a gammaherpesvirus that is closely related to herpesvirus saimiri (HVS) (28).
DNA sequence analysis of the entire 140.5 kbp of the KSHV genome reveals a number of cellular homologs which could possibly contribute to the pathogenesis associated with this virus (22, 28). These include a virus-encoded interleukin-6 IL-6 (vIL-6) (20, 21, 24), a vMIP1-α/β chemokine (20, 24), a Bcl-2 homolog (30), a viral IRF homolog (K9), a virus-encoded cyclin (11, 18), a virus-encoded G protein-coupled receptor (vGCR) (2), a virus-encoded FLICE inhibitory protein (v-FLIP) (34), and a neural cell adhesion molecule (N-CAM) homolog. The K9 open reading frame of KSHV was found to have overall amino acid identity of 13% to human ISGF3γ and ICSBP, with a conservation of the tryptophan-rich DNA binding region at the amino terminus (20). To investigate the role of K9, we have examined the functional consequence of K9 expression in IFN-mediated signal transduction. The expression of the K9 gene dramatically represses IFN-mediated gene regulation activity and also induces transformation of mouse fibroblasts. In addition, the K9 gene plays an important role in regulating the expression of some KSHV genes. These results demonstrate an elaborate molecular mimicry that has been developed by KSHV to antagonize the IFN-mediated cellular antiviral activity.
MATERIALS AND METHODS
Cell culture and transfection.
293 and NIH 3T3 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum. BJAB, BC-1, and BCBL-1 cells were grown in RPMI 1640 supplemented with 10% fetal calf serum. A calcium phosphate transfection was used for transient expression in 293 cells and for stable expression in NIH 3T3 cells.
Cloning of KSHV K9 from BCBL-1 cells.
The KSHV K9 open reading frame includes bp 86074 to 88164 of the published sequence of KSHV (28). A DNA fragment corresponding to the KSHV K9 was synthesized from BCBL-1 genomic DNA by PCR using primers containing a BamHI site at the 5′ end and an EcoRI site at the 3′ end for subsequent cloning. The 764-bp PCR-amplified DNA was digested with the restriction enzymes BamHI and EcoRI and subcloned into vector pcDNA3. DNA containing the KSHV K9 open reading frame was cloned in an antisense orientation into the NotI and BamHI sites of the tetracycline-inducible retroviral vector pBP JTR-1, kindly provided by Steven Reeves (25). In this construct, the antisense K9 gene is expressed from the cytomegalovirus early promoter, of which activity is regulated by the tetracycline treatment. The retroviral vector was electroporated into BCBL-1 cells, and these cells were selected for puromycin resistance for the next 5 weeks. For transient expression in 293 cells, KSHV K9 DNA was amplified by PCR and subcloned into the pFJ vector (33). The KSHV K9 gene was completely sequenced to verify 100% agreement with the original sequence using an ABI PRISM 377 automatic DNA sequencer.
Recombinant K9 protein and antibodies.
For purification of recombinant K9 protein from Escherichia coli, the K9 DNA fragment was amplified by PCR using primers containing BamHI- and SalI-recognition sequences at the ends and subcloned into BamHI and SalI cloning sites of the pQE-40 expression vector (Qiagen, San Diego, Calif.) with the potential of incorporating six histidines at the amino terminus. The lack of unwanted mutations was confirmed by direct DNA sequencing. When E. coli XL-1 blue containing plasmid pQE40-K9 reached an optical density at 600 nm of approximately 0.6, 1 mM isopropyl-β-d-thiogalactopyranoside was added, and cells were harvested 3 h after induction. Cells were solubilized with 6 M guanidine hydrochloride. Due to the presence of the affinity tail, His6-K9 protein was purified to virtual homogeneity in one step by Ni2+-chelate affinity chromatography. The purified recombinant His6-K9 protein was used to generate polyclonal antibody in New Zealand White rabbits. A Ni2+-chelate affinity column containing K9 protein was used to purify the antigen-specific antibodies. Antibody specific for K9 was eluted with high-pH solution (0.1 M triethylamine, pH 11.5).
Northern blot analysis.
Northern blot analysis was performed under standard conditions with random-labeled probes derived from vIL-6, K8, PAN/T1.1, small viral capsid antigen (sVCA), orf73, and cellular actin DNA. Total RNA was purified from BCBL-1 and BCBL-1/anti-K9 cells as instructed by the manufacturer (Qiagen), and 10 μg of total RNA was loaded in each lane. The filters were baked at 80°C for 2 h and then hybridized with radioactive probes.
Southern blot analysis.
Genomic DNA was digested overnight with restriction enzyme PstI. Digested DNA was separated on a 1% agarose gel, transferred to a nitrocellulose membrane, and subjected to a hybridization reaction. A labeled DNA fragment containing the vIL-6 or orf73 gene was used as a probe. Detection of DNA bands was performed with the protocol provided by the manufacturer (Boehringer Mannheim, Indianapolis, Ind.).
Luciferase assays.
NIH 3T3 cells were transfected by calcium phosphate protocol, and BCBL-1 cells were electroporated at 960 μF and 200 V. Cells were harvested 48 h after incubation with or without IFNs. All transfections included pGKβgal, which expresses β-galactosidase from a phosphoglucokinase promoter, and GAS-luc, GASmt-luc, GBP-ISRE-luc, or ISG15-ISRE-luc, described previously (16, 23, 36). Assays for luciferase or β-galactosidase activity were performed with a luminometer, using luciferase assay reagent or a β-galactosidase assay kit (Promega, Madison, Wis.). Luciferase values were normalized to β-galactosidase activity.
Immunoprecipitation and immunoblotting.
Cells were harvested and lysed with lysis buffer (0.3 M NaCl, 0.1% Nonidet P-40, 50 mM HEPES buffer [pH 8.0]) or radioimmunoprecipitation assay buffer (0.15 M NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris [pH 7.5]) containing 0.1 mM Na2VO3, 1 mM NaF, and protease inhibitors (leupeptin, aprotinin, phenylmethylsulfonyl fluoride, and bestatin). Immunoprecipitated proteins from cleared cell lysates were separated by SDS-polyacrylamide gel electrophoresis (PAGE), and detected by autoradiography of the dried gel slabs. For protein immunoblots, polypeptides in cell lysates corresponding to 105 cells were resolved in SDS-PAGE and transferred to a nitrocellulose membrane filter. Immunoblot detection was performed with a 1:2,000 dilution of primary antibody by enhanced chemiluminescence (Amersham).
Fluorescence-activated cell sorting (FACS) analysis.
Exponentially growing BCBL-1 cells were incubated with 50 μg of plasmid LXSG or LXSG-anti-vIL-6 in a 0.4-cm gap cuvette and given a 200-V and 960-μF charge from a Gene Pulser (Bio-Rad, Hercules, Calif.). After 48 h, green fluorescent cells were sorted out in a FACS Vantage (Becton Dickinson). Sorted cells were washed twice with phosphate-buffered saline and lysed with lysis buffer.
Assays for growth properties.
For serum dependence, 106 cells were seeded in 100-mm-diameter tissue culture dishes in DMEM plus 10% serum for 24 h. The cultures were washed four times with serum-free medium and transferred to DMEM with 0.1% serum. The cells were observed daily, and medium was changed every 4 days for 2 weeks. For assays for focus formation, 106 cells were plated in 100-mm-diameter tissue culture dishes and maintained with DMEM plus 10% serum changed every 4 days. At day 14, cells were photographed.
RESULTS
Identification of the KSHV K9 gene product.
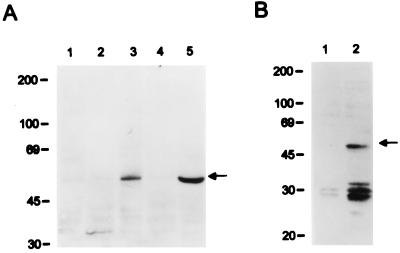
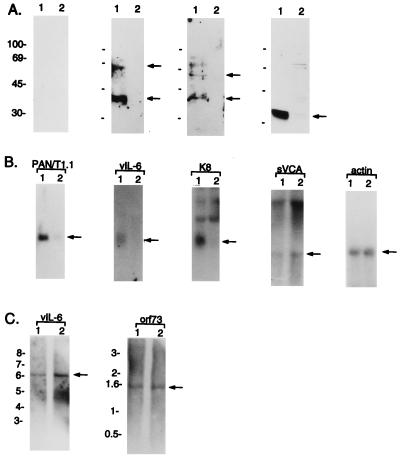
To demonstrate the expression of K9, we generated a rabbit polyclonal antibody against a purified bacterial His6-K9 fusion protein. To facilitate transient expression in 293 cells, an expression vector containing the KSHV K9 was constructed in plasmid pFJ containing the SRα-0 promoter (33). The anti-K9 antibody reacted specifically with a protein having an apparent molecular size of 50 kDa on immunoblots in 293 cells transfected with the K9 expression vector (Fig. 1A, lane 5). No such protein was detected in control 293 cells lacking the K9 gene. Lysates from BC-1 cells coinfected with KSHV and Epstein-Barr virus, BCBL- 1 cells infected with KSHV, and uninfected BJAB cells were used in an immunoblot assay with rabbit anti-K9 antibody. This experiment showed that a 50-kDa K9 protein was detected only in the BCBL-1 cells (Fig. 1A). In contrast, K9 was not detected from BC-1 cells and control BJAB cells. Previously, it has been shown that a K9 transcript was detected in BCP-1 cells, which are equivalent to BCBL-1 cells, but was not detected in BC-1 cells (20).
FIG. 1.
(A) Identification of KSHV K9 protein. Lysates from BJAB cells (lane 1), BC-1 cells (lane 2), BCBL-1 cells (lane 3), 293 cells transfected with the pFJ vector (lane 4), and 293 cells transfected with pFJ-K9 (lane 5) were used for immunoblot assay with anti-K9 antibody. (B) Construction of the NIH 3T3-K9 cell line. After selection with puromycin, lysates of NIH 3T3-babe cells (lane 1) and NIH 3T3-K9 cells (lane 2) were immunoblotted with anti-K9 antibody. Arrows indicate the K9 protein. Sizes are indicated in kilodaltons.
Repression of IFN-mediated signal transduction by KSHV K9.
To investigate the role of K9 in IFN-mediated signal transduction, the K9 reading frame was cloned into the retroviral expression vector pBabe-puro. NIH 3T3 cells were stably transfected with plasmid pBabe-puro or pBabe-K9 and then selected with puromycin (5 μg/ml). The 50-kDa K9 protein was detected by immunoblot analysis in NIH 3T3 cells stably transfected with pBabe-K9 (Fig. 1B). To examine the effect of K9 expression on transcriptional activation induced by IFNs, we measured the transcriptional activity of IFN-regulated promoters by using a luciferase reporter gene. Since type I IFN (IFN- α and IFN-β) and type II IFN (IFN-γ) act on overlapping but distinct sets of cis-acting elements (6, 7), we used three luciferase constructs that contained the different cis-acting elements: the GAS and the ISREs of the ISCBP15 promoter (ISG15-ISRE) and the guanylate binding protein (GBP-ISRE). The GAS element is regulated by IFN-γ, ISG15-ISRE is regulated by IFN-α/β, and GBP-ISRE is regulated by IFN-α/β and -γ (6, 7, 16, 23, 36).
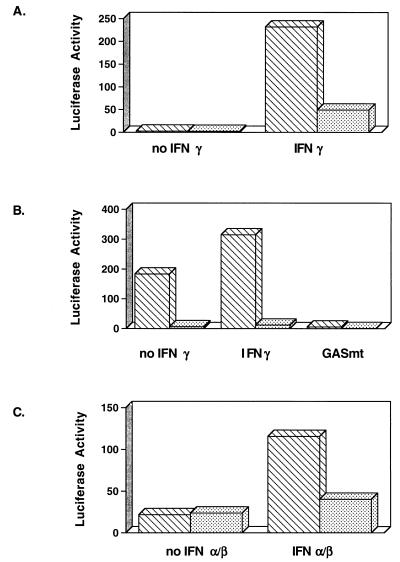
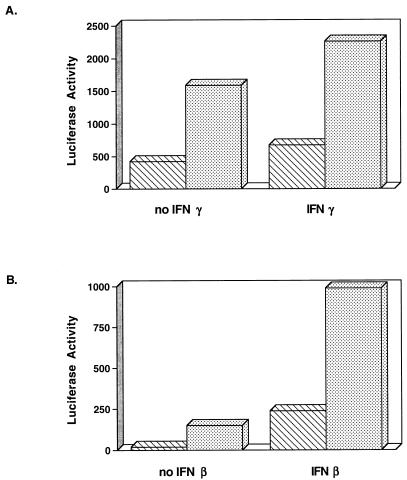
NIH 3T3-babe and NIH 3T3-K9 cells were transfected with a luciferase reporter plasmid and control β-galactosidase plasmid pGKβgal. After transfection, cells were incubated for 48 h in the presence or absence of IFNs. Luciferase activity was normalized for transfection efficiency by β-galactosidase activity. IFN-γ activated GBP-ISRE activity approximately 230-fold in NIH 3T3-babe cells, whereas the level of activation of GBP-ISRE by IFN-γ was drastically reduced in NIH 3T3-K9 cells (Fig. 2A). Unlike the level of GBP-ISRE activity the level of GAS activity was high in NIH 3T3-babe cells in the absence of IFN-γ, and IFN-γ treatment only weakly induced GAS activity in these cells (Fig. 2B). In contrast, the level of GAS activity in NIH 3T3-K9 cells was repressed approximately 200-fold in the absence of IFN-γ compared to that of NIH 3T3-babe cells (Fig. 2B). Additionally, IFN-γ treatment had almost no effect on GAS activity in NIH 3T3-K9 cells (Fig. 2B). No luciferase was detected in either cell line that was transfected with the mutant GAS element, which did not bind to the STAT1 transcriptional factor (6) (Fig. 2B). To determine the effect of K9 expression on type I IFN-α/β signal transduction, we examined the level of transcriptional activation of ISG15-ISRE by IFN-α/β treatment in NIH 3T3-babe and NIH 3T3-K9 cells. These experiments also showed that the level of transcriptional activation of ISG15-ISRE by IFN-α/β was dramatically reduced in NIH 3T3-K9 cells compared to NIH 3T3-babe cells (Fig. 2C). These results demonstrate that K9 expression in NIH 3T3 cells leads to marked repression in type I and II IFN-mediated signal transduction activity.
FIG. 2.
Expression of K9 represses IFN-mediated induction of GAS, GBP, and ISG15 promoter activity. NIH 3T3-babe (▧) and NIH 3T3-K9 (░⃞) cells were cotransfected with GBP-ISRE-luc (A), GAS-luc or GASmt-luc (B), or ISG15-ISRE-luc (C) together with pGKβgal, which expresses β-galactosidase from a phosphoglucokinase promoter. After transfection, cells were cultured in the presence or absence of mouse IFN-α/β or IFN-γ for 48 h. Assays for luciferase or β-galactosidase activity were performed as described in Materials and Methods. Values represent the average of three independent experiments.
Transformation of NIH 3T3 cells by the KSHV K9 expression.
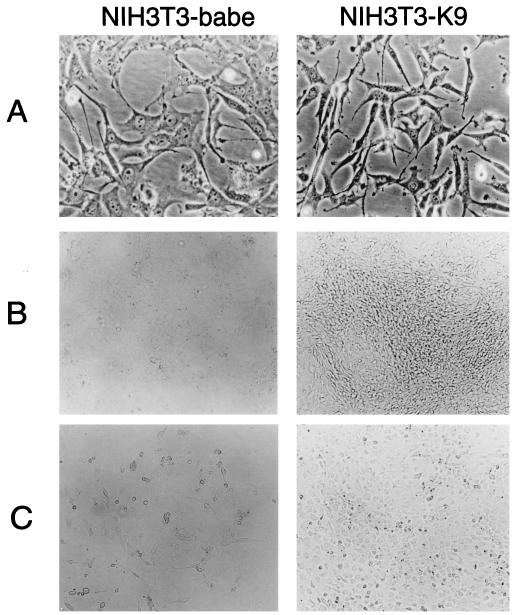
Cellular IRF-2 has been shown to function as a transcriptional repressor of IFN-mediated signal transduction (12). When IRF-2 was overexpressed in NIH 3T3 cells, cells became transformed and displayed enhanced tumorigenicity in nude mice (12). To investigate the potential transforming activity of the K9 gene, the growth properties of NIH 3T3-K9 cells were compared with those of NIH 3T3-babe cells. As shown in Fig. 3, the growth properties of NIH 3T3-K9 cells differed markedly from those of NIH 3T3-babe cells. NIH 3T3-K9 cells were considerably smaller than control NIH 3T3-babe cells (Fig. 3A). In focus-forming assays, NIH 3T3-K9 cells formed multiple foci which were recognizable even before cells reached confluence (Fig. 3B). The number of foci observed for NIH 3T3-K9 cells was over 1,000 per 100-mm-diameter tissue culture dish, while NIH 3T3-babe cells grew in flat monolayers and formed fewer than 50 foci (data not shown). Serum dependence of the cell lines was also tested over 14 days at 0.1% serum concentration. NIH 3T3-babe cells showed little, if any, increase in cell number at 0.1% serum concentration (Fig. 3C). In contrast, NIH 3T3-K9 cells continued to grow and reached confluence under the same conditions (Fig. 3C). Thus, the K9 gene of KSHV induces transformation of NIH 3T3 cells, resulting in morphologic changes, focus formation, and growth in reduced-serum conditions.
FIG. 3.
Transformation of NIH 3T3 cells by the KSHV K9 gene. Puromycin-resistant cells were obtained after transfection with the retroviral vector pBabe-puro or pBabe-K1. Puromycin-resistant cells were plated at 106 cells per 100-mm-diameter tissue culture dish. Preconfluent puromycin-resistant cells were photographed at a magnification of ×100 (A). After 14 days incubation, cells were photographed to show morphologic transformation at ×40 (B). For serum dependence (C), 106 cells were placed in 100-mm-diameter tissue culture dish, maintained in 0.1% serum for 14 days, and photographed at ×40.
Antisense K9 expression in BCBL-1 cells.
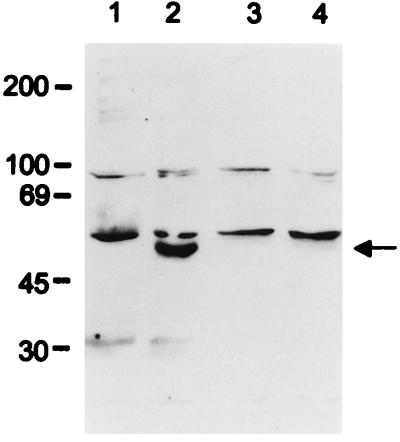
Since K9 was readily detected in BCBL-1 cells by immunoblot analysis, we examined the effect of K9 on IFN-regulated gene expression in these cells. We constructed a BCBL-1/anti-K9 cell line in which an antisense form of K9 gene was expressed. The full-length K9 gene was cloned into a tetracycline-inducible retroviral vector in an antisense orientation, which was then electroporated into BCBL-1 cells. After electroporation, puromycin-resistant BCBL-1 cells were selected in the presence of tetracycline to suppress the antisense K9 expression. Lysates of control BCBL- 1 and BCBL-1/anti-K9 cells were used for immunoblot assays with a rabbit anti-K9 antibody to measure the level of K9 expression. As shown in Fig. 4, K9 expression was drastically reduced in BCBL-1/anti-K9 cells in comparison with BCBL-1 cells. However, because of the leakiness of the tetracycline-inducible gene regulation, BCBL-1/anti-K9 cells showed a similar reduction of K9 expression in the presence or absence of tetracycline in the culture medium (Fig. 4). While slight morphologic changes were observed in BCBL-1/anti-K9 cells, no difference in growth rate between BCBL-1 cells and BCBL-1/anti-K9 cells was detected (data not shown).
FIG. 4.
Decreased K9 expression in BCBL-1/anti-K9 cells. Cell lysates were used for immunoblot assay with the anti-K9 antibody. Lane 1, control BJAB cells; lane 2, BCBL-1 cells; lane 3, BCBL-1/anti-K9 cells with tetracycline; lane 4, BCBL-1/anti-K9 cells without tetracycline. The arrow indicates K9 protein. Sizes are indicated in kilodaltons.
Increase of IFN-mediated transcriptional activity by the expression of antisense K9 in BCBL-1 cells.
To determine the effect of antisense K9 expression on IFN-regulated transcriptional activity in KSHV-infected B cells, BCBL-1 and BCBL-1/anti-K9 cells were electroporated with the luciferase reporter plasmids and the control β-galactosidase plasmid pGKβgal. The level of GAS and ISG15-ISRE activity of BCBL-1/anti-K9 cells was approximately four- to fivefold higher than that in the parental BCBL-1 cells in the presence or absence of IFN-γ stimulation (Fig. 5). These results demonstrated that the antisense K9 expression in BCBL-1 cells caused a significant increase of IFN-mediated transcriptional activity. Consistent with the results for NIH 3T3 cells, KSHV K9 functions as a repressor of IFN-mediated signal transduction in BCBL-1 cells.
FIG. 5.
Increased level of IFN-mediated transcriptional activity by decreased K9 expression. BCBL-1 (▧) and BCBL-1/anti-K9 (░⃞) cells were electroporated with GAS-luc (A) or ISG15-ISRE-luc (B) together with pGKβgal. After electroporation, cells were incubated with or without human IFN-β or IFN-γ for 48 h. Assays for luciferase or β-galactosidase activity were performed as described in Materials and Methods. Values represent the average of three independent experiments.
Downregulation of KSHV viral gene expression in BCBL-1/anti-K9 cells.
The detection of p27, p40, and p60 polypeptides in chemically stimulated BC-1 cells with KS-positive human sera has been described (19). We examined the expression of these polypeptides in BCBL-1 cells in which 1% of the culture displayed a spontaneous lytic cycle (27). Sera from patients with KS were used as the source of antibody for immunoblot analysis. KS-positive human sera detected two polypeptides, p40 and p60, in unstimulated BCBL-1 cells, while KS-negative normal human sera did not (Fig. 6A). Surprisingly, polypeptides p40 and p60 were not detected to an appreciable extent in BCBL-1/anti-K9 cells with the same KS-positive human sera as was used in BCBL-1 cells (Fig. 6A). Immunoblots with three additional KS-positive human sera exhibited similar results although with the different levels of reactivity against BCBL-1 cells (data not shown). We also measured the expression of KSHV vIL-6, which has been found previously in BC-1 and BCBL-1 cells (20). As was seen with p40 and p60, the amount of vIL-6 was dramatically decreased in BCBL-1/anti-K9 cells compared to BCBL-1 cells (Fig. 6A).
FIG. 6.
Downregulation of KSHV gene expressions in BCBL-1/anti-K9 cells. (A) Immunoblot assay of KSHV proteins. Lysates from BCBL-1 cells (lane 1) and BCBL-1/anti-K9 cells (lane 2) were immunoblotted with KS-negative normal human serum (a), KS-positive human serum 19 (b), KS-positive human serum 20 (c), or rabbit anti-vIL-6 antibody (d). Arrows indicate locations of the specific proteins. Sizes are indicated in kilodaltons. (B) Northern blot analysis of KSHV genes. Total RNA from BCBL-1 cells (lane 1) and BCBL-1/anti-K9 cells (lane 2) was separated on a 1% agarose gel, transferred to nitrocellulose, and hybridized with 32P-labeled PAN/T1.1, vIL-6, K8, sVCA, or cellular actin. Arrows indicate the location of the specific transcripts. (C) Southern blot analysis of KSHV genes. Purified genomic DNA of BCBL-1 (lane 1) or BCBL-1/anti-K9 cells (lane 2) was digested with restriction enzyme PstI. Labeled vIL-6 or orf73 probe was used for the hybridization. The vIL-6-positive band is expected to be a 6.3-kb DNA fragment, and the orf73-positive band is expected to be a 1.5-kb DNA fragment. Arrows indicate locations of the vIL-6- and orf73-specific bands. Sizes are indicated in kilobases.
To investigate further the downregulation of KSHV gene expression in BCBL-1/anti-K9 cells, we performed Northern blot analysis to measure the level of mRNA expression of several KSHV genes. Consistent with the results from the immunoblot assay, expression of the 0.8-kb mRNA of IL-6 was dramatically decreased in BCBL-1/anti-K9 cells compared to parental BCBL-1 cells (Fig. 6B). Since the identities of KSHV p40 and p60 are unknown, we measured the level of mRNA expression of other KSHV genes. Expression of the 0.7-kb mRNA of the K8 gene, which contains a putative purine binding motif (28), and the 1.1-kb PAN/T1.1 RNA, which has been shown to be a lytic cycle gene (32, 38), was also reduced in BCBL-1/anti-K9 cells compared to BCBL-1 cells (Fig. 6B). In contrast, the expression levels of sVCA and orf73 were not altered in BCBL-1/anti-K9 cells compared to BCBL-1 cells (Fig. 6B and data not shown). In addition, Southern blot analysis with the vIL-6 and orf73 genes as probes showed that the reduction of viral gene expression in BCBL-/anti-K9 cells was not due to the altered level of KSHV genomic DNA (Fig. 6C). Furthermore, PCR analysis with purified cellular genomic DNA confirmed the presence of KSHV DNA in BCBL-1/anti-K9 cells (data not shown).
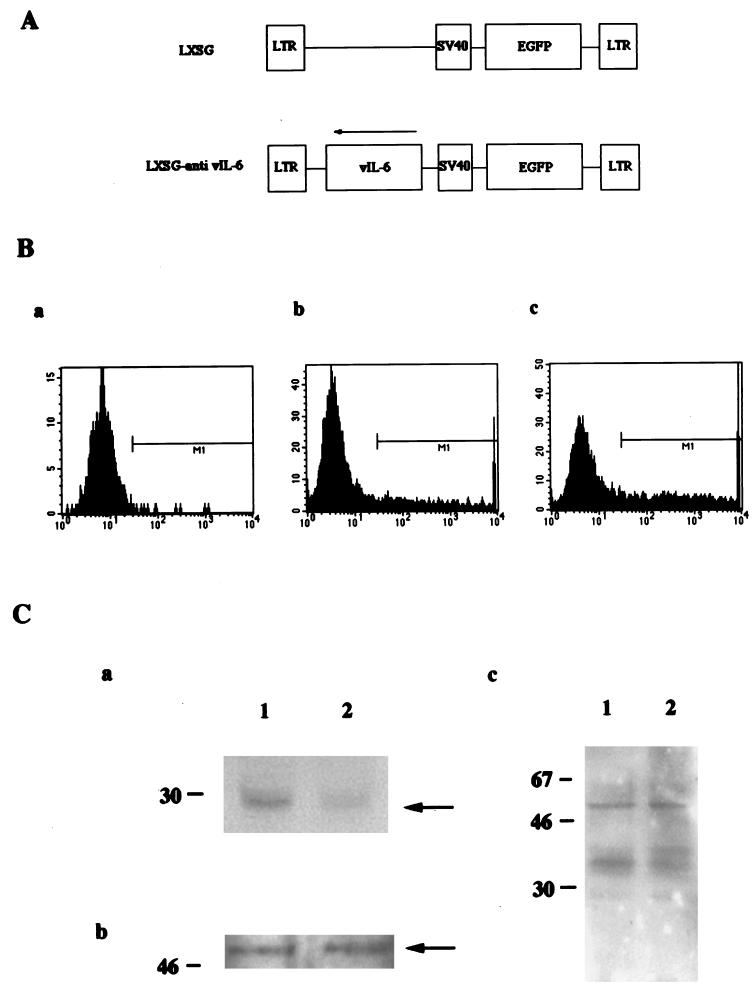
To examine the specific effect of the K9 antisense expression on KSHV expression, the vIL-6 gene was cloned into the retroviral vector LXSG in an antisense orientation. The retroviral vector LXSG, which contains the enhanced green fluorescence protein gene under the control of the simian virus 40 early promoter (Fig. 7A), has been used as a transient assay system for the expression of foreign genes (1). LXSG and LXSG-anti vIL-6 DNA were electroporated into BCBL-1 cells. FACS analysis showed that 15% of target BCBL-1 cells electroporated with LXSG, 35% of target BCBL-1 cells electroporated with LXSG-anti vIL-6, but less than 0.5% of BCBL-1 cells showed green fluorescence (Fig. 7B). Green fluorescent cells were sorted by FACS and lysed with lysis buffer. Equivalent amounts of proteins from sorted cells were used to examine the expression of vIL-6 by immunoblot analysis, which showed that the level of vIL-6 in BCBL-1/LXSG-anti-vIL-6 cells was lower than that in BCBL-1/LXSG cells (Fig. 7C). However, unlike BCBL-1/anti-K9 cells, similar levels of p40 and p60 polypeptides were detected in BCBL-1/LXSG-anti-vIL-6 cells compared to BCBL-1/LXSG cells (Fig. 7C). In addition, equivalent levels of K9 were detected in the two cell lines (Fig. 7C). This finding suggests that the reduction in the expression of several KSHV genes is likely due to the specific expression of the K9 antisense gene.
FIG. 7.
Reduced vIL-6 expression in BCBL-1 cells containing the vIL-6 antisense sequence. (A) Genetic organization of LXSG and LXSG-anti-vIL-6. LTR, long terminal repeat; SV40, simian virus 40. (B) FACS analysis of BCBL-1 cells electroporated with either LXSG or LXSG-anti-vIL-6. a, BCBL-1 (M1 < 0.5%); b, BCBL/LXSG (M1 = 15%); c, BCBL-1/LXSG-anti-vIL-6 (M1 = 35%). M1 indicates the gated cells for the green fluorescence. (C) Immunoblot assays. The same amounts of proteins from the sorted cells were used for immunoblot assay with anti-vIL-6 antibody (a), anti-K9 antibody (b), and human KS serum (c). Lane 1, BCBL-1/LXSG; lane 2, BCBL-1/LXSG-anti-vIL-6. Sizes are indicated in kilodaltons.
DISCUSSION
In this report, we have demonstrated that the K9 of KSHV encodes the first viral IRF (v-IRF). v-IRF functions as a repressor for cellular IFN-mediated signal transduction, it plays an important role in regulating the expression of several KSHV genes, and it has transforming activity in NIH 3T3 cells. Recently, we have shown that a recombinant HVS in which the STP oncogene of HVS was replaced with the K1 gene immortalized primary T lymphocytes to IL-2-independent growth and induced lymphoma in a common marmoset (17). KSHV also encodes a number of other gene products whose properties suggest a possible role in cell growth transformation. These include a G-protein-coupled receptor (2), a viral FLICE-inhibitory protein which can prevent apoptosis (34), and a virus-encoded cyclin (11, 18).
Transcriptional stimulation in response to IFNs is mediated by cellular IRFs (6, 7). These proteins are composed of a conserved DNA binding domain in the amino-terminal region and a divergent carboxyl-terminal region that serves as the regulatory domain (6, 7). The amino terminus of v-IRF is significantly homologous with the amino terminal DNA binding region of IRF, while the carboxyl terminus of v-IRF is divergent from the carboxyl transactivator/repressor region of IRF (20). KSHV v-IRF contains an additional 80 amino acids at the amino terminus which are not homologous with cellular IRFs. Interestingly, this region contains six repeats of a proline-rich P(X)2-3P motif, which matches well with the SH3 binding sequence (26, 37).
IRF-1 and IRF-2 exhibit opposite activities in IFN-mediated transcriptional activation, although they bind to the same ISRE sequence. IRF-1 activates the transcription of genes containing ISRE sites in their promoters, whereas IRF-2 represses the transcription of these genes (13). This finding suggests that IRF-2 antagonizes IRF-1 activity by competing with IRF-1 for binding to the promoter region. IRF-1 has also been suggested to function in a manner analogous to the tumor suppressor p53 in that it activates a set of genes whose products are required for the negative regulation of cell growth (12). Overexpression of IRF-2 induces transformation by antagonizing the cell growth-arresting activity of IRF-1 (12). By analogy to cellular IRF-2, the v-IRF of KSHV represses IFN-mediated transcriptional activation and induces cell growth transformation. This finding suggests that like IRF-2, v-IRF of KSHV may block IFN-mediated transcriptional activation by antagonizing IRF-1 activity.
ICSBP, which is expressed predominantly in macrophages and lymphocytes, represses transcriptional activation induced by type I and type II IFNs (8). ICSBP has been shown to form a complex with IRF-1 or IRF-2 in vivo and in vitro (8, 23). Multiple forms of interaction of ICSBP with the IRF family lead to transcriptional repression of IFN-regulated genes (3). Thus, identification of the cellular proteins which the v-IRF of KSHV interacts with will be important for understanding the mechanism of v-IRF action in repressing IFN-mediated signal transduction.
IFNs are a family of multifunctional cytokines that manifest antiviral activities. In fact, IFN is the first known cytokine. Although the detailed mechanism of the effects of IFNs against most viruses remains elusive, the steps of viral multiplication that are affected by IFNs have been identified for several families of viruses (35). For instance, IFNs have been shown to affect both the transactivation of immediate-early gene and the release of mature virions of herpes simplex virus (29, 31). While viruses are both inducers of IFN synthesis and the principal target of its action, it is not surprising that viruses have evolved a variety of mechanisms to counteract the inhibitory effects of IFN on viral replication (35). E1A of adenovirus inhibits IFN-induced signaling by downregulating the expression of STAT-1 and ISGF3γ (31), the terminal protein of hepatitis B virus also blocks the signaling by IFNs (9), EBNA-2 of Epstein-Barr virus inhibits IFN signaling by abolishing the induction of IFN-stimulated genes (15), and a major secreted protein (M-T7) of myxomavirus specifically binds IFN-γ and neutralizes its activity (35). Here, we describe an unique mechanism used by KSHV to antagonize IFN actions. Unlike other viruses, KSHV has developed an elaborate form of molecular mimicry by expressing a functional v-IRF gene which has significant homology with cellular IRFs. Thus, v-IRF may circumvent IFN-mediated functions by the host defenses by directly binding to cis-acting elements to repress the transcriptional activation or by sequestering cellular factors which are normally involved in IFN-regulated gene expression. We have found that v-IRF, in addition to having transcriptional repressor activity, plays an important role in KSHV gene expression. Further studies are required to understand the detailed role of v-IRF in the inhibition of IFN-mediated signal transduction and in the regulation of KSHV gene expression.
ACKNOWLEDGMENTS
We thank D. Ganem (BCBL-1 cells), P. Moore, Y. Chang (vIL-6 antibody), V. Garcia (orf26 antibody), G. Miller (sVCA antibody), and S. Reeves (pBP JTR-1 plasmid) for providing reagents. We especially thank L. Alexander and B. Means for critical reading of the manuscript. We also thank J. Newton for manuscript preparation.
This work was supported by Public Health Service grants CA31363 and RR00168.
ADDENDUM IN PROOF
After the manuscript was submitted, Gao et al. (S.-J. Gao, C. Boshoff, S. Jayachandra, R. A. Weiss, Y. Chang, and P. S. Moore, Oncogene 15:1979–1985, 1997) and Zimring et al. (J. C. Zimring, S. Goodbourn, and M. K. Offermann, J. Virol. 72:701–707, 1998) published similar results.
REFERENCES
- 1.Alexander L, Lee H, Rosenzweig M, Jung J U, Desrosiers R C. EGFP-containing vector system that facilitates stable and transient expression assays. BioTechniques. 1997;23:64–66. doi: 10.2144/97231bm12. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–350. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- 3.Bovolenta C, Driggers P H, Marks M S, Medin J A, Politis A D, Vogel S N, Levy D E, Sakaguchi K, Appella E, Coligan J E, Ozato K. Molecular interactions between interferon consensus sequence binding protein and members of the interferon regulatory factor family. Proc Natl Acad Sci USA. 1994;91:5046–5050. doi: 10.1073/pnas.91.11.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated Herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 6.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 7.David M. Transcription factors in interferon signaling. Pharmacol Ther. 1995;65:149–161. doi: 10.1016/0163-7258(94)00050-d. [DOI] [PubMed] [Google Scholar]
- 8.Driggers P H, Ennist D L, Gleason S L, Mak W, Marks M S, Levi B, Flanagan J R, Appella E, Ozato K. An interferon gamma-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc Natl Acad Sci USA. 1990;87:3743–3747. doi: 10.1073/pnas.87.10.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster G R, Ackrill A M, Goldin R D, Kerr I M, Thomas H C, Stark G R. Expression of the terminal protein region of hepatitis B virus inhibits cellular responses to interferons α and r and double-stranded RNA. Proc Natl Acad Sci USA. 1991;88:2888–2892. doi: 10.1073/pnas.88.7.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu X-Y, Schindler C, Improta T, Aebersold R, Darnell J E., Jr The proteins of ISGF-3, the interferon α-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci USA. 1992;89:7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godden-Kent D, Talbot S J, Boshoff C, Chang Y, Moore P, Weiss R A, Mittnacht S. The cyclin encoded by Kaposi’s sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada H, Kitagawa M, Tanaka N, Yamamoto H, Harada K, Ishihara M, Taniguchi T. Anti-oncogenic and oncogenic potentials of interferon regulatory factors-1 and -2. Science. 1993;259:971–974. doi: 10.1126/science.8438157. [DOI] [PubMed] [Google Scholar]
- 13.Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of the types 1 IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 14.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 15.Kanda K, Decker T, Aman P, Wahlstrom M, von Gabain A, Kallin B. The EBNA2-related resistance towards alpha interferon (IFN-α) in Burkitt’s lymphoma cells effects induction of IFN-induced genes but not the activation of transcription factor ISGF-3. Mol Cell Biol. 1992;12:4930–4936. doi: 10.1128/mcb.12.11.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanno Y, Kozak C A, Schindler C, Driggers P H, Ennist D L, Gleason S L, Darnell J E, Jr, Ozato K. The genomic structure of the murine ICSBP gene reveals the presence of the gamma interferon-responsive element, to which an ISGF3α subunit (or similar) molecule binds. Mol Cell Biol. 1993;13:3951–3963. doi: 10.1128/mcb.13.7.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H, Veazey R, Williams K, Li M, Guo J, Neipel F, Fleckenstein B, Lackner A A, Desrosiers R C, Jung J U. Deregulation of cell growth by the Kaposi’s sarcoma-associated herpesvirus K1 gene. Nat Med. 1998;4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- 18.Li M, Lee H, Yoon D-W, Albrecht J-C, Fleckenstein B, Neipel F, Jung J U. Kaposi’s sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi’s sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 21.Neipel F, Albrecht J-C, Ensser A, Huang Y-Q, Li J J, Friedman-Kien A E, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71:839–842. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity. J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson N, Marks M S, Driggers P H, Ozato K. Interferon consensus sequence-binding protein, a member of the interferon regulatory factor family, suppresses interferon-induced gene transcription. Mol Cell Biol. 1993;13:588–599. doi: 10.1128/mcb.13.1.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholas J, Ruvolo V R, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H-G, Hayward G S, Reitz M S. Kaposi’s sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 25.Paulus W, Baur I, Boyce F M, Breakefield X O, Reeves S A. Self-contained, tetracycline-regulated retroviral vector system for gene delivery to mammalian cells. J Virol. 1996;70:62–67. doi: 10.1128/jvi.70.1.62-67.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren R, Mayer B J, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1159–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 27.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Ganem D. Lytic growth of Kaposi’s sarcoma-associate herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 28.Russo J J, Bohenzxy R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi’s sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samuel C. Antiviral actions of interferon: interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 30.Sarid R, Sato T, Bohenzky R A, Russo J J, Chang Y. Kaposi’s sarcoma-associated herpesvirus encodes a functional Bcl-2 homologue. Nat Med. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 31.Sen G C, Ransohoff R M. Interferon-induced antiviral actions and their regulation. Adv Virus Res. 1992;42:57–102. doi: 10.1016/s0065-3527(08)60083-4. [DOI] [PubMed] [Google Scholar]
- 32.Sun R, Lin S-F, Gradoville L, Miller G. Polyadenylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takebe Y, Seiki M, Fujisawa J-I, Hoy P, Yokota K, Arai K-I, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J-L, Schröter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 35.Vilcek J, Sen G C. Interferons and other cytokines. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 375–399. [Google Scholar]
- 36.Wang I M, Blanco J C G, Tsai S Y, Tsai M-J, Ozato K. Interferon regulatory factors and TFIIB cooperatively regulate interferon-responsive promoter activity in vivo and in vitro. Mol Cell Biol. 1996;16:6313–6324. doi: 10.1128/mcb.16.11.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 38.Zhong W, Ganem D. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) J Virol. 1997;71:1207–1212. doi: 10.1128/jvi.71.2.1207-1212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]