Abstract
The Epstein-Barr virus (EBV) lytic switch gene, BZLF1, is tightly regulated in latently infected B cells. The BZLF1 gene promoter (Zp) contains several cis elements that have been previously shown to respond to inducers of the viral lytic cycle. These include four copies of an element referred to as the ZI domains and an element that contains a consensus CRE/AP-1 motif (ZII domain). In addition, Zp is autoregulated through two sites that bind the BZLF1 gene product Zta. The ZI domains have been shown to bind the ubiquitous cellular transcription factors Sp1 and Sp3 and/or the myocyte enhancer factor 2D (Liu et al., EMBO J. 16:143–153, 1997; Liu et al., Virology 228:9–16, 1997). Here we present a functional analysis of the ZII domain and show: (i) ATF-1 and ATF-2 appear to be the predominant cellular factors that bind to the CRE/AP-1 motif present in the ZII domain; and (ii) the region immediately upstream of the CRE/AP-1 motif contains a potent negative cis element, mutation of which results in a >10-fold increase in Zp activity. The negative cis element (ZIIR) in the ZII domain decreases both basal and induced Zp activity and thus is likely to play an important role in regulating reactivation of EBV. In addition, analysis of heterologous promoter constructs indicates that the function of ZIIR is context sensitive. Attempts to demonstrate a cellular factor binding to ZIIR have been unsuccessful, leaving unresolved the mechanism by which repression is mediated.
Epstein-Barr virus (EBV) is a lymphotropic human herpesvirus that latently infects B lymphocytes, resulting in a concomitant growth transformation of the infected cell. Infection is closely associated with several human cancers, including nasopharyngeal carcinoma and African Burkitt’s lymphoma and also plays a role in several lymphoproliferative diseases in immunocompromised individuals. In vitro, the transforming potential of EBV is evidenced by its ability to immortalize B lymphocytes to grow indefinitely in culture. Immortalization is achieved through the expression of a relatively small subset of EBV-encoded genes that serve to establish and maintain cellular transformation (for a review, see reference 28).
Propagation of EBV from host to host is dependent upon the activation of an estimated 100 or more viral genes, culminating in the production of infectious virions (28). While these genes remain quiescent during latency, a switch in the genetic program leading to the expression of viral replication-associated genes can be accomplished in vitro by treatment of latently infected B lymphocytes with various reagents, including phorbol esters, butyrate, Ca+2 ionophores, and anti-immunoglobulin (3, 14, 27, 35, 46, 50). Activation of the lytic cascade by cross-linking surface immunoglobulin or superinfection results initially in the expression of two viral genes, BZLF1 and BRLF1, which exhibit similar induction kinetics (maximal mRNA levels are reached between 2 and 4 h postinduction) (4, 17, 45). The BZLF1 gene product (referred to here as Zta but also called ZEBRA and EB1) has been shown to be a transcriptional activator (6, 8, 15, 20, 21, 32, 40, 47). Expression of Zta and Rta leads to the activation of early genes and ultimately to viral replication. Of all the viral transactivators examined, Zta is unique in that its expression alone can initiate the entire lytic cascade (9, 10, 25, 37), and regulation of Zta expression appears to be central to regulating entry into the lytic cycle.
Zta shares structural similarities with transcription factors of the basic leucine zipper family of proteins and is most closely related to proteins of the fos-jun extended family, and particularly Fos, with which it shares strong homology in the DNA binding domain (6, 15, 18, 22, 29, 31). Zta dimers bind to and activate transcription from AP-1 sites (15, 21, 47) as well as from specialized Z response elements present in the EBV lytic origins of DNA replication (32). In turn, Zta and AP-1-like sites present in the promoter region of BZLF1 play a critical role in the induction of Zta expression in response to anti-surface immunoglobulin antibodies, Ca2+ ionophores, or phorbol esters (7, 11, 19, 44, 47).
The BZLF1 promoter (Zp) exhibits very low basal activity which is potently upregulated by inducers of the viral lytic cycle (7, 11, 19, 44, 47). The region from bp −221 to +12 of Zp harbors the necessary cis elements for maintaining low basal activity and activation by lytic cycle-inducing agents (11, 19). Within this sequence, three distinct types of response elements have been defined (see Fig. 1). The first are A+T-rich sequences, termed ZI domains, four copies of which are interspersed in the promoter (ZIA-D). The second is represented by a unique element, ZII, which shares homology with consensus CRE/AP-1 binding sites (1, 12, 26, 30). The third is composed of two sites, termed ZIIIA and ZIIIB, which bind the BZLF1 gene product Zta (20). ZIIIA, but not ZIIIB, is an AP-1 response element (20). Induction of the BZLF1 gene appears to involve two steps: (i) initial activation of the promoter by inducers of the lytic cycle, mediated through the ZI and ZII domains, which results in low-level transcription of the BZLF1 gene; followed by (ii) autoactivation of the BZLF1 promoter, which is mediated through Zta binding to the ZIIIA and ZIIIB domains. It has previously been noted that Zta activation strongly synergizes with induction through the ZI and ZII domains (e.g., triggered by phorbol ester) (20). Thus, it has been proposed that the duration and magnitude of the initial signal may determine whether enough Zta is generated in an appropriate time interval to trigger the entire lytic cascade (20).
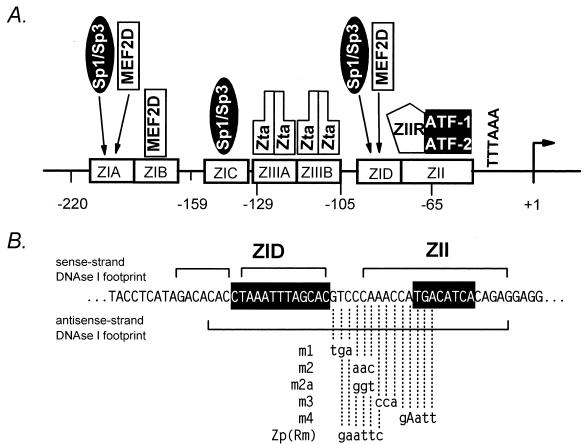
FIG. 1.
(A) Schematic illustration of the critical cis elements and known factors binding to these sites with the region from bp −221 to +14 of Zp. Binding of c-Jun and ATFa to the ZII CRE/AP-1 motif has also been reported (48). ZIIR, hypothetical cellular repressor binding to the negative cis element upstream of the CRE/AP-1 motif in the ZII domain (see text). (B) Nucleotide sequence of Zp in the region of the ZID and ZII domains. Shown are the regions protected from DNase I digestion with nuclear extract prepared from EBV-negative BL cell lines (19). Also summarized are the mutations that were introduced into the ZIIR element, with their designated names indicated to the left (see text).
The ZI domains in Zp have recently been shown to bind the ubiquitous cellular transcription factors Sp1 and Sp3 (5, 33) and/or the myocyte enhancer factor 2D (MEF2D) (5, 34). The ZIA and ZID domains can bind Sp1, Sp3, or MEF2D, while the ZIB domain binds only MEF2D and the ZIC domain binds only Sp1 and Sp3 (Fig. 1). Notably, we have previously shown that induction of the EBV lytic cycle by cross-linking surface immunoglobulin is inhibited by the immunosuppressants cyclosporin A and FK506 (23), and more recently we have shown that this sensitivity is dependent on the presence of the Zp MEF2 sites (ZIA, ZIB, or ZID) in conjunction with the ZII domain (34). In this paper, we dissect the ZII domain and identify two distinct cis elements within this region, the previously noted CREB/AP-1 motif (16, 20, 42, 48) and a previously unrecognized negative cis element. The latter cis element represses both basal and activated transcription from Zp and is likely to play an important role in regulating reactivation of EBV.
MATERIALS AND METHODS
Cell culture and transfections.
The EBV-negative Burkitt’s lymphoma B cell line DG75 was grown at 37°C in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, 100 U of penicillin, and 100 mg of streptomycin per ml. DG75 cells were transfected by DEAE-dextran and dimethyl sulfoxide (DMSO) shock, as previously described (5). Briefly, 107 cells per transfection were washed once with phosphate-buffered saline (PBS), and the cell pellet was resuspended in 0.5 ml of RPMI 1640 medium without serum. Cells were added to 0.5 ml of RPMI 1640 medium with DEAE-dextran (1 mg/ml) and 2 μg of plasmid DNA. After incubation at room temperature for 30 min, cells were subjected to DMSO shock (the addition of 0.5 ml of 20% DMSO for 2 min). Following washing with PBS, cells were resuspended in 10 ml of RPMI 1640 with 10% serum and cultured at 37°C in a 5% CO2 incubator. For induction by tetradecanoyl phorbol acetate (TPA), ionomycin, or both, the final concentrations of reagents were 20 ng/ml (TPA) and 1 μM (ionomycin). Overexpression of the catalytic subunit of protein kinase A (PKA) was achieved employing a Rous sarcoma virus long terminal repeat-driven expression vector, which has previously been described (36).
CAT and luciferase reporter gene assays.
Transfected cells were harvested 48 to 72 h posttransfection and washed once in PBS. For the chloramphenicol acetyltransferase (CAT) assay, cells were suspended in 100 μl of 0.25 M Tris-HCl (pH 7.5) and lysed by freeze-thawing three times. The cell lysate was collected after centrifugation, and the reporter gene assay was carried out as previously described (5, 24). The acetylated species of chloramphenicol were quantitated with a PhosphorImager (Molecular Dynamics). For assessing luciferase activity, cells were resuspended in cell lysis buffer, and the assay was performed according to the manufacturer’s protocol (Promega).
Construction of reporter constructs.
The −221ZpCAT reporter construct contains the BZLF1 promoter sequences from bp −221 to +12 and was constructed as described previously (5, 19). Artificial promoter constructs, 3×ZIB-ZII-βGCAT and 3×ZIC-ZII-βGCAT, were generated by cloning the indicated Zp elements upstream of the β-globin TATA box driving expression of the CAT reporter gene in the modified pGL2 vector (5). The 3×ZIB element was cloned into the AvaI-XbaI restriction sites, and the ZII element was cloned into the XbaI-BamHI sites. The sequences of the Zp elements were as follows: 3×ZIB, 5′-CCGGGCACCAGCTTATTTTAGACACTTCCACCAGCTTATTTTAGA CACTTCCACCAGCTTATTTTAGACACTTCT-3′; 3×ZIC, 5′-CCGGGCTCCTCCTCTTTTAGAAACTACTCCTCCTCTTTTAGAAACTACTCCTCCT CTTTTAGAAACTAT-3′; and 1×ZII, 5′-CATAGACGTCCCAAACCATGACATCACAGAGGAG-3′. Some of the artificial promoter constructs were also cloned into the pGL-2 luciferase reporter plasmid (Promega).
Mutations introduced into the ZII domain are summarized in Fig. 1, 2, and 6. Other mutations which are not shown in the figures were: 1×ZIIm2cm3, 5′-CATAGACGTCaacAACCATGgtATCACAGAGGAG-3′; and 3×ZIBm3, 5′-CCGGGCACCAGgggATTTTAGACACTTCCACCAGgggATTTTAGACACTTCCACCAGgggATTTTAGACACTTCT-3′ (lowercase letters indicate mutated residues). The pGL-2 luciferase vector containing the simian virus 40 (SV40) early promoter and the pGL-2 control vector containing the SV40 promoter and enhancer were purchased from Promega. The thymidine kinase promoter-luciferase plasmid was generously provided by David Leib (Washington University School of Medicine). The 3×ZIB-ZII with the spacer +4 was generated by cutting the construct with XbaI, filling in the 5′ overhangs with Klenow and dNTPs, and religating the construct. The 3×ZIB-ZII spacers +10 and +15 were generated by introducing the following linkers into the XbaI site: spacer +10, 5′-CTAGCGTTAC-3′; and spacer +15, 5′-CTAGCGTTACGACTC-3′. The structures of all the mutant and artificial promoters were confirmed by DNA sequencing.
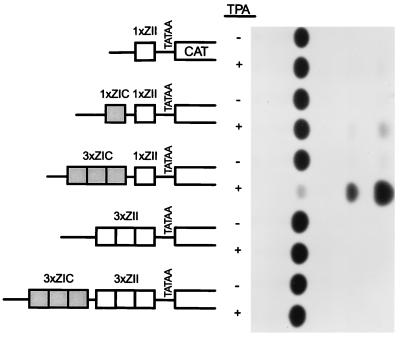
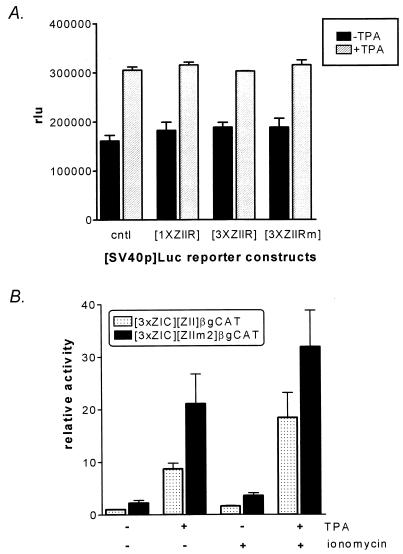
FIG. 2.
Activities of a series of artificial promoter constructs containing the ZIC and ZII domains, cloned upstream of the rabbit β-globin TATA box driving transcription of the CAT gene (5). A representative CAT assay is shown alongside the schematic illustrations of the artificial promoter constructs. The activities of the 1×ZII-βgCAT, 1×ZIC-1×ZII-βgCAT, and 3×ZIC-1×ZII-βgCAT reporter constructs have been previously published (5) and are shown here for comparison with the activities of the reporter constructs containing multiple copies of the ZII domain. The reporter constructs were transiently transfected into the EBV-negative BL cell line DG75, as described in Materials and Methods. As indicated, the activities of each construct were assayed in the absence and presence of TPA (final concentration, 10 ng/ml).
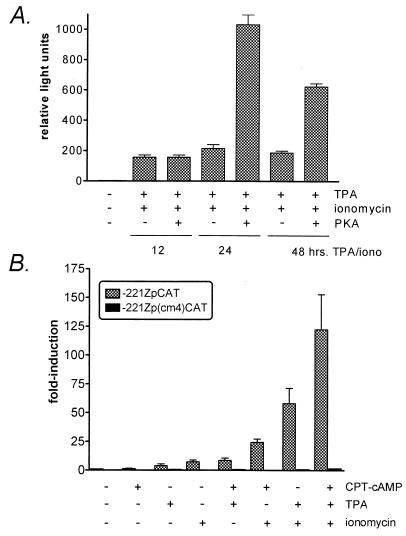
FIG. 6.
Analysis of cAMP inducibility of reporter constructs driven by Zp or the Zp(cm4) mutant. (A) DG75 cells were transiently transfected with the −221ZpLuciferase reporter construct along with a vector driving expression of the catalytic subunit of protein kinase A, as described in Materials and Methods. Transfected cells were treated for the indicated times with TPA (10 ng/ml) and ionomycin (1 μM), followed by assaying firefly luciferase activity on a Tuner Designs luminometer (Promega). The activities observed under each condition are given in relative light units. The data were compiled from six independent experiments and the standard errors of the means are shown. (B) DG75 cells were transiently transfected with either the −221ZpCAT or the equivalent reporter construct containing a mutation in the CRE/AP-1 motif [−221Zp(cm4)CAT] as described in Materials and Methods. Transfected cells were either not treated or treated for 48 h with the indicated reagents. The cells were harvested 48 h posttransfection and assayed for CAT activity. Fold induction relative to the activity of the uninduced −221ZpCAT reporter construct is shown. CPT-cAMP was used at a final concentration of 0.3 mM. TPA was added to a final concentration of 10 ng/ml, and ionomycin was added to a final concentration of 1 μM. The data are compiled from three independent experiments and standard errors of the means are shown.
EMSA.
DG75 nuclear extract was prepared as described previously (2, 13), and aliquots were stored at −70°C. Double-stranded oligonucleotide probes for electrophoretic mobility shift assays (EMSA) were labeled with T4 polynucleotide kinase and [γ-32P]ATP. EMSA reactions were performed in a total volume of 25 μl [20 mM Tris-HCl (pH 7.9), 10% glycerol, 0.1 M KCl, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, 1 μg of poly(dI-dC)(dI-dC), 5 μg of crude DG75 nuclear extract]. After incubation for 5 min at room temperature, 32P-labeled probe (25,000 cpm) was added, and the incubation was continued for an additional 25 min. The reaction mixtures were loaded onto a 4% nondenaturing polyacrylamide gel and electrophoresed in 0.5× Tris-borate-EDTA (1× Tris-borate-EDTA is 90 mM Tris, 64.6 mM boric acid, and 2.5 mM EDTA [pH 8.3]). Competition assays were carried out with unlabeled double-stranded DNA oligonucleotides that were added prior to addition of probe. Supershift experiments were performed with 1 μg of antibody against ATF-1, ATF-2, CREB-binding protein, c-Jun, or c-Fos (Santa Cruz Biotechnology, Inc.). The sense strand sequences of the oligonucleotides used in the EMSA were ZII, 5′-ACGTCCCAAACCATGACATCACAGAGGAG-3′ and ZIIcm3, 5′-ACGTCCCAAACCATGgtATCACAGAGGAG-3′.
DNase I footprinting.
DNase I footprinting was performed basically as previously described (19). Eighty micrograms of nuclear extract was incubated with 32P-labeled DNA in 10 mM Tris (pH 7.9), 0.5 mM EDTA, 0.5 mM dithiothreitol, 5% glycerol, and 2% polyvinylethanol with 1 μg of poly(dI-dC)(dI-dC) in a total volume of 50 μl. After incubation for 20 min at room temperature, DNase I was added, and the reaction was allowed to incubate for 30 s. Digestion was terminated by the addition of 120 μl of stop buffer (8 M urea, 0.5% sodium dodecyl sulfate, 5 mM EDTA), and the samples were subsequently extracted twice with phenol, twice with phenol-chloroform (1:1), and once with chloroform. The cleaved DNA was recovered by precipitation with ethanol, and the samples were separated on 8% denaturing (7 M urea) acrylamide gels as previously described (2).
RESULTS
The CRE/AP-1 site of the ZII domain is a positive cis element that is required for inducibility of Zp.
Previous limited mutagenesis of Zp provided evidence that the CRE/AP-1 motif in the ZII domain is essential for induction of Zp by known lytic cycle inducers (11, 19). We have recently reported that the activity of multimerized ZI domains (one to three copies) cloned into an artificial promoter construct containing the β-globin TATA box driving expression of the CAT reporter gene is greatly augmented by the presence of a single copy (1×) of the ZII domain (Fig. 2) (5, 7, 33, 34). However, when the ZII domain was multimerized (alone or in conjunction with three copies (3×) of the ZIC domain) and cloned upstream of a minimal promoter (β-globin TATA box) driving CAT gene expression (3×ZII-βGCAT or 3×ZIC-3×ZII-βGCAT), these reporter constructs failed to exhibit any detectable basal or phorbol ester-inducible activity. This observation suggested that the ZII domain may be composed of multiple cis elements, which can lead to either activation or repression of transcription.
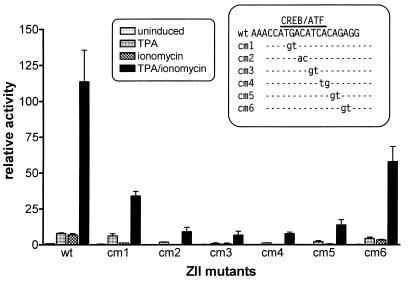
To further functionally dissect the ZII domain, we constructed a series of 2-bp mutations that span the CRE/AP-1 motif (see inset to Fig. 3). These mutations were then incorporated into an artificial promoter construct containing three copies of the ZIB domain linked to a single copy of the wild type (wt) or mutant ZII domain cloned upstream of the minimal β-globin promoter in the pGL-2 luciferase reporter plasmid (Promega). Notably, we have previously shown that the behavior of the 3×ZIB-1×ZII-βGCAT reporter construct closely mimics the behavior of the intact BZLF1 promoter (34). TPA and ionomycin inducibility of these reporter constructs was assessed in the EBV-negative Burkitt’s lymphoma cell line DG75 (Fig. 3). This analysis demonstrated that mutations that disrupted the core CRE/AP-1 motif severely attenuated TPA-and-ionomycin-inducible activity.
FIG. 3.
Impact of mutations in the CRE/AP-1 motif within the ZII domain on TPA and/or ionomycin inducibility of the 3×ZIB-1×ZII-βgLuciferase reporter construct. The artificial promoter construct, 3×ZIB-1×ZII-βgLuciferase, was generated as described in Materials and Methods. The mutations introduced into the ZII domain are shown in the inset. The EBV-negative BL cell line DG75 was transiently transfected, and cell lysates were collected 48 h posttransfection, as described in Materials and Methods. The final concentrations of TPA and ionomycin were 10 ng/ml and 1 μM, respectively. The fold inductions were calculated based on the activities of the uninduced wt construct. The data shown represented the results from two independent experiments, and the standard errors of the means are indicated.
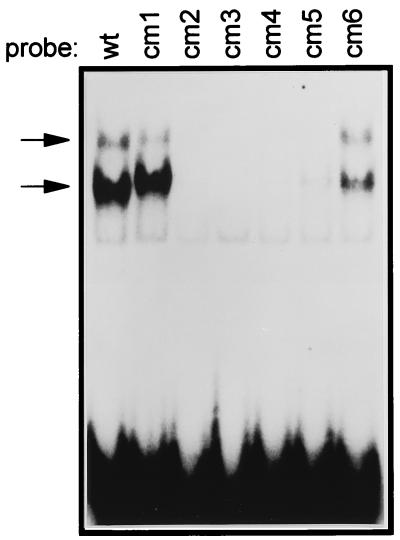
To extend this analysis, protein binding to the ZII domain was assessed by an EMSA employing crude nuclear extract prepared from the DG75 cell line (Fig. 4). Two major protein-DNA complexes were observed by EMSA with the wt ZII domain probe. These complexes were specific since they could be competed by the wt ZII domain probe but not by a ZII domain probe containing a mutation in the core of the CRE/AP-1 motif (ZIIcm4 mutant probe) (see Fig. 5A). Importantly, there was a direct correlation between the formation of the two major complexes observed by EMSA (Fig. 4) and the activities of the ZII mutants (Fig. 3). Thus, it appears that a functional CRE/AP-1 motif is critical for the activity of this artificial promoter construct.
FIG. 4.
EMSA with 32P-labeled probes containing either the wt ZII domain, or the indicated mutation in the ZII CRE/AP-1 motif. Crude nuclear extract from the DG75 cell line was used, as described in Materials and Methods. The nucleotide sequences of the ZII domain mutants are shown in the inset in Fig. 3. EMSA binding reaction conditions were as described in Materials and Methods. The specific complexes formed are indicated by arrows to the left of the gel.
FIG. 5.
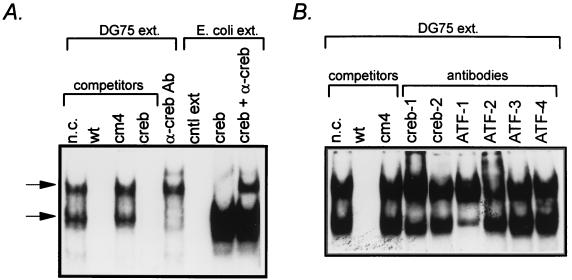
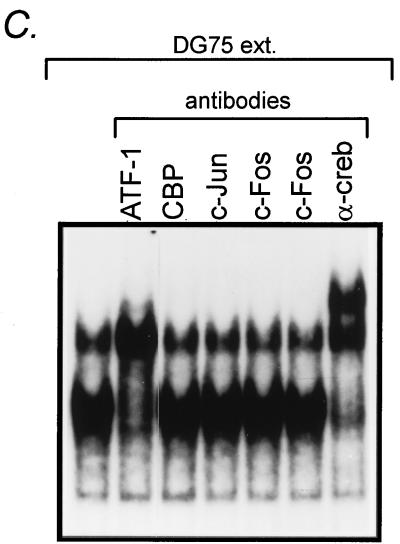
Analysis of cellular factors binding to the ZII domain. (A) Competition for binding to the ZII DNA probe with wt and cm4 mutant ZII domain oligonucleotides, as well as an oligonucleotide containing a consensus CREB binding site (100 ng of each cold oligonucleotide competitor was added to the EMSA binding reaction). In addition, the ability of a pan-CREB antibody (see text) to supershift the observed complexes is also shown, as well as binding or recombinant CREB protein. Conditions for the EMSA, antibody supershifts, and binding of recombinant CREB protein are described in Materials and Methods. (B and C) Antibody supershifts of specific complexes employing antibodies against specific CREB/ATF or AP-1 family members. For each supershift assay, 1 μg of antibody was added to the EMSA reaction as described in Materials and Methods. All antibodies were purchased from Santa Cruz Biotechnology, Inc.
ATF-1 and ATF-2 bind to the CRE/AP-1 site in the ZII domain.
To address what cellular factors are bound to the CRE/AP-1 motif in the ZII domain, we demonstrated that an oligonucleotide containing a consensus CRE site could compete for the formation of the complexes observed by EMSA with the wt ZII domain probe (Fig. 5A). In addition, when a pan-CREB rabbit polyclonal antibody (antiserum 244; raised against a peptide containing residues 128 to 162 of the human CREB-1 protein, a region that is well conserved in a number of CREB/ATF family members [49]), was incubated with the EMSA reaction, the lower complex disappeared and a faint supershift above the slower migrating complex was detectable (Fig. 5A). In other assays, this antibody resulted in a stronger supershift, as shown in Fig. 5C (α-creb). Incubation of the ZII domain probe with recombinant CREB-1 protein revealed the formation of a complex that migrated similarly to the lower complex observed by EMSA with DG75 nuclear extract (Fig. 5A). Addition of the pan-CREB rabbit polyclonal antibody to the EMSA reaction containing the recombinant CREB-1 protein resulted in a supershift that migrated at a position similar to that of the upper complex observed by EMSA with DG75 nuclear extract. Thus, it is possible that with DG75 nuclear extract, addition of the rabbit polyclonal antibody to the EMSA reaction results in a supershifted lower complex that comigrated with the upper complex, while a supershift of some or all of the upper complex may account for the appearance of the slower migrating supershifted complex.
To further address the question of what cellular factors bind to the CRE/AP-1 motif, we employed antibodies directed against specific CREB and AP-1 family members (Fig. 5B and C). Anti-ATF-1 antibody resulted in a strong supershift of the lower complex which, based on the increased band intensity (most apparent in Fig. 5C), appeared to comigrate with the upper complex. The anti-ATF-2 antibody appeared to supershift a portion of the top complex (Fig. 5B). In addition, a weak supershift of the lower complex by anti-CREB-1 antibody was observed by EMSA in some cases (Fig. 5B). However, antibodies directed against CREB-2, ATF-3, ATF-4, CREB-binding protein, c-Jun, or c-Fos did not appear to shift either the lower or upper complexes (Fig. 5B and C). Thus, it appears that ATF-1 and ATF-2 are the major cellular factors in DG75 nuclear extract that bind the ZII domain. Whether these factors bind as homodimers or are present as heterodimers with other, as yet unidentified CREB/ATF or AP-1 family members is unclear at this time.
The identification of CREB/ATF family members binding to the ZII domain raised the question of whether Zp is inducible by cAMP (39, 41). We initially assessed whether overexpression of the catalytic subunit of PKA could augment induction of Zp by TPA and ionomycin (Fig. 6A). While expression of the catalytic subunit of PKA had no effect on the level of activation of Zp after 12 h of TPA and ionomycin treatment, a fivefold enhancement was observed after 24 h of TPA and ionomycin induction (Fig. 6A). The augmentation of TPA and ionomycin induction of Zp was slightly lower (ca. threefold) after exposure of the cells to TPA and ionomycin for 48 h (Fig. 6A). In addition, we assessed cAMP induction of Zp employing the cAMP analog 8-chlorophenylthio-cAMP (CPT-cAMP). Addition of CPT-cAMP to DG75 cells transiently transfected with either a wt Zp (−221ZpCAT) reporter construct or the same reporter construct containing a mutation in the CRE/AP-1 motif −221Zp(cm4)CAT did not result in detectable induction of Zp activity (Fig. 6B). However, CPT-cAMP was able to augment induction of Zp by either TPA, ionomycin, or a combination of TPA and ionomycin (Fig. 6B). Furthermore, mutation of the CRE/AP-1 motif nearly completely abrogated induction of Zp by any combination of these reagents, underscoring the pivotal role this cis element plays in transcription initiation from Zp. These results indicate that cAMP alone is insufficient to activate transcription from Zp but can augment TPA and ionomycin induction.
Mutations upstream of the ZII domain CRE/AP-1 motif reveal a potent negative cis element.
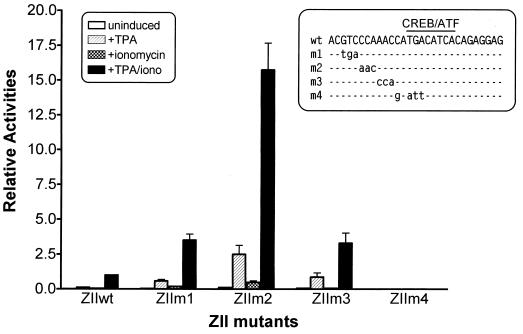
To identify other cis elements that might be present within the ZII domain, we introduced a series of 3-bp mutations (m1 through m3; see Fig. 7) upstream of the CRE/AP-1 motif. As described above, these mutations were cloned into the artificial promoter construct composed of three copies of the ZIB domain linked to a single copy of either the wt or mutated ZII domain upstream of the minimal β-globin TATA box. Surprisingly, all three mutations introduced upstream of the CRE/AP-1 motif strongly enhanced Zp inducibility with the m2 mutation having the largest impact (>15-fold enhancement of TPA-plus-ionomycin-induced activity; Fig. 7). To ensure that this was not the result of unwittingly generating a binding site for a transcriptional activator, a distinct mutation (m2a; Fig. 1B) was introduced into the ZII domain. The activity of the m2a mutant was nearly indistinguishable from that of the m2 mutant (data not shown). The m4 mutation, which disrupts the CRE/AP-1 motif (this mutation is the same as the previously reported MII mutation [19]), nearly completely abrogated inducibility of the artificial promoter (Fig. 7).
FIG. 7.
Impact of mutations in the region upstream of the CRE/AP-1 motif in the ZII domain on the induction by TPA and/or ionomycin. The mutations shown in the inset were introduced into the ZII domain in the context of the 3×ZIB-1×ZII-βgCAT reporter construct. The transfections were performed in DG75 cells, and the cell lysates were collected 48 to 72 h posttransfection for CAT assays. Their relative inductions of mutant 3×ZIB-ZII-CAT constructs are presented relative to the TPA-ionomycin-induced activity of the wt 3×ZIB-ZII. The data presented were compiled from four independent transfection and CAT assays, and standard errors of the means are shown.
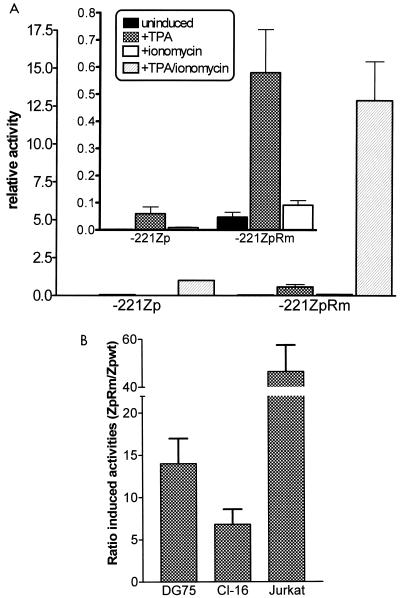
To determine whether the enhanced activity of the 3×ZIB-ZII-βgCAT reporter construct observed upon mutating the region upstream of the CRE/AP-1 was also observed in the context of the intact BZLF1 promoter, a 5-bp mutation (which introduced a diagnostic EcoRI site) was engineered into this region of Zp [Zp(Rm); Fig. 1B] in the context of the −221ZpCAT reporter construct. As observed with the 3×ZIB-ZIIm2-βgCAT reporter construct, introduction of a mutation in this region of Zp also dramatically increased promoter activity (>10-fold increase in TPA-plus-ionomycin-induced activity; see Fig. 8A). As shown in the inset of Fig. 8A, mutation of this negative cis element resulted in an increase in basal as well as TPA-, ionomycin-, and TPA-plus-ionomycin-induced Zp activities. The latter indicates that this cis element may play an important function in repressing Zp activity during latency. Based on the inhibition of Zp activity, we have named this cis element ZIIR for the ZII domain repressor.
FIG. 8.
Mutation of the ZIIR element in the context of the −221ZpCAT reporter construct. The mutation introduced into the ZIIR element (ZIIRm) is shown in Fig. 1. (A) The wt (−221ZpCAT) and mutant reporter construct (−221ZpRmCAT) were transiently transfected into the DG75 cell line, and CAT activity was assessed as described in Materials and Methods. Transfected cells were cultured in the presence or absence of TPA (20 ng/ml) and/or ionomycin (1 μM), as indicated. Activity is given relative to the TPA-plus-ionomycin-induced activity of the wt −221ZpCAT reporter construct. The inset shows the impact of the ZIIRm mutation on the basal, TPA-, and ionomycin-induced activities. The average induction by TPA plus ionomycin of the −221ZpCAT construct relative to the uninduced activity was 444-fold, while the average induction of the −221ZpRmCAT construct relative to the uninduced activity of the −221ZpCAT construct was 5,722-fold. The data presented represent four independent experiments, and the standard errors of the means are shown. (B) The activities of the wt (−221ZpCAT) and mutant (−221ZpRmCAT) reporter constructs were assessed in the EBV-positive BL cell line clone 16 (Cl-16) and the EBV-negative T-cell line Jurkat. Transfected cells were cultured in the presence of TPA (20 ng/ml) and ionomycin (1 μM). The ratios of the TPA-plus-ionomycin-induced activities of the mutant and wt reporter constructs are indicated. The data represent at least two independent experiments, and the standard errors of the means are shown.
To eliminate the possibility that the observed repression of Zp activity by the ZIIR cis element is unique to the EBV-negative DG75 BL cell line, we examined the activity of the −221ZpRm reporter construct in two other cell lines (the EBV-positive BL cell line clone 16 and the EBV-negative T cell line Jurkat). As shown in Fig. 8B, the −221ZpRm reporter construct was significantly more active than the unmutated reporter construct in both the clone 16 cell line (∼7-fold greater TPA-plus-ionomycin-induced activity) and the Jurkat cell line (∼45-fold greater TPA-plus-ionomycin-induced activity). Thus, the repressive function of the ZIIR cis element does not appear to be restricted to a specific cell type.
Repression of promoter activity by the ZIIR cis element is context sensitive.
To determine whether ZIIR can inhibit the activity of heterologous promoters, one and three copies of this cis element were cloned upstream of either the SV40 early promoter (Fig. 9A) or the herpes simplex virus thymidine kinase promoter (data not shown). In both cases, no repression of promoter activity was observed in the presence or absence of phorbol ester. As a negative control, three copies of the mutated ZIIR element (ZIIRm) were also cloned upstream of these heterologous promoters, and again no impact on promoter activity was observed (Fig. 9A and data not shown). Taken together, these data indicated that the position of ZIIR within a promoter may be critical for its function.
FIG. 9.
(A) Inability of the ZIIR element to repress transcription from the SV40 early promoter. One or three copies of the ZIIR element were cloned upstream of the enhancerless SV40 promoter as described in Materials and Methods. In addition, as a control, three copies of the mutated ZIIR element (ZIIRm; see Fig. 1) were also cloned upstream of the SV40 promoter. These reporter constructs were transiently transfected into the DG75 cell line, and uninduced and TPA induced activities were determined. The activities are presented as relative light units (rlu). The data represent the average of two independent experiments, and the standard errors of the means are shown. (B) Impact of mutating the ZIIR element in the context of the 3×ZIC-ZII-βgCAT reporter construct. The 3×ZIC-ZII-βgCAT reporter construct has been previously described (5). The data shown represent three independent assays, and the standard errors of the means are shown. The fold inductions were derived relative to the activity of the uninduced reporter construct.
To address the issue of whether the specific cellular factors bound upstream of ZIIR might affect the level of repression or activation observed, three copies of the ZIC domain from Zp were substituted for the three copies of the ZIB domain in the 3×ZI-ZII-βgCAT and 3×ZI-ZIIm2-βgCAT reporter constructs. We have previously reported that the ZIC element binds Sp1 and Sp3 but not MEF2D, while the ZIB element binds MEF2D but not Sp1 or Sp3 (summarized in Fig. 1A) (5, 33, 34). Mutation of ZIIR only resulted in a modest enhancement of promoter activity (<2-fold; Fig. 9B) when the activities of the 3×ZIC-ZII-βgCAT and 3×ZIC-ZIIm2-βgCAT reporter constructs were compared. Based on these results, it appears that the repression exerted by ZIIR is context sensitive.
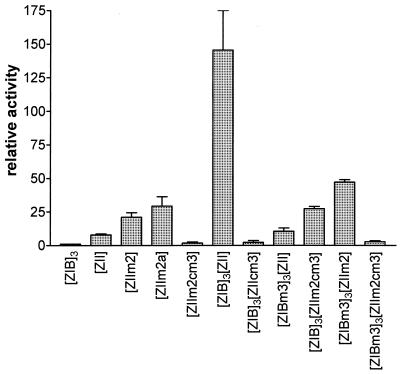
It is possible that ZIIR may be involved in modulating the synergy between MEF2D bound to the ZIB element and the factors bound to the ZII CRE/AP-1 motif. To address the issue of whether both a functional CRE/AP-1 motif and a functional MEF2D domain are required for ZIIR function, several artificial promoter constructs were generated containing various combinations of wt and mutated ZIB and ZII domains. These were transiently transfected into the DG75 cell line, and uninduced and TPA-plus-ionomycin-induced activities were examined (Fig. 10). Mutation of the ZIIR element in the context of an artificial promoter containing only a single copy of the ZII domain upstream of the β-globin TATA box (ZIIm2 and ZIIm2a; see Fig. 1B for mutations) resulted in a modest enhancement in activity relative to the unmutated ZII domain (Fig. 10). Mutation of the CRE/AP-1 motif in the context of the ZIIR mutation (ZIIm2cm3) abrogated the observed inducibility, demonstrating the requirement for a functional CRE/AP-1 motif (Fig. 10). Mutation of either the CRE/AP-1 motif or the MEF2D binding sites in the 3×ZIB-ZII-βgCAT reporter construct severely diminished inducible activity (see [ZIBm3]3[ZII] and [ZIB]3[ZIIcm3] in Fig. 10). Mutating ZIIR in the context of the artificial promoters containing either mutated MEF2D sites or a mutated CRE/AP-1 site resulted in diminished activation (3×ZIB-ZIIm2cm3 and 3×ZIBm3-ZIIm2; Fig. 10). Finally, mutation of ZIIR in the context of the artificial promoter in which the MEF2D sites and the CRE/AP-1 motif have been mutated resulted in little or no detectable activation (3×ZIBm3-ZIIm2cm3; Fig. 10). Thus, these results are consistent with the hypothesis that ZIIR functions to modulate the synergy between MEF2D and the factor(s) bound to the ZII CRE/AP-1 motif.
FIG. 10.
Derepression observed by mutating the ZIIR element requires a functional CRE/AP-1 motif and/or functional MEF2D binding sites. Artificial promoter constructs containing various combinations of wt and mutant ZIB and ZII domains were transfected into the DG75 cell line, followed by induction with TPA (10 ng/ml) and ionomycin (1.0 μM). The activities of the reporter constructs are shown relative to the activity of the 3×ZIB-βgCAT reporter construct, which was defined as 1.0. The activity of the 3×ZIB-1×ZIIm2-βgCAT reporter construct is not shown (relative activity, >2,000). The data presented were compiled from two independent experiments, and the standard errors of the means are shown.
The relative positions of the MEF2D and CRE/AP-1 elements on the DNA helix affect TPA and ionomycin inducibility and are independent of the ZIIR domain.
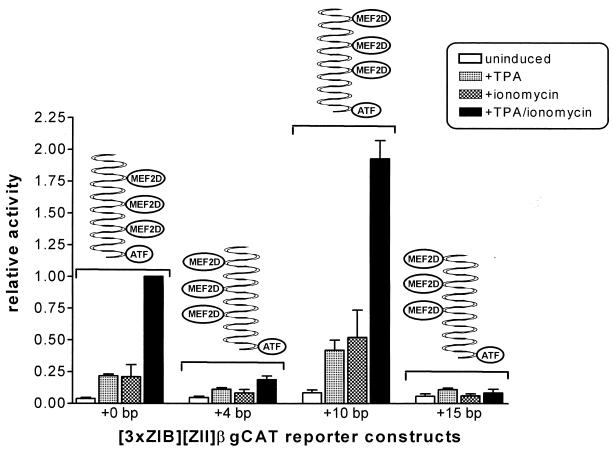
To determine whether the distance and orientation of the ZIB elements upstream of the ZII domain have an impact on activity and ZIIR function, several modified 3×ZIB-ZII-βgCAT and 3×ZIB-ZIIm2-βgCAT reporter constructs were generated. The distance between the ZII domain and the 3×ZIB cassette was increased by either 4, 10, or 15 bp. For both the 3×ZIB-ZII-βgCAT (Fig. 11) and the 3×ZIB-ZIIm2-βgCAT (data not shown) increasing the distance between the ZIB domains and the ZII domain by either 4 or 15 bp resulted in a dramatic reduction in the TPA, ionomycin, and TPA plus ionomycin inducibility. However, increasing the distance between these domains by 10 bp resulted in a slight enhancement in the inducibility of these constructs. This indicates that the phasing of the MEF2D and CRE/AP-1 cis elements on the DNA helix has a profound impact on the synergy observed. As depicted in Fig. 11, the original artificial promoter constructs were designed such that the cis elements were oriented on the same side of the helix (5).
FIG. 11.
Effect of alterations in the phasing of the MEF2D sites relative to the CRE/AP-1 motif. The 3×ZIB-ZII-βgCAT reporter construct and derivatives containing a spacer between the ZII domain and the ZIB domains of the indicated length were transiently transfected into the DG75 cell line as described in Materials and Methods. Transfected cells were either untreated or induced with TPA (10 ng/ml) and/or ionomycin (1.0 μM). The activities were calculated relative to the TPA-plus-ionomycin-induced activity of the parent reporter construct (no spacer), which was defined as 1.0. The data represent the average of four independent experiments, and standard errors of the means are shown.
Characterization of cellular protein binding to the ZII domain fails to identify a specific complex formed with the ZIIR element.
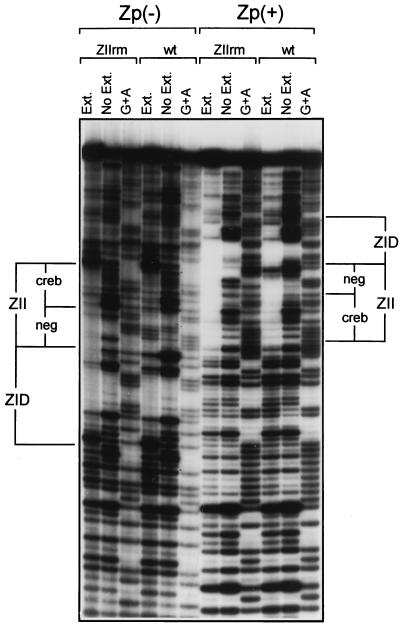
Based on the behavior of the ZIIR element, it seems likely that it functions through binding a cellular repressor. However, repeated attempts by EMSA have failed to identify a specific protein-DNA complex that is dependent on the ZIIR element. It should be noted that our original analysis of protein binding to Zp, by using DNase I footprinting with crude B-cell nuclear extracts, demonstrated at least partial protection of the ZIIR element (summarized in Fig. 1B) (19). Thus, to assess the possibility that the DNase I footprinting analyses detected binding of a cellular factor which cannot be readily identified by EMSA, DNase I footprinting employing both wt and mutant template and DG75 nuclear extract was carried out (Fig. 12). Surprisingly, mutation of ZIIR (ZIIrm) did not abrogate the protection of this region observed with DG75 nuclear extract (Fig. 12). This result suggests that the binding of MEF2D to the ZID domain and of CREB/ATF factors to the ZII domain is sufficient to prevent DNase I cleavage within the ZIIR element. Thus, neither EMSA nor DNase I footprinting analyses was able to provide direct evidence of cellular factor binding to ZIIR, leaving unresolved the mechanism by which this cis element functions.
FIG. 12.
DNase I protection analysis of the wt −221Zp and the ZIIRm mutant (−221ZpRm) employing crude nuclear extract prepared from the DG75 cell. Protection of both the sense strand [Zp(+)] and the antisense strand [Zp(−)] was analyzed, as described in Materials and Methods. The positions of the ZII and ZID domains are indicated. Ext., DG75 nuclear extract present; No Ext., nuclear extract absent.
DISCUSSION
In this report, we have dissected the functional elements within the ZII domain, a region previously identified as essential for Zp inducibility (5, 7, 11, 19, 20). A clearly recognizable CRE/AP-1 motif has been noted by a number of investigators, and in our original characterization of Zp we demonstrated that a mutation (MII) that disrupted this motif severely diminished TPA inducibility (19). More recently, two independent analyses of the ZII domain have been published (42, 48). Ruf and Rawlins (42) reported the characterization of a complex which they refer to as ZIIBC. This complex was reported to be composed of 26- and 36-kDa proteins and was shown to bind the ZII CRE/AP-1 motif. However, these investigators were unable, using antibody reagents against a wide panel of CREB and AP-1 family members, to identify the components of this complex. A detailed analysis of the factors binding to the ZII domain was also reported by Wang et al. (48), who identified 12 distinct DNA-protein complexes. These investigators were able to identify the presence of ATFa, ATF-1, ATF-2, and c-Jun in some of these complexes. In addition, they demonstrated that overexpression of ATF-1 led to activation of a Zp driven reporter construct. Notably, formation of some of the observed complexes was independent of the CRE/AP-1 motif, and may reflect binding to the ZIIR element.
In the analysis presented here, we identified two major complexes by EMSA and were able to demonstrate the presence of ATF-1, and perhaps CREB-1, in the lower complex and ATF-2 in the upper complex. Based on the results obtained by Wang et al. (48), it seems likely that the faster migrating complex we observe corresponds to the four closely migrating complexes which they refer to as group II, two of which they show contain ATF-1. Similarly, the upper complex we observed by EMSA likely correlates with the three closely migrating complexes that Wang et al. (48) refer to as group I, two of which they demonstrate contain ATF-2. Somewhat surprisingly, we failed to detect the apparently abundant complexes present in group IV. This may reflect differences in preparation of nuclear extracts or a failure to resolve these complexes from free probe in the gel. Notably, Wang et al. were not able to identify any of the cellular factors involved in forming the complexes present in either group III or group IV (fastest migrating complexes). At this point, it is unclear which cellular factor(s) is important for Zp function in vivo, although these investigators did demonstrate that overexpression of ATF-1 activated Zp (48) suggesting that ATF-1 may be important for Zp activity.
A number of negative cis elements have been identified in the region upstream of the BZLF1 gene transcription initiation site (38). Three binding sites for the cellular repressor YY1 have been identified in the region from bp −300 to −450 (38). Deletion of the two distal YY1 binding sites resulted in significant upregulation of promoter activity, while overexpression of YY1 was shown to downregulate Zp activity. In addition, five H1 motifs, which have been reported to function as negative cis elements, have been identified (43). Four of these map in the region from ca. bp −280 to −450 while the fifth maps just downstream of the ZII domain. Of the distal H1 motif, two overlap with identified YY1 binding sites (38). These investigators reported that binding to the distal H1 motifs was abrogated upon induction of the viral lytic cycle, suggesting that they are important for downregulating Zp activity during latency (43). Notably, with the exception of the H1 motif mapping downstream of the ZII domain, all the identified negative cis elements map upstream of bp −221. Thus, these distal negative cis elements cannot account for the extremely low basal activity exhibited by Zp driven reporter constructs containing sequences from bp −221 to +12.
In this paper, we have identified a potent negative cis element, ZIIR, which is likely to play an important role in downregulating transcription from Zp during latency. It is likely that ZIIR binds a cellular repressor, but to date we have failed to detect a specific protein complex binding to this domain. Alternatively, it is possible that one of the complexes that binds the CRE/AP-1 motif serves to repress transcription and that binding of this complex is sensitive to mutations in the region upstream of the CRE/AP-1 motif. If the latter is true, this complex either cannot be distinguished from the other CREB/ATF complexes observed by EMSA or cannot be detected by EMSA. DNase I footprinting of the ZpRm mutant indicated that the region of ZIIR is protected from DNase I digestion by B-cell nuclear extract, although methylation interference assays failed to identify any contacts within this region (data not shown). It is possible that in vivo footprinting may help illuminate this issue. In addition, generation of an EBV strain harboring the ZIIRm mutation will help provide definitive insight into the role of this negative cis element in regulating viral latency.
ACKNOWLEDGMENTS
We thank D. Leib and M. Montminy for recombinant CREB protein and the 244 anti-CREB antibody, respectively. We also thank members of the Speck lab, as well as David Leib, Skip Virgin, and members of their labs, for helpful discussions.
This research was supported by NIH grant CA52004 to S.H.S.
REFERENCES
- 1.Angel P, Imagawa M, Chiu R, Stein B, Imbra R J, Rahmsdort H J, Jonat C, Herrlich P, Karin M. Phorbol ester-inducible genes contain a common cis-element recognized by a TPA-modulated trans-acting factor. Cell. 1987;49:729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. 2nd ed. New York, N.Y: John Wiley and Sons; 1992. [Google Scholar]
- 3.Bauer G, Hofler P, zur Hausen H. Epstein-Barr virus induction by a serum factor. I. Induction and cooperation with additional inducers. Virology. 1982;121:184–194. doi: 10.1016/0042-6822(82)90128-3. [DOI] [PubMed] [Google Scholar]
- 4.Biggin M, Bodescot M, Perricaudet M, Farrell P J. Epstein-Barr virus gene expression in P3HR-1 superinfected Raji cells. J Virol. 1987;61:3120–3132. doi: 10.1128/jvi.61.10.3120-3132.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borras A M, Strominger J L, Speck S H. Characterization of the ZI domains in the Epstein-Barr virus BZLF1 gene promoter: role in phorbol ester induction. J Virol. 1996;70:3894–3901. doi: 10.1128/jvi.70.6.3894-3901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y-N, Dong D L-Y, Hayward G S, Hayward S D. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and dimerization domain that lacks the characteristic heptad leucine zipper motif. J Virol. 1990;64:3368–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatila T, Ho N, Liu P, Liu S, Mosialos G, Kieff E, Speck S H. The Epstein-Barr virus-induced Ca2+/calmodulin-dependent kinase type IV/Gr promotes a Ca2+-dependent switch from latency to viral replication. J Virol. 1997;71:6560–6567. doi: 10.1128/jvi.71.9.6560-6567.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 1986;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Countryman J, Jenson H, Seibel R, Wolf H, Miller G. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J Virol. 1987;61:3672–3679. doi: 10.1128/jvi.61.12.3672-3679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daibata M, Speck S H, Mulder C, Sairenji T. Regulation of the BZLF1 promoter of Epstein-Barr virus by second messengers in anti-immunoglobulin treated B cells. Virology. 1994;198:446–454. doi: 10.1006/viro.1994.1056. [DOI] [PubMed] [Google Scholar]
- 12.Delegeane A M, Ferland L H, Mellon P L. Tissue-specific enhancer of the human glycoprotein hormone α-subunit gene: dependence on cyclic AMP-inducible elements. Mol Cell Biol. 1987;7:3994–4002. doi: 10.1128/mcb.7.11.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dignam J D, Lebovitz M, Roeder R G. Accurate transcription by RNA polymerase II in soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faggioni A, Zompetta C, Grimaldi S, Barile G, Frati L, Lazdins J. Calcium modulation activates Epstein-Barr virus genome in latently infected cells. Science. 1986;254:1554–1556. doi: 10.1126/science.3012779. [DOI] [PubMed] [Google Scholar]
- 15.Farrell P J, Rowe D T, Rooney C M, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flamand L, Menezes J. Cyclic AMP-responsive element-dependent activation of Epstein-Barr virus Zebra promoter by human herpesvirus 6. J Virol. 1996;70:1784–1791. doi: 10.1128/jvi.70.3.1784-1791.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flemington E K, Goldfeld A E, Speck S H. Efficient transcription of the Epstein-Barr virus immediate-early BZLF1 and BRLF1 genes requires protein synthesis. J Virol. 1991;65:7073–7077. doi: 10.1128/jvi.65.12.7073-7077.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flemington E K, Borras A M, Lytle J P, Speck S H. Characterization of the Epstein-Barr virus BZLF1 protein transactivation domain. J Virol. 1992;66:922–929. doi: 10.1128/jvi.66.2.922-929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flemington E, Speck S H. Identification of phorbol ester response elements in the promoter of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1217–1226. doi: 10.1128/jvi.64.3.1217-1226.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flemington E, Speck S H. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1227–1232. doi: 10.1128/jvi.64.3.1227-1232.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemington E, Speck S H. Epstein-Barr virus BZLF1 trans activator induces the promoter of a cellular cognate gene, c-fos. J Virol. 1990;64:4549–4552. doi: 10.1128/jvi.64.9.4549-4552.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flemington E K, Speck S H. Evidence for coiled-coil dimer formation by an Epstein-Barr virus transactivator that lacks a heptad repeat of leucine residues. Proc Natl Acad Sci USA. 1990;87:9459–9463. doi: 10.1073/pnas.87.23.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldfeld A E, Liu P, Liu S, Flemington E K, Strominger J L, Speck S H. Cyclosporin A and FK506 block induction of the Epstein-Barr virus lytic cycle by anti-immunoglobulin. Virology. 1995;209:225–229. doi: 10.1006/viro.1995.1247. [DOI] [PubMed] [Google Scholar]
- 24.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyl-transferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grogan E, Jenson H, Countryman J, Heston L, Gradoville L, Miller G. Transfection of a rearranged viral DNA fragment, WZhet, stably converts latent Epstein-Barr viral infection to productive infection in lymphoid cells. Proc Natl Acad Sci USA. 1987;84:1332–1336. doi: 10.1073/pnas.84.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habener J. Cyclic AMP response element binding proteins: a cornucopia of transcription factors. Mol Endocrinol. 1990;4:1087–1094. doi: 10.1210/mend-4-8-1087. [DOI] [PubMed] [Google Scholar]
- 27.Kallin B, Luka J, Klein G. Immunochemical characterization of Epstein-Barr virus-associated early and late antigens in n-butyrate-treated P3HR-1 cells. J Virol. 1979;32:710–716. doi: 10.1128/jvi.32.3.710-716.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieff E, Liebovitz D. Epstein-Barr virus and its replication. In: Fields B, Knipe D, Chanock R, Hirsh M, Melnick J, Monath T, Roizman B, editors. Fields virology. New York, N.Y: Raven Press; 1990. pp. 1889–1920. [Google Scholar]
- 29.Kouzarides T, Packham G, Cook A, Farrell P. The BZLF1 protein of EBV has a coiled coil dimerisation domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene. 1991;6:195–204. [PubMed] [Google Scholar]
- 30.Lee W, Mitchell P, Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987;49:741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- 31.Lieberman P, Berk A. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J Virol. 1990;64:2560–2568. doi: 10.1128/jvi.64.6.2560-2568.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieberman P, Hardwick J M, Sample J, Hayward G S, Hayward S D. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol. 1990;64:1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S, Liu P, Borras A M, Chatila T, Speck S H. Cyclosporin A-sensitive induction of the Epstein-Barr virus lytic switch is mediated via a novel pathway involving a MEF2 family member. EMBO J. 1997;16:143–153. doi: 10.1093/emboj/16.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S, Borras A M, Liu P, Suske G, Speck S H. Binding of the ubiquitous cellular transcription factors Sp1 and Sp3 to the ZI domains in the Epstein-Barr virus lytic switch BZLF1 promoter. Virology. 1997;228:11–18. doi: 10.1006/viro.1996.8371. [DOI] [PubMed] [Google Scholar]
- 35.Luka J, Kallin B, Klein G. Induction of the Epstein-Barr virus (EBV) cycle in latently infected cells by n-butyrate. Virology. 1979;94:228–231. doi: 10.1016/0042-6822(79)90455-0. [DOI] [PubMed] [Google Scholar]
- 36.Maurer R A. Both forms of the cAMP-dependent protein kinase catalytic subunit can activate transcription of the PRL gene. J Biol Chem. 1989;264:6470–6873. [PubMed] [Google Scholar]
- 37.Miller G, Rabson M, Heston L. Epstein-Barr virus with heterogeneous DNA disrupts latency. J Virol. 1984;50:174–182. doi: 10.1128/jvi.50.1.174-182.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montalvo E A, Cottam M, Hill S, Wang Y-C J. YY1 binds to and regulates cis-acting negative elements in the Epstein-Barr virus BZLF1 promoter. J Virol. 1995;69:4158–4165. doi: 10.1128/jvi.69.7.4158-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montminy M R, Gonzalez G A, Yamamoto K K. Regulation of cAMP-inducible genes by CREB. Trends Neurosci. 1990;12:184–188. doi: 10.1016/0166-2236(90)90045-c. [DOI] [PubMed] [Google Scholar]
- 40.Packham G, Economou A, Rooney C M, Rowe D T, Farrell P J. Structure and function of the Epstein-Barr virus BZLF1 protein. J Virol. 1990;64:2110–2116. doi: 10.1128/jvi.64.5.2110-2116.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rehfuss R P, Walton K M, Loriaux M M, Goodman R H. The cAMP-regulated enhancer-binding protein ATF-1 activates transcription in response to cAMP-dependent protein kinase A. J Biol Chem. 1991;266:18431–18434. [PubMed] [Google Scholar]
- 42.Ruf I K, Rawlins D R. Identification and characterization of ZIIBC, a complex formed by cellular factors and the ZII site of the Epstein-Barr virus BZLF1 promoter. J Virol. 1995;69:7648–7657. doi: 10.1128/jvi.69.12.7648-7657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarzmann F, Prang N, Reichelt B, Rinkes B, Haist S, Marschall M, Wolf H. Negative cis-acting elements in the distal part of the promoter of Epstein-Barr virus trans-activator gene BZLF1. J Gen Virol. 1994;75:1999–2006. doi: 10.1099/0022-1317-75-8-1999. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu N, Takada K. Analysis of the BZLF1 promoter of Epstein-Barr virus: identification of an anti-immunoglobulin response sequence. J Virol. 1993;67:3240–3245. doi: 10.1128/jvi.67.6.3240-3245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tovey M, Lenoir G, Lours-Begon J. Activation of latent Epstein-Barr virus by antibody to human IgM. Nature (London) 1978;276:270–272. doi: 10.1038/276270a0. [DOI] [PubMed] [Google Scholar]
- 47.Urier G, Buisson M, Chambard P, Sergeant A. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 1989;8:1447–1453. doi: 10.1002/j.1460-2075.1989.tb03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y C, Huang J M, Montalvo E A. Characterization of proteins binding to the ZII element in the Epstein-Barr virus BZLF1 promoter: transactivation by ATF1. Virology. 1997;227:323–330. doi: 10.1006/viro.1996.8326. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto K K, Gonzalez G A, Menzel P, Rivier J, Montminy M R. Characterization of a bipartite activator domain in transcription factor CREB. Cell. 1990;60:611–617. doi: 10.1016/0092-8674(90)90664-z. [DOI] [PubMed] [Google Scholar]
- 50.zur Hausen H, O’Neil F, Freese U. Persisting oncogenic herpesvirus induced by the tumor promoter TPA. Nature (London) 1978;272:373–375. doi: 10.1038/272373a0. [DOI] [PubMed] [Google Scholar]