Supplemental Digital Content is Available in the Text.
Our study demonstrated that the excitatory Mas-related G protein–coupled receptor d plays a key role in mediating mechanical allodynia in a mouse model of painful diabetic neuropathy.
Keywords: Painful diabetic neuropathy, Neuropathic pain, Dorsal root ganglia, Single-cell RNA sequencing
Abstract
Painful diabetic neuropathy (PDN) is one of the most common and intractable complications of diabetes. Painful diabetic neuropathy is characterized by neuropathic pain accompanied by dorsal root ganglion (DRG) nociceptor hyperexcitability, axonal degeneration, and changes in cutaneous innervation. However, the complete molecular profile underlying the hyperexcitable cellular phenotype of DRG nociceptors in PDN has not been elucidated. This gap in our knowledge is a critical barrier to developing effective, mechanism-based, and disease-modifying therapeutic approaches that are urgently needed to relieve the symptoms of PDN. Using single-cell RNA sequencing of DRGs, we demonstrated an increased expression of the Mas-related G protein–coupled receptor d (Mrgprd) in a subpopulation of DRG neurons in the well-established high-fat diet (HFD) mouse model of PDN. Importantly, limiting Mrgprd signaling reversed mechanical allodynia in the HFD mouse model of PDN. Furthermore, in vivo calcium imaging allowed us to demonstrate that activation of Mrgprd-positive cutaneous afferents that persist in diabetic mice skin resulted in an increased intracellular calcium influx into DRG nociceptors that we assess in vivo as a readout of nociceptors hyperexcitability. Taken together, our data highlight a key role of Mrgprd-mediated DRG neuron excitability in the generation and maintenance of neuropathic pain in a mouse model of PDN. Hence, we propose Mrgprd as a promising and accessible target for developing effective therapeutics currently unavailable for treating neuropathic pain in PDN.
1. Introduction
Painful diabetic neuropathy (PDN) is an intractable complication affecting some 25% of diabetic patients.2,30 Painful diabetic neuropathy symptoms include neuropathic pain and small-fiber degeneration19,43,56,72 involving the degeneration of the axons of the nociceptive dorsal root ganglion (DRG) neurons that innervate the skin.44,74 Neuropathic pain associated with PDN is a debilitating affliction having a substantial impact on patients' quality of life and health care costs.17 Despite this prevalence and impact, current therapies for PDN are only partially effective.9,61,75 One critical barrier to developing novel and effective therapy for PDN is that the molecular mechanisms leading to neuropathic pain and to small-fiber degeneration are mostly unknown.
Neuropathic pain is associated with the hyperexcitability of neurons in pain pathways in the absence of appropriate stimuli.43,83,88 The cells responsible for this phenomenon include DRG nociceptors.43,83,88 Diabetic patients58 and animal models of PDN3,7 exhibit sensory neuron hyperexcitability, including spontaneous activity of DRG nociceptor axons.3,7,67 Consistent with these findings, our laboratory has shown that reducing the hyperexcitability of DRG nociceptors, identified by the sodium channel Nav1.8, which is expressed by 90% of nociceptors,69 reversed mechanical allodynia and small-fiber degeneration37 in the well-established high-fat diet (HFD) mouse model of PDN.56 It is known that in states of neuropathic pain, DRG nociceptors become hypersensitive to a variety of signaling molecules.10,27,38,48,53,79 However, the complete molecular profile underlying the hyperexcitable cellular phenotype of DRG nociceptors in PDN has not been elucidated. The identification of targets able to specifically modulate DRG nociceptor hyperexcitability could help in the further development of novel therapeutic approaches for PDN.
Because DRG neurons are a heterogeneous group of neurons,1,16,28,34,45,50,82 accurate and complete classification of the molecular properties of these neurons is imperative if we wish to understand which subtypes are involved in different pathological conditions such as PDN. Several groups have performed scRNA-seq of rodent and human sensory neurons16,28,45,68,82,87; however, single-cell RNA sequencing and the complete gene expression profile of molecularly distinct DRG cell types, along with the specific genes differentially expressed in each type, have not been addressed in PDN.
Using single-cell RNA sequencing of DRGs, we demonstrated an increased expression of the Mas-related G protein–coupled receptor d (Mrgprd) in a subpopulation of DRG neurons in the HFD mouse model of PDN. The role of Mrgprd-expressing neurons in mechanical nociception is well established in mice62,89 and in human.20,46 Mrgprd is an interesting target because it is a highly druggable, excitatory G protein–coupled receptor known to influence DRG neuron excitability to mechanical stimuli, expressed solely by the nociceptive neuronal population that extends out into the outermost layer of the skin.89 Indeed, here we demonstrated that limiting Mrgprd signaling reversed mechanical allodynia in the HFD mouse model of PDN. Furthermore, using in vivo calcium imaging, we demonstrated that activation of Mrgprd-positive cutaneous afferents that persist in diabetic mice skin resulted in an increased intracellular calcium influx into in DRG nociceptors that we assess in vivo as a readout of nociceptors hyperexcitability. Taken together, our data highlight a key role of Mrgprd-mediated DRG neuron excitability in the generation and maintenance of mechanical allodynia in a mouse model of PDN.
2. Materials and methods
2.1. Animals
All methods involving animals were approved by the Institutional Animal Care and Use Committee at Northwestern University. Animals were housed with food and water ad libitum on a 12-hour light cycle. We used the following mouse lines: C57/Bl6J (wild-type), Nav1.8-Cre;GCaMP6s, Nav1.8-Cre;Ai9, MrgprdΔEGFPf, Mrgprd-eGFP reporter mice (MrgprdΔEGFPf).89 Mrgprd-CreERT2 mice57 were crossed with hM4Di mice.64 The resulting Mrgprd-CreERT2;hM4Di mice were treated with tamoxifen 0.5 mg (Sigma-Aldrich, St. Louis, MO; T5648) given dissolved in corn oil via i.p. injection once per day starting postnatally day 10 (P10) through postnatally day 17 (P17). We observed that the metabolic profile of female mice on a HFD was extremely variable. Therefore, in this study, we only used male mice with the goal of reducing total mouse usage. Animals were given at least one week to drive recombination and reporter gene expression.
2.2. High-fat diet
Mice were fed a diet with a high fat content (42% fat) (Envigo TD88137, Envigo, Madison, WI), HFD for 10 weeks as previously described.25,37,51,56 Control mice were fed a regular diet (RD) containing 11% fat. After 10 weeks on RD or HFD, a glucose tolerance test was performed as described.37,56 Briefly, after fasting for 12 hours, mice were injected with a 45% d-glucose solution (2 mg glucose/g body weight). Animals were weighed on an electronic scale, and after fasting the animals for 12 hours, fasting blood glucose was measured using TrueTrack meter and TrueTrack glucose test strips. The mice were then injected with a 45% d-glucose solution (2 mg glucose/g body weight) and blood glucose was measured at 30, 60, and 120 minutes after injection (RD-Het n = 12 animals; RD-Homo n = 6 animals; HFD-Het n = 15 animals; HFD-Homo n = 15 animals). To compare “diabetic” vs “nondiabetic” HFD mice, we set the cutoff for diabetes (>178.41 mg/dL) at 2 SDs above the mean for glucose at 2 hours after glucose challenge, as determined from among wild-type littermate RD mice.37
2.2.1. Statistics
Blood glucose was analyzed using 1-way analysis of variance (ANOVA) followed by Tukey's test.
2.3. Isolation of dorsal root ganglion neurons and single-cell RNA sequencing using 10X Genomics platform
Lumbar DRG neurons from adult mice fed on a regular diet (n = 5 animals) and a high-fat diet (n = 5 animals) were isolated and dissociated following protocol established by Zeisel et al., 2018. A high viability single-cell suspension was prepared, and using the 10X Genomics chromium single cell kit v2, about 6000-8000 cells were recovered. Two rounds of the experiment (total n = 10 animals per group) were performed, and downstream cDNA synthesis, library preparation, and sequencing were performed according to the manufacturer's instruction. Illumina runs were demultiplexed and aligned using the 10X Genomics cell ranger pipeline. Dimensionality reduction, clustering, and differential expression of genes were done in Seurat v4.2.35 Raw matrix files for single-cell RNA sequencing were deposited in Dryad (https://doi.org/10.5061/dryad.9s4mw6mm5).
2.3.1. Statistics
We used the FindMarkers function in Seurat to perform a Wilcoxon rank sum test to identify the differentially expressed genes between RD and HFD.
2.4. RNAscope in situ hybridization
RNAscope in situ hybridization multiplex V2 was performed according to the manufacturer's instructions (Advanced Cell Diagnostics [ACD]). Dorsal root ganglions were isolated in an RNase-free manner, and the samples were then fixed in RNase-free 4% PFA for 24 hours. The samples were transferred to 30% sucrose for 24 hours and embedded in optimal cutting temperature (OCT). Twelve-micrometer sections were placed on SuperFrost Plus charged slides and stored at −20°C until ready to use. The slides were briefly washed in 1× PBS and followed by a 10-minute hydrogen peroxidase treatment at RT. The slides were then placed in a beaker containing 1× target retrieval solution that heated to 99 to 102°C for about 3 minutes. The slides were then cooled in DEPC-treated water and transferred to 100% ethanol for 3 minutes. The slides were completely air dried at RT, and hydrophobic barriers were drawn around the sections. The air-dried slides were placed on the HybEZ slide rack, and about 5 drops of RNAscope Protease III was added. The slide rack was placed onto a prewarmed humidity control tray and into the HybEZ oven at 40°C for 30 minutes. Probes Scn10a (C1, catalog 426011), neurofilament heavy chain (Nefh) (C2, catalog 443671), Mrgprd (C3, catalog 45692), Lpar3 (C1, catalog 43259), Trpv1 (C2, catalog 313331), human MRGPRD (C1, catalog 524871), and human TRPV1 (C2, catalog 415381) were used at the recommended concentration (C1:C2; 50:1). Probes were incubated for 2 hours at 40°C, and the slides were then stored in 5× saline sodium citrate solution. On day 2, AMP1, AMP2, and AMP3 were added sequentially with a 30-, 30-, and a 15-minute incubation period, respectively. Depending on the probe used, the appropriate HRP signals were developed. Briefly, 4 to 6 drops of HRP-C1 or HRP-C2 were added and incubated for 15 minutes at 40°C. This was followed by the addition of 1:100 dilution of TSA Plus fluorescein. The fluorophores were incubated at 40°C for 30 minutes, and this was followed by addition of the HRP blocker. Washes were performed using 1× wash buffer as recommended. The slides were then mounted using Vectashield mounting media containing DAPI.
2.4.1. Analysis
Tissue sections were analyzed by imaging the whole DRG using Olympus FV10i confocal microscope, and the images were processed using Fiji. Two blinded reviewers counted the positive cells for each subpopulation using the Cell Counter plugin and measured the Mrgprd mRNA expression as average intensity of Mrgprd+ dots per cell. Briefly, we first calculated the average background intensity (ABI) based on the integrated intensity of a background region in a selected area (ABI = IntDen background/area of selected background). Then we selected 10 dots per cell (2 cells per sample) and measured the area and integrated intensity of each dot. Lastly, we calculated the average intensity per single dot (AISD) using the formula: AISD = IntDen selected dots − ABI × area of selected dots/tot number of dots. Mean values of the counts from blinded reviewers were plotted and graphed.
2.4.2. Statistical analysis
Data were compared using unpaired t test. In all experiments (RD n = 3 animals; HFD n = 3 animals), values are expressed as mean ± SEM.
2.5. In vivo calcium imaging
Nav1.8-Cre;GCaMP6 animals were fed an RD or HFD diet for 10 weeks, then anesthetized by isoflurane and laminectomized, exposing the L4 DRG as described.52 The experimental setup and imaging were done as previously reported.52 Briefly, the mouse was positioned under the microscope by clamping the spinal column at L2 and L6; body temperature and isoflurane were constantly maintained and monitored throughout the imaging period. Silicone elastomer (World Precision Instruments) was used to cover the exposed DRG and surrounding tissue to avoid drying.52 A Coherent Chameleon-Ultra2 Ti:Sapphire laser was tuned to 920 nm, and GCaMP6s signal was collected by using a bandpass filter for the green channel (490-560 nm). Image acquisition was controlled using PrairieView software version 5.3. Images of the L4 DRG were acquired at 0.7 Hz, with a dwell time of 4 µs/pixel (pixel size 1.92 × 1.92 µm2), and a 10× air lens (Olympus UPLFLN U Plan Fluorite; 0.3 NA, 10 mm working distance). The scanned sample region was 981.36 × 981.36 µm2. Anesthesia was maintained using isoflurane (1.5%-2%) during imaging.
2.6. Intradermal β-alanine and capsaicin administration
Five microlitres of 100 mM β-alanine or 10 µM capsaicin was injected intradermally to the paw pad in hind paw of anesthetized mice in the set up described in the previous section. The needle was inserted into the paw for 10 seconds and any responses to the needle were not included in the analysis. After 10 seconds, β-alanine or capsaicin was released, and the neurons that responded to β-alanine or capsaicin was reported as the percentage of neurons that responded. Appropriate controls like saline and ethanol were used to rule out nonspecific responses because of needle injections.
2.7. Analysis of in vivo calcium imaging
Time series files were exported and further processed in Fiji (NIH). Any movement in the time series (on account of breathing) were adjusted using the template-matching plugin. Brightness and contrast were adjusted, and cells (region of interest [ROI]) were identified by looking for an increase in fluorescence during the stimulus application period, as previously reported.52 The identified cells were then carefully marked and a custom macro (where changes in [Ca2+]i were quantified by calculating the change in fluorescence for each ROI in each frame t of a time series using the formula: ΔF/F0 = (Ft − F0)/F0, where F0 = the average intensity during the baseline period before the application of the stimulus) was run followed by a Multi Measure Plugin to obtain the mean gray value of each ROI. Once the values were obtained, the ROIs that had a ΔF/F0 reading greater than 1 were included as a responding ROI/neuron (RD n = 7 animals, HFD n = 9 animals, average number of neurons imaged per DRG in RD = 197.5 ± 48.67 and HFD = 242.5 ± 69.16). To determine the percentage of responders, the total number of neurons imaged for each DRG was estimated by counting the number of neurons within a region of average density and extrapolating to the total DRG area.52 The area under the curve (AUC) was calculated for each responding ROI in GraphPad Prism 8.3, and the mean total peak area was calculated for each animal. The maximum amplitude was calculated by taking the maximum ΔF/F0 value for each responding ROI and the mean was calculated for each animal. Experimental procedures were designed to maximize robustness and minimize bias. Specifically, dynamic brush experiments were conducted using random experimental group assignments (diet and treatment). Investigators that performed in vivo calcium imagining experiments and endpoint analysis were blinded to the experimental conditions. We have experience with randomized allocation and blinded analysis using this mouse model with sequenced numbering of mice at weaning.37
2.7.1. Statistics
Percentage responders, AUC, and max amplitude were compared using a 2-tailed unpaired t test and Fisher exact test. Wilcoxon matched-pair signed rank test or paired t test was performed to compare saline and β-alanine and ethanol and capsaicin responses, respectively, in both experimental groups. Mann–Whitney test or unpaired t test was performed to compare effects of β-alanine and capsaicin, respectively, in RD v/s HFD conditions. Data are reported as mean ± SEM.
2.8. Detection of cutaneous innervation
Nav1.8-Cre;Ai9, MrgprdΔEGFPf, MrgprdΔEGFPf heterozygous, and homozygous animals were fed an RD or HFD for 10 weeks. The hind paws were harvested and fixed in Zamboni fixative for 24 hours, and then the overlying footpad skin was dissected, submerged in 30% sucrose solution for 24 hours, and embedded in Optimal Cutting Temperature Compound (OCT, Tissue-Tek). Thirty-micrometer sections were cut on a cryostat and counterstained by mounting solution with DAPI (Hardset, Vectashield).
2.8.1. Confocal analysis
Three separate sections from each animal were analyzed, composite Z-stack images of skin from the hind paw were imaged using Olympus FV10i, and the images were processed using Fiji. The epidermal–dermal junction was outlined by a blinded observer who also noted its length. Three blinded reviewers counted the nerves crossing this line using the Cell Counter plugin. Mean values of the counts from blinded reviewers were divided by the epidermal–dermal junction length to report IENF density.
2.8.2. Statistical analysis
Data were compared using 1-way ANOVA followed by Tukey test. In all experiments (RD n = 3-8 animals; HFD n = 3-7 animals), values are expressed as mean ± SEM.
2.9. Intraperitoneal injection with clozapine-N-oxide
Clozapine-N-oxide (CNO, 10 mg/kg; Sigma Aldrich, St. Louis, MO) in 200 μL of saline was injected with a 25-gauge needle to mice fed either an RD or HFD. Mice were tested for pain 1 and 4 hours after injection with either CNO (RD-CNO n = 6 mice; HFD-CNO n = 6 mice) or vehicle (RD-saline n = 7 mice; HFD-saline n = 6 mice).
2.10. von Frey
von Frey behavioral studies were performed as described. Briefly, mice were placed on a metal mesh floor and covered with a transparent plastic dome where they rested quietly after initial few minutes of exploration. Animals were habituated to this apparatus for 30 minutes a day, 2 days before behavioral testing. After acclimation, each filament was applied to 6 spots spaced across the glabrous side of the hind paw. Mechanical stimuli were applied with 7 filaments, each differing in the bending force delivered (10, 20, 40, 60, 80, 100, and 120 mN), but each fitting a flat tip and a fixed diameter of 0.2 mm. The force equivalence is 100 mN = 10.197 g. The filaments were tested in order of ascending force, with each filament delivered for 1 second in sequence from the first to the sixth spot, alternately from one paw to the other. The interstimulus interval was 10 to 15 seconds.37 The von Frey withdrawal threshold was defined as the force that evoked a minimum detectable withdrawal on 50% of the tests at the same force. Experimental procedures were designed to maximize robustness and minimize bias. Specifically, von Frey experiments were conducted using random experimental group assignments (diet and treatment). Investigators that performed von Frey tests and endpoint analysis were blinded to the experimental conditions. We have experience with randomized allocation and blinded analysis using this mouse model with sequenced numbering of mice at weaning.37
2.10.1. Statistical analysis
The incidence of foot withdrawal was expressed as a percentage of 6 applications of each filament as a function of force. A Hill equation was fitted to the function, relating the percentage of indentations eliciting a withdrawal to the force of indentation. From this equation, the threshold force was defined as the force corresponding to a 50% withdrawal rate.37 Data were compared by 1-way ANOVA followed by Kruskal–Wallis test (RD-Het n = 10 animals, RD-Homo n = 8 animals, HFD-Het n = 17 animals, and HFD-Homo n = 16 animals) and reported as mean ± SD.
2.11. Dynamic brush assay
Dynamic mechanical hypersensitivity was measured as previously described.15 Briefly, the animals were placed on a 7.25-mm spaced grid and covered with a transparent plastic dome where they rested quietly after initial few minutes of exploration. Animals were habituated to this apparatus for 45 minutes a day, 2 days before behavioral testing. The hind paw of the animals was lightly stroked in the heel to toe direction with a paintbrush (Cat # 06156-1004, Series4310L; Blick Art Supplies, Chicago, IL). Both the right and the left hind paw were brushed alternatively and scored with one minute of resting time between the brush strokes. We used a graded scoring system where the animal received a 0 score if the animal did not respond to the brush stimulus. Animals received a score of 1 if the animal lifted the paw off the mesh and placed it back on the mesh immediately. Animals received a score of 2 if there was sustained lifting (>2 seconds) or if there was flinching or licking of the stimulated paw. Experimental procedures were designed to maximize robustness and minimize bias. Specifically, dynamic brush experiments were conducted using random experimental group assignments (diet and treatment). Investigators who performed von Frey tests and endpoint analysis were blinded to the experimental conditions. We have experience with randomized allocation and blinded analysis using this mouse model with sequenced numbering of mice at weaning.37
2.11.1. Statistical analysis
Stroking was repeated a total of 10 times for each animal, and the scores were averaged. Data were compared by 1-way ANOVA followed by Kruskal–Wallis test (RD-Het n = 23 animals, RD-Homo n = 13 animals, HFD-Het n = 24 animals, and HFD-Homo n = 22 animals) and reported as mean ± SD.
3. Results
3.1. Single-cell transcriptomic profiling of dorsal root ganglion neurons in the HFD mouse model of painful diabetic neuropathy
To understand the mechanisms leading to DRG nociceptor excitability that underlies neuropathic pain in PDN, we used the well-established HFD mouse model of PDN, where mice are fed either a regular diet (RD) or a diet with a high content of fat (HFD) for about 10 weeks, during which time they develop mechanical allodynia and small-fiber degeneration.37,51 Our intention was to discover molecular markers responsible for the hyperexcitable cellular phenotype of DRG neurons as potential novel therapeutic targets. Hence, we identified the complete molecular profile of DRG neurons in their hyperexcitable state using an unbiased transcriptomic comparison of gene expression by lumbar DRG neurons from mice fed an HFD compared mice fed an RD. Dorsal root ganglion neurons are molecularly defined as distinct subtypes based on the expression of a set of molecular markers.16,28,45,82 To capture the full heterogeneity of DRG neurons and characterize changes in cell types and cell states, we performed single-cell RNA sequencing to understand the complete gene expression profile of molecularly distinct cell types together with the specific genes differentially expressed in each type in the HFD mouse model of PDN.
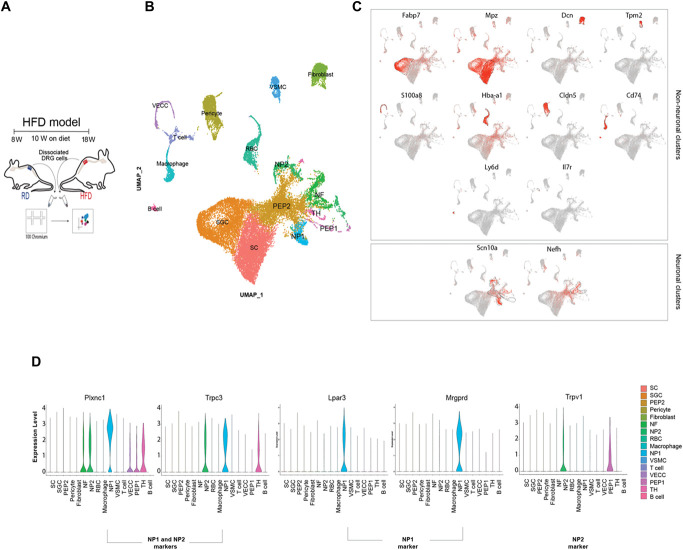
We prepared viable single-cell suspensions from lumbar DRGs in mice fed an RD or HFD and used the 10× platform for the single-cell capture and barcoding (Fig. 1A). We performed comparative clustering (n = 5 mice per group × 2 rounds; RD 6888 and 6693 cells; HFD 8567 and 6429 cells) and were able to clearly identify different cell types as well as conserved markers in the 2 conditions investigated. As expected, and in support of the literature on this topic,16,28,45,68,82,87 we were able to separate out neuronal and nonneuronal clusters. Within the neuronal clusters, we were able to identify distinct neuronal subtypes comparable with those described in the literature16,28,45,68,82,87 (Figs. 1B and C). In our comparative analysis of the single-cell data, we were particularly interested in the differential expression of GPCRs because they might represent important drug targets. Mrgprd is an interesting potential drug target because it is a G protein–coupled receptor known to influence DRG neuron excitability to mechanical stimuli in mice62,89 and in human.20,46 Additionally, Mrgprd is expressed by the nociceptive neuronal population that extends out into the outermost layer of the skin,89 making it a very accessible therapeutic target for PDN. We confirmed that in RD DRGs, Mrgprd is expressed in a subpopulation of neurons classified as nonpeptidergic type 1 neurons (NP1)82,87 (Fig. 1D). Raw matrix files for single-cell RNA sequencing were deposited in Dryad (https://doi.org/10.5061/dryad.9s4mw6mm5).
Figure 1.
Single-cell RNA-seq of lumbar DRG neurons in mice fed an RD or HFD. (A) Experimental model describing the 2 diet groups, isolation of DRG and single-cell capture and barcoding using the 10X Genomics platform. (B) Integrated UMAP plot visualizing neuronal (PEP, peptidergic; NF, neurofilament; NP, nonpeptidergic; TH, tyrosine hydroxylase) and nonneuronal clusters (VSMC, vascular smooth muscle cell; RBC, red blood cell; VECC, vascular endothelial capillary cell; SC, Schwann cell; SGC, satellite glial cells). (C) Feature plot indicating the expression of well-known nonneuronal markers and visualization of 2 broad neuronal markers—Nav1.8 and Nefh. (D) Violin plot showing expression of well-established markers used to identify the NP1 and NP2 subpopulation (n = 5 mice per group × 2 rounds; RD 6888 and 6693 cells; HFD 8567 and 6429 cells). DRG, dorsal root ganglion; HFD, high-fat diet; Nefh, neurofilament heavy chain; RD, regular diet. Lpar3, Lysophosphatidic Acid Receptor 3; Plxnc1, Plexin c1; Trpc3, Transient Receptor Potential Cation Channel subfamily C member 3; Trpv1, Transient Receptor Potential Cation Channel subfamily V member 1.
3.2. Overexpression of Mas-related G protein–coupled receptor d receptors in a subpopulation of dorsal root ganglion neurons in diabetic mice
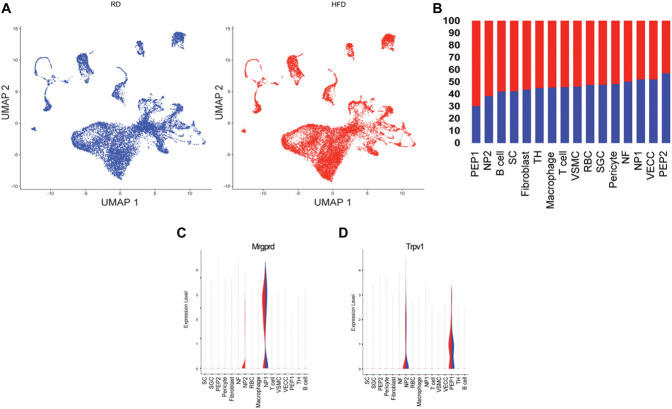
Aiming to further evaluate Mrgprd expression when comparing HFD and RD lumbar DRG, we performed a comparative analysis of the identified clusters (Fig. 2A). The relative proportion of cells in the neuronal clusters indicated an expansion in the peptidergic 1 (PEP1) and nonpeptidergic type 2 (NP2) clusters (Fig. 2B) in the HFD condition. When we compared RD and HFD DRGs, we observed that Mrgprd was significantly expressed in NP2 cluster (Fig. 2C, P-value of 0.002). The NP2 population expresses Trpv1, the receptor for capsaicin (Figs. 1D and 2D), and we observed that there were no differences in the expression of Trpv1 (Fig. 2D) in the NP2 population or in the PEP1 population when comparing RD and HFD conditions.
Figure 2.
Single-cell RNA-seq of lumbar DRG neurons reveals overexpression of Mrgprd in mice fed a HFD. (A) UMAP split by treatment (B) Percentage of cells within each neuronal cluster. (C) Violin plot indicating changes in gene expression of Mrgprd in the HFD condition. (D) Violin plot showing Trpv1 expression in the 2 diet conditions. (n = 5 mice per group × 2 rounds; RD 6888 and 6693 cells; HFD 8567 and 6429 cells). DRG, dorsal root ganglion; HFD, high-fat diet; Mrgprd, Mas-related G protein–coupled receptor d; RD, regular diet.
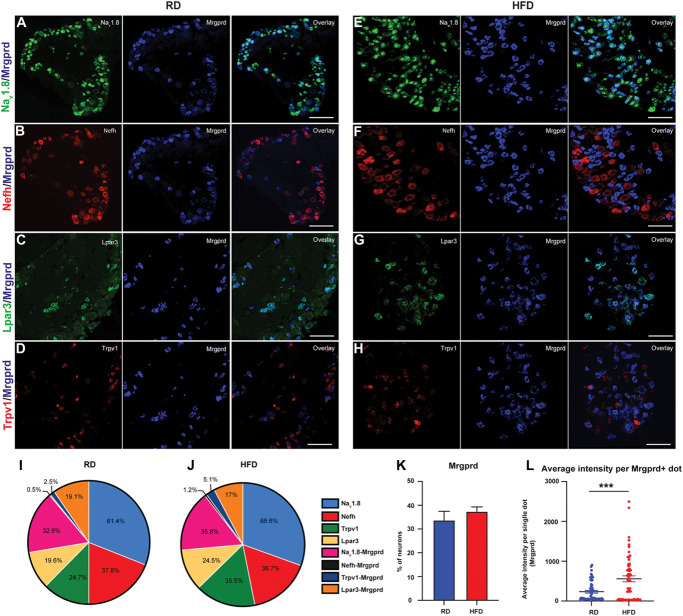
To validate the DRG subtypes identified with single-cell RNA seq, we performed RNA scope on frozen sections of lumbar DRG taken from mice fed RD or HFD. We used probes to detect markers characteristic of DRG subgroups. We know that Mrgprd-expressing neurons are a subpopulation of the Nav1.8 population as shown both in representative images (Figs. 3A and E). Additionally, both in RD and in HFD lumbar DRG, we verified the colocalization of Mrgprd and the Nefh expressed in NF DRG subpopulation (Figs. 3B and F). We also analyzed the expression of Mrgprd with lysophosphatidic acid receptor (Lpar3), expressed in NP1 class of neurons (Figs. 3C and G) and Trpv1, which is a marker of the NP2 population of DRG neurons82 (Figs. 3D and H). Quantification of DRG class of neurons, done in blind by 3 independent counters, showed no significant differences between the 2 diet conditions RD and HFD (Suppl Table 1, available at http://links.lww.com/PAIN/B957). In the pie chart (Figs. 3I and J), we show the percentage of expression of different neuron subtypes and their colocalization with other markers in both RD and HFD. Moreover, even if the quantification of Mrgprd-positive neurons in the 2 diet conditions showed no significant differences (Fig. 3K; RD 33.73 ± 3.76, n = 8 animals; HFD 37.41 ± 1.90, n = 7 animals), the quantification of Mrgprd mRNA as average intensity showed a robust difference among the 2 diet conditions (Fig. 3L; 2-tailed unpaired t test 325.4 ± 82.92, P = 0.0001; n = 60 dots and n = 3 animals in both RD and HFD).
Figure 3.
In situ hybridization validating the expression of Mrgprd (A–H) Nav1.8 (green), Nefh (red), Lpar3 (green), and Trpv1 (red) with Mrgprd (blue) from mice fed an RD or HFD. (I and J) Pie chart of each population along with the co-localization with other markers. In all experiments (RD n =3-4 mice and HFD n = 4-5 mice were imaged for each group). (K) Quantification of percentage of neurons expressing Mrgprd. (L) Quantification of Mrgprd mRNA as average intensity of Mrgprd+ dots per cell. Data compared using unpaired t tests. In all experiments (RD n = 8 mice and HFD n = 7 mice were imaged for each group), values are expressed as mean ± SEM. HFD, high-fat diet; Mrgprd, Mas-related G protein–coupled receptor d; Nefh, neurofilament heavy chain; RD, regular diet.
Given the molecular differences in neuronal subpopulations between mouse and human DRG,59,63,70,78 it was important to validate our potential candidate target using human DRGs. Using RNAscope, we validated the expression of MRGPRD in human DRGs from donor controls and patients with PDN provided to us by Anabios. We were able to confirm that MRGPRD is expressed in human DRGs both in control and in donors with PDN (Suppl Fig. 1A, ×10 magnification, available at http://links.lww.com/PAIN/B957). Additionally, as previously demonstrated,55,59,63,70,78 we confirmed that TRPV1 is more widely expressed in human DRGs as compared with mouse DRGs70 both in controls and in donors with PDN. Indeed, we observed that TRPV1 was expressed in most nociceptors and co-expressed with MRGPRD (Suppl Fig. 1A, available at http://links.lww.com/PAIN/B957, the inset shows a magnified image of a single neuron).
3.3. Mas-related G protein–coupled receptor d receptors are necessary for the establishment of static mechanical allodynia in diabetic mice
Using single-cell RNA sequencing, we have demonstrated an increased expression of the Mas-related G protein–coupled receptor d (Mrgprd) in a subpopulation of DRG neurons under diabetic conditions. To make a compelling case for Mrgprd receptors as a drug target for PDN, we wanted to demonstrate that manipulating these receptors reduces PDN symptoms. Hence, we examined the effects of reducing the expression of Mrgprd receptors in the development of pain hypersensitivity in the HFD mouse model of PDN using mice in which the Mrgprd receptor had been deleted and replaced with a gene for a fluorescent protein, EGFPf (MrgprdΔEGFPf).89 First, we induced PDN in MrgprdΔEGFPf mice by feeding them either an RD or HFD. Both MrgprdΔEGFPf heterozygous and homozygous mice fed HFD developed obesity (Suppl Fig. 2A, available at http://links.lww.com/PAIN/B957) and glucose intolerance (Suppl Fig. 2B and C, available at http://links.lww.com/PAIN/B957) like wild-type mice, demonstrating that Mrgprd receptor deletion does not alter their response to the changed metabolic profile prevailing in the HFD. We then tested both Het and Homo mice for mechanical allodynia (RD-Het n = 10 mice; RD-Homo n = 8 mice; HFD-Het n = 17 mice; HFD-Homo n = 16 mice) using von Frey withdrawal threshold measurements, as described elsewhere.6,37,51 Deletion of the Mrgprd receptors in RD-Homo did not change the withdrawal thresholds or result in mechanical allodynia compared with the heterozygous mice fed an RD (Fig. 4A; RD-Het 0.80 ± 0.93 v/s RD-Homo 0.70 ± 0.35, P = >0.99) indicating that Mrgprd deletion did not alter mechanical sensation in otherwise metabolically normal mice. We have previously shown that mice fed an HFD developed mechanical allodynia 6 weeks after commencement of the diet. In Het mice fed the HFD for 10 weeks, the withdrawal threshold was significantly reduced compared with RD-Het mice (RD-Het 0.80 ± 0.93 v/s HFD-Het 0.13 ± 0.24; P < 0.001), indicating mechanical allodynia in this group (Fig. 4A). We then determined whether deleting Mrgprd receptors would prevent the development of mechanical allodynia in HFD mice. We saw that there was a significant increase in the withdrawal threshold in HFD-Homo mice compared with HFD-Het (Fig. 4A; HFD-Het 0.13 ± 0.24 v/s HFD-Homo 0.33 ± 0.57, P = 0.03), indicating that deleting Mrgprd receptors prevented the establishment of static mechanical allodynia in this model of PDN.
Figure 4.
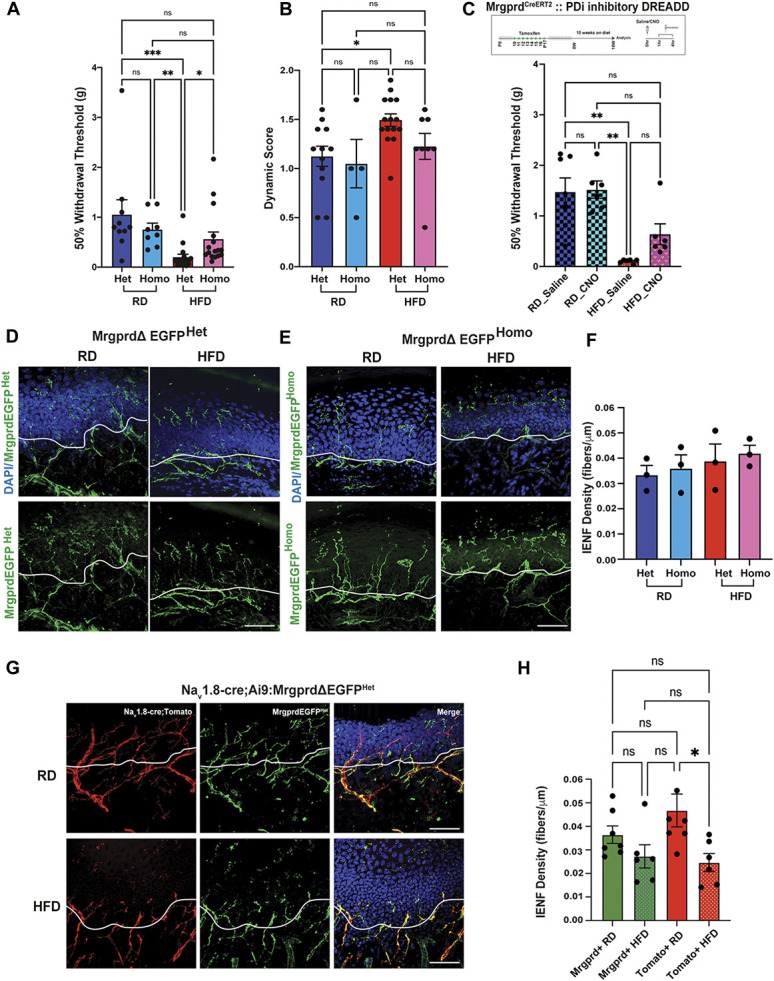
Mrgprd receptors are necessary for the establishment of static mechanical allodynia in diabetic mice. (A) von Frey testing of heterozygous (+/−) and homozygous (−/−) MrgprdΔEGFPf mice to evaluate static mechanical allodynia. Data were compared by 1-way ANOVA followed by Kruskal–Wallis test (RD-Het n = 10 animals, RD-Homo n = 8 animals, HFD-Het n = 17 animals, HFD-Homo n = 16 animals) and reported as mean ± SD. (B) Dynamic brush assay to evaluate the contribution of Mrgprd receptor to dynamic brush allodynia. Stroking was repeated for a total of 10 times for each animal and the scores were averaged. Data were compared by 1-way ANOVA followed by Kruskal–Wallis test (RD-Het n = 23 animals, RD-Homo n = 13 animals, HFD-Het n = 24 animals, HFD-Homo n = 22 animals) and reported as mean ± SD. (C) Schematic showing the generation of Mrgprd-CreERT2 crossed with hM4 DREADD mice and von Frey testing of RD and HFD Mrgprd-CreERT2;hM4Di mice to evaluate mechanical allodynia; RD saline n = 7 mice, RD CNO n = 6 mice, HFD saline n = 6 mice, HFD CNO n = 6 mice. ***P < 0.001, **P < 0.01, *P < 0.05. (D and E) Confocal micrographs of the glabrous skin of MrgprdΔEGFPf heterozygous and homozygous mice fed either an RD or HFD showing Mrgprd-expressing neuronal afferents labeled with GFP (green) and nuclear marker DAPI (blue). The white line demarcating the intraepidermal junction. (F) Quantification of the intraepidermal nerve fiber density in panels (D and E). (G) Confocal micrographs of the glabrous skin of Nav1.8-Cre;Ai9;MrgprdΔEGFPf mice fed either an RD or HFD showing Mrgprd-expressing afferents labeled with GFP (green), Nav1.8 afferents labelled with td-tomato (red), and nuclear marker DAPI (blue). The white line is demarcating the intraepidermal junction. (H) Quantification of the intraepidermal nerve fiber density in panel (G). Scale bar represents 50 μm. Data were compared using 1-way ANOVA followed by Tukey test. In all experiments (RD n = 7 animals; HFD n = 6 animals; 3 sections for each group), values are expressed as mean ± SEM, *P < 0.05. CNO, clozapine-N-oxide; HFD, high-fat diet; Mrgprd, Mas-related G protein–coupled receptor d; RD, regular diet.
To further characterize the pain phenotype, we tested for dynamic allodynia using the brush test15,21,54 in both Het and Homo mice fed either RD (RD-Het n = 23 mice; RD-Homo n = 13 mice) or HFD (HFD-Het n = 24 mice; HFD-Homo n = 22 mice) for 10 weeks. We observed that Het mice fed HFD (HFD-Het) developed dynamic allodynia at 10 weeks on diet (Fig. 4B; RD-Het 1.20 ± 0.33 v/s HFD-Het 1.45 ± 0.34 P = 0.04; RD-Homo 1.00 v/s HFD-Het P = 0.02). We observed that homozygous animals fed a HFD did not show an increase in the dynamic score compared with the HFD-Het mice (HFD-Het v/s HFD-Homo 1.20 ± 0.46 P = 0.18), demonstrating that Mrgprd receptors are necessary for the establishment of static but not brush (dynamic) mechanical allodynia in the HFD mouse model of PDN.
We next investigate the effects of modulating Mrgprd-positive DRG neuron excitability on PDN symptoms using a chemogenetic platform genetically introducing designer receptors exclusively activated by designer drug (DREADD) receptors into Mrgprd-positive DRG neurons. As previously described,37 we used an inhibitory DREADD receptor based on an engineered muscarinic acetylcholine receptor M4 (PDi), which works via activation of the inhibitory Gi/o protein pathway.4 Activation of this receptor with the small molecule agonist CNO or its metabolite clozapine inhibits neuronal activity (see Refs. 26,64,81 for review). We expressed inhibitory hM4 DREADD (PDi) receptors in Mrgprd-positive DRG neurons by crossing Mrgprd-CreERT2 mice57 with a mouse line that enables the conditional expression of DREADD receptors (hM4Di, Fig. 4C).64 Mrgprd is expressed more broadly in early development than in adulthood.47 Indeed, Liu et al. showed that tamoxifen treatment of MrgprdCreERT2 mice at early embryonic stages (E16.5-E17.5), labels Mrgprd-expressing neurons along with nonpeptidergic neurons expressing other Mrgpr genes, such as MrgprA3 and Mrgprb4.57 Aiming to express inhibitory hM4 DREADD (PDi) receptors in Mrgprd-positive nonpeptidergic nociceptors (NP1) specifically and not in nonpeptidergic neurons expressing other Mrgpr genes, such as Mrgpra3 and Mrgprb4, we have used a protocol that provided postnatal (P10-P17) tamoxifen treatment57 (Fig. 4C).
Next, we induced PDN in the Mrgprd-CreERT2; hM4Di mice by feeding them either an RD or HFD. As expected, we observed that HFD Mrgprd-CreERT2; hM4Di mice had significantly lower baseline withdrawal threshold for mechanical stimulation compared with animals on RD (Fig. 4C; RD 1.25 ± 0.38 v/s HFD 0.11 ± 0.04, P < 0.01). We then evaluated the consequences of reducing Mrgprd-positive DRG neuron excitability on mechanical allodynia in the HFD model by von Frey analysis 1 hour after a single intraperitoneal injection of either saline or CNO (10 mg/kg) (RD saline n = 7 mice, RD CN0 n = 6 mice, HFD saline n = 6 mice, HFD CNO n = 6 mice). We saw no changes in the withdrawal threshold when animals fed an RD were injected with either saline or CNO (Fig. 4C; RD-saline 1.48 ± 0.73, P = 0.58; n = 7 mice; RD-CNO 1.52 ± 0.42, P = 0.69; n = 6 mice) or when HFD animals were treated with CNO (Fig. 4C; HFD-saline 0.10 ± 0.04, P = 0.22; HFD-CNO 0.64 ± 0.51, P = 0.03). These data indicate that specifically modulating Mrgprd-positive DRG neuron (NP1) excitability via Gi-coupled GPCRs is not sufficient to reverse mechanical allodynia in the HFD mouse model of PDN.
3.4. Mas-related G protein–coupled receptor d–positive cutaneous afferents persist in diabetic mice skin
Using confocal microscopy, we examined skin innervation in both MrgprdΔEGFP heterozygous and MrgprdΔEGFP homozygous mice fed an RD (n = 3 mice) or a HFD (n = 3 mice) for 10 weeks (Figs. 4D and E). We observed that Mrgprd-positive cutaneous afferents normally crossed the intraepidermal junction and innervated the outer layer of skin even in the HFD condition, with no statistically significant difference in the intraepidermal nerve fiber (IENF) density, which is expressed as the number of nerves crossing the epidermal–dermal junction as a function of length (RD-Het 0.03 ± 0.004, HFD-Het 0.04 ± 0.007, RD-Homo 0.04 ± 0.005, HFD-homo 0.04 ± 0.003) between these group of mice (Fig. 4F). Skin innervation was normal in MrgprdΔEGFP heterozygous and MrgprdΔEGFP homozygous mice fed an RD or HFD (Figs. 4D–F), demonstrating that Mrgprd deletion did not interfere with normal neurite outgrowth. Moreover, we measured the number of nerve terminal branches in epidermis of both MrgprdΔEGFP heterozygous and homozygous mice, and we did not detect any significant difference (Suppl Fig. 3, available at http://links.lww.com/PAIN/B957, 1-way ANOVA followed by Brown–Forsythe test).
Previously, our laboratory has shown that there is a loss of the Nav1.8-positive intraepidermal nerve fibers in the HFD condition.37 We elected to use Nav1.8-Cre;Ai9;MrgprdΔEGFP heterozygous mice, in which in the same mouse strain Nav1.8-positive fibers are genetically labelled in red and Mrgprd-positive fibers in green. Using these mice, we investigated the skin innervation comparing mice fed either RD or HFD (Fig. 4G). In line with our previous results,37 we observed a significant decrease in IENF density in the Nav1.8-positive (td-tomato) afferents in the HFD condition (Nav1.8-positive (tomato) fibers RD-0.04 ± 0.007, n = 7, Nav1.8-positive (tomato)fibers HFD-0.02 ± 0.004, P < 0.022, n = 6) (Figs. 4G and H). However, the quantification of IENF density of Mrgprd-positive cutaneous afferents showed no differences between the 2 diet conditions (Mrgprd-positive fibers RD-0.04 ± 0.004, n = 7; Mrgprd-positive fibers HFD-0.03 ± 0.005, HFD n = 6) (Figs. 4G and H). Values are expressed as mean ± SEM. The quantification was done in blind by 3 independent counters. Given that these Mrgprd receptors remain in the outer layers of the skin, they become a remarkably interesting and accessible target for an antagonist that could be applied topically to the skin.
3.5. Activation of Mas-related G protein–coupled receptor d at the level of cutaneous afferents produces increased intracellular calcium concentration in dorsal root ganglion nociceptors in diabetic mice
The behavioral experiments demonstrated that Mrgprd receptors are necessary for the establishment of mechanical allodynia in the HFD mouse model of PDN. Using this mouse model, previous work in our laboratory37 had shown that DRG neurons become hyperexcitable. Moreover, reducing DRG nociceptor hyperexcitability with DREADD receptor technology, prevented and reversed neuropathic pain and neuronal calcium overload.37 Mrgprd is an excitatory receptor; therefore, we hypothesized that MrgprD signaling contributes to DRG nociceptor hyperexcitability underlying mechanical allodynia in the HFD model of PDN. To test this hypothesis, we used in vivo calcium imaging to assess nociceptor hyperexcitability by measuring changes in internal calcium concentration [Ca2+]i in these neurons. Given the cellular diversity and functional heterogeneity of DRG neurons,16,28,45,82 we selectively monitored [Ca2+]i in vivo in Nav1.8-positive DRG nociceptors by expressing the [Ca2+]i indicator protein GCaMP612 in these neurons (Nav1.8-Cre mice77 crossed with conditional reporter GCaMP6 mice; Ai96flox/flox;RCL-GCaMP6s12). Mrgprd is expressed in a subpopulation of Nav1.8-positive DRG neurons16,28,45,68,82,87 and, at the transcriptional level, we found no detectable differences in the expression of Nav1.8 within either of the 2 Mrgprd expressing clusters. Nav1.8-Cre;Ai96flox/flox;RCL-GCaMP6s mice were then fed either an RD or HFD for 10 weeks. Laminectomy was performed on anesthetized mice to expose the fourth lumbar (L4) DRG, which contains sensory neurons that innervate the paw.52
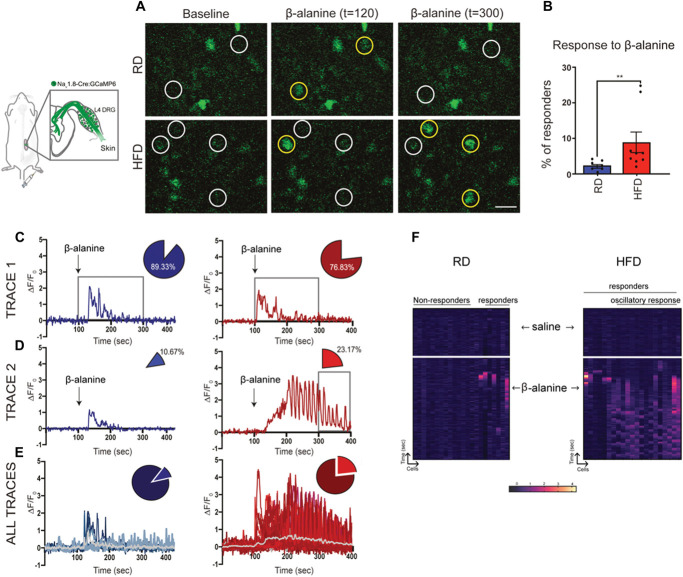
The in vivo physiological setup allowed us to examine DRG neuronal calcium signaling in real-time in response to paw manipulation with mechanical stimuli or application of drugs. Given that Mrgprd-positive fibers are remaining in the skin even in HFD condition (Figs. 4D–H), our calcium in vivo set up is ideal to check effect of activation of excitatory Mrgprd receptors expressed in these cutaneous afferents in the periphery. To functionally determine changes in Mrgprd expression, we used β-alanine, a known Mrgprd agonist,71 injected into the paw of mice fed either RD or HFD while monitoring calcium transients in vivo in the DRGs (RD: n = 7 mice; HFD: n = 9 mice; average number of neurons within the DRG imaged per animal RD = 197.5 ± 48.67; HFD 242.5 ± 69.16). The number and percentage of responding neurons along with the intensity of these responses were assessed with both vehicle saline injections (data not shown) and with β-alanine; representative images are shown in Figure 5A (magnified inset of the whole imaged DRG). Although neurons from both RD and HFD mice responded to β-alanine injections into the paw, we found an increase in the percentage of neurons that responded to the stimulus in HFD mice (Fig. 5B, RD 2.39 ± 0.51; HFD 8.88 ± 2.88, P = 0.008, white circles indicate neurons, and the yellow circles identify neurons that responded to the stimulus at a given time).
Figure 5.
Increased percentage of in vivo calcium responses to β-alanine in Nav1.8-expressing DRG neurons from mice fed a HFD. (A) Representative magnified images of whole DRG taken from RD (top) and HFD (bottom) mice showing neurons identified by white circles at baseline and during administration of β-alanine. A neuron that responds to the stimulus is identified by a yellow circle at t = 120 seconds and at t = 300 seconds. (B) Percentage of neurons responding to β-alanine; each black circle represents one animal. (C) Trace from a single neuron in RD (blue) and HFD (red) indicating change in fluorescence intensity (ΔF/F0) over time in seconds (sec). (D) Trace from a single neuron from RD and HFD in response to β-alanine injection. (C and D) Arrow indicates the addition of β-alanine injection at 100 seconds. The box shows the duration of response to β-alanine in both RD and HFD. The percentage of neurons responding within this duration is indicated by the pie chart. The percentage of neurons in the HFD continued to respond even after 300 seconds (HFD 23.17%; P = 0.021 vs RD 10.67%). (E) All responses from representative RD and HFD mice. The gray line indicates the average change in fluorescence intensity over time. (F) Heat map of 20 neurons shown as a function of time and their response to saline and β-alanine injections (arrows indicate the time of injection). Percentage responders were compared using a 2-tailed unpaired t test. Data reported as mean ± SEM. RD, n = 7 mice; HFD, n = 9 mice (average number of neurons within the DRG imaged per animal RD = 197.5 ± 48.67; HFD 242.5 ± 69.16). **P < 0.01. DRG, dorsal root ganglion; HFD, high-fat diet; RD, regular diet.
With our in vivo calcium imaging set up we were able to show that, among the DRG neurons taken from the HFD group, many responses were similar in amplitude to those seen in RD (Figs. 5C and E; RD 1.56 ± 0.14, HFD 1.78 ± 0.08, P = 0.19; RD, n = 7 mice; HFD, n = 9 mice; average number of neurons within the DRG imaged per animal: RD = 197.5 ± 48.67; HFD 242.5 ± 69.16). However, there was also a Mrgprd-expressing subpopulation of neurons from the HFD group in which activation of Mrgprd receptors with β-alanine injection in the paw produced marked long-lasting oscillatory calcium responses, something that was rarely observed in the RD group (Fig. 5D). Neurons in the RD group started to respond within 20 seconds after administration of β-alanine (t = 0-99 seconds of baseline reading, β-alanine injected at t = 100 seconds), and most of the calcium peaks were no longer detected beyond 300 seconds (Fig. 5D; RD 10.67%). Similarly, neurons in the HFD started to respond 20 seconds after the administration of β-alanine, but, interestingly, when compared with the RD group, about double the percentage of neurons in the HFD continued to respond even after 300 seconds (Fig. 5D; HFD 23.17% vs RD 10.67%, Fisher exact test, P = 0.021). Figure 5E shows all the responses from an individual animal as an illustration of the oscillatory calcium responses detected in the HFD group (gray trace indicates the average response).
The heat map visualization enables us to appreciate this oscillatory behavior of neurons in the HFD (Fig. 5F, 20 neurons from a representative animal fed an RD or HFD). So, while a population of β-alanine responsive neurons in the HFD responded with calcium transients similar in amplitude and duration to the neurons in the RD (Figs. 5C–F), a percentage of β-alanine–responsive neurons had a different profile, with oscillatory waves (Figs. 5C–F, RD = 3.17 ± 3.17, HFD = 17.99 ± 6.81, P = 0.07) and longer duration of response. Focusing on the Nav1.8-positive DRG neurons responding to β-alanine, we further determined their response to capsaicin (Suppl Fig. 4A-C, available at http://links.lww.com/PAIN/B957) and found no statistically significant differences when comparing RD and HFD groups. Taken together, these results indicate that activation of Mrgprd-positive cutaneous afferents that persist in diabetic mice skin produced DRG neuron hyperexcitability in diabetic mice.
4. Discussion
Our study demonstrated that excitatory Mrgprd receptors play a key role in the generation and maintenance of mechanical allodynia in PDN. Using single-cell RNA sequencing, we demonstrated increased Mrgprd expression in a subpopulation of nonpeptidergic DRG neurons in the HFD mouse model of PDN. Importantly, limiting Mrgprd signaling reversed static mechanical allodynia in the HFD mouse model of PDN demonstrating that Mrgprd receptors are necessary for the establishment of mechanical allodynia in this model of PDN. Furthermore, using in vivo calcium imaging, we demonstrated that activation of Mrgprd-positive cutaneous afferents that persist in diabetic mice produced increased intracellular calcium concentration in DRG neurons. Taken together, our data highlight a key role of Mrgprd-mediated DRG neuron excitability in the generation and maintenance of mechanical allodynia in a mouse model of PDN. Hence, we propose Mrgprd as a promising and accessible target for developing effective therapeutics currently unavailable for treating PDN.
Mrgprd is a promising therapeutic target. First, Mrgprd is a G protein–coupled receptor that has often been used as a druggable target.32,76,86 Second, Mrgprd is known to influence DRG neuron excitability in response to mechanical stimuli.57,60,84 Indeed, the role of Mrgprd both in mediating nonhistaminic itch and the excitability of polymodal nonpeptidergic nociceptors to mechanical in rodents has been clearly delineated.62 Similarly, in humans, the Mrgprd agonist β-alanine, generally used as supplement, can cause paresthesia and itch as side effect.18,46 Moreover, stimulation of Mrgprd receptor with intradermal β-alanine injection in healthy volunteers elicited not only itch but also burning and stinging sensation.46 Finally, Mrgprd is expressed by the nociceptive neuronal population that extends out into the outermost layer of the skin,62,89 making it a very accessible therapeutic target for PDN. From a translational perspective, an ideal therapy for PDN might involve Mrgprd-modifying drugs applied topically. Indeed, topical applications for treating PDN are very appealing as they should bypass drug side effects associated with systemic interventions. However, one limitation of our study is that we have used a genetic approach to demonstrate the role of Mrgprd in mediating neuropathic pain in the HFD mouse model of PDN as drugs for doing this are unavailable. The pharmacology of Mrgprd has not been extensively investigated, although an inverse agonist MU-684080 has been reported in the literature that might be an ideal candidate for examining models of PDN pain. Several peptides and small molecules have been proposed as ligands for Mrgprs.5,42,73 However, many of these ligands interact with multiple receptor types.39 Therefore, future screening for molecule Mrgprd antagonist or inverse agonist activity has to be performed using high-throughput electrophysiology or calcium image studies using human DRG neurons or nociceptors derived from human-induced pluripotent stem cells (iPSCs) if they are to be successfully translated to humans.65
Several groups have performed scRNA-seq of rodent DRG,16,28,45,68,82,87 facilitating their molecular characterization, allowing clustering of DRG neurons into distinct subtypes16,28,45,68,82,87 and delineating their developmental lineages.68 With this approach, we can now begin to describe gene expression changes within populations that accompany disease states. Indeed, scRNA profiling of DRG neurons has been performed in rodents and primates41 with chronic pain. Recently, single-cell/single-nucleus RNA sequencing has begun to reveal transcriptomic perturbations in DRG neurons after nerve injury.36,66 An increase in transcription of genes associated with cell death and alterations in pathways related to neuropathic pain was observed in the nonpeptidergic (NP) neuronal population. This heterogenous injury-induced transcriptional alteration within neuronal subtypes suggests that there are intrinsic differences in the genetic response to injury between subtypes of DRG neurons. Interestingly, a cluster expressing Atf3/Mrgprd appeared after 24 hours and transcriptomic changes within this cluster led to changes in neuronal phenotype within 2 days after injury,36 highlighting that distinct neuron types respond differently to injury and that injured Mrgprd-expressing neurons have particularly high reprogramming capabilities.
Characterization of DRG gene expression profiles has been extensively studied through bulk RNA sequencing in rodent models of PDN24,29,85 and from a limited number of donor control and with painful diabetic neuropathy31; however, to our knowledge, this is the first study using DRG single-cell RNA sequencing in a model of PDN. In the present study, we used single-cell RNA sequencing to demonstrate increased Mrgprd expression in a subpopulation of nonpeptidergic DRG neurons in the HFD mouse model of PDN. Interestingly in another clinically relevant rodent model of neuropathic pain, the chemotherapy-induced neuropathic pain caused by paclitaxel, Mrgprd mRNA translation was found to be significantly increased selectively in nociceptors.49 Our study also demonstrates that Mrgprd is significantly expressed in nonpeptidergic type 2 (NP2) DRG neurons in diabetic DRGs. The role of NP1 Mrgprd-expressing DRG neurons in mechanical nociception and to detect light punctate mechanical stimuli is well established in mice.11,40,46,62,89 The aberrant expression of Mrgprd receptors in NP2 subpopulation that in mice is dedicated to itch33 could have a role in mediating static mechanical allodynia in the HFD model of PDN. Understanding precisely which cell type mediates mechanical allodynia in PDN is of fundamental importance to our basic understanding of somatosensation and may provide an important way forward for identifying therapeutic targets in this disease. However, our study does not deconvolute the nature of DRG neurons subtypes expressing Mrgprd that are specifically associated with the occurrence of mechanical allodynia in our mouse model of PDN. Future studies designed to activate or silence selectively the NP1 or the NP2 subtype of nonpeptidergic neurons using specific markers for NP2 such as MrgprA3 and regents available including the MrgprA3GFP-cre mice33 combined with behavioral assessments are necessary to address these questions.
Numerous single-cell RNA-seq studies of human DRGs have demonstrated several significant molecular differences between rodent and human DRG neuron subtypes.55,59,63,70,78 Therefore, it is important to validate the molecular mechanisms underlying neuropathic pain discovered in mice using human tissue if they are to be successfully translated to humans.65 Using RNA scope, we confirmed MRGPRD expression in human DRGs from donor control and PDN donors (Suppl Fig. 1, available at http://links.lww.com/PAIN/B957). In this study, the power of this observation is limited by the extremely low number of patients; however, other studies have already demonstrated expression of MRGPRD in human DRG.63 Further transcriptomic studies in a large cohort of donors with clinically characterized PDN are needed to explore specific gene expression profile in diverse sensory phenotypes.
One of the hallmarks of PDN is small-fiber degeneration,19,43 particularly a “dying back” axonopathy that affects DRG nociceptor axons.44,74 Previously our laboratory has shown that there is a loss of the Nav1.8-positive intraepidermal nerve fibers in the HFD condition.37 Here we demonstrated that Mrgprd-positive cutaneous afferents normally cross the intraepidermal junction and innervate the outer layer of skin even in the HFD condition. It is possible that these cutaneous Mrgprd expressing afferents could be responsible for carrying mechanical allodynia in this model. Indeed, in the HFD model, we observed an increase in the percentage of neurons responding with calcium transients upon activation of Mrgprd receptors expressed in cutaneous nerve terminals by its known agonist β-alanine injected into the skin. Given that these Mrgprd receptor–expressing neurons remain in the outer layers of the skin, they represent an interesting and accessible target for an antagonist/inverse agonist that could be applied topically. However, one limitation of our study is that we were not able to establish if Mrgprd-positive afferents innervating the outermost layer of the skin even in the diabetic condition are nerves that are spared by degeneration or represent newly regenerating fibers. Several studies in rodent models of PDN13 and in patients with PDN have now demonstrated an increased regeneration of intraepidermal nerve fibers (IENFs).8,14 Indeed, increased regeneration of IENFs was able to distinguish between patients with painful and nonpainful diabetic neuropathy.14 Another study reports that increased regenerating IENFs were distinctly associated with ongoing burning pain in patients with diabetes.22 Recently, Kuner et al. have elegantly revealed the emergence of a form of chronic neuropathic pain that is driven by structural plasticity, abnormal terminal connectivity, and malfunction of nociceptors during reinnervation.23 Similar mechanisms could be at play in PDN. To answer this intriguing question, mouse genetic approaches could be applied in future studies. Indeed, we are currently exploring the status of Mrgprd-positive afferent and their regenerative properties in the skin of rodent models of PDN and in clinically well-characterized patients with PDN to enhance the translational validity of Mrgprd as a therapeutic target.
Regardless, our studies introduce the novel suggestion that Mrgprd plays a key role in the generation and maintenance of mechanical allodynia in PDN. Hence, we propose Mrgprd as promising accessible targets for developing effective therapeutics currently not available to treat neuropathic pain in diabetic neuropathy. Thus, these results have the potential for transforming the way neuropathic pain in diabetic neuropathy is treated, replacing the largely ineffective approaches that are currently available for patients afflicted with PDN.9 Furthermore, as demonstrated here, an unbiased approach combining single-cell transcriptomics and in vivo calcium imaging is a useful approach for revealing interesting targets that could be translated to produce more effective, disease-modifying therapies for patients suffering from PDN.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Appendix A. Supplemental digital content
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B957.
Supplementary Material
Acknowledgements
This work was supported by NIH R01 NS104295-01 and NIH HEAL initiative supplement R01 NS104295-01 (D.M.M.), NIH R01 AR077691-01 (D.M.M., R.J.M.), NIH R01AR064251 (R.J.M., A.M.M.), NIH P30AR079206 (A.M.M.), and NIH R01AR077019 (R.E.M.). The authors thank Dr. Rajeshwar Awatramani for helpful discussions. The authors thank the Northwestern University Sequencing Core Center for their assistance with the single-cell RNA sequencing (NIH S10OD025120).
Authors' contributions: D.S.G. performed single-cell RNA sequencing and analysis, in vivo calcium analysis, RNAscope in situ hybridization, pain behavioral studies, statistical analysis, figures, and manuscript revision. N.D.J. performed von Frey behavioral studies, immunohistochemical labeling, and statistical analysis, mouse breeding, administration of HFD, testing for diabetes, RNAscope in situ hybridization, evaluation by immunofluorescence of intraepidermal nerve fiber (IENF) density, figures, and manuscript revision. P.P. performed evaluation by immunofluorescence of intraepidermal nerve fiber (IENF) density, figures, analysis of RNAscope in situ hybridization, and manuscript revision. D.R. performed mouse breeding, administration of HFD, testing for diabetes and in vivo calcium surgery, and experimentation. R.E.M. set up and performed in vivo calcium imaging. A.M.M. aided in the analysis of the in vivo calcium imaging. D.M.M. supervised the project. D.M.M. and D.S.G. drafted the manuscript. N.S. aided in the analysis of the intraepidermal nerve fiber (IENF) density. D.M.M. and R.J.M. edited the manuscript. All authors read and approved the manuscript. All authors declare that they have no conflicting interests.
Data availability: The data that support the findings of this study are openly available as raw matrix files for single-cell RNA sequencing deposited in Dryad (https://doi.org/10.5061/dryad.9s4mw6mm5).
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.painjournalonline.com).
Contributor Information
Dale S. George, Email: dalesusan1@gmail.com.
Nirupa D. Jayaraj, Email: njayaraj@northwestern.edu.
Paola Pacifico, Email: paola.pacifico@northwestern.edu.
Dongjun Ren, Email: d-ren@northwestern.edu.
Nikhil Sriram, Email: nikhilsriram2023@u.northwestern.edu.
Rachel E. Miller, Email: rachel_miller@rush.edu.
Anne-Marie Malfait, Email: annemarie@annemariemalfait.com.
Richard J. Miller, Email: r-miller10@northwestern.edu.
References
- [1].Abraira VE, Ginty DD. The sensory neurons of touch. Neuron 2013;79:618–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Alam U, Sloan G, Tesfaye S. Treating pain in diabetic neuropathy: current and developmental drugs. Drugs 2020;80:363–84. [DOI] [PubMed] [Google Scholar]
- [3].Andersson DA, Gentry C, Light E, Vastani N, Vallortigara J, Bierhaus A, Fleming T, Bevan S. Methylglyoxal evokes pain by stimulating TRPA1. PLoS One 2013;8:e77986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci USA 2007;104:5163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bader M, Alenina N, Andrade-Navarro MA, Santos RA. MAS and its related G protein-coupled receptors, Mrgprs. Pharmacol Rev 2014;66:1080–105. [DOI] [PubMed] [Google Scholar]
- [6].Bhangoo SK, Ren D, Miller RJ, Chan DM, Ripsch MS, Weiss C, McGinnis C, White FA. CXCR4 chemokine receptor signaling mediates pain hypersensitivity in association with antiretroviral toxic neuropathy. Brain Behav Immun 2007;21:581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bierhaus A, Fleming T, Stoyanov S, Leffler A, Babes A, Neacsu C, Sauer SK, Eberhardt M, Schnölzer M, Lasitschka F, Neuhuber WL, Kichko TI, Konrade I, Elvert R, Mier W, Pirags V, Lukic IK, Morcos M, Dehmer T, Rabbani N, Thornalley PJ, Edelstein D, Nau C, Forbes J, Humpert PM, Schwaninger M, Ziegler D, Stern DM, Cooper ME, Haberkorn U, Brownlee M, Reeh PW, Nawroth PP. Methylglyoxal modification of Nav1.8 facilitates nociceptive neuron firing and causes hyperalgesia in diabetic neuropathy. Nat Med 2012;18:926–33. [DOI] [PubMed] [Google Scholar]
- [8].Bönhof GJ, Strom A, Püttgen S, Ringel B, Brüggemann J, Bódis K, Müssig K, Szendroedi J, Roden M, Ziegler D. Patterns of cutaneous nerve fibre loss and regeneration in type 2 diabetes with painful and painless polyneuropathy. Diabetologia 2017;60:2495–503. [DOI] [PubMed] [Google Scholar]
- [9].Bril V, England J, Franklin GM, Backonja M, Cohen J, Del Toro D, Feldman E, Iverson DJ, Perkins B, Russell JW, Zochodne D; American Academy of Neurology; American Association of Neuromuscular and Electrodiagnostic Medicine; American Academy of Physical Medicine and Rehabilitation. Evidence-based guideline: treatment of painful diabetic neuropathy: report of the American Academy of Neurology, the American Association of Neuromuscular and Electrodiagnostic Medicine, and the American Academy of Physical Medicine and Rehabilitation. Neurology 2011;76:1758–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cao DL, Zhang ZJ, Xie RG, Jiang BC, Ji RR, Gao YJ. Chemokine CXCL1 enhances inflammatory pain and increases NMDA receptor activity and COX-2 expression in spinal cord neurons via activation of CXCR2. Exp Neurol 2014;261:328–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cavanaugh DJ, Lee H, Lo L, Shields SD, Zylka MJ, Basbaum AI, Anderson DJ. Distinct subsets of unmyelinated primary sensory fibers mediate behavioral responses to noxious thermal and mechanical stimuli. Proc Natl Acad Sci U S A 2009;106:9075–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013;499:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cheng HT, Dauch JR, Hayes JM, Yanik BM, Feldman EL. Nerve growth factor/p38 signaling increases intraepidermal nerve fiber densities in painful neuropathy of type 2 diabetes. Neurobiol Dis 2012;45:280–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cheng HT, Dauch JR, Porzio MT, Yanik BM, Hsieh W, Smith AG, Singleton JR, Feldman EL. Increased axonal regeneration and swellings in intraepidermal nerve fibers characterize painful phenotypes of diabetic neuropathy. J Pain 2013;14:941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cheng L, Duan B, Huang T, Zhang Y, Chen Y, Britz O, Garcia-Campmany L, Ren X, Vong L, Lowell BB, Goulding M, Wang Y, Ma Q. Identification of spinal circuits involved in touch-evoked dynamic mechanical pain. Nat Neurosci 2017;20:804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chiu IM, Barrett LB, Williams EK, Strochlic DE, Lee S, Weyer AD, Lou S, Bryman GS, Roberson DP, Ghasemlou N, Piccoli C, Ahat E, Wang V, Cobos EJ, Stucky CL, Ma Q, Liberles SD, Woolf CJ. Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. eLife 2014;3:e04660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].daCosta DiBonaventura M, Cappelleri JC, Joshi AV. A longitudinal assessment of painful diabetic peripheral neuropathy on health status, productivity, and health care utilization and cost. Pain Med 2011;12:118–26. [DOI] [PubMed] [Google Scholar]
- [18].Décombaz J, Beaumont M, Vuichoud J, Bouisset F, Stellingwerff T. Effect of slow-release beta-alanine tablets on absorption kinetics and paresthesia. Amino Acids 2012;43:67–76. [DOI] [PubMed] [Google Scholar]
- [19].Divisova S, Vlckova E, Srotova I, Kincova S, Skorna M, Dusek L, Dubovy P, Bednarik J. Intraepidermal nerve-fibre density as a biomarker of the course of neuropathy in patients with type 2 diabetes mellitus. Diabetic Med 2016;33:650–4. [DOI] [PubMed] [Google Scholar]
- [20].Dong X, Han S, Zylka MJ, Simon MI, Anderson DJ. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell 2001;106:619–32. [DOI] [PubMed] [Google Scholar]
- [21].Duan B, Cheng L, Bourane S, Britz O, Padilla C, Garcia-Campmany L, Krashes M, Knowlton W, Velasquez T, Ren X, Ross S, Lowell BB, Wang Y, Goulding M, Ma Q. Identification of spinal circuits transmitting and gating mechanical pain. Cell 2014;159:1417–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Galosi E, La Cesa S, Di Stefano G, Karlsson P, Fasolino A, Leone C, Biasiotta A, Cruccu G, Truini A. A pain in the skin. Regenerating nerve sprouts are distinctly associated with ongoing burning pain in patients with diabetes. Eur J Pain 2018;22:1727–34. [DOI] [PubMed] [Google Scholar]
- [23].Gangadharan V, Zheng H, Taberner FJ, Landry J, Nees TA, Pistolic J, Agarwal N, Männich D, Benes V, Helmstaedter M, Ommer B, Lechner SG, Kuner T, Kuner R. Neuropathic pain caused by miswiring and abnormal end organ targeting. Nature 2022;606:137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gao Z, Feng Y, Ju H. The different dynamic changes of nerve growth factor in the dorsal horn and dorsal root ganglion leads to hyperalgesia and allodynia in diabetic neuropathic pain. Pain Physician 2017;20:E551–61. [PubMed] [Google Scholar]
- [25].George DS, Hackelberg S, Jayaraj ND, Ren D, Edassery SL, Rathwell C, Miller RE, Malfait AM, Savas JN, Miller RJ, Menichella DM. Mitochondrial calcium uniporter deletion prevents painful diabetic neuropathy by restoring mitochondrial morphology and dynamics. PAIN 2022;163:560–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gomez JL, Bonaventura J, Lesniak W, Mathews WB, Sysa-Shah P, Rodriguez LA, Ellis RJ, Richie CT, Harvey BK, Dannals RF, Pomper MG, Bonci A, Michaelides M. Chemogenetics revealed: DREADD occupancy and activation via converted clozapine. Science 2017;357:503–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gosselin RD, Dansereau MA, Pohl M, Kitabgi P, Beaudet N, Sarret P, Mélik Parsadaniantz S. Chemokine network in the nervous system: a new target for pain relief. Curr Med Chem 2008;15:2866–75. [DOI] [PubMed] [Google Scholar]
- [28].Goswami SC, Mishra SK, Maric D, Kaszas K, Gonnella GL, Clokie SJ, Kominsky HD, Gross JR, Keller JM, Mannes AJ, Hoon MA, Iadarola MJ. Molecular signatures of mouse TRPV1-lineage neurons revealed by RNA-Seq transcriptome analysis. J Pain 2014;15:1338–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Guo K, Eid SA, Elzinga SE, Pacut C, Feldman EL, Hur J. Genome-wide profiling of DNA methylation and gene expression identifies candidate genes for human diabetic neuropathy. Clin Epigenetics 2020;12:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gylfadottir SS, Christensen DH, Nicolaisen SK, Andersen H, Callaghan BC, Itani M, Khan KS, Kristensen AG, Nielsen JS, Sindrup SH, Andersen NT, Jensen TS, Thomsen RW, Finnerup NB. Diabetic polyneuropathy and pain, prevalence, and patient characteristics: a cross-sectional questionnaire study of 5,514 patients with recently diagnosed type 2 diabetes. PAIN 2020;161:574–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hall BE, Macdonald E, Cassidy M, Yun S, Sapio MR, Ray P, Doty M, Nara P, Burton MD, Shiers S, Ray-Chaudhury A, Mannes AJ, Price TJ, Iadarola MJ, Kulkarni AB. Transcriptomic analysis of human sensory neurons in painful diabetic neuropathy reveals inflammation and neuronal loss. Sci Rep 2022;12:4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Halls ML, Davenport AP, Summers RJ. Editorial: recent advances in G protein-coupled receptor signalling: impact of intracellular location, environment and biased agonism. Front Pharmacol 2021;12:707393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Han L, Ma C, Liu Q, Weng HJ, Cui Y, Tang Z, Kim Y, Nie H, Qu L, Patel KN, Li Z, McNeil B, He S, Guan Y, Xiao B, Lamotte RH, Dong X. A subpopulation of nociceptors specifically linked to itch. Nat Neurosci 2013;16:174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Handler A, Ginty DD. The mechanosensory neurons of touch and their mechanisms of activation. Nat Rev Neurosci 2021;22:521–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hao Y, Hao S, Andersen-Nissen E, Mauck WM, III, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hu G, Huang K, Hu Y, Du G, Xue Z, Zhu X, Fan G. Single-cell RNA-seq reveals distinct injury responses in different types of DRG sensory neurons. Sci Rep 2016;6:31851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jayaraj ND, Bhattacharyya BJ, Belmadani AA, Ren D, Rathwell CA, Hackelberg S, Hopkins BE, Gupta HR, Miller RJ, Menichella DM. Reducing CXCR4-mediated nociceptor hyperexcitability reverses painful diabetic neuropathy. J Clin Invest 2018;128:2205–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ji RR, Xu ZZ, Gao YJ. Emerging targets in neuroinflammation-driven chronic pain. Nat Rev Drug Discov 2014;13:533–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Klein A, Solinski HJ, Malewicz NM, Ieong HF, Sypek EI, Shimada SG, Hartke TV, Wooten M, Wu G, Dong X, Hoon MA, LaMotte RH, Ringkamp M. Pruriception and neuronal coding in nociceptor subtypes in human and nonhuman primates. Elife 2021;10:e64506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kupari J, Ernfors P. Molecular taxonomy of nociceptors and pruriceptors. PAIN 2023;164:1245–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kupari J, Usoskin D, Parisien M, Lou D, Hu Y, Fatt M, Lönnerberg P, Spångberg M, Eriksson B, Barkas N, Kharchenko PV, Loré K, Khoury S, Diatchenko L, Ernfors P. Single cell transcriptomics of primate sensory neurons identifies cell types associated with chronic pain. Nat Commun 2021;12:1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lansu K, Karpiak J, Liu J, Huang XP, McCorvy JD, Kroeze WK, Che T, Nagase H, Carroll FI, Jin J, Shoichet BK, Roth BL. In silico design of novel probes for the atypical opioid receptor MRGPRX2. Nat Chem Biol 2017;13:529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Latremoliere A, Woolf CJ. Central sensitization: a generator of pain hypersensitivity by central neural plasticity. J Pain 2009;10:895–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lauria G, Devigili G. Skin biopsy as a diagnostic tool in peripheral neuropathy. Nat Clin Pract Neurol 2007;3:546–57. [DOI] [PubMed] [Google Scholar]
- [45].Li CL, Li KC, Wu D, Chen Y, Luo H, Zhao JR, Wang SS, Sun MM, Lu YJ, Zhong YQ, Hu XY, Hou R, Zhou BB, Bao L, Xiao HS, Zhang X. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res 2016;26:967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liu Q, Sikand P, Ma C, Tang Z, Han L, Li Z, Sun S, LaMotte RH, Dong X. Mechanisms of itch evoked by beta-alanine. J Neurosci 2012;32:14532–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liu Y, Yang FC, Okuda T, Dong X, Zylka MJ, Chen CL, Anderson DJ, Kuner R, Ma Q. Mechanisms of compartmentalized expression of Mrg class G-protein-coupled sensory receptors. J Neurosci 2008;28:125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ma W, Quirion R. Inflammatory mediators modulating the transient receptor potential vanilloid 1 receptor: therapeutic targets to treat inflammatory and neuropathic pain. Expert Opin Ther Targets 2007;11:307–20. [DOI] [PubMed] [Google Scholar]
- [49].Megat S, Ray PR, Moy JK, Lou TF, Barragán-Iglesias P, Li Y, Pradhan G, Wanghzou A, Ahmad A, Burton MD, North RY, Dougherty PM, Khoutorsky A, Sonenberg N, Webster KR, Dussor G, Campbell ZT, Price TJ. Nociceptor translational profiling reveals the ragulator-rag GTPase complex as a critical generator of neuropathic pain. J Neurosci 2019;39:393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Meltzer S, Santiago C, Sharma N, Ginty DD. The cellular and molecular basis of somatosensory neuron development. Neuron 2021;109:3736–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Menichella DM, Abdelhak B, Ren D, Shum A, Frietag C, Miller RJ. CXCR4 chemokine receptor signaling mediates pain in diabetic neuropathy. Mol Pain 2014;10:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Miller RE, Kim YS, Tran PB, Ishihara S, Dong X, Miller RJ, Malfait AM. Visualization of peripheral neuron sensitization in a surgical mouse model of osteoarthritis by in vivo calcium imaging. Arthritis Rheumatol 2018;70:88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Miller RJ, Jung H, Bhangoo SK, White FA. Cytokine and chemokine regulation of sensory neuron function. Handb Exp Pharmacol 2009;194:417–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Murthy SE, Loud MC, Daou I, Marshall KL, Schwaller F, Kühnemund J, Francisco AG, Keenan WT, Dubin AE, Lewin GR, Patapoutian A. The mechanosensitive ion channel Piezo2 mediates sensitivity to mechanical pain in mice. Sci Transl Med 2018;10:eaat9897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Nguyen MQ, von Buchholtz LJ, Reker AN, Ryba NJ, Davidson S. Single-nucleus transcriptomic analysis of human dorsal root ganglion neurons. Elife 2021;10:e71752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Obrosova IG, Ilnytska O, Lyzogubov VV, Pavlov IA, Mashtalir N, Nadler JL, Drel VR. High-fat diet induced neuropathy of pre-diabetes and obesity: effects of “healthy” diet and aldose reductase inhibition. Diabetes 2007;56:2598–608. [DOI] [PubMed] [Google Scholar]
- [57].Olson W, Abdus-Saboor I, Cui L, Burdge J, Raabe T, Ma M, Luo W. Sparse genetic tracing reveals regionally specific functional organization of mammalian nociceptors. Elife 2017;6:e29507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ørstavik K, Namer B, Schmidt R, Schmelz M, Hilliges M, Weidner C, Carr RW, Handwerker H, Jørum E, Torebjörk HE. Abnormal function of C-fibers in patients with diabetic neuropathy. J Neurosci 2006;26:11287–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Price T, Torck A, Ray P, Hassler S, Hoffman J, Boitano S, Dussor G, Vagner J. RNA-seq based transcriptome profiling of human and mouse dorsal root ganglion reveals a potential role for Protease Activated Receptor 3 (PAR3) in pain processing. FASEB J 2016;30. doi: 10.1096/fasebj.30.1_supplement.710.6 [DOI] [Google Scholar]
- [60].Qu L, Fan N, Ma C, Wang T, Han L, Fu K, Wang Y, Shimada SG, Dong X, LaMotte RH. Enhanced excitability of MRGPRA3- and MRGPRD-positive nociceptors in a model of inflammatory itch and pain. Brain 2014;137:1039–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Quilici S, Chancellor J, Löthgren M, Simon D, Said G, Le TK, Garcia-Cebrian A, Monz B. Meta-analysis of duloxetine vs. pregabalin and gabapentin in the treatment of diabetic peripheral neuropathic pain. BMC Neurol 2009;9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rau KK, McIlwrath SL, Wang H, Lawson JJ, Jankowski MP, Zylka MJ, Anderson DJ, Koerber HR. Mrgprd enhances excitability in specific populations of cutaneous murine polymodal nociceptors. J Neurosci 2009;29:8612–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ray P, Torck A, Quigley L, Wangzhou A, Neiman M, Rao C, Lam T, Kim JY, Kim TH, Zhang MQ, Dussor G, Price TJ. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. PAIN 2018;159:1325–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ray RS, Corcoran AE, Brust RD, Kim JC, Richerson GB, Nattie E, Dymecki SM. Impaired respiratory and body temperature control upon acute serotonergic neuron inhibition. Science 2011;333:637–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Renthal W, Chamessian A, Curatolo M, Davidson S, Burton M, Dib-Hajj S, Dougherty PM, Ebert AD, Gereau RW, IV, Ghetti A, Gold MS, Hoben G, Menichella DM, Mercier P, Ray WZ, Salvemini D, Seal RP, Waxman S, Woolf CJ, Stucky CL, Price TJ. Human cells and networks of pain: transforming pain target identification and therapeutic development. Neuron 2021;109:1426–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Renthal W, Tochitsky I, Yang L, Cheng YC, Li E, Kawaguchi R, Geschwind DH, Woolf CJ. Transcriptional reprogramming of distinct peripheral sensory neuron subtypes after axonal injury. Neuron 2020;108:128–44.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Serra J, Duan WR, Locke C, Solà R, Liu W, Nothaft W. Effects of a T-type calcium channel blocker, ABT-639, on spontaneous activity in C-nociceptors in patients with painful diabetic neuropathy: a randomized controlled trial. PAIN 2015;156:2175–83. [DOI] [PubMed] [Google Scholar]
- [68].Sharma N, Flaherty K, Lezgiyeva K, Wagner DE, Klein AM, Ginty DD. The emergence of transcriptional identity in somatosensory neurons. Nature 2020;577:392–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Shields SD, Ahn HS, Yang Y, Han C, Seal RP, Wood JN, Waxman SG, Dib-Hajj SD. Nav1.8 expression is not restricted to nociceptors in mouse peripheral nervous system. PAIN 2012;153:2017–30. [DOI] [PubMed] [Google Scholar]
- [70].Shiers S, Klein RM, Price TJ. Quantitative differences in neuronal subpopulations between mouse and human dorsal root ganglia demonstrated with RNAscope in situ hybridization. PAIN 2020;161:2410–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Shinohara T, Harada M, Ogi K, Maruyama M, Fujii R, Tanaka H, Fukusumi S, Komatsu H, Hosoya M, Noguchi Y, Watanabe T, Moriya T, Itoh Y, Hinuma S. Identification of a G protein-coupled receptor specifically responsive to beta-alanine. J Biol Chem 2004;279:23559–64. [DOI] [PubMed] [Google Scholar]
- [72].Shun CT, Chang YC, Wu HP, Hsieh SC, Lin WM, Lin YH, Tai TY, Hsieh ST. Skin denervation in type 2 diabetes: correlations with diabetic duration and functional impairments. Brain 2004;127:1593–605. [DOI] [PubMed] [Google Scholar]
- [73].Solinski HJ, Gudermann T, Breit A. Pharmacology and signaling of MAS-related G protein-coupled receptors. Pharmacol Rev 2014;66:570–97. [DOI] [PubMed] [Google Scholar]
- [74].Sommer C, Lauria G. Skin biopsy in the management of peripheral neuropathy. Lancet Neurol 2007;6:632–42. [DOI] [PubMed] [Google Scholar]
- [75].Spallone V, Lacerenza M, Rossi A, Sicuteri R, Marchettini P. Painful diabetic polyneuropathy: approach to diagnosis and management. Clin J Pain 2012;28:726–43. [DOI] [PubMed] [Google Scholar]
- [76].Sriram K, Insel PAG. Protein-coupled receptors as targets for approved drugs: how many targets and how many drugs? Mol Pharmacol 2018;93:251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Stirling LC, Forlani G, Baker MD, Wood JN, Matthews EA, Dickenson AH, Nassar MA. Nociceptor-specific gene deletion using heterozygous NaV1.8-Cre recombinase mice. PAIN 2005;113:27–36. [DOI] [PubMed] [Google Scholar]
- [78].Tavares-Ferreira D, Shiers S, Ray PR, Wangzhou A, Jeevakumar V, Sankaranarayanan I, Cervantes AM, Reese JC, Chamessian A, Copits BA, Dougherty PM, Gereau RW 4th, Burton MD, Dussor G, Price TJ. Spatial transcriptomics of dorsal root ganglia identifies molecular signatures of human nociceptors. Sci Transl Med 2022; 16:14. doi: 10.1126/scitranslmed.abj8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Turner MD, Nedjai B, Hurst T, Pennington DJ. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta 2014;1843:2563–82. [DOI] [PubMed] [Google Scholar]
- [80].Uno M, Nishimura S, Fukuchi K, Kaneta Y, Oda Y, Komori H, Takeda S, Haga T, Agatsuma T, Nara F. Identification of physiologically active substances as novel ligands for MRGPRD. J Biomed Biotechnol 2012;2012:816159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Urban DJ, Roth BL. DREADDs (designer receptors exclusively activated by designer drugs): chemogenetic tools with therapeutic utility. Annu Rev Pharmacol Toxicol 2015;55:399–417. [DOI] [PubMed] [Google Scholar]
- [82].Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 2015;18:145–53. [DOI] [PubMed] [Google Scholar]
- [83].von Hehn CA, Baron R, Woolf CJ. Deconstructing the neuropathic pain phenotype to reveal neural mechanisms. Neuron 2012;73:638–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wang C, Gu L, Ruan Y, Geng X, Xu M, Yang N, Yu L, Jiang Y, Zhu C, Yang Y, Zhou Y, Guan X, Luo W, Liu Q, Dong X, Yu G, Lan L, Tang Z. Facilitation of MrgprD by TRP-A1 promotes neuropathic pain. FASEB J 2019;33:1360–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Yamazaki S, Yamaji T, Murai N, Yamamoto H, Matsuda T, Price RD, Matsuoka N. FK1706, a novel non-immunosuppressive immunophilin ligand, modifies gene expression in the dorsal root ganglia during painful diabetic neuropathy. Neurol Res 2012;34:469–77. [DOI] [PubMed] [Google Scholar]
- [86].Yang D, Zhou Q, Labroska V, Qin S, Darbalaei S, Wu Y, Yuliantie E, Xie L, Tao H, Cheng J, Liu Q, Zhao S, Shui W, Jiang Y, Wang MW. G protein-coupled receptors: structure- and function-based drug discovery. Signal Transduct Target Ther 2021;6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Zeisel A, Hochgerner H, Lonnerberg P, Johnsson A, Memic F, van der Zwan J, Haring M, Braun E, Borm LE, La Manno G, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S. Molecular architecture of the mouse nervous system. Cell 2018;174:999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zhang JM, Song XJ, LaMotte RH. Enhanced excitability of sensory neurons in rats with cutaneous hyperalgesia produced by chronic compression of the dorsal root ganglion. J Neurophysiol 1999;82:3359–66. [DOI] [PubMed] [Google Scholar]
- [89].Zylka MJ, Rice FL, Anderson DJ. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron 2005;45:17–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B957.