Abstract
Hepatitis delta virus (HDV) replicates by RNA-dependent RNA synthesis according to a double rolling circle model. Also synthesized during replication is a 0.8-kb, polyadenylated mRNA encoding the hepatitis delta antigen (HDAg). It has been proposed that this mRNA species represents the initial product of HDV RNA replication; subsequent production of genomic-length HDV RNA relies on suppression of the HDV RNA polyadenylation signal by HDAg. However, this model was based on studies which required the use of an HDV cDNA copy to initiate HDV RNA replication in cell culture, thus introducing an artificial requirement for DNA-dependent RNA synthesis. We have now used an HDV cDNA-free RNA transfection system and a method that we developed to detect specifically the mRNA species transcribed from the HDV RNA template. We established that this polyadenylated mRNA is 0.8 kb in length and its 5′ end begins at nucleotide 1631. Surprisingly, kinetic studies showed that this mRNA continued to be synthesized even late in the viral replication cycle and that the mRNA and the genomic-length RNA increased in parallel, even in the presence of HDAg. Thus, a switch from production of the HDAg mRNA to the full-length HDV RNA does not occur in this system, and suppression of the polyadenylation site by HDAg may not significantly regulate the synthesis of the HDAg mRNA, as previously proposed. These findings reveal novel insights into the mechanism of HDV RNA replication. A new model of HDV RNA replication and transcription is proposed.
Hepatitis delta virus (HDV) is an unusual subviral pathogen associated with fulminant and chronic hepatitis. HDV depends on hepatitis B virus coinfection to form infectious virions, since it uses hepatitis B surface antigen to form its viral envelope (25). It contains a single-stranded circular RNA genome of 1.7 kb (14, 22, 31), which can replicate in the absence of hepatitis B virus (4, 7). Three genotypes of HDV strains which differ by as much as 35% in nucleotide sequence (1) and may also differ in pathogenicity have been described. HDV replicates through a double rolling circle mechanism, generating multiple-length genomic and antigenomic-sense HDV RNAs, which are processed into monomeric circular HDV genomic and antigenomic RNAs (4, 15, 29). This replication process is presumably carried out by cellular RNA polymerase II (pol II), since it is inhibited by α-amanitin (21).
Also detected in HDV-infected liver tissue and some cultured cells replicating HDV RNA is a polyadenylated, 800-nucleotide (nt) antigenomic-sense HDV RNA (4, 8) that contains the open reading frame (ORF) for a protein termed hepatitis delta antigen (HDAg). This is the only protein produced by HDV but is usually composed of two forms: the small form (24 kDa) is required for viral RNA replication (13), while the large form (27 kDa) is a dominant negative inhibitor of HDV replication (3) and is required for virion assembly (2, 27). The production of large HDAg is the result of an RNA editing event during HDV replication (18), which extends the HDAg ORF for an additional 19 amino acids. This editing event is reported to occur at a late stage in the viral replication cycle, such that when the large form of HDAg is synthesized, HDV RNA replication will stop and virus assembly will begin.
The nature of the 0.8-kb mRNA has been controversial, since it is very difficult to detect in most cells replicating HDV RNA. It was proposed that this mRNA represents the initial product of HDV RNA replication; upon reaching the HDV RNA polyadenylation signal, the nascent transcript is polyadenylated and released as the HDAg-encoding mRNA (8, 10). The HDAg synthesized from this mRNA, in turn, inhibits the polyadenylation signal, thus allowing subsequent rounds of RNA replication to proceed beyond the polyadenylation signal, producing genomic-size (1.7-kb) HDV RNA (10). This model thus explains the paucity of the 0.8-kb mRNA species (15). Indeed, experimental evidence has shown that HDAg (both the small and large forms) can negatively regulate the HDV polyadenylation signal within an HDV cDNA construct (9, 10).
There is, however, a major conceptual flaw in this model. The mRNA for the large HDAg will not be synthesized under this scenario since RNA editing does not occur until later in infection, when a large amount of small HDAg has already been produced. This question has been difficult to address because the amount of the 0.8-kb RNA is very low or even undetectable in HDV-infected human or chimpanzee liver tissues or in cell cultures and transgenic mice (24) which actively replicate HDV RNA. A further complication is that HDV RNA replication has always been studied by using an HDV cDNA transfection approach (for a review, see reference 15). We have previously shown that the HDV cDNA contains several bidirectional promoters (19). Thus, the study of RNA transcription or replication from HDV RNA in these systems is compromised by the potential initiation of transcription from the cryptic promoters within the HDV cDNA. So far, the only successful HDV RNA transfection system reported is the transfection of HDV RNA into cells stably expressing an HDAg-encoding mRNA (7). Again, this artificial cDNA-based mRNA has complicated the study of authentic HDAg-encoding mRNA synthesis.
In this study, we developed an RNA transfection approach which was totally devoid of HDV cDNA and could detect specifically HDAg-encoding mRNA transcribed from HDV RNA. Using this approach, we found that HDV mRNA was abundantly synthesized throughout the HDV life cycle. Thus, contrary to the current model of HDV RNA transcription and replication (8, 10, 15, 16), there was no detectable inhibition of HDV mRNA synthesis by HDAg. Furthermore, the initiation point of the 0.8-kb mRNA may differ from the initiation point of HDV genomic RNA, again contradicting the current model. These findings provide novel insights into the mechanism of HDV RNA replication. A new model of HDV RNA replication and transcription is proposed.
MATERIALS AND METHODS
Cell culture and transfection.
Tsδ3 cells, which were derived from a temperature-sensitive hamster cell line (30) and stably express the small HDAg from an integrated cDNA copy of the HDAg-encoding mRNA under the cytomegalovirus promoter (11), were cultured at 33°C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 IU of penicillin per ml, and 100 mg of streptomycin per ml. H1δ9 cells, which contain an integrated cDNA for trimer HDV RNA (20), were cultured at 37°C in the same medium. Huh7 cells (23) were cultured at 37°C in DMEM supplemented with 10% fetal bovine serum, 100 IU of penicillin per ml, 100 mg of streptomycin per ml, 2 mM l-glutamate, and 1% nonessential amino acids (complete DMEM). All transfections were performed by using the DOTAP (Boehringer Mannheim) method according to the protocol provided by the manufacturer. Briefly, 1 day prior to transfection, Tsδ3 or Huh7 cells were seeded onto 60-mm-diameter dishes. On the following day, the cells were refreshed with 5 ml of the appropriate culture medium before transfection. Cells were transfected with 5 μg of plasmid cDNA (for Huh7 cells) or RNA (for Tsδ3 cells) in 0.15 ml of transfection mixture or 10 μg of RNA (for Huh7 cells) in 0.3 ml of transfection mixture. Following incubation overnight at 33°C (Tsδ3) or 37°C (Huh7), the culture medium was replaced with fresh medium and the cells were further incubated for an additional 1 to 5 days. For experiments involving the use of cycloheximide, 10 μg of cycloheximide per ml was added to the culture medium 1 or 2 days posttransfection and cells were incubated for a further 2 days.
Vectors and plasmid construction.
Plasmid PX9-I/II, which expresses an mRNA encoding the genotype I/II chimeric HDAg under the T7 promoter, was developed from plasmid PX9, which expresses an mRNA encoding the HDAg of the American isolate of genotype I. PX9 contains the pT7-3 plasmid backbone and HDV nt 21 through 658 (reading through nt 0) inserted in the BamHI-PstI site. To construct plasmid PX9-I/II, the EcoRI (in the multiple cloning site)-StuI (at HDV nt 1334) fragment from the plasmid PX9 was replaced with the corresponding fragment from plasmid 63 of an HDV genotype II cDNA clone (17). Thus, genotype I nt 21 to 1334 (reading through nt 0) were replaced with the corresponding genotype II nt 1663 to 1334. pKS/HDVD2, which contains a dimer HDV cDNA in a plasmid derived from plasmid pRC/CMV (12), was used for HDV cDNA transfections.
In vitro transcription.
Genomic HDV RNA (1.9 kb), which contains the entire HDV genome plus approximately 200 additional nt of HDV sequence, was transcribed from pKS/HDV1.9 (12) with T7 MEGAscript (Ambion) after linearization by EcoRV digestion. Antigenomic HDV RNA (1.9 kb) was transcribed from pKS/HDV1.9 with SP6 MEGAscript (Ambion) after linearization by SnaBI digestion. Capped, polyadenylated mRNA for small HDAg was transcribed from PX9 or PX9-I/II (see above) with T7 mMESSAGE mMACHINE (Ambion) after linearization by HindIII digestion.
Northern blot analysis.
Total RNA was extracted from transfected Tsδ3 and Huh7 cells or from H1δ9 cells by the guanidinium thiocyanate method (5). Polyadenylated RNA was isolated with an oligo(dT)-cellulose column (Sigma) according to the standard method (28). The RNA was digested with RQ1 DNase (Promega), treated with formaldehyde, electrophoresed through formaldehyde-containing 1.2% agarose gels, blotted onto a nitrocellulose membrane (Hybond C extra; Amersham), and probed with 32P-UTP-labeled HDV strand-specific riboprobes. Riboprobes for detecting HDV RNA were transcribed with T7 RNA polymerase (Promega) from plasmid S18 (to detect genomic HDV RNA) or S29 (to detect antigenomic HDV RNA), following linearization with EcoRV digestion (22). For the analysis of the H1δ9 HDV mRNA, various genomic-sense oligonucleotides (Table 1) were end labeled with 32P-ATP and T4 polynucleotide kinase (New England Biolabs). To detect newly synthesized HDAg mRNA in Huh7 cells transfected with genotype I HDV RNA (1.9 kb) and the chimeric genotype I/II mRNA, blots were probed with 32P-end-labeled oligonucleotide 1565, specific for the American isolate of genotype I HDV (22). The protocol for Northern blotting using oligonucleotide probes was adapted from that of Geliebter et al. (6). The membranes were prehybridized for 2 h at 55°C in 7% sodium dodecyl sulfate (SDS)–20 mM sodium phosphate (pH 7.0)–5× Denhardt’s solution–5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–100 μg of salmon sperm DNA per ml and hybridized overnight at 55°C in the same solution containing 10% dextran sulfate and 2 × 106 to 3 × 106 cpm of radiolabeled probe per ml. Blots were washed with 3× SSC–10 mM sodium phosphate (pH 7.0)–0.5× Denhardt’s solution–5% SDS for 1 min at room temperature followed by 1 h at 55°C. Northern blots probed with full-length HDV riboprobes were hybridized and washed as described previously (12). RNA extracted from H1δ9 cells, which express and replicate HDV RNA from an integrated cDNA trimer, was used as a positive control in all Northern blots. After autoradiography, computer images were generated by using Adobe Photoshop, version 3.0, and Canvas, version 5.0.
TABLE 1.
Sequences of primers used for Northern blotting and primer extension
| Oligonucleotide | Sequence |
|---|---|
| 63 | CGGTAAAGAGCATTGGAACGTCGGAGA |
| 303 | AATCACCTCCAGAGGACCCCTTCAGCGAAC |
| 468 | GAGTGAGGCTTATCCCGGGG |
| 676 | TTTCTTACCTGATGGCCGGC |
| 902 | CCCGAAGAGGAAAGAAGGACGCGAGACGCA |
| 1484 | TCTTCTTTGTCTTCCGGAGGTCTCTCTCG |
| 1565 | CCCCGCGGTCTTTCCTTCTTTCGGACC |
| 1634 | TACTCTTTTCTGTAAAGAGGAGACTGCTGG |
Western blot analysis.
Protein was extracted from transfected Huh7 cells according to the standard method (28). After denaturation by boiling in 2× sample buffer (100 mM Tris-HCl [pH 6.8], 200 mM dithiothreitol, 4% SDS, 0.2% bromophenol blue, 20% glycerol), 40 μg of protein from each sample was loaded onto a SDS–12.5% polyacrylamide minigel. The gel was electrophoresed for 60 to 90 min at 150 V. Proteins were then transferred to a nitrocellulose membrane (Hybond C extra; Amersham). Small and large HDAg were detected by the ECL (enhanced chemiluminescence) Western blot detection system (Amersham), using a rabbit polyclonal antibody against both forms of HDAg, and visualized by autoradiography.
Primer extension analysis.
RNA extracted from Tsδ3 and Huh7 cells (as described above) and the appropriate 32P-end-labeled oligonucleotides were coprecipitated in 0.3 M sodium acetate and 2.5 volumes of ethanol. The pellet was vacuum dried and resuspended in 8 μl of Tris-EDTA (pH 7.6) followed by the addition of 2 μl of 5× hybridization buffer (1.25 M KCl, 50 mM Tris-HCl [pH 7.4], 5 mM EDTA). The samples were incubated at 50°C for 1 h, followed by the addition of 40 μl of reverse transcription mix (25 mM KCl, 50 mM Tris-HCl [pH 7.5], 10 mM dithiothreitol, 3.5 mM MgCl2, 0.5 mM deoxynucleoside triphosphates, 100 μg of bovine serum albumin per ml, 20 U of avian myeloblastosis virus reverse transcriptase [Boehringer Mannheim]) and further incubation for 1 h at 37°C. Reaction mixtures were ethanol precipitated and dried as above, and the pellet was resuspended in 4 μl of double-distilled H2O. After the addition of 2 μl of sequencing gel loading buffer (98% formamide, 10 mM EDTA [pH 8.0], 0.025% xylene cyanol FF, 0.025% bromophenol blue), the samples were heat denatured, stored on ice, and loaded onto a 6% polyacrylamide gel containing 8 M urea. A dideoxy sequence (Amersham) generated from plasmid pKS/HDV1.9 primed by the same oligonucleotide used in the primer extension was used as a nucleotide sequence marker.
RESULTS
Heterogeneity of HDAg-encoding mRNA in cDNA-based HDV replication systems.
To characterize the mechanism of synthesis of the HDAg-encoding mRNA, we first attempted to study this mRNA in the various reported cDNA-based HDV replication systems in cell culture. Three systems were studied: Huh7 cells transiently transfected with a plasmid expressing a genomic dimer RNA of HDV, Tsδ3 cells, which stably express HDAg from an integrated cDNA copy of HDAg-encoding mRNA under the cytomegalovirus promoter (11), and H1δ9 cells, which stably express an antigenomic HDV trimer RNA from an integrated cDNA copy (20). Previously it has been very difficult to detect the 0.8-kb mRNA species in any of these systems. Based on the report that cycloheximide (CHX) treatment can inhibit mRNA translation and thereby stabilize some mRNA species (26), we treated these cell lines with 10 μg of CHX per ml. Figure 1 shows that a polyadenylated subgenomic HDV RNA of antigenomic sense was detected in various amounts in all three systems. However, these putative mRNAs surprisingly ran at different positions in denaturing agarose gels. The subgenomic mRNA in Huh7 cells transiently transfected with an HDV cDNA was barely detectable but appeared to have an electrophoretic mobility (Fig. 1, lane 1) similar to that of the previously characterized mRNA identified in cDNA-transfected COS7 cells (8). The mRNA was polyadenylated, and its 5′ end was mapped to nt 1631 (data not shown). The amount of this mRNA was not increased by treatment with CHX (lane 2). In H1δ9 cells, a significantly larger subgenomic RNA was detected. This RNA was polyadenylated and, unlike the mRNA from cDNA-transfected Huh7 cells, was stabilized by the addition of CHX (Fig. 1; compare lane 5 and lane 7). The mRNA expressed in Tsδ3 cells had an electrophoretic mobility between those of the mRNA species detected in cDNA-transfected Huh7 cells and H1δ9 cells. This mRNA was also polyadenylated and was most remarkably stabilized by CHX (Fig. 1; compare lane 9 and lane 11). These results indicated that the HDV subgenomic mRNA species synthesized in the three systems differed not only in amount but also in size and structure, since they differed in the ability to be stabilized by CHX.
FIG. 1.
Identification of subgenomic HDAg mRNAs in three cell culture systems. RNA was isolated from H1δ9, Tsδ3, and Huh7 cells transiently transfected with plasmid pKS/HDVD2, which expresses an HDV RNA dimer of genomic sense, at day 4 posttransfection. Some cells were treated with CHX (10 μg/ml) for 48 h before harvest. H1δ9 and Tsδ3 cells were separated into poly(A)+ and poly(A)− fractions. Northern blot of HDV antigenomic sense RNA was performed with a 32P-labeled 1.7-kb HDV genomic-sense RNA as a probe. The following antigenomic HDV RNAs are indicated: the 1.7-kb antigenomic HDV RNA (monomer), the H1δ9 HDAg endogenous mRNA (A), the Tsδ3 endogenous HDAg mRNA (B), and the HDAg mRNA in transiently transfected Huh7 cells (C). T, total unfractionated RNA.
To understand the basis for the mRNA heterogeneity in these systems, we performed primer extension studies using primer 1484 (Fig. 2A) to determine the initiation points of these mRNAs. The subgenomic mRNA produced in Tsδ3 cells from the cDNA construct used to establish HDAg expression in these cells (11) is predicted to begin 53 nt before the start codon for HDAg. Primer extension analysis confirmed this prediction (data not shown). We also performed primer extension on RNA extracted from Tsδ3 cells transfected with a 1.9-kb genomic sense HDV RNA to determine whether a second subgenomic mRNA species expressed from the HDV cDNA or replicating HDV RNA could be detected. No such primer-extended product was detected (data not shown). This result suggested that in Tsδ3 cells, which synthesize an mRNA from an integrated cDNA copy (11), HDV mRNA production from replicating HDV RNA is severely restricted.
FIG. 2.
Determination of the structure of the H1δ9 subgenomic mRNA. 32P-end-labeled oligonucleotides representing various regions of the HDV genome (A) were used to probe total RNA from CHX-treated (as described in the legend to Fig. 1) and untreated H1δ9 cells (B). (C) Oligonucleotide 63 was used for primer extension analysis of poly(A)-selected RNA from CHX-treated and untreated H1δ9 cells. Lanes 5 and 6, poly(A)− and poly(A)+ RNA, respectively, from CHX-untreated H1δ9 cells; lanes 7 and 8, poly(A)− and poly(A)+ RNAs, respectively, from CHX-treated H1δ9 cells. A dideoxy sequence generated from plasmid pKS/HDV1.9 by using oligonucleotide 63 served as a nucleotide sequence marker (lanes 1 to 4).
The subgenomic mRNA species in H1δ9 cells was much larger than the 0.8-kb mRNA species and was stabilized by CHX treatment (Fig. 1, lane 7). This mRNA was detectable with a full-length genomic RNA probe (S29) (Fig. 2B). To characterize this mRNA species, we first used oligonucleotide probes complementary to various regions of the HDV antigenome (Fig. 2A) to determine the origin of the additional HDV sequences seen in this mRNA. Oligonucleotide 1484, which is complementary to antigenomic HDV RNA in the HDAg ORF, bound to both monomer-length HDV RNA and the mRNA (Fig. 2B). Oligonucleotide 902, which hybridizes to antigenomic HDV RNA in the region between the poly(A) addition site and the ribozyme cleavage site, bound to monomer-length antigenomic HDV RNA but not to the subgenomic mRNA, indicating that the 3′ end of this mRNA was not extended beyond the polyadenylation signal reported for the 0.8-kb mRNA (8). However, oligonucleotides 1634 and 63, which are complementary to antigenomic HDV RNA sequence upstream of the previously characterized HDAg mRNA, bound to the H1δ9 mRNA, indicating that the 5′ untranslated region of this mRNA was longer than the previously described HDAg-encoding mRNA (8). Oligonucleotides 303, 468, and 676 did not bind to the mRNA; therefore, its 5′ end lay somewhere between nt 63 and 303. Primer extension analysis of H1δ9 RNA using oligonucleotide 63 revealed a band corresponding to a 5′ end located at nt 158; this band was detectable only in the polyadenylated RNA from CHX-treated H1δ9 cells (Fig. 2C, lane 8). This result indicated that this mRNA was initiated from an aberrant site not previously reported. No primer-extended product corresponding to an RNA species with a 5′ end at nt 1631 was detected even in CHX-treated samples (data not shown).
These results indicated that the HDV subgenomic mRNA species detected in various cDNA transfection systems have different origins of transcription. Therefore, these transcripts may represent aberrant transcription from cryptic promoters within the HDV cDNA. In all cases, the reported 0.8-kb RNA species was not, or was only barely, detected. Thus, the presence of HDV cDNA appears to interfere with the synthesis of the authentic subgenomic mRNA species, probably because of the presence of potent promoters in the HDV cDNA (19). This phenomenon may have led to the previous interpretations that the presence of HDAg inhibited the synthesis of the 0.8-kb RNA during HDV RNA replication (9, 10).
Abundance of an HDV RNA-templated mRNA in a cDNA-free transfection system.
The results presented above suggested that cDNA-initiated transcription may interfere with the synthesis of the HDAg-encoding mRNA. To study the authentic mRNA synthesis of HDV RNA, we therefore adapted an RNA-only transfection system to detect the HDAg mRNA produced from the replicating HDV genome. Briefly, in vitro-transcribed genomic HDV RNA (1.9 kb, slightly longer than the monomer-size RNA) was cotransfected with an in vitro-transcribed, capped, and polyadenylated HDAg-encoding mRNA into Huh7 cells. This approach led to robust HDV RNA replication in the transfected cells, as evidenced by the detection of antigenomic-sense HDV monomer RNA (Fig. 3B, lane 5). A subgenomic RNA species was also detected; however, it could not be distinguished from the mRNA used for transfection (Fig. 3B, lane 5). To distinguish between the mRNA produced from the genomic HDV template in the transfected cells and the exogenous mRNA used for transfection, we used for transfection a chimeric HDAg mRNA that contains sequences from both genotype I and genotype II, whereas the genomic RNA (1.9 kb) used for transfection was genotype I (Fig. 3A). This chimeric mRNA supported HDV RNA replication (Fig. 3B, lane 6). Since the oligonucleotide probe used for Northern blotting was specific for genotype I (nt 1565 to 1591), it detected the genotype I mRNA (100% complementarity to nt 1565 to 1591) (lane 3) but not the genotype I/II chimeric mRNA (55% complementarity to nt 1565 to 1591) (lane 4). Therefore, only the HDAg mRNA produced from the genomic RNA (genotype I) as a result of HDV RNA replication, not the transfected chimeric mRNA, would be detected (Fig. 3A; compare lanes 5 and 6). This probe also did not hybridize to the HDV RNA in H1δ9 cells (lane 1), which express the Italian isolate of genotype I HDV (20), since the Italian and American isolates of genotype I HDV also diverge in the region probed (74% homology in nt 1565 to 1591). This finding further indicates the specificity of this oligonucleotide probe. The result showed that a distinct 0.8-kb mRNA of antigenomic sense, which represents the RNA derived from the HDV monomer RNA, was synthesized at detectable levels (lane 6). The amount of this RNA was far greater than that seen in any cDNA transfection system reported so far. It is polyadenylated, whereas the genomic-size RNA does not contain poly(A) (Fig. 3C). Primer extension analysis of the polyadenylated RNA mapped the 5′ end to nucleotide 1631 (Fig. 3D, lane 6), indicating that this RNA-templated HDAg mRNA was initiated from the same site as the previously reported mRNA in a cDNA transfection system (8). Significantly, no corresponding primer-extended product was detected in the poly(A)-deficient fraction (Fig. 3D, lane 5), which represents all of the monomer-size RNA, even though this fraction contained at least 10 times more HDV-specific RNAs. Instead, a prominent band at nt 1646 was detected. This result suggests that the HDV genomic RNA may be initiated from a different site from the 0.8-kb mRNA species. As a control, this primer did not generate any primer extension products on the in vitro-transcribed chimeric mRNA used for transfection (Fig. 3D, lane 7).
FIG. 3.
Detection of the 0.8-kb subgenomic mRNA in the cDNA-free HDV RNA transfection system. (A) Schematic diagram of the mRNAs used for transfection and the oligonucleotide probe used for detection. (B) Total RNA harvested from Huh7 cells transfected with in vitro-transcribed mRNA I alone (lane 3), chimeric mRNA I/II alone (lane 4), mRNA I plus in vitro-transcribed 1.9-kb genomic HDV RNA (lane 5), or mRNA I/II plus 1.9-kb genomic HDV RNA (lane 6) was analyzed by Northern blotting. 32P-end-labeled oligonucleotide 1565, which is specific for the American isolate of HDV genotype I (22), was used as a probe. Total RNA extracted from H1δ9 cells and from mock-transfected Huh7 cells (lane 2) served as controls. (C) RNA isolated from Huh7 cells 4 days after transfection with 1.9-kb HDV RNA plus mRNA I/II was separated into poly(A)+ and poly(A)− fractions and analyzed as for panel B. (D) Primer extension analysis of the poly(A)− RNA (lane 5), poly(A)+ RNA (lane 6), and in vitro-transcribed mRNA I/II (lane 7), using oligonucleotide 1565 as the probe. A dideoxy sequence generated from plasmid pKS/HDV1.9 by using the same oligonucleotide served as a nucleotide sequence marker (lanes 1 to 4).
The HDAg mRNA and HDV monomer are synthesized in parallel in the presence of small HDAg.
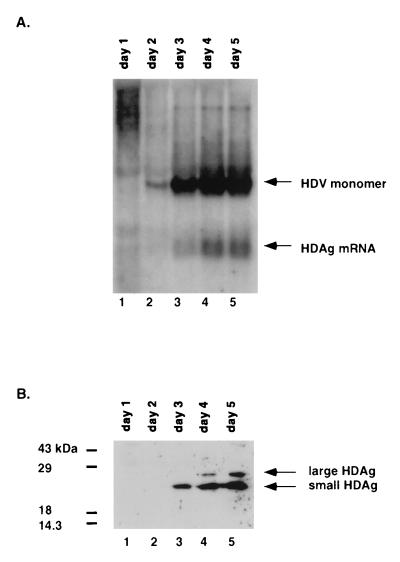
The experiments described above demonstrated the feasibility of this RNA-based transfection system for the study of de novo RNA-templated synthesis of the full-length antigenomic HDV RNA and HDAg mRNA. We next investigated the kinetics of the synthesis of these two RNA species. Previous studies indicated that HDAg could inhibit the production of the 0.8-kb HDAg-encoding mRNA in an HDV cDNA-transfected system, probably as a result of the inhibition of the polyadenylation signal in HDV cDNA or RNA (10). This inhibition was seen with both the large and small HDAg (9). Based on these studies, Hsieh and Taylor proposed that the first round of transcription on the HDV RNA template would produce the HDAg mRNA; small HDAg translated from this mRNA would then suppress polyadenylation during subsequent rounds of RNA synthesis, allowing the production of 1.7-kb antigenomic RNA (10). If this model is correct, it is predicted that the 0.8-kb mRNA would be synthesized first, followed by synthesis of the 1.7-kb monomer RNA, and that the 0.8-kb mRNA would be synthesized only early in the viral replication cycle. To test this hypothesis, we analyzed HDV RNA and HDAg from days 1 through 5 in the RNA transfection system described above. The results showed that the monomer RNA was detectable starting at day 2 and increased in amount through day 4 (Fig. 4A). Particularly, there was a significant increase between day 2 and day 3. Interestingly, the 0.8-kb mRNA species started to be detectable at day 3, and there was a particularly significant increase between days 3 and 4. There was a slight decrease in both the monomer RNA and the 0.8-kb mRNA from days 4 to 5. Thus, the mRNA and monomer RNA increased in parallel. The relative ratio between the monomer RNA and mRNA was determined by phosphorimager analysis and was found to remain roughly the same (approximately 3 in molar ratio except on day 2, when the amount of HDAg mRNA was too small to be reliably determined) throughout the 5-day period. The amount of HDAg in these cells was determined by immunoblotting using a polyclonal antibody against HDAg. Figure 4B shows that the small HDAg could be detected by day 3 and increased through day 5. By day 4, the large HDAg became detectable as a result of RNA editing, consistent with previous findings (18). It is notable that despite the presence of a large amount of HDAg at days 3 to 4, the amount of 0.8-kb mRNA continued to increase, particularly between days 3 and 4. This observation gave further support to the conclusion that the 0.8-kb mRNA species detected represents newly synthesized RNA but not merely stabilization of mRNA, since the template for the mRNA encoding the large HDAg will not be generated until an RNA editing event occurs late in infection (18). The accumulation of large HDAg at days 4 and 5 may explain the slight decrease in the amounts of both monomer RNA and the 0.8-kb mRNA at day 5, consistent with the previous finding that the large HDAg inhibits HDV RNA synthesis (3). These data reveal several important phenomena which contradict the current model for HDV mRNA synthesis (10, 15, 16). First, the mRNA and antigenomic monomer RNA are produced not sequentially but rather in parallel, suggesting that there is not a switch from mRNA synthesis to monomer RNA synthesis. Second, both species are still produced in the presence of HDAg, suggesting that HDAg does not inhibit the production of its own mRNA.
FIG. 4.
Kinetics of the synthesis of antigenomic HDV RNA and HDAg. Total RNA was isolated from Huh7 cells transfected with 1.9-kb HDV genomic RNA plus mRNA I/II on days 1 through 5 posttransfection and analyzed by Northern blotting using 32P-end-labeled oligonucleotide 1565 as a probe (A). Arrows indicate the 1.7-kb antigenomic HDV monomer and the 0.8-kb HDAg mRNA. (B) Protein isolated from a similar experiment was analyzed by Western blotting with a rabbit polyclonal antibody against HDAg. Arrows indicate the large (27-kDa) and small (24-kDa) forms of HDAg.
DISCUSSION
We describe in this report a novel system for the unequivocal detection of an HDAg-encoding mRNA transcribed from an HDV RNA. In this cDNA-free RNA transfection system, abundant synthesis of a 0.8-kb, polyadenylated, HDAg-encoding mRNA occurs during the HDV replication cycle. Kinetic studies of the production of antigenomic-sense HDV RNA in this system demonstrated that the amount of the RNA-templated HDAg-encoding mRNA increases in parallel with the monomeric-length (1.7 kb) antigenomic HDV RNA from days 1 to 4 posttransfection, followed by a slight decrease in both species from days 4 to 5 posttransfection. The ratio of the 1.7- and 0.8-kb antigenomic HDV RNAs remains the same throughout the replication cycle, despite increasing levels of HDAg. The 5′ end of the RNA-templated HDAg mRNA was mapped to nt 1631 by primer extension analysis. We did not detect a similar primer-extended band in the poly(A)-deficient fraction of RNA from cells replicating HDV but instead detected another major band upstream of the initiation site for HDAg mRNA synthesis, at nt 1646.
The above findings lead to several conclusions regarding the mechanism of HDV RNA replication and mRNA synthesis that contradict the current model of HDV replication (Fig. 5A). The current model (10, 15, 16) states that the 0.8-kb mRNA represents the initial product of HDV RNA replication, which is terminated by polyadenylation. Once HDAg is synthesized, polyadenylation will be inhibited during subsequent rounds of RNA synthesis, allowing elongation of the RNA transcript into multimeric HDV RNA. Our studies here provide three critical pieces of evidence contradicting this model. First, the HDV RNA polyadenylation signal does not appear to be suppressed by HDAg in cell culture. This conclusion is supported by our finding that HDAg mRNA synthesis continues to increase despite increasing levels of HDAg. Second, the HDAg mRNA may not be synthesized as the product of initiation of HDV RNA replication. This conclusion is supported by the finding that the 0.8-kb HDAg mRNA increases in parallel with the 1.7-kb antigenomic HDV RNA; if the HDAg mRNA were the initial product of HDV RNA replication, the mRNA would be predicted to appear only early in the HDV replication cycle. Our findings are in direct contrast to this prediction. Third, the mRNA and poly(A)− RNA (including HDV monomer RNA) may have different initiation points. This conclusion is suggested by the absence of a primer extension product at nt 1631 in poly(A)-deficient RNA from cells containing replicating HDV RNA. Instead, a major primer extension product at nt 1646 was detected in this fraction. This nucleotide is located at the site complementary to nt 1631 in the rod-like HDV RNA structure (14, 22, 31). Interestingly, this complementary region encompassing both nt 1631 and nt 1646 in the HDV cDNA has been shown to have a bidirectional promoter activity, suggesting that nt 1646 may serve as a signal for initiation of RNA synthesis (in the same orientation as the HDAg mRNA) (19). Although we cannot rigorously rule out the possibility that the product of initiation of RNA replication in the poly(A)− fraction is quickly degraded, the fact remains that no evidence can be provided to support a single initiation site for both the 0.8-kb HDAg mRNA and the 1.7-kb antigenomic HDV RNA.
FIG. 5.
Proposed models of HDV RNA transcription and replication. (A) The previously accepted model of HDV RNA transcription and replication (10, 15, 16). The initial product of replication from the genomic HDV RNA template is the 0.8-kb HDAg-encoding mRNA (arrow 1). HDAg produced from this mRNA suppresses the HDV polyadenylation signal, allowing synthesis of multimeric RNA (arrow 2), which is processed into full-length antigenomic HDV RNA (arrow 3). Subsequent rounds of replication bypass the polyadenylation signal due to the presence of HDAg and directly synthesize full-length antigenomic HDV RNA (arrow 4). (B) The proposed new model for HDV RNA transcription and replication presented in this report. The syntheses of 0.8-kb mRNA (a) and 1.7-kb monomer RNA (b) are independent and occur in parallel.
The current model for HDV RNA replication and transcription was developed from some critical assumptions regarding the mechanism of HDV transcription and replication, only a few of which are supported by experimental evidence. Essential to this model is the conclusion that HDAg suppresses the HDV RNA polyadenylation signal, originally reached by Hsieh and Taylor, who examined the effect of HDAg expression on an HDV cDNA polyadenylation signal (10). While expression of HDAg did appear to suppress the HDV cDNA polyadenylation signal, there was not a corresponding increase in readthrough transcripts (10). Therefore, HDAg may have functioned in these experiments to suppress pol II transcription from the DNA promoter. Indeed, preliminary evidence showed that HDAg has a capacity to inhibit DNA-templated pol II transcription (17a). Subsequent examination of the effects of HDAg on the HDV cDNA polyadenylation by Hsieh et al. did reveal an increase in readthrough transcripts in the presence of HDAg (9). However, reexamination of the data in this experiment revealed that there was still abundant polyadenylated transcript in the presence of HDAg. Thus, the observed effects of HDAg in inhibiting polyadenylation may have been associated with DNA-templated transcription. Furthermore, these studies (9, 10) showed that HDAg inhibited polyadenylation only when the HDV cDNA included sequences covering almost the full 1.7-kb genome. This result was interpreted to mean that a folded-back rod-like structure of HDV RNA was required for HDAg suppression of polyadenylation (9, 10, 16); however, this cDNA structure may also produce endogenous promoter activity in the HDV cDNA (19). Our studies presented here showed that when HDV RNA replication was initiated from an RNA template, HDAg did not suppress the synthesis of the HDAg mRNA, although we cannot entirely rule out the possibility that HDAg can partially inhibit HDAg mRNA polyadenylation. Regardless, the parallel increase in the 0.8-kb mRNA, the 1.7-kb antigenomic HDV RNA, and the small form of HDAg between days 2 and 3 posttransfection argues that HDV RNA replication is not dependent on the suppression of mRNA transcription by HDAg. Therefore, HDV RNA replication and subgenomic mRNA transcription may occur independently and in parallel throughout the viral life cycle (Fig. 5B). This new view of HDV RNA transcription and replication thus solves a puzzling question left unanswered by the old model of HDV RNA transcription, namely, how can the mRNA for large HDAg be synthesized late in infection if mRNA synthesis has already been shut off by the small HDAg? However, a different question arises as to how the polyadenylation signal is bypassed during RNA replication. Determining the answer to this question will require additional experimentation.
Another critical element of the current HDV replication model is the hypothesis that the sites of initiation of both mRNA transcription and genome replication are the same. This assumption has not been supported by experimental evidence so far. Because transcription of a functional mRNA for the expression of HDAg appears to be critical for replication in natural HDV life cycle (13), any studies that attempt to link the inhibition of mRNA initiation with inhibition of genome replication may not be able to prove that these two processes represent the same event. Recently, Wu et al. found that mutations at the top of the genomic HDV RNA rod (in the region of the putative RNA promoter for HDAg mRNA synthesis) which severely restricted HDV RNA replication could be rescued by small HDAg provided in trans (32). This finding suggests that sites critical for initiation of mRNA transcription may not be critical for initiation of replication. This is consistent with our data showing that the HDAg mRNA and poly(A)− HDV RNA have different primer extension products and thus may have different initiation points.
Finally, RNA pol II is hypothesized to be responsible for transcription of both the HDAg mRNA and the 1.7-kb antigenomic HDV RNA. This hypothesis is based on the finding that 1 μg of α-amanitin per ml could inhibit HDV RNA-templated transcription in vitro H1δ9 nuclear extract; however, it is possible that this was a DNA-templated transcription event, since the same authors were unable to inhibit HDV transcription with α-amanitin from an exogenous HDV RNA template added to HepG2 cells (21). Further, the relative sensitivities of synthesis of the various HDV RNA species (mRNA; 1.7-kb genomic and antigenomic RNAs) to α-amanitin have not been assessed. Thus, it is not clear whether mRNA transcription and genomic RNA replication of HDV are carried out by the same polymerase and regulated by the same mechanism. Our findings described here suggest the tantalizing possibility that the 0.8-kb HDAg mRNA and the 1.7-kb antigenomic RNA may be separately initiated but coordinately regulated. This possibility will require further studies.
ACKNOWLEDGMENTS
We thank C.-M. Lee (Chang-Gung Hospital, Kaoshiung, Taiwan) for his generous gift of the HDV genotype II clones and K.-S. Jeng for the tutoring and HDV plasmids on which this work depended.
L.E.M. is supported by a scholarship from the Life and Health Insurance Medical Research Fund. M.M.C.L. is an Investigator of the HHMI.
REFERENCES
- 1.Casey J L, Brown T L, Colan E J, Wignall F S, Gerin J L. A genotype of hepatitis D virus that occurs in northern South America. Proc Natl Acad Sci USA. 1993;90:9016–9020. doi: 10.1073/pnas.90.19.9016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang F-L, Chen P-J, Tu S-J, Wang C-J, Chen D-S. The large form of hepatitis δ antigen is crucial for assembly of hepatitis δ virus. Proc Natl Acad Sci USA. 1991;88:8490–8494. doi: 10.1073/pnas.88.19.8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chao M, Hsieh S-Y, Taylor J. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J Virol. 1990;64:5066–5069. doi: 10.1128/jvi.64.10.5066-5069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen P-J, Kalpana G, Goldberg J, Mason W, Werner B, Gerin J L, Taylor J. Structure and replication of the genome of hepatitis delta virus. Proc Natl Acad Sci USA. 1986;83:8774–8778. doi: 10.1073/pnas.83.22.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Geliebter J, Zeff R A, Schulze D H, Pease L R, Weiss E H, Mellor A L, Flavell R A, Nathenson S G. Interaction between Kb and Q4 gene sequences generates the Kbm6 mutation. Mol Cell Biol. 1986;6:645–652. doi: 10.1128/mcb.6.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glenn J S, Taylor J M, White J M. In vitro-synthesized hepatitis delta virus RNA initiates genome replication in cultured cells. J Virol. 1990;64:3104–3107. doi: 10.1128/jvi.64.6.3104-3107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsieh S-Y, Chao M, Coates L, Taylor J. Hepatitis delta virus genome replication: a polyadenylated mRNA for delta antigen. J Virol. 1990;64:3192–3198. doi: 10.1128/jvi.64.7.3192-3198.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh S-Y, Yang P-Y, Ou J T, Chu C M, Liaw Y F. Polyadenylation of the mRNA of hepatitis delta virus is dependent on the structure of the nascent RNA and regulated by the small or large delta antigen. Nucleic Acids Res. 1994;22:391–396. doi: 10.1093/nar/22.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh S-Y, Taylor J M. Regulation of polyadenylation of hepatitis delta virus antigenomic RNA. J Virol. 1991;65:6438–6446. doi: 10.1128/jvi.65.12.6438-6446.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang S B, Jeng K-S, Lai M M C. Studies of functional roles of hepatitis delta antigen in delta virus RNA replication. In: Dinter-Gottlieb G, editor. The unique hepatitis delta virus. R. G. Austin, Tex: Landes Company; 1995. pp. 95–109. [Google Scholar]
- 12.Jeng K-S, Daniel A, Lai M M C. A pseudoknot ribozyme structure is active in vivo and required for hepatitis delta virus RNA replication. J Virol. 1996;70:2403–2410. doi: 10.1128/jvi.70.4.2403-2410.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo M Y-P, Chao M, Taylor J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J Virol. 1989;63:1945–1950. doi: 10.1128/jvi.63.5.1945-1950.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo M Y-P, Goldberg J, Coates L, Mason W, Gerin G, Taylor J. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and application. J Virol. 1988;62:1855–1861. doi: 10.1128/jvi.62.6.1855-1861.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai M M C. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 16.Lazinski D W, Taylor J M. Recent developments in hepatitis delta virus research. Adv Virus Res. 1994;43:187–231. doi: 10.1016/s0065-3527(08)60049-4. [DOI] [PubMed] [Google Scholar]
- 17.Lee C-M, Changchien C S, Chung J C, Liaw Y F. Characterization of a new genotype II hepatitis delta virus from Taiwan. J Med Virol. 1996;49:145–154. doi: 10.1002/(SICI)1096-9071(199606)49:2<145::AID-JMV12>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 17a.Lo, K., and M. M. C. Lai. Unpublished data.
- 18.Luo G, Chao M, Hsieh S Y, Sureau C, Nishikura K, Taylor J. A specific base transition occurs on replicating hepatitis delta virus RNA. J Virol. 1990;64:1021–1027. doi: 10.1128/jvi.64.3.1021-1027.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macnaughton T B, Beard M R, Chao M, Gowans E J, Lai M M C. Endogenous promoters can direct the transcription of hepatitis delta virus RNA from a recircularized cDNA template. Virology. 1993;196:629–636. doi: 10.1006/viro.1993.1519. [DOI] [PubMed] [Google Scholar]
- 20.Macnaughton T B, Gowans E J, Jilbert A R, Burrell C J. Hepatitis delta virus RNA, protein synthesis and associated cytotoxicity in a stably transfected cell line. Virology. 1990;177:692–698. doi: 10.1016/0042-6822(90)90535-y. [DOI] [PubMed] [Google Scholar]
- 21.Macnaughton T B, Gowans E J, McNamara S P, Burrell C J. Hepatitis δ antigen is necessary for access of hepatitis δ virus RNA to the cell transcriptional machinery but is not part of the transcriptional complex. Virology. 1991;184:387–390. doi: 10.1016/0042-6822(91)90855-6. [DOI] [PubMed] [Google Scholar]
- 22.Makino S, Chang M F, Shieh C K, Kamahora T, Vannier D M, Govindarajan S, Lai M M C. Molecular cloning and sequencing of a human hepatitis delta virus RNA. Nature (London) 1987;329:343–346. doi: 10.1038/329343a0. [DOI] [PubMed] [Google Scholar]
- 23.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cell lines with differentiated function in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 24.Polo J M, Jeng K-S, Lim B, Govindarajan S, Hofman F, Sangiorgi F, Lai M M C. Transgenic mice support replication of hepatitis delta virus RNA in multiple tissues, particularly in skeletal muscle. J Virol. 1995;69:4880–4887. doi: 10.1128/jvi.69.8.4880-4887.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rizzetto M, Hoyer B, Canese M G, Shih J W-K, Purcell R H, Gerin J L. Delta agent: association of δ antigen with hepatitis B surface antigen and RNA in serum of δ-infected chimpanzees. Proc Natl Acad Sci USA. 1980;77:6124–6128. doi: 10.1073/pnas.77.10.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryu W-S, Bayer M, Taylor J. Assembly of hepatitis delta virus particles. J Virol. 1992;66:2310–2315. doi: 10.1128/jvi.66.4.2310-2315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 29.Taylor J, Mason W, Summers J, Golberg J, Aldrich C, Coates L, Gerin J, Gowans E. Replication of human hepatitis delta virus in primary cultures of woodchuck hepatocytes. J Virol. 1987;61:2891–2895. doi: 10.1128/jvi.61.9.2891-2895.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang E H, Tjian R. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- 31.Wang K S, Choo Q L, Weiner A J, Ou J H, Najarian R C, Thayer R M, Mullenbach G T, Denniston K J, Gerin J L, Houghton M. Structure, sequence, and expression of the hepatitis delta viral genome. Nature (London) 1986;323:508–514. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- 32.Wu T-T, Netter H J, Lazinski D W, Taylor J M. Effects of nucleotide changes on the ability of hepatitis delta virus to transcribe, process, and accumulate unit-length, circular RNA. J Virol. 1997;71:5408–5414. doi: 10.1128/jvi.71.7.5408-5414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]