Abstract
Schizophrenia (SCZ) is a debilitating neuropsychiatric disorder with high heritability and complex inheritance. In the past decade, successful identification of numerous susceptibility loci has provided useful insights into the molecular etiology of SCZ. However, applications of these findings to clinical classification and diagnosis, risk prediction, or intervention for SCZ have been limited, and elucidating the underlying genomic and molecular mechanisms of SCZ is still challenging. More recently, multiple Omics technologies – genomics, transcriptomics, epigenomics, proteomics, metabolomics, connectomics, and gut microbiomics – have all been applied to examine different aspects of SCZ pathogenesis. Integration of multi-Omics data has thus emerged as an approach to provide a more comprehensive view of biological complexity, which is vital to enable translation into assessments and interventions of clinical benefit to individuals with SCZ. In this review, we provide a broad survey of the single-omics studies of SCZ, summarize the advantages and challenges of different Omics technologies, and then focus on studies in which multiple omics data are integrated to unravel the complex pathophysiology of SCZ. We believe that integration of multi-Omics technologies would provide a roadmap to create a more comprehensive picture of interactions involved in the complex pathogenesis of SCZ, constitute a rich resource for elucidating the potential molecular mechanisms of the illness, and eventually improve clinical assessments and interventions of SCZ to address clinical translational questions from bench to bedside.
INTRODUCTION
Schizophrenia (SCZ) [1] affects ~1% of adults and poses for a huge health care burden worldwide [2]. Although SCZ results from changes in brain function, the underlying biological mechanisms are largely unknown [3]. In the past decade, new technologies combined with big data analytics have provided greater insights on its genetic architecture; this knowledge gained has exerted a profound impact on SCZ research. However, these advances have hitherto led to limited clinical applications [4].
Omic technologies aims at the collective characterization and quantification of pools of biological molecules at different levels (e.g., DNA variants, RNA transcript, epigenetic markers, proteins, metabolites, gut microbiota, and brain imaging) [5]. Multi-Omic data may provide system-level views with fine resolution, and identify key molecules, processes and events as potential targets for intervention through various integrative analytic approaches. Multi-Omic integration has already been used to identify potential treatment targets for SCZ symptoms [6]. Compared to individual-omics studies, multi-Omics can provide insights on disease mechanisms at multiple interacting levels, from genetic variation to molecular and cellular dysfunction [7].
Previous reviews have focused on the potential contribution of each single omic type, but few have comprehensively addressed the potential importance and value of integrative strategies. In this state-of-the-art review, we provide a broad survey of the individual-omics studies of SCZ, summarize the advantages and challenges for each, and then focus on studies in which multiple omics data have been integrated to unravel the complex pathophysiology of SCZ. We hope this will contribute towards the development of multi-Omic disease classification and risk prediction, and facilitate the translation of SCZ research from bench to bedside (Fig. 1).
Fig. 1. Overall strategy for Schizophrenia Omics research.
Genomics is highlighted in the center as most integrative omics approaches are genome-first. Pairwise interactions are shown by lines connecting different omics.
INDIVIDUAL-OMICS STUDIES OF SCHIZOPHRENIA
Genomics, transcriptomics, and epigenomics
As the heritability of SCZ is ~80% [8], genomics has provided the first powerful weapon to uncover disease mechanisms for SCZ [1, 9]. The most replicable genes from genome-wide association studies (GWAS), which focus on common variants, are ion-channel genes (e.g. CACNA2D2), neurotransmitter-related enzymes and receptors (e.g. DRD2), and genes in the major histocompatibility complex (MHC) regions [10, 11]. In parallel, investigations of rare or de novo mutations and copy number variations have revealed risk burden at the gene-set level (e.g. synaptic transmission networks) in SCZ development [12–14]. With the goal of using common variants to distinguish patients from controls, the polygenic risk score (PRS) was first developed for SCZ, and later became popularized across other complex phenotypes [15]. Recent efforts from large biobanks provide new hope for constructing actionable PRS that can be applied to personalize clinical intervention or management of SCZ symptoms [16]. Meanwhile, the studies of endophenotypes provide connections between genetic factors and disease onset [17], helping to identify causal mechanisms [18], and offering opportunities for early diagnosis and intervention [19].
Transcriptomics aims to study quantitative expression changes (transcription abundance) or qualitative changes (e.g. novel transcripts) of all genes and their isoforms. Gene expression is spatiotemporally dynamic, and the most important tissue for SCZ research has been the postmortem brain [20]. Earlier microarray-based mRNA expression analysis highlighted the involvement of DISC1 and GABA-A receptor beta-2 in the diseased brain; however, crucial questions of SCZ pathophysiology were not addressed [21]. The PsychENCODE project recently published the most detailed transcriptome study on the normal human brain and its dysregulation in neuropsychiatric disease patients [22, 23]. For SCZ, they found 4821 differentially expressed genes and 3803 isoforms; they also built a disease-specific co-expression network and trajectory [24]. In addition to dissecting the brain into different regions, new big data findings have also been augmented by single-cell and single-nucleic RNA sequencing (RNA-seq) data [25]. One study using single-cell RNA-seq found that different cell types have biologically distinct roles in SCZ [26]. The depiction of differences at the cellular level (i.e., GABAergic, glutamatergic, and glial neurons) will provide more meaningful biological insight into the pathogenesis of SCZ development [27].
Another mechanism that influences gene expression is epigenetics, which encompasses reversible but potentially transgenerational DNA changes (e.g., DNA methylation, histone modification, and MicroRNAs) without altering the underlying genetic sequences [28, 29]. Genome-wide epigenetic studies have been widely applied in SCZ research. Large public repositories (e.g., ENCODE, Roadmap, and FANTOM) have generated and accumulated context-specific epigenome maps corresponding to multiple tissues and cells at various developmental stages from humans and mice [30–32]. In another paper by PsychENCODE, DGCR5 long noncoding RNA was found to regulate the expression of several SCZ-related genes [33]. Paul et al. reconfirmed their finding of global hypomethylation in maternal immune activation-exposed mice and identified new pathways linked to neurodevelopmental conditions [34]. Girhar et al. found that neuronal H3K4me3 and H3K27ac histones were significantly overrepresented in SCZ [35].
Proteomics and metabolomics
Analysis at the protein and metabolite levels may reflect influences of environmental, developmental, and genetic factors on disease states [36, 37]. Both proteomic and metabolomic investigations of SCZ have shown differentially expressed proteins (e.g., NEFL, GNB1, and GLUL) [38] and amino acids (e.g., glutamate and glutamine) [39, 40] in glutamatergic signaling, corroborating the hypothesis that glutamatergic neurotransmission dysfunction is a central contributor to the pathogenesis of SCZ. Moreover, accumulating proteomic and metabolomic studies have indicated that altered energy metabolism matches the occurrence of impaired glucose tolerance and metabolic syndrome in SCZ patients [41–43].
Biomarkers are of highest clinical interest for comparative proteomics and metabolomics analysis, and central nervous system (CNS) activity has been shown influence gene expression in the peripheral blood of SCZ patients [44, 45]. More than 70% of potential proteomic biomarkers are involved in inflammatory responses [46]. Several studies have observed an association between the symptoms of SCZ and the cytokine expression levels in plasma [47, 48]. Saliva and sweat are other attractive body fluids for finding biomarkers, since their collection is non-invasive. Some studies have focused on differences in the saliva and sweat proteome between SCZ patients and controls [49, 50]. Several potential metabolomic biomarker signatures have been linked to both disease progression and treatment effectiveness in SCZ [51]. A recent review summarized results from 107 metabolomic studies on psychosis, and reported differentially expressed metabolites (e.g., N-acetylcysteine [NAA], lactate, and creatine) as potential biomarkers in two or more independent studies [52].
Gut microbiomics
Rapid 16S RNA sequencing technologies for the study of microbial populations have found that an imbalance in intestinal flora may reduce protectants and increase neurotoxin and inflammatory mediators, causing neuronal and synaptic damage which may contribute to the development of SCZ [53]. Recently, researchers found differential components in the fecal lactobacilli and oropharyngeal microbiome between first-episode SCZ patients and controls [54, 55], which is hypothesized to facilitate CNS inflammation related to SCZ [56]. Additionally, germ-free mice receiving SCZ microbiome fecal transplants were shown to have lower glutamate and higher glutamine and GABA, and displayed SCZ-relevant behaviors [57], suggesting that intestinal flora may influence neurotransmitters and pathogenesis of SCZ [58].
Differences in gut microbiota between individuals can be used to develop microbiota-based diagnostics for SCZ. A large number of studies have shown that the specific microbial panel enabled discriminating medication-free patients with SCZ from controls [59, 60]. Moreover, recent literature suggest that certain characteristics of the microbiome may be associated with the severity of psychiatric symptoms (negative symptoms and sleep) [61, 62]. Although no study has yet examined the relationship between the microbiota and cognition in SCZ, seropositivity for C. albicans has been associated with reduced memory abilities and overall cognition [63]. Recent work has shown that in some individuals, up to 70% of the drug transformation activity that occurs in their bodies can be ascribed to bacterial enzymes [64]. Hence, gut microbiota can be a major source of interpersonal variation in both drug efficacy and tolerability for SCZ patients.
Connectomics
The “connectome” describes human brain network of elements and connections [65], and connectomics is an approach to understand SCZ as a brain network disorder [66, 67]. Converging evidence suggests an abnormal connectome organization in SCZ patients and their relatives [67]. In line with white matter (WM) connectivity reductions reported from early to chronic stages of SCZ [68], structural connectome reductions affect the whole brain and span prominent associations and commissural WM pathways [69]. However, WM abnormalities vary in severity across different WM tract regions [70]. This aids in the development of tools to identify individuals at risk of transitioning to psychosis [71]. By means of WM fiber integrity measures to assess brain network architecture, a few findings have suggested that a disruption of rich-club organization and functional dynamics may reflect an early feature of SCZ pathophysiology [72]. A possible link between cortical myelination in the prefrontal cortex and structural connectome architecture in SCZ has been established [73]. Structural connectome impairments were more severe in SCZ subjects of clinical stages following the first episode than in first episode patients [74, 75]. The fragility of hubs to disconnection shows a significant association with the acceleration of gray matter loss in SCZ [76], suggesting that fragile prefrontal hub connections and topological volatility likely act as evolutionary influences on brain networks. Additionally, the disturbed connectome is involved in cognitive dysfunction in SCZ [76]. Structural and functional connectomes may be useful in clinical applications, having been shown to perform well in predicting treatment response [77]. Some multisite studies have successfully discriminated SCZ patients with relatively high accuracy using connectomic profiles [68, 78].
The most notable issue is that the specificity of the disrupted connectome to SCZ has not been addressed. A multiscale neuroscience framework may underlie alterations observed at the connectome level in SCZ [79]. Magnetic resonance imaging (MRI) studies have identified genetic contributions to structural [80] and functional [81] connectome deficits for SCZ. Moreover, studies targeting glial pathology may offer a unified gliocentric model to better understand the complex neural substrates underlying the disturbed connectome of SCZ [82].
Summary of individual-omics studies
Coverage of SCZ individual-omics studies in this review is purposefully brief, since each of them have been reviewed more in-depth by other groups (see Supplementary Table 1 for detail; also including Omics not mentioned here [e.g., Pharmacogenomics, Cellulomics, and Phenomics]). Those selected findings in each section above are based on the authors’ prior knowledge in psychiatry research and/or on consistent highlights from previous literature reviews. When piecing different aspects of omic evidences together, we arrive at the following summary: (1) SCZ is determined by the joint effects of environmental exposures and variation at multiple molecular/neural levels [83–85]; (2) the accumulated evidences highlights three different pathways (i.e., dopaminergic, glutamatergic, and inflammatory) as pivotal to understanding SCZ [86–88]; (3) as the main player and driver of SCZ research in the last decade, genomics (i.e., study of genome-wide variants and sequences) has not only revealed novel genes to understand disease etiology, but also generate resources and anchoring points (for other omics data) for translational medicine [89–91]. Nevertheless, a few unsolved questions still remain when restricting to genomics or other single-omics design alone. A major obstacle is the lack of clear annotation of noncoding GWAS hits. The integration of static genetic data and other dynamic omics data is necessary to define better biomarkers for clinical management of SCZ. In addition, the regulatory networks involved in SCZ development remain unclear [91], and the understanding of human microbiota and signaling for microbe-host interactions in SCZ is still in its infancy. These limitations and unsolved translational questions have propelled the application of multi-Omics approaches in SCZ research.
MULTI-OMICS STUDIES OF SCHIZOPHRENIA
As a multifactorial disorder, SCZ is likely to involve multiple gene regulatory networks that operate across different contextual levels [92]. Novel approaches included system genetics and integrative omics may be necessary for fully characterizing such complex etiology and mechanisms [93, 94], and enable translation to clinically actionable disease classification and management in mental health [91]. However, omics data from multiple sources are heterogeneous in formats and structures, and their integration has been facilitated by the creation of public data repositories and statistical tools (examples in Table 1). When designing a real multi-Omics study, we also need to take a few assumptions into consideration before collecting and analyzing data. One assumption corresponds to how multiple levels of molecular variation contribute to disease etiology (i.e., linear vs nonlinear/interactive) [95]. Another assumption deals with the initial focus of the investigation (i.e., genome-, phenotype-, or environment-first) [96]. We hypothesize the effects of different Omics data on SCZ are nonlinear and interactive, but genome-first approach provides an easier direction to disentangle the “Chicken or Egg” complexity (Fig. 1). To have a better view on where SCZ multi-Omics studies currently stand, we performed a systematic search using PubMed (see Supplementary Method) and identified 83 relevant publications listed in Supplementary Table 2. As we expected, the majority of the studies (75 out of 83) center around genomics data (i.e., genome-first). In the following sections, we summarize them according to four different categories; specifically, we highlight eight representative findings by integrative Omics in Fig. 2.
Table 1.
Resources and tools for Omics studies.
| Category | Type | Database/tool | Short description | URLs |
|---|---|---|---|---|
| Database | Proteomics | AlloMAPS | Allosteric signaling and mutations in proteins | http://allomaps.bii.a-star.edu.sg |
| Transcriptomics | APAatlas | Human alternative polyadenylation | https://hanlab.uth.edu/apa/ | |
| Transcriptomics | Coexpedia | Gene co expression data mapped to medical subject headings (MeSH). | http://www.coexpedia.org | |
| Genomics/Transcriptomics | GTEx | A comprehensive resource for gene expression and eQTL | http://www.gtexportal.org/home/ | |
| Genomics | PGC | GWAS summary statistics for neuropsychiatric diseases | https://www.med.unc.edu/pgc/ | |
| Multi-Omics | UK Biobank | GWAS genotypes, Brain MRI and EMR data | https://www.ukbiobank.ac.uk/ | |
| Genomics/Phenomics | All of US | GWAS genotypes, whole genome sequences, and EMR data | https://allofus.nih.gov/ | |
| Genomics | denovo-db | Human de novo gene variants detected by parent-child sequencing | http://denovo-db.gs.washington.edu | |
| Genomics | EnhancerAtlas | Enhancers in nine species | http://www.enhanceratlas.org/ | |
| Genomics | Gephebase | Genotype-phenotype relationships in eukaryotes | www.gephebase.org | |
| Genomics/Connectomics | ENIGMA | Genotypes and brain MRI data | http://enigma.ini.usc.edu/ | |
| Epigenomics | GIRD | Gene Transcription Regulation Database | http://gtrd.biouml.org | |
| Genomics | GWAS Central | GWAS datasets | https://www.gwascentral.org/index | |
| Genomics | Polygenic score catalog | database of polygenic scores | https://www.pgscatalog.org/ | |
| Epigenomics | HEDD | Human Enhancer Disease Database | http://zdzlab.einstein.yu.edu/1/hedd.php | |
| Genomics/Transcriptomics | m6AVar | Human variants affecting m6A sites | http://m6avar.renlab.org/ | |
| Transcriptomics/Connectomics | BrainSpan Atlas | Microarray and RNA-seq expression and brain imaging data | https://www.brainspan.org/static/home | |
| Genomics/Epigenomics | MGA | Mass Genome Annotation | http://ccg.vital-it.ch/mga/ | |
| Multi-Omics | mutLBSgeneDB | Mutations in Ligand Binding Sites Gene DataBase | http://www.zhaobioinfo.org/mutLBSgeneDB/ | |
| Multi-Omics | TSEA-DB | Tissue specificity of GWAS traits and phenotypes | https://bioinfo.uth.edu/TSEADB/ | |
| Transcriptomics | SCPortalen | Human and mouse single-cell centric database | http://single-cell.clst.riken.jp/ | |
| Multi-Omics | QTTbase | Quantitative Trait Loci across human phenotypes | http://mulinlab.org/qtlbase/index.html | |
| Transcriptomics | EVLncRNAs | Experimentally Validated IncRNAs including disease indications | http://biophy.dzu.edu.cn/EVLncRNAs/ | |
| Multi-Omics | CommonMind | Chromatin accessibility, gene expression | http://commonmind.org/WP | |
| Transcriptomics/Connectomics | Brain Map | Gene expression, in situ hybridization, MRI | http://human.brain-map.org/ | |
| Epigenomics | ReMap | Transcription factor ChlP-seq data | http://remap.cisreg.eu | |
| Transcriptomics/Epigenomics | spatialDB | Spatially resolved transcriptome | https://www.spatialomics.org/SpatialDB | |
| Multi-Omics | BrainCode | Integrative analysis of the human neuronal genome, transcriptome | http://www.humanbraincode.org/ | |
| Multi-Omics | Allen Brain Map | Multiple multi-Omics data for brain research | https://portal.brain-map.org/ | |
| Transcriptomics/Cellulomics | Human Cell Atlas | gene expression changes in single cell | https://www.humancellatlas.org/ | |
| Transcriptomics | SyntDB | IncRNAs and their evolutionary relationships in primates | http://syntdb.amu.edu.pl/ | |
| Epigenomics | mirTrans | Cell-specific transcriptional information for human miRNAs | http://mcube.nju.edu.cn/jwang/lab/soft/mirtrans/ | |
| Epigenomics | CFEA | Cell-free epigenome atlas | http://www.bio-data.cn/CFEA | |
| Genomics/Transcriptomics | BrainSeq Consortium | Gene expression, eQTLs | http://eqtl.brainseq.org/ | |
| Epigenomics | CistromeDB | ChlP-Seq and DNase-Seq data in human and mouse | http://cistrome.org/db | |
| Epigenomics | miRandola | Extracellular and circulating noncoding RNAs | http://mirandola.iit.cnr.it/ | |
| Epigenomics | EWAS Data Hub | DNA methylation array data and metadata | https://bigd.big.ac.cn/ewas/datahub | |
| Epigenomics | 3DIV | 3D-genome Interaction Viewer and database for Hi-C and pcHi-c data | http://kobic.kr/3div | |
| Multi-Omics | PsychENCODE | Chromatin accessibility, epigenetic modifiers, gene expression, QTLs | http://psychencode.org/ | |
| Epigenomics | BOCA | the open chromatin by ATAC-seq assay in different brain regions and cell types | http://icahn.mssm.edu/boca | |
| Transcriptomics/Pharmacogenomics | Connectivity Map | Transcriptional responses to chemical, genetic, and disease perturbation | https://portals.broadinstitute.org/cmap/ | |
| Transcriptomics/Proteomics | Differential NET | Differential protein-protein interactions in human tissues | http://netbio.bgu.ac.il/diffnet | |
| Genomics/Proteomics | ActiveDriverDB | Genome variation mapped against post-translational modifications | https://activedriverdb.org/ | |
| Proteomics | ADReCS-Target | Adverse Drug Reactions linked to proteins, genes and genetic variants | http://bioinf.xmu.edu.cn/ADReCS-Target | |
| Proteomics | PhaSepDB | Phase separation related proteins | http://db.phasep.pro/ | |
| Proteomics | DisNor | Protein interaction networks linking disease genes | http://disnor.uniroma2.it/ | |
| Proteomics | Tabloid Proteome | Protein associations inferred from Mass Spectrometry | http://iomics.ugent.be/tabloidproteome | |
| Proteomics/Metabolomics | pathDIP | Pathway data integration and analysis portal | http://ophid.utoronto.ca/pathDIP | |
| Transcriptomics/Proteomics | ProteomicsDB | Mass spectrometry of the human proteome | https://www.ProteomicsDB.org | |
| Proteomics/Metabolomics | Exposome-Explorer | Biomarkers of exposure to disease risk factors | http://exposome-explorer.iarc.fr | |
| Genomics | ExAC browser | Exome Aggregation Consortium sequence data | http://exac.broadinstitute.org | |
| Genomics | PopHuman | Population genomics-oriented genome browser | http://pophuman.uab.cat | |
| Proteomics | Membranome | A database of single-pass membrane proteins | http://membranome.org/ | |
| Proteomics | KEGG | integrated database to understand high-level functions | http://www.genome.ad.jp/kegg/ | |
| Metabolomics | METLIN | A metabolite mass spectral database | https://metlin.scripps.edu | |
| Metabolomics | HMDB | Human metabolome database | http://www.hmdb.ca | |
| Multi-Omics | jMorp | Multi-Omics dataset of 1000 healthy Japanese people | https://jmorp.megabank.tohoku.ac.jp/ | |
| Metabolomics | LIPID MAPS | in depth knowledge of lipid structure and function | http://www.lipidmaps.org/ | |
| Microbiomics | gutMEGA | Human gut metagenomics data | http://gutmega.omicsbio.info/ | |
| Microbiomics | gutMDisorder | Dysbiosis of the gut microbiota | http://bio-annotation.cn/gutMDisorder | |
| Connectomics | Human Connectome Project | Multimodal imaging data across the lifespan | https://www.humanconnectome.org/ | |
| Tools | Genomics/Phenomics | MRBase | Mendelian randomization using summary data from genome-wide association studies | http://www.mrbase.org |
| Genomics | CausalDB | Predicted causal variants from GWAS | http://mulinlab.org/causaldb | |
| Genomics/Epigenomics | FATHMM | prediction of pathogenic point mutations | http://fathmm.biocompute.org.uk | |
| Genomics/Epigenomics | Fgwas | a command line tool for integrating functional genomic information into a genome-wide association study | https://github.com/joepickrell/fgwas | |
| Genomics/Transcriptomics | TWAS | Imputed gene expression and perform TWAS | https://github.com/hakyimlab/PrediXcan | |
| Multi-Omics | PINSPIus | Omics data integration and disease subtyping | https://cran.r-project.org/web/packages/PINSPIus/ | |
| Genomics/Epigenomics | GARFIELD | a functional enrichment analysis approach described the GWAS analysis | https://www.ebi.ac.uk/birney-srv/GARFIELD/ | |
| Transcriptomics | WGCNA | Weighted gene co expression network analysis to build gene modules | https://horvath.genetics.ucla.edu/html/CoexpressionNetwork/RpackagesWGCNA/ | |
| Multi-Omics | ENLOC | integrative genetic association analysis of molecular QTL data and GWAS data | https://github.com/xqwen/integrative | |
| Genomics/Transcriptomics | eCaviar | Co-localization of eQTLs and trait-associated loci | https://github.com/fhormoz/caviar | |
| Genomics/Transcriptomics/ Epigenomics | SLDP | a method for looking for a directional effect of a signed functional annotation on a heritable trait using GWAS summary statistics. | https://github.com/yakirr/sldp | |
| Multi-Omics | Integrated systems genetics toolkit | Annotate gene function using multi-omics datasets | https://www.systems-genetics.org/ | |
| Genomics/Epigenomics | PAINTOR | Enrichment analysis of global trait-associated variants within annotations | https://github.com/gkichaev/PAINTOR_V3.0 | |
| Genomics/Epigenomics | GoShifter | a method to determine enrichment of annotations in GWAS significant loci. | https://github.com/immunogenomics/goshifter | |
| Genomics | PRSice | Calculate and apply polygenic risk scores analyses | https://www.prsice.info/ | |
| Genomics/Transcriptomics | PrediXcan | an approach to estimate the genetic association between disease risk and the local gene expression of thousands of genes. | http://twas-hub.org | |
| Genomics/Transcriptomics | SMR | Test for pleiotropic association between gene expression and complex trait | https://cnsgenomics.com/software/smr/ | |
| Multi-Omics | PsychENCODE-DSPN | Integrative omic analysis by deep learning | https://github.com/gersteinlab/PsychENCODE-DSPN | |
| Genomics/Transcriptomics | MESC | Estimate complex trait heritability mediated by assayed gene expression levels | https://github.com/douglasyao/mesc | |
| Transcriptomics | SCRNA-tools | designed for analyzing scRNA-seq data | https://www.scrna-tools.org/ | |
| Connectomics | nilearn | machine learning for neuroimaging in python | https://github.com/nilearn/nilearn | |
| Transcriptomics | Networknalyst | comprehensive gene expression analysis, meta-analysis & network biology | https://www.networkanalyst.ca/ | |
| Genomics/Epigenomics | PleioPred | a framework that leverages pleiotropy and functional annotations in genetic risk prediction | https://github.com/yiminghu/PleioPred | |
| Epigenomics | MethSurv | perform multivariable survival analysis using DNA methylation data | https://biit.cs.ut.ee/methsurv/ | |
| Genomics/Epigenomics | GREGOR | a tool built to evaluate global enrichment of trait-associated variants in experimentally annotated epigenomic regulatory features. | https://genome.sph.umich.edu/wiki/GREGOR | |
| Epigenomics | eFORGE | Histone labeling | https://eforge.altiusinstitute.org | |
| Proteomics | IMP | functional context of gene-gene networks | http://imp.princeton.edu/ | |
| Microbiomics | MetaNetX | Automated Model Construction and Genome Annotation for Large-Scale Metabolic Networks | https://www.metanetx.org/ | |
| Microbiomics | microbiomeDB | Mining and analyzing microbiome data | http://microbiomeDB.org |
Database and tools handling ≥3 types of omics data are denoted as ‘Multi-Omics’.
Fig. 2. Examples of integrative Omics studies on Schizophrenia.
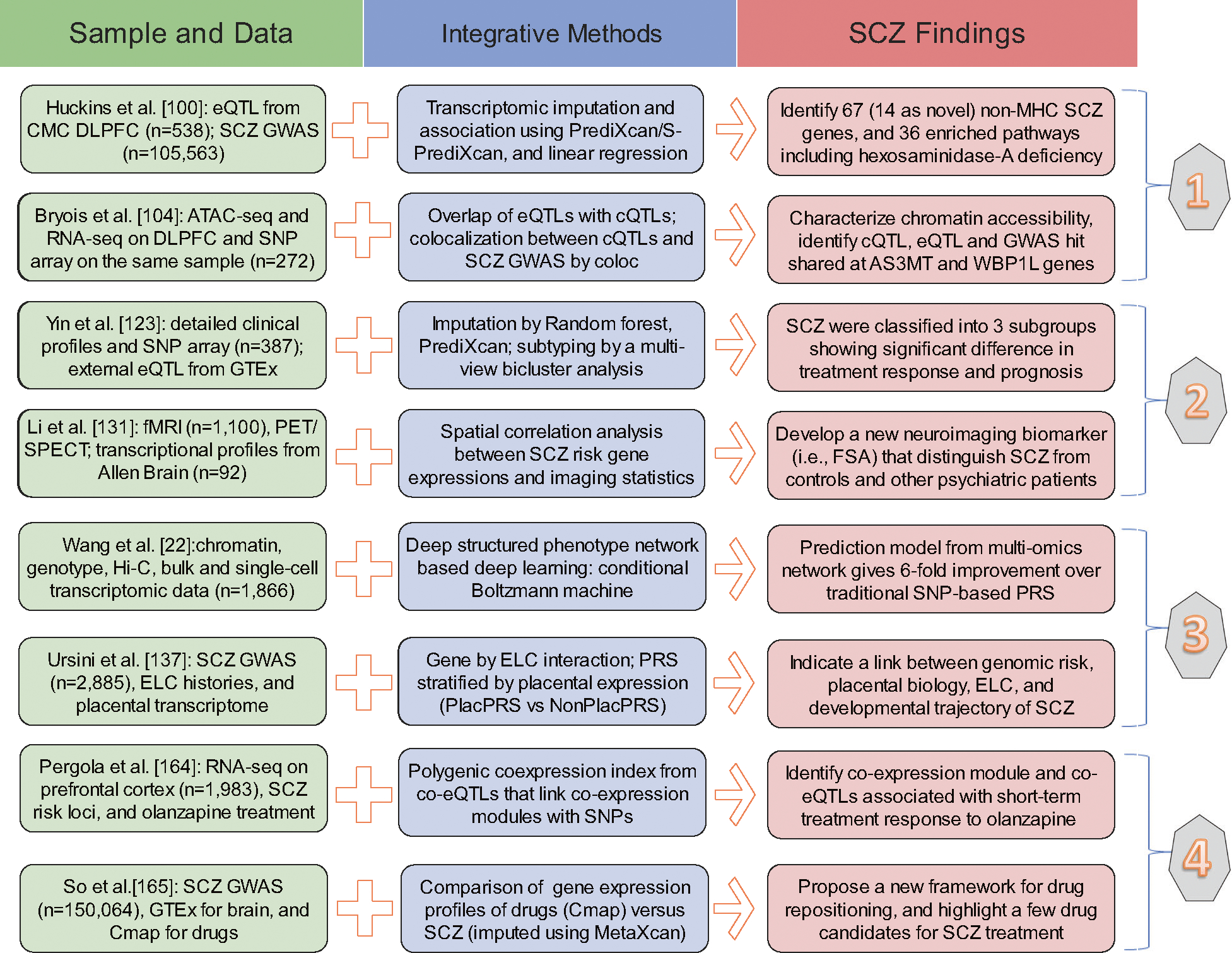
Eight recent reports were included as representative (two examples in each of those four categories) to demonstrate the usefulness of integrative Omics studies on schizophrenia (SCZ) research for pathogenesis (1), disease classification (2), risk prediction (3), and precise intervention (4). For each study, we briefly summarized the Omics data and the integrative methods used, and then highlighted the key SCZ findings identified by their application. eQTL expression quantitative trait locus, CMC CommonMind Consortium, DLPFC dorsolateral prefrontal cortex, GWAS genome-wide association study, MHC the major histocompatibility complex, cQTL chromatin quantitative trait locus, fMRI functional magnetic resonance imaging, PET positron emission tomography, SPECT single photon emission computed tomography, FSA functional striatal abnormalities, PRS polygenic risk score, ELC early-life complications, co-eQTL co-expression quantitative trait loci, Cmap The Connectivity Map.
Discovering and understanding the pathogenesis of SCZ
First, the combination of genomics and transcriptomics will lead to a better understanding of the associated noncoding SNPs and characterization of the molecular biology of the disease states. One recent study sequenced the polyA+ transcriptomes from the prefrontal cortex of 495 individuals and found widespread expression quantitative trait loci (eQTLs) that were also independently replicated [97]. Another study found that a few noncoding SNPs can disrupt transcription factor binding and change gene expression (POLR2A and CTCF) in human brain tissues [98]. Around 20% of SCZ-associated loci have variants that could contribute to altered gene expression and liability [99]. Most recently, PrediXcan model trained from CMC dorsolateral prefrontal cortex (DLPFC) omics data was applied to identify 413 genetic associations (67 non-MHC genes) with SCZ across 13 brain regions [100].
Second, genetic variations can regulate various cellular functions including DNA methylation and chromatin structure. The overlap between GWAS loci and differentially methylated sites can further annotate the extended genomic regions identified by SCZ GWAS and highlight potential regulatory variation causally involved in SCZ through co-localization [101]. Recently, many studies have focused on integrating methylome-wide association study (MWAS) results with GWAS findings and found that SCZ-associated DNA methylation differences overlapped with genetic susceptibility loci [101–103]. In parallel, the integration of GWAS and ATAC-seq of brain samples comprehensively identifies active gene regulatory elements in a brain region relevant to SCZ and quantifies how genetic variation alters functions, such as SNPs that modulate chromatin accessibility (i.e., chromatin QTL) [104]. From the perspective of gene regulation, transcription factor 4 (TCF4) binding sites were found in 39 of the 108 GWAS loci published by the Psychiatric Genomic Consortium (PGC) SCZ workgroup, showing their importance as regulators of neural genes and suggesting their functional interactions with potential relevance for SCZ [105, 106].
Third, the analysis of protein quantitative trait loci (pQTLs) can help elucidate causal pathways for genetic variants associated with neurological phenotypes and prioritize candidate targets for therapeutic intervention. Several studies have integrated pQTLs derived from human brain tissue and large-scale SCZ GWAS so as to provide a comprehensive functional annotation for all SCZ loci [107], and prioritized differentially expressed protein-coding genes between SCZ cases and controls [108]. These results may serve as an encyclopedia of SCZ susceptibility SNPs and offer holistic guides for post-GWAS functional experiments. Squires et al. summarized studies that combined rare variants of Regulators of G protein signaling (RGS) with bioinformatics and proteomic tools, finding that rare variants in functionally sensitive regions of RGS proteins could confer profound change-of-function phenotypes and lead to diseases, especially SCZ [109]. Metabolomics can also be used to bridge genotype to phenotype. For instance, a linkage disequilibrium (LD) score regression study, which calculated genetic correlations between SCZ and 172 medical, psychiatric, personality, and metabolomic phenotypes, has found a potential link between rare cases of SCZ (with 22q11.2 deletions) and serum citrate [110].
Fourth, human genetic profiles have been shown to influence overall gut microbiota composition [111], which may explain putative roles of some SCZ-associated genetic loci in this illness. Recently, some studies have combined 16S sequencing and omics to determine the effect of inflammatory bowel disease and dysregulated cytokines [112, 113]. It is now possible to identify the microbes present in the human body (membership) and their relative abundance using genomics; characterize their genetic potential (or gene pool) using metagenomics; and describe their ongoing functions using transcriptomics, proteomics, and metabolomics. However, current studies of microbiomes still mainly focus on intestinal disease; hence, further investigations using multi-Omics and including measures of the microbiomes on SCZ are needed.
Lastly, combining connectomics studies and genetics has not been commonly done for SCZ. Gurung and Prata systematically reviewed the effect of SCZ and bipolar disorder GWAS risk genes on the structure and function of the brain, including structural connectivity (ankyrin-G [ANK3] and zinc finger protein 804 A [ZNF804A]) and functional connectivity during executive tasks (calcium voltage-gated channel subunit alpha1C [CACNA1C] and ZNF804A) [114]. Imaging genetics aims to explain the genetic mechanisms behind brain changes associated with SCZ [115], but there have been only a few connectomics-genetics studies, indicating the effects of SNPs from ZNF804A, cholinergic receptor, muscarinic 3 (CHRM3), and D-amino acid oxidase activator (DAOA) on the connectome [116–118]. These findings suggest involvement of brain hubs (e.g. precuneus) and global graph metrics in SCZ genetic architecture.
Disease subtyping and clinical classification for SCZ
The ambiguity of SCZ classification causes difficulty for precise treatment plans and also hinders identifications of underlying disease mechanisms [90]. One possible solution is to improve subtyping and stratification within this disease [119]. However, subtyping based solely on clinical symptoms is almost an impossible task as the latest DSM-V has eliminated subtyping definitions previously present in the DSM-IV. Genetic information is still one of the five key elements for the on-going Research Domain Criteria [120] promoted by the National Institute of Mental Health in the United States [121]. However, there are several important limitations (i.e., unknown functional roles and high dimensionality) regarding a purely SNP-based approach. Integration across multi-Omics data may provide new clues towards a biologically based subtype classification for SCZ.
PsychENCODE investigators were among the first to examine how GWAS hits are distributed across transcriptomic or epigenomic systems in brain tissues, and identified different regulatory networks and expression subtypes of SCZ [23, 24]. Integration of genomics data with cell type-specific functional genomics annotation or data is also useful to distinguish SCZ from healthy controls or other mental disorders, as shown by a few single-cell Omics studies [26, 122]. Clustering on high-dimensional data is the most straightforward approach to derive molecular subtypes of complex diseases. A recent study proposed an analytic framework capable of identifying complex disease subgroups by leveraging both GWAS-predicted gene expression levels and clinical data using a multiview biclustering analysis [123], with predicted SCZ subgroups having different prognoses and treatment responses. This approach connects SNPs to genes via their effects on expression and is more biologically relevant and interpretable than a pure SNP-based analyses. Recent innovations also include overlaying (i.e., enrichment) different omics data onto the same expression network by unsupervised consensus clustering [124].
Alternatively, information borrowed from molecular endophenotypes can also be used to gain classification knowledge for SCZ. The Consortium on the Genetics of Schizophrenia (COGS) has conducted candidate gene and linkage analyses of many SCZ-related endophenotypes [125, 126]. These new perspectives can supplement traditional analyses of SCZ diagnosis and provide additional biological insights into the disease. Integration of GWAS with the peripheral blood proteome further sheds light on the functional role of these risk loci, and helps to determine how these associations map onto particular endophenotypes that could be useful for classifying SCZ. A recent study conducted enrichment analyses in peripheral blood samples and ascertained the overlap between proteomic findings and genetic loci identified in GWAS, finding that complement factors C3 and C4-related molecules were associated with an increased risk of SCZ [44]. Cheng et al. [127] utilized the latest pQTL data of blood proteins and GWAS data from the PGC, finding a significant association between insulin-like growth factor-binding protein 6 and SCZ. These results suggest that GWAS might benefit from integration with post-proteomics workflows to further classify and subtype SCZ.
Metabolic biomarkers correlate with exposures [128] and/or biological outcomes, and are therefore easily affected by the external environment. Integration of genetic studies with metabolomics may reduce the confounding of other factors. Several disease-loci associations have been identified in clinical settings by conducting a GWAS with metabolomics in blood serum samples [129]. A recent study conducted a two-sample Mendelian randomization analysis to assess the causal effects of 486 human serum metabolites on 5 major psychiatric disorders, and found that 2-methoxyacetaminophen sulfate had a robust effect on SCZ and identified a significant association between the glycine, serine, and threonine metabolism pathways and SCZ [130]. These results reveal that metabolomics is a new strategy for endophenotyping and early diagnosis of SCZ.
In the field of connectomics, a neuroimaging biomarker for SCZ identification, prognosis and subtyping has been developed based on functional striatal abnormalities (intrinsic and extrinsic connectome), recapitulating the distribution of dopamine and the expression profiles of polygenic risk in SCZ [131]. The conceptualization of ways to utilize genetic, epigenetic, and neuroimaging data in diagnosis and prediction of SCZ has gained ground [132, 133]. Disease subtyping and classification could also benefit from studies identifying brain markers of SCZ through combining connectomics with genomics analyses.
Disease risk prediction for SCZ
Risk prediction and stratification is another important task for clinical management of SCZ [90]. Although PRS was invented and first applied to SCZ [15], genotype data alone is not sufficient for predicting SCZ risk for individual patients [89]. Genomic risk and molecular endophenotypes are gaining increasing attention to refine risk models. Integration of genetics and other omics data together can increase the precision and sensitivity for SCZ risk stratification. A few new tools have been developed to handle new computation problems across different data resources, such as Omic Kriging and conditional deep Bolzmann machine [134, 135]. For applications in SCZ, Ayalew et al. [136] first used a convergent functional genomics approach, which borrowed information from the literature and tissue-specific gene expression to identify candidate genes with higher functional impact to achieve better predictive ability in independent cohorts. Ursini et al. found that PRSs for individuals with early-life complications (i.e., indication by placental gene expression) had more than five times greater risk than those without the complications [137]. Based on PsychENCODE integrative omics data, Wang et al. [22] improved the accuracy of the predictive model by 6-fold by adding expression data to additive genotype data, and the proposed machine learning algorithm ‘DSPN’ could further incorporate microRNA epigenetic data and neuroimaging data.
In addition to genomic risk scoring, a few other nongenetic predictors (i.e., childhood trauma and substance abuse) are also considered promising in developing risk prediction models for SCZ [138]. Another nonnegligible factor is family history of SCZ, which usually has an independent influence other than shared genetic effects [139]. Although an optimal risk stratification model for SCZ is not available yet, the emergence of the biggest-ever biobanks and electronic medical record (EMR) databases (e.g. UK Biobank and All of US in Table 1) is accelerating the effort [140, 141], from which detailed data of disease phenotypes, epidemiology, drug exposure, and genetics can be obtained for better views of disease onset and progression. Recently a cross-disorder PRS-pheWAS study that combined PGC and UK Biobank data together has shown strong correlation (or even possible causation) between schizophrenia genetic liability and psychological health, lifestyle, and socio-demographic factors [142]. Meanwhile, advanced statistics and deep learning algorithms designed for large-scale data with deep phenotyping and multi-Omics data allow better prediction of SCZ risk. On one hand, other versions of PRS calculation (e.g., polygenic transcriptome risk score [PTRS], and PleioPred) that leverage Omics annotation or phenome-wide correlations can further increase the accuracy of polygenic risk prediction in complex diseases [143, 144]. On the other hand, multivariate gene-environment interactions can be modeled and estimated by generalized mixed models. For instance, StructLMM is a new method to investigate gene-environment interaction in a high-throughput manner (i.e., multiple genes and environmental factors can be analyzed together) [145]. Taken together, the intriguing advancements in both resources and methodologies warrant better applications of omics data in future disease subtyping and risk prediction for SCZ.
Proteomic and metabolomic data can be used for mapping early biochemical changes in diseases, hence these data may offer an opportunity to develop predictive biomarkers for SCZ. SCZ-related biochemical processes, which can be traced in the cerebrospinal fluids (CSF) of prodromal patients, could serve as risk prediction indices [146]. When integrating lipid concentration levels with transcriptomic expressions from human prefrontal cortex, Yu et al. [147] found the relevance of lipidome organization and changes to SCZ progression. In addition, the disturbance of microbial metabolism may influence neurotransmitters in early developmental stages and contribute to subsequent onset of many human diseases [58]. For example, the pivotal roles of microbiome in the development and progression of colorectal cancer [148] and fibromyalgia [149] have been confirmed. Hence, integration of metabolome and microbiome may also help identify potential biomarkers for early diagnosis of SCZ.
In spite of these new opportunities, we are also facing the challenge of PRS (e.g., at DNA, RNA, or protein level) portability across different ethnic groups. Though heterogeneity of effect sizes and directions of GWAS hits across ancestral populations are relatively small for SCZ (European vs East Asian vs African), polygenic risk models trained in one population have reduced performance in the other population [11, 150, 151]. These differences may be attributable to differences in allele frequencies, LD structure, and varying interactions with other environmental risk factors in different populations [150]. This also caution us to match target populations with reference populations when applying risk models consisting of omics predictors.
A recent study combined PRS from genomics data with fMRI data in the Human Connectome Project, and identified connectivity associations serving as neural phenotype for understanding SCZ pathogenies and future risk prediction [81]. In addition, the integrative perspective involving gene-brain-environment relationships has also been proven valuable in investigating human behavioral health and mental diseases [85]. Future endeavors applying multi-Omics methods in SCZ may give us unprecedented power to study its neural mechanisms and identify new diagnostic and predictive tools.
Precise intervention for SCZ
SCZ is a heterogeneous syndrome with various clinical features (including psychosis, social, and mood symptoms) in individual patients, hence the management stratified by subtypes of different patients or risk stratification groups (e.g. those derived by methods in previous sections on “Disease subtyping and clinical classification for SCZ” and “Disease risk prediction for SCZ”) will also lead to personalized interventions and promisingly increase therapeutic success [90, 152]. Though psychological and social support is also used to manage SCZ symptoms, we limit the precise intervention here to antipsychotic drugs as it is still the main treatment option and has been mostly studied by genetics or omics approaches [84].
Pharmacogenomics that links genetic variants to antipsychotics response (e.g., drug metabolism genes CYP2C9, SLC22A1, and ABCB1) or to their adverse reactions (e.g. HLA genes and DRD2) are widely performed in research labs or clinics [153], and its progress has been already reviewed by other groups (see Supplementary Table 1). We highlight here a few representative findings from unbiased and hypothesis-free screening at genome-wide scale. Pardiñas et al. performed the first GWAS of clozapine metabolite plasma concentrations and identified CYP* and UGT* genes that may help in the clinical management of patients with treatment-resistant schizophrenia [154]. Another group studied clozapine-induced agranulocytosis/granulocytopenia (CIAG) by GWAS and identified HLA-B*59:01 as a risk factor for CIAG in the Japanese population [155]. With collaboration through Chinese Antipsychotics Pharmacogenomics Consortium, Yu et al. [156] identified five novel genes in association with response to different antipsychotics (e.g., olanzapine, risperidone, and aripiprazole) by two-stage GWAS; and Wang et al. [157] found rare genetic variations in glutamatergic or NMDA neurotransmission are implicated in short-term antipsychotic medication efficacy by whole exome sequencing. Compared to GWAS on SCZ itself, genome-wide pharmacogenomics studies are usually based on much smaller sample size (due to restricted drug medications) and will need more resources.
In parallel, other types of omics data have also been used to identify biomarkers underlying treatment response to antipsychotics. For instance, Readhead et al. [158] used human induced pluripotent stem cell (hiPSC)-based models and proved transcriptomic-based drug screening is helpful for SCZ drug discovery. Findings from epigenetics in antipsychotic response suggest that pharmacoepigenetic marker (e.g. histone deacetylases) can be promising new target to improve schizophrenia treatment [152, 159]. Proteomics and metabolomics studies have repeatedly demonstrated the significance of lipid metabolites’ quantities and/or alterations in schizophrenia patients receiving different antipsychotics [160, 161]. Another recent paper identified a number of SCZ-associated bacterial species representing potential microbial targets for future treatment [59], further consolidating the possibility that a novel microbiomics-based precise intervention and potential prevention options is possible through regulating inflammatory processes and immune responses involved in gut-microbiome-host interaction [88]. As an example from the neuroimaging community, a prospective positron emission tomography (PET) study revealed a difference in dopaminergic function between responders and non-responders at first episode of psychosis, which suggested dopamine dysfunction before starting treatment is linked to likelihood of responding to antipsychotic treatment [162]. Nevertheless, most of the aforementioned studies are observational and lack of mechanism support for therapeutic management and drug development.
Integration of multi-Omics data is now providing new clues to understand differences with respect to antipsychotic treatment outcomes among SCZ patients (i.e., positive symptoms, negative symptoms, or cognitive impairment), and is also shortening the path for implementing a mechanism-based precision intervention. A study by Kauppi et al. [163] used protein interactome to map polygenic link between antipsychotic drug targets and schizophrenia risk genes, and found that risk genes (e.g., CHRN, PCDH, and HCN families) involved in schizophrenia pathophysiology are reliable targets for novel drugs to treat cognitive or negative symptoms of schizophrenia. Pergola et al. [164] used PGC SCZ loci and prefrontal cortex co-expression network to obtain polygenic co-expression index (PCI), and found that PCI is relevant to olanzapine response in SCZ patients with positive symptom domain. Another transcriptome-wide association study (TWAS) compared the difference between imputed transcriptome from SCZ GWAS with drug-induced gene expression profiles from the Connectivity Map (CMap) database and found repositioning candidates enriched for multiple antipsychotics [165]. In addition, Price et al. [166] recently conducted a survey of cortical development aiming at cell type-resolved transcriptomic and epigenomic changes in the context of SCZ and proposed that focusing on multiple-Omic changes (e.g. genomic and epigenomic regulation on cellular identity) illuminates an impressive scene for future SCZ research including drug therapy. Therefore, integrative Omics provide important insight for understanding the mechanism whereby DNA variation leads to complex trait variation and can be informative for drug discovery and personalized treatment of SCZ patients.
CONCLUSIONS AND FUTURE PERSPECTIVES
We summarized SCZ omics studies from the perspective of genetic mechanism, clinical classification, risk prediction, and precise intervention. Their applications in SCZ and other neuropsychiatric disorders propel the field of precision psychiatry [90]. Multi-Omics research of SCZ helps to explain the complex relationships between alterations at different levels and is the most comprehensive way to explain the occurrence and development of SCZ. A scribed to disease characteristics and existing resources in mental health [167], combination of functional genomics approaches (i.e., genomics, transcriptomics, epigenomics) are most-widely adopted in current SCZ research [91]. As nucleic acids omics can be directly transferred to protein data, proteomics or metabolomics integrated with genomics will also lead to better understanding of the complex pathophysiology and possible therapeutic strategies in SCZ [168]. However, it is important to bear in mind that multi-Omics integrations can also bring new caveats (e.g., method difference, lack of comparable replication, and over fitting) [95] and challenges (e.g., interpretation, computational resources, and standards to be established) to us [94]; and sometimes, the combination of heterogeneous multiview datasets can even have lower performance of risk stratification than clean single Omics data [169]. Analyses and interpretation need to be conducted in a comparative, contextualized, and coherent manner.
A few perspectives deserve more attention in the future. One is to develop deeper phenotyping from increasingly detailed EMR data in health systems (e.g. Mental Health Research Network [MHRN]) and longitudinal cohorts (e.g. UK Biobank and All of US). The abundance of clinical diagnoses, their proxies, and putative endophenotypes from these data can enable both splitting and clumping of psychiatric disorders [170]. When restricting SCZ patients into well-defined developmental groups, one can reduce phenotype heterogeneity to identify novel genes and to build tailored genomic risk prediction models. There is also a need to incorporate other environmental exposures in disease subtyping or risk prediction with multi-Omics data, since the inclusion of known and novel gene-environment signals will increase stratification and prediction power [145]. With the rapid accumulation of detailed EMR data, the integration of environmental exposures, omics data at different molecular levels, and detailed clinical information can systematically reveal the joint effects of nature (i.e., molecular characteristics and regulatory network) and nurture (i.e., environment) on SCZ occurrence and development. Another method is to collect larger-scale omics data within the same subject. Compared with current integrative approaches that leverage data from different individuals, data on the same ones will create a more holistic view of the molecular events that lead to SCZ phenotypes [94]. For instance, a mostly recent report verified the importance of Wnt signaling pathway in the pathogenesis of neuropsychiatric disorders, through multi-Omics analysis (i.e., genomic, transcriptomic, and epigenomic data from the same subjects) of pluripotent stem cells from patients with 15q13.3 microdeletion and matched controls [171]. Wnt signaling plays an important role in neuronal survival and brain development, hence may be targeted for potential intervention of SCZ in future [172]. Lastly, it is also essential to generate more genomic resources and findings for traditionally less-represented ethnic populations, as this will not only be necessary to reduce health disparities across groups [173], but also help to better fine-map disease causal gene and increase the portability of risk predictions with Omics data across diverse populations [174]. Once these perspectives developed further, we can then investigate SCZ by integrating multi-Omics data through two other approaches (phenotype- and environment-first) that complement the major genome-first approach [96].
Overall, since SCZ is a complex disease, having only one method or single data set makes it difficult to fully capture the dynamic characteristics of the development and progression of the disease, which is influenced by both genes and life circumstances. While there are still many challenges ahead, a better translation from bench to bedside will only be realized in the future with a combined effort from multidimensional omics data, larger sample sizes, deeper phenotyping, and more integrated models.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the Natural Science Foundation of China, Shaanxi Province Innovation Capability Support Project (81772033 and 2020KJXX-039 to FG), National Heart, Lung, and Blood Institute (R01HL141845 to LKW), National Natural Science Foundation of China/Research Grants Council of Hong Kong Joint Research Scheme (8141101084 to PCS), Hong Kong Innovation and Technology Bureau Funding to State Key Laboratories, and Henry Ford Hospital Mentored Scientist grant (A20067 to HG). We thank Mr. Yang Cao and Ms. Dongru Chen for their help about the figures.
Footnotes
CONFLICT OF INTEREST
The authors declare no competing interests.
ADDITIONAL INFORMATION
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41380-021-01201-2.
Reprints and permission information is available at http://www.nature.com/reprints
REFERENCES
- 1.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–7. [DOI] [PubMed] [Google Scholar]
- 2.McGrath J, Saha S, Chant D, Welham J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol Rev. 2008;30:67–76. [DOI] [PubMed] [Google Scholar]
- 3.Bray NJ, O’Donovan MC. The genetics of neuropsychiatric disorders. Brain Neurosci Adv. 2019;2:2398212818799271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhuo C, Hou W, Li G, Mao F, Li S, Lin X, et al. The genomics of schizophrenia: Shortcomings and solutions. Prog Neuropsychopharmacol Biol Psychiatry. 2019;93:71–6. [DOI] [PubMed] [Google Scholar]
- 5.Bayes-Genis A, Liu PP, Lanfear DE, de Boer RA, González A, Thum T, et al. Omics phenotyping in heart failure: the next frontier. Eur Heart J. 2020;41:3477–84. [DOI] [PubMed] [Google Scholar]
- 6.McCutcheon RA, Reis Marques T, Howes OD. Schizophrenia-an overview. JAMA Psychiatry. 2020;77:201–10. [DOI] [PubMed] [Google Scholar]
- 7.Yang TL, Shen H, Liu A, Dong SS, Zhang L, Deng FY, et al. A road map for understanding molecular and genetic determinants of osteoporosis. Nat Rev Endocrinol. 2020;16:91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait – evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–92. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Børglum AD, Breen G, et al. Psychiatric genomics: an update and an agenda. Am J Psychiatry. 2018;175:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam M, Chen CY, Li Z, Martin AR, Bryois J, Ma X, et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet. 2019;51:1670–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickie IB, Scott J, McGorry PD. Clinical staging for mental disorders: a new development in diagnostic practice in mental health. Med J Aust. 2013;198:461–2. [DOI] [PubMed] [Google Scholar]
- 17.Benes FM, Matzilevich D, Lim B. Defining cellular endophenotypes for schizophrenia and bipolar disorder using gene expression profiling (GEP) studies of postmortem hippocampus. Int J Neuropsychopharmacol. 2006;9:S84. [Google Scholar]
- 18.Toulopoulou T, Zhang X, Cherny S, Dickinson D, Berman KF, Straub RE, et al. Polygenic risk score increases schizophrenia liability through cognition-relevant pathways. Brain. 2019;142:471–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenwood TA, Lazzeroni LC, Maihofer AX, Swerdlow NR, Calkins ME, Freedman R, et al. Genome-wide Association of Endophenotypes for Schizophrenia From the Consortium on the Genetics of Schizophrenia (COGS) Study. JAMA Psychiatry. 2019;76:1274–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lein ES, Belgard TG, Hawrylycz M, Molnar Z. Transcriptomic perspectives on neocortical structure, development, evolution, and disease. Annu Rev Neurosci. 2017;40:629–52. [DOI] [PubMed] [Google Scholar]
- 21.Horvath S, Janka Z, Mirnics K. Analyzing schizophrenia by DNA microarrays. Biol Psychiatry. 2011;69:157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, et al. Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018;362:eaat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science. 2018;362:eaat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science. 2018;362:eaat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnaswami SR, Grindberg RV, Novotny M, Venepally P, Lacar B, Bhutani K, et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc. 2016;11:499–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skene NG, Bryois J, Bakken TE, Breen G, Crowley JJ, Gaspar HA, et al. Genetici dentification of brain cell types underlying schizophrenia. Nat Genet. 2018;50:825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavalli G, Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–99. [DOI] [PubMed] [Google Scholar]
- 29.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng Q, Wang K, Brunetti T, Xia Y, Jiao C, Dai R, et al. The DGCR5 long non-coding RNA may regulate expression of several schizophrenia-related genes. Sci Transl Med. 2018;10:eaat6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basil P, Li Q, Gui H, Hui TCK, Ling VHM, Wong CCY, et al. Prenatal immune activation alters the adult neural epigenome but can be partly stabilised by a n-3 polyunsaturated fatty acid diet. Transl Psychiatry. 2018;8:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Girdhar K, Hoffman GE, Jiang Y, Brown L, Kundakovic M, Hauberg ME, et al. Cell-specific histone modification maps in the human frontal lobe link schizophrenia risk to the neuronal epigenome. Nat Neurosci. 2018;21:1126–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blackstock WP, Weir MP. Proteomics: quantitative and physical mapping of cellular proteins. Trends Biotechnol. 1999;17:121–7. [DOI] [PubMed] [Google Scholar]
- 37.Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, Mclean JA. Untargeted metabolomics strategies—challenges and emerging directions. J Am Soc Mass Spectrom. 2016;27:1897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.English JA, Pennington K, Dunn MJ, Cotter DR. The neuroproteomics of schizophrenia. Biol Psychiatry. 2011;69:163–72. [DOI] [PubMed] [Google Scholar]
- 39.Koike S, Bundo M, Iwamoto K, Suga M, Kuwabara H, Ohashi Y, et al. A snapshot of plasma metabolites in first-episode schizophrenia: a capillary electrophoresis time-of-flight mass spectrometry study. Transl Psychiatry. 2014;4:e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plitman E, de la Fuente-Sandoval C, Reyes-Madrigal F, Chavez S, Gómez-Cruz G, León-Ortiz P, et al. Elevated myo-inositol, choline, and glutamate levels in the associative striatum of antipsychotic-naive patients with first-episode psychosis: a proton magnetic resonance spectroscopy study with implications for glial dysfunction. Schizophr Bull. 2016;42:415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Chen T, Sun L, Zhao Z, Qi X, Zhou K, et al. Potential metabolite markers of schizophrenia. Mol Psychiatry. 2013;18:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ijaz S, Bolea B, Davies S, Savović J, Richards A, Sullivan S, et al. Antipsychotic polypharmacy and metabolic syndrome in schizophrenia: a review of systematic reviews. BMC Psychiatry. 2018;18:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pillinger T, Beck K, Gobjila C, Donocik JG, Jauhar S, Howes OD. Impaired glucose homeostasis in first-episode schizophrenia: a systematic review and meta-analysis. JAMA Psychiatry. 2017;74:261–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Comes AL, Papiol S, Mueller T, Geyer PE, Mann M, Schulze TG. Proteomics for blood biomarker exploration of severe mental illness: pitfalls of the past and potential for the future. Transl Psychiatry. 2018;8:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabherwal S, English JA, Focking M, Cagney G, Cotter DR. Blood biomarker discovery in drug-free schizophrenia: the contributionof proteomics and multiplex immunoassays. Expert Rev Poteomics. 2016;13:1141–55. [DOI] [PubMed] [Google Scholar]
- 46.Chan MK, Guest PC, Levin Y, Umrania Y, Schwarz E, Bahn S, et al. Converging evidence of blood-based biomarkers for schizophrenia: an update. Int Rev Neurobiol. 2011;101:95–144. [DOI] [PubMed] [Google Scholar]
- 47.Chan MK, Krebs MO, Cox D, Guest PC, Yolken RH, Rahmoune H, et al. Development of a blood-based molecular biomarker test for identification of schizophrenia before disease onset. Transl Psychiatry. 2015;5:e601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dahan S, Bragazzi NL, Yogev A, Bar-Gad M, Barak V, Amital H, et al. The relationship between serum cytokine levels and degree of psychosis in patients with schizophrenia. Psychiatry Res. 2018;268:467–72. [DOI] [PubMed] [Google Scholar]
- 49.Iavarone F, Melis M, Platania G, Cabras T, Castagnola M. Characterization of salivary proteins of schizophrenic and bipolar disorder patients by top-down proteomics. J Proteom. 2014;103:15–22. [DOI] [PubMed] [Google Scholar]
- 50.Raiszadeh MM, Ross MM, Russo PS, Schaepper MA, Zhou W, Deng J, et al. Proteomic analysis of eccrine sweat: implications for the discovery of schizophrenia biomarker proteins. J Proteome Res. 2012;11:2127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davison J, O’Gorman A, Brennan L, Cotter DR. A systematic review of metabolite biomarkers of schizophrenia. Schizophr Res. 2018;195:32–50. [DOI] [PubMed] [Google Scholar]
- 52.Li C, Wang A, Wang C, Ramamurthy J, Zhang E, Guadagno E, et al. Metabolomics in patients with psychosis: a systematic review. Am J Med Genet B Neuropsychiatr Genet. 2018;177:580–8. [DOI] [PubMed] [Google Scholar]
- 53.Yuan XX, Kang YL, Zhuo CJ, Huang XF, Song XQ. The gut microbiota promotes the pathogenesis of schizophrenia via multiple pathways. Biochem Biophys Res Commun. 2019;512:373–80. [DOI] [PubMed] [Google Scholar]
- 54.Schwarz E, Maukonen J, Hyytiäinen T, Kieseppä T, Orešič M, Sabunciyan S, et al. Analysis of microbiota in first episode psychosis identifies preliminary associations with symptom severity and treatment response. Schizophr Res. 2018;192:398–403. [DOI] [PubMed] [Google Scholar]
- 55.Castro-Nallar E, Bendall ML, Pérez-Losada M, Sabuncyan S, Severance EG, Dickerson FB, et al. Composition, taxonomy and functional diversity of the oropharynx microbiome in individuals with schizophrenia and controls. PeerJ. 2015;3:e1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dickerson F, Severance E, Yolken R. The microbiome, immunity, and schizophrenia and bipolar disorder. Brain Behav Immun. 2017;62:46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng P, Zeng B, Liu M, Chen J, Pan J, Han Y, et al. The gut microbiome from patients with schizophrenia modulates the glutamate-glutamine-GABA cycle and schizophrenia-relevant behaviors in mice. Sci Adv. 2019;5:eaau8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res.2018;1693:128–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu F, Ju Y, Wang W, Wang Q, Guo R, Ma Q, et al. Metagenome-wide association of gut microbiome features for schizophrenia. Nat Commun. 2020;11:1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen Y, Xu J, Li Z, Huang Y, Yuan Y, Wang J, et al. Analysis of gut microbiota diversity and auxiliary diagnosis as a biomarker in patients with schizophrenia: a cross-sectional study. Schizophr Res. 2018;197:470–7. [DOI] [PubMed] [Google Scholar]
- 61.Schwarz E, Maukonen J, Hyytiäinen T, Kieseppä T, Orešič M, Sabunciyan S, et al. Analysis of microbiota in first episode psychosis identifies preliminary associations with symptom severity and treatment response. Schizophr Res. 2018;192:398–403. [DOI] [PubMed] [Google Scholar]
- 62.Nguyen TT, Kosciolek T, Eyler LT, Knight R, Jeste DV. Overview and systematic review of studies of microbiome in schizophrenia and bipolar disorder. J Psychatr Res. 2018;99:50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Severance EG, Gressitt KL, Stallings CR, Katsafanas E, Schweinfurth LA, Savage CL, et al. Candida albicans exposures, sex specificity and cognitive deficits in schizophrenia and bipolar disorder. NPJ Schizophr. 2016;2:16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science. 2019;363:eaat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sporns O, Tononi G, Kotter R. The human connectome: A structural description of the human brain. PLoS Comput Biol. 2005;1:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fornito A, Zalesky A, Breakspear M. The connectomics of brain disorders. Nat Rev Neurosci. 2015;16:159–72. [DOI] [PubMed] [Google Scholar]
- 67.Fornito A, Bullmore ET. Connectomics: a new paradigm for understanding brain disease. Eur Neuropsychopharmacol. 2015;25:733–48. [DOI] [PubMed] [Google Scholar]
- 68.Cheng H, Newman S, Goñi J, Kent JS, Howell J, Bolbecker A, et al. Nodal centrality of functional network in the differentiation of schizophrenia. Schizophr Res. 2015;168:345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oestreich LKL, Randeniya R, Garrido MI. White matter connectivity reductions in the pre-clinical continuum of psychosis: a connectome study. Hum Brain Mapp. 2019;40:529–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mamah D, Ji A, Rutlin J, Shimony JS. White matter integrity in schizophrenia and bipolar disorder: Tract- and voxel-based analyses of diffusion data from the Connectom scanner. Neuroimage Clin. 2019;21:101649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Collin G, Seidman LJ, Keshavan MS, Stone WS, Qi Z, Zhang T, et al. Functional connectome organization predicts conversion to psychosis in clinical high-risk youth from the SHARP program. Mol Psychiatry. 2020;25:2431–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cui LB, Wei Y, Xi YB, Griffa A, De Lange SC, Kahn RS, et al. Connectome-based patterns of first-episode medication-naive patients with schizophrenia. Schizophr Bull. 2019;45:1291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wei Y, Collin G, Mandl RCW, Cahn W, Keunen K, Schmidt R, et al. Cortical magnetization transfer abnormalities and connectome dysconnectivity in schizophrenia. Schizophr Res. 2018;192:172–8. [DOI] [PubMed] [Google Scholar]
- 74.Griffa A, Baumann PS, Klauser P, Mullier E, Cleusix M, Jenni R, et al. Brain connectivity alterations in early psychosis: from clinical to neuroimaging staging. Transl Psychiatry. 2019;9:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang S, Gong G, Zhong S, Duan J, Yin Z, Chang M, et al. Neurobiological commonalities and distinctions among 3 major psychiatric disorders: a graph theoretical analysis of the structural connectome. J Psychiatry Neurosci. 2020;45:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gollo LL, Roberts JA, Cropley VL, Di Biase MA, Pantelis C, Zalesky A, et al. Fragility and volatility of structural hubs in the human connectome. Nat Neurosci. 2018;21:1107–16. [DOI] [PubMed] [Google Scholar]
- 77.Zhao X, Tian L, Yan J, Yue W, Yan H, Zhang D. Abnormal rich-club organization associated with compromised cognitive function in patients with schizophrenia and their unaffected parents. Neurosci Bull. 2017;33:445–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cui LB, Liu L, Wang HN, Wang LX, Guo F, Xi YB, et al. Disease definition for schizophrenia by functional connectivity using radiomics strategy. Schizophr Bull. 2018;44:1053–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van den Heuvel MP, Scholtens LH, Kahn RS. Multiscale neuroscience of psychiatric disorders. Biol Psychiatry. 2019;86:512–22. [DOI] [PubMed] [Google Scholar]
- 80.Voineskos AN. Genetic underpinnings of white matter ‘connectivity’: heritability, risk, and heterogeneity in schizophrenia. Schizophr Res. 2015;161:50–60. [DOI] [PubMed] [Google Scholar]
- 81.Cao H, Zhou H, Cannon TD. Functional connectome-wide associations of schizophrenia polygenic risk. Mol Psychiatry 2020. 10.1038/s41380-020-0699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dietz AG, Goldman SA, Nedergaard M. Glial cells in schizophrenia: a unified hypothesis. Lancet Psychiatry. 2020;7:272–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Os J, Kenis G, Rutten BP. The environment and schizophrenia. Nature.2010;468:203–12. [DOI] [PubMed] [Google Scholar]
- 84.Owen MJ, Sawa A, Mortensen PB. Schizophrenia. Lancet. 2016;388:86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sprooten E, Franke B, Greven CU. The P-factor and its genomic and neural equivalents: an integrated perspective. Mol Psychiatry 2021. 10.1038/s41380-021-01031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCutcheon RA, Krystal JH, Howes OD. Dopamine and glutamate in schizophrenia: biology, symptoms and treatment. World Psychiatry. 2020;19:15–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Radhakrishnan R, Kaser M, Guloksuz S. The link between the immune system, environment, and psychosis. Schizophr Bull. 2017;43:693–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dickerson F, Severance E, Yolken R. The microbiome, immunity, and schizophrenia and bipolar disorder. Brain Behav Immun. 2017;62:46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Weinberger DR. Thinking about schizophrenia in an era of genomic medicine. Am J Psychiatry. 2019;176:12–20. [DOI] [PubMed] [Google Scholar]
- 90.Rees E, Owen MJ. Translating insights from neuropsychiatric genetics and genomics for precision psychiatry. Genome Med. 2020;12:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sullivan PF, Geschwind DH. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell. 2019;177:162–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15:34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karczewski KJ, Snyder MP. Integrative omics for health and disease. Nat Rev Genet. 2018;19:299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype-phenotype interactions. Nat Rev Genet. 2015;16:85–97. [DOI] [PubMed] [Google Scholar]
- 96.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol.2017;18:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jaffe AE, Straub RE, Shin JH, Tao R, Gao Y, Collado-Torres L. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat Neurosci. 2018;21:1117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huo Y, Li S, Liu J, Li X, Luo XJ. Functional genomics reveal gene regulatory mechanisms underlying schizophrenia risk. Nat Commun. 2019;10:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takata A, Ionita-Laza I, Gogos JA, Xu B, Karayiorgou M. De novo synonymous mutations in regulatory elements contribute to the genetic etiology of autism and schizophrenia. Neuron. 2016;89:940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huckins LM, Dobbyn A, Ruderfer DM, Hoffman G, Wang W, Pardiñas AF, et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet. 2019;51:659–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hannon E, Dempster E, Viana J, Burrage J, Smith AR, Macdonald R, et al. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016;17:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mendizabal I, Berto S, Usui N, Toriumi K, Chatterjee P, Douglas C, et al. Cell type-specific epigenetic links to schizophrenia risk in the brain. Genome Biol. 2019;20:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li M, Li Y, Qin H, Tubbs JD, Li M, Qiao C et al. Genome-wide DNA methylation analysis of peripheral blood cells derived from patients with first-episode schizophrenia in the Chinese Han population. Mol Psychiatry 2020. 10.1038/s41380-020-00968-0. [DOI] [PubMed] [Google Scholar]
- 104.Bryois J, Garrett ME, Song L, Safi A, Giusti-Rodriguez P, Johnson GD, et al. Evaluation of chromatin accessibility in prefrontal cortex of individuals with schizophrenia. Nat Commun. 2018;9:3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xia H, Jahr FM, Kim NK, Xie L, Shabalin AA, Bryois J, et al. Building a schizophrenia genetic network: transcription factor 4 regulates genes involved in neuronal development and schizophrenia risk. Hum Mol Genet. 2018;27:3246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kibinge NK, Relton CL, Gaunt TR, Richardson TG. Characterizing the causal pathway for genetic variants associated with neurological phenotypes using human brain-derived proteome data. Am J Hum Genet. 2020;106:885–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Niu HM, Yang P, Chen HH, Hao RH, Dong SS, Yao S, et al. Comprehensive functional annotation of susceptibility SNPs prioritized 10 genes for schizophrenia. Transl Psychiatry. 2019;9:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Squires KE, Montañez-Miranda C, Pandya RR, Torres MP, Hepler JR. Genetic analysis of rare human variants of regulators of G protein signaling proteins and their role in human physiology and disease. Pharmacol Rev. 2018;70:446–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Duncan LE, Shen H, Ballon JS, Hardy KV, Noordsy DL, Levinson DF. Genetic correlation profile of schizophrenia mirrors epidemiological results and suggests link between polygenic and rare variant (22q11.2) cases of schizophrenia. Schizophr Bull. 2018;44:1350–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Goodrich JK, Davenport ER, Clark AG, Ley RE. The relationship between the human genome and microbiome comes into view. Annu Rev Genet. 2017;51:413–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jansson JK, Baker ES. A multi-omic future for microbiome studies. Nat Microbiol.2016;1:16049. [DOI] [PubMed] [Google Scholar]
- 113.Gill T, Brooks SR, Rosenbaum JT, Asquith M, Colbert RA. Novel inter-omic analysis reveals relationships between diverse gut microbiota and host immune dysregulation in HLA-B27-induced experimental spondyloarthritis. Arthritis Rheumatol. 2019;71:1849–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gurung R, Prata DP. What is the impact of genome-wide supported risk variants for schizophrenia and bipolar disorder on brain structure and function? A systematic review. Psychol Med. 2015;45:2461–80. [DOI] [PubMed] [Google Scholar]
- 115.Le BD, Stein JL. Mapping causal pathways from genetics to neuropsychiatric disorders using genome-wide imaging genetics: current status and future directions. Psychiatry Clin Neurosci. 2019;73:357–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen X, Zhang Z, Zhang Q, Zhao W, Zhai J, Chen M, et al. Effect of rs1344706 in the ZNF804A gene on the brain network. Neuroimage Clin. 2018;17:1000–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang Q, Cheng W, Li M, Ren H, Hu X, Deng W, et al. The CHRM3 gene is implicated in abnormal thalamo-orbital frontal cortex functional connectivity in first-episode treatment-naive patients with schizophrenia. Psychol Med. 2016;46:1523–34. [DOI] [PubMed] [Google Scholar]
- 118.Liu A, Chen X, Wang ZJ, Xu Q, Appel-Cresswell S, McKeown MJ. A genetically informed, group FMRI connectivity modeling approach: application to schizophrenia. IEEE Trans Biomed Eng. 2014;61:946–56. [DOI] [PubMed] [Google Scholar]
- 119.Jablensky A Subtyping schizophrenia: implications for genetic research. Mol Psychiatry. 2006;11:815–36. [DOI] [PubMed] [Google Scholar]
- 120.Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci. 2010;13:327–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sanislow CA, Ferrante M, Pacheco J, Rudorfer MV, Morris SE. Advancing translational research using NIMH research domain criteria and computational methods. Neuron. 2019;101:779–82. [DOI] [PubMed] [Google Scholar]
- 122.Maynard KR, Collado-Torres L, Weber LM, Uytingco C, Barry BK, Williams SR, et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat Neurosci. 2021;24:425–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yin L, Chau CKL, Sham PC, So HC. Integrating clinical data and imputed transcriptome from GWAS to uncover complex disease subtypes: applications in psychiatry and cardiology. Am J Hum Genet. 2019;105:1193–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nguyen T, Tagett R, Diaz D, Draghici S. A novel approach for data integration and disease subtyping. Genome Res. 2017;27:2025–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Greenwood TA, Lazzeroni LC, Calkins ME, Freedman R, Green MF, Gur RE, et al. Genetic assessment of additional endophenotypes from the Consortium on the Genetics of Schizophrenia Family Study. Schizophr Res. 2016;170:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Greenwood TA, Shutes-David A, Tsuang DW. Endophenotypes in schizophrenia: digging deeper to identify genetic mechanisms. J Psychiatr Brain Sci. 2019;4:e190005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cheng S, Guan F, Ma M, Zhang L, Cheng B, Qi X, et al. An atlas of genetic correlations between psychiatric disorders and human blood plasma proteome. Eur Psychiatry. 2020;63:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Smith AM, King JJ, West PR, Ludwig MA, Donley ELR, Burrier RE, et al. Amino acid dysregulation metabotypes: potential biomarkers for diagnosis and individualized treatment for subtypes of autism spectrum disorder. Biol Psychiatry. 2019;85:345–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sethi S, Brietzke E. Omics-based biomarkers: application of metabolomics in neuropsychiatric disorders. Int J Neuropsychopharmacol. 2015;19:pyv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang J, Yan B, Zhao B, Fan Y, He X, Yang L, et al. Assessing the causal effects of human serum metabolites on 5 major psychiatric disorders. Schizophr Bull. 2020;46:804–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li A, Zalesky A, Yue W, Howes O, Yan H, Liu Y, et al. A neuroimaging biomarker for striatal dysfunction in schizophrenia. Nat Med. 2020;26:558–65. [DOI] [PubMed] [Google Scholar]
- 132.Su-Ping D, Dongdong L, Calhoun VD, Yu-Ping W. Predicting schizophrenia by fusing networks from SNPs, DNA methylation and fMRI data. Annu Int Conf IEEE Eng Med Biol Soc. 2016;2016:1447–50. [DOI] [PubMed] [Google Scholar]
- 133.Jiang W, King TZ, Turner JA. Imaging genetics towards a refined diagnosis of schizophrenia. Front Psychiatry. 2019;10:494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wheeler HE, Aquino-Michaels K, Gamazon ER, Trubetskoy VV, Dolan ME, Huang RS, et al. Poly-omic prediction of complex traits: OmicKriging. Genet Epidemiol. 2014;38:402–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Salakhutdinov R, Hinton G. An efficient learning procedure for deep Boltzmann machines. Neural Comput. 2012;24:1967–2006. [DOI] [PubMed] [Google Scholar]
- 136.Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ursini G, Punzi G, Chen Q, Marenco S, Robinson JF, Porcelli A, et al. Convergence of placenta biology and genetic risk for schizophrenia. Nat Med. 2018;24:792–801. [DOI] [PubMed] [Google Scholar]
- 138.Stilo SA, Murray RM. Non-genetic factors in schizophrenia. Curr Psychiatry Rep.2019;21:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lu Y, Pouget JG, Andreassen OA, Djurovic S, Esko T, Hultman CM, et al. Genetic risk scores and family history as predictors of schizophrenia in Nordic registers. Psychol Med. 2018;48:1201–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet. 2011;12:417–28. [DOI] [PubMed] [Google Scholar]
- 141.Investigators AURP. The “All of Us” research program. N Engl J Med. 2019;381:668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Leppert B, Millard LAC, Riglin L, Davey Smith G, Thapar A, Tilling K, et al. A cross-disorder PRS-pheWAS of 5 major psychiatric disorders in UK Biobank. PLoS Genet. 2020;16:e1008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liang Y, Pividori M, Manichaikul A, Palmer AA, Cox NJ, Wheeler H et al. Polygenic transcriptome risk scores improve portability of polygenic risk scores across ancestries. bioRxiv 2020. 10.1101/2020.11.12.373647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hu Y, Lu Q, Liu W, Zhang Y, Li M, Zhao H. Joint modeling of genetically correlated diseases and functional annotations increases accuracy of polygenic risk prediction. PLoS Genet. 2017;13:e1006836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moore R, Casale FP, Jan Bonder M, Horta D, Bios C, Franke L, et al. A linear mixed-model approach to study multivariate gene-environment interactions. Nat Genet. 2019;51:180–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huang JT, Leweke FM, Tsang TM, Koethe D, Kranaster L, Gerth CW, et al. CSF metabolic and proteomic profiles in patients prodromal for psychosis. PLoS ONE. 2007;2:e756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yu Q, He Z, Zubkov D, Huang S, Kurochkin I, Yang X, et al. Lipidome alterations in human prefrontal cortex during development, aging, and cognitive disorders. Mol Psychiatry. 2020;25:2952–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nugent JL, Mccoy AN, Addamo CJ, Jia W, Sandler RS, Keku TO. Altered tissue metabolites correlate with microbial dysbiosis in colorectal adenomas. J Proteome Res. 2014;13:1921–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Clos-Garcia M, Andrés-Marin N, Fernández-Eulate G, Abecia L, Lavín JL, van Liempd S, et al. Gut microbiome and serum metabolome analyses identify molecular biomarkers and altered glutamate metabolism in fibromyalgia. EBioMedicine. 2019;46:499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bigdeli TB, Genovese G, Georgakopoulos P, Meyers JL, Peterson RE, Iyegbe CO, et al. Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Mol Psychiatry. 2020;25:2455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Millan MJ, Andrieux A, Bartzokis G, Cadenhead K, Dazzan P, Fusar-Poli P, et al. Altering the course of schizophrenia: progress and perspectives. Nat Rev Drug Discov. 2016;15:485–515. [DOI] [PubMed] [Google Scholar]
- 153.Zubiaur P, Soria-Chacartegui P, Koller D, Navares-Gómez M, Ochoa D, Almenara S, et al. Impact of polymorphisms in transporter and metabolizing enzyme genes on olanzapine pharmacokinetics and safety in healthy volunteers. Biomed Pharmacother. 2021;133:111087. [DOI] [PubMed] [Google Scholar]
- 154.Pardiñas AF, Nalmpanti M, Pocklington AJ, Legge SE, Medway C, King A, et al. Pharmacogenomic variants and drug interactions identified through the genetic analysis of clozapine metabolism. Am J Psychiatry. 2019;176:477–86. [DOI] [PubMed] [Google Scholar]
- 155.Saito T, Ikeda M, Mushiroda T, Ozeki T, Kondo K, Shimasaki A, et al. Pharmacogenomic study of clozapine-induced agranulocytosis/granulocytopenia in a japanese population. Biol Psychiatry. 2016;80:636–42. [DOI] [PubMed] [Google Scholar]
- 156.Yu H, Yan H, Wang L, Li J, Tan L, Deng W, et al. Five novel loci associated with antipsychotic treatment response in patients with schizophrenia: a genome-wide association study. Lancet Psychiatry. 2018;5:327–38. [DOI] [PubMed] [Google Scholar]
- 157.Wang Q, Man Wu H, Yue W, Yan H, Zhang Y, Tan L, et al. Effect of damaging rare mutations in synapse-related gene sets on response to short-term antipsychotic medication in chinese patients with schizophrenia: a randomized clinical trial. JAMA Psychiatry. 2018;75:1261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Readhead B, Hartley BJ, Eastwood BJ, Collier DA, Evans D, Farias R, et al. Expression-based drug screening of neural progenitor cells from individuals with schizophrenia. Nat Commun. 2018;9:4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kurita M, Holloway T, García-Bea A, Kozlenkov A, Friedman AK, Moreno JL, et al.HDAC2 regulates atypical antipsychotic responses through the modulation of mGlu2 promoter activity. Nat Neurosci. 2012;15:1245–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhuo C, Hou W, Tian H, Wang L, Li R. Lipidomics of the brain, retina, and biofluids: from the biological landscape to potential clinical application in schizophrenia. Transl Psychiatry. 2020;10:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pillinger T, McCutcheon RA, Vano L, Mizuno Y, Arumuham A, Hindley G, et al. Comparative effects of 18 antipsychotics on metabolic function in patients with schizophrenia, predictors of metabolic dysregulation, and association with psychopathology: a systematic review and network meta-analysis. Lancet Psychiatry. 2020;7:64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jauhar S, Veronese M, Nour MM, Rogdaki M, Hathway P, Turkheimer FE, et al. Determinants of treatment response in first-episode psychosis: an (18)F-DOPA PET study. Mol Psychiatry. 2019;24:1502–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kauppi K, Rosenthal SB, Lo MT, Sanyal N, Jiang M, Abagyan R, et al. Revisiting antipsychotic drug actions through gene networks associated with schizophrenia. Am J Psychiatry. 2018;175:674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pergola G, Di Carlo P, Jaffe AE, Papalino M, Chen Q, Hyde TM, et al. Prefrontal coexpression of schizophrenia risk genes is associated with treatment response in patients. Biol Psychiatry. 2019;86:45–55. [DOI] [PubMed] [Google Scholar]
- 165.So HC, Chau CK, Chiu WT, Ho KS, Lo CP, Yim SH, et al. Analysis of genome-wide association data highlights candidates for drug repositioning in psychiatry. Nat Neurosci. 2017;20:1342–9. [DOI] [PubMed] [Google Scholar]
- 166.Price AJ, Jaffe AE, Weinberger DR. Cortical cellular diversity and development in schizophrenia. Mol Psychiatry. 2021;26:203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tenenbaum JD, Bhuvaneshwar K, Gagliardi JP, Fultz Hollis K, Jia P, Ma L, et al. Translational bioinformatics in mental health: open access data sources and computational biomarker discovery. Brief Bioinform. 2019;20:842–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Borgmann-Winter KE, Wang K, Bandyopadhyay S, Torshizi AD, Blair IA, Hahn CG. The proteome and its dynamics: a missing piece for integrative multi-omics in schizophrenia. Schizophr Res. 2020;217:148–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rappoport N, Shamir R. Multi-omic and multi-view clustering algorithms: review and cancer benchmark. Nucleic Acids Res. 2018;46:10546–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sanchez-Roige S, Palmer AA. Emerging phenotyping strategies will advance our understanding of psychiatric genetics. Nat Neurosci. 2020;23:475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang S, Zhang X, Purmann C, Ma S, Shrestha A, Davis KN, et al. Network effects of the 15q13.3 microdeletion on the transcriptome and epigenome in human-induced neurons. Biol Psychiatry. 2021;89:497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Topol A, Zhu S, Tran N, Simone A, Fang G, Brennand KJ, et al. Signaling in human induced pluripotent stem cell neural progenitor cells derived from four schizophrenia patients. Biol Psychiatry. 2015;78:e29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177:1080. [DOI] [PubMed] [Google Scholar]
- 174.Peterson RE, Kuchenbaecker K, Walters RK, Chen CY, Popejoy AB, Periyasamy S, et al. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell. 2019;179:589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


