Abstract
Influenza virus nucleoprotein (NP) is a critical factor in the viral infectious cycle in switching influenza virus RNA synthesis from transcription mode to replication mode. In this study, we investigated the interaction of NP with the viral polymerase protein complex. Using coimmunoprecipitation with monospecific or monoclonal antibodies, we observed that NP interacted with the RNP-free polymerase protein complex in influenza virus-infected cells. In addition, coexpression of the components of the polymerase protein complex (PB1, PB2, or PA) with NP either together or pairwise revealed that NP interacts with PB1 and PB2 but not PA. Interaction of NP with PB1 and PB2 was confirmed by both coimmunoprecipitation and histidine tagging of the NP-PB1 and NP-PB2 complexes. Further, it was observed that NP-PB2 interaction was rather labile and sensitive to dissociation in 0.1% sodium dodecyl sulfate and that the stability of NP-PB2 interaction was regulated by the sequences present at the COOH terminus of NP. Analysis of NP deletion mutants revealed that at least three regions of NP interacted independently with PB2. A detailed analysis of the COOH terminus of NP by mutation of serine-to-alanine (SA) residues either individually or together demonstrated that SA mutations in this region did not affect the binding of NP to PB2. However, some SA mutations at the COOH terminus drastically affected the functional activity of NP in an in vivo transcription-replication assay, whereas others exhibited a temperature-sensitive phenotype and still others had no effect on the transcription and replication of the viral RNA. These results suggest that a direct interaction of NP with polymerase proteins may be involved in regulating the switch of viral RNA synthesis from transcription to replication.
Influenza viruses encompass a major group of human and animal pathogens belonging to enveloped, segmented, negative-strand RNA viruses. Following infection of permissive cells, both the transcription and the replication of influenza virus RNAs occur in the cell nucleus by a virus-specific RNA-dependent RNA polymerase protein complex (18). Various biochemical and genetic analyses have shown that three polymerase proteins (PB1, PB2, and PA) interact with each other and function as a three-polymerase protein (3P) heterocomplex in both transcription and replication of viral RNAs (vRNAs) (17, 30). Three types of influenza virus-specific RNAs are synthesized in infected cells. (i) mRNAs, the product of transcription, possess at the 5′ end a capped 10- to 13-nucleotide sequence of nonviral origin derived from the newly synthesized host nuclear RNAs, lack 17 to 22 nucleotides from the 3′ end, but possess poly(A) sequences at the 3′ end. (ii) cRNAs and (iii) vRNAs of plus and minus polarity, respectively, are the products of replication (17, 30). cRNAs are complete complementary copies of vRNA segments and do not possess either the capped primer at the 5′ end or poly(A) sequences at the 3′ end and function as the template for synthesis of vRNA which is also a complete copy of the cRNA template.
For transcription of mRNA, influenza virus uses a unique strategy in the host nucleus (17, 18). PB2, a member of the 3P complex, recognizes the capped host RNAs and cleaves the 5′ cap containing 10 to 13 nucleotides at a specific site, which is used by PB1, another member of the same 3P complex, as a primer for chain elongation. PB1 possessing the conserved polymerase motifs (7) uses the 5′-capped primer for initiating and continuing mRNA synthesis by chain elongation with the vRNA as a template (17). Transcription of mRNA is terminated at a specific site approximately 17 to 22 nucleotides from the 5′ end of the template vRNA, and poly(A) sequences are added at the 3′ end of viral mRNA by stuttering of the 3P complex on the oligo(U) stretch of the vRNA template (13). cis-acting elements such as a panhandle structure and poly(U) stretch of the template vRNA appear to be critical in both transcription termination and poly(A) addition at the 3′ end of mRNA (13, 22).
Unlike mRNA transcription, vRNA replication leading to synthesis of the cRNA and vRNA uses an entirely different mechanism of RNA synthesis by the 3P complex since it requires both primer-independent initiation of RNA synthesis at the 5′ end and chain completion without premature termination and without poly(A) addition at the 3′ end. Therefore, in the infectious cycle, a switch from transcription to replication after the primary transcription of vRNA must take place for cRNA synthesis, the first step in vRNA replication. The mechanism of the switch from transcription to replication is unclear at present and appears to require both viral and host factors (38). Genetic and biochemical studies have demonstrated that viral nucleoprotein (NP) is a critical factor in switching RNA synthesis from transcription mode to replication mode and that the switch to replication mode fails to occur in the absence of soluble NP both in vitro and in virus-infected cells (17).
Influenza virus NP is a major structural protein in virus particles and has multiple functions in the viral infectious cycle. It is a basic protein rich in arginine with a net positive charge of +14 at pH 6.5 (20). In vitro it binds to RNA nonspecifically, yet in vivo NP binds only to complete cRNA (plus polarity) and vRNA (minus polarity), forming cRNP and vRNP, respectively, and does not bind to viral mRNA (plus polarity) possessing 5′ cap and 3′ poly(A) sequences (17). NP has a karyophilic signal(s) (31, 41) for nuclear translocation and along with the 3P complex plays a critical role in nuclear translocation of vRNP after uncoating of the infecting virus. vRNP also interacts with M1 protein, suggesting a possible interaction of NP with M1 that likely plays a critical role in the budding process of virus particles (4). NP is a phosphoprotein (14, 34, 35) that has been shown to undergo autophosphorylation and also to possess phosphorylation activity in vitro (11).
Although NP is critically required for switching vRNA synthesis from transcription to replication, the function of NP in replication of vRNA remains unclear. So far, two mechanisms have been postulated for the role of NP in the transcription-to-replication switch. (i) NP has been shown to be an antitermination factor. Therefore, NP may facilitate melting of the panhandle structure of the vRNA template (6) and somehow stop stuttering of the 3P complex on the oligo(U) stretch of the vRNA template and thereby prevent chain termination leading to completion of cRNA synthesis. However, transcripts initiated with the capped primer cannot be antiterminated in the presence of soluble NP (6), suggesting that both the initiation of RNA synthesis in the absence of the capped primer at the 5′ end and the antitermination at the 3′ end are coordinated and most likely occur concurrently by the same mechanism. (ii) NP binds to the nascent product RNA during synthesis to form cRNP or vRNP. This binding of NP to the newly synthesized RNA will somehow cause antitermination and permit reading through the poly(U) tract in the vRNA template. On the other hand, the presence of cap at the 5′ end would prevent NP from binding to the nascent mRNA product and thereby prevent antitermination of the capped mRNA transcript (17). However, neither of these hypotheses could explain how the primer-independent initiation, the first step in cRNA or vRNA synthesis, could occur in the presence of free NP. For this reason, we have examined the third possibility, that the free NP could in fact directly interact with one or more components of the 3P complex and thereby modify the polymerase protein complex from transcription mode to replication mode. This function of NP could occur in addition to its known antitermination effect. In this report, we have demonstrated that NP indeed binds to the components of the polymerase protein complex both in virus-infected cells and in cells coexpressing polymerase and NP proteins. We have further shown that NP interacts with PB1 and PB2 but not with PA and that multiple regions of NP bind to PB2, a protein involved in binding and cleaving the 5′-capped RNA primer which is used for the initiation of viral mRNA transcription. In addition, we show that the NP-PB2 interaction is labile and that the COOH terminus of NP provides a regulatory role affecting the stability of NP-PB2 interaction. The implication of these results in the regulation of transcription and replication of influenza virus RNA is discussed.
MATERIALS AND METHODS
Viruses and cells.
Influenza virus (A/WSN/33) was grown in MDCK cells (7). Recombinant vaccinia virus expressing T7 RNA polymerase (VTF7.3) was a gift from Bernard Moss, National Institute of Allergy and Infectious Diseases, Bethesda, Md. (10). HeLa and CV1 cells were used for growing vaccinia virus stock and determining infectivity titer (PFU), respectively. COS1 cells were used for influenza virus polymerase activity assay (7, 8) and for studying protein-protein interactions by coexpression.
Plasmids and mutants of the NP gene.
Plasmids pGEM PB1, pGEM PB2, pGEM PA, and pGEM NP were used as described before (8). Standard techniques were used for DNA manipulation (37). COOH-terminal deletion mutants were constructed by using restriction enzyme sites to remove DNA encoding different amino acids from the full-length NP containing 498 amino acids (aa) as follows: ΔC33, SacI; ΔC140, HindIII; and ΔC165, SphI. Plasmid pET17b (Novagen, Madison, Wis.) containing an 11-aa T7 tag was used at the NH2 terminus to express different parts of NP as T7-tagged NP fusion proteins, using different restriction enzyme sites in NP as follows: NP I, aa 1 to 161 (EcoRI to BamHI); NP II, aa 160 to 256 (BamHI to BglII); NP III, aa 255 to 341 (BglII to BglII); NP IV, aa 340 to 498 (BglII to EcoRI); and NP V, aa 340 to 465 (BglII to SacI).
For the desired mutation of specific amino acids, PCR amplification with different oligonucleotides was used for site-directed mutagenesis of the NP gene. The PCR product was double digested with different restriction enzymes and ligated into pGEM NP by three-way ligation. Individual clones of pGEM NP containing serine-to-alanine (SA) mutations at aa 486, 482, 478, 473, and 467 were confirmed by sequencing the entire PCR-amplified DNA to ensure that additional mutations were not introduced by PCR amplification.
Plasmid Ribo-CAT (23) was used in influenza virus transcription-replication assays as described before (8). Plasmid pRSET B (Invitrogen, San Diego, Calif.) was used to clone PB1, PB2, PA, and NP to express them as histidine-tagged proteins.
Infection and transfection.
For analysis of protein-protein interactions during influenza virus infection, MDCK cells were infected with influenza virus (A/WSN/33) at a multiplicity of infection (MOI) of 5, unabsorbed viruses were removed by washing, and cells were labeled for 1 h after 6 h postinfection (hpi). For component expression experiments, COS1 cells in 60-mm-diameter dishes were infected with vaccinia virus VTF7.3 at an MOI of 5 for 1 h and then transfected with pGEM plasmids carrying genes encoding NP, PB1, PB2, or PA alone or in combination, using Lipofectin-mediated transfection as described before (7, 8). Cells were labeled at 14 h posttransfection (hpt) for 1 h, and the cell lysates were used for immunoprecipitation.
For the transcription-replication assay, COS1 cells in 60-mm-diameter dishes were infected with VTF7.3 at an MOI of 5 for 1 h and then transfected with a mixture of plasmid pGEM NP (or mutant NP; 5 μg), pGEM PB1 and pGEM PB2 (2 μg of each), pGEM PA (0.5 μg), and Ribo-CAT (3 μg) DNA by Lipofectin-mediated transfection. At 24 hpt, cells were lysed by freezing and thawing and assayed for chloramphenicol acetyltransferase (CAT) activity as described before (7, 8).
Radiolabeling and preparation of the infected cell lysate.
For radiolabeling of proteins, influenza virus (A/WSN/33)-infected MDCK cells at 5 hpi or VTF7.3-infected COS1 cells at 13 hpt were washed with phosphate-buffered saline containing 0.01% CaCl2 and 0.01% MgCl2, incubated in methionine and cysteine-free medium for 1 h at 37°C, and then labeled for 1 h in the same medium containing 50 μCi of Express 35S (New England Nuclear, Boston, Mass.) per ml. After labeling, cell monolayers were washed and scraped in cold phosphate-buffered saline and pelleted by centrifugation. Influenza virus-infected MDCK cells were lysed and separated into cytoplasmic and nuclear fractions as described previously (1). Nuclear fractions were then resuspended in 1 ml of TNE buffer (10 mM Tris HCl [pH 7.5], 20 mM NaCl, 2 mM EDTA) and lysed by sonication. Both cytoplasmic and nuclear fractions were diluted to 4 ml by adding TNE suspension buffer. The vRNP complex was removed from the cytoplasmic and nuclear fractions by centrifugation for 3 h at 48,000 rpm in an SW55 Ti rotor, yielding the supernatants (RNP-free lysate) and pellet (RNP) (6, 9). The RNP-free lysates from cytoplasmic and nuclear fractions were divided into two parts; one part was treated with RNases A (1 mg/ml) and T1 (1,000 U/ml) at 37°C for 30 min, and the other part remained untreated.
For preparing the lysates of His6-tagged proteins, the transfected COS1 cells were suspended in buffer A (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 2 mM imidazole, 1% Nonidet P-40) and lysed by sonication. The cell lysate was centrifuged at 12,000 rpm for 5 min at 4°C, and the supernatant was used for binding to TALON resin, a cobalt-immobilized metal affinity chromatography resin (Clontech, Palo Alto, Calif.).
Immunoprecipitation, His tag purification, and Western blotting.
For immunoprecipitation of labeled proteins in influenza virus-infected cells, both RNase-treated and untreated RNP-free supernatants of cytoplasmic and nuclear fractions were adjusted to 1× radioimmunoprecipitation assay (RIPA) buffer containing 10 mM Tris HCl (pH 7.5), 2 mM EDTA, 100 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 1% aprotinin, and 5 mg of bovine serum albumin per ml. Both samples were divided into five parts; each part was immunoprecipitated with either monospecific anti-PB1, anti-PB2, and anti-PA or monoclonal anti-NP antibodies or with normal serum (8). The immunoprecipitated complex was further washed with RIPA buffer containing 500 mM NaCl and finally in RIPA buffer without bovine serum albumin. The immunoprecipitate was analyzed by sodium dodecyl sulfate-polyacrylamide gel (8%) electrophoresis (SDS-PAGE).
For immunoprecipitation of cell lysates after individual component expression or coexpression, the labeled transfected COS1 cells were lysed in 1× RIPA buffer by sonication, and the lysate was clarified by centrifugation at 12,000 rpm at 4°C for 10 min. The lysate was then divided into two parts; one part was immunoprecipitated with monoclonal anti-NP antibodies, and other part was immunoprecipitated with monospecific anti-PB1, anti-PB2, or anti-PA antibodies according to the component proteins used for coexpression with NP (8). The immunoprecipitated complex was further washed and analyzed as described above.
When plasmid pET17b was used to express different parts of NP, anti-T7 tag antibody (Novagen) was used for immunoprecipitation. The immunoprecipitated sample was analyzed in SDS-polyacrylamide gels containing 10% polyacrylamide in the top half and 15% polyacrylamide in the bottom half.
For purification and analysis of His6-tagged proteins, lysates from cells transfected with pRSET PB1, pRSET PB2, pRSET PA, or pRSET NP were incubated with TALON metal affinity resin (Clontech) for 2 h at 4°C and washed with buffer A containing 500 mM NaCl and 5 mM imidazole. Finally, the resin-bound proteins were eluted, analyzed by SDS-PAGE (8% gel), and Western blotted, and portions of the same blot were probed with anti-PB1, anti-PB2, or anti-PA antibodies, depending on which proteins were expressed or coexpressed. Anti-WSN antibodies were used for probing NP in Western blot analysis. The blot was developed with Western blot-chemiluminescence agent (NEN).
RESULTS
Interaction of NP with the RNP-free polymerase protein complex in influenza virus-infected cells.
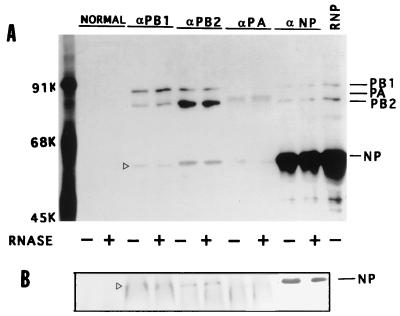
First we wanted to determine if viral NP interacted directly with the viral polymerase protein complex in influenza virus-infected cells. Accordingly, we used coimmunoprecipitation to identify which if any of the influenza virus polymerase protein(s) interacted with NP during viral infection. Since NP is the major structural component of RNP and the 3P complex is associated with RNP (27), it could be assumed that coimmunoprecipitation of any polymerase protein(s) and NP might be due to specific binding of both NP and the 3P complex with the vRNA, forming the vRNP-polymerase protein complex in virus-infected cells. To overcome the problem, we used the strategy described previously for removal of the RNP complex (6, 9). Briefly, MDCK cells were infected with influenza virus (A/WSN/33) at an MOI of 5 and labeled from 6 to 7 hpi. The labeled cells were fractionated into cytoplasm and nuclear fractions (1), and RNPs were pelleted from each fraction by ultracentrifugation (6, 9). Furthermore, since RNAs in the influenza virus RNP are susceptible to degradation by RNase treatment (15), any RNP (or RNA) contamination in the supernatant was further eliminated by treating the supernatant with RNases A and T1. Accordingly, the RNP-free supernatants from both cytoplasmic and nuclear fractions were divided into two equal aliquots which were either RNase treated or untreated. Finally, equal amounts of RNase-treated and untreated samples from cytoplasmic and nuclear fractions were immunoprecipitated with nonimmune serum or anti-PB1, anti-PB2, anti-PA, or anti-NP antibodies. As shown in Fig. 1A, anti-PB1, anti-PB2, and anti-PA antibodies immunoprecipitated the specific polymerase protein as well as the other members of the polymerase protein complex. It was shown previously that PA forms a less stable complex with either PB1 or PB2 and usually dissociates during immunoprecipitation (1, 9, 39). In addition, each of the antipolymerase antibodies also coimmunoprecipitated a 56-kDa protein. Conversely, anti-NP monoclonal antibodies immunoprecipitated NP as well as PB1 and PB2 but not PA. Results were similar with and without RNase treatment. To determine if the 56-kDa band which was coimmunoprecipitated with anti-PB1, anti-PB2, and anti-PA antibodies was NP, the relevant portion of the same gel was blotted and probed with anti-WSN antibodies. It should be noted that rabbit anti-WSN antibodies but not mouse monoclonal anti-NP antibodies could detect NP in Western blots. The results (Fig. 1B) show that the 56-kDa band that coimmunoprecipitated with anti-PB1 and anti-PB2 antibodies was NP, but this was not the case for the band coimmunoprecipitated by anti-PA antibodies. This protein brought down by anti-PA antibodies might be the 60-kDa protein which reacts nonspecifically with anti-PA antibodies as has been observed previously (1). Results from nuclear fractions were similar to those observed for the cytoplasmic fractions (data not shown). These results demonstrated that a fraction of RNP-free NP interacted with the 3P complex in influenza virus-infected cells. It should be noted that immunoprecipitation was carried out in the absence of SDS (0.1%) in RIPA buffer, as 0.1% SDS caused dissociation of the NP-polymerase protein complex and as a result the NP-polymerase protein complex could not be coimmunoprecipitated by either anti-polymerase or anti-NP antibodies (data not shown).
FIG. 1.
Presence of the NP-polymerase protein complex in influenza virus-infected cells. MDCK cells were infected with influenza virus (A/WSN/33) at an MOI of 5 and labeled with Express 35S (NEN) at 6 hpt for 1 h. Cells were lysed and fractionated into cytoplasmic and nuclear fractions, and vRNPs were removed from both fractions by ultracentrifugation (1, 6). Aliquots of RNP-free supernatants from the cytoplasmic fraction were treated with (+) or without (−) RNase. Each fraction was divided into five parts and immunoprecipitated with either normal serum or anti-PB1 anti-PB2, anti-PA, or anti-NP antibodies in the absence of SDS as noted in Materials and Methods. The RNP pellet was dissolved and immunoprecipitated with a mixture of anti-PB1, anti-PB2, and anti-NP antibodies. The immunoprecipitated complexes were separated by SDS-PAGE (8% gel) and autoradiographed (A). The gel in panel A was transferred to a membrane, and the relevant portion of the membrane was probed with anti-WSN antibodies and detected by chemiluminescence (B). The position of NP is shown with an open arrowhead. Similar results were obtained from the RNP-free nuclear supernatant (data not shown).
Interaction of NP with the viral polymerase protein complex by component expression.
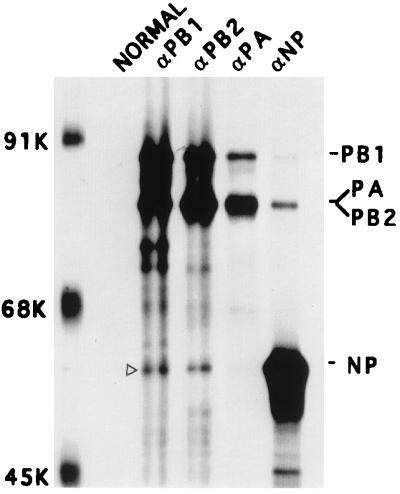
Although in influenza virus-infected cells the coimmunoprecipitation results with antipolymerase or anti-NP antibodies were similar with and without RNase treatment of the RNP-free polymerase protein complex, the presence of vRNA (or vRNP) in the infected cell lysate could not be completely ruled out. To eliminate the problem of vRNA (or vRNP) contamination, we used an expression system of individual components from cDNA (7, 8). Accordingly, we expressed three polymerase proteins and NP together, using the T7-vaccinia virus expression system. COS1 cells were infected with VTF7.3 at an MOI of 5 for 1 h and then transfected with pGEM PB1, pGEM PB2, pGEM PA, and pGEM NP DNAs. At 14 hpt, the transfected cells were labeled for 1 h and lysed in 1× RIPA buffer without SDS by sonication. The cell lysate was divided into five parts, and each part was immunoprecipitated by either normal serum or any one of the anti-PB1, anti-PB2, anti-PA, or anti-NP antibodies. Results (Fig. 2) show that the three polymerase proteins formed the 3P heterocomplex as expected and that PA was rather unstable in the 3P complex as seen earlier (1, 9, 17, 39). Furthermore, anti-PB1 and anti-PB2 but not anti-PA antibodies coimmunoprecipitated NP. Conversely, anti-NP antibodies coimmunoprecipitated PB1 and PB2. Although in this experiment PA and PB2 migrated to the same position, the band immunoprecipitated by NP was PB2 and not PA as shown by Western blotting (data not shown) and also as shown later (Fig. 3). These results demonstrated that the influenza virus NP interacted directly with polymerase protein PB1 and PB2 but not with PA in the absence of any vRNA or other viral components.
FIG. 2.
Interaction of NP with the polymerase protein complex in coexpressing cells. COS1 cells were infected with VTF7.3 at an MOI of 5 and transfected with a mixture of pGEM PB1, pGEM PB2 (3 μg of each DNA), pGEM PA (2 μg of DNA), and pGEM NP (1 μg of DNA). At 14 hpt, cells were labeled with Express 35S for 1 h, lysed, and divided into five parts. Each part was immunoprecipitated with either normal serum or anti-PB1, anti-PB2, anti-PA, or anti-NP antibodies. The immunoprecipitated complex was analyzed by SDS-PAGE (8% gel). The open arrowhead shows the position of NP.
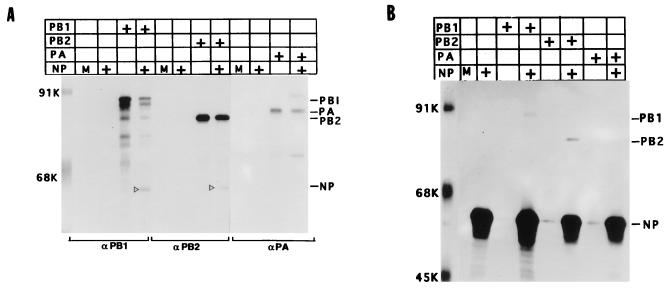
FIG. 3.
Interaction of NP with PB1 and PB2 in coexpressing cells. COS1 cells were infected with VTF7.3 at an MOI of 5 and transfected alone with pGEM NP (1 μg), pGEM PB1 (3 μg), pGEM PB2 (3 μg), or pGEM PA (2 μg) or cotransfected pairwise as indicated (+). At 14 hpt, cells were labeled with Express 35S for 1 h and lysed. The lysate was divided into two parts; one part was immunoprecipitated with anti-NP (B), and the other was immunoprecipitated with either anti-PB1, anti-PB2, or anti-PA antibodies (A). The immunoprecipitated complex was separated by SDS-PAGE (8% gel) and autoradiographed. M, mock transfected with pGEM 3; +, DNA used for transfection. The open arrowhead shows the position of NP.
Since NP interacted with two of the polymerase proteins (PB1 and PB2) in the presence of the whole polymerase protein complex, we wanted to determine whether 3P complex formation was required for interaction of PB1 and PB2 with NP or whether NP can interact with the individual polymerase proteins in the absence of 3P complex formation. We therefore coexpressed NP pairwise with PB1, PB2, or PA. Accordingly, COS1 cells were infected with VTF7.3 and then either mock transfected with pGEM 3, transfected individually with pGEM NP, pGEM PB1, pGEM PB2, or pGEM PA, or cotransfected pairwise with pGEM NP and one of the three plasmids expressing polymerase proteins. At 14 hpt, cells were labeled and lysed in RIPA buffer without SDS as described in Materials and Methods. The lysates were divided into two parts; one part was immunoprecipitated with either anti-PB1, anti-PB2, or anti-PA antibodies, depending on the P protein expressed, and the other part was immunoprecipitated by anti-NP antibodies. As shown in Fig. 3A, when PB1 or PB2 was coexpressed with NP and immunoprecipitated with either anti-PB1 or anti-PB2 antibodies, NP was coimmunoprecipitated along with either PB1 or PB2. But when PA was coexpressed with NP and immunoprecipitated with anti-PA antibodies, only PA and not NP was immunoprecipitated. However, it should be noted that since PA was expressed in lower amounts and formed a less stable complex even with other polymerase proteins, the formation of minor amounts of PA-NP complexes cannot be completely ruled out. Conversely, anti-NP antibodies immunoprecipitated NP as well as PB1 or PB2 but not PA from the lysates of coexpressing cells (Fig. 3B). These results also show that the interaction between NP and PB1 or PB2 was specific, as anti-PB1, anti-PB2, and anti-PA antibodies did not cross-react with NP (Fig. 3A). Likewise, anti-NP antibodies did not cross-react with either PB1, PB2, or PA (Fig. 3B).
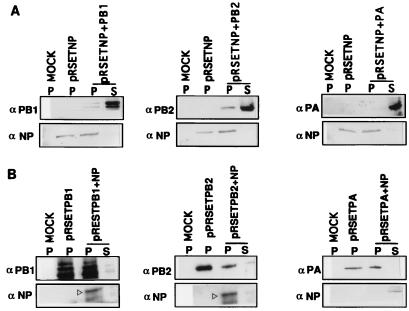
To demonstrate further that the interaction of PB1 and PB2 with NP was specific, we expressed PB1, PB2, and PA with a His6 tag at the NH2 terminus either individually or with NP. Conversely, we also expressed His6-tagged NP alone or with PB1, PB2, or PA. Accordingly, COS1 cells were infected with VTF7.3 and transfected with pGEM NP either alone or with pRSET PB1, pRSET PB2, or pRSET PA DNA. Alternatively, cells were transfected with pRSET NP DNA alone or with pGEM PB1, pGEM PB2, or pGEM PA DNA as described in Materials and Methods. At 14 hpt, cells were lysed and His6-tagged protein was purified by using TALON resin, a cobalt-based metal affinity resin, as described in Materials and Methods. The TALON-bound proteins were eluted, analyzed by SDS-PAGE (8% gel), and blotted to a membrane. Relevant portions of the blot were probed with either anti-WSN, anti-PB1, anti-PB2, or anti-PA antibodies. These results demonstrated that His-tagged NP formed complexes with PB1 or PB2 but not with PA (Fig. 4A). Conversely, His-tagged PB1 and PB2 but not PA interacted with NP (Fig. 4B). Analysis of supernatants (unbound) showed that both PA and NP were present when coexpressed but did not interact with each other. Taken together, these results from two independent experimental approaches using coimmunoprecipitation as well as His-tagged proteins demonstrate that PB1 and PB2 but not PA formed complexes with NP both in virus-infected cells and in cells coexpressing these proteins. These results further demonstrated that NP can interact with either PB1 or PB2 independently in the absence of 3P heterocomplex formation.
FIG. 4.
Copurification of the polymerase-NP protein complex, using either His-tagged NP or His-tagged PB1, PB2, or PA. COS1 cells in 60-mm-diameter dishes were infected with VTF7.3 at an MOI of 5 and transfected with pRSET NP (2 μg) alone or with pGEM PB1 (4 μg), pGEM PB2 (4 μg), or pGEM PA (2 μg) (A). In another set, VTF7.3-infected COS1 cells were transfected with pRSET PB1, (4 μg), pRSET PB2 (4 μg), or pRSET PA (2 μg) alone or with pGEM NP (2 μg). At 14 hpt cells were lysed as described in Materials and Methods. The lysate was incubated with TALON beads (Clontech) for 2 h with shaking in 4°C. The beads were then washed as described in Materials and Methods. TALON bead-bound (P) and unbound (S) proteins were analyzed by SDS-PAGE (8% gel) and Western blotted, and respective portions were probed with either anti-PB1, anti-PB2, anti-PA, or anti-WSN antibodies and developed in chemiluminescence solution. Open arrowheads show the positions of NP.
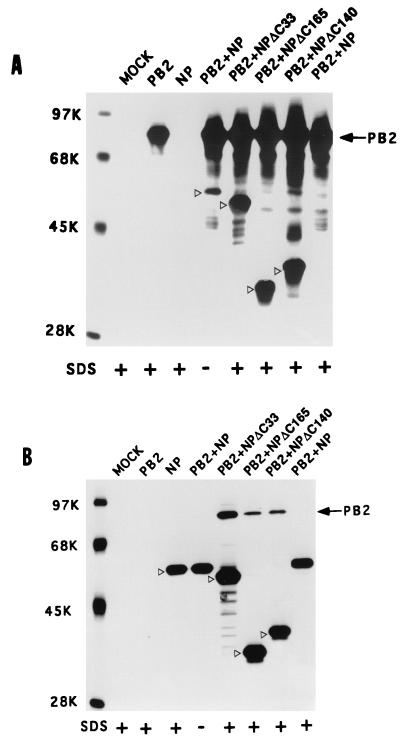
Deletion from the COOH terminus of NP enhances its binding to PB2.
Since NP-PB2 interaction is likely to affect PB2 function in switching from transcription to replication, we wanted to investigate the NP-PB2 interaction in further detail. To determine the regions of NP interacting with PB2, initially we made a number of COOH-terminal deletion mutants of NP. These NP deletion mutants were cotransfected with PB2. Accordingly, COS1 cells were infected with VT7.3 at an MOI of 5 and transfected with either pGEM NP or COOH-terminus deletion mutants of NP along with pGEM PB2. At 14 hpt, cells were labeled and lysed as described in Materials and Methods. The clarified lysate was divided into two parts; one part was immunoprecipitated with anti-NP antibodies, and the other was immunoprecipitated with anti-PB2 antibodies. As shown in Fig. 5A, anti-PB2 antibodies coimmunoprecipitated the wild-type (WT) NP as well as COOH-terminus deletion mutants of NP (Fig. 5A, lanes PB2+NP and PB2). Likewise, antibodies against NP coimmunoprecipitated PB2 along with NP deletion mutants (Fig. 5B, lanes PB2+NP and NP). As noted earlier, in the presence of 0.1% SDS in RIPA buffer, the complex between WT NP and WT PB2 became dissociated and could not be coimmunoprecipitated with either antibody (Fig. 5A and B, lanes PB2+NP, SDS+). But when 33 aa were deleted from the COOH terminus of NP (NPΔ33), PB2 and NP interacted with each other two to three times more in the absence of SDS (data not shown), and more importantly, a major fraction of the NP-PB2 complex was resistant to 0.1% SDS in RIPA buffer and in washing buffer (Fig. 5A and B, lanes PB2+NPΔC33, SDS+). Mutants with further deletion from the COOH terminus of NP (ΔC140 and ΔC165) behaved similarly to the NPΔC33 mutant (Fig. 5A, lanes PB2+NPΔC140 and PB2+NPΔC165; Fig. 5B, lanes PB2+NPΔC140 and PB2+NPΔC165). These results showed that the COOH terminus of NP affects the stability of NP-PB2 complex formation.
FIG. 5.
COOH-terminal deletion NP mutants bind strongly to PB2. COS1 cells in a 60-mm-diameter dish were infected with VTF7.3 at an MOI of 5 and transfected with pGEM PB2 and pGEM NP or NP mutants (2 μg of each). At 14 hpt, cells were labeled with Express 35S for 1 h, lysed by sonication, and clarified. The lysate was divided into two parts. One part was immunoprecipitated with anti-PB2 antibodies (A), and the other part was immunoprecipitated with anti-NP antibodies (B). The immunoprecipitated complex was analyzed by SDS-PAGE (8% gel). Positions of WT and mutant NP are shown with open arrowheads. +, immunoprecipitation and washing in the presence of 0.1% SDS.
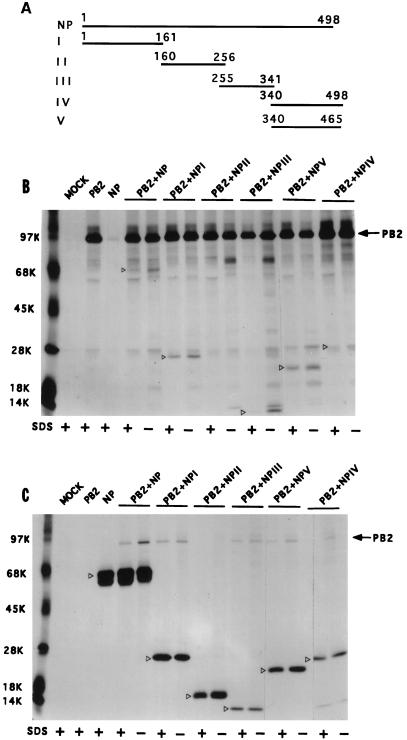
Multiple regions of NP interact independently with PB2.
To further dissect NP-PB2 interaction, the NP cDNA was digested with appropriate restriction enzymes into four fragments (NP I to NP IV [Fig. 6A]) as stated in Materials and Methods. In addition, we constructed NP V, which is same as NP IV except that it lacks the last 33 aa. Each NP fragment was cloned into a pET expression system with a T7 tag at the NH2 terminus in the proper translation frame.
FIG. 6.
Interaction of different NP fragments with PB2 protein. (A) Schematic diagram of different parts of NP expressed in pET vector. Numbers on the lines indicate amino acid residues of NP. These constructions were made by using appropriate restriction sites as stated in Materials and Methods. (B and C) Interactions of various parts of NP with PB2. COS1 cells were infected with VTF7.3 at an MOI of 5 and transfected with pET NP (or pET NP mutants) along with pGEM PB2 as described in Materials and Methods. At 14 hpt, cells were labeled with Express 35S, lysed, divided into two parts, and immunoprecipitated with either anti-PB2 (B) or T7 tag antibodies for NP mutants (C). The immunoprecipitated samples were analyzed by SDS-PAGE (10% [top half] and 15% [bottom half] polyacrylamide). Open arrowheads show the positions of WT and mutant NP.
The WT and NP deletion mutants (I to V) were coexpressed with PB2, and NP-PB2 complex formation was assayed by coimmunoprecipitation using either anti-PB2 antibodies (Fig. 6B) or anti-T7 tag antibodies (Fig. 6C) in the presence or absence of 0.1% SDS. Results show that NP I, NP III, NP IV, and NP V interacted with PB2 but NP II (aa 160 to 256) failed to form a complex with PB2 by immunoprecipitation using either anti-PB2 or anti-T7 tag antibodies. Complex formation could be demonstrated both in the presence and in the absence of SDS (0.1%), and more complex was present in the absence of SDS (0.1%) as expected. These results show that multiple regions of NP (I, III, and IV) can independently interact with PB2. These results also demonstrated the specificity of the coimmunoprecipitation, as NP II failed to form complex with PB2 although NP II was expressed well (Fig. 6C).
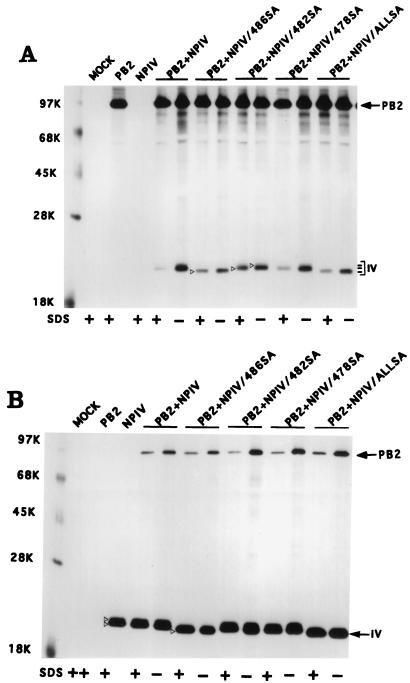
Mutational analysis of the COOH terminus of NP.
The COOH-terminal region of NP appears to regulate the stability of NP-PB2 interaction, as deletion of the last 33 aa of NP made the NP-PB2 complex stable to 0.1% SDS (Fig. 5). We therefore decided to determine the function of this COOH region by mutational analysis. Since NP is known to undergo phosphorylation and since serine is the only phosphoamino acid found in NP (14, 34, 35), we decided to mutate the five serine residues present in the last 33-aa sequence of NP either individually or together and determine their effect on NP-PB2 binding. Accordingly, NP mutants individually possessing 486SA, 482SA, or 478SA mutation and an NP mutant possessing these three SA mutations as well as 473SA and 467SA mutations were constructed by site-specific mutagenesis, and the effect of these mutations on the interaction of the NP IV fragment with PB2 was investigated. NP IV-PB2 interactions were analyzed by using the pET expression system for NP IV and NP IV SA mutants and the T7 expression system for PB2. Upon cotransfection, anti-T7 tag antibodies were used for immunoprecipitation of NP IV and NP IV mutants, whereas anti-PB2 antibodies were used for immunoprecipitation of PB2. Results (Fig. 7) show that SA mutations, either individually or together, in the NP IV fragment did not affect their binding to PB2 or the stability of the NP IV-PB2 complex in the presence of SDS. However, the 486SA mutation affected its migration in the gel. The mutant 486SA, when present alone or combined with others, migrated faster than the WT or other NP IV mutants (Fig. 7A). Whether this migration behavior was due to the effect of mutation on the structure of the polypeptide or due to its effect on phosphorylation remains to be determined.
FIG. 7.
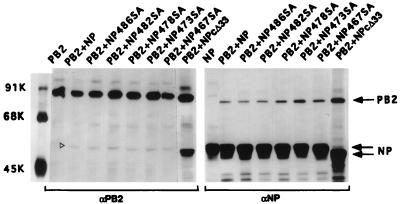
Effects of SA mutations on the binding of NP IV to PB2. COS1 cells were infected with VTF7.3 at an MOI of 5 and then transfected with either pET NP IV or pET NP IV mutants along with pGEM PB2 DNA. Cells were labeled with Express 35S at 14 hpt for 1 h, lysed in the absence of SDS, and divided into four parts. Two parts were adjusted to 0.1% SDS (+). One part from each preparation was immunoprecipitated with anti-PB2 (A) or with anti-T7 tag antibodies for NP (B). The immunoprecipitated complex was analyzed by SDS-PAGE (10% [top half] and 15% [bottom half] polyacrylamide). Open arrowheads show the positions of NP IV mutants. Three lines on the right indicate the positions of different NP IV mutant proteins (open arrowhead) in the gel. ALLSA, all SA mutations (467SA, 473SA, 478SA, 482SA, and 486SA) combined.
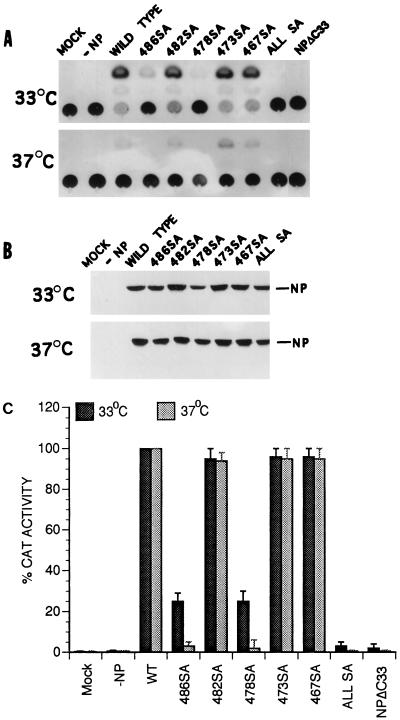
To determine if the SA mutations at the COOH terminus of NP affected its function in the influenza virus transcription-replication assay in vivo, we incorporated the SA mutations in the context of whole NP and used a modified Ribo-CAT system for assaying transcription and replication as described earlier (7, 8). In this system, the active polymerase protein complex is reconstituted in vivo and NP is required for replication and amplification of Ribo-CAT RNA under the control of the influenza virus RNA promoter. Since many of the single-base mutations are known to cause temperature-sensitive (ts) lesions (17, 21, 24, 25), we also wanted to determine the effect of SA mutations on temperature sensitivity. Since temperature sensitivity for influenza virus is usually determined at 33°C versus 39.5 to 42°C, we first investigated the effects at 33, 37, and 39.5°C in the in vivo transcription-replication assay using the WT proteins. We found that the vaccinia virus expression system used for the influenza virus transcription-replication assay was highly sensitive at 39.5°C and that little or no CAT expression was detected even for the WT viral proteins at 39.5°C (data not shown). More surprisingly, we observed that CAT expression was at least threefold higher at 33°C than at 37°C (Fig. 8A). We therefore analyzed both the WT and SA NP mutants at 33 and 37°C in the transcription-replication assay using the Ribo-CAT system (8). Results were compared by using the CAT activity of the WT NP at 33 and 37°C as 100% (Fig. 8C). Results show that NPΔC33 and all SA NP mutants combined together expressed little CAT activity at either 33 or 37°C and therefore were essentially nonfunctional at both temperatures. On the other hand, 482SA, 473SA, and 467SA NP mutants behaved like the WT protein at both temperatures. However, mutants 486SA and 478SA exhibited an intermediate phenotype: CAT activity was reduced to 20 to 30% of the WT level at 33°C but was essentially undetectable at 37°C, i.e., was highly ts (Fig. 8C). Western assay of the same lysate showed that essentially similar amounts of NP proteins were synthesized at both temperatures for the WT and mutant NP proteins (Fig. 8B). Since some NP SA mutations affected CAT activity in the in vivo transcription-replication assay, we wanted to determine if these SA mutations affected binding or stability of NP-PB2 interaction in the context of whole NP. Although the SA mutations in fragment NP IV did not affect its binding to PB2 (Fig. 7), these mutations may behave differently in the context of whole NP. Accordingly, NP-PB2 interaction was analyzed by coexpression of SA NP mutants and PB2. Results showed that NP-PB2 interactions of these SA NP mutants were essentially the same as for WT NP in the absence of 0.1% SDS (Fig. 9) and that the complex dissociated in the presence of 0.1% SDS in RIPA buffer (data not shown). Therefore, these SA mutations at the COOH terminus of NP did not affect the formation or stability of the NP-PB2 complex.
FIG. 8.
In vivo polymerase activity of NP mutants. COS1 cells in 60-mm-diameter dishes were infected with VTF7.3 at an MOI of 5 and transfected with DNA containing pGEM PB1 (3 μg), pGEM PB2 (3 μg), pGEM PA2 (0.5 μg), Ribo-CAT (3 μg), and pGEM NP (or NP mutant) (5 μg) in duplicate plates. One set of plates was kept at 33°C, and other set was kept at 37°C. At 24 hpt, cells were lysed and assayed for CAT activity (A). Parts of the same lysates were analyzed by SDS-PAGE (8% gel), Western blotted on a membrane, and probed with anti-WSN antibodies. The membrane was developed by chemiluminescence reagent (B). The CAT activities of WT and mutant NP proteins in panel A were quantified and compared, using the activity of the WT NP at 33 or 37°C as 100% (C).
FIG. 9.
Interaction of SA mutants of NP with PB2. COS1 cells were infected with VTF7.3 at an MOI of 5 and cotransfected with either pGEM NP or mutant NP (1 μg) along with pGEM PB2 (3 μg). At 14 hpt, cells were labeled with Express 35S for 1 h, lysed, and divided into two parts; one part was immunoprecipitated with anti-NP antibodies, and the other part was immunoprecipitated with anti-PB2 antibodies in the absence of SDS. The immunoprecipitated complex was analyzed by SDS-PAGE (8% gel).
DISCUSSION
In the influenza virus infectious cycle, NP plays a critical role in switching the transcription of viral mRNA to the replication of cRNA and vRNA. Studies with ts mutants have shown that different ts NP mutants can affect both cRNA and vRNA synthesis independently (17, 19). Biochemical studies have further shown that RNP-free soluble NP is required for switching mRNA to cRNA synthesis in vitro (6, 17). NP interacts with viral RNA (2, 16) as well as with itself, forming an oligomer (36), and with cellular proteins, (4, 32, 33, 41). It may also interact with other viral proteins such as M1 and possibly NS1 (4, 26), although M1-NP and M1-NS1 complexes have not been directly demonstrated. In this report, we have shown that NP can interact directly with the polymerase protein complex both in virus-infected cells and in cells coexpressing NP and polymerase proteins. We have further shown that NP can interact with PB1 and PB2 independently. Using two independent approaches, we have demonstrated that the interaction of NP with PB1 and PB2 is specific. Experiments reported here have further shown that the NP-PB2 complex is rather labile and therefore likely to be dynamic in nature. We have further shown that the sequences at the COOH terminus of NP regulate the strength and stability of NP-PB2 interaction since the deletion of 33 aa at the COOH terminus increases the amount of NP-PB2 complex formation and renders the NP-PB2 complex resistant to 0.1% SDS. However, how these COOH-terminal sequences affect NP-PB2 interaction is unclear. One possibility is that the COOH terminus may cover up and thereby mask some of the interacting domains and that removal of the COOH-terminal sequences would expose the interacting surface(s) of NP. Alternatively, removal of the COOH sequence could cause structural alteration of NP leading to exposure of the interacting domain(s). Since NP is a phosphoprotein (14, 34, 35) and since phosphorylation/dephosphorylation is known to affect many biological functions, including the interaction among proteins, it is likely that the state of NP phosphorylation regulates the affinity and stability of NP-PB2 interaction in virus-infected cells. Therefore, removal of the COOH sequences may have affected the phosphorylation of NP, which may be responsible for regulating the affinity of NP-PB2 interaction. We are now in the process of determining if deletion of COOH sequences affects phosphorylation of NP and if NP phosphorylation affects NP-PB2 interaction as well as transcription and replication of vRNA. It should be further noted that only a minor fraction of NP and PB2 formed complexes with each other either in virus-infected cells or in cells coexpressing WT proteins.
WSN ts 56 virus has a 314SN mutation in NP (21) and exhibits temperature sensitivity in the transcription-to-replication switch affecting cRNA synthesis (17, 19). It is not known if this ts effect is due to phosphorylation, as another mutation (332AT) in the same region (ts 81 in fowl plague virus) also exhibits temperature sensitivity (25). The role of either of these mutations on the phosphorylation of NP has not been determined. Recently, an SA mutation at position 3 of NP (3SA NP) of A/Victoria/3/75 virus was shown to partially affect phosphorylation and CAT activity (∼50% of the WT level) in an in vivo transcription-replication assay (3). However, these authors did not check for the temperature sensitivity of the 3SA NP mutant; furthermore, two viruses, A/WSN/33 and A/Swine/Cambridge/1/35, do not have a serine residue at position 3 of NP. Therefore, the sites of phosphorylation in NP and the role of phosphorylation in NP functions remain to be determined.
Our data show that multiple regions of NP interact with PB2. The COOH terminus (aa 340 to 498) of NP contains a PB2 binding site as well as a sequence regulating the NP-PB2 interaction in the last 33 aa of NP. NP II (aa 161 to 256) does not bind to PB2. An RNA binding region of NP has been identified within NH2-terminal aa 1 to 180 (2, 16), which overlap with the NP I region encompassing aa 1 to 161. However, further fine mapping of both RNA binding and PB2 binding regions will be needed to determine if there is any true overlap between these two functions.
As indicated earlier, the critical question as to how vRNA synthesis is switched from transcription mode to replication mode in the infectious cycle remains unexplained. It is likely that the 3P complex, template, and/or the product RNA become modified by the viral and cellular factors. Although RNP-free soluble NP has been shown to be involved in switching RNA synthesis from transcription to replication and in the synthesis of both cRNA and vRNA, the mode of NP function in these steps remains unclear. As mentioned earlier, the proposed antitermination effect due to NP binding to the product RNA or melting effect of NP on the panhandle structure of the template RNA cannot explain the efficient cap-independent initiation required for cRNA or vRNA synthesis. It is therefore possible that the observed binding of NP to PB1 and PB2 reported here facilitates cap-independent initiation in causing the transcription-to-replication switch. NP binding to PB2 may affect either the cap recognition or the cap cleavage function of PB2, thus reducing the availability of 5′-capped primers required for initiation of mRNA. Further, the binding of NP to PB1 may facilitate efficient cap-independent initiation and elongation. Antitermination (6) and processivity (12) functions of NP would permit more efficient chain completion. The recent observation that PB1 alone or PB1 and PA can permit synthesis of vRNA (28, 29, 40) may also support this hypothesis since all of these experiments were carried out with cell lines expressing NP along with PB1 and/or PA. It should also be noted that a ts defect in NP (ts 81) was extragenically suppressed by a ts defect in PB2 (24), suggesting a possible interaction between NP and PB2. Furthermore, a report that three anti-NP monoclonal antibodies interfered with the influenza virus RNA synthesis in vitro (5) would also suggest a possible interaction between NP and the polymerase protein complex. Therefore, further analysis of NP-PB1 as well as NP-PB2 interactions would help in defining the function of NP in this critical step of vRNA synthesis, the switch to replication from transcription including primer-independent initiation of RNA synthesis.
Finally, we have shown that an in vivo transcription-replication assay using CAT reporter protein can be used to analyze the temperature sensitivity of mutant proteins. It will be interesting to determine if the temperature sensitivity of the in vivo transcription-replication assay correlates with the ts phenotype of the infectious virus. If so, such an assay could be used as a screening procedure for selecting ts mutants which can be rescued by reverse genetics (23) for further analysis.
ACKNOWLEDGMENTS
This work was partially supported by NIH/NIAID grants AI16348, AI41681, and 5-T32-AI07323.
We thank Eleanor Berlin for typing the manuscript.
REFERENCES
- 1.Akkina R K, Chambers T M, Londo D R, Nayak D P. Intracellular localization of the viral polymerase proteins in cells infected with influenza virus and cells expressing PB1 protein from cloned cDNA. J Virol. 1987;61:2217–2224. doi: 10.1128/jvi.61.7.2217-2224.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albo C, Valencia A, Portela A. Identification of an RNA binding region within the N-terminal third of the influenza A virus nucleoprotein. J Virol. 1995;69:3799–3806. doi: 10.1128/jvi.69.6.3799-3806.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrese M, Portela A. Serine 3 is critical for phosphorylation at the N-terminal end of the nucleoprotein of influenza virus A/Victoria/3/75. J Virol. 1996;70:3385–3391. doi: 10.1128/jvi.70.6.3385-3391.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avalos R T, Yu Z, Nayak D P. Association of influenza virus NP and M1 proteins with cellular cytoskeletal elements in influenza virus-infected cells. J Virol. 1997;71:2947–2958. doi: 10.1128/jvi.71.4.2947-2958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barcena J, Ochoa M, De La Luna S, Melero J A, Nieto A, Ortin J, Portela A. Monoclonal antibodies against influenza virus PB2 and NP polypeptides interfere with the initiation step of viral mRNA synthesis in vitro. J Virol. 1994;68:6900–6909. doi: 10.1128/jvi.68.11.6900-6909.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaton A R, Krug R M. Transcription antitermination during influenza viral template RNA synthesis requires the nucleocapsid protein and the absence of a 5′ capped end. Proc Natl Acad Sci USA. 1986;83:6282–6286. doi: 10.1073/pnas.83.17.6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biswas S K, Nayak D P. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J Virol. 1994;68:1819–1826. doi: 10.1128/jvi.68.3.1819-1826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas S K, Nayak D P. Influenza virus polymerase basic protein 1 interacts with influenza virus polymerase basic protein 2 at multiple sites. J Virol. 1996;70:6716–6722. doi: 10.1128/jvi.70.10.6716-6722.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Detjen B M, St. Angelo C, Katze M G, Krug R M. The three influenza virus polymerase (P) proteins not associated with viral nucleocapsids in the infected cell are in the form of a complex. J Virol. 1987;61:16–22. doi: 10.1128/jvi.61.1.16-22.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuerst T R, Earl A L, Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galarza J M, Sowa A, Hill V M, Skorko R, Summers D F. Influenza A virus NP protein expressed in insect cells by a recombinant baculovirus is associated with a protein kinase activity and possesses single-stranded RNA binding activity. Virus Res. 1992;24:91–106. doi: 10.1016/0168-1702(92)90033-6. [DOI] [PubMed] [Google Scholar]
- 12.Honda A, Ueda K, Nagata K, Ishihama A. RNA polymerase of influenza virus: role of NP in RNA chain elongation. J Biochem. 1988;104:1021–1026. doi: 10.1093/oxfordjournals.jbchem.a122569. [DOI] [PubMed] [Google Scholar]
- 13.Hsu M-T, Parvin J D, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci USA. 1987;84:8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kistner O, Muller K, Schottissek C. Differential phosphorylation of the nucleoprotein of influenza A viruses. J Gen Virol. 1989;70:2421–2431. doi: 10.1099/0022-1317-70-9-2421. [DOI] [PubMed] [Google Scholar]
- 15.Klumpp K, Ruigrok R W H, Baudin F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi M, Toyoda T, Adyshev D M, Azuma Y, Ishihama A. Molecular dissection of influenza virus nucleoprotein: deletion mapping of the RNA binding domain. J Virol. 1994;68:8433–8436. doi: 10.1128/jvi.68.12.8433-8436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krug R M, Aloso-Caplen F V, Julkunon I, Katze M G. Expression and replication of the influenza virus genome. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 89–152. [Google Scholar]
- 18.Krug R M, St. Angelo C, Broni B, Shapiro G. Transcription and replication of influenza virion RNA in the nucleus of infected cells. Cold Spring Harbor Symp Quant Biol. 1987;52:353–358. doi: 10.1101/sqb.1987.052.01.040. [DOI] [PubMed] [Google Scholar]
- 19.Krug R M, Ueda M, Palese P. Temperature-sensitive mutants of influenza WSN virus defective in virus-specific RNA synthesis. J Virol. 1975;16:790–796. doi: 10.1128/jvi.16.4.790-796.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb R. Genes and proteins of the influenza viruses. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. pp. 1–87. [Google Scholar]
- 21.Li R, Palese P, Krystal M. Complementation and analysis of an NP mutant of influenza virus. Virus Res. 1989;12:97–112. doi: 10.1016/0168-1702(89)90057-9. [DOI] [PubMed] [Google Scholar]
- 22.Luo G X, Luytjes W, Enami M, Palese P. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J Virol. 1991;65:2861–2867. doi: 10.1128/jvi.65.6.2861-2867.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luytjes W, Krystal M, Enami M, Parvin J D, Palese P. Amplification, expression and packaging of a foreign gene by influenza virus. Cell. 1989;59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 24.Mandler J, Muller K, Scholtissek C. Mutants and revertants of an avian influenza A virus with temperature-sensitive defects in the nucleoprotein and PB2. Virology. 1991;181:512–519. doi: 10.1016/0042-6822(91)90883-d. [DOI] [PubMed] [Google Scholar]
- 25.Mandler J, Scholtissek C. Localization of the temperature-sensitive defect in the nucleoprotein of an influenza A/FPV/Rostock/34 virus. Virus Res. 1989;12:113–122. doi: 10.1016/0168-1702(89)90058-0. [DOI] [PubMed] [Google Scholar]
- 26.Marion R M M, Zurcher T, de la Luna S, Ortin J. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J Gen Virol. 1997;78:2447–2451. doi: 10.1099/0022-1317-78-10-2447. [DOI] [PubMed] [Google Scholar]
- 27.Murti K G, Webster R G, Jones I M. Localization of RNA polymerases of influenza viral ribonucleoproteins by immunogold labeling. Virology. 1988;164:562–566. doi: 10.1016/0042-6822(88)90574-0. [DOI] [PubMed] [Google Scholar]
- 28.Nakagawa Y, Kimura N, Toyoda T, Mizumoto K, Ishihama A, Oda K, Nakada S. The RNA polymerase PB2 subunit is not required for replication of influenza virus genome but is involved in capped mRNA synthesis. J Virol. 1995;69:728–733. doi: 10.1128/jvi.69.2.728-733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakagawa Y, Oda K, Nakada S. The PB1 subunit alone can catalyze cRNA synthesis and the PA subunit in addition to the PB1 subunit is required for viral RNA synthesis in replication of the influenza virus genome. J Virol. 1996;70:6390–6394. doi: 10.1128/jvi.70.9.6390-6394.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nayak D P. Influenza virus infections. In: Dulbecco R, editor. Encyclopedia of human biology. Vol. 5. New York, N.Y: Academic Press; 1997. pp. 67–80. [Google Scholar]
- 31.Neuman G, Castrucci M R, Kawaoka Y. Nuclear import and export of influenza virus nucleoprotein. J Virol. 1997;71:9690–9700. doi: 10.1128/jvi.71.12.9690-9700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Neill R E, Jaskunas R, Blobel G, Palese P, Moroianu J. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J Biol Chem. 1995;270:22701–22704. doi: 10.1074/jbc.270.39.22701. [DOI] [PubMed] [Google Scholar]
- 33.O’Neill R E, Palese P. NP1-1, a human homologue of SRP1, interacts with influenza virus nucleoprotein. Virology. 1994;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 34.Petri T, Dimmock N J. Phosphorylation of influenza virus nucleoprotein in vivo. J Gen Virol. 1981;57:185–190. doi: 10.1099/0022-1317-57-1-185. [DOI] [PubMed] [Google Scholar]
- 35.Privalsky M L, Penhoet E E. The structure and synthesis of influenza virus phosphoproteins. J Biol Chem. 1981;256:5368–5376. [PubMed] [Google Scholar]
- 36.Prokudina-Kantorovich E N, Seemenova N P. Intracellular oligomerization of influenza virus nucleoprotein. Virology. 1996;223:51–56. doi: 10.1006/viro.1996.0454. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Shimizu K, Handa H, Nakada S, Nagata K. Regulation of influenza virus RNA polymerase activity by cellular and viral factors. Nucleic Acids Res. 1994;22:5047–5053. doi: 10.1093/nar/22.23.5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.St. Angelo C, Smith G E, Summers M D, Krug R M. Two of the three influenza viral polymerase proteins expressed by using baculovirus vectors form a complex in insect cells. J Virol. 1987;61:361–365. doi: 10.1128/jvi.61.2.361-365.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toyoda T, Kobayashi M, Nakada S, Ishihama A. Molecular dissection of influenza virus RNA polymerase: PB1 subunit alone is able to catalyze RNA synthesis. Virus Genes. 1996;12:155–163. doi: 10.1007/BF00572954. [DOI] [PubMed] [Google Scholar]
- 41.Wang P, Palese P, O’Neill R E. The NPI-1 NPI-3 (karyopherin α) binding site on the influenza A virus nucleoprotein NP is a nonconventional nuclear localization signal. J Virol. 1997;71:1850–1856. doi: 10.1128/jvi.71.3.1850-1856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]