Abstract
Virtually all of our present understanding of endogenous murine leukemia viruses (MLVs) is based on studies with inbred mice. To develop a better understanding of the interaction between endogenous retroviruses and their hosts, we have carried out a systematic investigation of endogenous nonecotropic MLVs in wild mice. Species studied included four major subspecies of Mus musculus (M. m. castaneus, M. m. musculus, M. m. molossinus, and M. m. domesticus) as well as four common inbred laboratory strains (AKR/J, HRS/J, C3H/HeJ, and C57BL/6J). We determined the detailed distribution of nonecotropic proviruses in the mice by using both env- and long terminal repeat (LTR)-derived oligonucleotide probes specific for the three different groups of endogenous MLVs. The analysis indicated that proviruses that react with all of the specific probes are present in most wild mouse DNAs tested, in numbers varying from 1 or 2 to more than 50. Although in common inbred laboratory strains the linkage of group-specific sequences in env and the LTR of the proviruses is strict, proviruses which combine env and the LTR sequences from different groups were commonly observed in the wild-mouse subspecies. The “recombinant” nonecotropic proviruses in the mouse genomes were amplified by PCR, and their genetic and recombinant natures were determined. These proviruses showed extended genetic variation and provide a valuable probe for study of the evolutionary relationship between MLVs and the murine hosts.
Retroviruses are the only group of viruses known to have a “fossil” record. All mammals and birds, and probably most other vertebrates as well, have been subjected to retrovirus infection at some time in their evolutionary history, since endogenous elements closely related to known retroviruses have been found as abundant germ line elements in DNAs of all species examined (2, 7). The best-studied endogenous proviruses are those in mice, and it has been estimated that as much as 0.5% of the mouse genome consists of such elements. In inbred laboratory strains of mice, at least eight different groups of such endogenous elements have been identified. Those with close exogenous relatives include type B proviruses related to mammary tumor virus and type C proviruses related to murine leukemia virus (MLV) (2, 7).
The endogenous type C-related MLVs are a large and well-characterized group among the known endogenous proviruses. They are divided into two major groups, ecotropic and nonecotropic viruses, as classified by their potential host cell range, a property dictated by the surface (SU) protein encoded by the viral env gene. Ecotropic viruses can infect only mouse cells and are present in only one to five copies in common laboratory mouse strains (22, 26, 35). Nonecotropic viruses are subdivided into three groups, xenotropic, polytropic, and modified polytropic viruses, and are present in about 20 copies each in the genome of inbred mice (15, 26, 43). The endogenous nonecotropic MLV proviruses are useful for understanding the host-retrovirus interaction because they are abundant and highly polymorphic in the mouse genome (7). Our previous studies have shown that each nonecotropic provirus shares a set of polymorphisms in the env and long terminal repeat (LTR) regions that distinguish it from all the other groups (8, 42). Most usefully, the polymorphisms allowed us to develop a set of oligonucleotide probes that unambiguously detect all members of the nonecotropic groups in the mouse genome (11, 43). By using these group-specific probes, we could demonstrate several aspects of the proviruses, including their chromosomal locations in the common laboratory strains (3, 13–15, 44).
The study of nonecotropic viruses can also contribute to an understanding of the generation of oncogenic viruses in mice. The progeny of several of the nonecotropic proviruses can recombine with exogenous or other endogenous MLVs to give rise to oncogenic variants such as mink cell focus-forming (MCF) virus in certain laboratory mouse strains (6, 16, 33, 36, 41, 46). An endogenous xenotropic virus (Bxv-1) is the primary LTR donor for the MCF virus (17). A genetic exchange also occurs in the 5′ portion of the env gene encoding SU. This exchange usually involves the substitution of polytropic env sequences into an ecotropic virus background.
Endogenous nonecotropic proviruses are also found in wild-mouse species. Previous studies demonstrated that the env sequences of the proviruses are widely distributed in the subgenus Mus, especially Mus musculus species that are progenitors of common inbred laboratory strains (4, 21, 22, 26, 47, 50). These findings suggest that these germ line sequences were acquired independently in different wild mice and have remained largely segregated in the M. musculus species (26). Thus, detailed analysis of the endogenous nonecotropic proviruses in wild mice will allow us to evaluate the association between MLVs and the murine host during their evolutionary history.
In this study, we investigated endogenous nonecotropic MLVs in wild mice, including four major subspecies of M. musculus (M. m. castaneus, M. m. musculus, M. m. molossinus, and M. m. domesticus), as well as four common laboratory strains. We could demonstrate the detailed distribution of three groups of nonecotropic proviruses in the wild mice by using both env- and LTR-specific oligonucleotide probes. Moreover, we also examined the existence and significance of recombinant forms of nonecotropic proviruses in the mice. In this paper, we report an extensive polymorphism of the nonecotropic MLV proviruses and a possible evolutionary relationship between MLVs and wild mice.
MATERIALS AND METHODS
Mouse DNAs.
In addition to four common inbred laboratory strains (AKR/J, HRS/J, C3H/HeJ, and C57BL/6J), DNAs from four major subspecies of M. musculus were used in this study. The inbred wild-mouse strains used were CAST/Ei (M. m. castaneus), CZECH II/Ei (M. m. musculus), MOLC/Rk, MOLF/Ei, and MOLG/Dn (M. m. molossinus), and WSB/Ei and ZALENDE/Ei (M. m. domesticus). Strain WSB was separated from strain CLA, which was generated from wild mice trapped on a farm in Maryland. M. m. domesticus ZALENDE is originally from Europe (Switzerland) and was formerly classified as M. poschinavirus. DNA samples from these strains were obtained from the Mouse DNA Resource of The Jackson Laboratory, Bar Harbor, Maine.
Dried gel hybridization and oligonucleotide probes.
Hybridization in dried agarose gels (unblotting) was described previously (45). Briefly, genomic DNA digested with appropriate restriction enzymes was electrophoresed in a 0.8% agarose gel. After being stained with ethidium bromide (EtBr), the DNA in the gel was denatured. After being dried, the gel was hybridized for 16 h with a 5′-32P-labeled oligonucleotide probe (0.5 × 106 cpm/ml). The dried gel was then washed, briefly air dried, and exposed to X-ray film for 1 to 5 days with an intensifying screen at −70°C. The sequences of the oligonucleotide probes specific for each nonecotropic provirus env and LTR region (env: JS-4, JS-5, and JS-6/10; LTR: Pltr, Mltr, and Xltr) and the hybridization temperatures are described elsewhere (11, 43). The sequence of the oligonucleotide probe specific for recombinant provirus and the hybridization temperature are 5′-TTG AAC TCT GGC CAA GGG TGA C-3′ and 58°C (KT-45). A detailed protocol is available on request.
Synthetic oligonucleotide primers and PCR analysis.
To detect recombinant forms of nonecotropic MLVs in mouse genomes by PCR, we used six amplification primers. The locations of the amplification primers are indicated in Fig. 1A. The nucleotide sequences of the primers are as follows: XS-1, 5′-ACG GTC TCT ATG GTA CCT GG-3′; XA-3, 5′-ACT TTT CCA GAA ACT GTT GC-3′; PS-1, 5′-CTA TAG TCC CTG AGA CTG CC-3′; PA-2, 5′-CAC TGA CGT CTG AGA GCC AT-3′; mPS-1.1, 5′-GCA GCA TCT ATA CAA CCT AG-3′; and mPA-2, 5′-TCT ATC GGG GCT TCT GTG TC-3′.
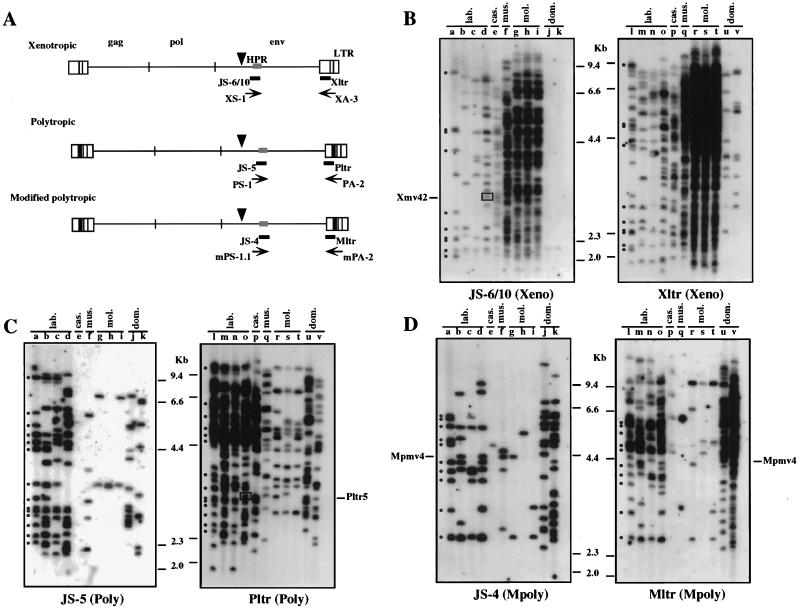
FIG. 1.
Distribution of nonecotropic proviruses in wild mice. (A) Locations of the group-specific PCR primers and probes. The structures of endogenous nonecotropic proviruses are shown. The approximate positions of the gag, pol, and env genes and the relative sizes of the LTRs are also indicated. The black boxes within the LTRs of the polytropic and modified polytropic viruses corresponded to the 190-bp inserted sequence (24). Arrowheads show conserved PvuII sites in the env gene in nonecotropic provirus genomes. (B to D) Unblotting analysis of PvuII-digested mouse DNAs was performed with xenotropic oligonucleotide probes (JS-6/10, env; Xltr, LTR) (B), polytropic oligonucleotide probes (JS-5, env; Pltr, LTR) (C), and modified polytropic oligonucleotide probes (JS-4, env; Mltr, LTR) (D). Lanes: a and l, AKR/J; b and m, HRS/J; c and n, C3H/HeJ; d and o, C57BL/6J (lab.); e and p, CAST/Ei (M. m. castaneus) (cas.); f and q, CZECH II/Ei (M. m. musculus) (mus.); g and r, MOLC/Rk (M. m. molossinus); h and s, MOLF/Ei (M. m. molossinus); i and t, MOLG/Dn (M. m. molossinus) (mol.); j and u, WSB/Ei (M. m. domesticus); k and v, ZALENDE/Ei (M. m. domesticus) (dom.). Known provirus loci that comigrate with wild-mouse fragments are shown on the side. Boxed bands indicate a comigrating fragment between two different probes. The approximate positions of molecular markers are also shown. Identically sized bands detected with both env and LTR probes are indicated by dots in lanes a and l.
The PCR was performed in a total volume of 50 μl containing 0.5 μg of genomic DNA, 50 pmol each of sense and antisense primers, and 2.5 U of Thermus aquaticus DNA polymerase (Taq polymerase; Perkin-Elmer Cetus, Norwalk, Conn.). The reaction mixtures for amplification were incubated at 94, 60, and 72°C for 1, 2, and 2 min, respectively. The cycle was repeated 30 times in a programmable cyclic reactor (ERICOMP, San Diego, Calif.). After amplification, the products were analyzed in a 0.8% agarose gel, stained with EtBr, and visualized by UV fluorescence. To denature the DNA before blotting, the gel was soaked in 0.5 M NaOH–1.5 M NaCl for 10 min, washed twice with H2O, and neutralized in 1.0 M Tris-HCl (pH 8.0)–1.5 M NaCl for 10 min. The DNA was transferred to a nylon membrane. After cross-linking of DNA, the membrane was analyzed by Southern blot hybridization with a 32P-labeled oligonucleotide as described previously (13).
Cloning and sequencing analysis.
The endogenous proviruses detected by PCR were cloned into pUC or pCR2.1 (Invitrogen Co., Purchase, N.Y.) vectors. DNA sequences were determined by the double-stranded dideoxy-chain termination method (40) with the Sequenase version 2.0 kit (United States Biochemical Co., Cleveland, Ohio). After sequencing, the sequence data were aligned by using the algorithm of Needleman and Wunsch (34) as implemented in the PILEUP program in the Genetics Computer Group program (9).
Nucleotide sequence accession numbers.
The provirus sequences reported in this study have been deposited in GenBank under accession no. AF017518 to AF017531.
RESULTS
Strain distribution of nonecotropic proviruses in wild mice.
In previous studies, distributions of xenotropic and polytropic env sequences in wild mice were detected using env-reactive probes (4, 25, 26). These studies suggested that xenotropic and polytropic env sequences were acquired independently in different wild-mouse subspecies. However, the polytropic probe used in these studies was nonspecific and reacted with too many fragments to allow unambiguous identification of individual proviruses (38). Furthermore, it is clear that not all MLV proviruses can be detected with group-specific probes from the env region (12, 29). Thus, to fully understand the distribution of nonecotropic MLVs in wild mice, we first hybridized appropriately digested wild-mouse DNA to both env- and LTR-specific oligonucleotide probes known to be highly specific for each group of nonecotropic proviruses. The locations of the probes are shown in Fig. 1A. Their exact sequences are described elsewhere (11, 43). Genomic DNAs were digested with PvuII and hybridized with the specific probes. The restriction enzyme PvuII was chosen to match the earlier proviral mapping efforts (11, 13–15). PvuII was preferred to other enzymes because the presence of conserved PvuII sites in the 5′ portion of known proviruses, in addition to the conserved site in env (Fig. 1A), results in a wide range of provirus-host junction fragment sizes.
As shown in Fig. 1, wild-mouse species contain numerous proviruses that are reactive with the group-specific probes but that differ greatly in number from one species to another and from the number in inbred strains. As we previously reported (11), most of the proviruses in inbred laboratory strains detected with env probes (lanes a to d) were also detected with the specific LTR probes (lanes l to o), although the latter hybridized to about twice as many fragments in total, consistent with the detection of 5′ as well as 3′ junction fragments. As an example, such fragments in DNA from the AKR/J mouse are indicated by dots in lanes a and l (Fig. 1).
The pattern of hybridization of the wild-mouse strains with the specific probes was quite different. Although almost all the strains had at least one provirus that reacted with each of the probes, the numbers of those proviruses were usually very different. Furthermore, in a number of cases, little or no correlation between fragments was detected with probes that recognize the same provirus in inbred laboratory strains. By using the xenotropic env (JS-6/10) probe, more than 60 fragments were detected in the DNAs from M. m. musculus and M. m. molossinus subspecies (Fig. 1B, lanes f to i) whereas only a faint band was demonstrated in each M. m. domesticus DNA (lanes j and k). Between 8 and 26 fragments were observed in the laboratory strains and M. m. castaneus DNAs (lanes a to e). These results are consistent with the distribution of xenotropic env sequences observed in previous studies (26). In contrast, by using the xenotropic LTR (Xltr) probe, about 10 bands were found in M. m. domesticus (lanes u and v). A similar pattern was observed with the polytropic probes (Fig. 1C). Although nearly 30 fragments were detected by the polytropic LTR (Pltr) probe in M. m. castaneus DNA (lane p), no specific fragment was detected by the polytropic env (JS-5) probe (lane e). Further, despite the detection of 14 to 23 polytropic LTR-reactive fragments in the DNAs of M. m. musculus and M. m. molossinus subspecies (lanes q to t), only a few polytropic env-reactive fragments were found in the DNAs (lanes f to i). By using the modified polytropic env (JS-4) probe, about 10 to 20 fragments were detected in the DNAs from four laboratory strains and M. m. domesticus (Fig. 1D, lanes a to d, j, and k). In contrast, only a few JS-4-reactive bands were seen in M. m. castaneus, M. m. musculus, and M. m. molossinus DNAs (lanes e to i). Furthermore, the faint bands detected by the modified polytropic LTR (Mltr) probe in M. m. castaneus and M. m. musculus DNAs did not show any correlation to the fragments that hybridized to the JS-4 probe (lanes e and q).
We tabulated the numbers of each group of provirus present in these strains (Table 1). The three endogenous nonecotropic groups showed a differential distribution in the M. musculus subspecies. Xenotropic sequences were distributed mainly in M. m. musculus and M. m. molossinus subspecies, while the polytropic and modified polytropic fragments were found predominantly in M. m. domesticus subspecies. In the laboratory strains of mice, the numbers of the fragments reactive with the env and LTR probes were well correlated, with approximately twice as many fragments being detected by the LTR-specific probes. In the wild mice, however, the env- and LTR-reactive fragments were not necessarily correlated. The lack of correlation could be due to sequence polymorphism in the proviruses, to internal deletions, or to recombination among the different groups. Consistent with recombination was the comigration of some of the xenotropic env-reactive fragments in M. m. castaneus DNA with the fragments detected with the polytropic LTR probe (Fig. 1B, lane e; Fig. 1C, lane p). In addition, a fragment detected with the polytropic LTR probe in the C57BL/6J strain (Fig. 1C, lane o, box) comigrated with a fragment that reacted with the xenotropic env probe, JS-6/10 (Fig. 1B, lane d, box). These fragments were identified and named Pltr5 and Xmv42, respectively, in our previous study and had been interpreted to be a possible recombinant between xenotropic and polytropic proviruses (11, 13).
TABLE 1.
Distribution of nonecotropic proviral env and LTR sequences in wild mice
| Strain | Subspecies | No. of reactive fragmentsa
|
||||||
|---|---|---|---|---|---|---|---|---|
| Xenotropic
|
Polytropic
|
Modified polytropic
|
Type A Xeno/Poly (KT-45) | |||||
| env | LTR | env | LTR | env | LTR | |||
| AKR/J | 11 | 25 | 17 | 38 | 11 | 24 | 0 | |
| HRS/J | 8 | 19 | 21 | 46 | 9 | 23 | 0 | |
| C3H/HeJ | 8 | 16 | 15 | 34 | 7 | 17 | 0 | |
| C57BL/6J | 18 | 36 | 22 | 46 | 10 | 26 | 1 | |
| CAST/Ei | M. m. castaneus | 26 | 35 | 0 | 30 | 1 | 5 | 15 |
| CZECH II/Ei | M. m. musculus | >60 | >100 | 5 | 23 | 4 | 1 | 0 |
| MOLC/Rk | M. m. molossinus | >60 | >100 | 2 | 14 | 2 | 6 | 4 |
| MOLF/Ei | M. m. molossinus | >60 | >100 | 2 | 14 | 1 | 3 | 5 |
| MOLG/Dn | M. m. molossinus | >60 | >100 | 2 | 14 | 2 | 5 | 4 |
| WSB/Ei | M. m. domesticus | 1 | 9 | 11 | 39 | 15 | 35 | 0 |
| ZALENDE/Ei | M. m. domesticus | 1 | 11 | 8 | 23 | 23 | >50 | 0 |
Approximate numbers of junction fragments as determined by counting the bands in the unblots shown in Fig. 1 and 7. Exact determinations require genetic analysis as well (11, 15). Results for fragments whose hybridization patterns were unlinked relative to inbred laboratory strains are shown in boldface type.
Detection of recombinant forms of nonecotropic proviruses in wild-mouse DNAs.
The distribution of the nonecotropic fragments suggested the existence of intragroup polymorphisms or recombinant proviruses among different groups of nonecotropic viruses in wild mice. To investigate this genetic variation, we searched directly for recombinant forms of nonecotropic MLVs in the wild mice by PCR. Sense and antisense primers were designed to prime at the hypervariable proline-rich (HPR) region of SU and the U3 region of the LTR, respectively (Fig. 1A). Because each standard nonecotropic group shares a set of polymorphisms in these regions and has strict linkage in the sequences (8, 43), members of these groups could be amplified only by corresponding primer pairs. Proviruses that are recombinant, relative to the standard ones, would yield product only when amplified with “mismatched” pairs. In the remainder of this paper, we refer to such proviruses as “recombinant,” for convenience of nomenclature. It should be kept in mind, however, that we have no way to tell whether they or the proviruses originally defined in inbred strains are the parental or recombinant forms.
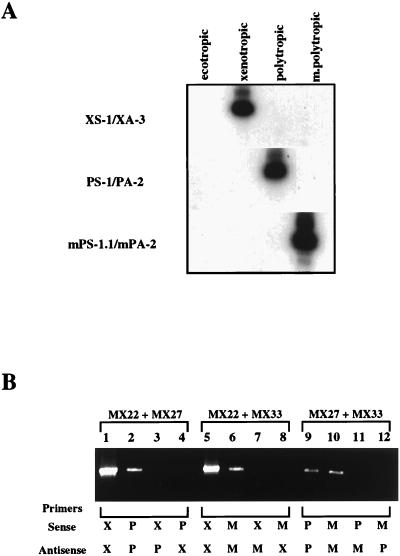
First, we investigated the specificity of the primers. As shown in Fig. 2A, under our amplification conditions, each matched primer pair could amplify only a provirus of the corresponding group. Furthermore, none of the proviruses was detectable with any mismatched combination of the sense and antisense primers (Fig. 2B).
FIG. 2.
Group-specific PCR primers for endogenous nonecotropic proviruses and specificity of the PCRs. (A) Specificity of the primer pairs. A Southern blot analysis of PCRs with each matched primer pair is shown. Each ecotropic (MX14) and nonecotropic (xenotropic, MX22; polytropic, MX27; modified polytropic, MX33) (42) proviral clone was used for the templates of the PCR. Primer pairs for each PCR are indicated on the left. (B) Specificity of the PCR. EtBr staining of the gels is shown. PCRs were performed in the presence of two different clones of nonecotropic proviruses with each combination of the primers. Templates: lanes 1 to 4, MX22 + MX27; lanes 5 to 8, MX22 + MX33; lanes 9 to 12, MX27 + MX33. Primer pair: lanes 1 and 5, XS-1/XA-3; lanes 2 and 9, PS-1/PA-2; lanes 6 and 10, mPS-1.1/mPA-2; lane 3, XS-1/PA-2; lane 4, PS-1/XA-3; lane 7, XS-1/mPA-2; lane 8, mPS-1.1/XA-3; lane 11, PS-1/mPA-2; lane 12, mPS-1.1/PA-2.
One concern in the use of PCR to search for recombinants is that the reaction itself might generate recombinant DNA molecules. Such recombination results when an incomplete product from one template hybridizes to another template and becomes a primer for amplification. Such artifactual template-primer pairs could be formed during the amplification of MLV sequences, since portions of env and the U3 regions of different groups of nonecotropic virus are highly conserved. To test for such an artifact, we performed PCR by using each primer pair in the presence of two different proviruses. Figure 2B shows the results of this experiment. An amplification product was observed only when a matching primer pair was present in the reaction mixture. Despite the high degree of sequence identity among the different proviruses, we could not find any products generated by PCR-mediated recombination. These results verified that under the conditions used here the oligonucleotide primers exhibited the desired degree of sequence specificity and that the PCRs themselves did not generate recombinant products.
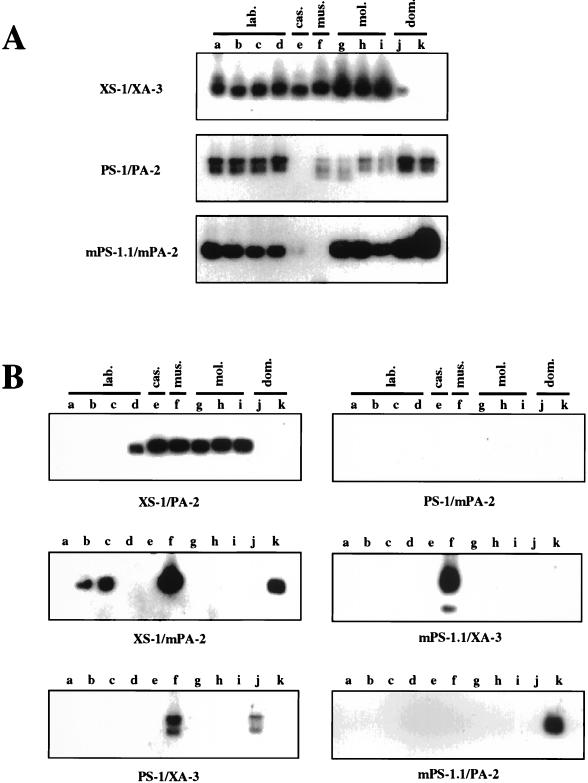
We first used the primer pairs to look for typical nonecotropic endogenous MLVs in the mouse DNAs. As shown in Fig. 3A, not all wild-mouse DNAs yielded the expected fragments. The xenotropic primer pair (XS-1/XA-3) could amplify fragments in the DNAs from four laboratory strains, M. m. castaneus, M. m. musculus, and M. m. molossinus (lanes a to i), but weak or no signals were found with M. m. domesticus DNAs (lanes j and k). Furthermore, no product was detected in DNA from M. m. castaneus with the polytropic primer pair (PS-1/PA-2) (lane e), whereas the modified polytropic primer pair (mPS-1.1/mPA-2) gave faint and no signals in M. m. castaneus and M. m. musculus DNAs, respectively (lanes e and f). These results were consistent with those obtained from the unblotting analysis in Fig. 1, where env- and LTR-reactive fragments did not show any correlation of polytropic and modified polytropic sequences, suggesting that they were present in different proviruses. Furthermore, only one xenotropic env-reactive fragment, which did not react with the LTR probe, was demonstrated in M. m. domesticus DNAs. These results also confirmed the specificity of these primers.
FIG. 3.
Detection of endogenous proviruses in genomic DNAs. Each group of nonecotropic proviruses (A) and recombinant forms of nonecotropic proviruses (B) were detected by PCR. The PCR products were analyzed by Southern blot hybridization with a 32P-labeled oligonucleotide probe (see Materials and Methods). The primer pairs used for the PCRs are indicated on the left (A) or at the bottom (B). Lanes: a, AKR/J; b, HRS/J; c, C3H/HeJ; d, C57BL/6J; e, CAST/Ei (M. m. castaneus); f, CZECH II/Ei (M. m. musculus); g, MOLC/Rk (M. m. molossinus); h, MOLF/Ei (M. m. molossinus); i, MOLG/Dn (M. m. molossinus); j, WSB/Ei (M. m. domesticus); k, ZALENDE/Ei (M. m. domesticus).
We next used combinations of the sense and antisense primers to test for the presence of recombinant forms of nonecotropic proviruses in the mouse DNAs. For example, a combination of xenotropic sense (XS-1) and polytropic antisense (PA-2) primers was used to look for recombinant proviruses with a xenotropic-type HPR region in SU and a polytropic-type U3 region in the LTR. As shown in Fig. 3B, possible recombinant forms were detected in at least one of the DNAs with all combinations of primers except polytropic-type env/modified polytropic (mPoly) LTR. Proviruses with xenotropic-type env and polytropic LTR sequences (Xeno/Poly) were detected in one laboratory strain (C57BL/6J) and all wild-mouse subspecies except M. m. domesticus (XS-1/PA-2; lanes d to i). This result was consistent with the previous detection of possible Xeno/Poly recombinant proviruses in C57BL/6J and M. m. castaneus DNAs (11, 13) (Fig. 1). Possible Xeno/mPoly recombinants were detected in two laboratory strains (HRS/J and C3H/HeJ), M. m. musculus, and one M. m. domesticus strain (ZALENDE/Ei) (XS-1/mPA-2; lanes b, c, f, and k). Fragments of two different sizes from Poly/Xeno recombinant proviruses analogous to class I MCF viruses were amplified from the DNA of M. m. musculus and one M. m. domesticus strain (WSB/Ei) (PS-1/XA-3; lanes f and j). Interestingly, each fragment detected in the WSB/Ei strain was slightly larger than that in M. m. musculus. Further, recombinant proviruses with modified polytropic-type env sequences (mPoly/Xeno and mPoly/Poly) were detected in the DNAs from M. m. musculus and a M. m. domesticus strain (ZALENDE/Ei), respectively (mPS-1.1/XA-3, lane f; mPS-1.1/PA-2, lane k).
Sequence of the env regions of recombinant proviruses.
To analyze the genetic nature of the possible recombinant endogenous MLVs detected in the PCR analysis, we examined at least five clones from each PCR product. The sequence analysis revealed the amplified fragments to be nonecotropic MLV-related proviruses containing sequences similar to two different groups in portions of the HPR and the U3 regions.
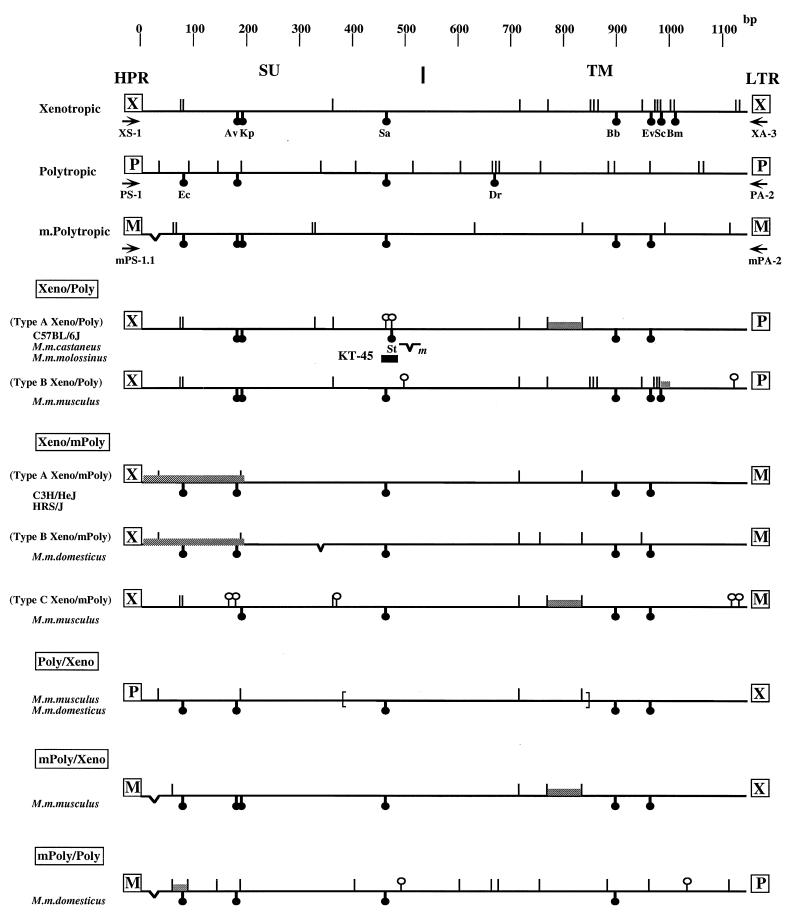
Figure 4 shows, in schematic form, the genetic structures of the env regions of the sequenced proviruses. The env sequences of the proviruses detected here were closely related to each other, because the regions we sequenced (the 3′ half of the SU and the transmembrane [TM] regions) are highly conserved in nonecotropic viruses, with the exception of the HPR region. However, characteristic sequences from at least two different groups were present in the env regions of the recombinant proviruses.
FIG. 4.
Schematic representation of env structures of recombinant proviruses. The nucleotide sequences of recombinant proviruses are compared with those of each nonecotropic provirus. Sequences of NZB (37) and CWM (32) were used for standard xenotropic viruses and MX27 and MX33 (42) were used for polytropic and modified polytropic viruses, respectively. Symbols: (|) nucleotide differences from consensus nonecotropic provirus sequence; (○|) unique nucleotide differences from consensus nonecotropic provirus sequence in recombinant proviruses; (☻) unique restriction enzyme site: Ec, EcoRII; Av, AvrII; Kp, KpnI; Sa, SacI; St, StyI; Dr, DraI; Ev, EcoRV; Sc, ScaI; Bm, BsmI; (V) deletion. The shaded areas indicate the possible recombinant region in each provirus. The location of the KT-45 hybridization probe is indicated. m in the type A Xeno/Poly recombinant virus indicates the deletion site in the clone from M. m. molossinus. The deletion region in the Poly/Xeno recombinant proviruses are shown by brackets. The boundary of SU and the TM region is also shown at the top. Arrows indicate PCR primers.
Unique structural features of the recombinant proviruses in the env region are as follows. First, the Xeno/Poly recombinants could be subdivided into two different types. Proviruses from C57BL/6J, M. m. castaneus, and M. m. molossinus were very similar to one another and are referred to as type A. These were different from those from the type B Xeno/Poly recombinant of M. m. musculus (Fig. 4). The type A Xeno/Poly recombinant proviruses contained xenotropic sequences up to the 3′ half of the TM region, while the type B proviruses were almost identical to xenotropic proviruses except in the 3′ quarter of the TM region. Thus, there appear to have been at least two distinct crossover events to generate recombinant viruses of this type. In addition, several clones from M. m. molossinus contained a 2-bp deletion near the end of the SU region, resulting in a frameshift and the introduction of a stop codon just upstream of the boundary of the SU and TM regions. Two unique nucleotide changes in the type A Xeno/Poly recombinant proviruses are worth noting. Near the end of the SU region, a SacI recognition site conserved in all groups of nonecotropic proviruses was missing from the type A recombinant proviruses and a nucleotide change introduced an additional StyI site in the region just 5 bp downstream from the absent SacI site (Fig. 4). These unique nucleotide changes allowed us to differentiate these type A Xeno/Poly recombinant proviruses from others; detailed analyses are given below.
The Xeno/mPoly recombinant proviruses could be subdivided into three types (Fig. 4). Although env sequences of type A (from C3H/HeJ and HRS/J strains) and type B (from an M. m. domesticus strain) recombinant proviruses were very similar to each other, the U3 region of the type A proviruses was distinct from that of the type B proviruses (see below). In contrast, env sequence of type C recombinant proviruses from M. m. musculus was distinct from those of the type A and type B recombinants (Fig. 4).
Interestingly, both type A and B proviruses contained polytropic sequences in first 200-bp region of their sequences (Fig. 4), although the xenotropic sense primer primed their amplification. This reactivity could not have been due to nonspecific hybridization of the xenotropic primer to polytropic sequence, because in this case, the primer pair, XS-1/PA-2, would have detected products in all mouse DNAs we examined except M. m. castaneus. Further, Poly/mPoly recombinant proviruses would also have been detected in the DNAs in which the Xeno/mPoly recombinant proviruses were observed. However, we could not detect any such proviruses by PCR analysis (Fig. 3B, XS-1/PA-2 and PS-1/mPA-2), indicating that the env sequence with characteristics of both xenotropic and polytropic viruses in the HPR region does exist in the mouse DNAs. Alternatively, it is possible that there is a crossover between xenotropic and polytropic proviruses just downstream of the primer binding site.
The Poly/Xeno recombinant proviruses detected in two different subspecies of wild mice had similar structural features (Fig. 4). Despite the primers used for their amplification, these proviruses were most closely related to modified polytropic provirus in all sequenced portions of the env gene. They resembled MCF viruses in that they encompassed polytropic env and xenotropic LTR sequences. In MCF viruses, however, some portions of the TM region are occupied by ecotropic sequences (18, 23, 39, 46, 49). Thus, the Poly/Xeno recombinant proviruses cannot be related to MCF viruses. The small fragments detected by PCR analysis in each strain of mice (Fig. 3B, PS-1/XA-3) represented the same type of recombinant proviruses lacking large parts of the env region (parentheses in Fig. 4).
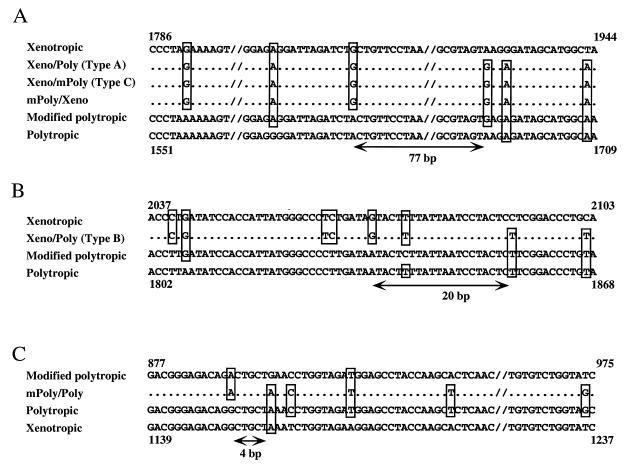
Possible crossover regions between different groups in the recombinant proviruses are shown in Fig. 5. They were in various locations in the different groups and ranged in size from 4 to 77 bp of identical sequences.
FIG. 5.
Possible recombinant regions in recombinant proviruses. (A) Sequences of possible recombination sites of type A Xeno/Poly, type C Xeno/mPoly, and mPoly/Xeno recombinant proviruses. (B and C) Recombination sites of type B Xeno/mPoly (B) and mPoly/Poly (C) recombinant proviruses. Nonecotropic provirus sequences are shown at the top and bottom. Only bases differing from those of at least one nonecotropic provirus are shown. The deduced recombination sites are indicated by arrows and correspond to the region indicated in Fig. 4. Sources of sequence data: xenotropic, NZB (37); polytropic, MX27 (42); modified polytropic, MX33 (42).
U3 regions of recombinant proviruses.
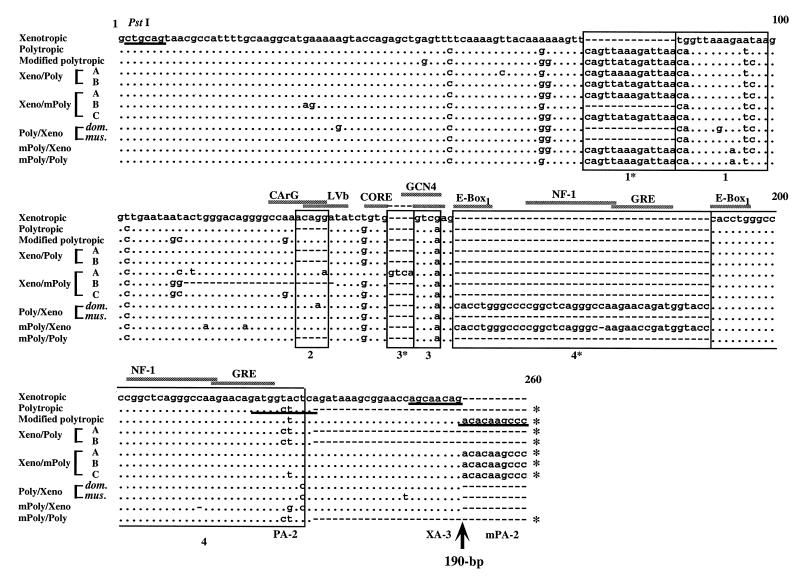
Sequence analyses were also performed on the U3 regions of the PCR-amplified recombinant proviruses. In Fig. 6, the sequences are compared with those of nonecotropic proviruses from inbred mice. Each typical nonecotropic provirus from inbred mice contained unique structures distinguishing it from the others. For example, polytropic and modified polytropic proviruses contained a 14-bp duplication of region 1 (1 and 1*) and region 2 was absent from the polytropic provirus sequence. Further, in polytropic and modified polytropic proviruses, a unique 190-bp insertion was present just downstream of region 4 (the core enhancer region) (Fig. 6) (24, 42).
FIG. 6.
U3 sequences of recombinant proviruses. The nucleotide sequences of recombinant proviruses are compared with analogous regions of xenotropic (NZB) (37), polytropic (MX27) (42), and modified polytropic (MX33) (42) proviruses. The sequence of the NZB xenotropic virus was used as a standard sequence. Dots indicate nucleotide identity. Dashes indicate the absence of a nucleotide. Direct repeats and unique sequences present in the proviruses are boxed; these regions are designated 1*, 1, 2, 3*, 3, 4*, and 4. Potential enhancer sequence regions are also indicated by the shaded bar. The position of the 190-bp insertion is shown by the arrow. Proviruses with the 190-bp insertion are indicated by asterisks on the 3′ end of the sequences. Primer sequences are underlined. The conserved PstI site is also shown.
The U3 sequences of several recombinant proviruses showed additional variation relative to the standard nonecotropic proviruses. Although the type B Xeno/Poly recombinant proviruses had a polytropic U3 sequence, regions 1* and 1 were exactly the same as those in the modified polytropic provirus. Conversely, the U3 sequences of type A Xeno/mPoly recombinant provirus were very similar to those of the modified polytropic provirus, although region 1* was identical to that of the polytropic provirus. Further, the type A Xeno/mPoly recombinant provirus had a unique 4-bp insertion creating a direct repeat (3* and 3) and a possible new enhancer binding sequence for GCN4 (CAGTCA) (Fig. 6) (20). The U3 region of the type B Xeno/mPoly recombinant provirus contained another unique feature. In spite of modified polytropic sequence, this type of provirus lacked region 1*, a characteristic feature of xenotropic proviruses. Furthermore, this type of recombinant provirus had a 23-bp deletion in the core enhancer region, resulting in the deletion of region 2 (Fig. 6).
The sequences of Poly/Xeno recombinant proviruses were virtually identical to that of the xenotropic provirus. However, these recombinant proviruses contained a modified polytropic region 1 (Fig. 6). Further, the provirus from the M. m. domesticus strain contained a 39-bp duplication (region 4*) in the core enhancer sequence region. This duplicated sequence was probably responsible for the differences in the length of PCR products found in the M. m. musculus and M. m. domesticus DNAs (Fig. 3B, PS-1/XA-3).
The U3 sequence of the mPoly/Xeno recombinant provirus contained a 14-bp direct repeat (region 1* and 1) on a base of xenotropic sequence. Further, a 38-bp direct repeat sequence (region 4*) was also found in the core enhancer region (Fig. 6), and repeats within this region often arise during replication of MCF viruses in mice (46). PCR analysis also confirmed that the mPoly/Xeno recombinant provirus, like xenotropic proviruses, does not have the 190-bp insertion (Fig. 6).
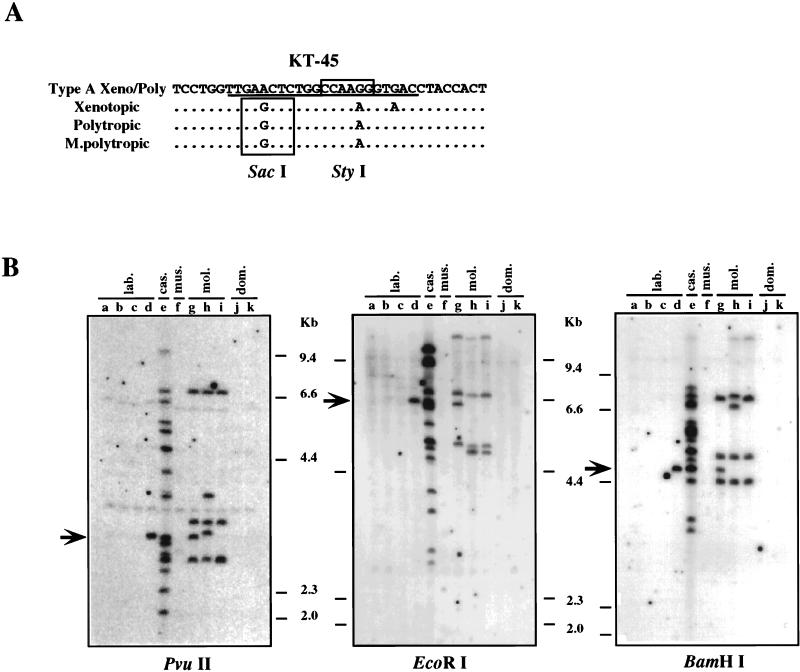
Distribution of the type A Xeno/Poly recombinant proviruses.
The unblotting and PCR analyses indicated that the type A Xeno/Poly recombinant provirus is widely distributed in M. m. castaneus and M. m. molossinus. To verify this possibility and determine the distribution of this type of recombinant provirus in more detail, we designed a specific probe, KT-45, by using a unique nucleotide sequence in the SU region (Fig. 4), resulting in two or three nucleotide differences from other nonecotropic proviruses (Fig. 7A). Initially, unblotting was performed on PvuII-digested mouse DNAs. As shown in Fig. 7B, reactive fragments were found in the C57BL/6J strain of inbred mice and the M. m. castaneus and M. m. molossinus subspecies. This distribution was consistent with the presence of the type A Xeno/Poly recombinant proviruses in the mice, indicating that KT-45 detected the type A Xeno/Poly recombinant proviruses specifically. Further, 15 fragments were found in M. m. castaneus DNA (Fig. 7B, lanes e; Table 1), and the fragments corresponded to almost half of the bands detected in the DNA by the Pltr probe (Fig. 1C, lane p; Table 1). Comparison of the numbers of the fragments detected by the Pltr and KT-45 probes in M. m. castaneus DNA implies that all or almost all the fragments detected by the polytropic LTR probe in M. m. castaneus DNA belonged to proviruses of this type. By contrast, this probe detected 4 or 5 fragments of the 14 or so Pltr-reactive fragments in the DNAs from M. m. molossinus strains (compare Fig. 7B, lanes g and h with Fig. 1C, lanes g to i, and Table 1), indicating that about one-third of the proviruses containing polytropic LTRs in the M. m. molossinus DNAs were type A Xeno/Poly recombinant proviruses. Another one-third were probably polytropic proviruses, since two polytropic env-reactive bands were found in each M. m. molossinus DNA (Fig. 1C, lanes r to t; Table 1). The remaining KT-45-reactive bands may be associated with different env sequences or may represent solo LTRs.
FIG. 7.
Distribution of type A Xeno/Poly recombinant provirus. (A) Specific oligonucleotide probe for the type A Xeno/Poly recombinant proviruses. The nucleotide sequence of the type A Xeno/Poly recombinant provirus is compared with analogous regions of xenotropic (NZB) (37), polytropic (MX27) (42), and modified polytropic (MX33) (42) proviruses. The sequence of the specific probe, KT-45, is underlined. Unique restriction recognition sites are boxed. (B) Detection of type A Xeno/Poly recombinant proviruses. Unblotting was performed by using PvuII-, EcoRI-, or BamHI-digested genomic DNAs. Lanes: a, AKR/J; b, HRS/J; c, C3H/HeJ; d, C57BL/6J; e, CAST/Ei (M. m. castaneus); f, CZECH II/Ei (M. m. musculus); g, MOLC/Rk (M. m. molossinus); h, MOLF/Ei (M. m. molossinus); i, MOLG/Dn (M. m. molossinus); j, WSB/Ei (M. m. domesticus); k, ZALENDE/Ei (M. m. domesticus). The approximate positions of molecular markers are shown on the right. Arrows indicate comigrating bands among different subspecies of mice.
All but one of the inbred strains contained no proviruses reactive with the KT-45 probe (Fig. 7B, lanes a to d). The exception was a single fragment found in the C57BL/6J strain, which seemed to comigrate with a band detected in M. m. castaneus and one M. m. molossinus DNA (Fig. 7B, lanes d, e, and g, arrow). The presence of a band of identical mobility in EcoRI- and BamHI-digested DNAs (Fig. 7B, lanes d, e, and g, arrow) confirms the sharing of this provirus among the three strains. As noted above, this fragment was identical to Xmv42 (11, 13) (Fig. 1B and C, boxed band). In our previous study, we reported that the Xmv42 is located on chromosome 11 and is closely linked to a modified polytropic provirus locus, Mpmv4 (15). As shown in Fig. 1D, the Mpmv4 fragment was also shared between the C57BL/6J strain and an M. m. molossinus strain (MOLC/Rk). However, Mpmv4 was not found in the M. m. castaneus DNA (Fig. 1D).
We next investigated the SU sequence of the type A Xeno/Poly recombinant provirus to predict the potential receptor usage of the recombinant proviruses. The host range of MLVs is specified by two variable regions (VRA and VRB) in the 5′ portion of SU, upstream of the HPR region (1). Type A Xeno/Poly recombinant provirus clones that encompass those regions were obtained from M. m. castaneus DNA by using an env sense primer that hybridizes to all groups of nonecotropic proviruses and the PA-2 antisense primer. Sequence analysis revealed that the nucleotide and deduced amino acid sequences of this region were virtually identical to that of xenotropic virus (data not shown), implying that the type A Xeno/Poly recombinant provirus should have a xenotropic receptor binding capacity. This observation is also consistent with the fact that infectious viruses classed as xenotropic have been isolated from these mice (27).
DISCUSSION
Distribution of nonecotropic proviruses in wild mice.
Endogenous nonecotropic MLV proviruses are stable genetic elements that are polymorphic in mice. The proviruses were fixed recently in the Mus germ line but are certainly older than ecotropic MLVs, which are neither widely distributed nor greatly amplified in mice (2, 7). Thus, the nonecotropic MLV proviruses should lead to a better understanding of the association between retroviruses and their host during evolutionary history. We describe here the analysis of polymorphism of the nonecotropic MLV proviruses in mice. Such analysis is a key step to develop an understanding of the virus-host association.
In this study, extended polymorphism of nonecotropic MLV proviruses was observed in inbred mice (AKR/J, HRS/J, C3H/HeJ, and C57BL/6J) and four major subspecies of M. musculus. The hybridization analysis with both env- and LTR-specific oligonucleotide probes for each of the nonecotropic groups allowed us to determine the detailed distribution of the proviruses. This analysis indicated that although the nonecotropic provirus sequences are widely distributed in the wild-mouse subspecies, each group shows a differential distribution. Xenotropic sequences were found mainly in M. m. musculus and M. m. molossinus, while the polytropic and modified polytropic fragments were found predominantly in M. m. domesticus. Together with previous observations that a species of Mus, M. spretus, has only polytropic env-like sequences (4, 26; our unpublished data), these observations imply that each nonecotropic provirus might have been integrated selectively into specific subspecies around the time of subspeciation. Alternatively, the different types of virus might have been able to infect only certain subspecies. In fact, it is known that wild-mouse species have shown a greater variability than inbred strains in susceptibility to the different host range groups of MLVs (25, 27, 31).
We also examined the correlation between env and LTR sequences of the proviruses. Although the linkage of those sequences in xenotropic, polytropic, and modified polytropic proviruses is relatively strict in common laboratory strains (Fig. 1) (11, 42, 43), in some wild mice there were exceptions to the rule. For example, the env and LTR sequences of xenotropic provirus did not correlate at all in M. m. domesticus (Fig. 1B). Furthermore, both polytropic and modified polytropic sequences appeared to be unlinked in some Asian wild mice (Fig. 1C and D). These observations could be explained by genetic features of the proviruses such as internal deletions, intragroup polymorphisms, or recombination with sequences of other groups. Although proviruses that show deletion or heterogeneity of the env sequences have been found in some inbred laboratory strains (12, 29, 30), the large numbers of proviruses that lack linkage between the env and LTR sequences in the wild-mouse subspecies suggested that the nonecotropic MLV proviruses show extensive genetic variation in wild mice that was not revealed in the laboratory strains.
Recombinant form of nonecotropic proviruses in wild mice.
By mixing and matching specific primers in a single PCR assay, we could detect several forms of proviruses in the mouse genomes which reacted as if they were recombinant relative to the standard proviruses found in inbred laboratory mice (Fig. 3B). We refer to these nonstandard proviruses as recombinant, by comparison to the proviruses in laboratory strains, although there is no way to judge, by sequence analysis alone, which types are precursors and which are recombinants. The sequence analyses of the env and LTR regions revealed that each recombinant provirus shows unique structural features not found in typical nonecotropic proviruses (Fig. 4 and 6). In some recombinant proviruses, possible crossover regions between different groups were found in the env genes, one of which was in three different types of recombinant proviruses (Fig. 5), implying a common origin. Interestingly, the possible recombinant regions we found here, just downstream from the HPR region and in the middle of the TM region, correspond to the recombinant sites observed in MCF viruses (5, 10, 17–19, 23, 46). These regions might contain “hot spots” for recombination between different viruses in mice or might be selected by the generation of replication-competent viruses.
In addition to recombinational differences, a number of distinctive sequence alternatives were observed in the recombinant relative to the standard nonecotropic proviruses. In the U3 region, the polytropic and modified polytropic proviruses have a complete 14-bp duplication of region 1 and the sequence of the modified polytropic provirus contains a few additional nucleotide changes. In contrast, the U3 region of the xenotropic provirus does not have this duplication and also lacks a 190-bp insertion found in the other two types (Fig. 6). The recombinant proviruses, however, did not follow these rules. For example, in the proviruses based on the modified polytropic U3 structure, the type A Xeno/mPoly recombinant provirus contained a polytropic-type region 1 whereas the type B Xeno/mPoly provirus lacked region 1* (Fig. 6). The mPoly/Xeno recombinant provirus contained a duplicated region 1 in a xenotropic U3 structure. Furthermore, region 1 of the Poly/Xeno recombinant proviruses was of the modified polytropic type. This observation might have useful implications for the evolutionary relationship among MLVs. With this possibility in mind, we are further analyzing the polymorphism of nonecotropic proviruses in other Mus species, including M. spretus and M. hortulanus (unpublished data).
The extensive genetic variation, both recombinational and mutational, among the proviruses of different subspecies of mice, as well as the polymorphism of insertion sites, shows that those proviruses are not static insertions in the germ line. Rather, their evolution must have involved extensive periods of replication as viruses separating insertions into the germ line. Indeed, both the recombinational and mutational events are similar to those seen during the generation of recombinant MCF viruses in some inbred strains of mice (10, 18, 19, 23, 28, 46).
Another interesting aspect of these proviruses was observed in the type A Xeno/Poly recombinant provirus. One recombinant provirus of this type was found in one laboratory strain and two subspecies of M. musculus (Fig. 4), indicating a common inheritance among these mice. Unique nucleotide changes in the SU region allowed us to generate a specific probe to fully characterize the recombinant proviruses in the mouse genomes (Fig. 7). We found that this type of provirus is present only in M. m. castaneus and M. m. molossinus subspecies and that all or almost all proviruses detected by the polytropic LTR probe in M. m. castaneus DNA were type A Xeno/Poly recombinant provirus (Fig. 1C). Interestingly, it has been demonstrated that M. m. castaneus shows resistance to polytropic virus infection in vitro, most probably because of a mutation of the gene for the polytropic receptor (31). Consistent with this observation, no polytropic env-containing sequence and only one modified polytropic env-containing sequence was present in the DNA sample from the subspecies (Fig. 1C and D). It is possible that the recombinant polytropic virus containing the xenotropic env sequence was selected by its ability to replicate in M. m. castaneus. Indeed, sequence analysis of the SU region of the recombinant implies that it encodes a xenotropic host range. It is believed that, unlike inbred laboratory mice, Asian wild mice can be genetically infected with xenotropic MLVs (27).
It is now clear that the inbred laboratory strains of mice were generated by interbreeding of a small number of wild mice, including M. m. molossinus and M. m. domesticus (47, 50). In fact, it has been shown that the Y chromosome of inbred strains was derived from either M. m. molossinus or M. m. domesticus (48). On the other hand, the M. m. molossinus subspecies is known to be a natural hybrid between M. m. castaneus and M. m. musculus in East Asia (47, 50). Considering this interpretation of the origin of M. m. molossinus and inbred mouse strains, it could be true that M. m. castaneus was the source of the type A Xeno/Poly recombinant proviruses in these mice. The distribution of the recombinant proviruses indicated that M. m. molossinus inherited the recombinant provirus from M. m. castaneus by interbreeding because, despite the relatively small number of samples tested, the M. m. molossinus subspecies contained multiple proviral genes and at least three of these represented shared proviral integrations with M. m. castaneus. Furthermore, it is also clear that the provirus, Xmv42, in the C57BL/6J strain was inherited from M. m. molossinus mice because the locus in the C57BL/6J strain comigrated with those of M. m. castaneus and an M. m. molossinus strain but only the M. m. molossinus shared a modified polytropic provirus locus (Mpmv4) linked with the recombinant proviruses in the C57BL/6J strain (Fig. 1D).
The proviruses detected in this study should provide valuable genetic markers for the evolutionary study of retroviruses and their murine host.
ACKNOWLEDGMENTS
We are grateful to Jonathan P. Stoye and Wayne N. Frankel for helpful comments and to Mary Bostic-Fitzgerald for preparing the manuscript.
This work was supported by National Cancer Institute award R35CA44385 to J.M.C. and a Leukemia Society of America Special Fellowship to K.T. J.M.C. was a Research Professor of the American Cancer Society.
REFERENCES
- 1.Battini J-L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 3.Bowes C, Li T, Frankel W N, Danciger M, Coffin J M, Applebury M L, Farber D B. Localization of a retroviral element within the rd gene coding for the b-subunit of cGMP-phosphodiesterase. Proc Natl Acad Sci USA. 1993;90:2955–2959. doi: 10.1073/pnas.90.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ch’ang L-Y, Yank W K, Myer F E, Koh C K, Boone L R. Specific sequence deletions in two classes of murine leukemia virus-related proviruses in the mouse genome. Virology. 1989;168:245–255. doi: 10.1016/0042-6822(89)90264-x. [DOI] [PubMed] [Google Scholar]
- 5.Chattopadhyay S K, Lander M R, Gupta S, Rands E, Lowy D R. Origin of mink cytopathic focus-forming (MCF) viruses: comparison with ecotropic and xenotropic murine leukemia virus genomes. Virology. 1981;113:465–483. doi: 10.1016/0042-6822(81)90175-6. [DOI] [PubMed] [Google Scholar]
- 6.Cloyd M W, Hartley J W, Rowe W P. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med. 1980;151:462–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin J M. Retrovirus variation and evolution. In: Cooper G M, Temin R G, Sugden B, editors. The DNA provirus. Washington, D.C: American Society for Microbiology; 1995. pp. 221–244. [Google Scholar]
- 8.Coffin J M, Stoye J P, Frankel W N. Genetics of endogenous murine leukemia viruses. Ann N Y Acad Sci. 1989;567:39–49. doi: 10.1111/j.1749-6632.1989.tb16457.x. [DOI] [PubMed] [Google Scholar]
- 9.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elder J H, Gautsch J W, Jensen F C, Lerner R A, Hartley J W, Rowe W P. Biochemical evidence that MCF murine leukemia viruses are envelope (env) gene recombinants. Proc Natl Acad Sci USA. 1977;74:4676–4680. doi: 10.1073/pnas.74.10.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frankel W N, Coffin J M. Endogenous nonecotropic proviruses mapped with oligonucleotide probes from the long terminal repeat region. Mamm Genome. 1994;5:275–281. doi: 10.1007/BF00389541. [DOI] [PubMed] [Google Scholar]
- 12.Frankel W N, Lee B K, Stoye J P, Coffin J M, Eicher E M. Characterization of the endogenous nonecotropic murine leukemia viruses of NZB/B1NJ and SM/J inbred strains. Mamm Genome. 1992;2:110–122. doi: 10.1007/BF00353859. [DOI] [PubMed] [Google Scholar]
- 13.Frankel W N, Stoye J P, Taylor B A, Coffin J M. Genetic analysis of endogenous xenotropic murine leukemia viruses: association with two common mouse mutations and the viral restriction locus Fv-1. J Virol. 1989;63:1763–1774. doi: 10.1128/jvi.63.4.1763-1774.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frankel W N, Stoye J P, Taylor B A, Coffin J M. Genetic identification of endogenous polytropic proviruses by using recombinant inbred mice. J Virol. 1989;63:3810–3821. doi: 10.1128/jvi.63.9.3810-3821.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankel W N, Stoye J P, Taylor B A, Coffin J M. A linkage map of endogenous murine leukemia proviruses. Genetics. 1990;124:221–236. doi: 10.1093/genetics/124.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green N, Hiai H, Elder J H, Schwartz R S, Khiroya R H, Thomas C Y, Tsichlis P N, Coffin J M. Expression of leukemogenic recombinant viruses associated with a recessive gene in HRS/J mice. J Exp Med. 1980;152:249–264. doi: 10.1084/jem.152.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoggan M D, O’Neill R R, Kozak C A. Nonecotropic murine leukemia viruses in BALB/c and NFS/N mice: characterization of the BALB/c Bxv-1 provirus and the single NFS endogenous xenotrope. J Virol. 1986;60:980–986. doi: 10.1128/jvi.60.3.980-986.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holland C A, Hartley J W, Rowe W P, Hopkins N. At least four viral genes contribute to the leukemogenicity of murine retrovirus MCF 247 in AKR mice. J Virol. 1985;53:158–165. doi: 10.1128/jvi.53.1.158-165.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holland C A, Wozney J, Hopkins N. Nucleotide sequence of the gp70 gene of murine retrovirus MCF 247. J Virol. 1983;47:413–420. doi: 10.1128/jvi.47.3.413-420.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hope I A, Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987;6:2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inaguma Y, Miyashita N, Moriwaki K, Huai W C, Mei-Lei J, Zinqiao H, Ikeda H. Acquisition of two endogenous ecotropic murine leukemia viruses in distinct Asian wild mouse populations. J Virol. 1991;65:1796–1802. doi: 10.1128/jvi.65.4.1796-1802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins N A, Copeland N G, Taylor B A, Lee B K. Organization, distribution and stability of endogenous ecotropic murine leukemia virus DNA in chromosomes of Mus musculus. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan A S. Nucleotide sequence analysis establishes the role of endogenous murine leukemia virus DNA segments in formation of recombinant mink cell focus-forming murine leukemia viruses. J Virol. 1984;50:864–871. doi: 10.1128/jvi.50.3.864-871.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan A S, Martin M A. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc Natl Acad Sci USA. 1983;80:2699–2703. doi: 10.1073/pnas.80.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak C A. Susceptibility of wild mouse cells to exogenous infection with xenotropic leukemia viruses: control by a single dominant locus on chromosome 1. J Virol. 1985;55:690–695. doi: 10.1128/jvi.55.3.690-695.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak C A, O’Neill R R. Diverse wild mouse origins of xenotropic, mink-cell focus-forming, and two types of ecotropic proviral genes. J Virol. 1987;61:3082–3088. doi: 10.1128/jvi.61.10.3082-3088.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak C A, Ruscetti S. Retroviruses in rodents. In: Levy J A, editor. Retroviridae. New York, N.Y: Plenum Press; 1992. pp. 405–487. [Google Scholar]
- 28.Laigret F, Repaske R, Boulukos K, Rabson A B, Khan A S. Potential progenitor sequences of mink cell focus-forming (MCF) murine leukemia viruses: ecotropic, xenotropic, and MCF-related viral RNAs are detected concurrently in thymus tissues of AKR mice. J Virol. 1988;62:376–386. doi: 10.1128/jvi.62.2.376-386.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamont C, Culp P, Talbott R L, Phillips T R, Trauger R J, Frankel W N, Wilson M C, Coffin J M, Elder J H. Characterization of endogenous and recombinant proviral elements of a highly tumorigenic AKR cell line. J Virol. 1991;65:4619–4628. doi: 10.1128/jvi.65.9.4619-4628.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levy D E, Lerner R A, Wilson M C. Normal expression of polymorphic endogenous retroviral RNA containing segments identical to mink cell focus-forming virus. J Virol. 1985;56:691–700. doi: 10.1128/jvi.56.3.691-700.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyu M S, Kozak C A. Genetic basis for resistance to polytropic murine leukemia viruses in the wild mouse species Mus castaneus. J Virol. 1996;70:830–833. doi: 10.1128/jvi.70.2.830-833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massey A C, Lawrenz S S, Innes D J, Thomas C Y. Origins of enhancer sequences of recombinant murine leukemia viruses from spontaneous B- and T-cell lymphomas of CWD mice. J Virol. 1994;68:3773–3783. doi: 10.1128/jvi.68.6.3773-3783.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mucenski M L, Taylor B A, Copeland N G, Jenkins N A. Characterization of somatically acquired ecotropic and mink cell focus-forming viruses in lymphomas of AKXD recombinant inbred mice. J Virol. 1987;61:2929–2933. doi: 10.1128/jvi.61.9.2929-2933.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 35.O’Brien S J, Moore J L, Martin M A, Womack J E. Evidence for the horizontal acquisition of murine AKR virogenes by recent horizontal infection of the germ line. J Exp Med. 1982;155:1120–1123. doi: 10.1084/jem.155.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Donnell P V, Stockert E, Obata Y, Old L J. Leukemogenic properties of AKR dualtropic (MCF) viruses: amplification of murine leukemia virus-related antigens on thymocytes and acceleration of leukemia development in AKR mice. Virology. 1981;112:468–563. doi: 10.1016/0042-6822(81)90301-9. [DOI] [PubMed] [Google Scholar]
- 37.O’Neill R R, Buckler C E, Theodore T S, Martin M A, Repaske R. Envelope and long terminal repeat sequences of a cloned infectious NZB xenotropic murine leukemia virus. J Virol. 1985;53:100–106. doi: 10.1128/jvi.53.1.100-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Neill R R, Khan A S, Hoggan M D, Hartley J W, Martin M A, Repaske R. Specific hybridization probes demonstrate fewer xenotropic than mink cell focus-forming murine leukemia virus env-related sequences in DNAs from inbred laboratory mice. J Virol. 1986;58:359–366. doi: 10.1128/jvi.58.2.359-366.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quint W, Boelens W, Wezenbeek P V, Cuypers T, Maandag E R, Selten G, Berns A. Generation of AKR mink cell focus-forming viruses: a conserved single-copy xenotropic-like provirus provides recombinant long terminal repeat sequences. J Virol. 1984;50:432–438. doi: 10.1128/jvi.50.2.432-438.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silver J. Role of mink cell focus-inducing virus in leukemias induced by Friend ecotropic virus. J Virol. 1984;50:872–877. doi: 10.1128/jvi.50.3.872-877.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoye J P, Coffin J M. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol. 1987;61:2659–2669. doi: 10.1128/jvi.61.9.2659-2669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoye J P, Coffin J M. Polymorphism of murine endogenous proviruses revealed by using virus class-specific oligonucleotide probes. J Virol. 1988;62:168–175. doi: 10.1128/jvi.62.1.168-175.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoye J P, Fenner S, Greenoak G E, Moran C, Coffin J M. Role of endogenous retroviruses as mutagens: the hairless mutation of mice. Cell. 1988;54:383–391. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- 45.Stoye J P, Frankel W N, Coffin J M. DNA hybridization in dried gels with fragmented probes: an improvement over blotting techniques. Technique. 1991;3:123–128. [Google Scholar]
- 46.Stoye J P, Moroni C, Coffin J. Virological events leading to spontaneous AKR thymomas. J Virol. 1991;65:1273–1285. doi: 10.1128/jvi.65.3.1273-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor B A. Recombinant inbred strains: use in gene mapping. In: Morse III H C, editor. Origins of inbred mice. New York, N.Y: Academic Press, Inc.; 1978. pp. 423–438. [Google Scholar]
- 48.Tucker P K, Lee B K, Lundrigan B L, Eicher E M. Geographic origin of the Y chromosomes in “old” inbred strains of mice. Mamm Genome. 1992;3:254–261. doi: 10.1007/BF00292153. [DOI] [PubMed] [Google Scholar]
- 49.Vogt M, Haggblom C, Swift S, Haas M. Envelope gene and long terminal repeat determine the different biological properties of Rauscher, Friend, and Moloney mink cell focus-inducing viruses. J Virol. 1985;55:184–192. doi: 10.1128/jvi.55.1.184-192.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yonekawa H, Gotoh O, Tagashira Y, Matsushima Y, Shi L I, Cho W S, Miyashita N, Moriwaki K. A hybrid origin of Japanese mice “Mus musculus molossinus.”. Curr Top Microbiol Immunol. 1986;127:62–67. [PubMed] [Google Scholar]