Abstract
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of Coronavirus Disease 2019 (COVID-19). The disease has a wide range of clinical manifestations, from asymptomatic to severe. Ancestral contribution, sex, immune response, and genetic factors influence the presentation of the disease. The objective of the present study was to validate these genetic variants in patients with severe COVID-19 who died and in survivor patients. Methods: Single nucleotide variants (SNVs) in six genes: ATPase plasma membrane Ca2+ transporting 2 (ATP2B2), transmembrane serine protease 2 (TMPRSS2), dedicator of cytokinesis 2 (DOCK2), (interferon alpha and beta receptor subunit 2) IFNAR2, tumor necrosis factor receptor superfamily, member 1A (TNFRSF1A), and tumor necrosis factor receptor superfamily, member 1B (TNFRSF1B), were explored in two groups: the first consisted of severe COVID-19-related patients (familial cases from 58 families, n = 130), and the second group of unrelated severe COVID-19 patients (n = 1045). In each study group, death was evaluated as the outcome.
Results
In non-related patients with severe COVID-19, carriers of GG genotype (rs2289274) in the ATP2B2 gene showed a high-risk probability of non-surviving (OR = 1.43). Survival analysis to 75 days indicates that carriers of GG have a higher risk than GA or AA genotypes (p = 0.0059). The haplotype GG (rs2289273-rs2289274) in ATP2B2 was found to be associated with a high risk of death in severe non-related COVID-19 patients. No significant associations were found between severe COVID-19-related patients and SNVs in ATP2B2, TMPRSS2, DOCK2, IFNAR2, TNFRSF1A, or TNFRSF1B.
Conclusions
Unrelated patients with severe COVID-19 that carry the GG genotype (rs2289274) in ATP2B2 showed a high death risk. Survival analysis to 75 days indicates that carriers of GG have a higher risk of non-survival compared to GA or AA genotypes.
Keywords: COVID-19, Genetic susceptibility, SARS-CoV-2, ATP2B2
Highlights
-
•
Patients with COVID-19 carriers rs2289274/ATP2B2 (GG) had a high risk of non-surviving.
-
•
The haplotype in ATP2B2 was associated with the risk of death in severe COVID-19.
-
•
No significant associations were found between severe COVID-19-related patients.
Abbreviations
- BMI
Body Mass Index
- CDC
Centers for Disease Control and Prevention
- GWAS
Genome-Wide Association Studies
- HWE
Hardy–Weinberg Equilibrium
- IMV
Invasive Mechanical Ventilation
- IQR
Interquartile Range
- LD
Linkage Disequilibrium
- SAH
Systemic Arterial Hypertension
- SNVs
Single Nucleotide Variations
- T2D
Type 2 Diabetes
1. Introduction
Infectious diseases result from the host's exposure to an agent in a specific environment. The host capable of being infected by an infectious agent possesses inherent factors such as ancestral contribution, sex, immune response, and genetic factors that become significant during the infection [1].
People infected by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) show heterogeneity in the clinical manifestations [2]. Also, subjects who developed the Coronavirus Disease 2019 (COVID-19) can present asymptomatic or severe disease with pneumonia, respiratory failure, acute respiratory distress syndrome (ARDS), and sepsis [3].
The global mortality rate among hospitalized COVID-19 patients is around 18 %. Older age and comorbidities (obesity, smoking, chronic lung disease, diabetes, hypertension, dyslipidemia, or heart disease) were identified as risk factors for death among COVID-19 patients [3]. The variation in mortality rates between populations and ethnicities is a strong indicator of the modulatory effect of host genetics on its pathogenesis [4]. In early 2020 in the United States, multiple members of a family showed a high case-fatality rate [5]. The authors proposed that family clusters could be a key to identifying host factors that predispose to a severe disease course or identify populations with increased risk of death [6].
Single Nucleotide Variants (SNVs) are more frequent genetic variations in the human genome. Studies on genetic predisposition have identified SNVs associated with the risk of COVID-19 in genes involved in the immune response like TLR3, TLR7, IFNAR2, and IFNAR1 [2].
Genome-wide association studies (GWAS) are one of the tools used to find SNVs among other genetic variations associated with specific diseases. Numerous research groups carried out GWAS in diverse populations during the COVID-19 pandemic to identify risk variants [[7], [8], [9], [10], [11], [12], [13]]. The case-control family design can determine familial risks, estimate the risk (penetrance) for measured genotypes, stratify risks by family history, and study modifiers in genetically susceptible individuals [14]. Identifying SNVs associated with increased risk in COVID-19 phenotypes may help to explain variability in the presentation of SARS-CoV-2 infection. This is also a key component for personalized disease management.
Through a literature search in GWAS and genetic association studies, we selected SNVs linked with COVID-19 or related to severity, mortality, and minor allele frequency (MAF) in the Latin American population greater than 10 % (Table 1).
Table 1.
SNVs selected to be evaluated in this study.
| Gene | SNV | MA | MAF, 1000 Genomes | MAF, CEU | Type or location | OR | p-value | Ancestry | Outcome/Association | Ref |
|---|---|---|---|---|---|---|---|---|---|---|
| IFNAR2 | rs2834158 | T | 0.39 | 0.32 | Intron variant | 1.38 | 0.027 | Mex | Death | 15 |
| rs1051393 | G | 0.39 | 0.32 | Missense variant | 1.3 | 0.041 | ||||
| rs3153 | A | 0.35 | 0.29 | Intron variant | 1.37 | 0.019 | ||||
|
TMPRSS2 |
rs12329760 |
T |
0.26 |
0.22 |
Missense variant |
NR |
0.04 |
Eu, Afr, As, and LA |
Severity |
16 |
| ATP2B2 | rs2289273 | A | 0.14 | 0.13 | Synonumous variant | 0.29 | 3.96E-05 | Chinese | SARS-CoV-2 infection | 8 |
| rs2289274 | A | 0.27 | 0.29 | 0.31 | 5.28E-05 | |||||
| TNFRS1B | rs3397 | C | 0.59 | 0.41 | 3′ UTR variant | NR | <0.01 | Mex | Low PaO2/FiO2 levels in severe COVID-19 | 17 |
| rs1061622 | G | 0.19 | 0.23 | Missense variant | NR | <0.05 | High sTNFR1 levels in severe COVID-19 |
MA; minor allele, MAF; Minor allele frequency, CEU; CEPH/CEU (Centre d'Etude du Polymorphisme Humain - Utah), NR; Not reported, Mex; Mexican-mestizo, Eu; European; Afr; African, As; South Asia, LA; Latin America, OR; Odds ratio, Ref; reference.
The objective of the present study was to validate these genetic variants in patients with severe COVID-19 who died compared to survivor patients. We explored these variants in unrelated (case-control study) and biologically related (familial cases) subjects.
2. Materials and methods
2.1. Study population
The cross-sectional study included COVID-19 patients hospitalized at the Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas (INER) in Mexico. The period of recruitment was from August 2020 to December 2021. The patients included did not have a previous history of vaccination and did not have a prior SARS-CoV-2 infection that would require hospitalization. Characteristics of patients included were described previously [15,16]. We studied two groups of severe COVID-19 patients: the first was COVID-19-related patients (familial cases from 58 families, n = 130); these patients were defined as familial subjects related in the first degree (horizontal or vertical consanguinity), including sib pairs, father, or mother, and at least one son or daughter. The second group included severe COVID-19 patients (n = 1045), not biologically related among them or to the subjects in the group of familial cases. In both groups, mortality was evaluated as the outcome. Exclusion criteria were described previously [17].
We recollected each patient's demographics, clinical data, signs, symptoms, and outcomes in the clinical record. The study was approved by the Ethics in Research Committee of the same institution (protocol number B11-23) and adhered to the principles of the Declaration of Helsinki. The approval of informed consent was obtained from the patients when possible or from the responsible family member.
Genotyping.
The DNA was extracted using a method described previously [17]. We evaluated eleven SNVs in six genes as follows; rs2289273, rs2289274 (ATPase plasma membrane Ca2+ transporting 2, ATP2B2); rs12329760 (transmembrane serine protease 2, TMPRSS2); rs60200309 (dedicator of cytokinesis 2, DOCK2); rs1051393, rs3153, rs2834158 (interferon alpha and beta receptor subunit 2, IFNAR2); rs767455, rs1800693 (tumor necrosis factor receptor superfamily, member 1A, TNFRSF1A) and (tumor necrosis factor receptor superfamily, member 1B, TNFRSF1B) in both groups.
The alleles were determined through real-time PCR (PCR) using the StepOnePlus equipment (Applied Biosystems, Foster City, CA, USA). We used TaqMan probes predesigned for the rs2289273 (C_15880954_10), rs2289274 (C_15881002_10), rs12329760 (C_25622353_20), rs60200309 (C_90065524_10), rs1051393 (C_2443247_30), rs3153 (C_9479908_10), rs2834158 (C_16072683_20), rs767455 (C___2298465_20), rs1800693 (C___2645714_10), rs1061622 (C___8861232_20) and rs3397 (C___8861228_20) (Applied Biosystems, Foster City, CA, USA). The PCR assay conditions were described previously [17].
2.2. Statistical analysis
Normal distribution was assessed employing the Kolmogorov-Smirnov test. Continuous data are shown as the median and interquartile range (IQR), and categorical data are shown as frequencies in percentage. The Hardy–Weinberg equilibrium (HWE) was calculated for the eleven polymorphisms in unrelated COVID-19 patients. We used the U-Mann-Whitney test and Fisher's exact test for the comparisons.
Subsequently, for each study group, we analyzed mortality as an outcome. The genotypes and alleles were analyzed using Epidat software v. 3.1 [18]; genetic models were also applied. For SNVs with a statistically significant association, linkage disequilibrium (LD) analysis in ATP2B2, IFNAR2, TNFRSF1A, and TNFRSF1B genes was performed with Haploview software v. 4.1 [19]. We used the Kaplan-Meier method to evaluate the survival probability according to the genotypes. We considered it statistically significant if p < 0.05.
3. Results
3.1. Characteristics of COVID-19 patients
The group of COVID-19-related patients (familial cases) had fewer men than the unrelated group (55 % vs. 65 %, p = 0.02) and were younger (54 vs. 58 years, respectively, p = 0.03). Body mass index (BMI) did not show statistical differences, nor did the requirement of invasive mechanical ventilation (IMV) (both p = 0.08); however, days with IMV were lower in related patients in comparison with unrelated individuals (7 vs. 9 days, respectively, p = 0.02). Day of symptom onset, PaO2/FiO2, and proportion of deaths were similar among the study groups (Table 2).
Table 2.
Characteristics in related and unrelated severe COVID-19 patients.
| Variable | Related (n = 130) | Unrelated (n = 1045) | p-value |
|---|---|---|---|
| Male (n, %) | 72 (55.4) | 682 (65.3) | 0.02 |
| Age (years) | 54 (46–62) | 58 (47–68) | 0.03 |
| BMI (kg/m2) | 30 (27–33) | 28 (26–33) | 0.08 |
| IMV requirement (n, %) | 81 (62.3) | 729 (69.8) | 0.08 |
| IMV (days) | 7 (0–18) | 9 (0–21) | 0.02 |
| Days of symptom onset | 9 (7–12) | 9 (7–13) | 0.25 |
| PaO2/FiO2(mm/Hg) | 156 (91–211) | 160 (123–203) | 0.05 |
| Deaths (n, %) | 40 (30.8) | 290 (27.8) | 0.68 |
Continuous data are presented as median (interquartile range, IQR) and categorical data as frequency (percentage, %). The statistical tests employed for the comparisons were U-Mann-Whitney and Fisher's exact test. BMI, Body Mass Index; IMV, invasive mechanical ventilation.
The most frequent comorbidities (>20 %) in both groups were type 2 diabetes (T2D), tobacco smoking, systemic arterial hypertension (SAH), and obesity. There were no significant differences in comorbidities between both groups, except for obesity (51 % vs. 41 %, p < 0.001). On the other hand, the most frequent signs (>20 %) in both groups were fever, headache, arthralgias, myalgia, dyspnea, cough, and sore throat (Supplementary Table 1).
3.2. SNVs associated with risk of death in unrelated patients with severe COVID-19
Our research group previously evaluated rs1051393, rs3153, rs2834158 (IFNAR2) [20]; rs767455, rs1800693 (TNFRSF1A) and rs1061622, rs3397 (TNFRSF1B) [15]; therefore, these variants were not analyzed in this study for unrelated patients. We evaluated unrelated patients' SNVs in ATP2B2, TMPRSS2, and DOCK2 genes. The rs2289274 in ATP2B2 and rs60200309 in DOCK2 did not comply with the HWE (p = 2.1E-7 and p = 1.1E-10, respectively).
The comparison of unrelated patients’ non-survivors and survivors showed that the subjects who died were younger and had lower BMI in comparison to the survivors. 92.4 % of non-survivor patients required IMV and more days with this type of assisted mechanical ventilation. The PaO2/FiO2 at admission to the hospital was lower in non-survivor patients (Supplementary Table 2).
Regarding the genetic association study, the codominant model showed no association between rs2289273 (ATP2B2), rs1232976 (TMPRSS2), rs60200309 (DOCK2), and risk of death in COVID-19. Allele frequencies in these SNVs were similar between survivors and non-survivor patients (Supplementary Table 3).
We found the rs2289274 (ATP2B2) to be significantly different distributed with the G allele associated with an increased risk (p = 0.018, OR 1.33, 95 % CI 1.05–1.68) in no survivors’ severe COVID-19 patients. The allele A was associated with a decreased risk in the same comparison (p = 0.018, OR 0.75, 95 % CI 0.59–0.94), similar to the codominant model, with the GA genotype (p = 0.044, OR 0.68, 95 % CI 0.50–0.94). When applied the full-genotype model, the association found was stronger (p = 0.015) with the GG genotype associated with risk (OR = 1.43, 95 % CI 1.08–1.91) of death in severe COVID-19 patients (Table 3).
Table 3.
Allele and genotype association analysis for ATP2B2 (rs2289274) in unrelated patients.
| Non-survivors (n = 290) | Survivors (n = 708) | p-value | OR (95 % CI) | |
|---|---|---|---|---|
| Allele | ||||
| G | 459 (79.1) | 1048 (74.0) | 1.33 (1.05–1.68) | |
| A | 121 (20.8) | 368 (25.9) | 0.018 | 0.75 (0.59–0.94) |
| Codominant model | ||||
| GG | 193 (66.6) | 411 (58.1) | ||
| GA | 73 (25.2) | 226 (31.9) | 0.044 | 0.68 (0.50–0.94) |
| AA | 24 (8.3) | 71 (10) | 0.71 (0.43–1.17) | |
| Full genotype | ||||
| GG | 193 (66.6) | 411 (58.1) | 0.015 | 1.43 (1.08–1.91) |
| GA | 73 (25.2) | 226 (31.9) | 0.041 | 0.71 (0.52–0.97) |
| AA | 24 (8.3) | 71 (10) | 0.460 | 0.80 (0.49–1.31) |
*p-value obtained using X2 test. The number of parentheses is a percentage. OR odds ratio; CI, confidence interval. A p-value <0.05 is considered as significant.
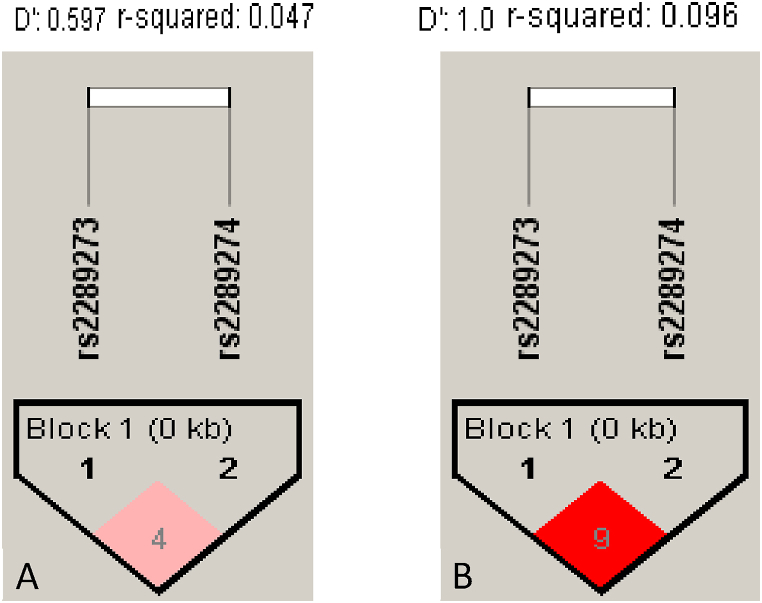
The haplotypes GG and GA (rs2289273-rs2289274) in ATP2B2 showed a significant association with high and low mortality risk, respectively (OR = 1.36 and OR = 0.74) (Table 4). However, a low LD was found in unrelated patients with COVID-19 severe (D’ = 0.59/r2 = 0.047). (Fig. 1A).
Table 4.
Association of haplotypes rs2289273-rs2289274 in ATP2B2 with mortality risk among unrelated patients.
| Haplotypes | Non-survivors n = 290 | Survivors n = 708 | p-value | OR (95 % CI) |
|---|---|---|---|---|
| GG | 454 (78.2) | 1027 (72.5) | 0.007 | 1.36 (1.08–1.71) |
| GA | 106 (18.2) | 326 (23.0) | 0.002 | 0.74 (0.58–0.95) |
| AA | 15 (2.6) | 42 (2.9) | 0.670 | – |
| AG | 5 (0.8) | 21 (1.5) | 1.400 | – |
*p-value obtained using X2 test. The number of parentheses is a percentage. OR, odds ratio; CI, confidence interval. A p-value <0.05 is considered significant.
Fig. 1.
Linkage disequilibrium (LD) of SNVs in the ATP2B2 gene. A) Unrelated patients (r2 = 0.047). B) Related patients (r2 = 0.096). LD was calculated using Haploview software [18].
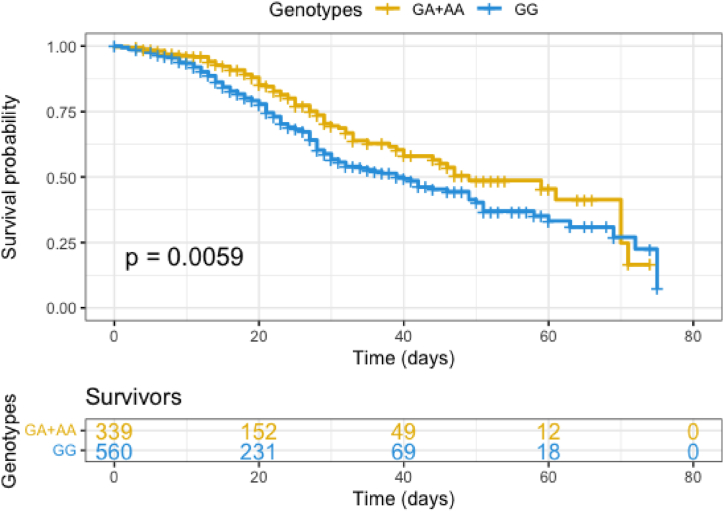
Survival analysis for 75 days in unrelated patients with COVID-19 severe by genotype (rs2289274/ATP2B2) indicated that the risk of death is lower in carriers of genotypes GA or AA in comparison to homozygous for the G allele (p = 0.0059, OR = 1.45, 95 % CI 1.11–1.89) (Fig. 2). The Kaplan–Meier survival curves cross around day 75, indicating a non-proportional hazard [21]; therefore, we used the log-rank test (p = 0.012).
Fig. 2.
Kaplan-Meier curve plots survival probability for unrelated patients with severe COVID-19 according to rs2289274 GA + AA or GG genotypes in ATP2B2.
3.3. Association of SNVs with risk of death in related COVID-19 patients
We evaluated eleven SNVs in ATP2B2, TMPRSS2, DOCK2, IFNAR2, TNFRSF1A and TNFRSF1B genes. In the transmission disequilibrium test (TDT), individuals with COVID-19 who survived were classified as unaffected, while non-survivor patients were identified as the index case. Individuals without information were classified as unknowns, which limited the analysis (Supplementary Figure 1, 2).
The comparison of demographic and clinical variables of related patients survivors and non-survivors showed male predominance in both groups (60 % and 53.5 %, respectively). The non-survivor group was older and had a higher BMI than the survivors' group; however, these variables did not show significant differences. 95.0 % of non-survivor patients required IMV and more days with this requirement. The PaO2/FiO2 at admission to the hospital was lower in non-survivor patients (Supplementary Table 4).
No significant differences between non-survivors and survivors were found in the subjects' genotype and allele frequencies (Supplementary Table 5). A tendency was observed in the codominant model for rs1232976 in TMPRSS2 with a higher frequency of the CT genotype in survivors vs. non-survivors (22.2 vs. 10.1, p = 0.08). In the same way, the genotype TC (rs767455) in TNFRSF1A showed a trend with a higher frequency in non-survivor patients (56.7 vs. 37.3, p = 0.06). LD for ATP2B2 was higher in related patients (D’ = 1) (Fig. 1B) in comparison with unrelated patients (D’ = 0.59); however, in related patients, the haplotypes obtained were not found to be associated with increased mortality risk between groups (Supplementary Table 6). The haplotype analysis for SNVs in IFNAR2, TNFRSF1A, and TNFRSF1B did not show significant differences in related patients. The LD between a particular locus pair depends on local recombination rates and the frequency in the study population [22]. The LD in related patients presented high values for SNVs in IFNAR2 but low ones for SNVs in TNFRSF1A and TNFRSF1B genes (Supplementary Figure 3).
4. Discussion
Several loci were evaluated in related and unrelated patients with severe COVID-19 using a candidate gene approach based on previous literature reviews on GWAS. Genetic variants in ATP2B2, TMPRSS2, DOCK2, IFNAR2, TNFRSF1A, and TNFRSF1B have been reported to be associated with susceptibility to COVID-19 and its severity, principally in Caucasian, Afro-American, and Asiatic populations [8,[15], [20], [23],24].
The populations included in this study were similar to those reported in the literature. According to the Centers for Disease Control and Prevention (CDC) in the USA, the risk of death from COVID-19 increases after the age of 40, and the risk is 25 times higher in those aged 50–64 years [25]. In our study, there was a higher percentage of men, and the age was 54 years for the related group and 58 years for unrelated patients.
Based on cohort studies and case series, overweight (BMI >25 kg/m2 but <30 kg/m2) suggestive among medical conditions of higher risk, and obesity (BMI ≥30 kg/m2) were more likely to develop into severe disease [26]. Our study did not find BMI significantly different between the analyzed groups. However, a high proportion of patients with pre-obesity (25.0–29.9 kg/m2) and obesity class I (30.0–34.9 kg/m2) [27] were observed in the family-related group. It is well known that families share habits, and these may be contributing to obesity, including metabolic syndrome, T2D, and SAH. These conditions are the most common complications of COVID-19, often co-occurring in the same patient [28].
The familial and unrelated groups did not differ in terms of comorbidities, and the prevalence of T2D, SAH, chronic lung diseases, ischemic heart disease, and smoking in both groups was similar to that reported in other studies [28]. Regarding signs and symptoms, the related patients showed more pronounced symptoms characterized by headache, myalgia, arthralgia, dyspnea, and anosmia, showing statistically significant differences between these signs and those in the unrelated group.
More of the 62 % of patients included in the study required IMV; related patients required lower days with this intervention than unrelated subjects; however, the PaO2/FiO2 at admission was higher in unrelated patients, and the percentage of survivors was greater in this group when compared to familial cases.
In the Mexican mestizo population, it has been shown that IFNAR2 variants are associated with mortality risk in unrelated patients with severe COVID-19. Additionally, increased levels of soluble IFNAR2 were found in survivors of COVID-19 compared to non-survivors. As for haplotype analysis, six haplotypes were reported in unrelated patients, one associated with risk of death and the other was found to be protective [20]. For the related patients with severe COVID-19, we analyzed three SNVs (rs3153, rs1051393, rs2834158) and reported three haplotypes with high LD (D’ = 0.88) but no significant association. The association of IFNAR2 loci with COVID-19 severity has been reported in different analyses [11,29,30]. The interferon-signaling pathway is essential in regulating immune response during viral infections. The rs1051393 missense variant led to a change of phenylalanine to valine. This variant is in the signal peptide region, affecting the IFNAR2 protein trafficking to the membrane [15]. However, in related patients with severe COVID-19, their association could not be demonstrated.
Other genetic variants relevant to COVID-19 are in the genes TNFRSF1A and TNFRSF1B. TNF-α pathway activation is through the receptors TNFR1 and TNFR2 [31]. High levels of soluble TNF-α have been reported in COVID-19 patients requiring intensive care compared to those hospitalized outside the intensive care unit [32]. In Mexican patients with severe COVID-19, rs1800693, rs767455 (TNFRSF1A), and rs1061622, rs3397 (TNFRSF1B) variants were associated with the risk of requiring IMV. However, in related patients with severe COVID-19, the rs1800693 and rs767455 were not associated with mortality risk. Previously, it was reported that the intron variant rs1800693 showed frequencies of TC + CC genotype significantly different between severe and mild in COVID-19 patients [33]. Among the best-known mechanisms that justify the relevance of the study of intronic variants is that they can affect the canonical donor/acceptor splice sites and modify the Indels and the spacing of the splicing motifs [34,35].
On the other hand, although the rs767455 variant was not associated with plasma levels of sTNF, sTNFR1, and sTNFR2, it was shown to be in high linkage of disequilibrium with rs1800693 (D’ = 0.94, r2 = 0.87) [15]. Finally, the TNFRSF1B gene variants rs1061622 and rs3397 were not associated with COVID-19 mortality risk in related patients. Methionine is changed to arginine in the rs1061622 variant, dramatically reducing the ability to activate NF-κB. In parallel, proinflammatory target genes of NF-κB, such as IL6 and IL8, are downregulated. All this acts through the TNFR-2 receptor, associated with pulmonary affections like cystic fibrosis and severe pulmonary disorder [36].
The rs3397 variant in the TNFRSF1B gene is in the 3′-untranslated region. Possible changes caused by rs3397 were investigated by in silico analysis of miRNA binding efficiency in genes potentially involved in COVID-19 pathogenesis. The rs3397 variant was reported to decrease the binding of has-miR-3126-5p, as well as increase the binding of has-miR-5581-5p, and cleave has-miR-122-3p all involved in COVID-19 susceptibility [37]. Previously, this variant was associated with higher levels of PaO2/FiO2 in COVID-19 patients carrying the TT genotype in contrast to TC + CC in the Mexican population, which is relevant as it may be a marker of COVID-19 severity [20]. However, we did not find an association with mortality risk in related patients, with low linkage disequilibrium values compared to what we previously reported in unrelated patients.
The rs12329769 in TMPRSS2 in a recessive genetic model (CC + CT vs. TT) has been associated with severe COVID-19 and high levels of inflammatory markers such as C-reactive protein (CRP), ferritin, LDH, and D-Dimer [23]. The role of TMPRSS2 in cell entry of SARS-CoV-2 could explain the association with COVID-19 susceptibility found in some studies [38]. The missense variant rs12329769 (Val197Met) is in the coding part of the gene. Some bioinformatics in silico analysis had predicted that this change could reduce enzyme stability and activity, reducing SARS-CoV-2 activation and, therefore, host cell entrance [39]. Our study did not find rs12329760 to be associated with mortality risk. The same result was reported for the rs12329760 variant and COVID-19 severity in Indonesian and German populations [40].
The rs60200309 (DOCK2) was reported to be associated with severe COVID-19 in patients less than 65 years old in a Japanese cohort [28]. DOCK2 downregulation inhibits macrophage migration in humans and hamsters infected with SARS-CoV-2 while decreasing type I IFN gene expression and increasing IFNG, IL6, and CCL5 [41]. We did not identify significant differences regarding this SNV. This underlines the importance of studying COVID-19 host genetics in non-Asian populations and the impact of population-specific risk alleles on various host genetic backgrounds. Although the minor allele frequency (MAF) was 0.10 in the mestizo-Mexican compared with 0.12 in the Japanese population, the variant was not associated with mortality risk.
We showed in unrelated patients with severe COVID-19 that rs2289274 (GG) in ATP2B2 is associated with an increased risk of death. Moreover, carriers of the GG haplotype (rs2289273-rs2289274) in ATP2B2 maintained the association with mortality risk in unrelated patients with severe COVID-19.
In unrelated patients, the LD is low compared to related patients. Survival analysis showed that the presence of the GG genotype is associated with a low probability of survival.
The SNVs rs2289273 and rs2289274 located in loci 3p25.3 in ATP2B2 were associated with susceptibility in asymptomatic COVID-19 patients [8]. Both are synonymous variants that change the G allele by A with an MAF of 0.14 and 0.28, respectively. For the rs2289274, the A-to-G nucleotide transition caused a glycine-to-serine substitution at a highly conserved amino acid position [42].
The ATP2B2 gene encodes a plasma membrane calcium transporter (ATPa2), a member of the P-type primary ion transport ATPase family distinguished by the production of an aspartyl phosphate intermediate during the reaction cycle. Despite extremely high concentration gradients, these enzymes play a crucial role in intracellular calcium homeostasis by removing bivalent calcium ions from eukaryotic cells [43]. The synonymous variant rs2289274 has been associated with hearing loss and COVID-19 in the Chinese population [8,44]. A study on murine model genes showed that those responsible for Ca2+ homeostasis, such as ATP2A3 and ATP2B2, are underregulated due to the presence of envelope (E) protein of SARS-CoV-2 [45].
We recognized the limitation of a relatively small sample size in related groups included in the study and the family structure. Since we have used multiplex families in this analysis, the combined TDT is only valid as a linkage test, not as an association test. The sib-TDT that uses unaffected siblings is of value in late-onset diseases. However, for complex diseases with low penetrance, the power is low [46]. Another limitation in unrelated patients was that we only included patients with severe forms of COVID-19 because the recruitment center was a third-level reference hospital, so we could not compare our findings with the disease's moderate, mild, or asymptomatic forms. However, we demonstrated in unrelated patients with severe COVID-19 SNVs in ATP2B2 that they are associated with a high risk of no survival.
5. Conclusion
Unrelated severe COVID-19 patients that carry the rs2289274 GG genotype in ATP2B2 showed a high mortality risk.
Survival analysis to 75 days shows that carriers of GG have a higher risk of non-survival compared to GA or AA genotypes.
Moreover, the haplotype GG (rs2289273-rs2289274) in ATP2B2 is associated with a high risk of non-survival in severe non-related COVID-19 cases.
Funding
This work was supported by the budget allocated to research (HLA Laboratory) from the Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas (INER).
Institutional review board statement
The study was conducted following the Declaration of Helsinki and approved by the Institutional Ethics Committee of Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas.
Informed consent statement
Informed consent was obtained from all subjects involved in the study.
Data availability statement
SUB13915955. Available in: https://www.ncbi.nlm.nih.gov/clinvar/?term=%22HLA%20Laboratory%2C%20Instituto%20Nacional%20de%20Enfermedades%20Respiratorias%20Ismael%20Cosio%20Villegas%22[submitter]+AND+%22ATP2B2%22 [gene].
CRediT authorship contribution statement
María Fernanda López-Bielma: Writing – original draft, Software, Methodology. Ramcés Falfán-Valencia: Writing – review & editing, Funding acquisition, Conceptualization. Aurelio Fierro-Piña: Methodology, Formal analysis. Edgar Abarca-Rojano: Validation, Supervision. Elizabeth Córdoba-Lanus: Writing – review & editing, Validation. Ingrid Fricke-Galindo: Supervision, Investigation. Priscila Romero-Villaseñor: Visualization, Investigation, Data curation. Ivette Buendía-Roldán: Writing – review & editing, Resources. Leslie Chávez-Galán: Writing – review & editing, Resources. María Esther Jaime-Capetillo: Visualization, Validation, Supervision. Gloria Pérez-Rubio: Writing – original draft, Software, Project administration, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
M.F.L.-B. had the support of the Consejo Nacional de Humanidades, Ciencias y Tecnologías (CONAHCYT) CVU:1165930 for the postgraduate degree in Master of Health Sciences at the Escuela Superior de Medicina of Instituto Politécnico Nacional.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e29493.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Shelton J.F., et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat. Genet. 2021;53(6):801–808. doi: 10.1038/s41588-021-00854-7. Jun. [DOI] [PubMed] [Google Scholar]
- 2.da Silva Torres M.K., et al. The complexity of SARS-CoV-2 infection and the COVID-19 pandemic. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.789882. Frontiers Media S.A., Feb. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grima P., Guido M., Zizza A. Clinical characteristics and risk factors associated with COVID-19 mortality in a non-Intensive Care Unit. J Prev Med Hyg. Mar. 2023;64(1):E3–E8. doi: 10.15167/2421-4248/jpmh2023.64.1.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiappelli F. CoViD-19 susceptibility. Bioinformation. Jul. 2020;16(7):501–504. doi: 10.6026/97320630016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D.D., Goel A. Estimating case fatality rates of COVID-19. Lancet Infect. Dis. 2020;20(7):773–774. doi: 10.1016/S1473-3099(20)30234-6. Lancet Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowe A., Chang D.D., Creek G. Multiple fatalities in a family cluster of COVID-19 with acute respiratory distress syndrome. Ochsner J. 2020;20(2):134–138. doi: 10.31486/toj.20.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellinghaus D., et al. Genomewide association study of severe covid-19 with respiratory failure. N. Engl. J. Med. 2020;383(16):1522–1534. doi: 10.1056/NEJMoa2020283. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan J., et al. Genome-wide association study of SARS-CoV-2 infection in Chinese population. Eur. J. Clin. Microbiol. Infect. Dis. 2022;41(9):1155–1163. doi: 10.1007/s10096-022-04478-5. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y., et al. Genome-wide association study of COVID-19 severity among the Chinese population. Cell Discov. 2021;7(1) doi: 10.1038/s41421-021-00318-6. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y., et al. Integrative genomics analysis reveals a 21q22.11 locus contributing risk to COVID-19. Hum. Mol. Genet. 2021;30(13):1247–1258. doi: 10.1093/hmg/ddab125. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pairo-Castineira E., et al. Genetic mechanisms of critical illness in COVID-19. Nature. Mar. 2021;591(7848):92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 12.The Severe Covid-19 GWAS Group Genome association study of severe covid-19 with respiratory failure. N. Engl. J. Med. 2020;383(16):1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F., et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 2020;6(1) doi: 10.1038/s41421-020-00231-4. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopper J.L., Bishop D.T., Easton D.F. Population-based family studies in genetic epidemiology. Lancet. Oct. 15, 2005;366(9494):1397–1406. doi: 10.1016/S0140-6736(05)67570-8. Elsevier B.V. [DOI] [PubMed] [Google Scholar]
- 15.Fricke-Galindo I., et al. TNFRSF1B and TNF variants are associated with differences in levels of soluble tumor necrosis factor receptors in patients with severe COVID-19. JID (J. Infect. Dis.) 2022;226(5):778–787. doi: 10.1093/infdis/jiac101. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez-Rubio G., Silva-Zolezzi I., Fernández-López J.C., et al. Genetic variants in IL6R and ADAM19 are associated with COPD severity in a Mexican mestizo population. COPD. 2016;13(5):610–615. doi: 10.3109/15412555.2016.1161017. [DOI] [PubMed] [Google Scholar]
- 17.Valencia-Pérez Rea D., Falfán-Valencia R., Fricke-Galindo I., Buendía-Roldán I., Chávez-Galán L., Nava-Quiroz K.J., Alanis-Ponce J., Pérez-Rubio G. The rs16969968 tobacco smoking-related single-nucleotide variant is associated with clinical markers in patients with severe COVID-19. Int. J. Mol. Sci. 2023;24(12):9811. doi: 10.3390/ijms24129811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Junta de Galicia and and Pan Americana Health Organization . Santiago de Compostela; España: 2006. EPIDAT 3.1 EPIDEMIOLOGICAL ANALYSIS FROM TABULATED DATA. [Google Scholar]
- 19.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. Jan. [DOI] [PubMed] [Google Scholar]
- 20.Fricke-Galindo I., et al. IFNAR2 relevance in the clinical outcome of individuals with severe COVID-19. Front. Immunol. 2022;13(Jul) doi: 10.3389/fimmu.2022.949413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazra A., Gogtay N. Biostatistics series module 9: survival analysis. Indian J. Dermatol. 2017;62(3):251–257. doi: 10.4103/ijd.IJD_201_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slatkin M. Linkage disequilibrium--understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 2008;9(6):477–485. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horowitz J.E., Kosmicki J.A., Damask A., et al. Genome-wide analysis provides genetic evidence that ACE2 influences COVID-19 risk and yields risk scores associated with severe disease. Nat. Genet. 2022;54(4):382–392. doi: 10.1038/s41588-021-01006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Namkoong H., et al. DOCK2 is involved in the host genetics and biology of severe COVID-19. Nature. 2022;609(7928):754–760. doi: 10.1038/s41586-022-05163-5. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Centers for Disease Control and Prevention, Interim Clinical Guidance for Management of Patients with Confirmed Coronavirus Disease (COVID-19).
- 26.Yang J., Hu J., Zhu C. Obesity aggravates COVID-19: a systematic review and meta-analysis. J. Med. Virol. 2021;93(1):257–261. doi: 10.1002/jmv.26237. John Wiley and Sons Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization, “‘A healthy lifestyle - WHO recommendations,’” “A healthy lifestyle - WHO recommendations”, Available: https://www.who.int/europe/news-room/fact-sheets/item/a-healthy-lifestyle---who-recommendations.
- 28.Richardson S., et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. JAMA, J. Am. Med. Assoc. 2020;323(20):2052–2059. doi: 10.1001/jama.2020.6775. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu D., Yang J., Feng B., Lu W., Zhao C., Li L. Mendelian randomization analysis identified genes pleiotropically associated with the risk and prognosis of COVID-19. J. Infect. 2021;82(1):126–132. doi: 10.1016/j.jinf.2020.11.031. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smieszek S.P., Polymeropoulos V.M., Xiao C., Polymeropoulos C.M., Polymeropoulos M.H. Loss-of-function mutations in IFNAR2 in COVID-19 severe infection susceptibility. J. Glob. Antimicrob. Resist. 2021;26:239–240. doi: 10.1016/j.jgar.2021.06.005. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gough P., Myles I.A. Tumor necrosis factor receptors: pleiotropic signaling complexes and their differential effects. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.585880. Frontiers Media S.A., Nov. 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortaz E., et al. Increased serum levels of soluble TNF-α receptor is associated with ICU mortality in COVID-19 patients. Front. Immunol. 2021;12(Apr) doi: 10.3389/fimmu.2021.592727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palacios Y., et al. Severe COVID-19 patients show an increase in soluble TNFR1 and ADAM17, with a relationship to mortality. Int. J. Mol. Sci. 2021;22(16) doi: 10.3390/ijms22168423. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellingford J.M., Ahn J.W., Bagnall R.D., et al. Recommendations for clinical interpretation of variants found in non-coding regions of the genome. Genome Med. 2022;14(1):73. doi: 10.1186/s13073-022-01073-3. Published 2022 Jul 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lomelin D., Jorgenson E., Risch N. Human genetic variation recognizes functional elements in noncoding sequence. Genome Res. 2010;20(3):311–319. doi: 10.1101/gr.094151.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Till A., et al. The Met-196 → Arg variation of human Tumor Necrosis Factor Receptor 2 (TNFR2) affects TNF-α-induced apoptosis by impaired NF-κB signaling and target gene expression. J. Biol. Chem. 2005;280(7):5994–6004. doi: 10.1074/jbc.M411541200. Feb. [DOI] [PubMed] [Google Scholar]
- 37.Karakas Celik S., Cakmak Genc G., Dursun A. A bioinformatic approach to investigating cytokine genes and their receptor variants in relation to COVID-19 progression. Int. J. Immunogenet. 2021;48(2):211–218. doi: 10.1111/iji.12522. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yaghoobi A., Lord J.S., Rezaiezadeh J.S., Yekaninejad M.S., Amini M., Izadi P. TMPRSS2 polymorphism (rs12329760) and the severity of the COVID-19 in Iranian population. PLoS One. 2023;18(2 February) doi: 10.1371/journal.pone.0281750. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paniri A., Hosseini M.M., Akhavan-Niaki H. First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. J. Biomol. Struct. Dyn. 2020:1–18. doi: 10.1080/07391102.2020.1767690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wulandari L., et al. Initial study on TMPRSS2 p.Val160Met genetic variant in COVID-19 patients. Hum. Genom. 2021;15(1) doi: 10.1186/s40246-021-00330-7. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferreira L.C., Gomes C.E.M., Rodrigues-Neto J.F., Jeronimo S.M.B. Genome-wide association studies of COVID-19: connecting the dots. Infect. Genet. Evol. 2022;106 doi: 10.1016/j.meegid.2022.105379. Elsevier B.V., Dec. 01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.V. A. Street, J. W. Mckee-Johnson, R. C. Fonseca, B. L. Tempel, and K. Noben-Trauth, “2 National Institutes of Health, National Institute on Deafness and Other Communication Disorders, Section on Murine Genetics,” 2085. [Online]. Available: http://genetics.nature.com.
- 43.Ficarella R., et al. A functional study of plasma-membrane calcium-pump isoform 2 mutants causing digenic deafness. 2006. www.pnas.orgcgidoi10.1073pnas.0609775104 [Online]. Available: [DOI] [PMC free article] [PubMed]
- 44.Wang S.L., et al. Gene-gene interaction of GJB2, SOD2, and CAT on occupational noise-induced hearing loss in Chinese han population. Biomed. Environ. Sci. 2014;27(12):965–968. doi: 10.3967/bes2014.131. Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- 45.Honrubia J.M., et al. SARS-CoV-2-Mediated lung edema and replication are diminished by cystic fibrosis transmembrane conductance regulator modulators. mBio. Jan. 2023;14(1) doi: 10.1128/mbio.03136-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cox A., et al. 1999. Combined Sib-TDT and TDT Provide Evidence for Linkage of the Interleukin-1 Gene Cluster to Erosive Rheumatoid Arthritis. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SUB13915955. Available in: https://www.ncbi.nlm.nih.gov/clinvar/?term=%22HLA%20Laboratory%2C%20Instituto%20Nacional%20de%20Enfermedades%20Respiratorias%20Ismael%20Cosio%20Villegas%22[submitter]+AND+%22ATP2B2%22 [gene].