Abstract
A series of vectors with heterologous genes was constructed from HSRV1, an infectious clone of human foamy virus (HFV), and transfected into baby hamster kidney cells to generate stably transfected vector cell lines. Two cis-acting sequences were required to achieve efficient rescue by helper virus. The first element was located at the 5′ end upstream of position 1274 of the proviral DNA. Interestingly, a mutation in the leader sequence which decreased the ability to dimerize in vitro inhibited transfer by helper HFV. A second element that was important for vector transfer was located in the pol gene between positions 5638 and 6317. Constructs lacking this element were only poorly transferred by helper HFV, even though their RNA was produced in the vector cell lines. This finding rules out the possibility that the observed lack of transfer was due to RNA instability. A minimal vector containing only these two elements could be successfully delivered by helper HFV, confirming that all essential cis-acting sequences were present. The presence of a sequence described as a second polypurine tract in HFV was not necessary for transfer. Our data identified the minimal sequence requirements for HFV vector transfer for the development of useful vector systems.
The packaging of genomic RNA into retroviral particles is a complex and highly specific process involving interactions between viral structural proteins and cis-acting sequences in the viral RNA, called psi (Ψ) or encapsidation signals (29). The packaging sequences for several retroviruses have been mapped. For Moloney murine leukemia virus (30), spleen necrosis virus (46), bovine leukemia virus (20), and type D retroviruses (44), these essential cis sequences are in close proximity to the 5′ end in the untranslated leader region of the genomic RNA, next to the splice donor site. In Moloney murine leukemia virus, psi can extend into the gag open reading frame (ORF) (5, 31). A more complex situation is found in retroviruses such as Rous sarcoma virus (RSV) and human immunodeficiency virus type 1 (HIV-1). For RSV, sequences upstream of the major splice donor site are involved in packaging, raising the question of how the spliced transcripts are distinguished from the genomic RNA in the encapsidation process (3, 21, 35). Direct-repeat sequences flanking the v-src gene enhance packaging by exerting an influence on several steps of the viral life cycle (41, 42). For HIV-1, sequences involved in packaging are located between the major splice donor site and the gag ORF (1, 12, 28). However, additional sequences in the env ORF enhance the efficiency of the process (7, 22, 37).
Secondary structures for psi sequences have been predicted, and a stem-loop structure common to a variety of retroviruses has been described (2, 17, 23, 47). Controversy exists as to whether dimerization plays an important role in RNA packaging (3, 6, 13, 26, 34); this idea is supported by the fact that the dimer linkage structure (DLS) of retroviruses, which mediates stable and noncovalent intermolecular linkage of the RNA monomers, coincides with the psi sequence.
Foamy viruses are a subfamily of retroviruses which may constitute good candidate vectors for gene delivery because of their broad tropism, their benign nature, and their greater packaging potential compared with that of other retroviruses (8, 18, 38, 40). However, the packaging sequence(s) has not been determined in detail. Recently, Russell and Miller (38) reported the construction of human foamy virus (HFV)-based vectors carrying genes for neomycin phosphotransferase and alkaline phosphatase. These constructs, with deletions in the gag, pol, and env ORFs, could be successfully transferred to recipient cells when cotransfected with wild-type helper HFV DNA, implying that the deleted sequences were not involved in transfer.
To investigate which sequences are necessary for gene delivery, a series of HFV-based vectors with deletions in the viral genome were constructed and stably transfected cell lines were generated. After transfection with a plasmid carrying helper HFV, it was found that sequences in the pol gene were needed, in addition to the leader region, to allow efficient transfer by helper HFV.
MATERIALS AND METHODS
Recombinant DNA.
Previously described laboratory procedures were followed for the manipulation of DNA, plasmid preparation, and cloning (39). The constructs used in the transfer experiments are depicted in Fig. 1 and were subjected to restriction enzyme digestion for verification of their construction. All constructs were derived from HSRV1 (36), an infectious molecular clone of HFV with a deletion in the U3 region. The positions cited refer to the proviral DNA. The start site of transcription (+1) of viral RNA coincides with position 778 of HSRV1.
FIG. 1.
Constructs used in the packaging experiments with respect to the pFOV-7 helper virus. Numbers indicate the proviral DNA of HSRV1. The U3 region is 777 bp long; thus, the start site of transcription (+1) of viral RNA coincides with position 778. IRES-hygror indicates the gene encoding hygromycin resistance under the control of the poliovirus IRES. SV40-gusA denotes the gusA gene under the control of the SV40 early promoter. Solid lines indicate HSRV1 sequences present in the vector constructs. Transfer was assayed by GUS (vector) and β-Gal (helper HFV) expression per milliliter. Values shown are the means of at least three independent experiments. The ratios of numbers of GUS-positive colonies to numbers of β-Gal-positive colonies are given in percentages. The asterisk in plasmid pIH-1, BamHI denotes the site of mutation.
The construction of plasmid pHSG11 has been described elsewhere (8). In this plasmid, the accessory bel genes are replaced by the gusA reporter gene, coding for β-glucuronidase (GUS), under the control of the simian virus 40 (SV40) early promoter (SV40-gusA cassette). To generate plasmids expressing hygromycin resistance, the HindIII/ClaI fragment of pBabehygro (32) was inserted into plasmid pPBS behind the poliovirus internal ribosomal entry site (IRES). This IRES-hygror cassette was then inserted as an SpeI/SalI fragment into plasmid pHSG-11 digested with SpeI and NheI (the SalI and NheI sites being blunt ended) to replace sequences between positions 5894 and 9250. The resulting plasmid was called pIH-7 (Fig. 1). Vector pIH-1 was generated by insertion of the SpeI fragment from positions 5895 to 6927 into the SpeI site of pIH-7.
Constructs pIH-2, pIH-3, and pIH-4, which have internal deletions in the leader and gag ORF, were generated with primer pairs P1-P2, P1-P3, and P1-P4, respectively. These were used to amplify by PCR sequences from a KpnI site of plasmid pHSRV1 50 bp before the beginning of the viral U3 region up to positions 1551, 1274, and 1173, respectively. The oligonucleotides used are listed in Table 1 with nonviral sequences underlined. The amplicons were then used to replace the sequences up to the second BglII site at position 2153 of plasmid pHSG11. The BstXI fragment of this plasmid consisting of the 3′ pol sequences from position 6011 to the beginning of the SV40-gusA cassette was also exchanged for the equivalent fragment of pIH-1, which contains the IRES-hygror cassette. Thus, plasmids pIH-2, pIH-3, and pIH-4 had internal deletions of proviral sequences from positions 1551 to 2153, 1274 to 2153, and 1173 to 2153, respectively. Plasmid pIH-1, BamHI resulted from mutant M32 (16), which represents the insertion of a BamHI linker into the Klenow fragment-treated AvrII site at position 1180 in the leader region of plasmid pIH-1.
TABLE 1.
Nucleotide sequences of the oligonucleotides useda
| Name | Sequence | Position on HSRV1 |
|---|---|---|
| P1 | 5′ GACTGGTACCCCTGTAAACAATGCTGG 3′ | −50 |
| P2 | 5′ GACTAGATCTCCAAACCTCAATGGACCTTCAG 3′ | 1551 |
| P3 | 5′ GACTAGATCTCCAGAGCTTCAACATCAAGTTC 3′ | 1274 |
| P4 | 5′ GACTAGATCTATAATTTACAAATAAACCCGAC 3′ | 1173 |
| P5 | 5′ GGATCCATTTAAATCTCTCAATGTACTC 3′ | 5869 |
| P6 | 5′ GACTACTAGTCTCGAGGTCTCAAAGAAGCAGGCC 3′ | 6372 |
| P7 | 5′ GACTACTAGTCTCGAGACCAGGAACGAGAGGAGG 3′ | 6318 |
| P8 | 5′ GTCAGCTAGCGAATTCCAGAATACAATGG 3′ | 6766 |
| P9 | 5′ AGTCGCTAGCATATGCATCCACTTAGAAT 3′ | 7786 |
| P10 | 5′ GATCTCGAGATTTAAATTAATTAACCAAAGTG 3′ | 6011 |
| P11 | 5′ TTGGTTAATTAATTTAAATCTCGA 3′ | |
| P12 | 5′ GACTTAATTAAACAACTAGGACGCTGTC 3′ | 5638 |
| P13 | 5′ GACTCTAGACCAGGAACGAGAGGAGG 3′ | 6317 |
| P14 | 5′ GACTATTTAAATATTAGTTTATAGTCCCATTG 3′ | 4678 |
| P15 | 5′ GACTTAATTAATGACCCTGTAATAATTG 3′ | 5428 |
| P20 | 5′ GACAATTGGCGCCCAACGTGGGGC 3′ | 1118 |
| P21 | 5′ CCGGTGTTGTAACAGGGAACACTCC 3′ | 2255 |
| P22 | 5′ GACTAAGCTTCCAGTTCATCTCTAGTCA 3′ | 1655 |
Nonviral sequences are underlined. P11 is a nonviral sequence.
Constructs pIH-5 and pIH-6 encode proviral DNA up to positions 6372 and 6318, respectively. Sequences including positions 5869 to 6372 and 5869 to 6318 were amplified with primer pairs P5-P6 and P5-P7, and the respective amplicons were digested with SpeI and cloned into the SpeI site of plasmid pIH-7 as described above. Plasmid pIH-8 was generated from pIH-7 by deletion of the viral sequences from the SwaI site at position 2817. Construct pIH-9 was generated by PCR amplification of HSRV1 sequences from positions 6766 to 7786 with primer pair P8-P9 and insertion of the amplicon as an NheI fragment into the SpeI site of plasmid pIH-7.
Vector pIH-10 represents plasmid pIH-6 with an internal deletion from positions 1551 to 6011 of a BstXI site. It was constructed by replacing sequences of pIH-2 with annealed oligonucleotides P10 and P11, introducing SwaI and PacI sites for cloning. The BstXI fragment was then exchanged for the IRES-hygror cassette of plasmid pIH-6 plus proviral sequences up to position 6318. To generate plasmid pIH-11, sequences from positions 5638 to 6317 were amplified with primer pair P12-P13 and cloned as a PacI/XbaI fragment into vector pIH-10 digested with PacI and SpeI. This mutant has an internal deletion from positions 1551 to 5638. Plasmid pIH-12 originated from plasmid pIH-10 but includes additional pol sequences from positions 4678 to 5428 of pHSRV1. The amplicon generated from primer pair P14-P15 was cloned as a SwaI/PacI fragment into pIH-10.
To monitor the transfer of plasmids pIH-2 and pIH-3 by helper HFV, total cellular DNA from the recipient cells was extracted with a DNA extraction kit (Qiagen). About 0.5 μg of DNA was PCR amplified between positions 1118 and 2255 with primer pair P20-P21. The generated fragments were gel purified and sequenced on an automated sequencer with AmpliTaqFS and the ABI310 sequence analysis system (Perkin-Elmer).
Cells and DNA transfections.
Stably transfected vector cell lines were generated with Lipofectin (GIBCO BRL) from BHK/Bel-1 cells, which constitutively express the viral transactivator Bel-1 (8). They were selected in Dulbecco’s modified Eagle’s medium containing 5% fetal calf serum, 0.5 mg of G-418 (GIBCO BRL) per ml, and 400 μg of hygromycin (Sigma) per ml. The selection medium was changed daily until colonies of hygromycin-resistant cells were observed. These were subsequently pooled, and stable transfection was verified by staining for GUS expression.
BHLL cells (9) were used as recipient cells in the transfer experiments. They were cultured in Dulbecco’s modified Eagle’s medium supplemented with 5% fetal calf serum. These cells carry the lacZ gene under the transcriptional control of the HFV long terminal repeat (LTR). Infection by helper HFV expressing the viral transactivator Bel-1 was monitored by 5-bromo-4-chloro-3-indolyl-β-d- galactopyranoside (X-Gal) staining, whereas the vector was quantified by 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-GlcA) staining for GUS expression.
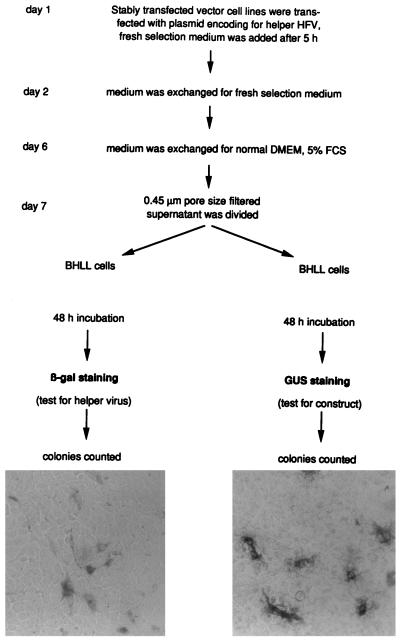
To produce helper HFV, 4 × 105 cells of the vector cell lines were transfected in the presence of Lipofectamine (GIBCO BRL) with 5 μg of DNA from plasmid pFOV-7 (40). The selection medium was changed daily but was replaced with nonselective medium on the day before the supernatant was harvested (Fig. 2).
FIG. 2.
Schematic presentation of the protocol followed to test for transfer. DMEM, Dulbecco’s modified Eagle’s medium; FCS, fetal calf serum.
Transfer of the construct by helper HFV.
On day 7 posttransfection with plasmid pFOV-7, supernatant fluid collected from the stably transfected vector cell lines was assayed for the presence of the helper virus and the construct (Fig. 2). The supernatant was filtered through a 0.45-μm-pore-size membrane (Acrodisc), and 1, 0.1, or 0.01 ml in duplicate was plated onto BHLL cells. One replicate was assayed for the presence of helper HFV by counting β-galactosidase (β-Gal)-positive foci, and the other was assayed for GUS expression to determine the transfer of the construct.
Staining for GUS and β-Gal activities.
Staining for GUS and β-Gal expression has been described elsewhere (8, 9).
RNase protection assay.
To generate the antisense probe, sequences from positions 1118 to 1655 of proviral DNA were amplified by PCR with primer pair P20-P22 and cloned as a MunI/HindIII fragment into plasmid pSPT18 (Boehringer Mannheim Biochemicals) digested with EcoRI/HindIII. The plasmid was digested with SspI at position 1392 of HSRV1 and transcribed with SP6 polymerase (Boehringer-Mannheim) in the presence of 32P. This procedure resulted in a radiolabelled riboprobe 272 nucleotides (nt) long. Of those, 263 nt were complementary to full-length helper HFV from positions 1393 to 1655 or 159 nt (from positions 1393 to 1551) for the transcripts with internal deletions. Total cellular RNA was extracted from stably transfected cells with the RNeasy kit (Qiagen), and 10 μg was ethanol precipitated along with 4 × 105 cpm of gel-purified riboprobe. RNase protection assays were carried out with an RPA II kit (Ambion). Plasmid pBR322 digested with MspI provided a size marker for electrophoresis. A linearized plasmid for the human β-actin gene (Ambion) generated a radiolabelled transcript of 218 nt, 127 nt of which were complementary to cellular RNA to provide an internal control.
RESULTS
A series of HFV-based vectors was constructed; each construct carried the IRES-hygror cassette and the SV40-gusA cassette (Fig. 1). To investigate the transfer of the constructs by replication-competent helper HFV, the vectors were stably transfected into BHK/Bel-1 cells (8). The derived vector cell lines were then transfected with plasmid pFOV-7 carrying replication-competent helper HFV (38). The supernatant was plated on recipient BHLL cells (9) 1 week after transfection (Fig. 2), when the HFV-induced cytopathic effect of the vector producing-cell lines was maximal. Since BHLL cells carry the gene for β-Gal under the control of the HFV LTR, helper virus infection was monitored by the addition of X-Gal, while vector transfer was assayed by incubation with X-GlcA.
Delineation of sequences in the leader and gag ORF required for transfer of HFV vectors.
Plasmid pIH-1 generated a vector with sequences from positions 1 to 6927 of proviral DNA. When BHK/Bel-1 cells stably expressing this construct were transfected with helper virus DNA and the supernatant was plated on BHLL cells, vector-transduced, GUS-positive foci approximated 10% of β-Gal-positive foci resulting from infection by helper HFV (Fig. 1). This result demonstrated that the cis-acting sequences essential for transfer were present. The criterion for transfer adopted was that the construct titer should constitute at least 1% of the helper virus titer. Constructs with a titer of less than 1% that of helper HFV were described as being poorly transferred. To investigate the contribution of the 5′ end of the genome, mutants pIH-2, pIH-3, and pIH-4, with internal deletions of proviral sequences from positions 1551 to 2153, 1274 to 2153, and 1173 to 2153, respectively, were constructed. Transfer was observed for mutants with deletions in gag (pIH-2 and, to a lesser extent, pIH-3). However, no transfer was observed for construct pIH-4, in which part of the leader region was also deleted (Fig. 1), suggesting that sequences crucial for transfer lie upstream of position 1274.
Retrovirus genomes frequently recombine, and it was theoretically possible that transfer could be accounted for by the regeneration of a packaging signal by recombination between helper HFV and vector genomes. To investigate this possibility, vector cell lines expressing deletion mutants pIH-2, pIH-3, and pIH-4 were transfected with plasmid pFOV-7, and the supernatant was added to BHK/Bel-1 cells to enable the synthesis of vector-derived RNA and the subsequent expression of the IRES-hygror cassette. Following incubation in selection medium, no hygromycin-resistant colonies were detected for construct pIH-4, only for constructs pIH-2 and pIH-3. Although the supernatant must have represented a mixture of viral particles encoding the construct and helper HFV, no cytopathic effect appeared in the hygromycin-resistant cells. These cells stained blue when tested for GUS expression, indicating the presence of the construct (data not shown).
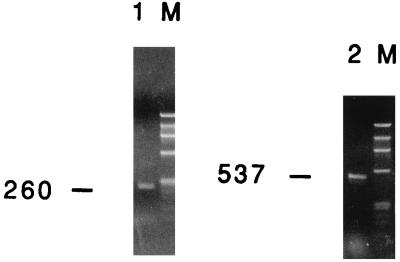
Cellular DNA was extracted and subjected to PCR with primer pair P20-P21 (Table 1). For helper virus, a fragment of 1,138 bp was expected, whereas deletion mutants pIH-2 and pIH-3 were expected to provide fragments of 537 and 260 bp, respectively. When plasmids pIH-2 and pIH-3 were mixed and PCR was performed, we found that the longer PCR product, from pIH-2, was less abundantly produced. We were, however, able to detect the PCR product from plasmid pIH-2 if this plasmid was present at 10% the amount of pIH-3. Control DNA from uninfected cells gave no signal (data not shown); no band corresponding to the size of full-length helper virus was observed with DNA from the hygromycin-resistant cells, whereas deletion mutant fragments of the expected sizes were readily seen (Fig. 3). DNA sequencing confirmed the transfer of original deletion mutants pIH-2 and pIH-3.
FIG. 3.
Assay for recombination between construct pIH-2 or pIH-3 and helper HFV used in the transfer experiments. Vector cell lines for each construct were transfected with helper HFV DNA, and the supernatant was used to infect BHK/Bel-1 cells. This procedure allowed transcription from the LTR of the constructs and subsequent expression of the IRES-hygror cassette. Primer pair P20-P21 was used to amplify DNA from hygromycin-resistant colonies. Due to the sizes of their internal deletions, PCR fragments of 537 and 260 bp would be expected if plasmids pIH-2 and pIH-3 were transferred without recombination. Helper HFV would generate a fragment of 1,138 bp. Lanes: 1, PCR product of cells exposed to helper HFV and construct pIH-3; 2, PCR product of cells exposed to helper HFV and construct pIH-2 (in both cases, no fragment corresponding to helper HFV could be detected); M, molecular weight marker φX174 cut with HaeIII. The sizes of the bands are, from top to bottom (in base pairs): 1,353, 1,078, 872, 603, 310, 281, 271, 234, 194, 118, and 72.
The DLS consists of several sites which direct the dimerization of HFV RNA in vitro and is located in the region containing the primer binding site, the leader region, and the start of the gag gene. Mutations in a palindromic sequence of 10 nt in site II (SII) within the leader region led to a decreased ability of RNA to dimerize in vitro (16). To investigate the functional relationship of this in vitro finding to the transfer by helper HFV, we constructed a plasmid (pIH-1, BamHI) in which the palindrome was extended by the insertion of a BamHI linker at position 1180 of the proviral DNA. The RNA of this mutant dimerizes poorly in vitro (16). Figure 1 shows that the transfer of this mutant by helper HFV was also greatly reduced, suggesting the importance of an intact DLS for transfer.
Additional sequences in the pol ORF are required for the transfer of HFV vectors.
Plasmids pIH-5 and pIH-6 were constructed to include proviral sequences from positions 1 to 6372 and 1 to 6318, respectively. These constructs could be transferred (Fig. 1). Plasmid pIH-6 lacks a sequence, described as a second polypurine tract (PPT), which might play a role in reverse transcription (25, 43). However, this sequence was dispensable for the transfer of vector genomes. In contrast to these plasmids, plasmid pIH-7, with sequences up to position 5894, was found to be poorly transferred by helper HFV, constituting less than 1% of the values obtained for helper HFV (Fig. 1). We generated two other constructs, pIH-8 and pIH-9. Plasmid pIH-8 includes HSRV1 sequences from positions 1 to 2817. Plasmid pIH-9 includes proviral sequences from positions 1 to 7786 but has an internal deletion from positions 5893 to 6766. Both constructs behaved in a manner similar to that of plasmid pIH-7, indicating that they also lacked important sequences.
Construction of an HFV vector containing minimal cis-acting sequences.
To examine the possibility that further sequences in the gag or pol ORF were necessary for transfer by HFV, an internal deletion in the proviral sequences from positions 1551 to 6011 of plasmid pIH-6 was constructed. The resulting construct, pIH-10, was poorly transferred, whereas plasmid pIH-11, containing HSRV1 sequences up to position 6317 and having an internal deletion from positions 1551 to 5638 of the proviral DNA, was efficiently transferred. Another construct, pIH-12, which resulted from insertion of proviral sequences from positions 4678 to 5428 into construct pIH-10, was only poorly transferred (Fig. 1). These results demonstrate the importance of the pol sequences after position 5638 of HSRV1, which cannot be substituted by other upstream sequences. In the viral RNA, this position corresponds to position 4861. These results are in keeping with the findings of Russell and Miller (38) and, furthermore, define for the first time the minimal cis-acting sequences required for vector transfer.
Stability of vector RNA.
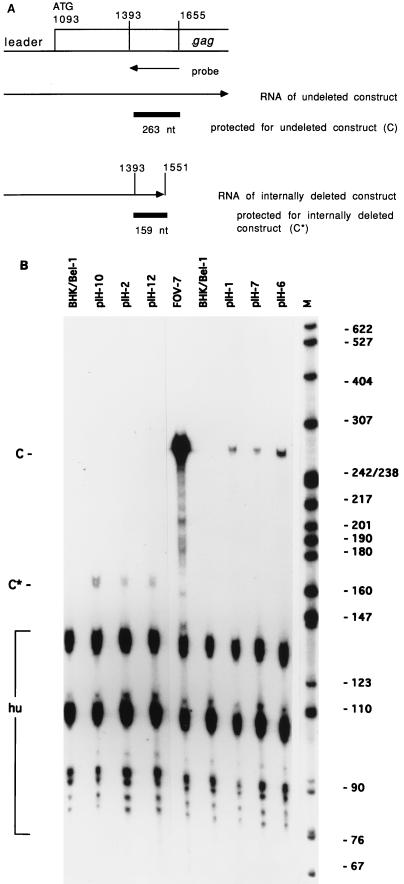
We addressed the question of whether the vector cell lines produced the same amounts of RNA from transferable and nontransferable constructs to exclude the possibility that the lack of transfer resulted from an instability in the vector cell lines of RNAs lacking the pol sequences. To investigate this question, an RNase protection assay was performed. The riboprobe was generated from positions 1393 to 1655 of the proviral sequences (Fig. 4A), representing positions 616 to 878 of HFV RNA, and hybridized only to unspliced RNA, since the major splice donor site of HFV lies at position 51 (33). Cellular RNA was extracted from a set of vector cell lines generated from three transferable and three poorly transferable constructs. These two groups of constructs included examples with or without an internal deletion (Fig. 4B).
FIG. 4.
RNase protection assay. (A) Localization of the riboprobe in the gag ORF. Numbers indicate the proviral DNA of HSRV1. The expected sizes of the protected RNA fragments for constructs with and without internal deletions protected by the riboprobe are indicated. nt, nucleotides. (B) RNA from stably transfected vector cell lines was compared with RNA from BHK/Bel-1 cells and RNA from BHK/Bel-1 cells producing helper HFV (FOV-7). Vectors pIH-10, pIH-2, and pIH-12 yielded a protected fragment of 159 nt (C*). Vectors pIH-1, pIH-7, and pIH-6 yielded a protected fragment of 263 nt (C). A fragment (127 nt) of human β-actin RNA (hu) served as an internal control. M, molecular weight marker pBR322 cut with MspI. The sizes of the DNA fragments are indicated in nucleotides.
Mutants with internal deletions comprised the poorly transferable constructs pIH-10 (Fig. 4B, lane 2) and pIH-12 (lane 4) as well as the transferable construct pIH-2 (lane 3). Mutants without internal deletions included the transferable constructs pIH-1 (Fig. 4B, lane 7) and pIH-6 (lane 9) as well as the poorly transferable construct pIH-7 (lane 8). RNAs from untreated BHK/Bel-1 cells (Fig. 4B, lanes 1 and 6) and BHK/Bel-1 cells transfected with plasmid pFOV-7 (lane 5) were included as controls. For constructs pIH-10, pIH-2 and pIH-12, with internal deletions from proviral position 1551 (RNA, 774), a protected band of 159 nt was seen (Fig. 4B, lanes 2, 3, and 4, C*), whereas helper HFV and constructs pIH-1, pIH-7 and pIH-6, without an internal deletion, resulted in the protection of a fragment of 263 nt (lanes 7, 8, and 9, C). A 127-nt fragment of the human β-actin gene served as an internal control.
Some variations in RNA production were noted with, for example, constructs pIH-10 (Fig. 4B, lane 2) and pIH-2 (lane 3) as well as constructs pIH-7 (lane 8) and pIH-6 (lane 9). These variations may be accounted for by the fact that the vector cell lines represented pooled populations of transfected cells and, thus, included cells with different integration and subsequent transcription events. However, since construct pIH-10, being more abundant than construct pIH-2, was still only poorly transferred (Fig. 1), the variations in RNA synthesis cannot account for the different transfer efficiencies of the constructs. Viral RNA from control BHK/Bel-1 cells transfected with plasmid pFOV-7 gave a much stronger signal (Fig. 4B, lane 5) than constructs in the vector cell lines. This result suggests that the RNA synthesis of helper HFV also exceeded the synthesis of the constructs in the respective vector cell lines. The reason for this finding is not known, but it probably explains why each deletion mutant was transferred at only a fraction of the amount observed for helper HFV. Several fragments smaller than the human β-actin gene fragment were observed. They were, however, not present if hybridization was carried out with a riboprobe for construct RNA only (data not shown). Furthermore, since these bands also appeared in BHK/Bel-1 cells without construct RNA (Fig. 4B, lanes 1 and 6), they must be specific for the β-actin probe. A possible explanation for their presence could be that mismatches occurred when the human β-actin gene and the hamster equivalent in the BHK/Bel-1 cells hybridized. Mismatches are likely to result in higher sensitivity to RNase treatment and the generation of smaller protected species.
DISCUSSION
The successful use of vectors based on replication-competent and replication-incompetent HFV to deliver heterologous genes has been reported (8, 18, 38, 40). However, these vectors still encode large parts of viral sequences, limiting the space available for the incorporation of foreign DNA. To exploit further foamy viruses as vectors for gene delivery, we investigated what sequences in the viral genome were necessary for the construction of a minimal HFV-based vector. Because the titer of viruses released from foamy virus-infected cells is low, we used an indirect approach to monitor whether helper HFV could transfer constructs carrying reporter genes. The experiments described here, therefore, cannot strictly distinguish among the impairment of nucleoplasmic transport, the packaging process, and subsequent steps of the replication cycle after the infection of recipient cells (e.g., reverse transcription and integration).
Constructs carrying the genes hygror and gusA were introduced into the viral genome, and stably expressing vector cell lines were generated. After transfection of these vector cell lines with helper HFV, the supernatant was assayed for the packaged construct by infection of recipient cells and staining for the reporter gene. From these experiments it was evident that functional HFV vectors must contain two distinct regions of the viral genome. The first of these two regions, located at the 5′ end of the proviral genome upstream of position 1274 (RNA, 497), by analogy to other retroviruses probably constitutes a common DLS-encapsidation packaging signal (6, 10, 15, 20). Three sites (SI, SII, and SIII) in the leader region are important for full dimerization of HFV RNA in vitro (16). A deletion mutant (pIH-4) lacking SII and SIII was not a functional vector. However, deletion mutants pIH-2 and pIH-3, both of which include the complete DLS, could be transferred (Fig. 1), as proved by sequence analysis after transfer to recipient cells. The observation that plasmid pIH-1, BamHI, carrying a mutation in the SII palindrome which impaires dimerization in vitro, also inhibited transfer by helper HFV indicates that these two functional sites are closely linked. The splice donor site of HFV is located at position 51 of viral RNA (33). Therefore, the putative packaging sequence in the leader region of viral RNA is present only on unspliced genomic RNA.
The second region needed for efficient transfer by helper HFV lies in the pol ORF between positions 5638 and 6317 of proviral DNA (4861 and 5540, respectively, of viral RNA). Deletion mutants which lacked these sequences could not be transferred, whereas constructs which included these sequences constituted functional vectors (Fig. 1). It has been reported that the pol ORF of foamy viruses possesses a second central PPT, a feature shared only by the lentiviruses (reviewed in reference 24), and a dual mode of initiation of plus-strand DNA synthesis has been suggested (25, 43). This is the case for HIV-1, for which a PPT duplication has been described (45), and mutation of this central PPT reduces the infectivity of the virus (19). In HSRV1, the central PPT is located at positions 6340 to 6351. However, since pIH-6 and pIH-11 could be efficiently transferred in the absence of this sequence, its influence in the context of replication-defective vectors is, at most, minor.
Additional env sequences which are required to produce vectors based on HIV-1 have been described, although subsequent investigations could not define a specific sequence (7, 22, 37). For RSV, direct-repeat sequences flanking the v-src gene are also essential cis-acting sequences (41, 42). The situation with HFV seems to be similar in that two sequences from different parts of the genome must be present simultanously to enable efficient transfer by helper HFV. The mechanism by which the region in pol exerts its influence is unclear, but the RNase protection assay carried out showed that a potential instability or a low rate of synthesis of RNA lacking this region was not the explanation. Like other complex retroviruses (14), HFV has a transcriptional transactivator. In order to allow cytoplasmic expression of unspliced and singly spliced mRNAs encoding the structural proteins, some complex retroviruses, such as HIV-1, require a posttranscriptional transactivator (Rev) and the presence of a specific recognition element (Rev-responsive element) within these mRNAs. No Rev- or Rev-responsive element-like functions are required for HFV gene expression (27). Moreover, it was shown recently that the Pol protein of HFV is not required for packaging (4). These findings beg the question of how and where the unspliced genomic RNA of HFV is recognized and packaged. A possible function of the sequences in the pol ORF is that they work in a manner similar to that of a constitutive transport element, as described for Mason-Pfizer monkey virus (11), allowing unspliced RNA to effectively reach the cytoplasm. So far, we have not investigated whether the sequences in the pol ORF can act as a constitutive transport element or whether they are responsible for other functions.
Recently, it became clear that the life cycle of foamy viruses is different from that of other retroviruses and that it follows a pathway not unlike that of hepadnaviruses (48). The detailed contributions of the pol sequences to the replication process in general and to packaging in particular remain to be determined. However, with the identification of the minimal cis-acting sequences described here and the construction of a useful vector (pIH-11) with only about 3.6 kb of proviral sequences remaining for the delivery of heterologous genes, it will be possible to fully exploit the potential packaging capacity of HFV as a vector.
ACKNOWLEDGMENTS
We are grateful to the EC and to the Wellcome Trust for funding this study and to the Jefferiss Research Trust for laboratory support.
Plasmid pPBS with the poliovirus IRES was provided by R. Vile of the ICRF Unit, Hammersmith Hospital, London, United Kingdom. We thank Albert Stühler for helpful discussion.
REFERENCES
- 1.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alford R L, Honda S, Lawrence C B, Belmont J W. RNA secondary structure analysis of the packaging signal for Moloney murine leukemia virus. Virology. 1991;183:611–619. doi: 10.1016/0042-6822(91)90990-s. [DOI] [PubMed] [Google Scholar]
- 3.Aronoff R, Hajjar A M, Linial M L. Avian retroviral RNA encapsidation: reexamination of functional 5′ RNA sequences and the role of nucleocapsid Cys-His motifs. J Virol. 1993;67:178–188. doi: 10.1128/jvi.67.1.178-188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin D W, Linial M L. The roles of Pol and Env in the assembly pathway of human foamy virus. J Virol. 1998;72:3658–3665. doi: 10.1128/jvi.72.5.3658-3665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender M A, Palmer T D, Gelinas R E, Miller A D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J Virol. 1987;61:1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkowitz R D, Hammarskjöld M-L, Helga-Maria C, Rekosh D, Goff S P. 5′ Regions of HIV-1 are not sufficient for encapsidation: implications for the HIV-1 packaging signal. Virology. 1995;212:718–723. doi: 10.1006/viro.1995.1530. [DOI] [PubMed] [Google Scholar]
- 8.Bieniasz P D, Erlwein O, Aguzzi A, Rethwilm A, McClure M O. Gene transfer using replication-defective human foamy virus vectors. Virology. 1997;235:65–72. doi: 10.1006/viro.1997.8658. [DOI] [PubMed] [Google Scholar]
- 9.Bieniasz P D, Rethwilm A, Pitman R, Christie I, McClure M O. A comparative study of higher primate foamy viruses including a new virus from a gorilla. Virology. 1995;207:217–228. doi: 10.1006/viro.1995.1068. [DOI] [PubMed] [Google Scholar]
- 10.Bieth E, Gabus C, Darlix J-L. A study of the dimer formation of Rous sarcoma virus RNA and of its effect on viral protein synthesis in vitro. Nucleic Acids Res. 1990;18:119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K-T, Rekosh D, Hammarskjöld M-L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchschacher G L, Jr, Panganiban A T. Human immunodeficiency virus vectors for inducible expression of foreign genes. J Virol. 1992;66:2731–2739. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen B R. Human immunodeficiency virus as a prototypic complex retrovirus. J Virol. 1992;65:1053–1056. doi: 10.1128/jvi.65.3.1053-1056.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darlix J-L, Gabus C, Nugyere M-T, Clavel F, Barre-Sinoussi F. cis Elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 16.Erlwein O, Cain D, Fischer N, Rethwilm A, McClure M O. Identification of sites that act together to direct dimerization of human foamy virus RNA in vitro. Virology. 1997;229:251–258. doi: 10.1006/viro.1997.8438. [DOI] [PubMed] [Google Scholar]
- 17.Harrison G P, Hunter E, Lever A M. Secondary structure model of the Mason-Pfizer monkey virus 5′ leader sequence: identification of a structural motif common to a variety of retroviruses. J Virol. 1995;69:2175–2186. doi: 10.1128/jvi.69.4.2175-2186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirata R K, Miller A D, Andrews R G, Russell D W. Transduction of hematopoietic cells by foamy virus vectors. Blood. 1996;88:3654–3661. [PubMed] [Google Scholar]
- 19.Hungnes O, Tjotta E, Grinde B. Mutations in the central polypurine tract of HIV-1 result in delayed replication. Virology. 1992;190:440–442. doi: 10.1016/0042-6822(92)91230-r. [DOI] [PubMed] [Google Scholar]
- 20.Katoh I, Yasunaga T, Yoshinaka Y. Bovine leukemia virus RNA sequences involved in dimerization and specific gag protein binding: close relation to the packaging sites of avian, murine, and human retroviruses. J Virol. 1993;67:1830–1839. doi: 10.1128/jvi.67.4.1830-1839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katz R A, Terry R W, Skalka A M. A conserved cis-acting sequence in the 5′ leader of avian sarcoma virus RNA is required for packaging. J Virol. 1986;59:163–167. doi: 10.1128/jvi.59.1.163-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaye J F, Richardson J H, Lever A M. cis-Acting sequences involved in human immunodeficiency virus type 1 RNA packaging. J Virol. 1995;69:6588–6592. doi: 10.1128/jvi.69.10.6588-6592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konings D A M, Nash M A, Maizel J V, Arlinghaus R B. Novel GACG-hairpin pair motif in the 5′ untranslated region of type C retroviruses related to murine leukemia virus. J Virol. 1992;66:632–640. doi: 10.1128/jvi.66.2.632-640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kupiec J-J, Sonigo P. Reverse transcriptase jumps and gaps. J Gen Virol. 1996;77:1987–1991. doi: 10.1099/0022-1317-77-9-1987. [DOI] [PubMed] [Google Scholar]
- 25.Kupiec J J, Tobaly-Tapiero J, Canivet M, Santillana H M, Flügel R M, Peries J, Emanoil-Ravier R. Evidence for a gapped linear duplex DNA intermediate in the replicative cycle of human and simian spumaviruses. Nucleic Acids Res. 1988;16:9557–9565. doi: 10.1093/nar/16.20.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laughrea M, Jette L, Mak J, Kleiman L, Liang C, Wainberg M A. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J Virol. 1997;71:3397–3406. doi: 10.1128/jvi.71.5.3397-3406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee A H, Lee H Y, Sung Y C. The gene expression of human foamy virus does not require a post-transcriptional transactivator. Virology. 1994;204:409–413. doi: 10.1006/viro.1994.1545. [DOI] [PubMed] [Google Scholar]
- 28.Lever A M L, Göttlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linial M L, Miller A D. Retroviral RNA packaging: sequence requirements and implications. In: Swanstrom R, Vogt P K, editors. Retroviruses: strategies of replication. New York, N.Y: Springer-Verlag; 1990. pp. 125–152. [DOI] [PubMed] [Google Scholar]
- 30.Mann R, Mulligan R C, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 31.Mansky L M, Krueger A E, Temin H M. The bovine leukemia virus encapsidation signal is discontinuous and extends into the 5′ end of the gag gene. J Virol. 1995;69:3282–3289. doi: 10.1128/jvi.69.6.3282-3289.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors and a complementary helper free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muranyi W, Flügel R M. Analysis of splicing pattern of human spumatetrovirus by polymerase chain reaction reveals complex RNA structures. J Virol. 1991;65:727–735. doi: 10.1128/jvi.65.2.727-735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paillart J-C, Berthoux L, Ottmann M, Darlix J-L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pugatsch T, Stacey D W. Identification of a sequence likely to be required for avian retroviral packaging. Virology. 1983;128:505–511. doi: 10.1016/0042-6822(83)90279-9. [DOI] [PubMed] [Google Scholar]
- 36.Rethwilm A, Baunach G, Netzer K-O, Maurer B, Borisch B, ter Meulen V. Infectious DNA of the human spumaretrovirus. Nucleic Acids Res. 1990;18:733–738. doi: 10.1093/nar/18.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richardson J H, Child L A, Lever A M L. Packaging of human immunodeficiency virus type 1 RNA requires cis-acting sequences outside the 5′ leader region. J Virol. 1993;67:3997–4005. doi: 10.1128/jvi.67.7.3997-4005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell D W, Miller A D. Foamy virus vectors. J Virol. 1996;70:217–222. doi: 10.1128/jvi.70.1.217-222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Schmidt M, Rethwilm A. Replicating foamy virus-based vectors directing high level expression of foreign genes. Virology. 1995;210:167–178. doi: 10.1006/viro.1995.1328. [DOI] [PubMed] [Google Scholar]
- 41.Simpson S B, Zhang L, Craven R C, Stoltzfus C M. Rous sarcoma virus direct repeat cis elements exert effects at several points in the virus life cycle. J Virol. 1997;71:9150–9156. doi: 10.1128/jvi.71.12.9150-9156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorge J, Ricci W, Hughes S H. cis-Acting RNA packaging locus in the 115-nucleotide direct repeat of Rous sarcoma virus. J Virol. 1983;48:667–675. doi: 10.1128/jvi.48.3.667-675.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tobaly-Tapiero J, Kupiec J J, Santillana H M, Canivet M, Peries J, Emanoil-Ravier R. Further characterization of the gapped DNA intermediates of human spumavirus: evidence for a dual role in initiation of plus-strand DNA synthesis. J Gen Virol. 1991;72:606–608. doi: 10.1099/0022-1317-72-3-605. [DOI] [PubMed] [Google Scholar]
- 44.Vile R G, Ali M, Hunter E, McClure M O. Identification of a generalised packaging sequence for D-type retroviruses and generation of a D-type retroviral vector. Virology. 1992;189:786–791. doi: 10.1016/0042-6822(92)90607-q. [DOI] [PubMed] [Google Scholar]
- 45.Wain-Hobson S, Sonigo P, Danos O, Cole S, Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985;40:9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe S, Temin H M. Encapsidation sequences for spleen necrosis virus, an avian retrovirus, are between the 5′ long terminal repeat and the start of the gag gene. Proc Natl Acad Sci USA. 1982;79:5986–5990. doi: 10.1073/pnas.79.19.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Temin H M. A double hairpin structure is necessary for the efficient encapsidation of spleen necrosis virus retroviral RNA. EMBO J. 1994;13:713–726. doi: 10.1002/j.1460-2075.1994.tb06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu S, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. The human foamy virus replication pathway is distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]