Abstract
Tumor dissemination into the surrounding stroma is the initial step in a metastatic cascade. Invasion into stroma is a non-autonomous process for cancer, and its progression depends upon the stage of cancer, as well as the cells residing in the stroma. However, a systems framework to understand how stromal fibroblasts resist, collude, or aid cancer invasion has been lacking, limiting our understanding of the role of stromal biology in cancer metastasis. We and others have shown that gene perturbation in stromal fibroblasts can modulate cancer invasion into the stroma, highlighting the active role stroma plays in regulating its own invasion. However, cancer invasion into stroma is a complex higher-order process and consists of various sub-phenotypes that together can result in an invasion. Stromal invasion exhibits a diversity of modalities in vivo. It is not well understood if these diverse modalities are correlated, or they emanate from distinct mechanisms and if stromal biology could regulate these characteristics. These characteristics include the extent of invasion, formation, and persistence of invasive forks by cancer as opposed to a collective frontal invasion, the persistence of invading velocity by leader cells at the tip of invasive forks, etc. We posit that quantifying distinct aspects of collective invasion can provide useful suggestions about the plausible mechanisms regulating these processes, including whether the process is regulated by mechanics or by intercellular communication between stromal cells and cancer. Here, we have identified the sub-characteristics of invasion, which might be indicative of broader mechanisms regulating these processes, developed methods to quantify these metrics, and demonstrated that perturbation of stromal genes can modulate distinct aspects of collective invasion. Our results highlight that the genetic state of stromal fibroblasts can regulate complex phenomena involved in cancer dissemination and suggest that collective cancer invasion into stroma is an outcome of the complex interplay between cancer and stromal fibroblasts.
Keywords: Stromal invasion, Collective invasion, Cancer-stroma interaction
Introduction
Cancer metastasis is a complex phenomenon consisting of overlapping processes occurring over many physiological scales [1]. Dissemination into the surrounding stroma is one of the earlier steps of a primary tumor as it begins its journey towards metastasis. Epithelial tumor cells were considered to acquire a mesenchymal state due to acquired mutations, after which they would invade into the stroma and escape into the vasculature for distant tissues, creating secondary metastatic nodes [2, 3]. Recent findings have added considerable complexity to the dogma, with initial dissemination being considered as a combination of epithelial and mesenchymal states, and contesting evidence showing that cancer cells can micrometastasize as cell-clusters [4–6], or need to transition into a mesenchymal state to metastasize [7–9]. In addition, there is an increased appreciation of the role of stroma in regulating cancer phenotypes, with activated fibroblasts now increasingly being recognized to promote cancer growth and metastasis [10–14]. With cancer being the primary source of genetic diversity in this milieu, the role of the genetic state of the stromal cells in regulating dissemination has escaped focus. Indeed, because stromal fibroblasts do not acquire mutations by uncontrolled growth, they could be attractive gene targets with less probability to acquire drug resistance or escape surveillance.
Stromal invasion of cancer is a complex process, comprised of many different sub-processes which together form the phenotype of collective invasion. Since each sub-process may be dictated by distinct mechanisms, it is non-trivial to predict the effect of stromal genetic state on cancer invasion. The collective invasion may entail a frontal movement of epithelial cancer cells into the stroma, or conversely, the formation of invasive forks which forge into the stromal barrier while the larger tumor mass remains relatively non-invasive. Leader cells which form the leading edge of the invasive forks may undergo epithelial to mesenchymal transition (EMT), and disseminate into and through the stroma without maintaining cell–cell contacts with the remaining fork. These, and many such other sub-characteristics, if measured, can provide useful insights into the plausible mechanisms regulating these processes but are often overlooked for the broader phenotype of total levels of metastasis. Because cancer continuously evolves, owing to acquired mutations as well as a high proliferation rate, minor aspects of the invasion, if selected for, can have a profound effect on metastasis.
We and others have previously shown that stromal genetic state can regulate the overall phenotype of collective cancer invasion. It is, however, not known if gene expression changes in human stromal fibroblasts could influence the complex collective invasion characteristics, and if modulating the stromal genetic state could allow secondary control of tumor dissemination. Here, we demonstrate that gene perturbation in pathways that we previously found to be important for decreased stromal resistance to cancer in the human stroma can produce a diverse set of sub-phenotypes in cancer invasion.
Using a reductive and multi-throughput model of melanoma stromal dissemination consisting of A375 malignant melanoma cells, and BJ skin fibroblasts, we found that gene silencing in BJ could alter A375 collective invasion characteristics beyond the overall extent of invasion we previously reported. In this report, we have identified and developed methods to quantify various sub characteristics which are inherent in the collective invasion of cancer into the stroma and found that melanoma invasion could present a diversity of responses. These diverse responses suggest a stroma-cancer cross talk, or modulation of stromal mechanics by gene silencing, which could lead to the emergence of new patterns of collective melanoma invasion. Our work underscores our previous findings that the stromal genetic state plays a key role in permitting or resisting tumor dissemination, as well as in regulating key aspects of invasion presenting new avenues to target tumor metastasis by modulating stromal biology.
Results
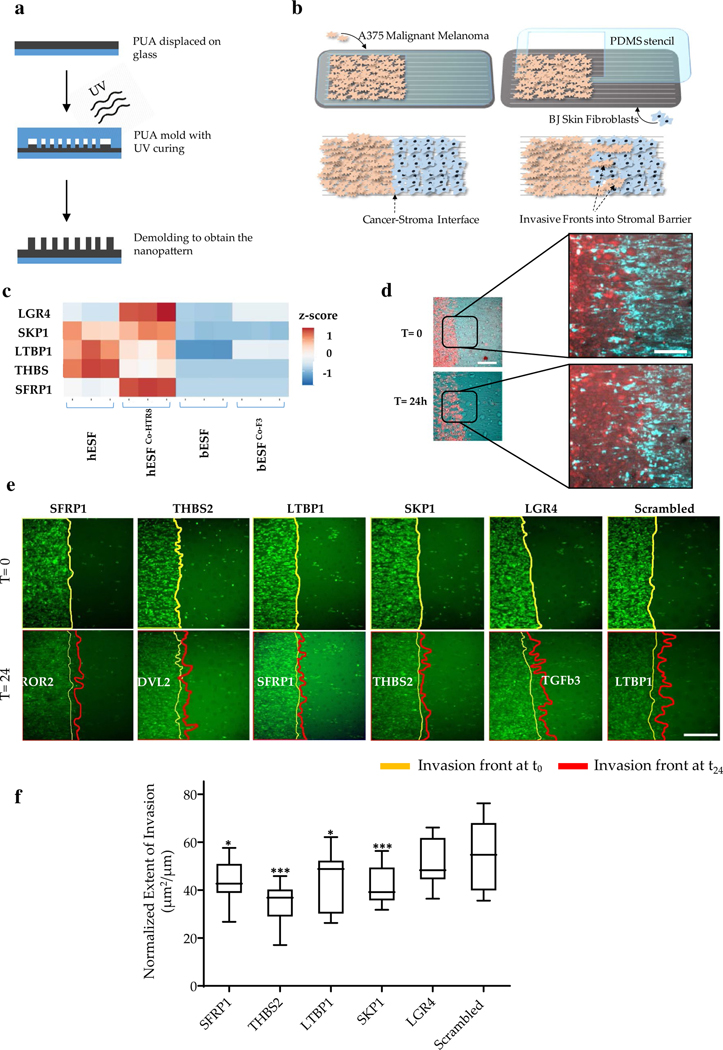
To study the effect of stromal fibroblast genes on cancer invasion, we chose a combination of A375 malignant melanoma cell line, and BJ, untransformed skin fibroblasts, and studied their invasion on our nanopatterned stromal invasion platform reported earlier [18]. Nanopatterned substrates mimicking extracellular ultrastructure in various tissues were fabricated using capillary force lithography [1, 2] (Fig. 1a). A375 cells were labeled with Cell Tracker green, seeded on the nanostructure in the region remaining accessible in PDMS (polydimethylsiloxane) stencil on nanofabricated polyurethane substrates [3, 4] (Fig. 1b). Thereafter, the stencil was removed, and unlabeled BJ stromal cells were introduced to occupy the empty space created by the stencil, thereby creating two juxtaposed monolayers of either cell types [19] (Fig. 1b). Using live-cell microscopy, we observed the invasive characteristics of A375 cells into the BJ stromal fibroblast monolayer (Fig. 1e). This platform presents a reductive model to measure stromal invasion in multi-throughput; however, it does not present all the complexities present in a 3D cancer microenvironment. We have shown that the nanopatterned invasion assay can allow sensitive and quantitative discrimination between malignant and non-malignant stromal invasion [4].
Fig. 1.
Multiplexed stromal gene knockdown to test melanoma invasion into skin stromal fibroblasts. a Schematic showing the fabrication workflow of the collective stromal invasion device substrate; PUA polyurethane; b Schematic showing assay setup and experimental workflow consisting of patterned seeding of labeled invasive A375 cells using stencils, and subsequent seeding of stromal BJ fibroblasts, and observing formation and penetration of invasive fronts; c Heatmap showing relative expression of selected genes with high expression in human endometrial stromal fibroblasts (hESFs) compared to bovine endometrial stromal fibroblasts (bESFs) with or without co-culture of the species-specific trophoblasts (HTR8, and F3, respectively); Heatmap shows the Z-score; d Representative image showing an invasive A375 front into the BJ stromal fibroblast monolayer at time 0 h and 24 h, scale bar = 200 μm. Inset shows a magnified view of the juxtaposed A375 (red) and BJ monolayer, scale bar = 100 μm; e Time-lapse images showing initial (yellow line, 0 h) and final (red line, 24 h) position of the invasive A375 fronts into the stromal BJ monolayer, wherein BJ cells are transduced with siRNA listed above the panels; f Quantified extent of A375 frontal invasion across different stromal BJ conditions listed in e. The end of boxes refer to as upper and lower quartile, the horizontal bar refers to the mean, and the end of lines refer to the lowest or highest data (*p value < 0.05, **p value < 0.005, ***p value < 0.0005)
We had previously described that bovine fibroblasts resist collective invasion by downregulation of Wnt and TGFb signaling, implicating key genes regulating the process. Both these pathways have been previously described to be important in fibroblast mediated cancer metastasis [20–23]. We sought to test if alteration of stromal gene expression could alter collective invasion in more complex ways than manifested by a broader total invasion metric. Towards this, we tested new targets within the Wnt and TGFb signaling pathways differentially expressed between bovine and human fibroblasts (endometrial), augmenting our previously tested list of targets [18]. Using RNA Sequencing data we previously reported, we identified genes present in non-canonical Wnt signaling and TGFβ pathway, which were upregulated in human endometrial stromal cells (hESF), with or without co-culture with their respective trophoblasts (HTR8) compared to bovine endometrial stromal cells (bESF), with or without co-culture with their respective trophoblasts (F3). (Fig. 1c). These genes included LGR4 encoding a G-protein coupled receptor and a key Wnt activator which also regulates breast cancer and squamous cell carcinoma metastasis [24, 25], SKP1 encoding S-phase kinase-associated protein 1, an essential component of the SCF ubiquitin ligase complex which regulates degradation of various SMADs and other factors [26]; LTBP1 encoding a latent TGFb1 binding protein promoting EMT transition [27]; THBS2 encoding thrombospondin 2, a cell–cell and cell–matrix adhesion protein and also a biomarker for colorectal cancer [6]; and SFRP1 encoding the secreted frizzled-related protein 1, with an elevated level in cancer stroma influencing stromal-to epithelial signaling [5]. The objective of this study was to create quantitative measures for the diversity of tumor dissemination phenotypes which could meaningfully indicate plausible mechanisms guiding those individual metrics.
Characterization of diverse modes of stromal invasion
We first tested the effect of gene knockdown on the total extent of invasion. We seeded A375 cells, and BJ cells after siRNA mediated gene knockdown 48 h after siRNA transfection to maximize the effect of gene knockdown. A complete monolayer was observed for each condition allowing a juxtaposed interaction of A375 and BJ collective fronts (Fig. 1d). The initial and final area occupied by A375 cells were segmented, difference calculated and normalized by the initial length of the interface between A375 and BJ monolayers to calculate the extent of cancer invasion over a unit interface length (Fig. 1e, f). We found that all stromal knockdowns, except LGR4, resulted in a significant reduction of stromal invasion compared to scrambled control (Fig. 1e, f). However, morphogenetic observation of the invasive fronts showed that even disruption of the same protein network through its various components resulted in different invasive characteristics (Fig. 1e). We, therefore, sought to further test the sub-characteristics of this invasive process.
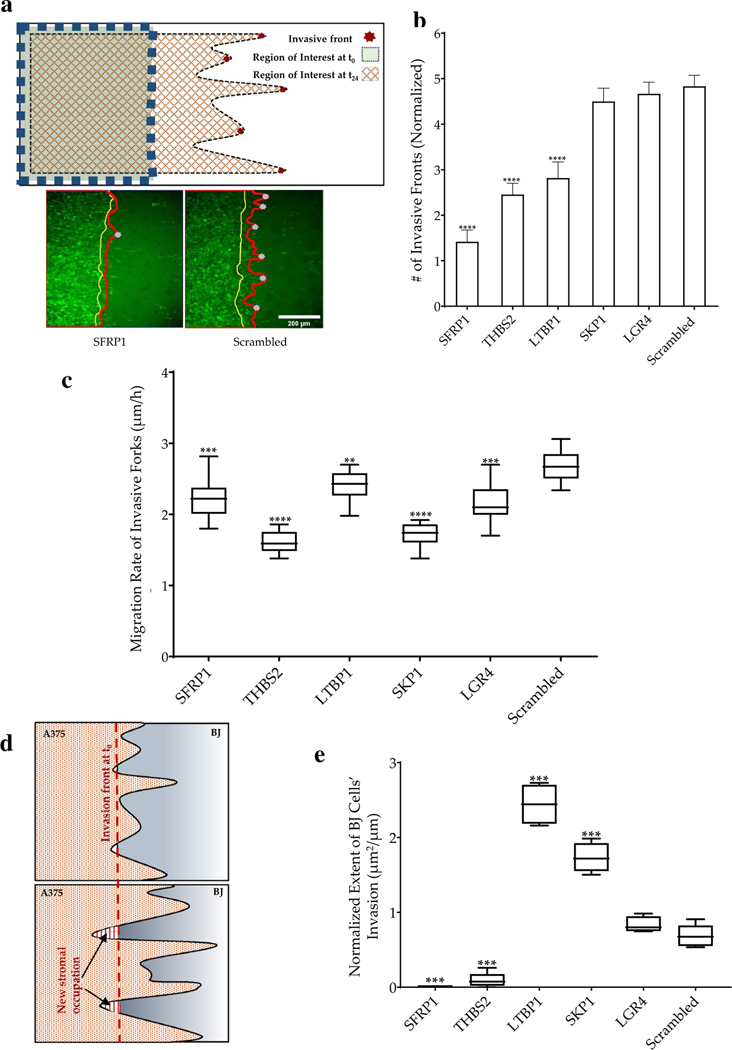
Collective migration of epithelial cells can contextually, on anisotropic surfaces, results in the formation of invasive fronts consisting of the leader (or pioneer cells) which are highly migratory, followed by follower cells that divide rapidly to maintain cell–cell contact as the front advances [28]. Invasive fronts have been described in vivo in disseminating cancer populations [29, 30]. We tested if gene knockdown in stroma could change the number of invasive fronts that get initiated in the melanoma monolayers (Fig. 2a, b). We found that knockdown of SFRP1, THBS2, and LTBP1 significantly reduced the number of invasive fronts in a unit length of melanoma-stroma interface. Interestingly, although SKP1 knockdown had resulted in significant repression of total invasion, it did not result in the reduction of invasive front formation by the invading melanoma cells (Fig. 2b). It is notable that change in expression of a single gene could have such a dramatic effect on the pattern of collective movement from different cell types, but these observations also could suggest plausible mechanisms at play wherein the stromal contribution to cancer invasion is important.
Fig. 2.
Phenotypic characterization of the invasive fronts. a Schematic showing formation of invasive fronts and identification of the leader cells in the invasive fronts; and images of invading A375 cells showing a representative example of the interface at time 0 (yellow line), and after 24 h (red line); Also shown below are examples of invasive forks penetrating BJ cells transduced with siRNA targeting SFRP1, or control; gray dots depict the tip of the invasive fronts; Scale bar = 200 μm; b Number of invasive fronts penetrating into BJ cells with different siRNA knockdowns. c Rate of penetration of invasive forks in all conditions listed in b. d Schematic showing new area occupied by stromal cells after 24 h (indicated by arrows). e The normalized extent of BJ cells’ invasion of the cancer cells. The end of boxes refer to as upper and lower quartile, the horizontal bar refers to the mean, and the end of lines refer to the lowest or highest data. Statistical significance was calculated for each siRNA as an unpaired t-test against the control Scr. (*p value < 0.05, **p value < 0.005, ***p value < 0.0005, ****p value < 0.0001). In each of the above figures, gene name refers to the siRNA knockdown of the specific gene
Although the formation of invasive fronts is formulated as an autonomous cancer phenomenon, wherein cancer cells assume phenotypes of highly migratory leader cells and less migratory follower cells; our data demonstrate that stromal genetic state could modulate collective cancer invasion. While an overall reduction in the extent of invasion (Fig. 1f) may be an outcome of multiple plausible mechanisms, reduction in the invasive fronts indicates either a heightened barrier formation within the stroma which invasive forks are unable to breach, or secretion of factors which limit specification of leader cells within cancer, or mechanical stromal response to nascent fork formations. We further measured the rate of invasive fork growth, suggesting penetration of invasive fronts, and found that all knockdown conditions significantly reduced the rate of fork penetration, indicating a strong influence of stromal fibroblasts in regulating collective cancer penetration (Fig. 2c). This reduction may be induced by inhibition of attractive chemokine signals from the stroma in the experimental conditions. LGR4, for example, is a key activator of Wnt signaling necessary for stabilization and nuclear translocation of b-catenin. LGR4 knockdown could result in the stabilization of b-catenin destruction complex and, therefore, reduction in gene products responsible for inducing migration, including ADAM10, DKK1, and DKK4, BMP4, and various metalloproteinases [31]. Similarly, SFRP1 is a secreted frizzled-related protein, and maybe pro-invasive for invading melanoma cells, and its knockdown could induce a modest reduction in cancer migration. Knockdown of thrombospondin in the stromal fibroblast appeared to have the most drastic effect, both in the overall reduction in the extent of invasion, as well as in regulating cancer penetration. Secretome analysis of cancer-associated fibroblasts has revealed THBS2 gene products to be present and a key to regulating cancer metastasis [32], and increasing evidence suggests a THBS2 mediated interaction between cancer and stromal fibroblasts [33]. We also observed that the stromal fibroblasts themselves could push the invading melanoma cells in certain locations, while in other locations, the invading cells continued to advance invasive forks. We, therefore, quantified the new territory occupied by the competing stromal fibroblasts after 24 h, competitively from the invading melanoma cells (Fig. 2d). We surprisingly found that while knockdown of SFRP1 and THBS2 had resulted in a significantly lower new area occupied by BJ cells vs control, knockdown of LTPB1 and SKP1 indeed resulted in significantly higher new aerial occupation by BJ cells into the A375 territory (Fig. 2e). Because all these conditions had resulted in a significant reduction in the extent of A375 invasion, these data suggested that while SFPR1 and THBS2 knockdown resisted the overall growth of invasion, knockdown of LTBP1 and SKP1 accompanied a combination of push and pull of the invasive forks (Fig. 2e).
Velocities of leader cells exhibit stromal influence on regulating the temporal spread of cancer
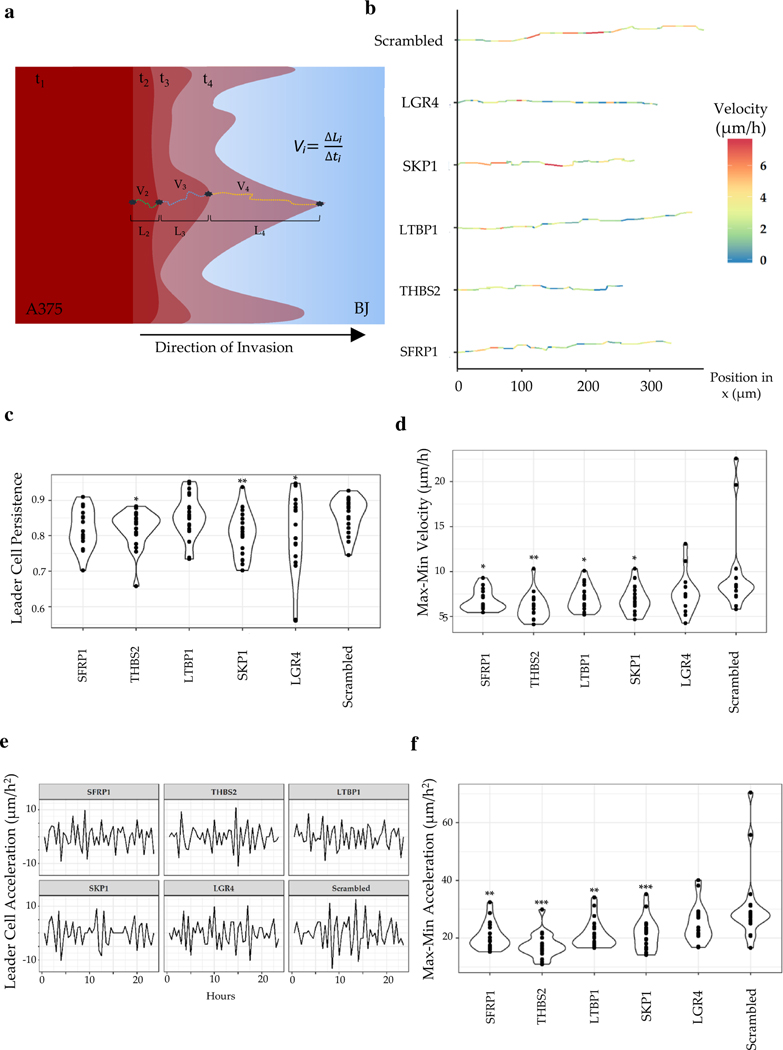
Our data showed that stromal state could influence the fate of invasive fronts formed by the collectively invading cancer cells. Penetration into the stroma is a key first step into the metastatic cascade. We, therefore, sought to further characterize the leader cells penetration into the stroma. We identified the granular instantaneous velocity of the leader cells into the stromal monolayer. The anisotropic arrangement of nano grooves rendered the movement of invasive fronts highly directional. Using time-lapse resolved fluorescence data in the invasive fronts representing the mean total movement for each gene knockdown, we computed their instantaneous velocities (Fig. 3a). Specifically, first, we identified the directional movement for each identified invasive front. Thereafter, we identified the invasive fork most closely matching the mean movement and measured the instantaneous velocity by computing the directional displacement every hour (Fig. 3a, b).
Fig. 3.
Phenotypic characterization of the speed and the path of the leader cells. a Schematic showing calculation of instantaneous velocity and migration rate of the tip of invasive forks calculated from acquired time-lapse images; ti: time-stamp, Vi: velocity, Li: length traversed by the leader cell in the i-th interval; b average trajectory of a representative leader cell for each condition; instantaneous velocity is color coded; c Violin plot showing persistence of movement of the leader cells; each dot refers to an instance of an invasive fork; d Difference in the maximal, and minimal velocity of penetrating A375 leader cells in the different stromal background; e Representative instantaneous acceleration of leader cells plotted over time. f The difference in the maximal and the minimal acceleration of penetrating A375 leader cells in the different stromal background. Statistical significance was calculated for each siRNA as an unpaired t test against the control Scr. (*p value < 0.05, **p value < 0.005, ***p value < 0.0005)
Interestingly, we found that while for the scrambled control, the velocity fluctuated between medium and high, for all conditions but SKP1 knockdown, the velocity fluctuated between low and high (Fig. 3b). Closer observation revealed that although in the early phase of the invasion, when the invasive forks are probably getting established, the rate of penetration is moderately high, eventually is curtailed by stroma in most experimental conditions. These data suggest an active stromal response to cancer invasion, which is activated when the stromal breach is sensed. This response is, notably, absent for scrambled control, indicating that ELI specific gene inhibition could increase the responsiveness of stromal fibroblasts.
Notably, the instantaneous velocity of the leader cells showed regular fluctuation of movement for all conditions (Fig. 3b). This observation may indicate a null expectation of cellular movement, which may occur through periodic steps and pause, or a continuous stromal-cancer interaction which gives way to allow cancer to move forward. We calculated the directional persistence of A375 leader cells in different stromal background, calculated as the extent of continuous directional movement into the stroma vs. in other random directions, and found that knockdown of THBS2, SKP1, and LGR4 significantly reduced the persistence of penetrative fork growth compared to the scrambled control condition (Fig. 3c). These data indicate differences from the more global metric of the extent of areal invasion (Fig. 1f), suggesting that while knockdown of SFRP1 and LTBP1 may limit A375 invasion, they may not resist a persistently penetrating fork into the stroma. In contrast, LGR4 knockdown significantly resisted the persistence of penetration, although the areal extent of total invasion was not affected. These data indicate differences in mechanisms guiding stromal-dependent invasion between LGR4 and other knockdowns, suggesting that LGR4 knockdown may confer a more resistive stromal force, which specifically targets invasive breach into the collective stromal monolayer (Fig. 3c). A distinct calculation of the difference between the peak velocities of A375 leader cells in each stromal condition revealed that while in the control conditions, there were instances of rapid fluctuations in velocities, for knockdown conditions, the penetrating velocities of the leading cells were controlled in stricter limits (Fig. 3d). This calculation suggests a plausible reduction in stromal assistive engagement upon siRNA mediated downregulation of Wnt signaling. Estimation of the instantaneous acceleration of leader cells revealed that whereas after an initial low acceleration, the control stromal monolayer resulted in high fluctuation of acceleration of the fork penetration, for other conditions, we did not see any sharp increase in the fluctuations of the acceleration itself (Fig. 3e). Although it is difficult to speculate the mechanisms driving such diverse phenotypes, we posit that a non-varying persistence suggests an equilibrium between stromal fibroblasts and A375, possibly due to mechanical changes in the BJ stroma due to gene knockdown. A highly varying persistence shows more non-equilibrium dynamics, possibly due to continuous secretory signaling between cancer and stroma and an underdamped force generation feedback between the stroma and invasive cells (Fig. 3f).
Finally, our data and analyses show that collective invasion is constituted of various sub characteristics, which could provide finer suggestions to the possible mechanisms driving the dissemination of tumor into the stroma.
Discussion
Stromal effect on cancer invasion has been a subject of intense research interest in the last few years, with accumulating evidence suggesting that cancer can collude with an activated stromal compartment to advance towards metastasis. Recent findings have indicated that tumor dissemination into the stroma is a complex process and may involve multiple mechanisms including a complete mesenchymal transition of epithelial cancer cells, dissemination as epithelial clusters, a hybrid response, dissemination by invading fronts wherein migratory leader cells are followed by proliferative follower cells, etc. We considered that these mechanisms, which seem conflicting, may all co-exist and may be an outcome of the interplay between the tumor and the stromal microenvironment, with additional inputs from the non-stromal tumor microenvironment. This necessitates identifying and quantifying the sub-characteristics of tumor invasion, test if these characteristics are influenced by the stromal biology, and if the stromal influence is correlative for all the invasive sub-phenotypes. We used a bioengineered platform we have previously published [18, 19], which, although does not incorporate all the physiological aspects present in the cancer microenvironment [1], nevertheless allows a multi throughput and reductive modeling of the stromal invasion process in a physiological setting. We performed these experiments on untransformed skin fibroblast cell line, BJ, observing the invasion of malignant A375 melanoma cells into a monolayer of BJ, which had been subjected to siRNA-based knockdown of a battery of genes.
We found that the collective invasion of A375 showed a marked diversity of behavior in response to BJ cells in different knockdown conditions, suggesting that stromal signaling could profoundly affect cancer invasion sub-phenotype. It is remarkable that a change in expression of a single gene could have such a dramatic effect on the pattern of collective movement from different cell types. These observations point to either a complex interplay of secreted molecules between stroma and cancer which are affected by Wnt and TGFβ signaling or that gene knockdown changes stromal mechanics, resulting in complex pattern formation of stromal invasion. We found that while regulation of TGFβ secondary response (by knockdown of SKP1 and LTBP1) within the fibroblasts could significantly alter the overall extent of melanoma invasion, inhibition of Wnt crosstalk (by knockdown of SFRP1 and LGR4) did not result in large changes in stromal resistance, but instead altered the migration rate of invasive forks. Interestingly, SFRP1 and THBS2 knockdown affected stromal resistance by directly resisting the growth of melanoma invasion. In contrast, LTBP1 and SKP1 knockdown showed a combination of push and pull of invasive fronts, with the stromal fibroblasts moving into previously occupied melanoma area. Wnt and TGFβ signaling has been extensively reported to be crucial for cancer stem cell maintenance, as well as cancer growth and metastasis, but its role in stromal-cross talk with cancer is not well understood. Our data indicate that while TGFβ resulted in a pro-fibrotic response in the stroma, Wnt-mediated communication is necessary for continuous penetrative invasion of melanoma within the stromal monolayer.
We posit that further mathematical modeling and simulation of simple mechanical and signaling interactions may show the emergence of the diversity of invasive behavior shown in this work. In conclusion, our work details various sub-phenotypes that together constitute cancer dissemination into the stroma and highlights that stromal state could regulate these phenotypes distinctly. Further, our work suggests plausible mechanisms that may drive these subphenotypes, which could be targeted specifically to limit cancer dissemination into the stromal surrounding.
Materials and methods
Cell culture
Human skin fibroblast (BJ) cells were grown in Eagle’s Minimum Essential Medium (EMEM) with 10% of fetal bovine serum (FBS) and 1% antibiotic/antimycotic, for 72 h before siRNA transfection. The human melanoma cells (A375) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% of fetal bovine serum (FBS) and 1% antibiotic/antimycotic for 48 h before labeling and culturing on the nanopatterns.
Fabrication of nanotextured mold
Nanopatterns were prepared using electron-beam lithography on a silicon wafer with a layer of photoresist applied using the spin coating. The sub-micron parallel grooves were formed using the deep reactive ion etcher (STS ICP Etcher) after developing the photoresist. Adding the UV curable polyurethane (PUA) dropwise on the prepared silicon master and a layer of polyethylene terephthalate (PET) film made it ready to be cured by UV (n = 200–400 nm, 100 mJ/cm2) for 1 min. Then the mold was peeled off and overnight UV exposure was performed to finalize the unreacted acrylate group and have the PUA mold with a thickness of 50 μm.
Fabrication of nanostructure substrate
Nanostructured substrates were prepared using capillary force lithography as previously described [15, 16]. Specifically, a PET film containing the PUA patterns was used as a replica mold to create a secondary PUA substrate. Glass coverslips were cleaned for 1 h using 0.1 M NaOH, washed with DIH2O and dried. Primer was applied using a paintbrush (phosphoric acrylate and propylene glycol monomethyl ether acetate, ratio 1:10), and coverslip baked for 20–30 min at 70 °C. 200 ml of PUA precursor was dispensed dropwise, and the replicate mold was placed on top, after which the combination was cured under UV (ν = 250–400 nm, 100 mJ/cm2) for 1 min. The mold was then peeled off, and the remaining semi-polymerized substrate was further polymerized under UV overnight, thereby terminating residual active acrylate groups.
Stromal invasion assay
A PDMS (polydimethylsiloxane) mold, which was fabricated by a stereolithographic plastic mold, was used for cell patterning. The monomer and crosslinker (with a ratio of 10:1) were mixed and cured at 80 °C for 4 h and then cast in the pre-designed mold to form a PDMS stencil. The nanopatterned substrate was coated with Laminin (25 μg/ml). After washing with isopropanol and drying with N2 steam, the stencil was placed on the substrate. To remove air and bubbles under the stencil, the device was kept in vacuum. The labeled A375 cells were seeded at a density of 5*105 cells and attached to the surface overnight. The stencil was removed using the tweezers. The unlabeled stromal cells were seeded at a density of 5 × 105 to fill and attach that area which was covered by the stencil before. After incubation for 5 h, the unattached cells were removed by washing off. Then the plate was ready to be mounted on the stage of the live-cell microscopy.
Small-interfering RNA (siRNA) transfection
siRNA transfection in a 24 well plate was performed with Lipofectamine RNAimax. Cells with 60% confluency were transfected with 50 nmol of siRNA per well. After incubation overnight, the transfection mixture was removed, and the normal growth medium was added. Imaging was performed after 48 h of transfection. All siRNAs were obtained from IDT (Table 1).
Table 1.
List of siRNA with the sequences
| siRNA | siRNA Duplex Name | Sequence 5′ to 3′ |
|---|---|---|
| SFRP1 | Hs.Ri.SFRP1.13.1 | AAG GUU UUA AAA CAG UCU |
| THBS2 | Hs.Ri.THBS2.13.2 | CGA AUG CAG AGA AUA UUA |
| LTBP1 | Hs.Ri.LTBP1.13.3 | GGA AUU GCA AGU CCU CUG |
| SKP1 | Hs.Ri.SKP1.13.1 | CCA UCA UGA AUG CAA GAU |
| LGR4 | Hs.Ri.LGR4.13.3 | UUG CUU UGG UCC AAU CAG |
Time-lapse microscopy
After allowing the cells to expand freely in the culture plate, time-lapse images were recorded. The plate was mounted on the stage of a live cell imaging microscope (Zeiss Observer Axio Z1), which was equipped with an ORCA Flash 4.0 CMOS camera. We used an EC Plan-Apochromat 10x/0.8 WD-0.55 objective for imaging cells for 24 h, with a time interval of 30 min each. Images were acquired using ZEN 2012 Software while using Definite Focus 2 from Zeiss.
Image analysis and cell tracking
We tracked the cells’ migration, velocity, displacement, and the invasion fronts using the manual tracker and Region of Interest (ROI) panels in the Fiji software package [17]. The Instantaneous velocity of the invading fronts and the rate of penetration of invasive forks were measured by averaging the displacement observed within each 30-min time interval. The length of the invasion was defined by measuring the horizontal distance of the initial point of one cell and its final location. Because of nanofabricated substrates, the movement of cells was largely unidirectional. By tracing the ROI of A375 cells for the initial time point and the final one, the invasion fronts were defined. For each ROI, the number of invasion fronts was manually counted and analyzed.
Acceleration, persistence and velocity changes calculation
The leader cells velocity changes within their trajectory, their acceleration and persistence were calculated by a custom script in R. The velocity changes have been indicated by a color code in their positions over time. The persistence of movement of leader cells is defined as the ratio of total displacement in the direction of invasion divided by the total path length.
Gene expression heat map
Gene expression in human and bovine endothelial stromal fibroblasts with and without co-culture with trophoblasts was calculated in terms of their transcripts per million (TPM) values. Z scores were calculated by normalizing TPM values for each gene independently. Selected genes were thought to be involved in the non-canonical Wnt, and TGFb pathways were selected for analysis and represented in the heat map. All RNA Sequencing data was obtained from our previous report [18].
Statistics analysis
Statistical analysis was performed using Student’s unpaired t-test to compare each condition with Scrambled. In addition, ANOVA and significance of (adjusted) p-value 0.05, followed by Dunnett’s test results, are provided in the supplementary.
Supplementary Material
Funding
This research was funded by the UConn Health Startup fund provided by the UConn Health Dental School and Biomedical Engineering Department, as well as by National Cancer Institute, USA funded U54 sub-contract to UConn Health: 1U54CA209992–02.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s13577-020-00427-6) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest Authors declare no conflict of interest.
References
- 1.Suhail Y, Cain MP, Vanaja K, Kurywchak PA, Levchenko A, Kalluri R. Systems biology of cancer metastasis. Cell Syst. 2019;9(2):109–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168(4):670–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Eaton EN, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525(7568):256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qutaish MQ, Zhou Z, Prabhu D, Liu Y, Busso MR, Izadnegahdar D, et al. Cryo-imaging and software platform for analysis of molecular MR imaging of micrometastases. Int J Biomed Imaging. 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155(7):1639–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padmanaban V, Krol I, Suhail Y, Szczerba BM, Aceto N, Bader JS, et al. E-cadherin is required for metastasis in multiple models of breast cancer. Nature. 2019;573(7774):439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia D, Jolly MK, Tripathi SC, Den Hollander P, Huang B, Lu M, et al. Distinguishing mechanisms underlying EMT tristability. Cancer Converg. 2017;1(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jolly MK, Tripathi SC, Jia D, Mooney SM, Celiktas M, Hanash SM, et al. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget. 2016;7(19):27067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shamir ER, Pappalardo E, Jorgens DM, Coutinho K, Tsai W-T, Aziz K, et al. Twist1-induced dissemination preserves epithelial identity and requires E-cadherin. J Cell Biol. 2014;204(5):839–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shangguan C, Gan G, Zhang J, Wu J, Miao Y, Zhang M, et al. Cancer-associated fibroblasts enhance tumor 18F-FDG uptake and contribute to the intratumor heterogeneity of PET-CT. Theranostics. 2018;8(5):1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clocchiatti A, Ghosh S, Procopio M-G, Mazzeo L, Bordignon P, Ostano P, et al. Androgen receptor functions as transcriptional repressor of cancer-associated fibroblast activation. J Clin Investig. 2018;128(12):5531–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulet CR, Bernard G, Tremblay S, Chabaud S, Bolduc S, Pouliot F. Exosomes induce fibroblast differentiation into Cancer-associated fibroblasts through TGFβ signaling. Mol Cancer Res. 2018;16(7):1196–204. [DOI] [PubMed] [Google Scholar]
- 13.Ding S-M, Lu A-L, Zhang W, Zhou L, Xie H-Y, Zheng S-S, et al. The role of cancer-associated fibroblast MRC-5 in pancreatic cancer. J Cancer. 2018;9(3):614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen M, Xiang R, Wen Y, Xu G, Wang C, Luo S, et al. A whole-cell tumor vaccine modified to express fibroblast activation protein induces antitumor immunity against both tumor cells and cancer-associated fibroblasts. Sci Rep. 2015;5(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahn EH, Kim Y, An SS, Afzal J, Lee S, Kwak M, et al. Spatial control of adult stem cell fate using nanotopographic cues. Biomaterials. 2014;35(8):2401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim D-H, Smith RR, Kim P, Ahn EH, Kim H-N, Marbán E, et al. Nanopatterned cardiac cell patches promote stem cell niche formation and myocardial regeneration. Integr Biol. 2012;4(9):1019–33. [DOI] [PubMed] [Google Scholar]
- 17.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kshitiz G, Afzal J, Maziarz JD, Hamidzadeh A, Liang C, Erkenbrack EM, et al. Evolution of placental invasion and cancer metastasis are causally linked. Nat Ecol Evol. 2019;3(12):1743–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kshitiz DA, Afzal J, Suhail Y, Ahn EH, Goyal R, Hubbi ME, et al. Control of the interface between heterotypic cell populations reveals the mechanism of intercellular transfer of signaling proteins. Integr Biol Quant Biosci Nano Macro. 2015;7(3):364–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren Y, Jia H-H, Xu Y-Q, Zhou X, Zhao X-H, Wang Y-F, et al. Paracrine and epigenetic control of CAF-induced metastasis: the role of HOTAIR stimulated by TGF-ss1 secretion. Mol Cancer. 2018;17(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batlle E, Massague J. Transforming growth factor-beta signaling in immunity and cancer. Immunity. 2019;50(4):924–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin P, Wang W, Zhang Z, Bai Y, Gao J, Zhao C. Wnt signaling in human and mouse breast cancer: focusing on Wnt ligands, receptors and antagonists. Cancer Sci. 2018;109(11):3368–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karagiannis GS, Poutahidis T, Erdman SE, Kirsch R, Riddell RH, Diamandis EP. Cancer-associated fibroblasts drive the progression of metastasis through both paracrine and mechanical pressure on cancer tissue. Mol Cancer Res. 2012;10(11):1403–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue Z, Yuan Z, Zeng L, Wang Y, Lai L, Li J, et al. LGR4 modulates breast cancer initiation, metastasis, and cancer stem cells. FASEB J. 2018;32(5):2422–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Song Y, Ling Z, Li Y, Ren X, Yang J, et al. R-spondin 2-LGR4 system regulates growth, migration and invasion, epithelial-mesenchymal transition and stem-like properties of tongue squamous cell carcinoma via Wnt/β-catenin signaling. EBioMedicine. 2019;44:275–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan CH. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandramouli A, Simundza J, Pinderhughes A, Cowin P. Choreographing metastasis to the tune of LTBP. J Mammary Gland Biol Neoplasia. 2011;16(2):67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J, Kim D-H, Shah SR, Kim H-N, Kshitiz G, Kim P, et al. Switch-like enhancement of epithelial-mesenchymal transition by YAP through feedback regulation of WT1 and Rho-family GTPases. Nat Commun. 2019;10(1):2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol. 2012;14(8):777–83. [DOI] [PubMed] [Google Scholar]
- 30.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10(7):445–57. [DOI] [PubMed] [Google Scholar]
- 31.Herbst A, Jurinovic V, Krebs S, Thieme SE, Blum H, Göke B, et al. Comprehensive analysis of β-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/β-catenin signaling. BMC Genomics. 2014;15(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bronisz A, Godlewski J, Wallace JA, Merchant AS, Nowicki MO, Mathsyaraja H, et al. Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol. 2011;14(2):159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.del Pozo MY, Park D, Ramachandran A, Ombrato L, Calvo F, Chakravarty P, et al. Mesenchymal cancer cell-stroma crosstalk promotes niche activation, epithelial reversion, and metastatic colonization. Cell Rep. 2015;13(11):2456–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.