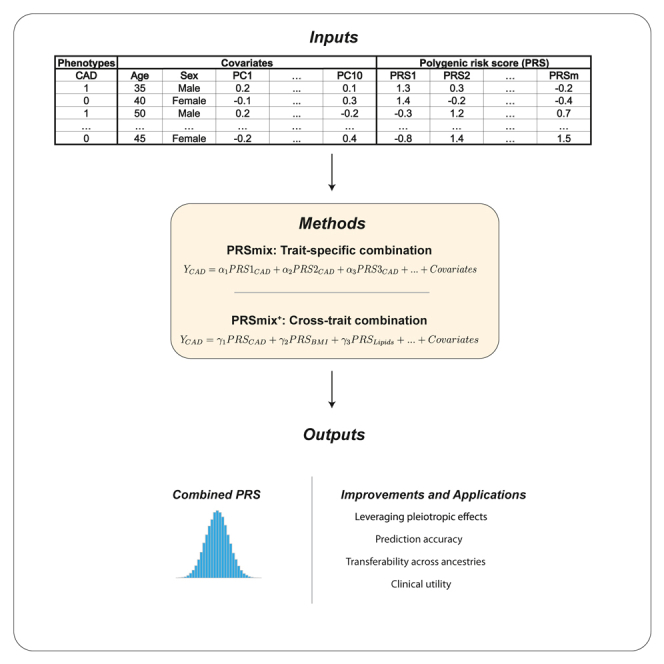
Summary
Polygenic risk scores (PRSs) are an emerging tool to predict the clinical phenotypes and outcomes of individuals. We propose PRSmix, a framework that leverages the PRS corpus of a target trait to improve prediction accuracy, and PRSmix+, which incorporates genetically correlated traits to better capture the human genetic architecture for 47 and 32 diseases/traits in European and South Asian ancestries, respectively. PRSmix demonstrated a mean prediction accuracy improvement of 1.20-fold (95% confidence interval [CI], [1.10; 1.3]; p = 9.17 × 10−5) and 1.19-fold (95% CI, [1.11; 1.27]; p = 1.92 × 10−6), and PRSmix+ improved the prediction accuracy by 1.72-fold (95% CI, [1.40; 2.04]; p = 7.58 × 10−6) and 1.42-fold (95% CI, [1.25; 1.59]; p = 8.01 × 10−7) in European and South Asian ancestries, respectively. Compared to the previously cross-trait-combination methods with scores from pre-defined correlated traits, we demonstrated that our method improved prediction accuracy for coronary artery disease up to 3.27-fold (95% CI, [2.1; 4.44]; p value after false discovery rate (FDR) correction = 2.6 × 10−4). Our method provides a comprehensive framework to benchmark and leverage the combined power of PRS for maximal performance in a desired target population.
Keywords: integrative, combination, PRS, clinical utility, cross ancestry, South Asian
Graphical abstract
Highlights
-
•
PRSmix pipeline integrates multiple PRSs for a target population
-
•
PRSmix improved PRS prediction accuracy and portability across ancestries
-
•
PRSmix does not require pre-defined traits for the combination
-
•
PRSmix outperforms previous combination methods
Polygenic risk score (PRS) demonstrated a utility to predict the clinical phenotypes and outcomes of individuals. Truong et al. developed a PRSmix pipeline to combine multiple PRSs for a target population without the need for pre-defined traits. The method demonstrated better performance compared to combination frameworks with pre-defined traits.
Introduction
Thousands of polygenic risk scores (PRSs) have been developed to predict an individual’s genetic propensity to diverse phenotypes.1 PRSs are generated when risk alleles for distinct phenotypes are weighted by their effect size estimates and summed.2 Risk alleles included in PRS have traditionally been identified from genome-wide association study (GWAS) results conducted on a training dataset, which are weighted and aggregated to derive a PRS to predict distinct phenotypes. The association between PRS and the phenotype of interest is subsequently evaluated in a test dataset that is non-overlapping with the training dataset.3
Most PRS have been developed in specific cohorts that may vary in terms of population demographics, admixture, environment, and SNP availability. Limited validation of many PRSs outside of the training datasets and poor transferability of PRSs to other populations may limit their clinical utility. However, pooling of data from individual PRSs generated and validated in diverse cohorts has the potential to improve the predictive ability of PRSs across diverse populations. The Polygenic Score (PGS) Catalog is a publicly available repository that archives SNP effect sizes for PRS estimation. The SNP effect sizes were developed from various methods (e.g., P + T,4 LDpred,5,6 PRS-CS7) to obtain the highest prediction accuracy in the studied dataset. PRS metadata enable researchers to replicate PRSs in independent cohorts and aggregate SNP effects to refine PRSs and enhance the accuracy and generalizability in broader populations.8 However, optimizing PRS performance requires methodological approaches to adjust GWAS estimate effect sizes that take into account correlated SNPs (i.e., linkage disequilibrium) and refine the PRS for the target population.4,5,7,9,10,11,12 Furthermore, numerous scores are often present for single traits with varied validation metrics in non-overlapping cohorts. There is a lack of standardized approaches combining PRSs from this growing corpus to enhance prediction accuracy and generalizability while minimizing bias for a target cohort.8,11,13 Additionally, recent studies have selected scores based on prior knowledge of clinical risk factors to the main traits.8,14,15 However, this strategy may neglect important information from other traits. Our study leverages the diversity of traits analyzed across cohorts and PRS methodologies.
To address these issues, we sought to (1) validate previously developed PRSs in two geographically and ancestrally distinct cohorts, the All of Us (AoU) Research Program and the Genes & Health (G&H) cohort, and (2) present and evaluate new methods for combining previously calculated PRSs to maximize performance beyond all best-performing published PRSs. To aggregate the genetic information across different sources, we proposed PRSmix, a framework to combine the PRS from the same trait with the outcome trait. Previous studies highlighted the effect of pleiotropic information on a trait’s genetic architecture.14,16 Therefore, we proposed PRSmix+ to additionally combine PRSs from other genetically correlated traits to further improve the PRS for a given trait.
To assess the prediction improvement, we performed PRSmix and PRSmix+ for 47 traits in European ancestry and 32 traits in South Asian ancestry. We evaluated (1) the relative improvement of the proposed framework over the best-performing pre-existing PRS for each trait; (2) the efficient training sample sizes required to improve the PRS; (3) the predictive improvement in six groups, namely anthropometrics, blood counts, cancer, cardiometabolic, biochemistry, and other conditions; and (4) the clinical utility and pleiotropic effect of the newly built PRS for coronary artery disease (CAD). Overall, we show that PRSmix and PRSmix+ significantly improved prediction accuracy. An R package for preprocessing and harmonizing the SNP effects from the PGS Catalog as well as assessing and combining the scores was developed to facilitate the combining of pre-existing PRSs for both ancestry-specific and cross-ancestry contexts using the totality of published PRSs. The development of this framework has the potential to improve precision health by improving the generalizability in the application of PRSs.17
Results
Overview of methods
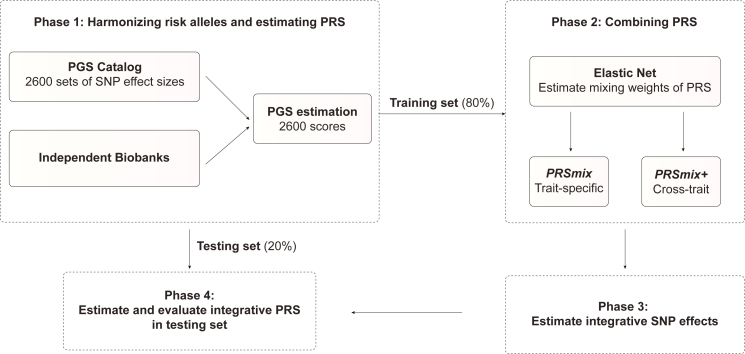
A single PRS may only reflect genetic effects captured in the discovery dataset of a single study that may be only a part of the total genetic effects underlying the trait of interest. Therefore, we harmonized and combined multiple sets of PRSs to establish a new set of scores, which gather information across studies and traits. Our approach leveraged multiple well-powered PRSs to improve prediction accuracy and is detailed in Figure 1.
Figure 1.
The framework of the trait-specific and cross-trait PRS integration
In phase 1, we obtained the SNP effects from the PGS Catalog and then harmonized the effect alleles as the alternative alleles in the independent cohorts. In each independent biobank (AoU, G&H), we estimated the PRS and split the data into training (80%) and testing (20%) datasets. In phase 2, in the training dataset, we trained the Elastic Net model with high-power scores to estimate the mixing weights for the PRSs. The training phase could include PRSs from traits corresponding to outcomes (PRSmix) or all traits (PRSmix+). The training was adjusted for age, sex, and 10 principal components (PCs). In phase 3, we adjusted the per-allele effect sizes from each single PRS by multiplying with the corresponding mixing weights obtained in the training phase. The final per-allele effect sizes are estimated as the weighted sum of the SNP effects across different single scores. In phase 4, we evaluated the re-estimated per-allele effect sizes in the testing dataset.
Our combination frameworks leveraged the PGS Catalog18 as the resource of SNP effects to estimate single PRSs. To avoid overfitting, we used AoU and G&H cohorts (see STAR Methods) due to non-overlapping samples from the original GWAS. We randomly divided the target cohort into a training set (80%) and a testing set (20%). We selected traits from the PGS Catalog that have the highest number of PRSs. For the stability of the linear combination, we curated binary traits with a prevalence >2% in the target cohort. Continuous traits were assessed using incremental R2, which is estimated as the difference between the full model of PRS and covariates (age, sex, and 10 principal components [PCs]) and the null model of only covariates. For binary traits, the prediction accuracy was converted to liability R2 with disease prevalence approximated as the prevalence in the corresponding cohort.
To combine the scores, we employed Elastic Net19 to construct linear combinations of the PRS. We proposed two combination frameworks: (1) PRSmix combines the scores developed from the same outcome trait, and (2) PRSmix+ combines all the high-power scores across other traits. Trait-specific combinations, PRSmix, can leverage the PRSs developed from different studies and methods to more fully capture the genetic effects underlying the traits. It has also been shown that complex traits are determined by genes with pleiotropic effects.14 Therefore, we additionally proposed a cross-trait combination, PRSmix+, to make use of pleiotropic effects and further improve prediction accuracy.
First, we evaluated the improvement for each method, defined as the fold ratio of the method compared to the prediction accuracy of the best single PRS. For a fair comparison with the proposed framework, we selected the best-performing PRS from the set of traits matched with the outcome trait from the training set and evaluated by incremental R2 in the testing set. First, we performed simulations to assess the improvement with various heritabilities and training sample sizes. We estimated the slope of improvement of prediction accuracy by increasing training sample sizes for various heritabilities.
Next, we applied the proposed frameworks in two distinct cohorts: (1) the AoU program, in which 47 traits were tested in US residents of European ancestry; and (2) the G&H cohort, in which 32 traits were tested in British South Asian ancestry (Table S1). In each cohort, we compared the improvement of our proposed framework with the single best score from the PGS Catalog. We estimated the averaged fold ratio as a measure of the improvement of prediction accuracy by our approach compared to the best single score from PGS Catalog. We also classified the traits into six categories as anthropometrics, blood counts, cancer, cardiometabolic, biochemistry, and other conditions (Tables S2 and S3). Cancer traits were not considered in the younger G&H cohort due to their low prevalence (<2%). We then present additional detailed analyses for CAD focused on clinical utility improvements relative to existing PRSs.
Simulations were used to evaluate the combination frameworks
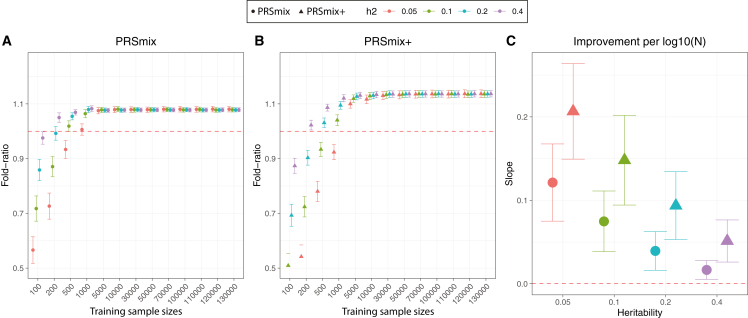
To compare the performance of PRSmix and PRSmix+ against the best single PRS and evaluate the sample sizes needed for training the mixing weights, we performed simulations with real genotypes of European ancestry in the UK Biobank given the large sample sizes available (Figure 2). Briefly, we randomly split 7,000 individuals as a testing dataset mimicking the testing size of 20% of real data. In the remaining dataset, we used 200,000 individuals for GWAS to estimate the SNP effect sizes for PRS calculations. Finally, with the rest of the data, we randomly selected different sample sizes as the training sample to evaluate the sample sizes needed to train the mixing weights. To assess the improvement of PRS performance, we computed the fold ratio of prediction accuracy R2 between PRSmix and PRSmix+ against the best-performing single simulated PRS.
Figure 2.
Simulations to demonstrate the predictive improvement of PRSmix and PRSmix+
(A and B) The points and triangles represent the mean fold ratio of R2 between (A) PRSmix and (B) PRSmix+, respectively, versus the best single PRS.
(C) The improvement per logarithm with base 10 of sample size for various heritabilities was represented as a slope of a linear regression of fold ratio ∼log10(N). In simulations, the correlation within simulated trait-specific PRSs was 0.8, and the correlation between trait-specific and correlated PRSs was 0.4 (see STAR Methods). The whiskers demonstrate CIs across 200 replications. The dashed red lines represent the reference for fold ratio equal to 1 for (A) and (B), and equal to 0 for (C).
Our results showed that the trait-specific combination, PRSmix, showed no improvement with the training sample smaller than 500 for most of the traits. Our simulations illustrated that traits with low heritability required a larger sample size to achieve an improvement compared to traits with high heritability (Figures 2A and 2B). PRSmix demonstrated a better performance compared to the best single PRS with training sample sizes from Ntraining = 200 samples for the high heritable trait (h2 = 0.4) to Ntraining = 5,000 samples for the low heritable trait (h2 = 0.05) (Figures 2A and 2B). We observed that PRSmix demonstrated a saturation of improvement from Ntraining = 5,000. PRSmix+ demonstrated negligible further improvement when the training sample size was increased from 5,000 but maintained consistent improvement relative to PRSmix and the best single PRS. Moreover, we observed that traits with higher heritability or higher best prediction accuracy of a single PRS demonstrated a smaller improvement compared to traits with a smaller heritability (Figure 2C).
Combining trait-specific PRSs improves prediction accuracy (PRSmix)
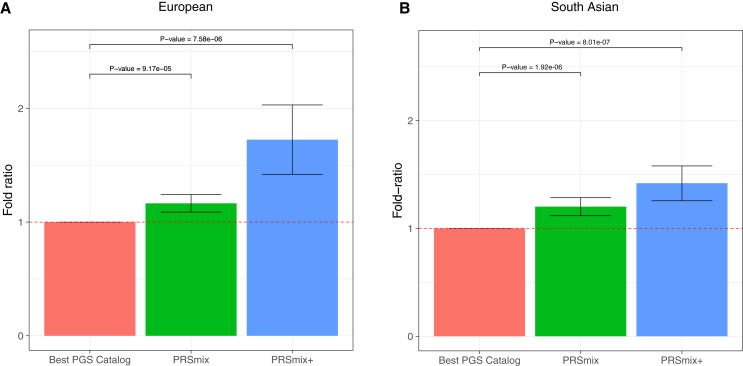
To determine if a trait-specific combination, namely PRSmix, would improve the accuracy of PRS prediction, we used whole-genome sequencing data from European ancestry participants in the AoU Research Program, and imputed G&H participants of South Asian ancestry. We randomly split the independent cohorts into training (80%) and testing sets (20%). The training set was used to train the weights of each PRS, referred as mixing weights, which indicates how much each PRS explains the phenotypic variance in the training set, and the PRS accuracies were evaluated in the testing set (Figure 1). We curated 47 traits and 32 traits in the AoU and G&H cohorts, respectively. For binary traits, we removed traits with a prevalence of smaller than 2% (see STAR Methods; Table S1). Traits with the best-performance trait-specific single PRS that showed a lack of power were also removed. Overall, we observed a significant improvement compared to one using a two-tailed paired t test with PRSmix. PRSmix significantly improves the prediction accuracy compared to the best PRS estimated from the PGS Catalog. PRSmix improved 1.20-fold (95% CI, [1.10; 1.3]; p = 9.17 × 10−5) and 1.19-fold (95% CI, [1.11; 1.27]; p = 1.92 × 10−6) compared to the best PRS from PGS Catalog for European ancestry and South Asian ancestry, respectively.
In European ancestry, we observed the greatest improvement of PRSmix against the best single PRS for rheumatoid arthritis of 3.36-fold. Furthermore, in South Asian ancestry, we observed that PRSmix of CAD had the best improvement of 2.32-fold compared to the best-performance single PRS. Details of the prediction accuracy are shown in Figures S1 and S2 and Tables S2 and S3. This was consistent with findings in simulations since traits with a lower single PRS performance demonstrated a better improvement with the combination strategy. Additionally, the number of final features with non-zero weight in the model is provided in Table S4. On average, PRSmix incorporated 10 PRSs in both European and South Asian ancestry, and PRSmix+ incorporated 69 and 32 PRSs in European and South Asian ancestry, respectively.
Cross-trait combination further improved PRS accuracy and highlighted the contribution of pleiotropic effects (PRSmix+)
We next assessed the contribution of pleiotropic effects from cross-trait PRSs to determine if these would further improve the combination framework (PRSmix+) by including high-power PRSs from within 2,600 PRSs in the PGS Catalog. To evaluate the power of PRS and improve computational efficiency, we employed the theoretic power and variance of incremental R2 for continuous traits and liability R2 for binary traits (see STAR Methods). We observed that PRSmix+ further improved the prediction accuracy compared to the best PGS Catalog in European ancestry (Figure 3A) and South Asian ancestry (Figure 3B). We observed an improvement of 1.72-fold (95% CI, [1.40; 2.04]; p = 7.58 × 10−6) and 1.42-fold (95% CI, [1.25; 1.59]; p = 8.01 × 10−7) higher compared to the best PGS Catalog for European ancestry and South Asian ancestry, respectively. PRSmix+ significantly improved the prediction accuracy compared to PRSmix in both European and South Asian ancestry with 1.46-fold (95% CI, [1.17; 1.75]; p = 0.002) and 1.19-fold (95% CI, [1.07; 1.32]; p = 0.001), respectively (Figure S3). PRSmix yielded an equivalent number of non-zero mixing weights between European ancestry (median = 8; interquartile range [IQR] = [5; 12]) and South Asian ancestry (median = 8; IQR = [3; 14]). However, PRSmix+ demonstrated a higher number of non-zero weights in European ancestry (median = 55; IQR = [30; 76]) compared to South Asian ancestry (median = 32; IQR = [11; 49]). The median absolute mixing weights were similar between European ancestry and South Asian ancestry (Figure S4). We note that most of the best PRSs across traits were developed from 2021 onward. The details of the most recent trait-specific PRSs, PRSs with largest sample sizes, best PRSs being compared, and score with highest weights for European ancestry and South Asian ancestry are provided in Tables S5 and S6, respectively.
Figure 3.
Comparison of PRSmix and PRSmix+ versus the best PGS Catalog in European and South Asian ancestries
The relative improvement compared to the best single PRS was assessed in (A) the European ancestry in the AoU cohort and (B) South Asian ancestry in the G&H cohort. PRSmix combines trait-specific PRSs and PRSmix+ combines additional PRSs from other traits. The best PGS Catalog score was selected by the best-performance trait-specific score in the training sample and evaluated in the testing sample. The prediction accuracy (R2) was calculated as incremental R2, which is a difference of R2 between the model with PRS and covariates including age, sex, and 10 PCs versus the base model with only covariates. Prediction accuracy for binary traits was assessed with liability-R2 where disease prevalence was approximately estimated as a proportion of cases in the testing set. The bars represent the ratio of prediction accuracy of PRSmix and PRSmix+ versus the best PRS from the PGS Catalog across 47 traits and 32 traits in AoU and G&H cohorts, respectively, and the whiskers demonstrate 95% CIs. p values for significance difference of the fold ratio from 1 using a two-tailed paired t test.
Consistent with our simulation results, a smaller improvement was observed for traits with a higher baseline prediction accuracy from PGS Catalog (Figure S5), noting that the baseline prediction accuracy depends on the heritability and genetic architecture (i.e., polygenicity). In contrast, more improvement was observed for traits with lower heritability, thus lower prediction accuracy, when comparing the single best PRS (Figure 1C).
We next transferred the PRS weights trained in European ancestry on South Asian ancestry to assess the transferability of the linear combination across ancestries, and we observed a better performance compared to the best score from PGS Catalog for most of the traits and a slightly lower performance for A1c, asthma, platelet, rheumatoid arthritis, red cell distribution width (RDW), and triglyceride (Table S7). However, performing a linear combination with the matched ancestry still demonstrated a better performance than using transferred weights.
We also compared PRSmix and PRSmix+ using all scores or only ancestry-matched scores from PGS Catalog (Figure S6). We observed that combining using all scores improved the prediction accuracy better than combining only ancestry-matched scores to the target population. With PRSmix, using all scores improved the prediction accuracy 1.2-fold (95% CI, [1.09; 1.31]; p = 0.0002) compared to using ancestry-matched scores. With PRSmix+, using all scores improved the prediction accuracy 1.12-fold (95% CI, [1.07; 1.18]; p = 2.14 × 10−5) compared to using ancestry-matched scores (Figure S6).
Prediction accuracy and predictive improvement across various types of traits
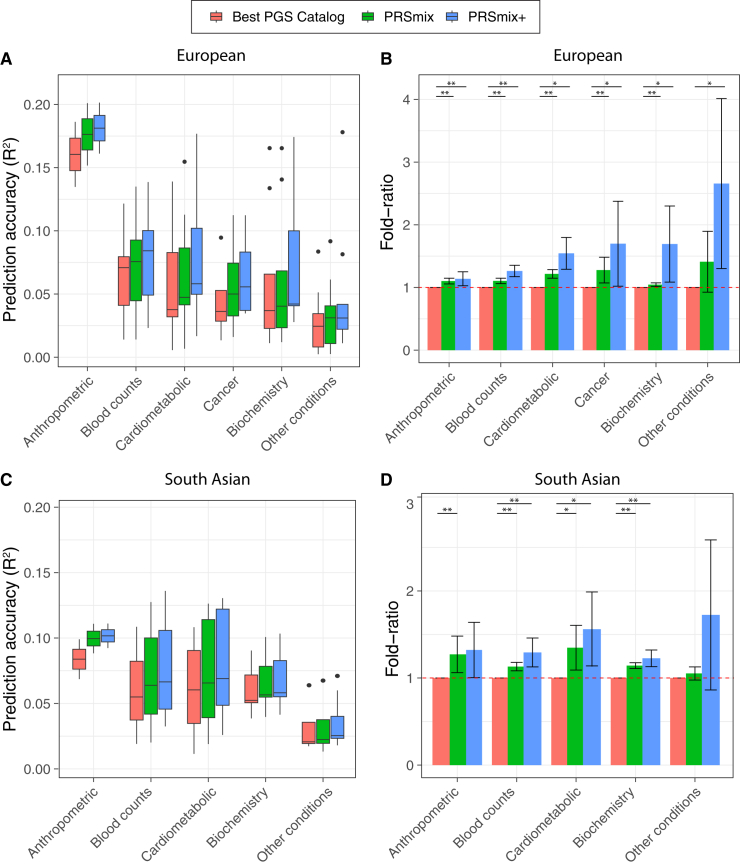
We next compared PRSmix and PRSmix+ with the best PRS estimated from the PGS Catalog across six categories, including anthropometrics, blood counts, cancer, cardiometabolic, biochemistry, and other conditions (see STAR Methods). PRSmix demonstrates a higher prediction accuracy across all types of traits in both European and South Asian ancestries (Figure 4). We observed a similar trend in the predictive performance of PRSmix+ across different types of traits. In European ancestry, the smallest improvement with PRSmix+ was in anthropometric traits of 1.14-fold (95% CI, [1.03; 1.25]; p = 0.01) while other conditions (including depression, asthma, migraine, current smoker, hypothyroid, osteoporosis, glaucoma, rheumatoid arthritis, and gout) obtained the highest mean predictive improvement but also with high variance of 2.66-fold (95% CI, [1.30; 4.01]; p = 0.01) (Table S8). In South Asian ancestry, the mean predictive improvement was highest in the other conditions (including asthma, migraine, current smoker, and rheumatoid arthritis) type at 2.10-fold (95% CI, [0.787; 3.405]; p = 0.1). Biochemistry demonstrated the smallest improvement of 1.23-fold (95% CI, [1.15; 1.31]; p = 5.8 × 10−9). We note that PRSmix and PRSmix+ improve prediction accuracy for all traits (Tables S2 and S3). The large variance could be due to the wide range of improvement and the small number of traits in each subtype.
Figure 4.
Prediction accuracy and improvement across various types of traits in the European and South Asian ancestry
We classified the traits into six main categories for European ancestry in the AoU cohort and five categories for South Asian ancestry in the G&H cohort due to the low prevalence of cancer traits in G&H. The prediction accuracies (A and C) are estimated as incremental R2 and liability R2 for continuous traits and binary traits, respectively. The relative improvements (B and D) are estimated as the fold ratio between the prediction accuracies of PRSmix and PRSmix+ against the best PGS Catalog. The order on the axis followed the decrease in the prediction accuracy of PRSmix+. The boxplots in (A) and (C) show the first to the third quartile of prediction accuracies for 47 traits and 32 traits in European and South Asian ancestries, respectively. The whiskers reflect the maximum and minimum values within the 1.5 × IQR for each group. The bars in (B) and (D) represent the mean prediction accuracy across the traits in that group and the whiskers demonstrate 95% CIs. The red dashed lines in (B) and (D) represent the ratio equal to 1 as a reference for comparison with the best PGS Catalog score. The asterisks indicate p values: ∗p < 0.05 and ∗∗p < 0.05/number of traits in each type with a two-tailed paired t test.
Comparison with previous combination methods
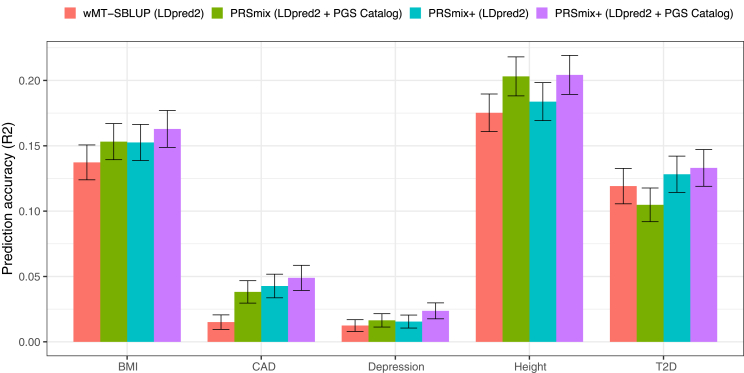
There have been several studies proposed to incorporate multiple traits to improve prediction accuracy of the target trait.8,15,20 For example, the weighted index for multi-trait summary statistics best linear unbiased predictor (wMT-SBLUP)15 created a weighted index for correlated PRSs and required the input sample sizes, genetic correlation, and heritability across all pairs of traits from GWAS summary statistics to be determined. Krapohl et al.20 and Albiñana et al.13 combined PRSs using scores estimated from LDpred2.5 Here, we benchmarked PRSmix and PRSmix+ against wMT-SBLUP using summary statistics and a combination of PRSs developed by LDpred2 with a pre-defined set of correlated traits to the main outcomes and an extension of scores generated by different methods from PGS Catalog (Figure 5).
Figure 5.
Benchmarking previous methods with PRSmix and PRSmix+
LDpred2-auto was used as the baseline method to input in the methods. Five traits from Maier et al.15 and 26 publicly available GWASs for European ancestry were curated. The components of each combination method are denoted in parentheses. wMT-SBLUP was conducted with the input of sample sizes from the GWAS summary statistics and heritabilities and genetic correlation between all pairs of traits using LD score regression. PRSmix (LDpred2 + PGS Catalog) combined target trait-specific scores within 26 scores and PGS Catalog. Elastic Net (LDpred2) was performed using Elastic Net with all scores from 26 traits generated with LDpred2-auto. PRSmix+ (LDpred2 + PGS Catalog) was conducted using 26 scores from LDpred2-auto and scores from all traits obtained from PGS Catalog. Incremental R2 and liability R2 were used for continuous traits and binary traits, respectively. The whiskers demonstrate 95% CIs of mean prediction accuracy. CAD, coronary artery disease; T2D, type 2 diabetes.
We first observed that integrating scores by Elastic Net with scores from pre-defined traits improved prediction accuracy compared to wMT-SBLUP ranging between 1.08-fold (95% CI, [1.03; 1.12]; p value after false discovery rate (FDR) correction = 0.36) for type 2 diabetes (T2D) and 2.87-fold (95% CI, [1.58; 4.15]; p = 0.006) for CAD (Tables S9 and S10). PRSmix demonstrated a similar performance with a combination of LDpred2 scores for BMI, CAD, and depression. PRSmix+, with scores from both pre-defined traits and PGS Catalog, demonstrated a consistent boost in prediction accuracy compared to wMT-SBLUP between 1.12-fold (95% CI, [1.02; 1.21]; p = 0.016) for T2D and 3.27-fold (95% CI, [2.1; 4.44]; p = 2.6 × 10−4) for CAD. PRSmix+ equipped with both LDpred2-auto and PGS Catalog scores also outperformed the Elastic Net combination of LDpred2 scores best observed with 1.6-fold (95% CI, [1.31; 1.89]; p = 1.1 × 10−4) for depression. Interestingly, height, a highly polygenic trait,21 similarly demonstrated the best performance under a trait-specific combination (PRSmix with trait-specific LDpred2-auto and PGS Catalog scores) and PRSmix+ equipped with both LDpred2-auto and PGS Catalog scores (Figure 5). Employing pleiotropic effects only provided a small improvement with height (Table S10). On the other hand, T2D demonstrated that all methods of cross-trait combinations provided a significant improvement over the trait-specific combination (Figure 5).
Clinical utility for CAD
To evaluate the utility of the proposed methods, we assessed the PRSmix and PRSmix+ for CAD, which is the leading cause of disability and premature death among adults.22,23,24 The single best CAD PRSs (PRSCAD) s from the PGS Catalog in the training set were from Koyama et al.25 and Tamlander et al.26 in European and South Asian ancestries, respectively (Figure S7 and S8). In the testing set, liability R2 with Koyama et al. for European ancestry was 0.03 (95% CI, [0.03; 0.04]; p < 2 × 10−16) and with Tamlander et al. for South Asian ancestry was 0.006 (95% CI, [0.003; 0.009]; p = 2.39 × 10−4) (Figure 6).
Figure 6.
Comparison of prediction accuracies with PRSmix, PRSmix+, and CAD PRS from PGS Catalog
PRSmix was computed as a linear combination of CAD PRS and PRSmix+ was computed as a linear combination of all significant PRSs obtained from the PGS Catalog. The PRSs were evaluated in the testing set with liability R2 in the (A) European ancestry from the AoU cohort and (B) South Asian ancestry from the G&H cohort. The bars indicate the mean prediction accuracy and the whiskers show 95% CIs.
Subsequently, we assessed the clinical utility of the integrative model with PRS and established QRISK3, clinical risk factors, including age, sex, total cholesterol, high-density lipoprotein cholesterol (HDL-C), systolic blood pressure, BMI, T2D, and current smoking status versus the traditional model with clinical risk factors (Figure 7; Table S11). In European ancestry, the CAD PRSmix+ integrative score improved the continuous net reclassification of 33% (95% CI, [22%; 44%]; p = 4.15 × 10−9) compared to PRSmix (30%; 95% CI, [20%; 44%]; p = 1.4 × 10−10) and the best PRS from the PGS Catalog (24%; 95% CI, [13%; 36%]; p = 2.05 × 10−5). In South Asian ancestry, the integrated score with PRSmix+ showed significant continuous net reclassification of 27% (95% CI, [16%; 39%]; p = 3.69 × 10−6) compared to PRSmix (23%; 95% CI, [11%; 34%]; p = 6.56 × 10−5) and the best PGS Catalog (11%; 95% CI, [−0.3%; 23%]; p = 0.05). Our results also demonstrated an improvement in net reclassification for models without clinical risk factors.
Figure 7.
Net reclassification improvement for CAD with the addition of PRSs to the baseline model in European and South Asian ancestries
The baseline model for risk prediction includes QRISK3, age, sex, total cholesterol, HDL-C, systolic blood pressure, BMI, T2D, and current smoking status. We compared the integrative models with PGS Catalog, PRSmix, and PRSmix+ in addition to clinical risk factors versus the baseline model with only factors. The points indicate the mean estimate for continuous net reclassification improvement (NRI) and the whiskers indicate 95% CIs estimated from 500 bootstraps. HDL-C, high-density lipoprotein cholesterol.
We assessed the incremental area under the curve (AUC) between the full model of PRS and covariates and the null model with only covariates (Table S12). PRSmix+ demonstrated an incremental AUC of 0.02 (95% CI, [0.018; 0.02]; p < 2.2 × 10−16) and 0.008 (95% CI, [0.007; 0.009]; p < 2.2 × 10−16) in European and South Asian ancestries, respectively. PRSmix obtained an incremental AUC of 0.016 (95% CI, [0.016; 0.017]; p < 2.2 × 10−16) and 0.006 (95% CI, [0.005; 0.007]; p < 2.2 × 10−16) in European and South Asian ancestries, respectively. The best PGS Catalog had the smallest incremental AUC of 0.012 (95% CI, [0.011; 0.013]; p < 2.2 × 10−16) and 0.003 (95% CI, [0.002; 0.003]; p < 2.2 × 10−16) in European and South Asian ancestries, respectively.
We also compared the risks for individuals in the top decile versus the remaining population (Table S13). For European ancestry, an increased risk with odds ratio (OR) per 1 SD of the best PGS Catalog, PRSmix, and PRSmix+ were 1.43 (95% CI, [1.30–1.57]; p < 2.2 × 10−16), 1.60 (95% CI, [1.45–1.76]; p < 2.2 × 10−16), and 1.74 (95% CI = [1.58; 1.91]; p < 2.2 × 10−16), respectively. The top decile of PRSmix+ compared to the remaining population demonstrated an increased risk of OR = 2.53 (95% CI, [1.96; 3.25]; p = 8.64 × 10−13). The top decile for the best PGS Catalog versus the remainder was OR = 1.67 (95% CI, [1.27; 2.19]; p = 2 × 10−4). For South Asian ancestry, an increased risk with OR per 1 SD of the best PGS Catalog, PRSmix, and PRSmix+ was 1.24 (95% CI, [1.13; 1.37]; p < 1.52 × 10−16), 1.39 (95% CI, [1.33; 1.46]; p < 2.2 × 10−16), 1.40 (95% CI, [1.27; 1.55]; p < 2.2 × 10−16), and 1.50 (95% CI = [1.36; 1.66]; p < 2.2 × 10−16), respectively. In South Asian ancestry, PRSmix+ demonstrated an OR of 2.34 (95% CI, [1.79; 3.05]; p = 4.22 × 10−10), and, with the best PGS Catalog, OR was 1.73 (95% CI, [1.30; 2.28]; p = 1.31 × 10−4) for the top decile versus the remaining population.
Moreover, we observed that there was a modest improvement for PRSmix from the training size of 5,000 in both European and South Asian ancestries (Figure S9), which aligned with our simulations (Figures 2A and 2B). Our results demonstrated the generalization of our combination methods across diverse ancestries to improve prediction accuracy. To obtain maximized prediction accuracy in real data, SNP heritability, polygenicity, and heterogeneity in the definition of the disease may influence the sample sizes need for combination. However, our simulation highlighted the most important information, including heritability and the genetic correlation between traits to combine. Improvements from the combination benefit from imposing the penalty by the ElasticNet on unimportant features across multiple scores in the combination. With PRSmix+, our empirical result with CAD showed that there was a modest improvement with training sample sizes larger than 5,000.
Finally, we conducted phenome-wide association studies (PheWASs) in AoU between PRSCAD with 1,815 phecodes to compare the pleiotropy of PRS and assess the relationship between CAD PRS and disease phenotypes given the inherent use of pleiotropy in development (Table S14). As expected, PRSmix+ had a stronger association for coronary atherosclerosis relative to the single best PRS from the PGS Catalog. PRSmix+ associations with cardiometabolic risk factors were significantly greater with averaged fold ratio = 1.10 (95% CI, [1.09–1.12]; p value with paired t test = 1.07 × 10−25) and 1.07 (95% CI, [1.05–1.081]; p = 4.8 × 10−13) for circulatory system and endocrine/metabolic system (Table S15). The PheWAS result for PRSmix+ aligned with the list of traits from the selected PRS (Table S14).
Discussion
In this paper, we propose a trait-specific framework (PRSmix), and cross-trait framework (PRSmix+) to leverage the combined power of existing scores. We performed and evaluated our method using the AoU and G&H cohorts showcasing a framework to develop the optimal PRS for a given trait in a target population leveraging all existing PRSs. Across 47 traits in the AoU cohort and 32 traits in the G&H cohort with either continuous traits or binary traits with prevalence >2%, we demonstrated substantial improvement in average prediction R2 by using a linear combination with Elastic Net with 5-fold cross-validation. The empiric observations are concordant with simulations. To our knowledge, there has been a number of emerging studies to combine PRS, but there is a limited number of frameworks that comprehensively evaluate, harmonize, and leverage the combination of these scores.8,27,28 Our studies permit several conclusions for the development, implementation, and transferability of PRS.
First, externally derived and validated PRSs are generally not the optimal PRS for a given cohort. Consistent with other risk predictors, recalibration within the ultimate target population improves performance.29 By leveraging the PGS Catalog, our work carefully harmonizes the risk alleles to estimate PRS across all scores and provides newly estimated per-allele SNP effects (provided to the PGS Catalog) to assist the interpretability of the models.
Second, previous studies selected an arbitrary training sample size to estimate the mixing weights, which may lead to a poor power of the combination frameworks and inaccurate estimate of sampling variance.10 We assessed the expected sample sizes to estimate the mixing weights via simulations and real data. Our results demonstrated that, while low heritability traits benefit the most, they require a greater training sample size.
Third, we leveraged all PRSs, including those not trained on the primary trait, to systematically optimize PRS for a target cohort. We showed that PRSmix improved the prediction by combining the scores matching the outcome trait. In addition, we showed that PRSmix+ was able to leverage the power of cross-traits, which highlighted the contribution of pleiotropic effects to enhance PRS performance. We leverage prior work demonstrating the effects of pleiotropy on complex traits.14,30,31 Our results demonstrated that South Asian ancestry required an equivalent number of trait-specific scores to combine similar to the European ancestry. Since the majority of scores in PGS Catalog were developed on European ancestry, PRSmix+ incorporated a higher number of PRSs in the European ancestry compared to the South Asian ancestry.
Fourth, we showed that using a linear combination in a matched ancestry still demonstrated a better performance than using transferred pre-trained weights from another ancestry. This indicates that methods that attempt to re-adjust PRS weights cross-ancestry are less effective for prediction than directly getting the weights from the matched ancestry. We showed that providing the linear combination model with all scores from all ancestries demonstrated a better predictive accuracy than using only ancestry-matched PRSs to the outcome trait. Additionally, the variability between the two cohorts would decrease prediction accuracy. There might be more contributing factors that influence the prediction accuracy, such as sample sizes of the PGS panel and polygenicity of the traits,6,32 ancestral consistency between discovery GWAS and linkage disequilibrium (LD) reference panels in PRS methods,33 ancestry proportions in the discovery GWAS,33 and cohort-specific contexts.34
Fifth, we demonstrated that our method outperformed previous methods combining scores. We showed that PRSmix+ outperformed wMT-SBLUP15 using a limited number of correlated traits. wMT-SBLUP required GWAS’s sample sizes, heritability, and genetic correlation between all traits. LDpred2-auto required GWAS summary statistics and initialized heritability and proportion of causal SNPs. Krapohl et al.20 and Abraham et al.8 proposed to use Elastic Net to combine the scores developed from summary statistics, and correlated traits were selected with prior knowledge. However, these strategies consider scores developed from particular methods using pre-defined summary statistics. Our framework utilizes all PRSs available in the PGS Catalog, which were optimized for their target traits. Additional summary statistics and PRSs could be added to further enhance the models. We let our Elastic Net model penalize the component PRSs without the need for prior knowledge. Elastic Net can select PRSs to include and efficiently handle multi-collinearity.35,36,37 Furthermore, PRSmix and PRSmix+ only required a set of SNP effects to estimate the PRSs and estimated the prediction accuracy to the target trait to select the best scores for the combination. Additionally, compared to the preselected traits for stroke by Abraham et al.,8 we also observed that our method could identify more related risk factors to include compared to previous work conducted on stroke such as usual walking pace, arthropathies, and lipoprotein(a) (Figure S10; Tables S16 and S17). Therefore, our method is more comprehensive in an unbiased way in terms of choosing the risk factors and traits to include with empirically improved performance. We demonstrated that T2D has a greater prediction accuracy when incorporating information from multiple traits. T2D is a common highly polygenic condition correlated with other cardiometabolic risk factors as well as social/lifestyle factors. Previous PRSs for T2D may not closely consider pleiotropic effects from correlated traits to improve PRS for T2D. PRS for T2D could still have room for improvements by further incorporating genetically correlated traits in the future. Furthermore, with a limited pre-defined list of correlated traits, we showed that cross-trait combination may give a similar performance to combining trait-specific scores in PGS Catalog (BMI, CAD, depression in Figure 5). Across different traits, we demonstrated that PRSmix+ and PRSmix has an overall better power compared to other methods. Cross-trait combination for height does not significantly improve prediction accuracy, whereas T2D demonstrated a higher accuracy with any cross-trait combination methods. Intuitively, height has been known as a well-established highly polygenic trait thanks to its enormous sample sizes in a recent study21 with well-powered scores from the PGS Catalog.
Sixth, greater performance is observed even for non-European ancestry groups underrepresented in GWAS and PRS studies. We empirically demonstrate the value of training and incorporating pleiotropy with all available PRSs to improve performance, including multiple metrics of clinical utility for CAD prediction in multiple ancestries. In South Asian ancestry, we observed that PRSmix and PRSmix+ demonstrated a significant improvement with the best improvement for CAD. Of note for CAD, the relative improvements in South Asian ancestry were higher than in European ancestry for PRSmix and equivalent for PRSmix+. Transferability of PRS has been shown to improve the clinical utility of PRS in non-European ancestry.17,38 Although the prediction accuracy for South Asian ancestry is still limited, our results highlighted the transferability of predictive improvement with PRSmix and PRSmix+ to South Asian ancestry. We anticipate that ongoing and future efforts to improve our understanding of the genetic architecture in non-European ancestries will further improve the transferability of PRS across ancestry.
Last, traits with low heritability or generally low-performing single PRS benefit the most from this approach, especially with PRSmix+, such as migraine in both European and South Asian ancestries. Additionally, our results showed that pleiotropic effects play an important role in understanding and improving prediction accuracies of complex traits. However, anthropometric traits, which are highly polygenic39 and have good predictive performance using the best PGS Catalog, also showed improvement with the combination framework in both European and South Asian ancestries.
Given that PRSmix+ outperformed PRSmix, one might consider whether there is a reason to use PRSmix instead of PRSmix+. We observed that, in cases of highly heritable traits or high performance with a single PRS, there was only marginal improvement of PRSmix+ over PRSmix. In this scenario, PRSmix could provide similar predictive performance while being less time consuming because trait-specific PRS inputs only are required. However, for traits with lower heritability, PRSmix+ shows a marked improvement over PRSmix and would be preferred. Wang et al.40 showed that the theoretical prediction accuracy of the target trait using the PRS from the correlated trait is a function of genetic correlation, heritability, number of genetic variants, and sample size. Future directions could include defining the minimum parameters required for the performance of the PRSmix+ model to improve on single trait-specific PRS.
In conclusion, our framework demonstrates that leveraging different PRSs either trait specific or cross-trait can substantially improve model stability and prediction accuracy beyond all existing PRSs for a target population. Importantly, we provide software to achieve this goal in independent cohorts.
Limitations of the study
Our work has several limitations. First, the majority of scores from PGS Catalog were developed in European ancestry populations. Further non-European SNP effects will likely improve the single PRS power, which may, in turn, also improve the prediction accuracy of our proposed methods. Second, the Elastic Net makes a strong assumption that the outcome trait depends on a linear association with the PRS and covariates. However, a recent study demonstrated that there is no statistical significance difference between linear and non-linear combinations for neuropsychiatric diseases.27 Third, we estimated the mixing weights for each single SNP as a mixing weight of the PRS. Future studies could consider linkage disequilibrium between the SNPs and functional annotations of each SNP. Fourth, our frameworks were conducted on binary traits with a prevalence >2%. Additional combination PRS models are emerging that seek to use pre-existing genotypic data from genetically related, but low-prevalence, conditions to improve the prediction accuracy of rare conditions.27 Fifth, the baseline demographic characteristics (i.e., age, sex, social economic status) in the target cohort might limit the validation and transferability of PRS.41 Although these factors were considered by using a subset of the target cohorts as training data, it is necessary to have PRS developed on similar baseline characteristics. Last, with the expanding of all biobanks, there might be no perfect distinction between the samples deriving PRS and the testing cohort, and future studies may consider the potential intersection of samples to train the linear combination.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Data | ||
| UK Biobank | Bycroft et al.42 | https://www.ukbiobank.ac.uk/ |
| All of Us Research Program | National Institute of Health | https://allofus.nih.gov/ |
| PGS Catalog | PGS Catalog | https://www.pgscatalog.org/ |
| Methods | ||
| R version 4.0.0 | v.4.0.0 | https://www.r-project.org/about.html |
| PRSmix (at publication) | v.1.0.0 | Zenodo: https://doi.org/10.5281/zenodo.10581592 Github: https://github.com/buutrg/PRSmix |
Resource availability
Lead contact
Further information and requests for resources should be directed to the lead contact, Pradeep Natarajan (pnatarajan@mgh.harvard.edu).
Materials availability
The study did not generate any new reagents or materials.
Data and code availability
-
•
UK Biobank individual-level data are available for request through the UK Biobank with application (https://www.ukbiobank.ac.uk). The All of Us individual-level data are available for request through the All of Us platform (https://www.researchallofus.org). The All of Us and Genes & Health individual-level data is a controlled access dataset and may be granted at https://www.researchallofus.org/ and https://www.genesandhealth.org/, respectively. Data can be accessed through the secure All of Us Researcher Workbench platform, which is a cloud-based analytic platform that was built on the Terra platform.57 Researchers gain access to the platform after they complete a 3-step process including registration, completion of ethics training, and attesting to a data use agreement attestation.58 All of Us uses a tiered approach based on what genomic data is accessible through the Controlled Tier, and includes both whole genome sequencing (WGS), genotyping array variant data in multiple formats, as well as variant annotations, access to computed ancestry, and quality reports.59 This study includes data on the 98,600 participants with (WGS) data in the All of Us v6 Curated Data Repository release. Participant data in this data release was collected between May 6, 2018 and April 1, 2021. This project is registered in the All of Us program under the workspace name “Polygenic risk score across diverse ancestries and biobanks. The code is provided at https://github.com/buutrg/PRSmix. The version at the publication is at https://doi.org/10.5281/zenodo.10581592. The published article includes all other data generated or analyzed during this study.
-
•
Individual-level genomic data and longitudinal phenotypic data from the UK Biobank, a large-scale population-based cohort with genotype and phenotype data in approximately 500,000 volunteer participants recruited from 2006 to 2010 was used. Baseline assessments were conducted at 22 assessment centers across the UK using touch screen questionnaire, computer assisted verbal interview, physical tests, and sample collection including for DNA (https://www.ukbiobank.ac.uk). The UK Biobank Application is 7089.
-
•
Polygenic risk scores were obtained from the Polygenic Score (PGS) Catalog,18 which is a publicly accessible resource cataloging published PRS, including the metadata. The metadata provides information describing the computational algorithms used to generate the score, and performance metrics to evaluate a PRS.18 At the time of this study, over 2,600 PRS were cataloged in the PGS Catalog (version July 18, 2022) designed to predict 538 distinct traits. The PGS Catalog is freely available at https://www.pgscatalog.org/. Our new scores are deposited in the PGS Catalog.
-
•
The weights from the PRSmix and PRSmix+ scores in this manuscript have been returned to the PGS Catalog. The R package to implement PRSmix and PRSmix+ in independent datasets is at https://github.com/buutrg/PRSmix.
Experimental model and study participant details
The All of Us Research Program is a longitudinal cohort continuously enrolling (starting May 2017) U.S. adults ages 18 years and older from across the United States, with an emphasis on promoting inclusion of diverse populations traditionally underrepresented in biomedical research, including gender and sexual minorities, racial and ethnic minorities, and participants with low levels of income and educational attainment.43 Participants in the program can opt-in to providing self-reported data, linking electronic health record data, and providing physical measurement and biospecimen data.44 Details about the All of Us study goals and protocols, including survey instrument development,45 participant recruitment, data collection, and data linkage and curation were previously described in detail.44,46
The Genes & Health biobank
Genes & Health is a community-based genetics study enrolling British South Asian, with an emphasis on British Bangladeshi (two-thirds) and British Pakistani (remaining) people, with a goal of recruiting at least 100,000 participants. Currently, over 52,000 participants have enrolled since 2015. All participants have consented for lifelong electronic health record access and genetic analysis. The study was approved by the London South East National Research Ethics Service Committee of the Health Research Authority. 97.4% of participants in Genes & Health are in the lowest two quintiles of the Index of Multiple Deprivation in the United Kingdom. The cohort is broadly representative of the background population with regard to age, but slightly over-sampled with females and those with medical problems since two-thirds of people were recruited in healthcare settings such as General Practitioner surgeries.47
A linear combination of scores
We proposed PRSmix to combine PRS of outcome traits and PRSmix+ to combine high-power PRS (defined in the following subsection) from all traits obtained from PGS Catalog. The linear combination was conducted by using an Elastic Net algorithm from the “glmnet” R package48 (version 4.1) to combine the estimated PRS. First, we randomly split the independent cohorts into 80% of training and 20% testing. The PRS in the training set was standardized with mean 0 and variance 1. Before conducting linear combination, we first evaluated the performance of each individual PRS by their power and pP-value (see below). Summary-based methods to estimate genetic correlation between traits require full GWAS summary statistics including marginal effect sizes and standard error whereas the PGS Catalog only provides the adjusted SNP effect sizes from various PRS methods. We employed the predictive R2 to estimate variance explained of PRS for the outcome trait to select the secondary traits. PRS demonstrating theoretically significant predictive R2 and high power were selected for combination. We selected high-power scores defined as power >0.95 with pP-value ≤ 0.05 for the combination with Elastic Net.
An Elastic Net algorithm was used with 5-fold cross-validation and default parameters to estimate the mixing weights of each PRS. The mixing weights were then divided by the corresponding original standard deviation of the PRS in the training set.
Where and is the mixing weight estimated from the Elastic Net and standard deviation of PRSi in the training set, respectively. is the adjusted mixing weight for PRSi. To derive the per-allele effect sizes from the combination framework, we multiplied the SNP effects with the corresponding adjusted mixing weights:
Where is the adjusted effect size of SNPj and is the original effect sizes of SNPj in PRSi. We set if SNPj is not in PRSi. The adjusted effect sizes were then utilized to calculate the final PRS.
The mixing weights for PGS Catalog scores for PRSmix and PRSmix+ in European ancestry are provided in Tables S16 and S17, respectively. For South Asian ancestry, the mixing weights for PRSmix and PRSmix+ in European ancestry are provided in Tables S18 and S19, respectively.
Power and variance of PRS accuracy
The prediction accuracy (R2) was calculated as incremental R2 which is a difference of R2 between the model with PRS and covariates including age, sex, and 10 PCs versus the base model with only covariates. Incremental R2 indicates the difference between the full model and the covariate-only model which isolated the explanatory power of PRS.2 Prediction accuracy for binary traits was assessed with liability R2 where disease prevalence was approximately estimated as a proportion of cases in the testing set.
We selected high-power PRS to conduct the combination by assessing the power and variance of prediction accuracy. The power of PRS can be estimated based on the power of the two-tailed test of association as follow3,49:
| (Equation 1) |
where is the Chi-squared distribution function, is the significance level, and is the non-centrality parameter which can be estimated as
| (Equation 2) |
where N, is the sample size and estimated prediction accuracy in the testing set, respectively. can be estimated as incremental or liability for continuous traits and binary traits, respectively. Briefly, incremental compared the difference in goodness-of-fit between a full model with PRS and covariates including age, sex, and first 10 PCs, and a null model with only covariates. Additionally, for binary traits, liability was estimated with the disease prevalence approximated as the prevalence in the samples. The theoretical variance and standard error of can be estimated as follow50,51,52:
| (Equation 3) |
Therefore, we can analytically estimate the confidence interval of prediction accuracy for each of the score.
To compare the improvement, for instance between PRSmix and the best PGS Catalog, we estimate the mean fold-ratio of R2 across different traits with its 95% confidence interval and evaluated the significance difference from 1 using a two-tailed paired t-test.
Simulations
We used UK Biobank European ancestry to conduct simulations for trait-specific and cross-trait combinations. Overall, we simulated 4 traits with heritability equal to 0.05, 0.1, 0.2, and 0.4. We randomly selected M = 1000 causal SNPs among 1.1 million HapMap3 variants with INFO >0.6, MAF >0.01 and pP-value Hardy-Weinberg equilibrium >10−7. We removed individuals with PC1 and PC2 > 3 standard deviation from the mean to remove outliers of the inferred genetic ancestry. We randomly remove one in a pair of related individuals with closer than 2nd degree. The genetic components were simulated as a linear combination of 6 PRSs where PRS1, PRS2, and PRS3 were considered trait-specific scores with genetic correlations equal to 0.8. PRS4, PRS5 and PRS6 are simulated as cross-trait scores with genetic correlation equal to 0.4. The SNP effects for PRSs are simulated by a multivariate normal distribution MVN(0, ) where is the covariance matrix between PRSs. The main diagonal contains the heritability of the traits as and the covariance between PRSs are simulated as where is the genetic correlation between PRSs (0.8 for trait-specific scores and 0.4 for cross-trait scores). The PRSs of the outcome are estimated by the weighted combination of PRS where the weights follow U(0,1). 7 phenotypes were simulated as where g is PRS and e is the residuals.
We split the simulated cohort into 3 data sets for: 1) GWAS (200,000 individuals – 60%) 2) training set (130,000 individuals – 38%): training the mixing weights with a linear combination and 3) testing set (7000 individuals – 2%): testing the combined PRS. We incorporated PRS1, PRS2 and PRS3 to assess the trait-specific PRSmix framework. We combined all 6 single PRS to evaluate the cross-trait PRSmix+ framework. We compared the fold-ratio of the R2 of the combined PRS to the R2 of best single PRS to assess the improvement of the combination strategy. To evaluate the improvement across different heritabilities, we estimated the slope of improvement per log10(N) increase of training sample sizes on the fold-ratio of predictive improvement.
Sample and genotyping quality control
The AoU data version 5 contains more than 700 million variants from whole genome sequencing.44 We curated European ancestry by predicted genetic ancestry with a probability >90% provided by AoU yielding 48,112 individuals in the AoU. For variant quality control beyond AoU central efforts, we further filtered SNPs to include MAF >0.001 which retained 12,416,130 SNPs. We performed a similar quality control for imputed genotype data for South Asian ancestry in the Genes & Health cohort with additional criteria of INFO score >0.6 and genotype missing rate <5%. Individuals with a missing rate >5% were removed. Eventually, 44,396 individuals and 8,935,207 SNPs remained in Genes & Health.
Clinical outcomes
Clinical phenotypes were curated using a combination of electronic health record data, direct physical measurements, and/or self-reported personal medical history data, from the All of Us v6 Data Release as detailed in Table S20. Individuals in the Genes and Health cohort were also curated with similar definitions based on ICD10, SNOMED and operation codes (Table S21). Traits with the best performing single trait-specific PRS with power <0.95 such as hemoglobin, sleep apnea, and depression were removed. Binary traits with a prevalence <2% were removed.
Assessment of clinical utility
We applied PRSmix and PRSmix+ for coronary artery disease as a clinical application. The phenotypic algorithm includes at least one ICD or CPT code below: ICD9 410x, 411x, 412x; ICD10 I22x, I23x, I24.1, I25.2 CPT 92920–92979 (PCI), 33533–33536, 33517–33523, 33510–33516 (CABG) or self-reported personal history of MI or CAD. CAD in Genes and Health cohort was defined with at least one ICD10 I22x, I23x, I24.1, I25 or operation codes K401, K402, K403, K404, K411, K451, K452, K453, K454, K455, K491, K492, K499, K502, K751, K752, K753, K754, K758, K759 or SNOMED codes 1755008, 22298006, 54329005, 57054005, 65547006, 70211005, 70422006, 73795002, 233838001, 304914007, 401303003, 401314000.
We calculated the QRISK3 10-year predicted risk for CAD using the R package QRISK3 v0.3.0. Definition of QRISK3 are described with Type 1 diabetes, Type 2 diabetes, chronic kidney diseases (stage 3,4,5), atrial fibrillation, systemic lupus erythematosus, migraine, rheumatoid arthritis, severe mental illness following.53
The category-free NRI was used to evaluate the clinical utility. NRI was calculated by adding the PRS to the baseline logistic model including age, sex, the first 10 principal components, and clinical risk factors. The clinical risk factors include total cholesterol, HDL-C, BMI, type 2 diabetes, and current smoking status or model includes only age, sex, and 10 principal components. NRI was calculated as the sum of NRI for cases and NRI for controls:
and estimate the proportion of cases that had higher or lower risk after classification with logistic regression, respectively. The confidence interval for NRI was estimated with 500 bootstraps. We also compared the risk increase between individuals in the top decile of PRS versus those remaining in the population. In addition to liability R2 to compare the PRS performance, we also used the incremental area under the curve (AUC) to compare the PRS. The incremental AUC was estimated as the difference between the AUC of models with the integrative score versus the model with only clinical variables. We also used the baseline model with QRISK354 which estimate the 10-year risk of ASCVD and adjusted for traditional risk factor described above.
wMT-SBLUP and linear combination of LDpred2-auto derived scores
LDpred2-auto: LDpred2 is a Bayesian method that computes the adjusted SNP effect sizes from GWAS summary statistics. LDpred2 utilizes the SNP effect sizes as prior and incorporates LD between markers to infer the posterior effect sizes. In our work, we implemented LDpred2-auto55 since this method can infer heritability and the proportion of causal variants. LDpred2-auto was conducted with 800 burn-in iterations and 500 iterations. The proportion of causal variants was initialized between 10−4 and 0.9. Furthermore, LDpred2-auto does not require a validation set, the SNP effect sizes were averaged between scores. We used 1,138,726 HapMap3 variants that overlapped with SNPs from whole-genome sequencing data in the All of Us cohort. The LD reference panel developed from European ancestry was provided by the LDpred2-tutorial.
wMT-SBLUP: wMT-SBLUP15 calculates the mixing weights of PRS using sample sizes from GWAS summary statistics, SNP-heritability and genetic correlation. We compared wMT-SBLUP with our method using 5 traits that were originally assessed with wMT-SBLUP including CAD, T2D, depression, height, and BMI. We curated 26 publicly available GWAS summary statistics (Table S22) and performed LDpred2-auto with quality controls suggested by Privé et al..5,55 We used LD score regression to estimate SNP-heritability and genetic correlation across 26 traits. For each of the 5 outcome traits, we selected correlated traits with pP-value of genetic correlation less than 0.05.
Elastic Net for linear combination: we also implemented linear combination by Elastic Net with the LDpred2-auto-derived PRSs for contributing traits since this strategy was proposed by several works.8,20,27 We selected scores with significant variance explained (pP-value<0.05) to the outcome trait and conducted Elastic Net using the glmnet R package.48
Phenome-wide association study
We obtained the list of 1815 phecodes from the PheWAS website (last accessed December 2022).56 The phecodes were based on ICD-9 and ICD-10 to classify individuals. PheWAS was conducted on European ancestry only in AoU. For each phecodes as the outcome, we conducted an association analysis using logistic regression on PRS and adjusted for age, sex, and first 10 PCs. The significance threshold for PheWAS was estimated as 2.75 x 10−5 (0.05/1815) after Bonferroni correction.
Quantification and statistical analysis
Analyses were performed on the AoU Researcher Workbench in Jupyter Notebook 14 using R version 4.0.0 programming language. Results are reported in compliance with the AoU Data and Statistics Dissemination Policy.
Additional resources
This study is not a clinical trial, there is no clinical registry number. There is no external sites that have been generated to support discussion or use of the information/data/material created by the manuscript.
Acknowledgments
We would like to thank Alkes L. Price for critical comments on this work. L.E.H. is supported by the National Human Genome Research Institute (K08HG012221). P.N. is supported by grants from NHGRI (U01HG011719), NHLBI (R01HL142711, R01HL127564, and R01HL151152), and Massachusetts General Hospital (Paul & Phyllis Fireman Endowed Chair in Vascular Medicine). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The AoU Research Program is supported by the National Institutes of Health, Office of the Director, Regional Medical Centers, 1 OT2 OD026549, 1 OT2 OD026554, 1 OT2 OD026557, 1 OT2 OD026556, 1 OT2 OD026550, 1 OT2 OD 026552, 1 OT2 OD026553, 1 OT2 OD026548, 1 OT2 OD026551, and 1 OT2 OD026555; IAA # AOD 16037; Federally Qualified Health Centers, HHSN 263201600085U; Data and Research Center, 5 U2C OD023196; Biobank, 1 U24 OD023121; The Participant Center, U24 OD023176; Participant Technology Systems Center, 1 U24 OD023163; Communications and Engagement, 3 OT2 OD023205 and 3 OT2 OD023206; and Community Partners, 1 OT2 OD025277, 3 OT2 OD025315, 1 OT2 OD025337, and 1 OT2 OD025276. In addition, the AoU Research Program would not be possible without the partnership of its participants.
G&H has recently been core funded by Wellcome (WT102627 and WT210561), the Medical Research Council (UK) (M009017), Higher Education Funding Council for England Catalyst, Barts Charity (845/1796), Health Data Research UK (for London substantive site), and research delivery support from the NHS National Institute for Health Research Clinical Research Network (North Thames). G&H has recently been funded by Alnylam Pharmaceuticals, Genomics PLC and a Life Sciences Industry Consortium of Bristol-Myers Squibb Company, GlaxoSmithKline Research and Development Limited, Maze Therapeutics Inc., Merck Sharp & Dohme LLC, Novo Nordisk A/S, Pfizer Inc., and Takeda Development Center Americas Inc.
We thank Social Action for Health, Centre of The Cell, members of our community advisory group, and staff who have recruited and collected data from volunteers. We thank the NIHR National Biosample Centre (UK Biocentre), the Social Genetic & Developmental Psychiatry Centre (King’s College London), Wellcome Sanger Institute, and Broad Institute for sample processing, genotyping, sequencing, and variant annotation. We thank Barts Health NHS Trust, NHS Clinical Commissioning Groups (City and Hackney, Waltham Forest, Tower Hamlets, Newham, Redbridge, Havering, Barking, and Dagenham), East London NHS Foundation Trust, Bradford Teaching Hospitals NHS Foundation Trust, Public Health England (especially David Wyllie), Discovery Data Service/Endeavour Health Charitable Trust (especially David Stables), and NHS Digital for GDPR-compliant data sharing backed by individual written informed consent.
Most of all, we thank all of the volunteers participating in the AoU Research Program and G&H.
Author contributions
B.T. and P.N. designed experiments. B.T. performed statistical analyses. B.T., L.H., and Y.R. had unrestricted access to the AoU dataset. L.H. assisted in processing AoU phenotype data. B.T., S.H.L., and P.N. provided guidance and feedback on analyses. B.T. and P.N. wrote the manuscript with assistance from all authors (Y.R., Q.Q.H., W.H., H.M., D.A.v.H., Y.W., A.R.M., and S.H.L.). All authors agreed to submit the manuscript, read and approved the final draft, and take full responsibility for its content.
Declaration of interests
P.N. reports grants from Allelica, Amgen, Apple, Boston Scientific, Genentech, and Novartis; is a consultant to Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Foresite Labs, HeartFlow, Novartis, Genentech, and GV; reports scientific advisory board membership with Esperion Therapeutics, Preciseli, and TenSixteen Bio; is a scientific co-founder of TenSixteen Bio; and reports spousal employment at Vertex Pharmaceuticals, all unrelated to the present work.
Published: March 19, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2024.100523.
Supplemental information
References
- 1.Catalog, P.G.S. PGS Catalog - the Polygenic Score Catalog. http://www.pgscatalog.org/.
- 2.Choi S.W., Mak T.S.-H., O’Reilly P.F. Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 2020;15:2759–2772. doi: 10.1038/s41596-020-0353-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi S.W., O’Reilly P. SA20 - PRSice 2: POLYGENIC RISK SCORE SOFTWARE (UPDATED) AND ITS APPLICATION TO CROSS-TRAIT ANALYSES. Eur. Neuropsychopharmacol. 2019;29:S832. doi: 10.1016/j.euroneuro.2017.08.092. [DOI] [Google Scholar]
- 5.Privé F., Arbel J., Vilhjálmsson B.J. LDpred2: better, faster, stronger. Bioinformatics. 2021;36:5424–5431. doi: 10.1093/bioinformatics/btaa1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vilhjálmsson B.J., Yang J., Finucane H.K., Gusev A., Lindström S., Ripke S., Genovese G., Loh P.-R., Bhatia G., Do R., et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am. J. Hum. Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge T., Chen C.-Y., Ni Y., Feng Y.-C.A., Smoller J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019;10:1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abraham G., Malik R., Yonova-Doing E., Salim A., Wang T., Danesh J., Butterworth A.S., Howson J.M.M., Inouye M., Dichgans M. Genomic risk score offers predictive performance comparable to clinical risk factors for ischaemic stroke. Nat. Commun. 2019;10:5819. doi: 10.1038/s41467-019-13848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung W., Chen J., Turman C., Lindstrom S., Zhu Z., Loh P.-R., Kraft P., Liang L. Efficient cross-trait penalized regression increases prediction accuracy in large cohorts using secondary phenotypes. Nat. Commun. 2019;10:569. doi: 10.1038/s41467-019-08535-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weissbrod O., Kanai M., Shi H., Gazal S., Peyrot W.J., Khera A.V., Okada Y., Biobank Japan Project. Martin A.R., Finucane H.K., Price A.L. Leveraging fine-mapping and multipopulation training data to improve cross-population polygenic risk scores. Nat. Genet. 2022;54:450–458. doi: 10.1038/s41588-022-01036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inouye M., Abraham G., Nelson C.P., Wood A.M., Sweeting M.J., Dudbridge F., Lai F.Y., Kaptoge S., Brozynska M., Wang T., et al. Genomic Risk Prediction of Coronary Artery Disease in 480,000 Adults: Implications for Primary Prevention. J. Am. Coll. Cardiol. 2018;72:1883–1893. doi: 10.1016/j.jacc.2018.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruan Y., Lin Y.-F., Feng Y.-C.A., Chen C.-Y., Lam M., Guo Z., Stanley Global Asia Initiatives. He L., Sawa A., Martin A.R., et al. Improving polygenic prediction in ancestrally diverse populations. Nat. Genet. 2022;54:573–580. doi: 10.1038/s41588-022-01054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albiñana C., Zhu Z., Schork A.J., Ingason A., Aschard H., Brikell I., Bulik C.M., Petersen L.V., Agerbo E., Grove J., et al. Multi-PGS enhances polygenic prediction by combining 937 polygenic scores. Nat. Commun. 2023;14:4702. doi: 10.1038/s41467-023-40330-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C., Yang C., Gelernter J., Zhao H. Improving genetic risk prediction by leveraging pleiotropy. Hum. Genet. 2014;133:639–650. doi: 10.1007/s00439-013-1401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maier R.M., Zhu Z., Lee S.H., Trzaskowski M., Ruderfer D.M., Stahl E.A., Ripke S., Wray N.R., Yang J., Visscher P.M., Robinson M.R. Improving genetic prediction by leveraging genetic correlations among human diseases and traits. Nat. Commun. 2018;9 doi: 10.1038/s41467-017-02769-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe K., Stringer S., Frei O., Umićević Mirkov M., de Leeuw C., Polderman T.J.C., van der Sluis S., Andreassen O.A., Neale B.M., Posthuma D. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019;51:1339–1348. doi: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 17.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert S.A., Gil L., Jupp S., Ritchie S.C., Xu Y., Buniello A., McMahon A., Abraham G., Chapman M., Parkinson H., et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat. Genet. 2021;53:420–425. doi: 10.1038/s41588-021-00783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buch G., Schulz A., Schmidtmann I., Strauch K., Wild P.S. A systematic review and evaluation of statistical methods for group variable selection. Stat. Med. 2023;42:331–352. doi: 10.1002/sim.9620. [DOI] [PubMed] [Google Scholar]
- 20.Krapohl E., Patel H., Newhouse S., Curtis C.J., von Stumm S., Dale P.S., Zabaneh D., Breen G., O’Reilly P.F., Plomin R. Multi-polygenic score approach to trait prediction. Mol. Psychiatry. 2018;23:1368–1374. doi: 10.1038/mp.2017.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yengo L., Vedantam S., Marouli E., Sidorenko J., Bartell E., Sakaue S., Graff M., Eliasen A.U., Jiang Y., Raghavan S., et al. A saturated map of common genetic variants associated with human height. Nature. 2022;610:704–712. doi: 10.1038/s41586-022-05275-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klarin D., Natarajan P. Clinical utility of polygenic risk scores for coronary artery disease. Nat. Rev. Cardiol. 2022;19:291–301. doi: 10.1038/s41569-021-00638-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heart Association Council on Epidemiology A. Circulation; 2022. Heart Disease and Stroke Statistics—2022 Update: A Report from the American Heart Association. [DOI] [PubMed] [Google Scholar]
- 24.Arnett D.K., Blumenthal R.S., Albert M.A., Buroker A.B., Goldberger Z.D., Hahn E.J., Himmelfarb C.D., Khera A., Lloyd-Jones D., McEvoy J.W., et al. 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;140:e596–e646. doi: 10.1161/CIR.0000000000000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koyama S., Ito K., Terao C., Akiyama M., Horikoshi M., Momozawa Y., Matsunaga H., Ieki H., Ozaki K., Onouchi Y., et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat. Genet. 2020;52:1169–1177. doi: 10.1038/s41588-020-0705-3. [DOI] [PubMed] [Google Scholar]
- 26.Tamlander M., Mars N., Pirinen M., FinnGen, Widén E., Widén E., Ripatti S. Integration of questionnaire-based risk factors improves polygenic risk scores for human coronary heart disease and type 2 diabetes. Commun. Biol. 2022;5:158. doi: 10.1038/s42003-021-02996-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albiñana C., Zhu Z., Schork A.J., Ingason A., Aschard H., Brikell I., Bulik C.M., et al. Multi-PGS Enhances Polygenic Prediction by Combining 937 Polygenic Scores. Nature Communications. 2023;14:4702. doi: 10.1038/s41467-023-40330-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., Zhan J., Jin J., Zhang J., Lu W., Zhao R., Ahearn T.U., Yu Z., O’Connell J., Jiang Y., et al. Novel methods for multi-ancestry polygenic prediction and their evaluations in 5.1 million individuals of diverse ancestry. bioRxiv. 2022 doi: 10.1101/2022.03.24.485519. Preprint at. [DOI] [Google Scholar]
- 29.Sud M., Sivaswamy A., Chu A., Austin P.C., Anderson T.J., Naimark D.M.J., Farkouh M.E., Lee D.S., Roifman I., Thanassoulis G., et al. Population-Based Recalibration of the Framingham Risk Score and Pooled Cohort Equations. J. Am. Coll. Cardiol. 2022;80:1330–1342. doi: 10.1016/j.jacc.2022.07.026. [DOI] [PubMed] [Google Scholar]
- 30.Carroll R.J., Bastarache L., Denny J.C. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30:2375–2376. doi: 10.1093/bioinformatics/btu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bastarache L., Denny J.C., Roden D.M. Phenome-Wide Association Studies. JAMA. 2022;327:75–76. doi: 10.1001/jama.2021.20356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Kanai M., Tan T., Kamariza M., Tsuo K., Yuan K., Zhou W., Okada Y., Huang H., et al. BioBank Japan Project Polygenic prediction across populations is influenced by ancestry, genetic architecture, and methodology. Cell Genom. 2023;3 doi: 10.1016/j.xgen.2023.100408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Namba S., Lopera E., Kerminen S., Tsuo K., Läll K., Kanai M., Zhou W., Wu K.-H., Favé M.J., et al. Global Biobank analyses provide lessons for developing polygenic risk scores across diverse cohorts. Cell Genom. 2023;3 doi: 10.1016/j.xgen.2022.100241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hou K., Xu Z., Ding Y., Harpak A., Pasaniuc B. Calibrated prediction intervals for polygenic scores across diverse contexts. medRxiv. 2023 doi: 10.1101/2023.07.24.23293056. Preprint at. [DOI] [Google Scholar]
- 35.Zou H., Hastie T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Series B Stat. Methodol. 2005;67:301–320. doi: 10.1111/j.1467-9868.2005.00503.x. [DOI] [Google Scholar]
- 36.Zhou D.-X. On grouping effect of elastic net. Stat. Probab. Lett. 2013;83:2108–2112. doi: 10.1016/j.spl.2013.05.014. [DOI] [Google Scholar]
- 37.Zou H., Hastie T. Addendum: Regularization and variable selection via the elastic net. J. R. Stat. Soc. Series B Stat. Methodol. 2005;67:768. doi: 10.1111/j.1467-9868.2005.00527.x. [DOI] [Google Scholar]
- 38.Wang M., Menon R., Mishra S., Patel A.P., Chaffin M., Tanneeru D., Deshmukh M., Mathew O., Apte S., Devanboo C.S., et al. Validation of a Genome-Wide Polygenic Score for Coronary Artery Disease in South Asians. J. Am. Coll. Cardiol. 2020;76:703–714. doi: 10.1016/j.jacc.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lloyd-Jones L.R., Zeng J., Sidorenko J., Yengo L., Moser G., Kemper K.E., Wang H., Zheng Z., Magi R., Esko T., et al. Improved polygenic prediction by Bayesian multiple regression on summary statistics. Nat. Commun. 2019;10:5086. doi: 10.1038/s41467-019-12653-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Tsuo K., Kanai M., Neale B.M., Martin A.R. Challenges and opportunities for developing more generalizable polygenic risk scores. Annu. Rev. Biomed. Data Sci. 2022;5:293–320. doi: 10.1146/annurev-biodatasci-111721-074830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mostafavi H., Harpak A., Agarwal I., Conley D., Pritchard J.K., Przeworski M. Variable prediction accuracy of polygenic scores within an ancestry group. Elife. 2020;9 doi: 10.7554/eLife.48376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mapes B.M., Foster C.S., Kusnoor S.V., Epelbaum M.I., AuYoung M., Jenkins G., Lopez-Class M., Richardson-Heron D., Elmi A., Surkan K., et al. Diversity and inclusion for the All of Us research program: A scoping review. PLoS One. 2020;15 doi: 10.1371/journal.pone.0234962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The “All of Us” Research Program. Denny J.C., Rutter J.L., Goldstein D.B., Philippakis A., Smoller J.W., Jenkins G., Dishman E. The "All of Us" Research Program. N. Engl. J. Med. 2019;381:668–676. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cronin R.M., Jerome R.N., Mapes B., Andrade R., Johnston R., Ayala J., Schlundt D., Bonnet K., Kripalani S., Goggins K., et al. Development of the initial surveys for the All of Us Research Program. Epidemiology. 2019;30:597–608. doi: 10.1097/EDE.0000000000001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.All of Us Research Program Protocol . 2020. All of Us Research Program | NIH.https://allofus.nih.gov/about/all-us-research-program-protocol [Google Scholar]
- 47.Finer S., Martin H.C., Khan A., Hunt K.A., MacLaughlin B., Ahmed Z., Ashcroft R., Durham C., MacArthur D.G., McCarthy M.I., et al. Cohort Profile: East London Genes & Health (ELGH), a community-based population genomics and health study in British Bangladeshi and British Pakistani people. Int. J. Epidemiol. 2020;49:20–21i. doi: 10.1093/ije/dyz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010;33:1–22. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S.H., Clark S., van der Werf J.H.J. Estimation of genomic prediction accuracy from reference populations with varying degrees of relationship. PLoS One. 2017;12 doi: 10.1371/journal.pone.0189775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wishart J., Kondo T., Elderton E.M. The mean and second moment coefficient of the multiple correlation coefficient, in samples from a normal population. Biometrika. 1931;22:353. doi: 10.2307/2332101. [DOI] [Google Scholar]
- 51.Stuart A., Ord K., Arnold S. Wiley; 2010. Kendall’s Advanced Theory of Statistics, Classical Inference and the Linear Model. [Google Scholar]
- 52.Momin M.M., Lee S., Wray N.R., Lee S.H. Significance tests for R2 of out-of-sample prediction using polygenic scores. Am. J. Hum. Genet. 2023;110:349–358. doi: 10.1016/j.ajhg.2023.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riveros-Mckay F., Weale M.E., Moore R., Selzam S., Krapohl E., Sivley R.M., Tarran W.A., Sørensen P., Lachapelle A.S., Griffiths J.A., et al. Integrated polygenic tool substantially enhances coronary artery disease prediction. Circ. Genom. Precis. Med. 2021;14 doi: 10.1161/CIRCGEN.120.003304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hippisley-Cox J., Coupland C., Brindle P. Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: prospective cohort study. BMJ. 2017;357:j2099. doi: 10.1136/bmj.j2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Privé F., Arbel J., Aschard H., Vilhjálmsson B.J. Identifying and correcting for misspecifications in GWAS summary statistics and polygenic scores. HGG Adv. 2022;3 doi: 10.1016/j.xhgg.2022.100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Denny J.C., Ritchie M.D., Basford M.A., Pulley J.M., Bastarache L., Brown-Gentry K., Wang D., Masys D.R., Roden D.M., Crawford D.C. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pereira F. Terra.Bio; 2020. Home.https://terra.bio/ [Google Scholar]
- 58.Researcher Workbench. https://www.researchallofus.org/workbench/.
- 59.Data Methods – All of Us Research Hub. https://www.researchallofus.org/data-tools/methods.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
UK Biobank individual-level data are available for request through the UK Biobank with application (https://www.ukbiobank.ac.uk). The All of Us individual-level data are available for request through the All of Us platform (https://www.researchallofus.org). The All of Us and Genes & Health individual-level data is a controlled access dataset and may be granted at https://www.researchallofus.org/ and https://www.genesandhealth.org/, respectively. Data can be accessed through the secure All of Us Researcher Workbench platform, which is a cloud-based analytic platform that was built on the Terra platform.57 Researchers gain access to the platform after they complete a 3-step process including registration, completion of ethics training, and attesting to a data use agreement attestation.58 All of Us uses a tiered approach based on what genomic data is accessible through the Controlled Tier, and includes both whole genome sequencing (WGS), genotyping array variant data in multiple formats, as well as variant annotations, access to computed ancestry, and quality reports.59 This study includes data on the 98,600 participants with (WGS) data in the All of Us v6 Curated Data Repository release. Participant data in this data release was collected between May 6, 2018 and April 1, 2021. This project is registered in the All of Us program under the workspace name “Polygenic risk score across diverse ancestries and biobanks. The code is provided at https://github.com/buutrg/PRSmix. The version at the publication is at https://doi.org/10.5281/zenodo.10581592. The published article includes all other data generated or analyzed during this study.
-
•
Individual-level genomic data and longitudinal phenotypic data from the UK Biobank, a large-scale population-based cohort with genotype and phenotype data in approximately 500,000 volunteer participants recruited from 2006 to 2010 was used. Baseline assessments were conducted at 22 assessment centers across the UK using touch screen questionnaire, computer assisted verbal interview, physical tests, and sample collection including for DNA (https://www.ukbiobank.ac.uk). The UK Biobank Application is 7089.
-
•
Polygenic risk scores were obtained from the Polygenic Score (PGS) Catalog,18 which is a publicly accessible resource cataloging published PRS, including the metadata. The metadata provides information describing the computational algorithms used to generate the score, and performance metrics to evaluate a PRS.18 At the time of this study, over 2,600 PRS were cataloged in the PGS Catalog (version July 18, 2022) designed to predict 538 distinct traits. The PGS Catalog is freely available at https://www.pgscatalog.org/. Our new scores are deposited in the PGS Catalog.
-
•
The weights from the PRSmix and PRSmix+ scores in this manuscript have been returned to the PGS Catalog. The R package to implement PRSmix and PRSmix+ in independent datasets is at https://github.com/buutrg/PRSmix.