Abstract
Infection with Kaposi’s sarcoma-associated herpesvirus (KSHV) is closely associated with Kaposi’s sarcoma (KS) and primary effusion lymphoma, with viral genomes present in a latent state in the majority of tumor cells. Here we describe a cluster of latently expressed viral genes whose mRNAs are generated from a common promoter. Two mRNAs in this region encode the latency-associated nuclear antigen, the product of open reading frame 73 (ORF73). The larger RNA, of 5.8 kb, is an unspliced transcript that includes ORF72 and -71 at its 3′ end; it initiates at nucleotides (nt) 127880 to 127886 from a promoter lacking recognizable TATA elements. A less abundant mRNA, of 5.4 kb, is a variant of this transcript, in which 336 nt of 5′ noncoding information has been removed by RNA splicing. A third, more abundant RNA is generated from the same promoter region via splicing from the common splice donor at nt 127813 to an acceptor 5′ to ORF72; this transcript is the presumed mRNA for ORF72, which encodes the viral cyclin D homolog. All three RNAs are 3′ coterminal. In situ hybridization analysis with probes that can detect all three transcripts shows that the RNAs are detectable in a large fraction of BCBL-1 cells prior to lytic induction and in >70% of KS spindle cells in primary KS tumors. This confirms that these transcripts are indeed latent RNAs and suggests a role for their products in viral persistence and/or KSHV-associated proliferation.
The genome of Kaposi’s sarcoma-associated herpesvirus (KSHV) (also known as human herpesvirus 8) was initially identified by representational difference analysis of Kaposi’s sarcoma (KS) tumor samples (3). It has since been detected in a variety of lymphoproliferative disorders, including body cavity lymphoma or primary effusion lymphoma (PEL) (1) and multicentric Castleman’s disease (31). The epidemiological evidence implicating KSHV as a causative agent for KS is strong. (i) KSHV DNA is detected in virtually all KS tumor biopsies from human immunodeficiency virus (HIV)-positive or HIV-negative patients (20, 35). (ii) Anti-KSHV seroreactivity is found in ≥80% of KS patients but in less than 6% of healthy blood donors in the United States (10, 12, 19). Seropositivity for KSHV precedes the onset of KS and correlates with increased KS risk, suggesting that rather than being a correlative marker KSHV is directly involved in KS pathogenesis (8). The KSHV-specific antibody response includes a strong response to a latency-associated nuclear antigen (LANA) (11, 13, 23), which is one of the proteins encoded by the KSHV latent messages identified in this study.
On the basis of the complete sequence of the 137-kbp unique region (L) and terminal repeat regions (H), KSHV is classified as a human gammaherpesvirus, the lymphotropic subgroup of the herpesvirus family (22, 29). All herpesviruses display two modes of replication: lytic replication, during which the host cell is destroyed and viral progeny are released, and latent replication, during which the viral genome persists but shows restricted gene expression and no release of viral progeny (reviewed in reference 28). KSHV conforms to this paradigm as well. In cultured B cells from PEL tumors, the virus genome persists as a circular episome during viral latency and is capable of reactivating and replicating in response to outside stimuli (18, 25, 26). Only a subset of viral genes are transcribed during KSHV latency (37). In the distantly related Epstein-Barr virus, the latency-associated genes are essential for episome maintenance and host cell transformation (reviewed in references 14 and 27). By analogy, important KSHV genes involved in growth deregulation and viral genomic persistence are likely to be found among those transcribed during viral latency.
By preparing labeled cDNA from KS tumors and annealing it to arrays of cloned viral DNA fragments, we previously identified KSHV-specific transcripts emanating from the region of open reading frame K12 (ORFK12) as the most abundant RNAs in latently infected cells (36, 37). In subsequent experiments, we (11) and others (30) have searched for additional regions of viral DNA likely to be transcribed in latency by probing Northern blots of RNA from uninduced and induced PEL cell lines with probes from different genomic regions, looking for genes that were preferentially expressed prior to lytic induction. This revealed that a region just to the right of ORFK12 is also expressed during latency; this region spans ORF71 to -73. One of the products of this region, that encoded by ORF73, has recently been identified as LANA, the immunodominant latent antigen initially detected serologically (11, 13, 23). Here we present a detailed analysis of the transcription of this region and show that separate mRNAs encoding LANA and the viral cyclin D homolog are generated from a common latency-specific promoter. Both RNAs are abundantly expressed in KS tumors as well as PEL cell lines. The finding that the viral cyclin is expressed as a latent gene in two neoplastic conditions suggests a role for this gene product in the pathogenesis of the abnormal proliferation observed in KSHV-associated diseases.
MATERIALS AND METHODS
Cell lines.
All cell lines were from the American Type Culture Collection. HeLa, CV-1, and 293 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin at 37°C, 5% CO2. LnCAP and BCBL-1 cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 0.05 mM 2-mercaptoethanol, 1 mM sodium bicarbonate, 2 mM l-glutamine, penicillin, and streptomycin at 37°C, 5% CO2.
Plasmids.
All DNA was from a KSHV lambda library derived from a KS lesion (37) unless otherwise indicated. pDD2 contains a 736-bp HindIII-BamHI fragment (nucleotides [nt] 122791 to 123527) comprising most of ORF72 in pBluescript II KS(+) (Stratagene). pDD41 contains a 1,763-bp NcoI fragment (nt 127609 to 129370) cloned into the NcoI site of pGL3basic (Promega), and pDD43 contains the same fragment in the opposite orientation. pDD83 is derived from pDD41 by internal NheI deletion, leaving a 552-bp NcoI-NheI fragment (nt 127609 to 128159), and pDD53 (nt 127607 to 127910) was derived from pDD41 by an internal SmaI deletion. pML10 contains ORF71 (nt 122145 to 122711).
Induction of viral replication and RNA isolation.
A total of 5 × 106 BCBL-1 cells were induced with 20 ng of phorbol-12-tetradecanoate-13-acetate (TPA)/ml, and RNA was isolated 48 h after TPA or mock treatment. Total cellular RNA was isolated with RNAzol (Tel-Test Inc., Friendswood, Tex.) and poly(A) enriched as previously described (16).
cDNA isolation.
A poly(dT)-primed cDNA library was constructed from induced and uninduced BCBL-1 cells in λZAP (Stratagene). Specific cDNA clones were isolated by hybridization with KSHV sequences from pDD2 (ORF72) and pML10 (ORF71), according to the manufacturer’s procedures.
Northern blotting and hybridization.
Northern blotting and hybridization have been described previously (16). Briefly, RNA from BCBL-1 cells was poly(A) enriched with the Oligotex mRNA system (Qiagen, Inc.), run on a 1% formaldehyde gel blotted to Hybond membrane (Amersham), and hybridized overnight at 65°C in Church buffer (5) (1% [wt/vol] bovine serum albumin, 1 mM EDTA, 0.5 M NaHPO4 [pH 7.2], 7% [wt/vol] sodium dodecyl sulfate), washed in a solution of 40 mM NaHPO4 (pH 7.2), 0.1% sodium dodecyl sulfate, and 1 mM EDTA, and exposed to film for 48 h. Probes were randomly labeled with [32P]dCTP by using the Redivue random priming kit (Amersham).
In situ hybridization.
In situ probes for the latent transcripts were derived from pDD2 by in vitro transcription and hybridized to Kaposi’s sarcoma tissue sections as described previously (32).
RT-PCR.
Total RNA (0.5 μg) was reverse transcribed for 1 h at 42°C with 25 pmol of primer 7308 (5′-GCATTCCCGGGGGCGCCATC; nt 127279 to 127298) in order to identify the 5.8-kb transcript untranslated region (UTR), or primer 7208 (5′-GAAAAGGAGTCTGCCGCGGCATAGC; nt 123188 to 123212) in order to identify the 1.7-kb transcript UTR, in a 20-μl reaction mixture containing 200 U of superscriptII reverse transcriptase and buffer (Gibco-BRL), 20 U of RNAsin (Promega), 5 mM dithiothreitol, and 125 μM (each) deoxynucleoside triphosphate (Pharmacia). Two microliters of this reaction mixture was then added to 95 μl containing 1 U of Taq polymerase and buffer (15 μM MgCl2; Perkin-Elmer), primer 7311 (5′-TCCTCGGGAAATCTGGTCT; nt 127297 to 127315) for the 5.8-kb UTR or primer 7208 and one of the following upstream primers: 1 (5′-TGTCTGAGAGCTCCCCCTTG; nt 127383 to 127401), 2 (5′-CTGGACTTGCCTAGGTAGCA; nt 127633 to 127614), 3 (5′-TTTCTCACGCCCGGATTATATATC; nt 127656 to 127633), 4 (5′-ATGGGTTATTGGCCGTTTCTG; nt 127677 to 127657), 5 (5′-TGTGTGGTGGTTTTCGAAAAAC; nt 127699 to 127678), 6 (5′-GGTGTACATGATTTGTGTTAAGG; nt 127722 to 127700), 7 (5′-AGCAGCAGCTTGGTCCGGCTG; nt 127844 to 127824), 8 (5′-TTGGAGGCAGCTGCGCCACGAAGC; nt 127883 to 127860), or 9 (5′-GCGGCGCCCGGGACAATC; nt 127919 to 127902). The amplification conditions were 30 cycles of 1 min at 94°C, 2 min at 58°C, and 2 min at 72°C, followed by 10 min at 72°C. All amplification products were cloned into pCR2.1 (Invitrogen) and sequenced.
5′ RACE.
5′ rapid amplification of cDNA ends (RACE) was performed on 10 μg of BCBL-1 RNA with primers 7311 and 7308 according to the manufacturer’s instructions (Boehringer Mannheim, Inc.). RACE products after two rounds of PCR under the conditions described above were cloned into pCR2.1 and sequenced.
S1 nuclease assay.
Hybridization (12 h) and S1 nuclease digestion (1 h) were performed at 28°C on 30 μg of total BCBL-1 RNA or yeast RNA by using 3′-32P-labeled oligonucleotide 7325 (5′-GACGTGACTGCTTCGTGGCGCAGCTGCCTCCAAATGATACACACAT; nt 127851 to 127896) and the S1-Kit (Ambion, Inc., Austin, Tex.) according to the manufacturer’s procedures.
Promoter assays.
Reporter DNA (2 μg) was transfected into 3 × 105 cells per 60-mm-diameter dish by using Lipofectamine (Gibco-BRL), and transfection efficiency was measured in each transfection by cotransfection of 500 ng of pSEAP encoding secreted alkaline phosphatase (SEAP) under control of the simian virus 40 (SV40) promoter-enhancer (Clontech). Transactivation was calculated as the mean of three separate experiments normalized to SEAP levels. SEAP and luciferase levels were determined by using chemiluminescent substrate in a Turner luminometer according to the manufacturer’s instructions.
RESULTS
ORF71, -72, and -73 are transcribed during KSHV latency.
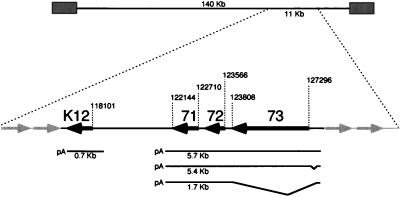
In KSHV the leftward-oriented ORF71, -72, and -73 are located immediately adjacent to each other, initiating at nucleotide positions 122710, 123566, and 127296, respectively (Fig. 1) (throughout this report nucleotide numbers are according to Russo et al. [29]). They are separated from the K12 locus by a 4-kb noncoding (intergenic) region. Just to the right of ORF73 lie the rightward-oriented ORFK14 (nt 127883 to 128929, encoding a putative OX-2 homolog) and ORF74 (nt 129371 to 130550, encoding a G-protein-coupled receptor homolog); these genes are transcribed in a bicistronic transcript that is regulated differently from the ORF71 to -73 transcription unit, being strongly upregulated during lytic growth (13a).
FIG. 1.
Location of clustered latency transcripts in KSHV genome. The numbers indicate nucleotide positions, based on the sequence of Russo et al. (29). The boldface arrows represent ORFK12, -71, -72, and -73.
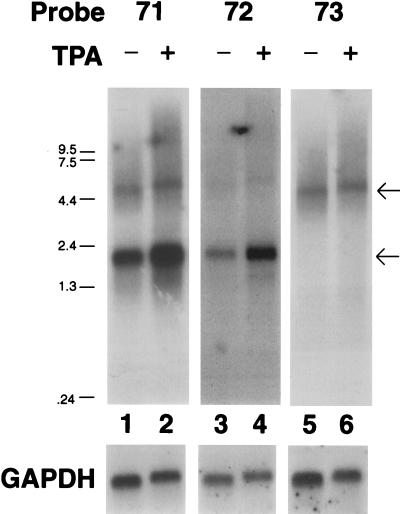
To characterize the transcription of ORF71, ORF72, and ORF73, RNA from the PEL cell line BCBL-1 was examined by Northern blotting with probes specific for the individual coding regions. BCBL-1 cells do not contain Epstein-Barr virus but harbor replication-competent KSHV. Upon treatment with TPA these cells undergo the complete program of KSHV gene expression, resulting, ultimately, in viral replication and the release of mature virions (26). Poly(A)+ RNA was isolated from BCBL-1 cells 48 h after TPA or mock treatment. Hybridization with a 921-bp BamHI-SacI DNA fragment (nt 126474 to 127394) representing the 5′ end of ORF73 identified a single transcript of approximately 5.8 kb. The levels of this transcript were similar in the presence or absence of TPA (Fig. 2, lanes 5 and 6), in marked contrast to typical KSHV lytic transcripts, which are strongly upregulated by TPA (37). Hybridization with a 737-bp BamHI-HindIII DNA fragment (nt 122791 to 123527) located within the coding region of ORF72 detected two transcripts of approximately 5.8 and 1.7 kb (Fig. 2, lanes 3 and 4). As with the ORF73 probe, the level of the 5.8-kb transcript was unaffected by TPA. The smaller, 1.7-kb transcript was modestly increased by TPA. However, this induction was small compared to that of typical KSHV lytic genes and varied in extent from experiment to experiment, typically in the range of two- to fourfold over the uninduced level (the samples shown in Fig. 2 show an induction ratio at the upper end of what we usually observe). To verify that all transcripts encompassing ORF72 had the same polarity, the blots were rehybridized with a single-stranded antisense RNA probe, which yielded an identical pattern while a sense probe detected no signal (data not shown). Hybridization with a 567-bp DNA fragment (nt 122145 to 122711) specific for ORF71 detected transcripts of 5.8 and 1.7 kb and no additional, smaller species (Fig. 2, lanes 1 and 2). The 5.8-kb transcript was again detected by this probe, which also showed it to be unaffected by TPA, while the 1.7-kb transcript was marginally upregulated. All filters contained equivalent amounts of poly(A) RNA, as evidenced by hybridization with a GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe (Fig. 2).
FIG. 2.
Analysis of LAT transcription. Shown is an autoradiogram of a Northern blot with probes specific for ORF71 (lanes 1 and 2), ORF72 (lanes 2 and 3), and ORF73 (lanes 5 and 6) of BCBL-1 RNA from TPA-induced (lanes 2, 4, and 6) or mock-treated (lanes 1, 3, and 5) cells. The blot was rehybridized with a GAPDH probe as a loading control. The arrows indicate the migration of the 5.8- and 1.7-kb messages.
The patterns of hybridization shown in Fig. 2 establish that the 5.8-kb RNA contains ORF-73, -72, and -71 and is the likely mRNA for ORF73, given that this ORF is its most 5′ coding region and that its AUG is in a favorable context for translational initiation (15). The 1.7-kb transcript spans ORF72 and -71 and is the presumed mRNA for ORF72, encoding the viral cyclin (2, 4, 17). No other messages were consistently identified with this assay, suggesting that the 5.8- and 1.7-kb messages represent the major latent messages in this region.
In situ hybridization analysis.
To exclude the possibility that the RNA hybridization signals described above emanated solely from the 2 to 4% of cells within the BCBL-1 population known to undergo spontaneous lytic replication, the BCBL-1 cells were analyzed by in situ hybridization with an ORF72-specific probe (which would detect all the transcripts from this gene cluster). Using an ORF72 antisense riboprobe, a signal was detected in >40% of the cells prior to TPA treatment (data not shown). It is likely that an even higher percentage of the cells express the RNAs at levels below the detection threshold of our in situ analysis, since virtually all nuclei in this population stain for LANA (12); in any case, the large number of positive cells prior to TPA induction excludes the possibility that these messages are only expressed in lytic infection.
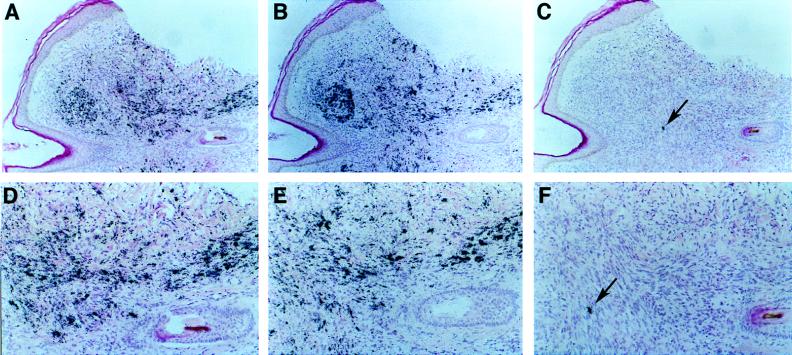
Next, by examining sections of primary KS tumors, we investigated the more important question of whether this cluster is expressed in latently infected KS tumor cells. As shown in Fig. 3A and D, the majority of spindle cells in KS express detectable transcripts spanning ORF72; this pattern is virtually identical to that observed with probes spanning ORFK12 (Fig. 3B and E), previously characterized as a latent RNA (32), and agrees well with similar recent findings of Davis et al. (6). However, the exposure time to detect a comparable signal for the ORF72 probe was twice as long, suggesting that these latency transcripts are much less abundant than the ORFK12 transcript (a finding that accords well with our earlier cDNA analysis [37]). By contrast, the hybridization signal of a well-characterized lytic message (nut-1) was restricted to a small subset of cells (Fig. 3C and F). No signal was detected after hybridization with an ORF72 sense probe (data not shown).
FIG. 3.
In situ hybridization of a KS tumor section. Shown are subjacent sections of a KS tumor from an HIV-negative patient hybridized with specific probes and counterstained with hematoxylin and eosin stain. Hybridization was done with an ORF72 probe (A and D), an ORFK12 probe (B and E), and a nut-1 probe (C and F). Exposure was for 7 days (A and D) and 3 days (B, E, C, and F). Magnification: ×47 (A to C) and ×96 (D to F). The arrow indicates a single cell positive for the lytic nut-1 RNA.
The 5.8- and 1.7-kb RNAs are coterminal.
The sizes and coding organizations of the RNAs suggested that their 3′ ends were likely to be coterminal. Accordingly, we isolated cDNA clones from a poly(dT)-primed cDNA library made from BCBL-1-derived RNA. The library was screened with probes specific for ORF71 and -72, and 15 independent clones were isolated and sequenced. All of the clones contained the entire ORF71 and terminated within or just upstream of ORF72; none revealed evidence of RNA splicing in this region. Fourteen of the 15 clones terminated 14 to 25 nt downstream of a consensus AAUAAA polyadenylation signal (nt 122093 to 122089) just 3′ of ORF71 (Table 1); 9 of the 15 terminated at a common nucleotide, 122070. (The remaining clone terminated just 1 nt after this signal.) This same poly(A) site was identified by Rainbow et al. (23) in a partial cDNA clone that extended into ORF73 and expressed a fragment of that coding region. As the next candidate poly(A) site is located 1,075 nt downstream, these results, coupled with the sizes of the transcripts, establish that the 5.8- and 1.7-kb RNAs are processed at this common poly(A) signal.
TABLE 1.
Characteristics of cDNA clones isolated by hybridization with probes specific for ORF72 and ORF71
| Probe | 3′ end (nt) | 5′ end (nt) | Size (bp) |
|---|---|---|---|
| 72 | 122064 | 123721 | 1,657 |
| 122070 | 123513 | 1,443 | |
| 122068 | 123735 | 1,667 | |
| 122070 | 123737 | 1,667 | |
| 122089 | 123068 | 1,440 | |
| 122070 | 123083 | 1,013 | |
| 122075 | 123126 | 1,051 | |
| 71 | 122070 | 123204 | 1,134 |
| 122070 | 122916 | 846 | |
| 122065 | 123026 | 961 | |
| 122070 | 123654 | 1,584 | |
| 122073 | 123513 | 1,440 | |
| 122070 | 123513 | 1,443 | |
| 122070 | 123730 | 1,660 | |
| 122070 | 123730 | 1,660 |
An additional ORF73 mRNA is derived by alternative splicing.
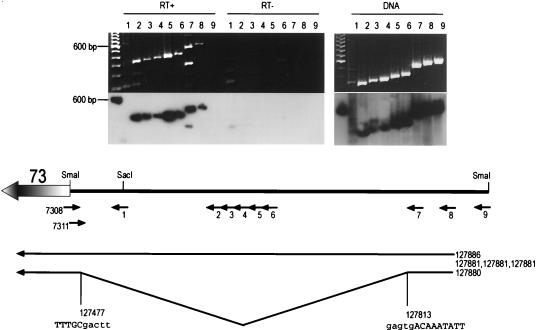
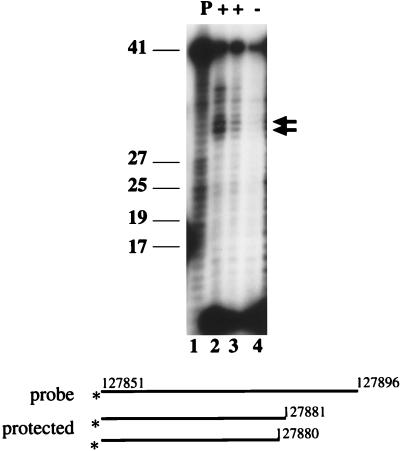
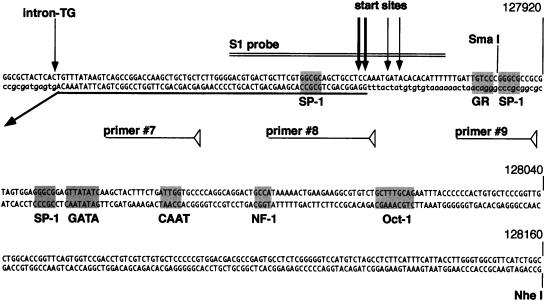
To determine the 5′ end of the ORF73 message, reverse transcription (RT)-PCR amplification of viral RNA was performed, using downstream primers within ORF73 (designated 7308 and 7311 [Fig. 4]) and various upstream primers (primers 1 to 9 [Fig. 4]). All primers up to and including primer 8, but not the more distal primer 9, yielded amplification products identical in size to the control PCR products derived by amplifying viral genomic DNA (Fig. 4, top). Amplification was dependent on reverse transcriptase, and the amplified product corresponded to the 5′ UTR region of ORF73, as determined by DNA hybridization (Fig. 4, lower gel panel) as well as cloning and sequencing (data not shown). This demonstrated the presence of an unspliced 5′ UTR and located the 5′ end of the RNA between primers 8 and 9. The exact transcription initiation site was determined by 5′ RACE, which yielded five clones terminating at nt 127881 (three clones), 127885, and 127886. This initiation site was independently confirmed by S1 nuclease protection (Fig. 5). BCBL-1 RNA, 30 and 15 μg, was hybridized at 28°C to a 46-mer oligonucleotide (7325; see Materials and Methods for the sequence) (Fig. 5, lane 1) spanning the region between primers 8 and 9. Following S1 nuclease digestion, fragments of 30, 31, 34, and 36 nt were protected (Fig. 5, lanes 2 and 3). No specific fragment was protected after hybridization and S1 nuclease digestion with nonspecific RNA (Fig. 5, lane 4). These bands are consistent with the RACE-predicted initiation sites within nt 127880 to 127886 and exclude the possibility that the ends mapped in the RACE reaction were artificially generated by premature termination by reverse transcriptase.
FIG. 4.
RT-PCR analysis of the ORF73 5′ UTR. BCBL-1 RNA or DNA was reverse transcribed with primer 7308 and PCR amplified with the primers indicated above the lanes. A one hundred-base-pair molecular size ladder is shown in the left lane of each gel. The 600-bp fragment hybridized to the polylinker sequence in the probe. Where indicated (RT−), reverse transcriptase was omitted from the reaction. An ethidium-bromide-stained gel (top) and an autoradiogram of the same gel hybridized with a probe specific for the UTR (lower gel panel) are shown. Note that in the RT+ panel two fragments are amplified in lane 7 and none in lane 9. The deduced structure of the 5′ UTR and the locations of primers (small arrows) are shown below the gels. The numbers on the right of the diagram denote the 5′ end and splice junction, as determined from RACE clones.
FIG. 5.
S1 nuclease protection analysis of the KSHV latency promoter. Shown is an autoradiogram of a 12% denaturing acrylamide gel. Lane 1 shows the input, undigested probe; lanes 2 to 4 show the protected fragments resulting from hybridization to 30 μg of BCBL RNA (lane 2), 15 μg of BCBL-1 RNA (lane 3), or 30 μg of yeast RNA (lane 4) followed by S1 nuclease digestion at 28°C. The arrows point to the two most prominent protected fragments, and size standards are indicated on the left. The asterisks denote the site of the radiolabel.
In addition to the full-length unspliced 5′ UTR, RT-PCR with primer 7 also amplified a smaller product (Fig. 4, RT+, lane 7). Cloning and sequencing identified this band as emanating from an alternatively spliced message lacking nt 127477 to 127813, which are framed by consensus donor and acceptor splice signals (Fig. 4). Notably, an identical splice has also been observed in a single partial cDNA clone bearing ORF73 coding sequences (23). The entire intron is located in the 5′ UTR of the RNA; hence, the alternative splice does not affect the coding information for ORF73. Based on the KSHV sequence, the exact size of the unspliced RNA is 5,814 nt and that of the spliced RNA is 5,478 nt, a difference that might be expected to be resolved by Northern blotting. However, our Northern blots detected only a single band in this region, with a size of ca. 5.8 kb (Fig. 2). Since small products should be amplified more efficiently than longer ones when common primers are employed, the lower copy number of the smaller RT-PCR product indicates that its spliced RNA template must be substantially less abundant than that of the cognate unspliced RNA. This presumably explains why it was not detected by Northern blotting. (The failure to detect this species by RT-PCR with primer 8 can also be explained: it is due to the fact that the start sites of the RNA map within the sequence of this primer, impairing annealing to the oligonucleotide.)
The 1.7-kb ORF72 mRNA is also derived by splicing from the latent promoter.
To determine whether the 1.7-kb latent transcript might also be derived by alternative splicing from a common pre-mRNA, RT-PCR was performed with downstream primers located within ORF72 (primers 7208 and 7207) and primer 7 upstream of the ORF73 alternative splice donor (Fig. 4). Four independent clones were isolated and sequenced; these revealed a 4,037-nt splice (nt 127813 to 123776) which removes the entire ORF73 coding region. This predicts a 1,777-nt transcript in which the same splice donor site (nt 127813) used in the ORF73 transcript is fused to an alternative splice acceptor (at nt 123776), some 209 nt 5′ to the AUG for ORF72 (encoding v-cyclin). The predicted size of this RNA is in excellent agreement with the size of the RNA observed by Northern blotting (Fig. 2).
Two hundred fifty base pairs upstream of the ORF73 start site suffice to drive reporter gene expression.
The region 5′ to the start sites of the 5.8-kb transcript was scanned for recognizable cis-acting elements characteristic of pol II promoters. No canonical TATA box was identified by using a variety of pattern-matching programs; the presence of multiple 5′ ends in the RNA is consistent with this finding, since TATA-less promoters typically display such degeneracy in their initiation sites. The region upstream of nt −30 contains numerous predicted transcription factor binding sites, including recognition elements for SP-1, CAAT, NF1, Oct-1 and GATA family member binding sites (Fig. 6).
FIG. 6.
Sequence and putative transcription factors surrounding the latent start site. Shown are the principal start sites (vertical arrows), the predicted binding sites for selected transcription factors (shaded), and the positions of primers used for PCR and S1 analysis. The boldfaced horizontal arrow indicates the 5′ region and splice site of the 5.4- and 1.7-kb transcripts. The boxed sequence represents the predicted ORFK14 coding region. Nucleotide positions are according to the sequence of Russo et al. Lowercase letters represent adjacent sequence not present in the mRNA.
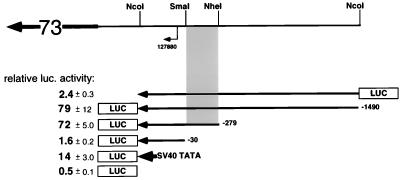
To see if this region could function as a promoter in a heterologous context, DNA fragments from the region were cloned into a promoterless luciferase reporter construct (pGL3basic) and assayed for promoter activity by transient transfection of 293 cells (Fig. 7). (293 cells were chosen because of evidence that they are at least semipermissive for KSHV infection [7, 24].) Luciferase activity was determined as the mean ± standard deviation of three independent transfections, each normalized for transfection efficiency with a SEAP reporter (pSEAP) as the internal standard. A 1,763-bp NcoI-NcoI fragment (pDD41; nt 127609 to 129370) with the ORF73 start site facing the luciferase gene resulted in a 79- ± 12-fold increase in luciferase expression, while the same DNA in the opposite direction (pDD43) did not register significant activity above background (2.4- ± 0.3-fold compared to 0.5- ± 0.1-fold for the promoterless vector alone). A reporter construct containing a shorter, 552-bp NcoI-NheI fragment (pDD83; nt 127609 to 128159) had essentially the same activity (72- ± 5-fold above background), while a 303-bp NcoI-SmaI fragment (pDD53; nt 127607 to 127910) extending only 30 bp upstream of the ORF73 transcription initiation site was inactive (1.6- ± 0.2-fold) (Fig. 7). For purposes of comparison, in the same series of transfections the SV40 minimal TATA and initiation element (without the SV40 enhancer, pGL3SV40) had an activity of 14 ± 3. Qualitatively similar results were obtained in other cell lines: LnCAP, which is semipermissive for KSHV infection, and HeLa and CV-1, which are nonpermissive (24). This localizes the minimal ORF73 promoter to a 250-bp region between nt 127910 and 128159 of KSHV (Fig. 7).
FIG. 7.
Promoter analysis of the KSHV latency promoter. Shown are fold transactivations (averages ± standard deviations) for selected reporter constructs and their locations relative to the latent start site. Nucleotide positions are according to the sequence of Russo et al. (29). The shaded area represents the minimal ORF73 promoter. The boldfaced arrow containing 73 represents the ORF73 transcript. luc., luciferase.
DISCUSSION
This report describes the characterization of 5.8-, 5.5-, and 1.7-kb latent KSHV mRNAs capable of encoding at least the products of ORF73 and -72. These messages are transcribed at low-to-moderate levels in the absence of TPA and are poorly inducible by that compound. This transcription unit is directly adjacent to another latently expressed region of the genome, that surrounding ORFK12. We do not know if the clustering of these latency-associated genes into a contiguous region of the genome has regulatory or evolutionary significance. In KS tumors, both the K12 region and ORF73-ORF72 RNAs are present in the majority of the spindle cells of the tumor. However, the regulation of the K12 locus differs from that of the ORF73-ORF72 cluster in that it is directed by independent promoter elements that display a high basal level of activity and strong upregulation by TPA. In fact, work by us (11, 37) and others (30) shows that the ORF73-ORF72 cluster is the only region of the genome that displays its characteristic pattern of significant basal activity and poor inducibility by TPA. We anticipate that additional latent genes remain to be discovered, but most of these will likely have more complicated transcriptional phenotypes than those described here: for example, low or variable basal expression with significant induction by TPA. Such genes can be confused with lytic-cycle genes when analyzed in biochemical experiments on cell populations; single-cell analysis by in situ hybridization or immunohistochemistry will likely be required for their proper characterization.
Although the RNA structures defined here identify the likely templates for translation of ORF73 and -72, we have been unable to identify monocistronic transcripts for ORF71, which encodes a viral protein posited to be involved in the prevention of apoptosis (34). No RNA of the proper size is evident in Northern blots with ORF71-specific probes (Fig. 2). It remains possible that the product of ORF71 is encoded by a rare, independently initiated monocistronic RNA that is below the detection threshold of our analytical procedures; alternatively, it may be that the 1.7-kb mRNA may be functionally bicistronic. Experimental tests of the latter possibility are under way.
The structures of the transcripts described here are strongly predictive of the expression of LANA and v-cyclin in PEL and KS tissues. (Indeed, LANA protein expression is known to occur in PEL cells.) The function of LANA is unknown, though its nuclear location and many acidic repeats suggest that it might be a modulator of transcription. Alternatively, it could serve an EBNA1-like function in viral plasmid maintenance and/or other as-yet-undiscovered functions. The identification of v-cyclin (encoded by ORF72) as the product of a latently expressed gene is particularly noteworthy, given the strong association of latent KSHV infection with several human neoplasms (KS and PEL). Others have shown that the KSHV cyclin gene can associate with and activate cdk6, overcome an Rb-mediated growth arrest, and cause the activity of cdk6 to become resistant to inhibition by known cdk inhibitors (9, 17, 33). Moreover, deregulated cellular D-type cyclins are strongly and specifically linked to several forms of human cancer (21). The finding that this gene is a part of the viral latency program, which in other gammaherpesviruses includes key genes affecting growth deregulation, strongly suggests an important role for its product in KSHV-related proliferative syndromes.
ACKNOWLEDGMENTS
M.L. and R.R. contributed equally to this work.
This work was supported by a grant from the National Institutes of Health. Rolf Renne is a Fellow of the Leukemia Society.
REFERENCES
- 1.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Nador R G, Bai F, Bohenzky R A, Russo J J, Moore P S, Chang Y, Knowles D M. Kaposi’s sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi’s sarcoma and malignant lymphoma. J Virol. 1996;70:8218–8223. doi: 10.1128/jvi.70.11.8218-8223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 4.Chang Y, Moore P S, Talbot S J, Boshoff C H, Zarkowska T, Godden K, Paterson H, Weiss R A, Mittnacht S. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. doi: 10.1038/382410a0. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 5.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis M A, Sturzl M A, Blasig C, Schreier A, Guo H G, Reitz M, Opalenik S R, Browning P J. Expression of human herpesvirus 8-encoded cyclin D in Kaposi’s sarcoma spindle cells. J Natl Cancer Inst. 1997;89:1868–1874. doi: 10.1093/jnci/89.24.1868. [DOI] [PubMed] [Google Scholar]
- 7.Foreman K E, Friborg J, Jr, Kong W P, Woffendin C, Polverini P J, Nickoloff B J, Nabel G J. Propagation of a human herpesvirus from AIDS-associated Kaposi’s sarcoma. N Engl J Med. 1997;336:163–171. doi: 10.1056/NEJM199701163360302. [DOI] [PubMed] [Google Scholar]
- 8.Gao S J, Kingsley L, Hoover D R, Spira T J, Rinaldo C R, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore P S. Seroconversion to antibodies against Kaposi’s sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi’s sarcoma. N Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 9.Godden-Kent D, Talbot S J, Boshoff C, Chang Y, Moore P, Weiss R A, Mittnacht S. The cyclin encoded by Kaposi’s sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kedes D H, Ganem D, Ameli N, Bacchetti P, Greenblatt R. The prevalence of serum antibody to human herpesvirus 8 (Kaposi sarcoma-associated herpesvirus) among HIV-seropositive and high-risk HIV-seronegative women. JAMA. 1997;277:478–481. [PubMed] [Google Scholar]
- 11.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi’s sarcoma-associated herpesvirus. J Clin Invest. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kedes D H, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. . (Erratum, 2:1041.) [DOI] [PubMed] [Google Scholar]
- 13.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss R A, Talbot S J. Identification of a major latent nuclear antigen LNA-1, in the human herpesvirus 8 genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- 13a.Kirshner, J., and D. Ganem. Unpublished data.
- 14.Klein G. Epstein-Barr virus strategy in normal and neoplastic B cells. Cell. 1994;77:791–793. doi: 10.1016/0092-8674(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 15.Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 16.Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi’s sarcoma-associated herpesvirus. Virology. 1997;236:147–154. doi: 10.1006/viro.1997.8713. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Lee H, Yoon D-W, Albrecht J-C, Fleckenstein B, Neipel F, Jung J U. Kaposi’s sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mesri E A, Cesarman E, Arvanitakis L, Rafii S, Moore M A, Posnett D N, Knowles D M, Asch A S. Human herpesvirus-8/Kaposi’s sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J Exp Med. 1996;183:2385–2390. doi: 10.1084/jem.183.5.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller G, Rigsby M O, Heston L, Grogan E, Sun R, Metroka C, Levy J A, Gao S J, Chang Y, Moore P. Antibodies to butyrate-inducible antigens of Kaposi’s sarcoma-associated herpesvirus in patients with HIV-1 infection. N Engl J Med. 1996;334:1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- 20.Moore P S, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi’s sarcoma in patients with and without HIV infection. N Engl J Med. 1995;332:1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 21.Motokura T, Bloom T, Kim H G, Juppner H, Ruderman J V, Kronenberg H M, Arnold A. A novel cyclin encoded by a bcl1-linked candidate oncogene. Nature. 1991;350:512–515. doi: 10.1038/350512a0. [DOI] [PubMed] [Google Scholar]
- 22.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S-J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol. 1996;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 27.Rickenson A B, Kief E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 28.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2221–2230. [Google Scholar]
- 29.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 32.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanton C, Mann D J, Fleckenstein B, Neipel F, Peters G, Jones N. Herpes viral cyclin/Cdk6 complexes evade inhibition by CDK inhibitor proteins. Nature. 1997;390:184–187. doi: 10.1038/36606. [DOI] [PubMed] [Google Scholar]
- 34.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 35.Whitby D, Howard M R, Tenant-Flowers M, Brink N S, Copas A, Boshoff C, Hatzioannou T, Suggett F E, Aldam D M, Denton A S, et al. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi’s sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 36.Zhong W, Ganem D. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) J Virol. 1997;71:1207–1212. doi: 10.1128/jvi.71.2.1207-1212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]