Abstract
The 25-kilodalton (25K), 12K, and 8K movement proteins of potato virus X are derived from overlapping open reading frames (ORFs). Using an in vivo complementation assay, we have shown that the 25K protein is expressed from a functionally monocistronic mRNA, whereas the 12K and 8K proteins are from a bicistronic mRNA. Translation of the 8K ORF is by leaky ribosome scanning through the 12K ORF.
The triple gene block (TGB) of potato virus X (PVX) encodes three proteins (25-kilodalton [25K], 12K, and 8K proteins) required for virus transport (5) and is a genetic module that is conserved among members of the Potexvirus, Hordeivirus, Furovirus, and Carlavirus genera (18, 21, 28). Several properties of the TGB proteins have been identified, but it is not known how all of these activities contribute to virus movement. For example, consistent with the predicted properties of a viral movement protein, the 25K protein of PVX influences plasmodesma gating (2) and the homologous 25K protein of foxtail mosaic potexvirus has ATPase and RNA binding activity (29). However, unlike some viral movement proteins affecting plasmodesma gating, the 25K protein of PVX accumulates in laminate inclusions and has not been localized to plasmodesmata (11). The 12K and 8K proteins of PVX were studied by using in vitro translation systems and may be membrane-associated proteins (27).
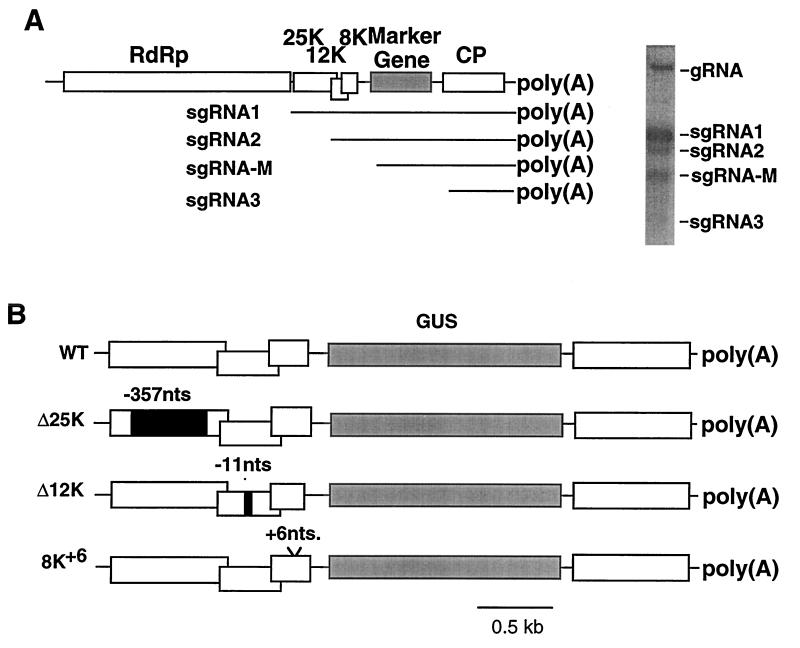
In vivo studies indicated that two separate subgenomic RNAs (sgRNAs) are required for expression of the entire TGB of barley stripe mosaic hordeivirus and beet necrotic yellow vein furovirus (16, 31). In both cases, expression of the first open reading frame (ORF) was from a functionally monocistronic mRNA, while expression of the two downstream ORFs was from a bicistronic mRNA (7, 32). In the case of PVX, the results of in vitro translation studies (12, 26) using synthesized RNAs indicated that two 3′ coterminal sgRNAs (sgRNA1 and sgRNA2) of 2.1 and 1.4 kb were necessary for translation of the entire TGB, while a third sgRNA of 0.9 kb (sgRNA3) was required for expression of the viral coat protein (CP; Fig. 1A).
FIG. 1.
(A) Diagrammatic representation of the PVX genome. Open boxes indicate the five viral ORFs, and the shaded box indicates an introduced marker gene (in this study, GUS or GFP). The proteins encoded by each ORF include the RNA-dependent RNA polymerase (RdRp), the TGB proteins (25K, 12K, and 8K), and CP. The sgRNAs of the TGB and CP genes are labelled sgRNA1, -2, and -3. The sgRNA-M is synthesized from a duplicated sgRNA promoter and is the mRNA for the marker protein. The Northern blot of RNA from infected tobacco protoplasts shows each of the predicted genomic and sgRNAs. (B) Diagrammatic representation of the 3′ region of the genomes of PVX.GUS (WT) and three mutant derivatives, Δ25K, Δ12K, and 8K+6. Black boxes within the TGB ORFs of the Δ25K and Δ12K viruses depict deletions of 357 and 11 nt, respectively. The 8K+6 mutant has an insertion of 6 nt.
However, in several studies of PVX (9, 10, 20) and of the related potexviruses, foxtail mosaic virus, white clover mosaic virus, and clover yellow mosaic virus (3, 6, 14), only two RNA species corresponding to sgRNA1 and sgRNA3 were observed in infected plants. These data suggest that the PVX sgRNA2 either accumulates to a very low level or is not required for complete translation of the TGB.
Here we describe an in vivo analysis of the PVX TGB translation strategy. We produced plants carrying TGB transgene constructs corresponding to possible mono-, bi-, and tricistronic mRNAs. The results of this study confirm the results of in vitro studies indicating that the 25K ORF is translated from a functionally monocistronic mRNA and that the 12K and 8K proteins are derived from a functionally bicistronic mRNA. Additionally, we present evidence that the 8K ORF is translated by leaky ribosome scanning through the 12K ORF.
Separate RNAs are required for expression of the PVX TGB proteins.
To determine the expression strategy of the PVX TGB, we produced transgenic tobacco expressing mono-, bi-, and tricistronic mRNAs mimicking TGB mRNAs that may be present in infected plants. Six binary plasmids containing PVX sequences were prepared for Agrobacterium-mediated transformation of tobacco leaf discs and are described in Table 1. To produce these constructs, the entire TGB and fragments of the TGB were amplified by PCR and inserted between the 35S promoter and octapine synthase terminator in the plasmid SLJ4D4 (19). Fragments containing the promoter, PVX ORF(s), and terminator were recovered from the SLJ4D4 plasmid by digestion with EcoRI and HindIII and were ligated to the EcoRI-HindIII-linearized binary plasmid SLJ7292 as described previously (1).
TABLE 1.
Cell-to-cell movement of mutant PVX.GUS viruses on transgenic tobacco
| Transgene constructa | No. of lines testedb | No. of transgenic linesc complementing mutant:
|
|||
|---|---|---|---|---|---|
| Δ25K | Δ12K | 8K+6 | |||
| TGB100 | 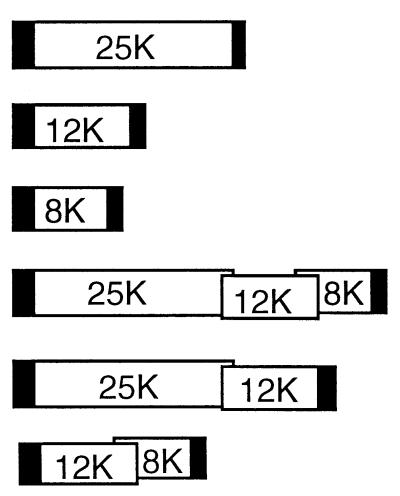 |
22 | 16 | —d | — |
| TGB200 | 9 | — | 3 | — | |
| TGB300 | 11 | — | — | 7 | |
| TGB400 | 21 | 21 | 0e | 0 | |
| TGB500 | 5 | 5 | 0 | — | |
| TGB600 | 6 | — | 4 | 3 | |
Dark boxes indicate promoter and terminator regions. 25K, 12K, and 8K are the names of the TGB ORFs of PVX.
Ten to 20 plants per line were tested by inoculation with each virus.
Number of lines complementing cell-to-cell movement of mutant viruses was measured by GUS assay at 5 dpi. Wild-type PVX.GUS moved from cell to cell in all lines tested.
—, lines not tested with specific viruses.
Lines yielding no positive results.
Expression of the TGB100, TGB300, TGB400, and TGB500 transgenes was analyzed by Western blotting using anti-25K and anti-8K sera (data not shown). Expression of the transgenes was also analyzed by Western or Northern blotting with selected T1 transgenic plants (data not shown). Accumulation of the 12K protein in the TGB200 and TGB600 lines was not analyzed by Western blotting because we did not have suitable antisera.
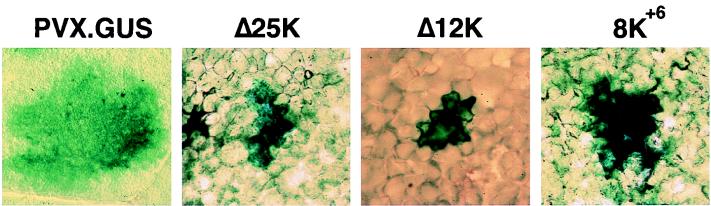
Two versions of PVX were used in these studies and were derived from PVX.GUS and PVX.GFP plasmids. These plasmids contain cDNA of PVX with either the β-glucuronidase (GUS) or green fluorescence protein (GFP) genes inserted adjacent to a duplicated sgRNA3 promoter (4, 9). Three PVX.GUS derivatives were prepared by PCR mutagenesis (17) and used initially to assess transgenic complementation of viral movement defects (Fig. 1B). The Δ25K mutant contains a deletion mutation extending between genomic nucleotides (nt) 4587 to 4945 as described previously (2), the Δ12K mutant contains a frameshift mutation resulting from the deletion of 11 nt between genomic nt 5240 to 5250, and the 8K+6 mutant has 6 nt (CGCGTA) inserted after nt 5542 in the PVX genome. The transgenic tobacco plants were inoculated with infectious transcripts (of the 8K+6 mutant) or with virions of Δ25K and Δ12K. The virions obtained from transgenic tobacco expressing the Δ25K and Δ12K viral genomes (1) were used in this study as a source of inoculum for approximately 1,000 plants that was produced more economically than by transcription of cDNA clones.
Leaves of T1 transgenic plants and nontransgenic plants inoculated with Δ25K, Δ12K, and 8K+6 were detached at 5 days postinoculation (dpi) and processed for histochemical assay of GUS, as described previously (13). By 5 dpi on nontransgenic plants, PVX.GUS foci were 10 to 15 cells in diameter (Fig. 2). However, the three mutant viruses were defective for cell-to-cell movement (Fig. 2), and in the inoculated leaves the GUS activity was restricted to initially infected cells.
FIG. 2.
Results of GUS histochemical analysis of tobacco leaves at 5 dpi with PVX.GUS, Δ25K, Δ12K, and 8K+6 mutant viruses. Expanding infection foci on leaves inoculated with PVX.GUS were photographed with a 1× objective and a Leica MZ12 microscope. The GUS activity in single infected cells on leaves inoculated with mutant viruses was photographed with a 20× objective and a Zeiss Axiophot microscope.
The cell-to-cell movement defects were complemented in trans when the mutant viruses were inoculated to plants expressing the individual wild-type TGB ORFs (Table 1). Thus, on the TGB100 lines expressing the 25K gene, there was complementation of the Δ25K mutant (16 of 22 lines tested positive); in the TGB200 lines expressing the 12K gene there was complementation of the Δ12K mutant (3 of 9 lines tested positive); and in the TGB300 lines expressing the 8K gene there was complementation of the 8K+6 mutant (7 of 11 lines tested positive; Table 1). These data confirm findings that the 25K protein is required for PVX cell-to-cell movement (2, 25) and demonstrate directly that the 12K and 8K proteins are also required for virus movement out of the initially infected cells.
There was also complementation of the TGB mutants inoculated to plants carrying bi- or tricistronic transgenes. However, on these lines, with one exception, complementation occurred only if the PVX mutation corresponded to the 5′-most ORF in the transgene (Table 1). Thus, the Δ25K mutant was complemented on TGB400 (21 of 21 lines tested positive) and TGB500 lines (5 of 5 lines tested positive). The 25K ORF was the 5′-most ORF in both of these transgenes. The Δ12K mutant was complemented on TGB600 lines (four of six lines tested positive), in which the 5′-most ORF encoded the 12K protein, but not on lines TGB400 or TGB500, in which the 12K ORF was internal. The one exception in which there was complementation of an internal ORF was with the 8K+6 mutant inoculated to TGB600 lines. In these lines, the 8K ORF was expressed as an internal ORF in the transgene RNA. Thus, there was expression of the 12K and 8K proteins from a bicistronic 12K/8K mRNA in the TGB600 lines but not from a tricistronic mRNA encoding the entire TGB in the TGB400 and TGB500 lines. These complementation data are therefore consistent with the previous in vitro studies (26), suggesting that the 2.1-kb sgRNA1 is functionally monocistronic for the 25K protein and that the 12K and 8K proteins are encoded in sgRNA2.
The translation strategy of the PVX 12K and 8K proteins from a bicistronic RNA.
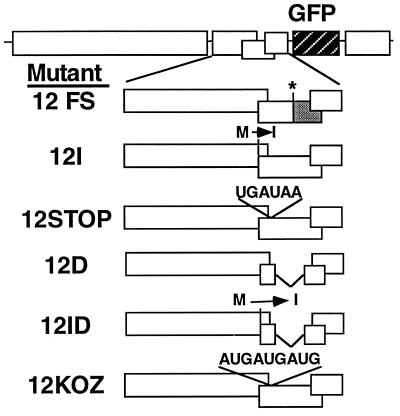
The complementation data (Table 1) show that the TGB600 mRNA served as a bicistronic mRNA for expression of the 12K and 8K proteins. If there is a corresponding bicistronic viral mRNA in PVX-infected cells, we predicted that certain types of mutations in the 12K ORF of inoculated PVX would act in cis to prevent translation of the 8K ORF. The type of 12K mutation that would prevent 8K protein expression depends on the mechanism by which the 8K ORF is translated from a bicistronic mRNA. Thus, six PVX.GFP mutants (Fig. 3) containing mutations in the 12K ORF were prepared by PCR mutagenesis (17). The 12FS mutant has a deletion mutation identical to that of Δ12K. In the 12I mutant, the AUG translation initiation codon was converted to an AUA codon by replacing the methionine codon with one for isoleucine. The 12STOP mutant contains the sequence C UAG UGA UAA in which the C replaces a U in the PVX and has the UGA UAA motif inserted after nt 5167. This motif introduced two stop codons 20 nt downstream of the 12K ORF initiation codon. The mutant 12KOZ contains the sequence GGGGACCAUGAUGAUGGCAG GG inserted at this same position. This sequence contains three translation initiation codons within a Kozak consensus sequence (23, 30). The 12D mutant lacks the coding sequence for the entire 12K ORF between nt 5170 to 5423, leaving intact only the first 20 nt, overlapping the 25K ORF, and the last 70 nt, of which 67 nt overlap the 8K ORF. The 12ID mutant contains a replacement of the translation initiation codon with a codon for isoleucine and a deletion of 20 nt within the 12K ORF.
FIG. 3.
Diagrammatic representation of PVX.GFP and six mutant derivatives. The 12FS mutation interferes with expression of a region of the 12K ORF, as indicated by the shaded box. Mutations converting a methionine start codon to an isoleucine codon are indicated above the 12K ORF of mutants 12I and 12ID. Nucleotide sequence insertions in the 12STOP and 12KOZ mutants are also indicated above the 12K ORF. Deletions in mutants 12D and 12ID are indicated by gaps in the 12K ORF.
The mutations in 12FS, 12I, 12ID, and 12STOP (Fig. 3) would each prevent translation of the 8K ORF if it was translated by ribosome frameshifting or by readthrough of the 12K ORF stop codon. The ribosomes either would not initiate translation of the 12K ORF or would disengage before reaching the 8K ORF. The 12D mutant would prevent internal entry of ribosomes translating the 8K ORF by removal of any sequences that could potentially function as an internal ribosome entry site. Finally, the 12KOZ mutant would prevent translation of the 8K ORF by leaky ribosome scanning because the mutation introduces several initiation codons at the start of the 12K ORF. All three of these initiation codons lie within a good translation context (22, 30) so that leaky ribosome scanning would be prevented.
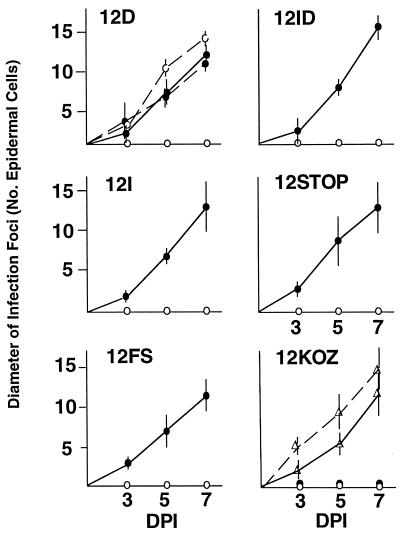
Each of these 12K mutations was introduced into the PVX.GFP background so that cell-to-cell movement could be monitored directly under UV illumination over a 7-day period (Fig. 4). On nontransgenic plants, each 12K mutant virus was restricted to the initially infected cells, as evidenced by GFP expression. However, all 12K mutant viruses, except 12KOZ, moved from cell to cell on TGB200 plants expressing the 12K protein. The phenotype of 12KOZ is likely due to an effect of the mutations on expression of the 8K ORF, because the cell-to-cell movement defect was complemented on TGB600 plants expressing both the 12K and 8K proteins. Therefore, we conclude that expression of the 8K protein in PVX is achieved by leaky ribosome scanning through the 12K ORF.
FIG. 4.
Time course analysis of expanding PVX infection foci, measured by using GFP as a visual marker to count the number of infected epidermal cells across an infection focus. The average diameters of 10 infection foci on an inoculated leaf of nontransgenic (○), TGB200 (•), and TGB600 (▵) tobacco plants were measured at 3, 5, and 7 dpi. Dotted lines in two panels indicate the cell-to-cell movement of wild-type PVX.GFP, while solid lines indicate the cell-to-cell movement of the mutant viruses named in each panel.
Leaky ribosome scanning plays a role in translation from downstream start codons in some positive-stranded RNA viruses and retroviruses (15, 24) and requires that the translation initiation codon of the first ORF is in a suboptimal sequence context (22). Efficient translation initiation from the 5′ end of the mRNA requires a purine at position −3 from the start codon and a guanine at position +4 from the start codon (8, 15). Conversely, a suboptimal context has a pyrimidine at position −3 from the start codon and position +4 does not contain a guanine. The translation initiation codon of the PVX 12K ORF is in an unfavorable context for translation initiation, with a pyrimidine (cytosine) at position −3 and a uridine at position +4. The initiation codons of the second ORF in the TGB of beet necrotic yellow vein furovirus and barley stripe mosaic hordeivirus are also in a suboptimal context, indicating that, like PVX, these viruses use leaky scanning as a control mechanism for translation of the third TGB ORF (7, 32).
The conserved linear arrangement of the genes within the TGBs of the hordeivirus, furovirus, and potexvirus families and the similar TGB expression strategies suggest that the mechanisms used for expression of the TGB proteins have functional significance. The most obvious functional reason for conservation of the TGB would concern the stoichiometry of TGB protein production. For example, by coupling expression of the 12K and 8K proteins from the same RNA it could be ensured that they are produced in relative amounts that result in the most efficient function. An indication that TGB protein function is dependent on relative levels is from the complementation of TGB mutations in beet necrotic yellow vein furovirus: the complementation of defective cell-to-cell movement occurred only if the 13K and 15K TGB proteins were expressed from the same bicistronic mRNA (7). The situation in PVX is not precisely the same because, as described here, cell-to-cell movement was complemented by expression of any of the TGB proteins from monocistronic transgenes. However, in none of our TGB transgenic lines was there complementation of long-distance movement defects in the inoculated virus (data not shown). Perhaps precise stoichiometry of the TGB proteins is required for PVX to enter or leave the vascular system.
Acknowledgments
The Sainsbury Laboratory is funded by the Gatsby Charitable Foundation.
We thank Tamas Dalmay, Kostya Kanyuka, Isabelle Malcuit, and Steven Rudd for comments on the draft script.
REFERENCES
- 1.Angell S M, Baulcombe D C. Consistent gene silencing in transgenic plants expressing a replicating potato virus X RNA. EMBO J. 1997;16:3675–3684. doi: 10.1093/emboj/16.12.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angell S M, Davies C, Baulcombe D C. Cell-to-cell movement of potato virus X is associated with a change in the size exclusion limit of plasmodesmata in trichome cells of Nicotiana clevelandii. Virology. 1996;215:197–201. doi: 10.1006/viro.1996.0046. [DOI] [PubMed] [Google Scholar]
- 3.Bancroft J B, Rouleau M, Johnston R, Prins L, Mackie G A. The entire nucleotide sequence of foxtail mosaic virus RNA. J Gen Virol. 1991;72:2173–2181. doi: 10.1099/0022-1317-72-9-2173. [DOI] [PubMed] [Google Scholar]
- 4.Baulcombe D C, Chapman S N, Santa Cruz S. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 1995;7:1045–1053. doi: 10.1046/j.1365-313x.1995.07061045.x. [DOI] [PubMed] [Google Scholar]
- 5.Beck D L, Guilford P J, Voot D M, Andersen M T, Forster R L S. Triple gene block proteins of white clover mosaic potexvirus are required for transport. Virology. 1991;183:695–702. doi: 10.1016/0042-6822(91)90998-q. [DOI] [PubMed] [Google Scholar]
- 6.Bendena W G, Bancroft J B, Mackie G A. Molecular cloning of clover yellow mosaic virus RNA: identification of coat protein coding sequences in vivo and in vitro. Virology. 1987;157:276–284. doi: 10.1016/0042-6822(87)90270-4. [DOI] [PubMed] [Google Scholar]
- 7.Bleykasten-Grosshans C, Guilley H, Bouzoubaa S, Richards K E, Jonard G. Independent expression of the first two triple gene block proteins of beet necrotic yellow vein virus complements virus defective in the corresponding gene but expression of the third protein inhibits viral cell-to-cell movement. Mol Plant-Microbe Interact. 1997;10:240–246. [Google Scholar]
- 8.Cavener D R, Ray S C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman S N, Kavanagh T A, Baulcombe D C. Potato virus X as a vector for gene expression in plants. Plant J. 1992;2:549–557. doi: 10.1046/j.1365-313x.1992.t01-24-00999.x. [DOI] [PubMed] [Google Scholar]
- 10.Davenport G F, Baulcombe D C. Mutation of the GKS motif of the RNA dependent RNA polymerase from potato virus X either disables or eliminates virus replication. J Gen Virol. 1997;78:1247–1251. doi: 10.1099/0022-1317-78-6-1247. [DOI] [PubMed] [Google Scholar]
- 11.Davies C, Hills G J, Baulcombe D C. Subcellular localisation of the 25kDa protein encoded by the triple gene block of potato virus X. Virology. 1993;197:166–175. doi: 10.1006/viro.1993.1577. [DOI] [PubMed] [Google Scholar]
- 12.Dolja V V, Grama D P, Morozov S Y, Atabekov J G. Potato virus X-related single- and double-stranded RNAs. Characterization and identification of terminal structures. FEBS Lett. 1987;214:308–312. [Google Scholar]
- 13.Dolja V V, McBride H J, Carrington J C. Tagging of plant potyvirus replication and movement by insertion of β-glucuronidase into the viral polyprotein. Proc Natl Acad Sci USA. 1992;89:10208–10212. doi: 10.1073/pnas.89.21.10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forster R L S, Beck D L, Guilford P J, Voot D M, Van Dolleweerd C J, Andersen M T. The coat protein of white clover mosaic potexvirus has a role in facilitating cell-to-cell transport in plants. Virology. 1992;191:480–484. doi: 10.1016/0042-6822(92)90215-b. [DOI] [PubMed] [Google Scholar]
- 15.Futterer J, Hohn T. Translation in plants—rules and exceptions. Plant Mol Biol. 1996;32:159–189. doi: 10.1007/BF00039382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmer D, Bouzoubaa S, Hehn A, Guilley H, Richards K, Jonard G. Efficient cell-to-cell movement of beet necrotic yellow vein virus requires 3′ proximal genes located on RNA2. Virology. 1992;189:40–47. doi: 10.1016/0042-6822(92)90679-j. [DOI] [PubMed] [Google Scholar]
- 17.Ho S, Hunt H D, Horton R M, Pullen K J, Peare L R. Site-directed mutagenesis by overlap extension using the PCR. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 18.Huisman M J, Linthorst H J M, Bol J F, Cornelissen B J C. The complete nucleotide sequence of potato virus X and its homologies at the amino acid level with various plus-stranded RNA viruses. J Gen Virol. 1988;69:1789–1798. doi: 10.1099/0022-1317-69-8-1789. [DOI] [PubMed] [Google Scholar]
- 19.Jones J D G, Shlumukov L, Carland F, English J J, Scofield S R, Bishop G, Harrison K. Effective vectors for transformation, expression of heterologous genes, and assaying transposon excision in transgenic plants. Transgenic Res. 1992;1:285–297. doi: 10.1007/BF02525170. [DOI] [PubMed] [Google Scholar]
- 20.Kim K H, Hemenway C. The 5′ nontranslated region of potato virus X RNA affects both genomic and subgenomic RNA synthesis. J Virol. 1996;70:5533–5540. doi: 10.1128/jvi.70.8.5533-5540.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 24.Lin C-G, Lo S J. Evidence for involvement of ribosomal leaky scanning mechanism in the translation of the hepatitis B virus pol gene from the viral pregenome RNA. Virology. 1992;188:342–352. doi: 10.1016/0042-6822(92)90763-f. [DOI] [PubMed] [Google Scholar]
- 25.Morozov S Y, Fedorkin O N, Juttner G, Schiemann J, Baulcombe D C, Atabekov J G. Complementation of a potato virus X mutant mediated by bombardment of plant tissues with cloned viral movement protein genes. J Gen Virol. 1997;78:2077–2083. doi: 10.1099/0022-1317-78-8-2077. [DOI] [PubMed] [Google Scholar]
- 26.Morozov S Y, Miroshnichenko N A, Solovyev A G, Fedorkin O N, Zelenina D A, Lukasheva L I, Karasev A V, Dolja V V, Atabekov J G. Expression strategy of the potato virus X triple gene block. J Gen Virol. 1991;72:2039–2042. doi: 10.1099/0022-1317-72-8-2039. [DOI] [PubMed] [Google Scholar]
- 27.Morozov S Y, Miroshnichenko N A, Zelenina D A, Fedorkin O N, Solovijev A G, Lukasheva L I, Atabekov J G. Expression of RNA transcripts of potato virus X full-length and subgenomic cDNAs. Biochimie. 1990;72:677–684. doi: 10.1016/0300-9084(90)90051-h. [DOI] [PubMed] [Google Scholar]
- 28.Petty I T D, Jackson A O. Mutational analysis of barley stripe mosaic virus RNA β. Virology. 1990;179:712–718. doi: 10.1016/0042-6822(90)90138-h. [DOI] [PubMed] [Google Scholar]
- 29.Rouleau M, Smith R J, Bancroft J B, Mackie G A. Purification, properties, and subcelular localization of foxtail mosaic potexvirus 26-kDa protein. Virology. 1994;204:254–265. doi: 10.1006/viro.1994.1530. [DOI] [PubMed] [Google Scholar]
- 30.Wakiyama M, Hirao I, Kumagai I, Miura K. Effect of tandem repeated AUG codons on translation efficiency of eukaryotic mRNA carrying a short leader sequence. Mol Gen Genet. 1993;238:59–64. doi: 10.1007/BF00279531. [DOI] [PubMed] [Google Scholar]
- 31.Weiland J J, Edwards M C. Evidence that the Aa gene of barley stripe mosaic virus encodes determinants of pathogenicity to oat (Avena sativa) Virology. 1994;201:116–126. doi: 10.1006/viro.1994.1271. [DOI] [PubMed] [Google Scholar]
- 32.Zhou H, Jackson A O. Expression of the barley stripe mosaic virus RNAβ “triple gene block.”. Virology. 1996;216:367–379. doi: 10.1006/viro.1996.0072. [DOI] [PubMed] [Google Scholar]