ABSTRACT
Functional genomics techniques, such as transposon insertion sequencing and RNA-sequencing, are key to studying relative differences in bacterial mutant fitness or gene expression under selective conditions. However, certain stress conditions, mutations, or antibiotics can directly interfere with DNA synthesis, resulting in systematic changes in local DNA copy numbers along the chromosome. This can lead to artifacts in sequencing-based functional genomics data when comparing antibiotic treatment to an unstressed control. Further, relative differences in gene-wise read counts may result from alterations in chromosomal replication dynamics, rather than selection or direct gene regulation. We term this artifact “chromosomal location bias” and implement a principled statistical approach to correct it by calculating local normalization factors along the chromosome. These normalization factors are then directly incorporated into statistical analyses using standard RNA-sequencing analysis methods without modifying the read counts themselves, preserving important information about the mean-variance relationship in the data. We illustrate the utility of this approach by generating and analyzing a ciprofloxacin-treated transposon insertion sequencing data set in Escherichia coli as a case study. We show that ciprofloxacin treatment generates chromosomal location bias in the resulting data, and we further demonstrate that failing to correct for this bias leads to false predictions of mutant drug sensitivity as measured by minimum inhibitory concentrations. We have developed an R package and user-friendly graphical Shiny application, ChromoCorrect, that detects and corrects for chromosomal bias in read count data, enabling the application of functional genomics technologies to the study of antibiotic stress.
IMPORTANCE
Altered gene dosage due to changes in DNA replication has been observed under a variety of stresses with a variety of experimental techniques. However, the implications of changes in gene dosage for sequencing-based functional genomics assays are rarely considered. We present a statistically principled approach to correcting for the effect of changes in gene dosage, enabling testing for differences in the fitness effects or regulation of individual genes in the presence of confounding differences in DNA copy number. We show that failing to correct for these effects can lead to incorrect predictions of resistance phenotype when applying functional genomics assays to investigate antibiotic stress, and we provide a user-friendly application to detect and correct for changes in DNA copy number.
KEYWORDS: data normalization, transposon insertion sequencing, RNA sequencing, chromosomal copy number, bacterial genomics, ciprofloxacin, median sliding window, data correction
INTRODUCTION
Functional genomics technologies, such as transposon insertion sequencing (TIS) (1) and RNA-sequencing (RNA-seq) (2), have emerged as effective, high-throughput methods for investigating gene function. Most analysis of functional genomics data relies on quantifying and comparing sequencing read counts, with results often expressed as relative (log) ratios of read counts between an experimental condition and control. Critically, the calculation of these log read count ratios depends on accurate normalization. Common normalization methods correct for differences in sequencing depth between experiments and assume this normalization factor is constant for all genes assayed. Here, we identify a phenomenon that violates these normalization assumptions resulting from treatments that affect DNA replication in bacteria, which we have coined chromosomal location bias (CLB). We present a new normalization technique and associated tool, named “ChromoCorrect,” that can be easily applied to any functional genomics data to identify and correct for CLB.
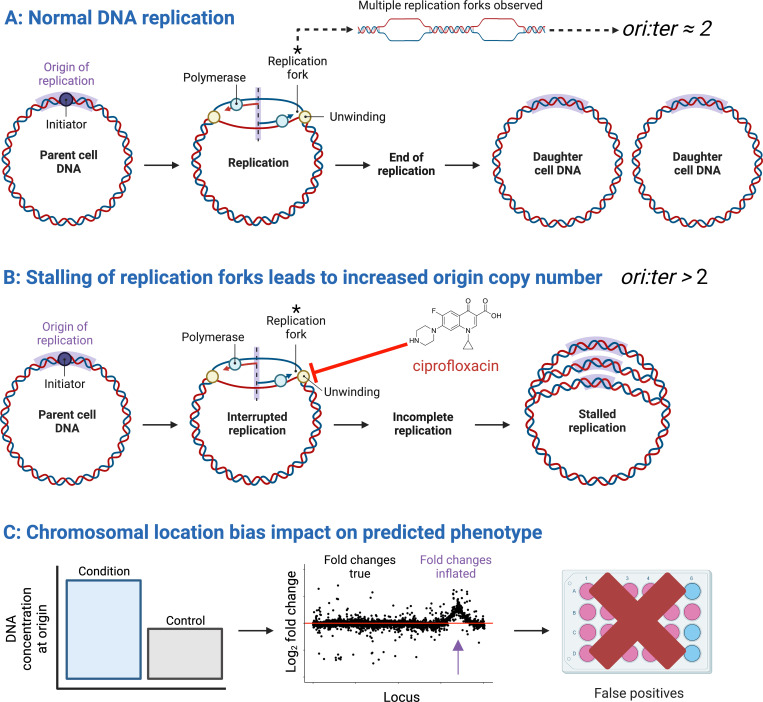
During normal exponential growth in bacteria, the time between cell divisions is significantly shorter than the time needed to complete chromosomal replication. This leads to cells containing more copies of DNA around the origin of replication (oriC) compared to the terminus (ter) and is due to the firing of multiple simultaneous replication forks (3) (Fig. 1A). In many sequencing-based assays, this difference in DNA copy numbers translates into higher read counts around the origin as compared to the terminus due to a higher availability of template nucleic acids. Under most conditions, the ratio of oriC-ter reads remains constant between a treatment and an untreated control and does not interfere with results. However, certain treatments specifically alter the oriC-ter ratio, such as exposure to the DNA gyrase-targeting antibiotic ciprofloxacin (Fig. 1B). This introduces large changes in read counts that primarily reflect changes in the DNA copy number near the origin rather than gene regulation (RNA-seq) or mutant fitness (TIS) (Fig. 1C). These distortions, or CLB, in turn can lead to incorrect predictions of drug sensitivity or resistance.
Fig 1.
Chromosomal location bias as a result of higher chromosomal copy number near the origin after ciprofloxacin treatment and the downstream effects on read counts. (A) Normal DNA replication producing two daughter cells. The origin of replication is colored purple. Multiple replication forks naturally lead to more origin (oriC) than terminus (ter) DNA. (B) Ciprofloxacin prevents DNA unwinding, stalling the replication fork. This produces a highly inflated relative concentration of DNA proximal to the origin. (C) The knock-on effects of increased DNA concentration around the origin on the read counts and fold changes, an observable peak at the origin in the relative log2FCs and potential false positive predictions of downstream drug sensitivity testing.
A comprehensive study by Slager et al. (4) showed that treatment with a range of antimicrobials modified both DNA and RNA copy numbers, resulting in altered oriC-ter ratios across the genomes for multiple bacterial species. In a subsequent review, Slager et al. (5) suggested that not normalizing for this effect in RNA-seq analyses may lead to an overestimation of differential gene expression. Previous studies have attempted to correct CLB in antibiotic-treated functional genomics data using local regression methods such as Lowess (6–8). Typically, local regression is performed on the raw read counts, with normalized counts produced to replace the raw reads for the differential analysis. However, most differential analysis tools for sequencing data rely on count models that assume counts of similar magnitude have similar variance (9, 10). Providing modified or transformed counts violates these assumptions and will lead to incorrect assessment of statistical significance. We often observe distortions in the local read count density spanning several orders of magnitude following antibiotic treatment, which would lead to concomitantly large distortions in resulting P-values. Hence, there is a clear need for a statistically sound methodology to properly address CLB in functional genomics data.
Here, we develop a statistically principled approach for correcting CLB using a local normalization factor rather than directly providing normalized counts. These local normalization factors can then be provided to differential analysis tools such as edgeR (9) or DESeq2 (10, 11) as offsets alongside the raw counts to correct for CLB within the statistical model. This preserves important features of the data needed for accurate calculation of P values, namely the mean-variance trend, while also producing fold-changes that have the CLB effect removed. Based on this method we have developed an application for identifying and correcting for CLB named “ChromoCorrect.” We have made our diagnostic and normalization procedure available as a graphical Shiny application that can be applied to any sequencing-based functional genomics assay. We apply ChromoCorrect to a data set we generated for this study that displays strong CLB: TIS output of an Escherichia coli K12 library challenged with ciprofloxacin. We confirm that ciprofloxacin produces the predicted large local distortions in read counts around the origin of replication, confounding the TIS counts such that they no longer accurately reflect the fitness of individual mutants. We show, using minimum inhibitory concentration (MIC) assays, that these distortions lead to incorrect predictions of mutant ciprofloxacin sensitivity and resistance. We also demonstrate that our normalization approach, after processing with ChromoCorrect, corrects these, rendering accurate fold changes that align well with independently determined mutant phenotypes.
RESULTS
Chromosomal location bias distorts a range of functional genomics data sets
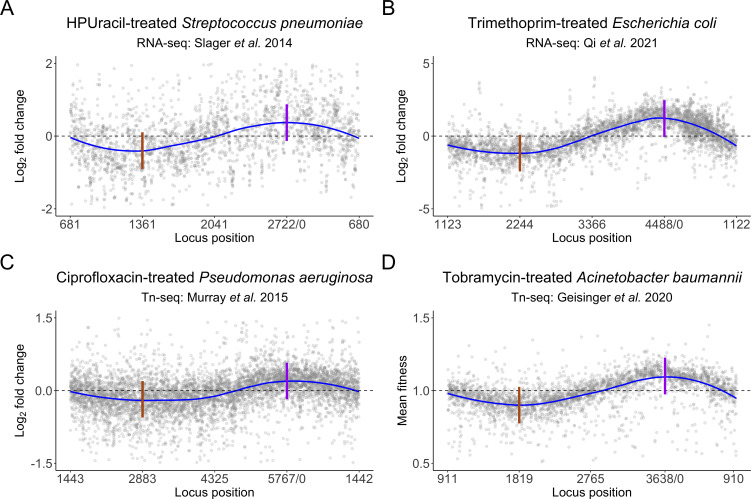
To illustrate the prevalence of CLB, we collected instances of CLB in existing RNA-seq and TIS data sets from four different antibiotic treatments across four different bacteria and plotted log2 fold changes along the chromosome (Fig. 2) (4, 7, 12, 13). In all cases examined, we see clear evidence of CLB, with a local increase in DNA copy number identified around the origin. Taken together, this supports that CLB is widespread in antibiotic-treated functional genomics data sets and that a robust method to detect and remove CLB is necessary to produce accurate predictions for downstream analysis.
Fig 2.
Read count log2 fold changes versus genome location plots displaying chromosomal location bias under varying conditions and organisms using different experimental techniques. Blue lines indicate trendlines over the individual gray points. Dashed gray lines indicate the expected trend line. Solid vertical lines indicate the terminus (brown) and origin of replication (purple). All experiments have decreased fold changes or fitness scores that dip at the terminus and increase at the origin. (A) RNA-Seq analysis of 6(p-Hydroxyphenylazo)-uracil-treated Streptococcus pneumoniae. (B) RNA-Seq analysis of trimethoprim-treated Escherichia coli. (C) Tn-seq analysis of ciprofloxacin-treated Pseudomonas aeruginosa. (D) Tn-seq analysis of tobramycin-treated Acinetobacter baumannii. Data sources: (A): Slager et al. (4), (B): Qi et al. (12), (C): Murray et al. (13), and (D): Geisinger et al. (7).
A principled normalization procedure to correct for chromosomal location bias
To correct CLB, we have developed a normalization procedure that produces offsets that can be directly incorporated into differential testing using packages such as edgeR (9) or DEseq2 (10) without modifying the input count data. These offsets can be thought of as a gene-specific normalization factor, which in this case includes a correction for the local read density across the chromosome.
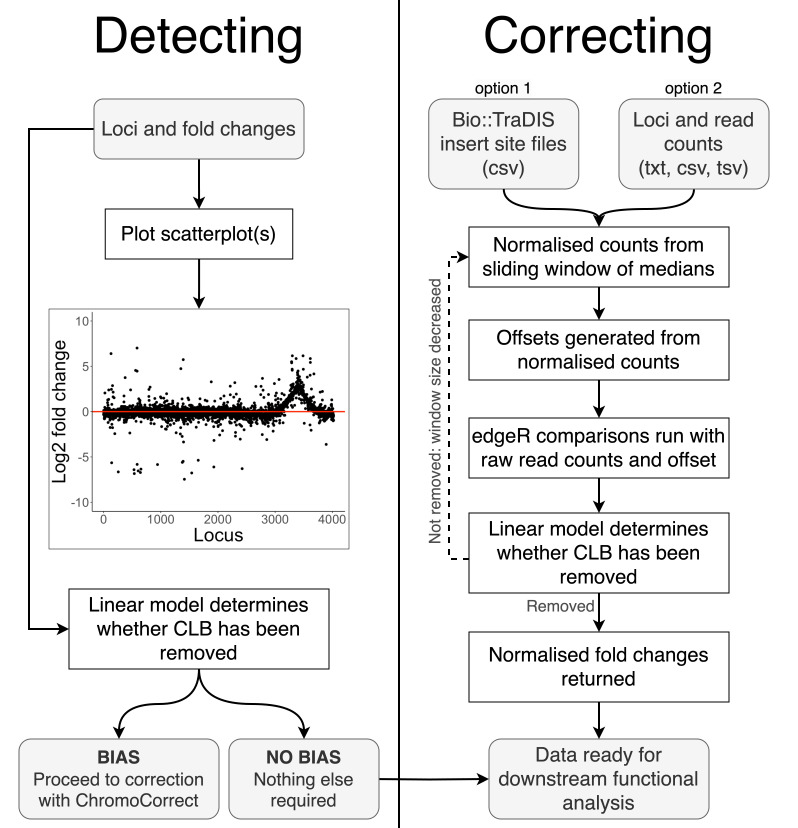
Our normalization procedure comprises three major steps as outlined in Fig. 3. First, we calculate the local median read depth along the chromosome using a sliding window over the local gene neighborhood. The number of flanking genes included in the sliding window starts at 500 and is dynamically determined from the data set by fitting a linear model and testing the slope and y-intercept of the fitted line. Each iteration reduces the window size by 100 loci if the window is not small enough to accurately fit the trendline until a minimum window size of 200 is reached. We have found that medians calculated from windows smaller than 200 loci can be unduly influenced by a small number of genes with particularly high or low read counts. Second, the sliding window analysis is used to calculate a gene read count normalized for local read density by dividing the actual read count for each gene by the ratio of the local to global average read counts. This normalized count is then used to derive a gene-wise normalization factor, additionally incorporating differences in effective library size between replicates using the trimmed mean of M-values (TMM) method (14) (see Materials and Methods). Finally, these offsets are provided to the edgeR (9) glmFit function alongside the raw counts for differential analysis, where they are directly incorporated into the statistical model for testing purposes. This procedure maintains the information contained in the raw counts, necessary for accurate statistical analysis, while correcting the resulting estimated log2FCs for local distortions in DNA copy number.
Fig 3.
Schematic of pipeline for detecting and correcting chromosomal location bias using ChromoCorrect. Gray boxes indicate user input or output, and white indicates automated steps. Detecting requires the log2 fold changes of each locus to plot a scatterplot of fold change by chromosomal position, which graphs trends in the fold changes for the user to visualize the pipeline’s assessment. The app and the R console display a message recommending normalization if chromosomal location bias is detected by a fitted linear model. Correcting requires read counts per locus in a txt, csv, or tsv file format, which are then normalized using a sliding window of medians with a default size of 500. An offset matrix is generated from the normalized counts to input along with the raw read counts into edgeR. A linear model is fitted again during correction to determine whether the default sliding window is small enough to capture the trend and repeats the normalization procedure with a smaller window otherwise. The corrected analysis is returned after the normalization is complete.
The ChromoCorrect app
The normalization techniques described here have been organized into an R Shiny app for easy use by researchers wanting to diagnose and normalize data affected by CLB. Instructions for installing and running the app can be found on Github (https://github.com/BarquistLab/ChromoCorrect/) or accessed online through ShinyApps (https://thecainlab.shinyapps.io/ChromoCorrect/).
Transposon insertion sequencing case study: ciprofloxacin
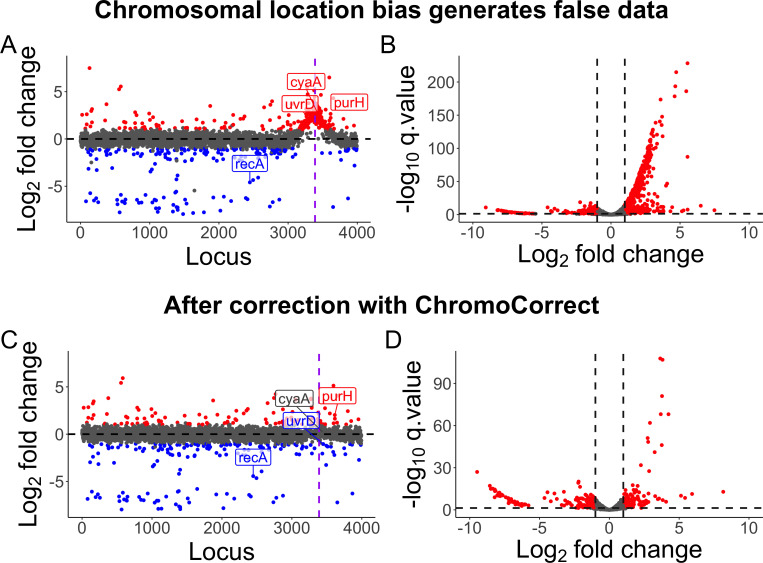
To illustrate the functionality of ChromoCorrect, we generated a data set applying the transposon-directed insertion-site sequencing (TraDIS) TIS technique to a dense library of E. coli transposon mutants exposed to ciprofloxacin compared to an untreated control. This data set was generated using E. coli K12 BW25113, the parental strain of the Keio collection (15). A TraDIS library of 350,000 unique Tn5 mutants (16) was challenged with a subinhibitory concentration of ciprofloxacin (1/2 MIC) with growth overnight. After analysis with the TraDIS toolkit (17), we confirmed ciprofloxacin as an inducer of exaggerated CLB as it displayed a distinctive peak of increased reads around the origin of replication (Fig. 4A). This peak reflects the expected increase in DNA copy number at the origin compared to the rest of the genome that ciprofloxacin induces as it targets topoisomerases and stalls the replication fork and DNA synthesis (4, 18). Around the peak, the observed inflation in transposon insertions occurred largely between loci 3023 and 3622, 300 loci on either side of the origin of replication (oriC is located between locus 3322, mnmG and locus 3323, mioC). This entire 600 locus region had an average log2FC of 1.5 (a fold change increase of 2.8 compared to the untreated control), whereas the rest of the genome had a more typical average log2FC of 0.1. Strikingly, this means that many of these 600 loci would meet the standard “2-fold” cutoff often used to prioritize genes for further investigation.
Fig 4.
Visualizing and diagnosing chromosomal location bias in a ciprofloxacin-treated transposon insertion sequencing data set. Each point represents a locus of the genome. The x-axis is the chromosomal location, and the y-axis is the log2FC from ciprofloxacin treated versus untreated comparisons. (A) Volcano plot before correction, showing a large skew of significant genes (red) to the right, representing an increased prevalence of these mutants compared to the no antibiotic control. (B) Locus by fold change scatterplot pre-normalization with loci plotted in chromosomal order. The dashed black line shows the expected trend of the data if not affected by CLB. The purple dotted line is the origin of replication, where a large peak of elevated read counts is seen. (C) Locus by fold change scatterplot post-normalization with no peak. (D) Volcano plot post-normalization with no skew. Normalization performed with a sliding median window size of 200. Some mutant examples from our phenotypic validation are labeled in the scatterplots. Blue genes represent significant genes with log2FCs ≤ −1, and red genes indicate significant genes with log2FCs ≥ 1.
After analysis with the TraDIS toolkit (17), the ciprofloxacin treatment TIS data yielded 754 genes with significant values (q-value < 0.05), with 468 having an absolute log2FC value over 1. Of these 468 genes, 391 (84%) of these were located between loci positions 3023 and 3623, despite this region representing only 15% of the genome. Another confirmation of this data bias was visualization by a volcano plot showing log2FC versus −log10 q-value (Fig. 4B). The fold change was skewed to the right, indicating a bias towards cells that appeared to have a higher frequency of insertions in many genes. After normalizing the data using ChromoCorrect with an automatically determined sliding window median of 200 genes, we identified 272 significant genes (a 64% reduction), with only 163 having an absolute log2FC value over 1 (a 65% reduction). The normalized volcano plot and locus by fold change scatterplot show the CLB has been removed (Fig. 3C and D).
Chromosomal location bias leads to incorrect predictions of ciprofloxacin sensitivity
Using TIS or other functional genomics techniques to identify entire gene suites involved in antibiotic stress tolerance is well-established (8, 13, 19). However, the possible presence of CLB in these data sets, if left uncorrected, may result in false positive predictions that are carried on into laboratory analysis. To demonstrate the implications of CLB for predictions of antibiotic resistance (or sensitivity), as well as to validate the use of ChromoCorrect to generate more biologically accurate predictions, we tested the phenotypes of various E. coli BW25113 single gene mutants from the Keio collection (15) that represent different types of potential errors that may arise because of CLB. In total, 11 mutants were tested for their ciprofloxacin resistance and sensitivity profile (between 5 and 20 ng/mL) via an MIC assay and compared to the wild type (WT) BW25113 resistance level (10 ng/mL). For this, we examined genes that exhibited dissimilar outputs before and after correction, particularly focusing on those that were identified as significant prior to correction but not afterward. The 11 mutants tested represented four distinct classes of genes (Table 1). The first class represented a positive control and included the known antibiotic resistance determinants acrB, involved in efflux (20, 21), and recA, involved in DNA recombination and repair (22, 23). As expected, both genes remained significant after correction with ChromoCorrect with a predicted sensitivity phenotype and were confirmed by the MIC assay. The second class included five genes with a predicted sensitivity phenotype before but not after applying ChromoCorrect. As predicted by ChromoCorrect, all five mutants displayed no change in MIC compared to WT, confirming that they were falsely designated as ciprofloxacin sensitivity genes prior to correction for CLB. The third class comprised three gene representatives predicted to be sensitive both before and after correction, and all showed a twofold increase (20 ng/mL) in ciprofloxacin MIC, confirming a true sensitivity phenotype. Finally, we examined genes with the most extreme mispredictions of phenotype in the absence of correction for CLB, those that shifted from predicted mutant resistance to predicted sensitivity after correction. There were eight genes whose log2FC went from positive to negative values, but only one (uvrD) met the phenotypic validation threshold of |log2FC| ≥ 1 before and after correction. The misprediction of ∆uvrD phenotype prior to correction was confirmed by increased ciprofloxacin sensitivity in our MIC assay. Neglecting to address this bias would have falsely classified uvrD as mediating ciprofloxacin sensitivity, not resistance. In summary, all mutants tested by MIC assay reflected the predicted phenotype after correction of CLB, including six instances where the analysis based on uncorrected data led to an incorrect prediction of resistance phenotype. This emphasizes the critical role of ChromoCorrect’s normalization in ensuring accurate and reliable gene fitness assessments.
TABLE 1.
Single gene E. coli BW25113 Keio knockouts validated in this study
| Predicted mutant phenotype | Gene | Function | TraDIS log2FC | Experimental mutant phenotype MIC50 (ng/mL) | |
|---|---|---|---|---|---|
| Before ChromoCorrect | After ChromoCorrect | ||||
| - | WT | - | - | - | 10 |
| Cip sensitive before and after | acrB | Multidrug efflux system protein | −0.89 | −0.83 | ≤5 |
| recA | DNA recombination and repair protein | −1.99 | −1.92 | ≤5 | |
| Cip resistant before and WT phenotype after | cyaA | Adenylate cyclase | 2.48 | 0.15 | 10 |
| ilvN | Acetolactate synthase 1 small subunit | 2.13 | −0.06 | 10 | |
| mtlD | Mannitol-1-phosphate dehydrogenase | 1.77 | 0.13 | 10 | |
| fdhE | Formate dehydrogenase formation protein | 1.66 | 0.13 | 10 | |
| pfkA | 6-phosphofructokinase I | 1.32 | −0.03 | 10 | |
| Cip resistant before and after | purH | Bifunctional transformylase/ cyclohydrolase | 2.50 | 1.64 | 20 |
| dgkA | Diacylglycerol kinase | 1.62 | 1.07 | 20 | |
| rpe | D-ribulose-5-phosphate 3-epimerase | 1.45 | 1.07 | 20 | |
| Cip resistant before and sensitive after | uvrD | DNA-dependent ATPase I and helicase II | 1.32 | −1.06 | ≤5 |
Use of the Shiny application
The app is split into two main tabs: detecting and correcting (Fig. 3). Detecting requires the upload of analyzed output files containing log2FC values to visualize any CLB, while the Correcting tab requires the upload of the read counts for the conditions affected by CLB and the associated no-stress control.
The first step within the app is to assess whether data sets that have undergone differential analysis are affected by CLB. This can be done using the Detecting tab. The user inputs one or more files containing locus tags and fold change information, and a locus by fold change scatterplot for each condition is generated. The user can cycle through the uploaded data sets in the drop-down menu in the sidebar to determine if any of the experimental conditions are affected by CLB. It is deemed present if the general trend of the fold changes is not flat and distributed around zero, as demonstrated in Fig. 3A. The “decision” box will contain red text suggesting correction if CLB is detected, or black text if not detected.
If the analysis produced CLB, the user can fix the issue with the second tab: Correcting. This tab requires the upload of a file with a locus_tag column and read counts, or the upload of read files produced by the tradis_gene_insert_sites script from Bio-TraDIS (17). The tab requires at least four insert site files or columns of read counts: two biological replicates of the experimental condition and two biological replicates of the associated control condition. Once the upload is complete, the user must define which condition specifies the control. The app will compute an edgeR comparison before and after normalization and produce two scatterplots of locus versus log fold change associated with the two analyses. The default window size of 500 is suitable for smooth trend lines. The window will automatically reduce in size if the data has not been normalized effectively due to any sharper trend lines present. The user can then export the corrected output, free from CLB.
DISCUSSION
This study addresses the fundamental issue of CLB, which we show impacts a wide variety of functional genomics analyses, resulting in false positives and negatives or incorrect interpretation of data. CLB arises from nucleic acid copy number fluctuations along the chromosome, typically around the origin and/or terminus, which can be exacerbated by replication-disrupting events like replication-targeted antibiotic treatment. To solve this problem, we introduce ChromoCorrect, a normalization tool that effectively corrects for CLB producing accurate log2FCs and significance values for each locus. Using a ciprofloxacin-treated TraDIS data set in E. coli, we demonstrated that CLB leads to incorrect predictions of antibiotic resistance phenotypes that can be corrected using ChromoCorrect.
Over the past decade, functional genomics techniques, like TIS and RNA-Seq, have been employed to comprehensively assess the effects of various selection pressures, such as antibiotic exposure, on microbial fitness and cellular responses (8, 13, 19, 24, 25). Here, we show that CLB can lead to incorrect prediction of phenotype using our own antibiotic-treated TIS data, as a cautionary tale for future studies. Our study retrospectively highlighted the prevalence of CLB in published functional genomics data sets across diverse species and under a range of conditions. These conditions include both treatment with direct DNA-targeting antimicrobials, such as fluoroquinolones, but also antimicrobials that have an indirect effect on DNA replication, like trimethoprim, HPUra, and tobramycin (Fig. 2) (4, 7, 12, 13). Trimethoprim targets the dihydrofolate reductase of the folate biosynthesis pathway and reduces the availability of tetrahydrofolate, a precursor to the essential DNA components thymidine and thymine (26) eventually leading to “thymineless death” (27). Thymine starvation stalls replication forks, initially leading to a transient increase in origin-proximal DNA before the replicating DNA is destabilized and degraded, leading to ultimate depletion of origin-proximal DNA (28). Similarly, the uracil analogue HPUra has an indirect effect on DNA replication by stalling replication forks (4). The mechanism by which tobramycin, a ribosome-targeting antibiotic, contributes to the observed CLB is not obvious but could represent another indirect or secondary effect. Our work suggests that CLB may be exerting a more significant influence on the interpretation of sequencing data than currently acknowledged and an important future study would be to comprehensively assess the prevalence of CLB across functional genomics data sets.
The four organisms we investigated span both Gram-negative and Gram-positive bacteria, indicating that CLB occurs in diverse species, and is likely relevant beyond these well-studied organisms. Our method bears conceptual similarity to peak-to-trough ratio (PTR) methods that use oriC-ter ratios to determine bacterial growth rates from genomic or metagenomic DNA sequencing data (29–31), which have been shown to accurately predict growth rates in a wide range of bacteria in pure culture. The success of these methods suggests that beyond antibiotic treatments that interfere with DNA replication, CLB may also affect the results of functional genomics comparison between bacterial cultures growing at different rates.
Biases arising from altered oriC-ter ratios in TIS data have been recognized previously and corrected for using local regression methods (6–8), though the origins of these biases have not been clearly described. Our contribution has been to highlight the prevalence of CLB in bacterial functional genomics data. A major advantage of ChromoCorrect is an ability to directly incorporate the local normalization factors as an offset into differential analysis tools such as edgeR (9) and DESeq2 (10). This preserves the mean-variance relationship in the underlying count data, which is important for accurate estimation of statistical significance. This approach was inspired by the transcript quantification package tximport that corrects for differences in isoform abundance during gene-level differential expression in eukaryotes (32). By facilitating the import of offsets into established RNA-seq analysis tools, ChromoCorrect can be used seamlessly with existing pipelines.
Although clearly a critical step in the analysis of functional genomics data, the interpretation of correcting for CLB requires care and depends on the technology analyzed. In the case of RNA-seq, CLB is reflective of a genuine increase in RNA synthesized from the origin-proximal region, and the primary danger is that this could be interpreted as a specific regulatory response rather than a direct result of antibiotic activity on DNA replication dynamics. In contrast for TIS experiments, CLB introduces artifacts that can lead to false predictions of phenotype. Since TIS uses transposon-flanking reads as a proxy for mutant abundance, and local distortions in DNA copy number will lead to a local distortion in template DNA abundance, mutants containing transposon insertions in the vicinity of the origin will appear to be more frequent in the population than they really are in data affected by CLB.
Our study highlights the importance of scrutinizing data for CLB to improve the reliability of conclusions drawn from functional genomics data. We recommend that future microbial functional genomic data sets with read counts produced, especially those that involve antibiotic exposure, be screened for the presence of CLB, and if so, to correct the data using ChromoCorrect before proceeding with time and labor-intensive biological interpretation and laboratory experiments.
MATERIALS AND METHODS
Software
Analyses were performed using R (version 4.0.3), R Studio (version 2022.07.2). The application was developed using RShiny (version 1.7.3).
TraDIS library ciprofloxacin challenging and sequencing
An E. coli K12 TraDIS library was generated as previously described (16) and challenged with subinhibitory ciprofloxacin (40 µg/mL) in 10 mL of Mueller Hinton cation-adjusted media. Genomic DNA was extracted using the DNeasy UltraClean Microbial Kit (Qiagen) according to the manufacturer’s instructions and was sequenced on an Illumina HiSeq2500 platform at the Wellcome Sanger Institute.
Identifying chromosomal location bias
The data were run through the Bio::TraDIS pipeline (17) using SMALT mapping and a minimum read count of 10. To identify whether CLB was present, the log2FCs were plotted in genome order, with locus on the x-axis and log2FC on the y-axis.
Generating normalized read counts and offsets with ChromoCorrect
For each condition, the read counts are obtained, and the first 1,000 genes are appended to the end of the file and the last 1,000 to the beginning to mimic a circular genome for the median sliding window function. Read counts of zero are excluded from the data set to remove their influence on the median. The medianFilter function from package FBN (version 1.5.1) is used to calculate a median for each locus based on an adjustable window size. We have found that a default window size of 500 is sufficient for many smoother trends, whilst sharps trends need to be computed with a smaller window size. A ratio for each point is calculated by dividing the locus’ median by the mean median of all loci. The normalized read count is obtained by dividing the raw read count by the ratio computed for each locus. The normalized counts are not used as a replacement for the raw read counts, instead, an offset data set is created. The offset is computed as the natural logarithm of the raw count, subtracted from the natural logarithm of the normalized count. An arbitrary pseudocount of 0.1 is added to both raw and normalized counts before log transforming to prevent the undefined log of 0 occurring. The effective library size is calculated using the edgeR package (version 3.32.1) calcNormFactors function applied to the normalized counts, multiplied by the column sums of the normalized counts. Following this, the offset values are adjusted to account for library size differences by subtracting the logarithm of the effective library size.
Comparisons using edgeR within ChromoCorrect
ChromoCorrect incorporates the offset along with the raw counts in edgeR to perform differential analysis and assess the normalization. Genes are filtered based on a minimum count threshold (default value of 10). Raw counts are put into a DGEList with groupings and scaleOffset() is used to offset the raw read counts. Library size normalization is not computed due to the inclusion of the offset, which produces a custom normalization factor per gene. Common negative binomial (estimateGLMCommonDisp()) and Bayes tagwise (estimateGLMTagwiseDisp()) dispersions for general linear models are calculated. A gene-wise negative binomial for general linear models (glmFit() and glmLRT()) is fit with contrasts to produce likelihood ratio tests per gene between the control and conditions, producing the log2 fold changes and adjusted P values. Following the analysis, ChromoCorrect generates summary statistics and diagnostic plots, automatically calculating the adjustment of the window size if bias has not been mitigated. The process repeats until a satisfactory result is achieved or a minimum window size of 200 is reached. The code produces scatterplots of the data before and after bias is removed for a visual reference.
Minimum inhibitory concentration assays
MIC assays were performed for the single gene knockouts to determine their breakpoint compared to WT cultures. Mutants were steaked from frozen onto Mueller Hinton (MH) agar plates and incubated overnight at 37˚C. Three single colonies of each mutant were inoculated into 5 mL of cation-adjusted MH broth (CAMHB) and grown overnight at 37˚C and 200 rpm shaking. A 1/100 dilution of the overnight cultures was made in 5 mL of fresh CAMHB and grown for 2.5 h until the exponential phase. MICs were performed with triplicate technical replicates in a 96-well plate with approximately 1 × 105 cells per 150 µL well and grown overnight at 37˚C with 200 rpm shaking. Cells were imaged after 16 h at OD600 on a PHERAstar plate reader (BMG Labtech). Wells were blanked and averaged within triplicates. MIC was determined as the lowest concentration that inhibited at least 50% of growth compared to the untreated mutant positive control.
ACKNOWLEDGMENTS
The authors would like to thank Julian Parkhill and the Wellcome Sanger Institute sequencing team for kindly providing the TIS sequencing data used in this publication. The authors would also like to thank Claire Maher, Hannah Lott, and Natasha Delgado for prototype testing of ChromoCorrect.
G.J.S. acknowledges the Helmholtz Information & Data Science Academy (HIDA) for providing financial support for a short-term research visit to the Helmholtz Institute for RNA-based Infection Research (HIRI), acknowledges an Australian Research Council-funded scholarship from project grant DE180100929, and acknowledges financial support from the Australasian Genomic Technologies Association. This project was partially funded by the Bavarian State Ministry for Science and the Arts through the research network bayresq.net to L.B. and Australian National Health and Medical Research Council (NHMRC) Project Grant 1159752 to A.K.C. and L.B. A.K.C. was supported by an Australian Research Council (ARC) Future Fellowship (FT220100152).
Contributor Information
Lars Barquist, Email: lars.barquist@helmholtz-hiri.de.
Amy K. Cain, Email: amy.cain@mq.edu.au.
Anthony Fodor, University of North Carolina at Charlotte, Charlotte, North Carolina, USA.
DATA AVAILABILITY
TraDIS sequencing reads were deposited in the European Nucleotide Archive (ENA) under study accession number PRJEB35059. Control conditions are biosamples SAMEA6429763 and SAMEA6429764, ciprofloxacin conditions are SAMEA6429767 and SAMEA6429768. RNA sequencing data in S. pneumoniae reported in Slager et al. (4) sourced from ENA study accession PRJNA235855. RNA-Seq data in P. aeruginosa reported in Murray et al. (13) sourced from ENA study accession PRJNA291292.
The ChromoCorrect code is publicly available and can be found on Github at https://github.com/BarquistLab/ChromoCorrect and as an online interface at https://thecainlab.shinyapps.io/ChromoCorrect/.
REFERENCES
- 1. Cain AK, Barquist L, Goodman AL, Paulsen IT, Parkhill J, van Opijnen T. 2020. A decade of advances in transposon-insertion sequencing. Nat Rev Genet 21:526–540. doi: 10.1038/s41576-020-0244-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Colgan AM, Cameron AD, Kröger C. 2017. If it transcribes, we can sequence it: mining the complexities of host–pathogen–environment interactions using RNA-seq. Curr Opin Microbiol 36:37–46. doi: 10.1016/j.mib.2017.01.010 [DOI] [PubMed] [Google Scholar]
- 3. Skarstad K, Boye E, Steen HB. 1986. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J 5:1711–1717. doi: 10.1002/j.1460-2075.1986.tb04415.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Slager J, Kjos M, Attaiech L, Veening JW. 2014. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell 157:395–406. doi: 10.1016/j.cell.2014.01.068 [DOI] [PubMed] [Google Scholar]
- 5. Slager J, Veening J-W. 2016. Hard-wired control of bacterial processes by chromosomal gene location. Trends Microbiol 24:788–800. doi: 10.1016/j.tim.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zomer A, Burghout P, Bootsma HJ, Hermans PWM, van Hijum SAFT. 2012. ESSENTIALS: software for rapid analysis of high throughput transposon insertion sequencing data. PLoS One 7:e43012. doi: 10.1371/journal.pone.0043012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geisinger E, Mortman NJ, Dai Y, Cokol M, Syal S, Farinha A, Fisher DG, Tang AY, Lazinski DW, Wood S, Anthony J, van Opijnen T, Isberg RR. 2020. Antibiotic susceptibility signatures identify potential antimicrobial targets in the Acinetobacter baumannii cell envelope. Nat Commun 11:4522. doi: 10.1038/s41467-020-18301-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2:e00315-10. doi: 10.1128/mBio.00315-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Nat Prec. doi: 10.1038/npre.2010.4282.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qi Q, Angermayr SA, Bollenbach T. 2021. Uncovering key metabolic determinants of the drug interactions between trimethoprim and erythromycin in Escherichia coli. Front Microbiol 12. doi: 10.3389/fmicb.2021.760017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murray JL, Kwon T, Marcotte EM, Whiteley M. 2015. Intrinsic antimicrobial resistance determinants in the superbug Pseudomonas aeruginosa . mBio 6:e01603-15. doi: 10.1128/mBio.01603-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:1–9. doi: 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K‐12 in‐frame, single‐gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maher C, Maharjan R, Sullivan G, Cain AK, Hassan KA. 2022. Breaching the barrier: genome-wide investigation into the role of a primary amine in promoting E. coli outer-membrane passage and growth inhibition by ampicillin. Microbiol Spectr 10:e0359322. doi: 10.1128/spectrum.03593-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barquist L, Mayho M, Cummins C, Cain AK, Boinett CJ, Page AJ, Langridge GC, Quail MA, Keane JA, Parkhill J. 2016. The TraDIS toolkit: sequencing and analysis for dense transposon mutant libraries. Bioinformatics 32:1109–1111. doi: 10.1093/bioinformatics/btw022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ojkic N, Lilja E, Direito S, Dawson A, Allen RJ, Waclaw B. 2020. A roadblock-and-kill mechanism of action model for the DNA-targeting antibiotic ciprofloxacin. Antimicrob Agents Chemother 64:e02487-19. doi: 10.1128/AAC.02487-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leshchiner D, Rosconi F, Sundaresh B, Rudmann E, Ramirez LMN, Nishimoto AT, Wood SJ, Jana B, Buján N, Li K, Gao J, Frank M, Reeve SM, Lee RE, Rock CO, Rosch JW, van Opijnen T. 2022. A genome-wide Atlas of antibiotic susceptibility targets and pathways to tolerance. Nat Commun 13:3165. doi: 10.1038/s41467-022-30967-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pakzad I, Zayyen Karin M, Taherikalani M, Boustanshenas M, Lari AR. 2013. Contribution of AcrAB efflux pump to ciprofloxacin resistance in Klebsiella pneumoniae isolated from burn patients. GMS Hyg Infect Control 8:Doc15. doi: 10.3205/dgkh000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nolivos S, Cayron J, Dedieu A, Page A, Delolme F, Lesterlin C. 2019. Role of AcrAB-TolC multidrug efflux pump in drug-resistance acquisition by plasmid transfer. Science 364:778–782. doi: 10.1126/science.aav6390 [DOI] [PubMed] [Google Scholar]
- 22. Lusetti SL, Cox MM. 2002. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem 71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940 [DOI] [PubMed] [Google Scholar]
- 23. Machuca J, Recacha E, Gallego-Mesa B, Diaz-Diaz S, Rojas-Granado G, García-Duque A, Docobo-Pérez F, Blázquez J, Rodríguez-Rojas A, Pascual A, Rodríguez-Martínez JM. 2021. Effect of RecA inactivation on quinolone susceptibility and the evolution of resistance in clinical isolates of Escherichia coli. J Antimicrob Chemother 76:338–344. doi: 10.1093/jac/dkaa448 [DOI] [PubMed] [Google Scholar]
- 24. Jana B, Cain AK, Doerrler WT, Boinett CJ, Fookes MC, Parkhill J, Guardabassi L. 2017. The secondary resistome of multidrug-resistant Klebsiella pneumoniae. Sci Rep 7:42483. doi: 10.1038/srep42483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boinett CJ, Cain AK, Hawkey J, Do Hoang NT, Khanh NNT, Thanh DP, Dordel J, Campbell JI, Lan NPH, Mayho M, Langridge GC, Hadfield J, Chau NVV, Thwaites GE, Parkhill J, Thomson NR, Holt KE, Baker S. 2019. Clinical and laboratory-induced colistin-resistance mechanisms in Acinetobacter baumannii. Microb Genom 5:e000246. doi: 10.1099/mgen.0.000246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sangurdekar DP, Zhang Z, Khodursky AB. 2011. The association of DNA damage response and nucleotide level modulation with the antibacterial mechanism of the anti-folate drug trimethoprim. BMC Genomics 12:583. doi: 10.1186/1471-2164-12-583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khodursky A, Guzmán EC, Hanawalt PC. 2015. Thymineless death lives on: new insights into a classic phenomenon. Annu Rev Microbiol 69:247–263. doi: 10.1146/annurev-micro-092412-155749 [DOI] [PubMed] [Google Scholar]
- 28. Sangurdekar DP, Hamann BL, Smirnov D, Srienc F, Hanawalt PC, Khodursky AB. 2010. Thymineless death is associated with loss of essential genetic information from the replication origin. Mol Microbiol 75:1455–1467. doi: 10.1111/j.1365-2958.2010.07072.x [DOI] [PubMed] [Google Scholar]
- 29. Korem T, Zeevi D, Suez J, Weinberger A, Avnit-Sagi T, Pompan-Lotan M, Matot E, Jona G, Harmelin A, Cohen N, Sirota-Madi A, Thaiss CA, Pevsner-Fischer M, Sorek R, Xavier R, Elinav E, Segal E. 2015. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349:1101–1106. doi: 10.1126/science.aac4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown CT, Olm MR, Thomas BC, Banfield JF. 2016. Measurement of bacterial replication rates in microbial communities. Nat Biotechnol 34:1256–1263. doi: 10.1038/nbt.3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Joseph TA, Chlenski P, Litman A, Korem T, Pe’er I. 2022. Accurate and robust inference of microbial growth dynamics from metagenomic sequencing reveals personalized growth rates. Genome Res 32:558–568. doi: 10.1101/gr.275533.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soneson C, Love MI, Robinson MD. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4:1521. doi: 10.12688/f1000research.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
TraDIS sequencing reads were deposited in the European Nucleotide Archive (ENA) under study accession number PRJEB35059. Control conditions are biosamples SAMEA6429763 and SAMEA6429764, ciprofloxacin conditions are SAMEA6429767 and SAMEA6429768. RNA sequencing data in S. pneumoniae reported in Slager et al. (4) sourced from ENA study accession PRJNA235855. RNA-Seq data in P. aeruginosa reported in Murray et al. (13) sourced from ENA study accession PRJNA291292.
The ChromoCorrect code is publicly available and can be found on Github at https://github.com/BarquistLab/ChromoCorrect and as an online interface at https://thecainlab.shinyapps.io/ChromoCorrect/.