ABSTRACT
Helicobacter pylori is a highly successful pathogen that poses a substantial threat to human health. However, the dynamic interaction between H. pylori and the human gastric epithelium has not been fully investigated. In this study, using dual RNA sequencing technology, we characterized a cytotoxin-associated gene A (cagA)-modulated bacterial adaption strategy by enhancing the expression of ATP-binding cassette transporter-related genes, metQ and HP_0888, upon coculturing with human gastric epithelial cells. We observed a general repression of electron transport-associated genes by cagA, leading to the activation of oxidative phosphorylation. Temporal profiling of host mRNA signatures revealed the downregulation of multiple splicing regulators due to bacterial infection, resulting in aberrant pre-mRNA splicing of functional genes involved in the cell cycle process in response to H. pylori infection. Moreover, we demonstrated a protective effect of gastric H. pylori colonization against chronic dextran sulfate sodium (DSS)-induced colitis. Mechanistically, we identified a cluster of propionic and butyric acid-producing bacteria, Muribaculaceae, selectively enriched in the colons of H. pylori-pre-colonized mice, which may contribute to the restoration of intestinal barrier function damaged by DSS treatment. Collectively, this study presents the first dual-transcriptome analysis of H. pylori during its dynamic interaction with gastric epithelial cells and provides new insights into strategies through which H. pylori promotes infection and pathogenesis in the human gastric epithelium.
IMPORTANCE
Simultaneous profiling of the dynamic interaction between Helicobacter pylori and the human gastric epithelium represents a novel strategy for identifying regulatory responses that drive pathogenesis. This study presents the first dual-transcriptome analysis of H. pylori when cocultured with gastric epithelial cells, revealing a bacterial adaptation strategy and a general repression of electron transportation-associated genes, both of which were modulated by cytotoxin-associated gene A (cagA). Temporal profiling of host mRNA signatures dissected the aberrant pre-mRNA splicing of functional genes involved in the cell cycle process in response to H. pylori infection. We demonstrated a protective effect of gastric H. pylori colonization against chronic DSS-induced colitis through both in vitro and in vivo experiments. These findings significantly enhance our understanding of how H. pylori promotes infection and pathogenesis in the human gastric epithelium and provide evidence to identify targets for antimicrobial therapies.
KEYWORDS: Helicobacter pylori, cytotoxin-associated genes A, dual RNA sequencing, ATP-binding cassette transporter, oxidative phosphorylation, alternative splicing, inflammatory bowel disease
INTRODUCTION
Helicobacter pylori, a Gram-negative and transmissible pathogen that colonizes the stomachs of almost half of the world’s population, poses a substantial threat to human health (1). The World Health Organization has classified H. pylori as a “high-priority” pathogen (2). According to the consensus of the 2015 Kyoto conference, all H. pylori-positive individuals should receive eradication therapy unless there are compelling contraindications (3). Patients infected with this pathogen typically exhibit a sustained immune response (chronic gastritis) that can progress to atrophic gastritis, intestinal metaplasia (IM), and, ultimately, gastric adenocarcinoma, according to Correa’s cascade (4). Studies have determined that several virulence factors contribute to the pathogenicity of H. pylori, especially the cytotoxin-associated gene A (cagA), which is delivered through bacterial type IV secretion and causes chronic inflammation and oncogenesis in gastric epithelial cells (5).
H. pylori was initially identified as a noninvasive microorganism adhering to the gastric mucosa and surviving in the gastric lumen. However, its facultative intracellular nature has been increasingly recognized by researchers in recent decades, with evidence revealing that the bacterium can invade, survive, and replicate in both epithelial cells and professional phagocytes in vitro and in vivo (6–8). A small proportion of H. pylori was found to colonize within parietal cells in the murine gastric epithelium (9). This ability to reside within gastric epithelial cells endows H. pylori with a selective survival advantage, allowing it to evade eradication by immunocytes and antibiotics that act extracellularly. Capurro et al. reported that H. pylori secretes vacuolating cytotoxin A (vacA), a virulence factor, to create an intracellular niche in vivo. This niche protects H. pylori from antibiotics and leads to infection recrudescence after therapy (10). We recently described an evasion strategy where H. pylori subverts autophagosomes into a pro-survival niche by disrupting lysosomal acidification and maturation, enabling the bacteria to survive within the host cell and establish persistent colonization (11, 12). However, to date, studies to systemically characterize how intracellular H. pylori reprograms host cell transcriptomes and how cagA contributes to colonization within the host cell are lacking.
The advent of dual RNA sequencing (RNA-seq) through probe-independent massively parallel cDNA sequencing now offers the opportunity for the comprehensive and simultaneous whole-genome transcriptional profiling of both the pathogen and infected host cells at high resolution (13–15). Using this technology, we infected a normal human gastric epithelial cell line (GES-1) with wild-type (WT) or cagA-mutated H. pylori strains to monitor the progression of H. pylori infection over time and simultaneously generated high-resolution transcriptome profiles of H. pylori and the human host cells. The depth of sequencing used allowed us to identify infection-specific host transcriptome alterations and characterize the adaption and pathogenesis strategies of the microorganism during infection. In addition, we performed in vitro and in vivo experiments to validate the findings obtained from the dual RNA-seq data set. Collectively, this study is the first to perform a dual-transcriptome analysis of H. pylori during dynamic interaction with the gastric epithelium and to provide new insights into the pathogenesis of H. pylori.
RESULTS
Identification of host-induced H. pylori-specific stress responses using dual RNA-seq
To explore the interaction between intracellular H. pylori and the human gastric epithelium, we first labeled H. pylori with fluorescein isothiocyanate (FITC) and determined the internalization efficiency of this microorganism by coculturing it with GES-1 cells for 3 or 24 h. We observed that most FITC-labeled H. pylori were stained by the antibody in Triton X-100-permeabilized GES-1 cells. However, in cells not treated with Triton X-100, these FITC-labeled bacteria could not be detected by the antibody (Fig. S1A), indicating that H. pylori can invade GES-1 cells as early as 3 h post coincubation. Gentamicin, an aminoglycoside antibiotic that poorly penetrates the eukaryotic cell membrane (16), was used to eliminate extracellular bacteria. We found that treatment with gentamicin (100 µg/mL) efficiently inhibited the biofilm formation (Fig. S1B) and bacterial growth (Fig. S1C) of multiple H. pylori strains, including TN2GF4, NCTC 11637, PMSS1, and 7.13. A recent study suggested that gentamicin exerts adverse effects, such as mitochondrial damage in cell culture (17). Thus, we examined whether gentamicin supplementation could induce reactive oxygen species and the associated DNA damage in GES-1 cells. Our results revealed that gentamicin at a concentration of 100 µg/mL had no adverse impact on the proliferation of GES-1 cell biofilm formation (Fig. S1D). Moreover, in contrast to carbonyl cyanide 3-chlorophenylhydrazone, we did not observe an increased production of mitochondrial superoxide in gentamicin-treated GES-1 cell biofilm formation (Fig. S1E). Similarly, the level of 8-hydroxy-2′-deoxyguanosine (8-OHdG), a hallmark of DNA damage, was not significantly increased in cells coincubated with gentamicin biofilm formation (Fig. S1E). These results collectively demonstrated that gentamicin at a concentration of 100 µg/mL effectively restricted the growth of H. pylori but had minimal adverse effects on the proliferation of GES-1 cells.
Next, we cocultured GES-1 cells with WT or △cagA H. pylori TN2GF4 strains at a multiplicity of infection (MOI) of 100 for 3 h. Subsequently, gentamicin (100 µg/mL) was used to eliminate extracellular bacteria. We monitored genome-scale transcriptomic events during the infection by performing dual RNA-seq on both infected and mock-infected GES-1 cells at time 0 (uninfected GES-1 cells and H. pylori before infection) and 12 and 24 h post-infection, with three biologically independent experiments per timepoint (Fig. 1A). The sequencing libraries yielded over 121 million total reads (median, 73,752,510 reads; range, 9,815,954 to 86,633,416 reads). After adapter trimming and removal of low-quality reads, we retained an average of 46 million clean reads per sample (median, 54,816,222 reads; range, 9,309,838 to 70,169,125 reads), of which approximately 28.4% and 71.6% of the total reads originated from the pathogen and the human genome, respectively (Fig. 1B). As anticipated, only 0.72% of human and 0.06% of H. pylori reads were mapped and counted as ribosomal RNAs, indicating the successful depletion of host and bacterial rRNAs. The high number of usable reads in each library indicated the reliability of our dual RNA-seq approach, including total RNA extraction, host and bacterial mRNA enrichment, and cDNA library preparation, which enabled us to determine different gene expression patterns in host-pathogen transcripts with high resolution.
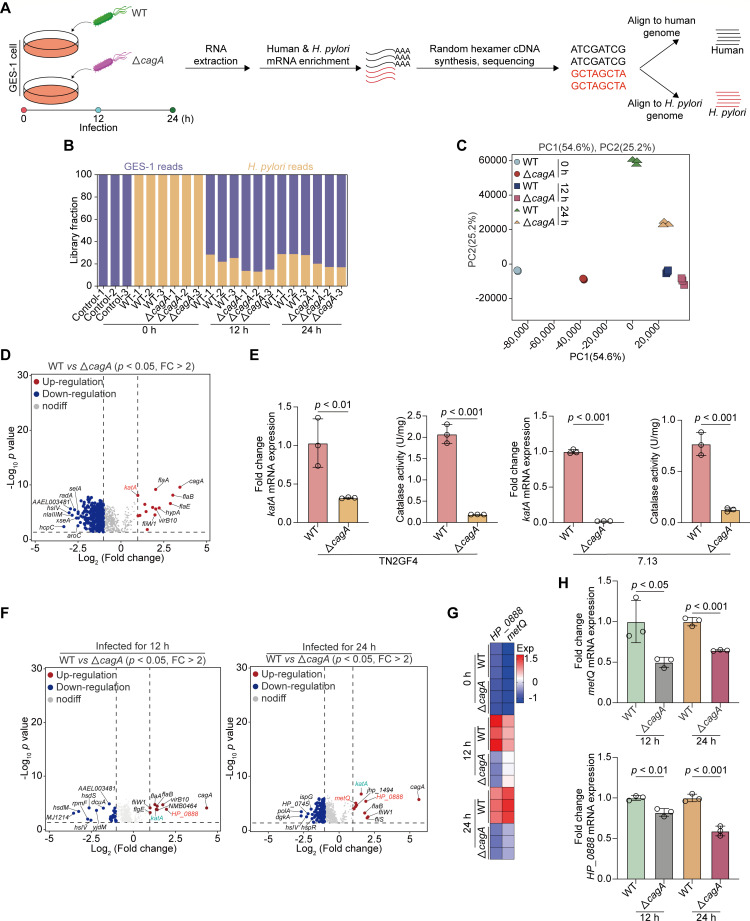
Fig 1.
Identification of host-induced H. pylori-specific stress responses using dual RNA-Seq. (A–D) GES-1 cells were infected with WT or cagA-mutated H. pylori TN2GF4 strains (MOI 100) for 3 h and then exposed to gentamicin (100 µg/mL) for 12 or 24 h to eliminate the extracellular bacteria as detailed in “supplemental Materials and Methods.” (A) Experimental workflow for dual RNA-seq RNA extraction, library preparation, and sequencing processing. (B) The proportions of reads aligned to H. pylori or GES-1 cells in each library. On average, there are 46 million reads per library: 71.6% of the reads aligned to the human genome and 28.4% to the H. pylori genome. (C) Principal component analysis (PCA) of the H. pylori transcriptome from different experiment conditions. (D) Volcano plot showing the DEGs obtained from WT H. pylori TN2GF4 strain compared with cagA-mutated TN2GF4 strain at time 0 using the DESEq2 toolkit. P value < 0.05 was considered statistically significant, Benjamini-Hochberg adjusted two-sided Wilcoxon test. (E) Total mRNA or proteins were extracted from WT or cagA-mutated H. pylori TN2GF4 (left) or 7.13 (right) strains. The expression levels of katA mRNA and catalase activity were determined. H. pylori 16S rRNA was used as the loading control for katA mRNA. (F) Volcano plot showing the DEGs obtained from WT H. pylori TN2GF4 strain compared with cagA-mutated TN2GF4 strain after coculturing with GES-1 cells for 12 (left) or 24 h (right). P value < 0.05 was considered statistically significant, Benjamini-Hochberg adjusted two-sided Wilcoxon test. (G) Heatmap showing the expression of ATP-binding cassette (ABC) transporter-related genes, metQ and HP_0888, in WT or cagA-mutated H. pylori TN2GF4 strains after coculturing with GES-1 cells for 0, 12, or 24 h. Color coding was based on normalized expression levels. (H) GES-1 cells were infected with WT or cagA-mutated H. pylori TN2GF4 strains (MOI 100) for 3 h and then exposed to gentamicin (100 µg/mL) for 12 or 24 h to eliminate the extracellular bacteria. The expression levels of metQ (upper) and HP_0888 (down) mRNA were determined. H. pylori 16S rRNA was used as the loading control. All the quantitative data were presented as means ± SD from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001.
We systematically profiled the transcriptional events of H. pylori at each timepoint to identify infection-specific gene expression, which may lead to adjustments in the virulence and fitness of the pathogen. PCA showed distinct global gene expression in WT and cagA-mutant H. pylori at different incubation periods, and the profiles of the three biological replicates were clustered together (Fig. 1C). To investigate the cagA-dependent transcriptome in H. pylori, we analyzed gene expression profiles in WT and △cagA H. pylori strains at time 0 using the differential expression analysis package DESeq2 with a twofold cutoff and a Benjamini-Hochberg false discovery rate of <0.05 as the threshold for inclusion. We identified 14 genes with significantly higher expression in WT H. pylori than in the cagA-mutant strain (Fig. 1D). Moreover, the expression level of cagA was the highest in WT H. pylori [Fold change (FC) = 10.9, P = 3.0E−10], indicating the successful mutation of the cagA factor. A set of genes related to the flagellar regulatory system, including flaB (FC = 8.24, P = 8.6E−09), flgE (FC = 7.47, P = 2.9E−07), flaA (FC = 4.12, P = 7.7E−10), fliW1 (FC = 4.00, P = 3.5E−05), fliS (FC = 2.93, P = 0.01), fliD (FC = 2.66, P = 4.1E−07), motB (FC = 2.17, P = 4.4E−05), and motA (FC = 2.02, P = 5.12E−05), was significantly downregulated in the cagA isogenic mutant strain. This finding is consistent with that of a previous study indicating that cagA is associated with bacterial motility (18). Moreover, we observed a marked reduction in the expression level of katA in the △cagA strain (FC = 2.0, P = 9.4E−09). The katA (catalase) gene of H. pylori encodes an antioxidant enzyme that protects the bacteria from oxidative stress (19). To verify whether the cagA mutation is responsible for the decreased expression of katA, we measured the katA mRNA level and catalase activity in two H. pylori strains, TN2GF4 and 7.13, with or without the cagA mutation. We observed that the levels of katA mRNA expression and catalase activity were both significantly decreased in △cagA strains (Fig. 1E), indicating that katA is transcriptionally inactivated in the absence of cagA expression in H. pylori. In contrast, we identified that a cluster of genes, including hcpC (Sel1 repeat family protein), nlaIIIM (DNA adenine methylase), and radA (DNA repair protein), were upregulated in the cagA isogenic mutant strain (Fig. 1D).
Next, we compared transcriptional diversity between WT and cagA-mutated strains after coculturing with GES-1 cells for 12 or 24 h. Our findings revealed that katA and flagellum-related genes were highly expressed in the transcriptome of the WT H. pylori strain (Fig. 1F). Moreover, compared with △cagA strains, HP_0888 and metQ were both identified to be strongly upregulated in the WT H. pylori strain upon coincubation with GES-1 cells for 12 (HP_0888: FC = 4.0, P = 1.8E−04; metQ: FC = 1.3, P = 9.8E−04) or 24 h (HP_0888: FC = 3.8, P = 4.9E−06; metQ: FC = 2.3, P = 1.1E−05, Fig. 1F). HP_0888 and metQ are both critical components of ABC transporters that mediate the uptake of host-provided nutrients into the cytoplasm of H. pylori for its proliferation and pathogenicity (20, 21). Heatmap analysis showed that such upregulation occurred after coculturing of H. pylori with GES-1 cells (Fig. 1G). Furthermore, we performed quantitative real-time PCR (qRT-PCR) and determined that the expression of HP_0888 and metQ was both significantly declined in cagA-mutated strains after coculturing them with GES-1 cells for 12 or 24 h (Fig. 1H). This finding indicated that ABC transporter genes, HP_0888 and metQ, were regulated by cagA in response to host-imposed stresses during H. pylori infection.
H. pylori cagA activated oxidative phosphorylation by disrupting electron transportation
To explore H. pylori cagA-reprogrammed host transcriptomic profiles, we performed PCA analysis and observed a clear separation in GES-1 cells infected with WT or cagA-mutated H. pylori strains for 12 or 24 h (Fig. 2A). Next, using DESeq2, we identified 176 DEGs (FC > 2 or <0.5, P < 0.05) in GES-1 cells infected with the H. pylori WT strain compared with the cagA-mutated strain after 12 h post-challenge. Using the same filter criteria, we profiled 222 DEGs whose expression was either positively or negatively regulated by H. pylori cagA after 24 h of infection. Subsequently, we analyzed these DEGs to predict their biological functions through gene set enrichment analysis (GSEA) and identified four biological processes, including oxidative phosphorylation, common to both timepoints (Fig. 2B and C). Oxidative phosphorylation, a crucial source of ATP generation, supports cell growth and the progress of intracellular metabolic pathways (22, 23). This process is accomplished by transporting electrons to a series of transmembrane proteins located in the mitochondrial inner membrane, known as the electron transport chain (ETC) (24). Impaired electron transport leads to the aberrant accumulation of reactive oxygen species (ROS). We identified genes responsible for cagA-driven oxidative stress and observed a pronounced reduction in genes related to the ETC in GES-1 cells infected with the WT H. pylori strain compared with their control counterparts (Fig. 2D). These included genes encoding the subunits of NADH: ubiquinone oxidoreductase (NDUFA10, NDUFS6, NDUFB2, and NDUFS7), cytochrome c oxidase (COX7B, COX8A, COX4I1, COX5B, and COX7C), ATP synthase (ATP6V0A4, ATP5MC2, ATP5F1B, and ATP5MC1), CYC1 (encoding cytochrome c), PPA2, and UQCRC1. However, this reduction was significantly reversed in △cagA-infected GES-1 cells. Furthermore, we performed qRT-PCR and verified that the mRNA expression of NDUFS6, NDUFA10, ATP6V0A4, NDUFS7, and NDUFB2 was significantly decreased in GES-1 cells infected with the WT H. pylori strain for 12 or 24 h. Consistently, the reduction in the expression of these genes was significantly mitigated in GES-1 cells co-coculturing with the △cagA H. pylori strain (Fig. 2E), indicating that cagA triggered oxidative stress by disrupting electron transportation in GES-1 cells.
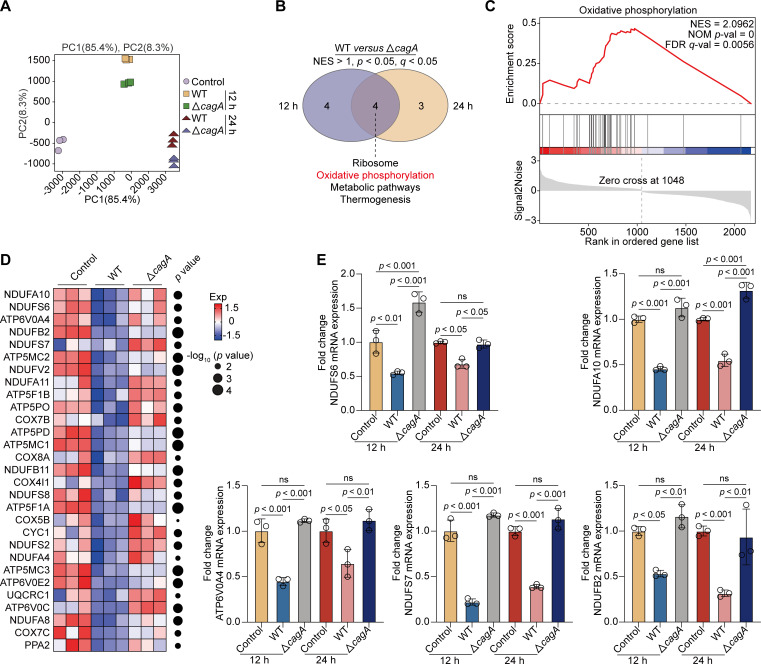
Fig 2.
H. pylori cagA activated oxidative phosphorylation by disrupting the electron transportation. (A–D) GES-1 cells were infected with WT or cagA-mutated H. pylori TN2GF4 strains (MOI 100) for 3 h and then exposed to gentamicin (100 µg/mL) for 12 or 24 h to eliminate the extracellular bacteria. (A) PCA diagram showing the host cell transcriptome from different experiment conditions. (B) Venn diagram showing the number of GSEA-enriched biological pathways in GES-1 cells infected with WT versus cagA-mutated H. pylori TN2GF4 strains for 12 (left) or 24 h (right), with four pathways including oxidative phosphorylation (red) being simultaneously enriched in both two timepoints. (C) GSEA enrichment plot showed that the “oxidative phosphorylation” pathway was enriched in GES-1 cells infected with WT versus cagA-mutated H. pylori TN2GF4 strains for 24 h. (D) Heatmap showing the expression of ETC-related genes in uninfected GES-1 cells or cells infected with WT or cagA-mutated H. pylori TN2GF4 strains for 24 h. Dot size indicates the −log10 transformed P values, color coding based on normalized expression levels. (E) GES-1 cells were infected with WT or cagA-mutated H. pylori TN2GF4 strains (MOI 100) for 3 h and then exposed to gentamicin (100 µg/mL) for 12 or 24 h to eliminate the extracellular bacteria. The mRNA expression of NDUFS6, NDUFA10, ATP6V0A4, NDUFS7, and NDUFB2 was determined. β-Actin was used as the loading control. All the quantitative data were presented as means ± SD from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001.
Repressed expression of multiple splicing factors in response to H. pylori infection
Next, we characterized H. pylori-driven transcriptomic events in GES-1 cells. To examine gene expression profiles correlated with H. pylori infection, we used the DESeq2 toolkit and identified 3,197 DEGs in GES-1 cells infected with H. pylori for 24 h compared with mock infection (FC > 2 or <0.5, P < 0.05). Of these, 1,967 genes were upregulated and 1,230 genes were downregulated in H. pylori-infected cells (Fig. 3A), suggesting altered transcriptional activity in response to H. pylori infection. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that DEGs downregulated in response to H. pylori infection were enriched in the spliceosome pathway (Fig. 3B). To further confirm the KEGG annotation results, we performed Gene Ontology (GO) analysis and determined that these downregulated genes were enriched in RNA/rRNA/ncRNA processing and ncRNA/rRNA/nucleic acid metabolic processes (Fig. 3C). Using the same filter criteria, we profiled 2,003 DEGs that were downregulated in △cagA-infected GES-1 cells compared with mock-infected cells (FC < 0.5, P < 0.05). KEGG analysis revealed that the DEGs downregulated in response to △cagA H. pylori strain infection were also enriched in the spliceosome pathway (Fig. 3D), suggesting that H. pylori-perturbed RNA splicing and RNA stability processes were not dependent on cagA expression.
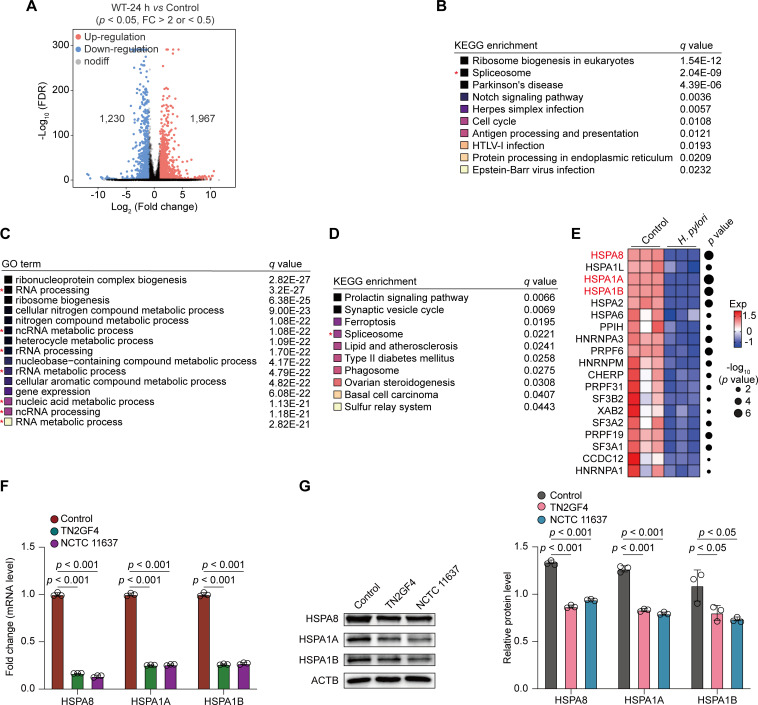
Fig 3.
Repressed expression of multiple splicing factors in response to H. pylori infection. (A–E) GES-1 cells were infected with WT or cagA-mutated H. pylori TN2GF4 strains (MOI 100) for 3 h and then exposed to gentamicin (100 µg/mL) for 24 h to eliminate the extracellular bacteria. (A) Volcano plot showing the DEGs obtained from GES-1 cells infected with WT H. pylori TN2GF4 strain for 24 h compared with mock infection (FC > 2 or <0.5, P value < 0.05). Of these, 1,967 DEGs were upregulated, whereas 1,230 DEGs were downregulated in H. pylori-infected cells. Benjamini-Hochberg adjusted two-sided Wilcoxon test. (B) KEGG annotation of the top 10 enriched biological pathways using the downregulated DEGs from WT H. pylori TN2GF4-infected GES-1 cells compared with mock infection after 24 h post-challenge. Colored squares represented the q value (black, small; yellow, big). (C) GO annotation of the top 15 enriched biological pathways using the downregulated DEGs from WT H. pylori TN2GF4-infected GES-1 cells compared with mock infection after 24 h post-challenge. Colored squares represented the q value (black, small; yellow, big). (D) KEGG annotation of the top 10 enriched biological pathways using the downregulated DEGs from cagA-mutated H. pylori TN2GF4-infected GES-1 cells compared with mock infection after 24 h post-challenge. Colored squares represented the q value (black, small; yellow, big). (E) Heatmap showing the expression of splicing regulators in uninfected GES-1 cells or cells infected with WT H. pylori TN2GF4 strains for 24 h. Dot size indicates the −log10 transformed P values, color coding based on normalized expression levels. (F and G) GES-1 cells were infected with H. pylori TN2GF4 or NCTC11637 strains (MOI 100) for 3 h and then exposed to gentamicin (100 µg/mL) for 24 h to eliminate the extracellular bacteria. The mRNA (F) and protein (G) expression of HSPA8, HSPA1A, and HSPA1B was determined in uninfected GES-1 cells or cells infected with H. pylori TN2GF4 or NCTC 11637 strains. β-Actin was used as the loading control. All the quantitative data were presented as means ± SD from three independent experiments. *P < 0.05 and ***P < 0.001.
We explored the transcriptional profile of splicing factors involved in the spliceosome pathway and identified that numerous well-established splicing regulators were downregulated in response to H. pylori infection, including heat shock proteins (HSPs: HSPA8, FC = 0.069, P = 1.84E−07; HSPA1L, FC = 0.302, P = 0.0028; HSPA1A, FC = 0.082, P = 1.51E−08; HSPA1B, FC = 0.115, P = 4.49E−07; HSPA2, FC = 0.278, P = 1.94E−05; and HSPA6, FC = 0.250, P = 0.018), splicing factor 3a Subunit 2 (SF3A2, FC = 0.599, P = 0.0066), splicing factor 3b subunit 2 (SF3B2, FC = 0.684, P = 0.043), heterogeneous nuclear ribonucleoprotein M (HNRNPM, FC = 0.44, P = 0.002), and heterogeneous nuclear ribonucleoprotein A3 (HNRNPA3, FC = 0.429, P = 2.6E−03) and heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1, FC = 0.766, P = 0.04, Fig. 3E). Consistent with the dual RNA-seq data, coculturing GES-1 cells with two H. pylori strains, TN2GF4 and NCTC 11637, revealed the downregulation of HSPA8, HSPA1A, and HSPA1B at both mRNA and protein levels compared with mock treatment (Fig. 3F and G), indicating that H. pylori disrupted RNA splicing and RNA stability by reducing the expression levels of several splicing factors in GES-1 cells.
H. pylori infection modulates mRNA splicing of functional genes involved in the cell cycle process
Because the DEGs were enriched in the spliceosome pathway, we quantified splicing changes caused by H. pylori infection in GES-1 cells using the rMATS algorithm (25). Five major modes of alternative splicing (AS) events, namely, alternative 5′ splice sites (A5SS), alternative 3′ splice sites (A3SS), mutually exclusive exons (MXE), retained intron (RI), and skipped exons (SE), have been described in metazoan organisms (26). Using an adjusted P value cutoff of <0.05, we identified 9,895 splicing events that significantly changed in H. pylori-infected GES-1 cells compared with the controls (Fig. 4A). Specifically, the majority of AS events were exon skipping (69.06%), with 3,062 of the remaining events categorized into other splicing modes, including 1,267 for MXE, 611 for A5SS, 801 for A3SS, and 383 for RI. Thus, these data support the involvement of H. pylori in mRNA splicing during coculturing with GES-1 cells, with cassette exons being the most frequent targets upon H. pylori challenge.
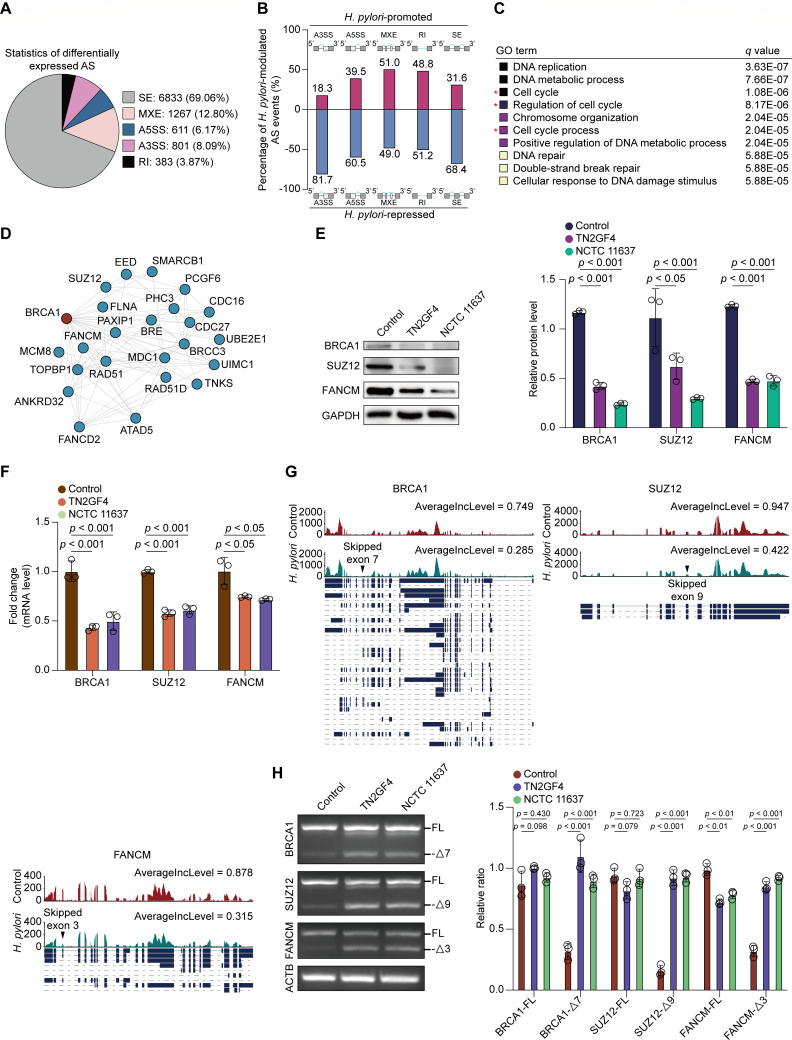
Fig 4.
H. pylori infection modulated mRNA splicing of functional genes that were involved in cell cycle process. (A–D) GES-1 cells were infected with WT H. pylori TN2GF4 strain (MOI 100) for 3 h and then exposed to gentamicin (100 µg/mL) for 24 h to eliminate the extracellular bacteria. (A) Pie charts showing the proportion of each type of significantly altered splicing events in H. pylori-infected GES-1 cells compared with the controls using the rMATS algorithm. (B) Percentage of H. pylori-promoted or repressed splicing events in GES-1 cells. (C) GO annotation of the top 10 enriched biological pathways using the functional genes affected by H. pylori-promoted splicing events. Colored squares represented the q value (black, small; yellow, big). (D) The protein-protein interaction network for functional genes involved in the cell cycle process based on the STRING database. (E and F) GES-1 cells were infected with H. pylori TN2GF4 or NCTC11637 strains (MOI 100) for 3 h and then exposed to gentamicin (100 µg/mL) for 24 h to eliminate the extracellular bacteria. (E) The protein levels of BRCA1, SUZ12, and FANCM were determined and quantified in uninfected GES-1 cells or cells infected with H. pylori TN2GF4 or NCTC 11637 strains for 24 h. GAPDH was used as the internal control. (F) The mRNA expression of BRCA1, SUZ12, and FANCM was determined in uninfected GES-1 cells or cells infected with H. pylori TN2GF4 or NCTC 11637 strains for 24 h. (G) Exon skipping in the seventh exon of BRCA1, the nineth exon of SUZ12, and the third exon of FANCM as visualized by the IGV software. Black arrowheads indicate splicing sites. (H) GES-1 cells were infected with H. pylori TN2GF4 or NCTC11637 strains (MOI 100) for 3 h and then exposed to gentamicin (100 µg/mL) for 24 h to eliminate the extracellular bacteria. RT-PCR analysis of alternative splicing patterns of the changed splicing genes in control and H. pylori-infected GES-1 cells. β-Actin was used as the internal control. The expression of full-length and exon-skipping isoforms of the three genes was quantified. All the quantitative data were presented as means ± SD from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001.
To gain deep insights into the spectrum of genes that were aberrantly spliced, we defined H. pylori-modulated AS events using a stringent filter criteria (IncLevelDiff ≥ 0.1 or ≤−0.1, adjusted P < 0.05, and splice junction read coverage ≥ 20). We identified 736 H. pylori-promoted and 1,294 H. pylori-repressed splicing events in 628 and 1,064 genes in GES-1 cells, respectively (Fig. 4B). Approximately 65.8% of H. pylori-modulated AS events was SE, followed by MXE (15.2%), A3SS (8.7%), A5SS (6.1%), and RI (4.2%). Next, we focused on the 628 genes affected by H. pylori-promoted AS events and predicted their molecular functions by performing GO term enrichment analysis. We determined that 106 of these genes, including breast cancer type 1 (BRCA1), SUZ12 polycomb repressive complex 2 subunit (SUZ12), and FA complementation group M (FANCM), were involved in the cell cycle process (Fig. 4C). The protein-protein interaction network constructed using the STRING database further confirmed extensive interactions among these genes (Fig. 4D). Subsequently, we validated the expression of BRCA1, SUZ12, and FANCM in H. pylori-infected GES-1 cells and observed significant abrogation of the mRNA and protein expression of these three genes in cells treated with H. pylori strains TN2GF4 and NCTC 11637 (Fig. 4E and F). The results of rMATS analysis revealed that H. pylori infection promoted the skipping of the seventh exon in BRCA1, the ninth exon in SUZ12, and the third exon in FANCM, as visualized using the Integrative Genomics Viewer program (Fig. 4G). To verify the aberrant splicing of these three genes, we performed semi-quantitative and real-time PCR experiments using specific primers for target genes. We observed that although the pre-mRNA levels of these three genes remained stable, the expression of the exon-skipping isoforms of BRCA1 (exon 7), SUZ12 (exon 9), and FANCM (exon 3) was significantly elevated upon H. pylori infection (Fig. 4H), indicating that H. pylori treatment led to aberrant pre-mRNA splicing of functional genes involved in the cell cycle process.
Gastric H. pylori colonization reduced the severity of chronic DSS-induced colitis
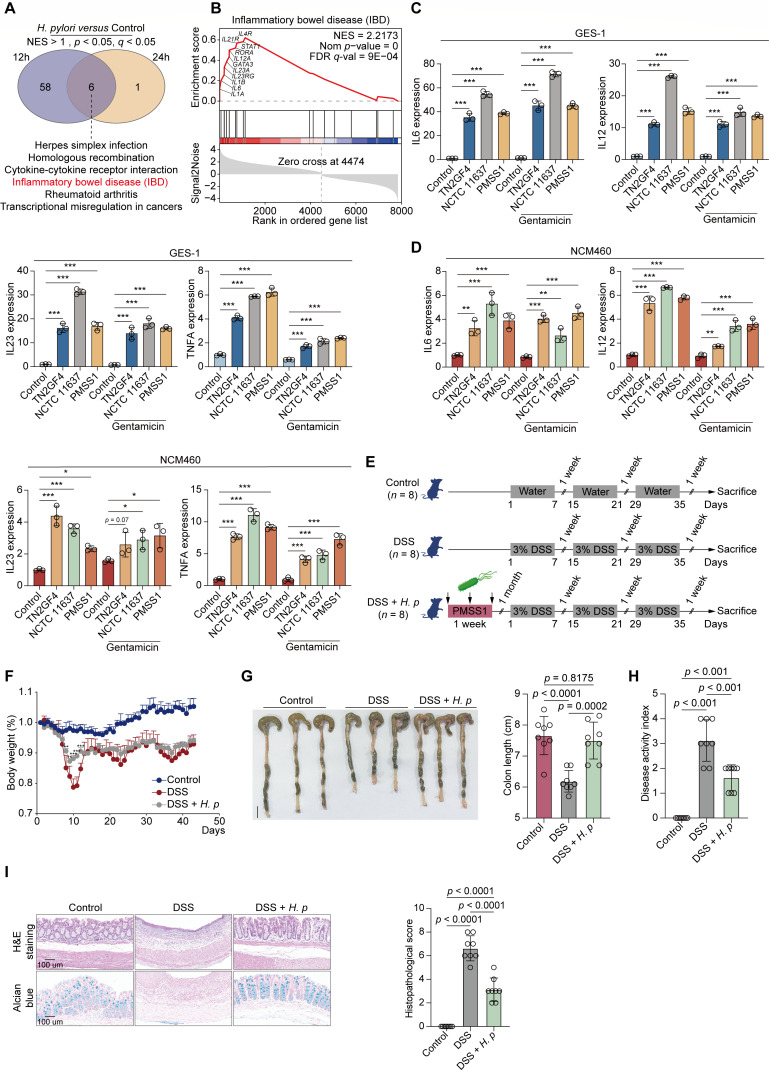
As mentioned above, we identified 3,197 DEGs in GES-1 cells infected with H. pylori for 24 h compared with mock-infected cells (FC > 2 or <0.5, P < 0.05). To further explore infection-specific transcriptome signatures, we profiled 1,831 DEGs in H. pylori-infected GES-1 cells at 12 h using the same filter criteria. Of these, 808 upregulated and 1,023 downregulated DEGs were identified in H. pylori-infected GES-1 cells compared with the controls. We then performed GSEA analysis to examine biological functions in H. pylori-infected GES-1 cells at 12 or 24 h post-challenge by importing these DEGs. We observed that a cluster of inflammatory bowel disease (IBD)-related cytokines and genes was strongly upregulated in GES-1 cells infected with H. pylori for 12 or 24 h (Fig. 5A and B). Consistent with the dual RNA-seq data, coculturing of GES-1 cells with three H. pylori strains, namely, TN2GF4, NCTC 11637, and PMSS1, resulted in significantly elevated expression of IBD-associated genes, including IL6, IL12, IL23, and TNFA, upon H. pylori infection, regardless of whether eliminating extracellular pathogens by gentamicin or not (Fig. 5C).
Fig 5.
Gastric H. pylori colonization alleviated the severity of chronic DSS-induced colitis. (A and B) GES-1 cells were infected with WT H. pylori TN2GF4 strain (MOI 100) for 3 h and then exposed to gentamicin (100 µg/mL) for 12 or 24 h to eliminate the extracellular bacteria. (A) Venn diagram showing the number of GSEA-enriched biological pathways in H. pylori-infected GES-1 cells compared with mock infection after 12 or 24 h challenge, with six pathways including IBD pathway (red) being simultaneously enriched in both two timepoints. (B) GSEA enrichment plot showed that the “IBD” pathway was enriched in H. pylori-infected GES-1 cells for 24 h compared with mock infection. IBD pathway-related genes that were upregulated in response to H. pylori infection were labeled. (C and D) GES-1 (C) or NCM460 (D) cells were infected with H. pylori TN2GF4, NCTC 11637, or PMSS1 strains (MOI 100) for 24 h with or without gentamicin (100 µg/mL) treatment. The mRNA expression of IL6, IL12, IL23, and TNFA were determined. β-Actin was used as the loading control. The quantitative data were presented as means ± SD from three independent experiments. (E–J) C57BL/6J mice were orally inoculated with H. pylori PMSS1 strain (n = 8 animals) or the vehicle (n = 8 animals) for 1 month, followed by the administration of three cycles of 3% DSS (7 days/cycle), each separated by 7 days of regular water. (E) Schematic overview of the experimental design. (F) The changes of mice body weight after DSS administration were monitored. Mean ± SD from eight mice in each group. (G) (Left) Representative photographs of mouse colon tissue from each group were presented. Scale bar = 1 cm. (Right) The colon length of each mice was recorded. (H) The DAI index per mice was evaluated. (I) The histological analysis of mice colon tissue was performed by hematoxylin and eosin (H&E) and alcian blue staining. Scale bar = 100 µm. Histological scores of the DSS-induced colitis were evaluated. The quantitative data were presented as means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001.
Previous epidemiological data have indicated an inverse association between H. pylori infection and the risk of IBD due to the ability of H. pylori to induce immune tolerance (27). However, detailed mechanisms through which H. pylori infection protects against IBD still remain unclear. To assess whether H. pylori infection can stimulate a similar IBD-associated immune response in a normal human intestinal epithelial cell line, we selected NCM460 to be cocultured with these H. pylori strains, such as TN2GF4, NCTC 11637, and PMSS1, with or without gentamicin administration. Consistently, we discovered that H. pylori coincubation substantially elevated the expression of IL6, IL12, IL23, and TNFA, regardless of the use of gentamicin to eliminate extracellular pathogens (Fig. 5D).
We further investigated whether H. pylori infection can efficiently protect against the development of IBD in an in vivo model. We used the mouse-adapted, cagPAI-positive H. pylori strain PMSS1 to colonize the stomach of mice and then assessed whether gastric H. pylori colonization can mitigate the severity of chronic colitis induced by 3.0% dextran sulfate sodium (DSS) (Fig. 5E). We observed severe clinical symptoms in all colitis mice that received DSS, including loss of body weight and stool consistency, and stool with occult blood. However, mice precolonized with H. pylori exhibited significantly less body weight than those treated with DSS alone (Fig. 5F). Likewise, H. pylori pretreatment substantially alleviated the shortening of colon length in DSS-treated mice (Fig. 5G). Disease activity index (DAI) scores were markedly decreased in mice pretreated with H. pylori compared with those treated with DSS alone (Fig. 5H). Moreover, we collected colon tissues for histological assessment by performing H&E and alcian blue staining and observed weakened intestinal epithelial destruction, limited inflammatory cell infiltration, and declined goblet cell loss in H. pylori precolonized mice compared with those administered DSS alone (Fig. 5I). Thus, these findings collectively demonstrated that gastric H. pylori colonization significantly upregulated the expression levels of IBD-associated genes, efficiently attenuated colonic inflammation, and mitigated the destruction of the colonic mucosal structure caused by DSS administration.
H. pylori-sustained Muribaculaceae abundance contributed to the restoration of intestinal barrier function damaged by DSS treatment
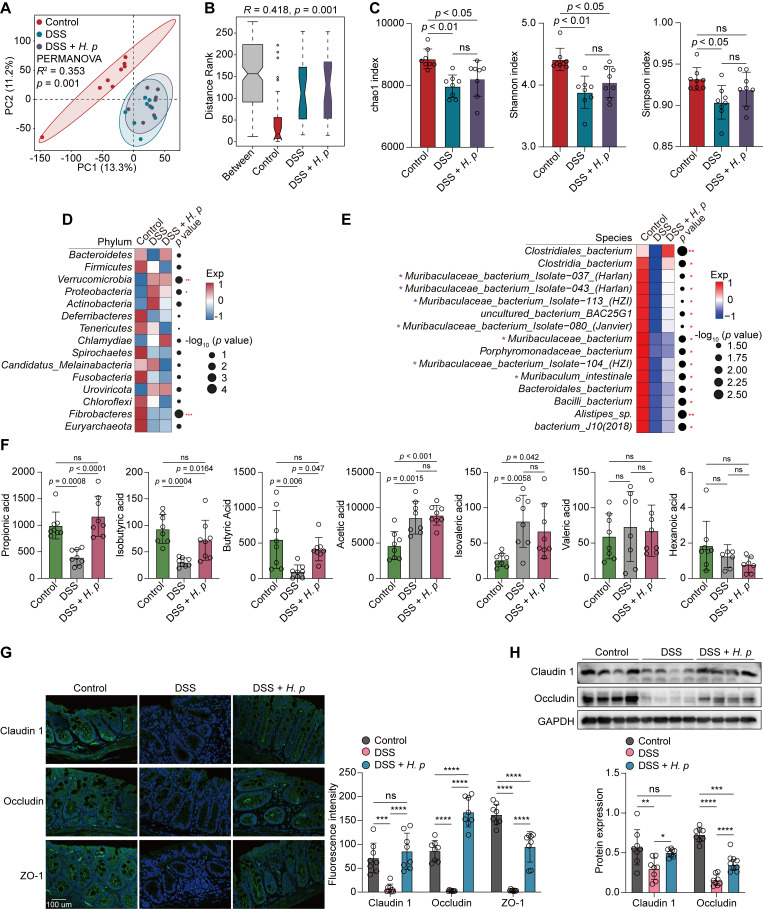
Disordered intestinal microbiota is the underlying pathogenic mechanism of IBD (28–30). To investigate whether intestinal flora is involved in H. pylori-alleviated colitis, we collected stool samples from these mice and subjected them to high-throughput shotgun metagenomic sequencing to profile their microbial community taxonomic composition. PCA at the species level revealed distinct structures of the intestinal microbiota among these three groups [R2 = 0.353, P = 0.001, permutational multivariate analysis of variance (PERMANOVA)] despite no marked structural changes between DSS alone and DSS and H. pylori cotreatment groups (Fig. 6A). This distinction was associated with differences in community composition, as determined by analysis of similarities (ANOSIM) analysis (R = 0.418, P = 0.001; Fig. 6B). This finding indicated a more heterogeneous community structure in DSS-treated and DSS and H. pylori-cotreated mice than in control mice. We further observed a decline in the α-diversity of the intestinal microbiota, as indicated by the chao1, Shannon, and Simpson indexes, in DSS-treated mice compared with their control counterparts. However, higher chao1, Shannon, and Simpson indexes were observed in H. pylori-pretreated mice compared with those treated with DSS alone, although the difference was not statistically significant (Fig. 6C). This finding suggested the recovery of the richness and diversity of the intestinal microbiota in H. pylori-precolonized mice.
Fig 6.
H. pylori-sustained Muribaculaceae abundance contributed to the restoration of intestinal barrier function damaged by DSS treatment. (A–H) C57BL/6J mice were orally inoculated with H. pylori PMSS1 strain (n = 8 animals) or the vehicle (n = 8 animals) for 1 month, followed by the administration of three cycles of 3% DSS (7 days/cycle), each separated by 7 days of regular water. (A) PCA diagram showing the β-diversity of mice fecal microbiota among the three groups at the species level. (B) ANOSIM test was applied to compare microbial structure dissimilarity between and within groups. Two-sided Wilcoxon rank-sum test. (C) The α-diversity of intestinal microbiota in the three groups at the species level was evaluated by chao1 (left), Shannon (middle), and Simpson (right) indices. (D and E) Analysis of the differences in the mice intestinal microbiota at the phylum (D) or species (E) levels. Dot size indicates the −log10 transformed P values, color coding based on normalized expression levels. Two-sided Wilcoxon rank-sum test. (F) The concentrations of seven types of short-chain fatty acid (SCFA) in mice colon tissue from each group were determined by absolute quantitative metabolomics. (G) The fluorescence intensities of Claudin 1, Occludin, and ZO-1 in the mice colon were determined by immunofluorescence staining. Scale bar = 100 µm. (H) The protein levels of Claudin 1 and Occludin in the mice colon were determined and quantified by western blotting. GAPDH was used as the internal control. All the quantitative data were presented as means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001.
Next, we performed metagenomic phylogenetic analysis (MetaPhlAn2) to examine taxonomic abundance in these fecal samples. We determined that at the phylum level, the abundance of Bacteroidetes was decreased in DSS-treated mice relative to the control mice, whereas its level was restored in H. pylori-pretreated mice (Fig. 6D). Conversely, the abundance of Proteobacteria was increased following DSS treatment, whereas H. pylori precolonization markedly limited its accumulation in the mouse colon (Fig. 6D). At the species level, we identified that the abundance of a group of bacteria belonging to the Muribaculaceae family within the Bacteroidetes phylum was significantly decreased in DSS-treated mice, whereas their abundance was markedly sustained after H. pylori precolonization (Fig. 6E). These microorganisms included Muribaculaceae_bacterium_Isolate−037_(Harlan) (P = 0.03), Muribaculaceae_bacterium_Isolate−043_(Harlan) (P = 0.04), Muribaculaceae_bacterium_Isolate−113_(HZI) (P = 0.04), Muribaculaceae_bacterium_Isolate−080_(Janvier) (P = 0.04), Muribaculaceae_bacterium (P = 0.02), Muribaculaceae_bacterium_Isolate−104_(HZI) (P = 0.02), and Muribaculum_intestinale (P = 0.02).
Muribaculaceae has been newly identified as a type of SCFA-producing bacteria (31), especially propionate and butyrate (32). Their abundance was markedly decreased in mice with fatty liver disease (33). To determine whether the restored abundance of Muribaculaceae was accompanied by increased SCFA levels in H. pylori-pretreated mouse colon, we conducted absolute quantitative metabolomic analysis to measure SCFA levels in mouse stool samples. As expected, we discovered significant reductions in propionic acid, isobutyric acid, and butyric acid levels in DSS-treated mice, whereas the levels of these three SCFAs were substantially restored upon gastric H. pylori colonization (Fig. 6F), indicating that H. pylori-elevated Muribaculaceae bacteria may contribute to the enrichment of propionic and butyric acids in the mouse colon. However, this phenomenon was not observed for other SCFA members (Fig. 6F), suggesting that Muribaculaceae bacteria were preferentially responsible for the production of propionic and butyric acids but not the other SCFAs.
SCFAs, including propionate and butyrate, produced by bacterial fermentation in the gut, have long been of interest to researchers for their capacity to maintain intestinal barrier integrity and homeostasis (34). To explore whether H. pylori precolonization could effectively repair the intestinal barrier damaged by DSS, we determined the expression levels of three tight junction proteins, namely, Claudin 1, Occludin, and ZO-1, which are directly responsible for intestinal barrier function (35). As shown in Fig. 6G, the immunofluorescence intensity of these three proteins was significantly decreased in the colon of DSS-treated mice relative to those from the control counterparts. However, such reduction was markedly reversed by H. pylori precolonization. Consistently, immunoblotting analysis confirmed that the decreased protein levels of Claudin 1 and Occludin in DSS-treated mice were significantly restored by H. pylori pretreatment (Fig. 6H), indicating that the intestinal barrier integrity damaged by DSS treatment was recovered upon H. pylori colonization.
DISCUSSION
The recent development of dual RNA-seq technology has enabled researchers to simultaneously profile RNA expression in host-pathogen systems, offering novel insights into complex host-pathogen interactions. In this study, by leveraging the advantages of this technology, we presented the first dual-transcriptome analysis of H. pylori and its human host, revealing the bacterium’s adaptation and pathogenesis mechanisms regulated by cagA during H. pylori infection of human gastric epithelial cells and exploring reprogrammed host transcriptomes in response to H. pylori colonization. The key events occurring during the interaction between H. pylori and host cells were further validated by performing in vitro and in vivo experiments using multiple standard bacterial strains.
H. pylori has a limited ability to use carbohydrates as a carbon source, forcing it to rely on exogenous amino acids and peptides provided by the host. The ABC transporter that mediates the import of these substrates into the cytoplasm of H. pylori is pivotal for bacterial pathogenicity and virulence (20). In this study, we discovered significantly elevated expression of ABC transporter-related genes, metQ and HP_0888, upon coculturing H. pylori with gastric epithelial cells. However, such upregulation was largely mitigated in the cagA-mutated strain, which suggests that cagA benefits intracellular H. pylori colonization by facilitating the uptake of host-provided nutrients. Moreover, we observed an inhibitory impact of H. pylori cagA on electron transportation, as evidenced by the general repression of a set of ETC-related genes. The disrupted oxidative phosphorylation process could lead to the accumulation of ROS in host cells, thereby causing DNA damage and chromosomal instability and thus offering a pathological potential for gastric carcinogenesis (36). These results enhance our understanding of the pathogenic mechanisms of cagA-positive H. pylori strains upon colonization in the human stomach.
As a fundamental mechanism responsible for protein polymorphism, AS is a central mode of genetic regulation in metazoan organisms (37). Previous studies have indicated a correlation between aberrant splicing and gastric diseases, especially adenocarcinoma (38, 39). Liu et al. recently analyzed AS events in H. pylori-negative gastric cancer patients using the TCGA data set and identified several survival-related AS events (40). However, a systematic profiling of AS events in response to H. pylori infection has yet to be conducted. In this study, we observed decreased expression of multiple splicing regulators, including HSPAs, HNRNPs, SF3B2, SF3A1, and SF3A2, in H. pylori-infected gastric epithelial cells. We globally analyzed H. pylori-regulated AS events using the rMATS algorithm, revealing that all common modes of AS occurred in H. pylori-infected GES-1 cells, with cassette exons being the most frequently targeted upon bacterial challenge. By profiling genes affected by H. pylori-promoted splicing events, we found that a substantial number of these genes were involved in the cell cycle process, which may partially explain the cell cycle arrest induced by H. pylori infection as reported in previous studies (41). Future studies are needed to determine whether H. pylori-induced aberrant splicing events are involved in the pathogenesis of this microorganism.
Numerous epidemiological studies have suggested a protective role for chronic H. pylori infection against IBD, with a lower H. pylori infection rate observed in patients with ulcerative colitis and Crohn’s disease than in those without IBD (42–44). Some researchers have attributed this protective effect to H. pylori-induced systemic immune tolerance or to medical therapies used by patients with IBD, including metronidazole (45–47). However, whether H. pylori-modulated immune response can affect the pathogenesis of IBD still remains debatable. Consistent with previous data, our results indicated that gastric H. pylori colonization significantly mitigated the severity of DSS-induced colitis, as evidenced by alleviated clinical symptoms in DSS-treated mice. Mechanistically, we are inspired by a recent review (30) and started to investigate whether H. pylori-altered intestinal microbiota was involved in protecting against DSS-induced colitis. Resultingly, we identified a cluster of propionic and butyric acid-producing bacteria, Muribaculaceae, selectively enriched in the colon of H. pylori-precolonized mice, which may contribute to the restoration of intestinal barrier function damaged by DSS treatment. Increased production of SCFAs is considered beneficial for health because these metabolites play a versatile role in supporting gut homeostasis (48). Thus, apart from H. pylori-induced immune tolerance, our findings indicate the potential involvement of the intestinal microbiota remodeled by H. pylori in protecting against IBD. Further elucidation of the mechanism underlying this protective effect can help in the development of effective strategies for the treatment and prevention of IBD.
In summary, this study presents a robust and systematic characterization of the interplay between H. pylori and human gastric epithelial cells, offering mechanistic insights into the bacterium’s adaptive and pathogenic strategies. These results enhance our understanding of how H. pylori promotes infection and pathogenesis in the human gastric epithelium and provide evidence to identify targets for antimicrobial therapies.
MATERIALS AND METHODS
Bacterial and cell culture
Wild-type and cagA-mutant H. pylori TN2GF4 strains, NCTC 11637 (ATCC43504), were obtained from the Department of Microbiology, The Chinese University of Hong Kong. The H. pylori strain PMSS1 and 7.13 wild-type and cagA-mutant strains were kind gifts from Dr. Chuan XIE (The First Affiliated Hospital of Nanchang University, Jiangxi, China). H. pylori was initially grown on horse blood agar plates (Columbia Blood Agar Base with DENT Selective Supplements by Oxoid) in an anaerobic jar with a microaerophilic environment for 5 days at 37°C. GES-1 (normal human gastric epithelial cell line) and NCM460 (normal human colon mucosal epithelial cell line) were from Dr. Wang Hong Ying (Chinese Academy of Medical Sciences, Beijing, China). GES-1 and NCM460 cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Thermo Fisher) supplemented with 10% fetal bovine serum (Gibco, Thermo Fisher) at 37°C in 5% CO2.
Reagents, antibodies, and commercial kits
Primary antibodies we used were anti-H. pylori (ab7788, Abcam), anti-HSPA8 (10654-1-AP, Proteintech), anti-HSPA1A (10995-1-AP, Proteintech), anti-HSPA1B (25405-1-AP, Proteintech), anti-BRCA1 (22362-1-AP, Proteintech), anti-SUZ12 (20366-1-AP, Proteintech), anti-FANCM (12954-1-AP, Proteintech), anti-ZO-1 (21773-1-AP, Proteintech), anti-Claudin 1 (13050-1-AP, Proteintech), anti-Occludin (66378-1-Ig, Proteintech), anti-β-Actin (4967S, Cell Signaling Technology), and anti-GAPDH (2118S, Cell Signaling Technology). Secondary antibodies used included anti-mouse conjugated to horseradish peroxidase (A2304, Sigma-Aldrich) and anti-rabbit conjugated to horseradish peroxidase (NA9340, GE Healthcare) for western blotting; anti-rabbit conjugated to Alexa Fluor 568 (A11011) and anti-mouse conjugated to Alexa Fluor 488 (A21202) were from Life Technology for confocal microscopy.
The Catalase Activity Assay Kit was purchased from Abcam (ab83464). DSS (160110) was from MP Biomedicals (Shanghai) Co. Ltd. MitoSox Red mitochondrial superoxide indicator was from Thermo Fisher Scientific (M3608). The 8-OHdG ELISA Kit was from Abcam (ab201734).
Statistical analysis
All data were expressed as the mean ± standard derivation. Differences between two groups were compared by the Mann-Whitney U test or Student’s t-test. Multiple group comparisons were made by the Kruskal-Wallis test or one-way analysis of variance followed by Tukey’s t-test. P values less than 0.05 were considered statistically significant. See other details in the supplementary experimental procedures.
ACKNOWLEDGMENTS
The authors would like to thank Professor Peng CHEN for intellectually contributing to the project. We also thank Gene Denovo technology for high-throughput sequencing and technical supports. Professional English language editing support was provided by AsiaEdit (asiaedit.com).
This work was supported by the National Natural Science Foundation of China (No. 81974070), Guangdong Basic and Applied Basic Research Foundation (2020A1515011063, 2023A1515011685, and 2023A1515030071), Guangdong Young Innovative Talents Foundation (2020KQNCX013), Shenzhen Science and Technology Program (JCYJ20210324131010027, JCYJ20220530154205011, and JCYJ20230807142314030), and Research Foundation of Shenzhen Hospital of Southern Medical University (PT2018GZR05 and PT2018GZR10).
W.G. conceived, designed, and managed the study. W.H., Z.Y.Z., and Z.Y.H. performed the data analysis and experiment validation. W.H. wrote the manuscript. All authors contributed intellectually to the project through discussion and critically reviewed the manuscript.
Contributor Information
Zhan Gang Xiao, Email: zhangangxiao@swmu.edu.cn.
Wei Gong, Email: gongwei@smu.edu.cn.
Emily K. Cope, Northern Arizona University, Flagstaff, Arizona, USA
ETHICS APPROVAL
This study was approved by the Ethic Committee of Southern Medical University (NYSZYYEC20190017).
DATA AVAILABILITY
The authors confirm that the data supporting the findings of this study are available within the article and the raw sequencing results can be accessed with the accession number GSE243405.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00206-24.
Identification of host-induced H. pylori-specific stress responses using Dual RNA-Seq, related to Fig. 1.
Supplemental materials and methods and legend to Fig. S1.
Primers.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Malfertheiner P, Camargo MC, El-Omar E, Liou JM, Peek R, Schulz C, Smith SI, Suerbaum S. 2023. Helicobacter pylori infection. Nat Rev Dis Primers 9:19. doi: 10.1038/s41572-023-00431-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- 3. Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N, Malfertheiner P, Kyoto Global Consensus C. 2015. Kyoto global consensus report on Helicobacter pylori gastritis. Gut 64:1353–1367. doi: 10.1136/gutjnl-2015-309252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Correa P. 1988. A human model of gastric carcinogenesis. Cancer Res 48:3554–3560. [PubMed] [Google Scholar]
- 5. Hatakeyama M. 2014. Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host Microbe 15:306–316. doi: 10.1016/j.chom.2014.02.008 [DOI] [PubMed] [Google Scholar]
- 6. Deen NS, Huang SJ, Gong L, Kwok T, Devenish RJ. 2013. The impact of autophagic processes on the intracellular fate of Helicobacter pylori: more tricks from an enigmatic pathogen? Autophagy 9:639–652. doi: 10.4161/auto.23782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu W, Chan H, Lu L, Wong KT, Wong SH, Li MX, Xiao ZG, Cho CH, Gin T, Chan MTV, Wu WKK, Zhang L. 2020. Autophagy in intracellular bacterial infection. Semin Cell Dev Biol 101:41–50. doi: 10.1016/j.semcdb.2019.07.014 [DOI] [PubMed] [Google Scholar]
- 8. Necchi V, Candusso ME, Tava F, Luinetti O, Ventura U, Fiocca R, Ricci V, Solcia E. 2007. Intracellular, intercellular, and stromal invasion of gastric mucosa, preneoplastic lesions, and cancer by Helicobacter pylori. Gastroenterology 132:1009–1023. doi: 10.1053/j.gastro.2007.01.049 [DOI] [PubMed] [Google Scholar]
- 9. Oh JD, Karam SM, Gordon JI. 2005. Intracellular Helicobacter pylori in gastric epithelial progenitors. Proc Natl Acad Sci USA 102:5186–5191. doi: 10.1073/pnas.0407657102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Capurro MI, Greenfield LK, Prashar A, Xia S, Abdullah M, Wong H, Zhong XZ, Bertaux-Skeirik N, Chakrabarti J, Siddiqui I, O’Brien C, Dong X, Robinson L, Peek RM, Philpott DJ, Zavros Y, Helmrath M, Jones NL. 2019. VacA generates a protective intracellular reservoir for Helicobacter pylori that is eliminated by activation of the lysosomal calcium channel TRPML1. Nat Microbiol 4:1411–1423. doi: 10.1038/s41564-019-0441-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang L, Hu W, Cho CH, Chan FK, Yu J, Fitzgerald JR, Cheung CK, Xiao ZG, Shen J, Li LF, Li MX, Wu JC, Ling TK, Chan JY, Ko H, Tse G, Ng SC, Yu S, Wang MH, Gin T, Ashktorab H, Smoot DT, Wong SH, Chan MT, Wu WK. 2018. Reduced lysosomal clearance of autophagosomes promotes survival and colonization of Helicobacter pylori. J Pathol 244:432–444. doi: 10.1002/path.5033 [DOI] [PubMed] [Google Scholar]
- 12. Hu W, Zhang L, Li MX, Shen J, Liu XD, Xiao ZG, Wu DL, Ho IHT, Wu JCY, Cheung CKY, Zhang YC, Lau AHY, Ashktorab H, Smoot DT, Fang EF, Chan MTV, Gin T, Gong W, Wu WKK, Cho CH. 2019. Vitamin D3 activates the autolysosomal degradation function against Helicobacter pylori through the PDIA3 receptor in gastric epithelial cells. Autophagy 15:707–725. doi: 10.1080/15548627.2018.1557835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J. 2016. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529:496–501. doi: 10.1038/nature16547 [DOI] [PubMed] [Google Scholar]
- 14. Nuss AM, Beckstette M, Pimenova M, Schmühl C, Opitz W, Pisano F, Heroven AK, Dersch P. 2017. Tissue dual RNA-seq allows fast discovery of infection-specific functions and riboregulators shaping host-pathogen transcriptomes. Proc Natl Acad Sci USA 114:E791–E800. doi: 10.1073/pnas.1613405114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanchez-Villamil JI, Tapia D, Khakhum N, Widen SG, Torres AG. 2022. Dual RNA-seq reveals a type 6 secretion system-dependent blockage of TNF-α signaling and BicA as a Burkholderia pseudomallei virulence factor important during gastrointestinal infection. Gut Microbes 14:2111950. doi: 10.1080/19490976.2022.2111950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phillips I, Eykyn S, King BA, Jenkins C, Warren CA, Shannon KP. 1977. The in vitro antibacterial activity of nine aminoglycosides and spectinomycin on clinical isolates of common Gram-negative bacteria. J Antimicrob Chemother 3:403–410. doi: 10.1093/jac/3.5.403 [DOI] [PubMed] [Google Scholar]
- 17. Elliott RL, Jiang XP. 2019. The adverse effect of gentamicin on cell metabolism in three cultured mammary cell lines: "are cell culture data skewed?”. PLoS One 14:e0214586. doi: 10.1371/journal.pone.0214586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Figura N, Trabalzini L, Mini R, Bernardini G, Scaloni A, Talamo F, Lusini P, Ferro E, Martelli P, Santucci A. 2004. Inactivation of Helicobacter pylori cagA gene affects motility. Helicobacter 9:185–193. doi: 10.1111/j.1083-4389.2004.00224.x [DOI] [PubMed] [Google Scholar]
- 19. Hong Y, Wang G, Maier RJ. 2007. A Helicobacter hepaticus catalase mutant is hypersensitive to oxidative stress and suffers increased DNA damage. J Med Microbiol 56:557–562. doi: 10.1099/jmm.0.46891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rahman MM, Machuca MA, Khan MF, Barlow CK, Schittenhelm RB, Roujeinikova A. 2019. Molecular basis of unexpected specificity of ABC transporter-associated substrate-binding protein DppA from Helicobacter pylori. J Bacteriol 201:e00400-19. doi: 10.1128/JB.00400-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nguyen PT, Lai JY, Kaiser JT, Rees DC. 2019. Structures of the Neisseria meningitides methionine-binding protein MetQ in substrate-free form and bound to l- and d-methionine isomers. Protein Sci 28:1750–1757. doi: 10.1002/pro.3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ashton TM, McKenna WG, Kunz-Schughart LA, Higgins GS. 2018. Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res 24:2482–2490. doi: 10.1158/1078-0432.CCR-17-3070 [DOI] [PubMed] [Google Scholar]
- 23. Harrington JS, Ryter SW, Plataki M, Price DR, Choi AMK. 2023. Mitochondria in health, disease, and aging. Physiol Rev 103:2349–2422. doi: 10.1152/physrev.00058.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilson DF. 2017. Oxidative phosphorylation: regulation and role in cellular and tissue metabolism. J Physiol 595:7023–7038. doi: 10.1113/JP273839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shen S, Park JW, Lu Z, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA 111:E5593–E5601. doi: 10.1073/pnas.1419161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rogalska ME, Vivori C, Valcárcel J. 2023. Regulation of pre-mRNA splicing: roles in physiology and disease, and therapeutic prospects. Nat Rev Genet 24:251–269. doi: 10.1038/s41576-022-00556-8 [DOI] [PubMed] [Google Scholar]
- 27. Rokkas T, Gisbert JP, Niv Y, O’Morain C. 2015. The association between Helicobacter pylori infection and inflammatory bowel disease based on meta-analysis. United European Gastroenterol J 3:539–550. doi: 10.1177/2050640615580889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Imdad A, Pandit NG, Zaman M, Minkoff NZ, Tanner-Smith EE, Gomez-Duarte OG, Acra S, Nicholson MR. 2023. Fecal transplantation for treatment of inflammatory bowel disease. Cochrane Database Syst Rev 4:CD012774. doi: 10.1002/14651858.CD012774.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adolph TE, Zhang J. 2022. Diet fuelling inflammatory bowel diseases: preclinical and clinical concepts. Gut 71:2574–2586. doi: 10.1136/gutjnl-2021-326575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bai X, Jiang L, Ruan G, Liu T, Yang H. 2022. Helicobacter pylori may participate in the development of inflammatory bowel disease by modulating the intestinal microbiota. Chin Med J 135:634–638. doi: 10.1097/CM9.0000000000002008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lagkouvardos I, Lesker TR, Hitch TCA, Gálvez EJC, Smit N, Neuhaus K, Wang J, Baines JF, Abt B, Stecher B, Overmann J, Strowig T, Clavel T. 2019. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome 7:28. doi: 10.1186/s40168-019-0637-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang J, Han L, Liu Z, Zhang W, Zhang L, Jing J, Gao A. 2023. Genus unclassified_Muribaculaceae and microbiota-derived butyrate and indole-3-propionic acid are involved in benzene-induced hematopoietic injury in mice. Chemosphere 313:137499. doi: 10.1016/j.chemosphere.2022.137499 [DOI] [PubMed] [Google Scholar]
- 33. Kannt A, Papada E, Kammermeier C, D’Auria G, Jiménez-Hernández N, Stephan M, Schwahn U, Madsen AN, Østergaard MV, Dedoussis G, Francino MP, MAST4HEALTH consortium . 2019. Mastiha (Pistacia lentiscus) improves gut microbiota diversity, hepatic steatosis, and disease activity in a biopsy-confirmed mouse model of advanced non-alcoholic steatohepatitis and fibrosis. Mol Nutr Food Res 63:e1900927. doi: 10.1002/mnfr.201900927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim CH. 2023. Complex regulatory effects of gut microbial short-chain fatty acids on immune tolerance and autoimmunity. Cell Mol Immunol 20:341–350. doi: 10.1038/s41423-023-00987-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chelakkot C, Ghim J, Ryu SH. 2018. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med 50:1–9. doi: 10.1038/s12276-018-0126-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu Y, Shi Y, Han R, Liu C, Qin X, Li P, Gu R. 2023. Signaling pathways of oxidative stress response: the potential therapeutic targets in gastric cancer. Front Immunol 14:1139589. doi: 10.3389/fimmu.2023.1139589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marasco LE, Kornblihtt AR. 2023. The physiology of alternative splicing. Nat Rev Mol Cell Biol 24:242–254. doi: 10.1038/s41580-022-00545-z [DOI] [PubMed] [Google Scholar]
- 38. Zhang Y, Ma S, Niu Q, Han Y, Liu X, Jiang J, Chen S, Lin H. 2020. Features of alternative splicing in stomach adenocarcinoma and their clinical implication: a research based on massive sequencing data. BMC Genomics 21:580. doi: 10.1186/s12864-020-06997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Y, Yuan Y. 2017. Alternative RNA splicing and gastric cancer. Mutat Res Rev Mutat Res 773:263–273. doi: 10.1016/j.mrrev.2016.07.011 [DOI] [PubMed] [Google Scholar]
- 40. Liu C, Hu C, Li Z, Feng J, Huang J, Yang B, Wen T. 2020. Systematic profiling of alternative splicing in Helicobacter pylori-negative gastric cancer and their clinical significance. Cancer Cell Int 20:279. doi: 10.1186/s12935-020-01368-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Naumann M, Sokolova O, Tegtmeyer N, Backert S. 2017. Helicobacter pylori: a paradigm pathogen for subverting host cell signal transmission. Trends Microbiol 25:316–328. doi: 10.1016/j.tim.2016.12.004 [DOI] [PubMed] [Google Scholar]
- 42. Sonnenberg A, Genta RM. 2012. Low prevalence of Helicobacter pylori infection among patients with inflammatory bowel disease. Aliment Pharmacol Ther 35:469–476. doi: 10.1111/j.1365-2036.2011.04969.x [DOI] [PubMed] [Google Scholar]
- 43. Yu Y, Zhu S, Li P, Min L, Zhang S. 2018. Helicobacter pylori infection and inflammatory bowel disease: a crosstalk between upper and lower digestive tract. Cell Death Dis 9:961. doi: 10.1038/s41419-018-0982-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Halme L, Rautelin H, Leidenius M, Kosunen TU. 1996. Inverse correlation between Helicobacter pylori infection and inflammatory bowel disease. J Clin Pathol 49:65–67. doi: 10.1136/jcp.49.1.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Higgins PDR, Johnson LA, Luther J, Zhang M, Sauder KL, Blanco LP, Kao JY. 2011. Prior Helicobacter pylori infection ameliorates Salmonella typhimurium-induced colitis: mucosal crosstalk between stomach and distal intestine. Inflamm Bowel Dis 17:1398–1408. doi: 10.1002/ibd.21489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang H, Dai Y, Liu Y, Wu T, Li J, Wang X, Wang W. 2018. Helicobacter pylori colonization protects against chronic experimental colitis by regulating TH17/Treg balance. Inflamm Bowel Dis 24:1481–1492. doi: 10.1093/ibd/izy107 [DOI] [PubMed] [Google Scholar]
- 47. Castaño-Rodríguez N, Kaakoush NO, Lee WS, Mitchell HM. 2017. Dual role of Helicobacter and Campylobacter species in IBD: a systematic review and meta-analysis. Gut 66:235–249. doi: 10.1136/gutjnl-2015-310545 [DOI] [PubMed] [Google Scholar]
- 48. Blaak EE, Canfora EE, Theis S, Frost G, Groen AK, Mithieux G, Nauta A, Scott K, Stahl B, van Harsselaar J, van Tol R, Vaughan EE, Verbeke K. 2020. Short chain fatty acids in human gut and metabolic health. Benef Microbes 11:411–455. doi: 10.3920/BM2020.0057 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of host-induced H. pylori-specific stress responses using Dual RNA-Seq, related to Fig. 1.
Supplemental materials and methods and legend to Fig. S1.
Primers.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and the raw sequencing results can be accessed with the accession number GSE243405.