ABSTRACT
Human adenoviruses (HAdVs) are causative agents of morbidity and mortality throughout the world. These double-stranded DNA viruses are phylogenetically classified into seven different species (A–G). HAdV-G52, originally isolated in 2008 from a patient presenting with gastroenteritis, is the sole human-derived member of species G. Phylogenetic analysis previously suggested that HAdV-G52 may have a simian origin, indicating a potential zoonotic spillover into humans. However, evidence of HAdV-G52 in either human or simian populations has not been reported since. Here, we describe the isolation and in vitro characterization of rhesus (rh)AdV-69, a novel simian AdV with clear evidence of recombination with HAdV-G52, from the stool of a rhesus macaque. Specifically, the rhAdV-69 hexon capsid protein is 100% identical to that of HAdV-G52, whereas the remainder of the genome is most similar to rhAdV-55, sharing 95.36% nucleic acid identity. A second recombination event with an unknown adenovirus (AdV) is evident at the short fiber gene. From the same sample, we also isolated a second, highly related recombinant AdV (rhAdV-68) that harbors a distinct hexon gene but nearly identical backbone compared to rhAdV-69. In vitro, rhAdV-68 and rhAdV-69 demonstrate comparable growth kinetics and tropisms in human cell lines, nonhuman cell lines, and human enteroids. Furthermore, we show that coinfection of highly related AdVs is not unique to this sample since we also isolated coinfecting rhAdVs from two additional rhesus macaque stool samples. Our data collectively contribute to elucidating the origins of HAdV-G52 and provide insights into the frequency of coinfections and subsequent recombination in AdV evolution.
IMPORTANCE
Understanding the host origins of adenoviruses (AdVs) is critical for public health as transmission of viruses from animals to humans can lead to emergent viruses. Recombination between animal and human AdVs can also produce emergent viruses. HAdV-G52 is the only human-derived member of the HAdV G species. It has been suggested that HAdV-G52 has a simian origin. Here, we isolated from a rhesus macaque, a novel rhAdV, rhAdV-69, that encodes a hexon protein that is 100% identical to that of HAdV-G52. This observation suggests that HAdV-G52 may indeed have a simian origin. We also isolated a highly related rhAdV, differing only in the hexon gene, from the same rhesus macaque stool sample as rhAdV-69, illustrating the potential for co-infection of closely related AdVs and recombination at the hexon gene. Furthermore, our study highlights the critical role of whole-genome sequencing in understanding AdV evolution and monitoring the emergence of pathogenic AdVs.
KEYWORDS: adenovirus, AdV, human adenovirus, HAdV, rhesus adenovirus, rhAdV
INTRODUCTION
Adenoviruses (AdVs) are nonenveloped, double-stranded DNA (dsDNA) viruses belonging to the Adenoviridae family. Since their discovery in the 1950s, over 100 human (H)AdV genotypes have been discovered and classified into seven species (A–G) based on genetic and biological features (1, 2). HAdVs cause a variety of mild-to-severe diseases affecting the eye and the respiratory, urinary, and gastrointestinal tracts (3). AdVs have also been isolated from other mammalian (e.g., nonhuman primate, NHP) and nonmammalian (e.g., avian) species. A subset of these non-HAdVs have been leveraged as candidate vaccine vectors, as well as models for AdV antiviral development and pathogenesis (4–6). However, there are few therapeutics and no publicly available vaccines to combat AdV infections.
AdV evolution via recombination is a challenge to preventing and treating AdV infections (7–10). Recombination commonly occurs at hypervariable “hotspots” under high immune pressure including capsid and immune evasion genes of dsDNA viruses (e.g., herpes simplex viruses 1 and 2 and papillomavirus) but can also occur throughout the genome in both nonstructural and structural genes (11–14). For HAdVs, hotspots are frequently the hexon capsid, penton, and fiber receptor genes, which are responsible for virion structure and attachment to epithelial cells (7, 9, 10, 15–17). Recombinant genotypes originating from two parental AdVs that infect the same host species, as well as pairs that infect different host species (zoonoses), have been reported with increasing frequency over the past two decades; several recombinants have also been associated with outbreaks, such as keratoconjunctivitis caused by HAdV-85 (18–29). Zoonotic AdV recombinants are particularly concerning in a community lacking pre-existing immunity, especially if the recombinant AdV exhibits enhanced pathogenicity and transmissibility (28, 30–33).
The recent increase in the number of known NHP-derived AdVs (over 210 reported in NCBI as of October 2023) has fueled concerns for pathogenic zoonotic AdVs due to their high genetic relatedness with HAdVs and ability to replicate in human cells (5, 28, 33–47). The majority of known NHP AdVs have been detected in the gastrointestinal tract, whereas most HAdVs affect the respiratory tract (species B and E) or eye (species B, D, and E) (3, 29, 39, 48). In contrast, only two HAdVs of the F species—HAdV-F40 and HAdV-F41—are frequently linked to human gastrointestinal infection (49, 50). HAdV-G52, the sole human-derived member of the G species, was isolated from the stool of a patient with gastroenteritis (51). However, HAdV-G52 has not been identified in any human sample since its report in 2007 (52, 53). Phylogenetic analysis suggests that HAdV-G52 may be of simian rather than human origin, and it is noteworthy that the isolation of HAdV-G52 utilized primary monkey kidney cells (51). The remaining members of the HAdV-G species are derived from NHPs and have not been reported in humans.
Although coinfection of replicating AdVs is a prerequisite for recombination, reports describing AdV coinfection are limited (54–56). Our understanding of coinfection is also currently limited to HAdV coinfections in the respiratory tract even though the gastrointestinal tract is also a likely site of coinfection due to persistent HAdV infection (57–61). One major limitation is that most current diagnostic methods cannot discern the presence of more than one AdV, especially if they are highly similar. Conventional, consensus PCR-based AdV screening methods that target a conserved locus in the genome may fail to detect multiple or recombinant AdVs (22, 23, 53, 62–66). In contrast, multiplex- and microarray-based assays can detect multiple AdV genotypes by leveraging a collection of genotype-specific probes targeting a single gene like hexon or penton. However, reliance on a single or few loci can preclude the identification of recombinant or novel HAdVs in the absence of whole-genome sequencing (22, 23, 43, 62, 67, 68). For instance, a number of HAdVs (e.g., HAdV-D53 and HAdV-D63) were originally misclassified on the basis of hexon-targeted neutralization and sequencing assays but were later reclassified following whole-genome sequencing (20, 23). Direct culturing is another strategy for detecting coinfection in clinical scenarios, but this method is not routinely interrogated for AdV coinfection. Altogether, limited investigation of AdV coinfection impedes a deeper understanding of the rate, mechanism, and functional impact of recombination on AdV biology.
In this study, we identified, isolated, and characterized two closely related, hexon-recombinant rhAdVs (rhAdV-68 and rhAdV-69) coinfecting the gastrointestinal tract of a single rhesus macaque. One of the rhAdVs possessed the hexon of HAdV-G52, suggesting a simian rather than human origin for HAdV-G52. Despite differences in hexon, rhAdV-68 and rhAdV-69 shared similar in vitro growth kinetics and tropisms. We also identified and isolated highly related rhAdVs coinfecting two additional rhesus macaques. Identification of an HAdV-G52 hexon recombinant rhAdV and multiple coinfections expands our understanding of AdV evolution and suggests that continual surveillance is critical for identifying novel and recombinant AdVs with unknown pathogenic potential.
RESULTS
Isolation of rhesus adenoviruses 68 and 69 in cell culture
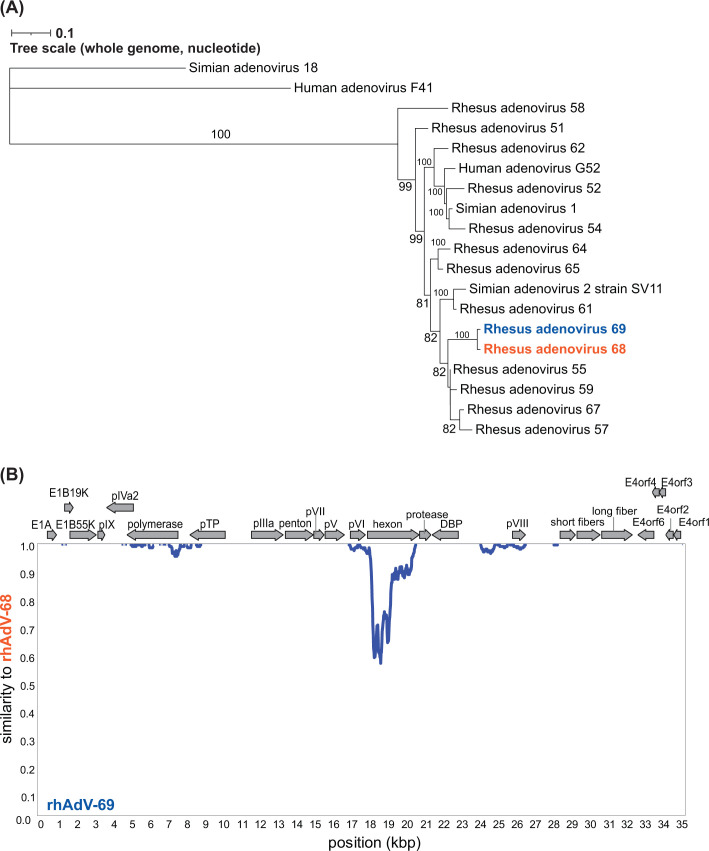
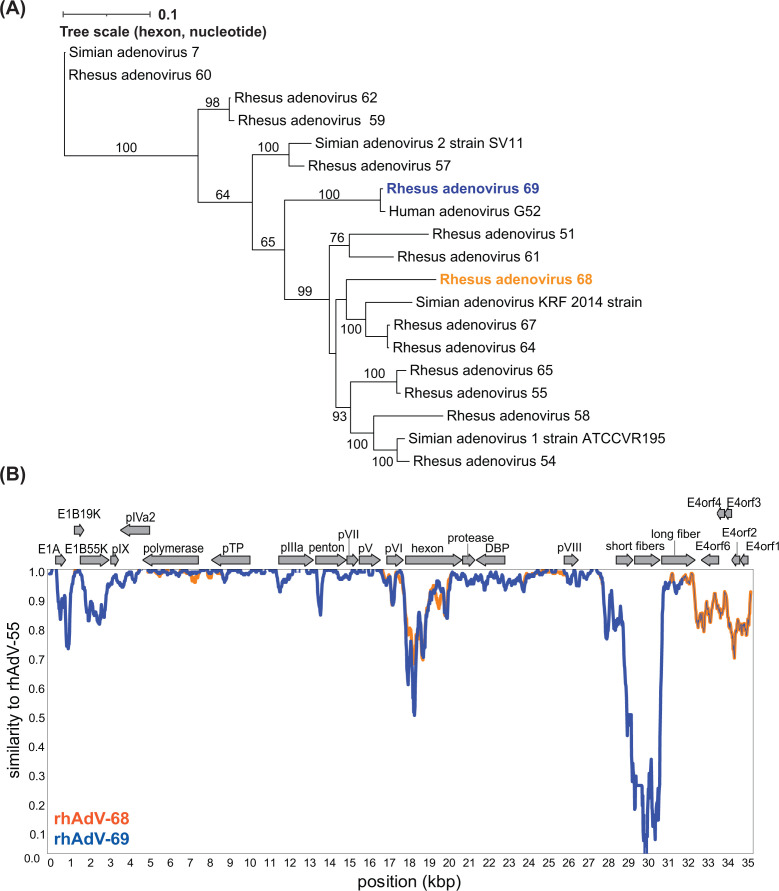
Previous high-throughput sequencing (HTS) analysis of the stool virome of rhesus macaques from a simian immunodeficiency virus (SIV) vaccine study identified several samples harboring sequences from novel AdVs (69). Our initial efforts to isolate novel AdVs from a single sample (sample 14581) using Vero E6 cells (African green monkey kidney epithelial cells) yielded cytopathic effects (CPEs) that could be passaged. After five passages, we performed one limiting dilution assay (LDA; Fig. S1A, passages 1–6). HTS of DNA extracted from whole lysates from two separate wells in the LDA identified two distinct, full-length rhAdVs genomes referred to as rhAdV-68 and rhAdV-69 in accordance with the current naming scheme for unclassified AdVs derived from rhesus macaque samples (1, 5). Phylogenetic analysis demonstrated that both rhAdV-68 and rhAdV-69 were most closely related to rhAdV-55 at 95.39% and 95.36% nucleotide identity (ni), respectively (Fig. 1A; Table S1). Furthermore, rhAdV-68 and -69 were nearly identical to each other, sharing 98.60% overall ni across the genome (Fig. 1A). The divergence occurred primarily in the hexon region (85.55% ni), while the backbones of the two viruses shared 99.74% ni (Fig. 1B). Phylogenetic analysis of the hexon demonstrated that the rhAdV-69 hexon is nearly identical to HAdV-G52 hexon at the nucleotide level (99.20% ni) and 100% identical at the amino acid level (Fig. 2A; Fig. S2A). In contrast, the hexon gene of rhAdV-68 is most similar to rhAdV-64 at the nucleotide and amino acid levels (89.30% and 95.10% identities, respectively; Fig. 2A; Fig. S2A).
Fig 1.
Identification of highly similar, coinfecting rhAdV-68 and rhAdV-69. (A) Whole genome maximum likelihood tree (unrooted) was generated using iqtree2 with 1,000 bootstraps and the TIM2 + F + R3 substitution model following multiple sequence alignment of nucleotide sequences with MAFFT. Bootstrap values greater than 60 are shown at nodes. (B) Similarity plot comparing rhAdV-68 and rhAdV-69 genomes using a 200 bp sliding window and 20 bp step size. Genome organization for both rhAdV-68 and rhAdV-69 is depicted above the similarity plot. Protein-coding regions are represented by colored arrows indicating the transcriptional orientation.
Fig 2.
Phylogenetic analysis of rhAdV-68 and rhAdV-69. (A) Hexon gene maximum likelihood trees (unrooted) were generated using iqtree2 with 1,000 bootstraps and the TIM2 + F + G4 substitution model following multiple sequence alignment of nucleotide sequences with MAFFT. Bootstrap values greater than 60 are shown at nodes. (B) Similarity plot comparing rhAdV-68 and rhAdV-69 genomes to the closest relative, rhAdV-55, using a 200 bp sliding window and 20 bp step size. Genome organization for the three rhAdVs is depicted above the similarity plot. Protein-coding regions are represented by colored arrows indicating the transcriptional orientation.
Comparison of rhAdV-68 and rhAdV-69 to their closest relative, rhAd V-55, identified high variability in several regions, including the early genes E1A, E1B, and E4 and the short fiber genes (Fig. 2B). The short fiber gene of rhAdV-68 and rhAdV-69 shared 55.36% amino acid identity in their closest relative, human mastadenovirus F (Fig. S2B). These data suggest that rhAdV-68 and rhAdV-69 are the result of rhAdV-55 undergoing at least two recombination events in the hexon and short fiber genes (Fig. S3A).
Virus purification by further limiting dilution assay
While rhAdV-69 was the only rhAdV in one culture (P6.2), the other culture (P6.1) of the LDA contained both rhAdV-68 and rhAdV-69 according to HTS. We confirmed the composition of rhAdV-68 and rhAdV-69 in each culture of the LDA with virus-specific Taqman quantitative PCR (qPCR) primers and probes targeting the hexon region (Fig. S1B). To physically separate rhAdV-68 and rhAdV-69, we performed a second LDA followed by two rounds of inoculation on Vero E6 cells to further amplify the rhAdVs (Fig. S1A). qPCR of the final passages (P9.1 and P9.2) confirmed that we had successfully separated rhAdV-68 and rhAdV-69 (Fig. S1B). In addition, whole-genome sequencing yielded no reads from the unique hexon region of rhAdV-68 detected in the rhAdV-69 culture and vice versa (Table S2). Sequencing also confirmed that the genome sequences from P6 to P9 were identical, indicating no new mutations were acquired for either rhAdV-68 or rhAdV-69 during the additional passages.
Comparative growth kinetics of rhAdV-68 and rhAdV-69
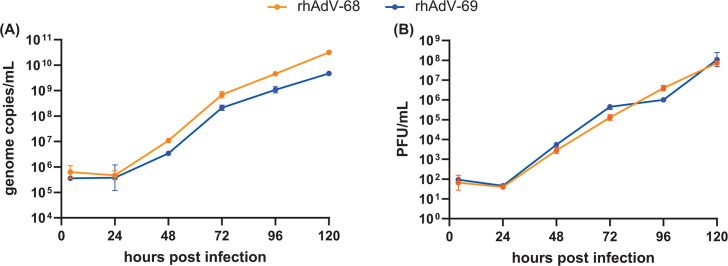
Hexon may play a role in viral attachment, entry, and replication of HAdVs (70). Given that rhAdV-68 and rhAdV-69 primarily differed at the hexon genes, we compared the viral growth kinetics of both viruses in Vero E6 cells (MOI = 0.01; Fig. 3). rhAdV-68 and rhAdV-69 exhibited similar multi-step growth kinetics, including an eclipse period of approximately 24 hours and similar peak viral titers {[rhAdV-68, 7.9 ± 0.1; rhAdV-69, 8.0 ± 0.4 log10[plaque forming unit (PFU/mL)]} and genome copy levels [rhAdV-68, 10.7 ± 0.1; rhAdV-69, 9.7 ± 0.1 log10(genome copies/mL)] at 120 hours post infection (HPI). Infection with either virus induced 100% CPE by 120 HPI. No significant differences in genome copy levels or viral titers were detected at any timepoint.
Fig 3.
Comparative growth kinetics of rhAdV-68 and rhAdV-69. Multi-step growth kinetics of rhAdV-68 and rhAdV-69 were acquired in Vero E6 cells infected with MOI = 0.01. Extracellular viral (A) genome copy number and (B) PFU were quantified with qPCR and plaque assay, respectively. Data from three biological replicates are shown as the geometric mean with error bars representing the geometric SD.
Comparative tropism of rhAdV-68 and rhAdV-69
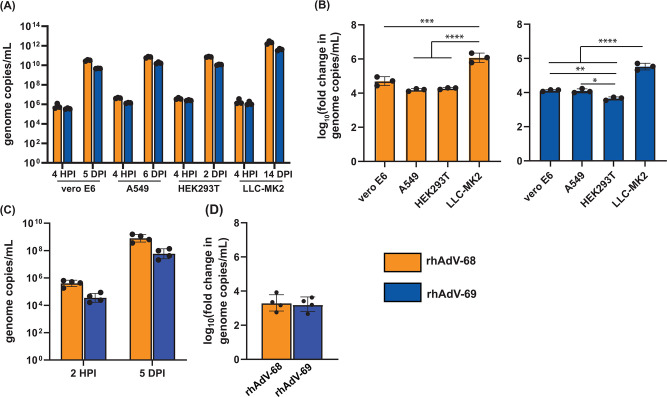
To determine if genetic differences influenced viral tropism, we next compared the tropisms of rhAdV-68 and rhAdV-69 in multiple immortalized cell lines and a human jejunal enteroid monolayer model lacking STAT1 (Fig. 4). We inoculated HEK293T (human embryonic kidney epithelia), A549 (human lung adenocarcinoma), and LLC-MK2 (rhesus macaque kidney epithelia) with rhAdV-68 or rhAdV-69 (MOI = 0.01) and then quantified viral genome copy levels at 4 HPI and once cultures exhibited 100% CPE (Fig. 4A). Each cell line supported rhAdV-68 and rhAdV-69 replication, with rhAdV-68 and rhAdV-69 replicating to similar levels for a given cell line. However, the extent to which rhAdV-68 and rhAdV-69 replicated in a specific cell line differed (Fig. 4B). Infection in LLC-MK2 cells produced the greatest log10 fold-change in viral genome copy levels for both rhAdV-68 (6.07 ± 0.28) and rhAdV-69 (5.51 ± 0.19). rhAdV-69 also replicated to a greater extent in A549 cells (4.10 ± 0.10) and Vero E6 cells (4.12 ± 0.05) than HEK293T cells (3.65 ± 0.10). In contrast, viral genome copy levels were similar for Vero E6, A549, and HEK293T cells infected with rhAdV-68 (Fig. 4B).
Fig 4.
Comparative tropism of rhAdV-68 and rhAdV-69 in cell lines and jejunal enteroids. Extracellular rhAdV-68 and rhAdV-69 genome copy levels acquired (A and B) after Vero E6, A549, HEK293T, and LLC-MK2 cells (MOI = 0.01) reached 100% CPE at 5 DPI, 6 DPI, 2 DPI, and 14 DPI, respectively; and (C and D) at 5 DPI in human STAT1-/- jejunal monolayer enteroids (MOI = 5). Log10 fold-change in viral copy number in (B) and (D) was calculated as the log-transformed ratio of genome copies per milliliter at the terminal timepoint vs 4 HPI. Data from three (A and B) or four (C and D) biological replicates are shown as the geometric mean with error bars representing the geometric SD. One-way ANOVA with multiple comparisons, *P-value ≤ 0.05; **P-value ≤ 0.01; ***P-value ≤ 0.001; ****P-value ≤ 0.001.
rhAdV-68 and rhAdV-69 also infected and replicated to similar levels in differentiated human STAT-/- jejunal enteroid monolayers by 120 HPI (MOI = 5; Fig. 4C). rhAdV-68 and rhAdV-69 genome copy levels also increased by similar extents (3.30 ± 0.49 and 3.22 ± 0.41 log10 fold-change, respectively) in the enteroid monolayers (Fig. 4D).
Evidence of coinfections in additional rhesus macaque stool samples
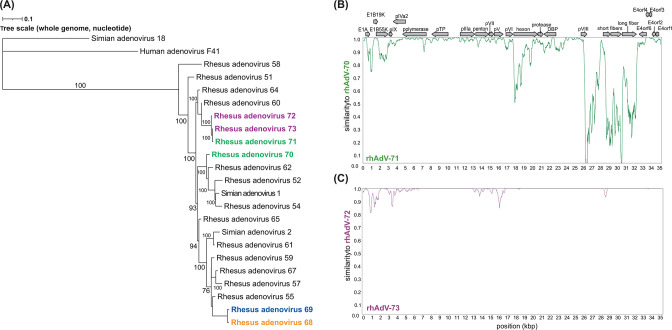
To our knowledge, gastrointestinal coinfection of closely related AdVs has not been previously reported in humans or NHPs. Therefore, we next examined two additional rhesus macaque stool samples (samples 14585 and 14587) containing rhAdVs sequences in our cohort to determine if rhAdV coinfection in the gastrointestinal tract is a more generalized phenomenon, and, if so, the relationship of the two viruses to each other (69). Using the culture approach followed by LDA described above, we successfully isolated and obtained full genome sequences for two pairs of coinfecting rhAdVs: rhAdV-70 and -71 from sample 14585 and rhAdV-72 and -73 from sample 14587 (Fig. 5; Fig. S4). Phylogenetic analysis indicated that rhAdV-71, -72, and -73 were most closely related to rhAdV-60 (99.1%–99.4% ni) and rhAdV-70 to rhAdV-62 (98.5% ni; Fig. 5A; Table S1). One coinfecting rhAdVs pair (rhAdV-70 and -71) displayed less than 85% genome-wide ni, reflecting two distinct virus species. The second pair (rhAdV-72 and -73) was highly similar (99.11%) with the regions of differences primarily localized to early and pV genes, suggesting recombination with other rhAdVs (rhAdV-64 and rhAdV-51, respectively; Fig. 5B and C; Fig. S3C). Further investigation suggested that rhAdV-70, -71, -72, and -73 all likely underwent other recombination events at the short and long fiber genes, as well as other nonstructural and structural genes like hexon, penton, and early E1A/B genes (Fig. S3B, C and S4).
Fig 5.
Genomic characterization of coinfecting rhAdV pairs rhAdV-70 and -71 and rhAdV-72 and -73. (A) Whole genome maximum likelihood tree (unrooted) was generated using iqtree2 with 1,000 bootstraps and the TIM2 + F + G4 substitution model following multiple sequence alignment of nucleotide sequences with MAFFT. Bootstrap values greater than 60 are shown at nodes. Coinfecting rhAdVs isolated from three separate rhesus macaque stool samples in this study are colored as follows: rhAdV-68 (orange) and -69 (blue) from sample 14581; rhAdV-70 and -71 from sample 14585 (green); and rhAdV-72 and -73 from sample 14587 (purple). Similarity plots comparing coinfecting rhAdVs from (B) sample 14585 and (C) sample 14587 using a 200 bp sliding window and 20 bp step size.
DISCUSSION
Phylogenetic analysis suggests that HAdV-G52 has a simian rather than human origin (51). However, evidence of HAdV-G52 in simian or human samples has not been reported since its initial discovery (51–53, 71). Here, we report the detection of a novel rhesus adenovirus genome that possesses the HAdV-G52 hexon gene, making this only the second time the HAdV-G52 hexon sequence has been described. The fact that the rhAdV-69 hexon is 100% identical to the HAdV-G52 hexon at the amino acid level and differs only by 21 nucleotides strongly supports the hypothesis that HAdV-G52 has a simian origin.
Within the sample in which we isolated rhAdV-69, we also obtained a second rhAdV (rhAdV-68) that predominantly differed only at the hexon gene in comparison to rhAdV-69, suggesting that rhAdV-68 and -69 are recombinants. We showed that these two coinfecting rhAdVs exhibited similar in vitro replication kinetics and tropisms. We also identified additional examples of rhAdV coinfection of two additional rhesus macaques. One of these samples harbored a pair of rhAdVs that differed only at limited loci, suggesting they are also related by recombination. Our findings collectively support the idea that the gastrointestinal tract can be a reservoir of multi-AdV coinfections, making it an ideal site for the evolution of AdV via recombination. Therefore, these results highlight the critical need for AdV surveillance in the gastrointestinal tract since high levels of recombination between coinfecting AdVs might lead to novel, pathogenic genotypes.
All the rhAdVs isolated in this study were recovered from immunocompromised rhesus macaques terminally infected with SIV (69). The presence and isolation of AdVs from these samples is not unexpected since it is well-established that immunocompromised individuals, such as those infected with HIV (SIV for NHPs) or undergoing organ transplantation, are at greater risk for both de novo AdV infection and expansion of persistent enteric AdVs (35, 69, 72–74). AdVs can persist in various mucosal sites of immunocompetent and immunocompromised individuals, such as within the gastrointestinal lamina propria (59, 61, 75, 76). However, there is limited data demonstrating coinfection in immunocompromised hosts, let alone in immunocompetent hosts or the gastrointestinal tract (54, 55). Instead, coinfection studies have primarily focused on pediatric populations with acute respiratory disease, wherein the coinfection rate may be as high as ~10% (56, 77). Failure to identify coinfections and recombinant AdVs is likely due to the use of low-resolution genotyping tools like PCR. Only recently have HTS technologies been employed in clinical settings to fully characterize HAdV infections (78, 79). Our findings highlight the value of HTS-based assays for characterizing HAdV infections since single-locus-based approaches would likely have been unable to reveal the coinfections identified in this study. Furthermore, the combination of virus culture and complete genome sequencing of purified viruses allowed us to identify regions of high variation in genomes that are otherwise >99% identical. Additional HTS-based surveillance studies determining the prevalence of HAdVs in the gastrointestinal tract of immunocompetent children and adults should also be performed to determine baseline levels of HAdV coinfection and recombination.
The recombinant rhAdVs isolated in this study demonstrated potential recombination events at well-documented genetic “hotspots”: hexon, fiber, and penton—the major capsid genes that mediate viral attachment and entry (7, 10). We also detected potential recombination at genes involved in viral replication (e.g., E1A and E1B19K) and encoding minor capsid components (e.g., pV and protease). There are few investigations of recombination at these genes, which are typically shielded from the host immune system and, thus, considered under less immune pressure (15, 16). Again, whole-genome sequencing provides insights into additional regions subject to recombination, suggesting there may be unrecognized and unanticipated biological pressures at these loci. Therefore, future investigations of how recombination at these genes affects the molecular properties of AdVs will likely reveal new insights into the host-AdV interactions.
Despite the differences between hexon and other genes, multi-step growth kinetics in Vero E6 cells and tropisms for rhAdV-68 and rhAdV-69 were similar. This suggests that neither virus has a significant competitive advantage over the other in vitro in the cells we tested. Future work using in vivo models is needed to examine the possible impacts of the hexon recombination events on virus fitness under immune pressures from the host. Like other simian-derived AdVs, rhAdV-68 and rhAdV-69 can replicate in both human and NHP cell lines, although the extent depends on the cell type (5, 36, 45). We observed the increase in replication in the rhesus kidney epithelial cell line LLC-MK2, suggesting that rhAdV-68 and rhAdV-69 may be adapted for replication in a simian host. rhAdV-68 and rhAdV-69 also demonstrated similar changes in replication levels in two human cell lines (human lung adenocarcinoma and kidney epithelia) and differentiated human jejunal enteroid monolayers lacking STAT1. Replication of AdVs in enteroid monolayers has been previously demonstrated only for HAdV-C5 and HAdV-F41 (80). The ability of rhAdV-68 and rhAdV-69 to replicate in human cells raises the possibility of zoonotic transmission. Even if these viruses themselves are of limited or no pathogenicity, their ability to infect humans could lead to further AdV recombination that generates more pathogenic AdVs. Apart from this, a potential benefit of their human tropism is the possibility that these rhAdVs may be suitable for AdV-vectored-based vaccine research if levels of pre-existing immunity in humans are low (5).
In contrast to the rhAdV-69 hexon gene, the short fiber gene shared by rhAdV-68 and rhAdV-69 is distinct from known fiber genes, with the closest relative being the short fiber of human mastadenovirus F (55.36% amino acid identity). This divergence is likely due to recombination between rhAdV-55 and an unknown AdV rather than viral mutation, which plays a minor role in AdV diversity compared to other dsDNA viruses (81–84). Therefore, rhAdV-68 and rhAdV-69 likely derived from recombination between an unknown AdV closely related to rhAdV-55 and either HAdV-G52 (for rhAdV-69) or an AdV related to rhAdV-64 (for rhAdV-68).
Future studies are needed to define the tissue tropism of these novel rhAdVs beyond the gastrointestinal tract. We suggest that these rhAdVs are derived from the gastrointestinal tract since they were recovered from stool samples and can grow in human intestinal organoids. However, HAdVs not associated with gastrointestinal disease or tropism have been detected in human fecal samples; therefore, we cannot rule out whether these rhAdVs infect other tissues and are merely passing through the gastrointestinal tract (57, 59, 74). Samples from other tissues from the infected animal were unavailable for us to investigate this possibility.
In conclusion, this study demonstrates the presence and characterization of highly related, recombinant, and coinfecting rhAdVs from the gastrointestinal tract of rhesus macaques using a set of bioinformatic and in vitro approaches. Our data provide insight into the origins of HAdV-G52, suggesting that it may indeed be of simian rather than human origin. Additionally, these data suggest that surveillance of gastrointestinal AdV infection will be critical for monitoring the emergence of novel, recombinant, and potentially pathogenic AdV genotypes.
MATERIALS AND METHODS
Cell culture
Vero E6 (African green monkey kidney epithelia), LLC-MK2 (rhesus macaque kidney epithelia), A549 (human lung adenocarcinoma), and HEK293T (human embryonic kidney epithelia) were maintained in Dulbecco’s modified Eagle medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), 10 mM HEPES buffer (Gibco), 100 U/mL penicillin streptomycin (100 U/mL, Gibco), 2 mM L-glutamine (Gibco), and 0.1 mM MEM nonessential amino acids (Gibco) at 37°C with 5% CO2. For virus infections, all cells were cultured in the same medium with FBS content reduced to 2%.
Adenovirus cultivation, isolation, and sequencing from stool samples
Virus isolation
Three rhesus macaques stools collected in a previous study were analyzed in this study (14581, 14585, and 14587) (69). rhAdV-68 and rhAdV-69 were isolated from sample 14581 according to the workflow presented in Fig. S1A. Briefly, confluent Vero E6 cells in a 96-well plate format were initially inoculated with 20 µL of stool filtrate (passage 1). Following six days of incubation, infections were subjected to three cycles of freeze-thaw. The resulting whole-cell lysates were via low-speed centrifugation (2000 rpm, 10 minutes), and the supernatant was passaged to a new confluent Vero E6 cell layer (passage 2). This was repeated for a total of five passages before performing two successive LDAs. For LDAs, 10 µL of a 10-fold serial dilution of clarified cell lysate supernatant was used to inoculate confluent layers of Vero E6 cells in 96-well formats (n = 8 wells per dilution). Clarified whole cell lysate was collected from the greatest dilution at which CPE was still observed, centrifuged, and then either frozen or immediately passaged to a new layer of confluent Vero-E6 cells for further passaging. A similar strategy was used to isolate coinfecting rhAdV pairs from sample 14585 (rhAdV-70 and -71) and sample 14587 (rhAdV-72 and -73).
Whole-genome sequencing
For passage 5, total nucleic acid was extracted from the clarified supernatant of whole cell lysate using the Direct-zol RNA Miniprep kit (Zymo Research) according to the manufacturer’s protocol. RNA was randomly amplified as previously described before library preparation with the NEBNext Ultra DNA prep Kit for Illumina (New England Biolabs) (85). An Agilent 2100 Bioanalyzer was used to quantify and assess the quality of libraries before sequencing on Illumina MiSeq (2 × 250 bp paired-end reads). Raw data were analyzed using the Chan Zuckerberg ID (CZID) platform (86). For passages 6 and 9, total DNA was extracted from the supernatant of whole cell lysate using the Qiagen DNeasy Blood and Tissue Kit (Qiagen) according to manufacture protocol. Libraries were constructed using the Illumina DNA Library prep kit (Illumina). Libraries were sequenced on Illumina NextSeq (2 × 150 bp paired-end reads) and analyzed with both the CZID platform (https://czid.org/) and a custom assembly pipeline described below (86).
Bioinformatic analyses
Viral assembly was manually verified using the following pipeline. Quality-filtered, trimmed (Trimgalore), and deduplicated (BBMap) reads failing to align to the human genome GRCh38 were used for de Bruijn-based assembly using metaSPAdes with BayesHammer error correction (87). Whole genomes were annotated with the Genome Annotation Transfer Utility tool (88). For phylogenetic analysis, sequences were aligned with MAFFT using progressive methods with default settings (89). Maximum likelihood trees were constructed using iqtree2 with 1,000 bootstrap replications (90). The substation model that minimized the Bayesian information criterion was chosen on a per-tree basis. Recombination analysis and visualization were performed with Simplot using the Kimura (2-parameter) distance model, maximum likelihood tree model, 2.0Ts/Tv ratio, 100 bootstrap replicates, and sliding window of 200 bp with a 20 bp step size (91). Variant analysis was performed according to GATK standards (92). In brief, raw reads were quality-filtered (Phred score >30), trimmed (adapter removal and 15 bp from the 5’ end), and then aligned to either the rhAdV-68 or rhAdV-69 genomes using BWA mem with seed length of 30 and default settings. Aligned reads were then deduplicated with Picard prior to variant calling with VarScan2. Searches for closest relatives were performed using BLASTn.
qPCR assay
Total DNA was extracted from the supernatant of whole-cell lysates using the Qiagen DNeasy Blood and Tissue Kit (Qiagen) according to manufacture protocol. qPCR was performed using TaqMan Fast Virus 1-Step Master Mix (Applied Biosystems) on ViiA 7 Real-Time PCR System in 96-well plate format. Standard curves using templates targeting the unique regions of the hexon genes of rhAdV-68 and rhAdV-69 were generated to quantify viral genome copy levels. TaqMan qPCR primers and probes (Integrated DNA Technologies) targeting unique regions of the hexon genes of rhAdV-68 and rhAdV-69 are provided below. All samples were measured as technical duplicates.
rhAdV-68 forward: 5′-GGGTACCGCGTACAATTCC-3′, rhAdV-68 reverse: 5′-ACTCTCAGCTTGTTGCTGTC-3′, rhAdV-68 probe: 5’-/56-FAM/AACCCTGCA/ZEN/GAATGGGAGGATACC/3IABkFQ/−3’ rhAdV-69, forward: 5′-TACTCCGGCACCGCTTA-3′ rhAdV-69, reverse: 5′-GTGCTTGGGCTCTCACTTT-3′ rhAdV-69, and probe: 5’-/56-FAM/TCCGTAGAG/ZEN/TGGCCGGATAACACT/3IABkFQ/−3’.
Plaque assay
Confluent Vero E6 cells in six-well plates were infected with 500 µL of 10-fold serial dilutions of rhAdV-68 or rhAdV-69 for 1 hour at 37°C with periodic gentle shaking. After incubation, cells were washed once with culture media without FBS. An overlay containing a 1:1 ratio of 1.2% SeaPlaque Agarose (Millipore) and 2× Modified Eagle Medium (Gibco) supplemented with 4% FBS was applied. After a minimum 10 days of incubation at 37°C, cells were fixed overnight with 4% formaldehyde and then stained with crystal violet for plaque counting.
Multi-step growth curve
Vero E6 cells were infected with MOI = 0.01 of rhAdV-68 or rhAdV-69 (titrated by plaque assay) for 1 hour at 37°C. After incubation, cells were washed once with culture media without FBS. The supernatant was collected and clarified (2,000 rpm, 10 minutes) at 4, 24, 48, 72, 96, and 120 HPI for quantification of the extracellular virus by plaque assay and qPCR. Fresh media were supplemented at each collection timepoint to account for the volume of supernatant that was collected. Samples were collected for three biological replicates.
Virus infection of cell lines
A549, HEK293T, and LLC-MK2 cells were inoculated with MOI = 0.01 of rhAdV-68 or rhAdV-69 for 1 hour at 37°C. After incubation, cells were washed once with culture media without FBS. Whole-cell lysates were collected at 4 HPI and once infections reached 100% CPE, which was 6 days for A549, 2 days for HEK293T, and 14 days for LLC-MK2. Fresh media were supplemented at each collection timepoint to account for the volume of supernatant that was collected. Samples were collected for three biological replicates. Whole-cell lysates were clarified via low-speed centrifugation (2,000 rpm, 10 minutes), and DNA was extracted from the resulting supernatant using the Qiagen DNeasy Blood and Tissue Kit (Qiagen) according to manufacture protocol. qPCR was used to quantify viral genome copies as described above. Log10 fold-change in viral genome copy level was calculated as the genome copies per milliliter at the terminal timepoint divided by that at 4 HPI.
Virus infection of enteroid cultures
The J2 human STAT1-/- enteroid cultures were generated as previously described (93). Cultures were purchased (TMC Digestive Diseases Center Gastrointestinal Experimental Model Systems Core), maintained, and passaged in Matrigel matrix (Corning, #354230) as multilobular three-dimensional (3D) cultures in 24-well plates supplemented with enteroid growth medium (WRNE, with Wnt, R-Spondin, and Noggin growth factors in 50% L-WRN cell conditioned medium). To generate confluent monolayers, the 3D enteroids were dissociated with TrypLE Express (Thermo, #12604–013), filtered with a 40 µm cell strainer (Corning, #431750), and seeded onto collagen IV-coated 96-well plates as monolayers.
Following culture in growth medium for 24 hours, seeded cells were plated in differentiation medium for 5 days before inoculation. For virus infection, monolayers (approximately 2 × 104 cells/well) were inoculated with rhAdVs at MOI = 5 for 2 hours at 37°C in 5% CO2. Monolayers were then washed twice with a complete medium without growth factors [CMGF(−)] to remove unbound viruses. Inoculated wells were cultured in a differentiation medium for five days. Samples were collected at 0 HPI (immediately after removal of unbound virus) and 5 DPI for DNA extraction and quantification of viral genome copy levels with qPCR. Each experiment was performed with four technical replicates in each condition.
Statistical analysis
All analyses were performed with GraphPad Prism (version 10). Comparisons between three or more groups were evaluated with one-way ANOVA with multiple comparisons. Comparisons between two groups were evaluated with t tests with Welch’s correction. *P-value ≤ 0.05; **P-value ≤ 0.01; ***P-value ≤ 0.001; ****P-value ≤ 0.0001.
ACKNOWLEDGMENTS
We would like to thank Dr. Adriana Kajon from the Lovelace Respiratory Research Institute; Dr. Matthew Weitzman from the University of Pennsylvania, Perelman School of Medicine; and Dr. Scott Handley from the Department of Pathology and Immunology, Washington University in Saint Louis for helpful suggestions and advice. We would also like to extend our thanks to MariaLynn Crosby and Jessica Hoisington-Lopez at the Washington University in Saint Louis DNA Sequencing Lab at the Center for Genome Sciences and Systems Biology for their technical support with our HTS experiments.
This work was supported by the National Institutes of Health grants T32GM139774 (awarded to A.N.P.) and RC2DK116713 (awarded to D.W.). The funders had no role in study design, data collection, and interpretation or the decision to submit the work for publication.
Contributor Information
David Wang, Email: davewang@wustl.edu.
Koenraad van Doorslaer, College of Agriculture & Life Sciences, University of Arizona, Tucson, Arizona, USA.
DATA AVAILABILITY
Complete genome sequences are available at GenBank for rhAdV-68 to rhAdV-73 (PP134848, PP134849, PP134850, PP134851, PP134852, and PP134853). Raw sequencing data have been deposited to the Sequence Read Archive as PRJNA1061574.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.00043-24.
Isolation and separation of rhAdV-68 and rhAdV-69.
Phylogenetic analysis of rhAdV-68 and rhAdV-69 hexon and short fiber genes.
Schematic overview of genomes of coinfecting rhAdVs.
Comparative analysis of rhAdV-70–rhAdV-73.
Genomic characterization of coinfecting rhAdVs.
Assessment of virus isolation using HTS.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Benkő M, Aoki K, Arnberg N, Davison AJ, Echavarría M, Hess M, Jones MS, Kaján GL, Kajon AE, Mittal SK, Podgorski II, San Martín C, Wadell G, Watanabe H, Harrach B, ICTV Report Consortium . 2022. ICTV virus taxonomy profile: Adenoviridae 2022. J Gen Virol 103:001721. doi: 10.1099/jgv.0.001721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. HAdV working group. Available from: http://hadvwg.gmu.edu. Retrieved 7 May 2023.
- 3. Ison MG, Hayden RT. 2016. Adenovirus. Microbiol Spectr 4. doi: 10.1128/microbiolspec.DMIH2-0020-2015 [DOI] [PubMed] [Google Scholar]
- 4. Wilson SS, Bromme BA, Holly MK, Wiens ME, Gounder AP, Sul Y, Smith JG. 2017. Alpha-defensin-dependent enhancement of enteric viral infection. PLoS Pathog 13:e1006446. doi: 10.1371/journal.ppat.1006446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abbink P, Kirilova M, Boyd M, Mercado N, Li Z, Nityanandam R, Nanayakkara O, Peterson R, Larocca RA, Aid M, Tartaglia L, Mutetwa T, Blass E, Jetton D, Maxfield LF, Borducchi EN, Badamchi-Zadeh A, Handley S, Zhao G, Virgin HW, Havenga MJ, Barouch DH. 2018. Rapid cloning of novel rhesus adenoviral vaccine vectors. J Virol 92:e01924-17. doi: 10.1128/JVI.01924-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tatsis N, Ertl HCJ. 2004. Adenoviruses as vaccine vectors. Mol Ther 10:616–629. doi: 10.1016/j.ymthe.2004.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lukashev AN, Ivanova OE, Eremeeva TP, Iggo RDY. 2008. Evidence of frequent recombination among human adenoviruses. J Gen Virol 89:380–388. doi: 10.1099/vir.0.83057-0 [DOI] [PubMed] [Google Scholar]
- 8. Ismail AM, Lee JS, Lee JY, Singh G, Dyer DW, Seto D, Chodosh J, Rajaiya J. 2018. Adenoviromics: mining the human adenovirus species D genome. Front Microbiol 9:2178. doi: 10.3389/fmicb.2018.02178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crawford-Miksza LK, Schnurr DP. 1996. Adenovirus serotype evolution is driven by illegitimate recombination in the hypervariable regions of the hexon protein. Virology 224:357–367. doi: 10.1006/viro.1996.0543 [DOI] [PubMed] [Google Scholar]
- 10. Robinson CM, Singh G, Lee JY, Dehghan S, Rajaiya J, Liu EB, Yousuf MA, Betensky RA, Jones MS, Dyer DW, Seto D, Chodosh J. 2013. Molecular evolution of human adenoviruses. Sci Rep 3:1812. doi: 10.1038/srep01812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Norberg P, Kasubi MJ, Haarr L, Bergström T, Liljeqvist J-A. 2007. Divergence and recombination of clinical herpes simplex virus type 2 isolates. J Virol 81:13158–13167. doi: 10.1128/JVI.01310-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koelle DM, Norberg P, Fitzgibbon MP, Russell RM, Greninger AL, Huang M-L, Stensland L, Jing L, Magaret AS, Diem K, Selke S, Xie H, Celum C, Lingappa JR, Jerome KR, Wald A, Johnston C. 2017. Worldwide circulation of HSV-2 × HSV-1 recombinant strains. Sci Rep 7:44084. doi: 10.1038/srep44084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Angulo M, Carvajal-Rodríguez A. 2007. Evidence of recombination within human alpha-papillomavirus. Virol J 4:33. doi: 10.1186/1743-422X-4-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murahwa AT, Tshabalala M, Williamson A-L. 2020. Recombination between high-risk human papillomaviruses and non-human primate papillomaviruses: evidence of ancient host switching among alphapapillomaviruses. J Mol Evol 88:453–462. doi: 10.1007/s00239-020-09946-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gonzalez G, Koyanagi KO, Aoki K, Kitaichi N, Ohno S, Kaneko H, Ishida S, Watanabe H. 2014. Intertypic modular exchanges of genomic segments by homologous recombination at universally conserved segments in human adenovirus species D. Gene 547:10–17. doi: 10.1016/j.gene.2014.04.018 [DOI] [PubMed] [Google Scholar]
- 16. Robinson CM, Rajaiya J, Walsh MP, Seto D, Dyer DW, Jones MS, Chodosh J. 2009. Computational analysis of human adenovirus type 22 provides evidence for recombination among species D human adenoviruses in the penton base gene. J Virol 83:8980–8985. doi: 10.1128/JVI.00786-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dhingra A, Hage E, Ganzenmueller T, Böttcher S, Hofmann J, Hamprecht K, Obermeier P, Rath B, Hausmann F, Dobner T, Heim A. 2019. Molecular evolution of human adenovirus (HAdV) species C. Sci Rep 9:1039. doi: 10.1038/s41598-018-37249-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hashimoto S, Gonzalez G, Harada S, Oosako H, Hanaoka N, Hinokuma R, Fujimoto T. 2018. Recombinant type human mastadenovirus D85 associated with epidemic keratoconjunctivitis since 2015 in Japan. J Med Virol 90:881–889. doi: 10.1002/jmv.25041 [DOI] [PubMed] [Google Scholar]
- 19. Walsh MP, Seto J, Jones MS, Chodosh J, Xu W, Seto D. 2010. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J Clin Microbiol 48:991–993. doi: 10.1128/JCM.01694-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaneko H, Aoki K, Ishida S, Ohno S, Kitaichi N, Ishiko H, Fujimoto T, Ikeda Y, Nakamura M, Gonzalez G, Koyanagi KO, Watanabe H, Suzutani T. 2011. Recombination analysis of intermediate human adenovirus type 53 in Japan by complete genome sequence. J Gen Virol 92:1251–1259. doi: 10.1099/vir.0.030361-0 [DOI] [PubMed] [Google Scholar]
- 21. Ji T, Li L, Li W, Zheng X, Ye X, Chen H, Zhou Q, Jia H, Chen B, Lin Z, Chen H, Huang S, Seto D, Chen L, Feng L. 2021. Emergence and characterization of a putative novel human adenovirus recombinant HAdV-C104 causing pneumonia in Southern China. Virus Evol 7:veab018. doi: 10.1093/ve/veab018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu EB, Wadford DA, Seto J, Vu M, Hudson NR, Thrasher L, Torres S, Dyer DW, Chodosh J, Seto D, Jones MS. 2012. Computational and serologic analysis of novel and known viruses in species human adenovirus D in which serology and genomics do not correlate. PLoS One 7:e33212. doi: 10.1371/journal.pone.0033212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singh G, Robinson CM, Dehghan S, Schmidt T, Seto D, Jones MS, Dyer DW, Chodosh J. 2012. Overreliance on the hexon gene, leading to misclassification of human adenoviruses. J Virol 86:4693–4695. doi: 10.1128/JVI.06969-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaneko H, Aoki K, Ohno S, Ishiko H, Fujimoto T, Kikuchi M, Harada S, Gonzalez G, Koyanagi KO, Watanabe H, Suzutani T. 2011. Complete genome analysis of a novel intertypic recombinant human adenovirus causing epidemic keratoconjunctivitis in Japan. J Clin Microbiol 49:484–490. doi: 10.1128/JCM.01044-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robinson CM, Singh G, Henquell C, Walsh MP, Peigue-Lafeuille H, Seto D, Jones MS, Dyer DW, Chodosh J. 2011. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology 409:141–147. doi: 10.1016/j.virol.2010.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsushima Y, Shimizu H, Phan TG, Ushijima H. 2011. Genomic characterization of a novel human adenovirus type 31 recombinant in the hexon gene. J Gen Virol 92:2770–2775. doi: 10.1099/vir.0.034744-0 [DOI] [PubMed] [Google Scholar]
- 27. Walsh MP, Chintakuntlawar A, Robinson CM, Madisch I, Harrach B, Hudson NR, Schnurr D, Heim A, Chodosh J, Seto D, Jones MS. 2009. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 4:e5635. doi: 10.1371/journal.pone.0005635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Borkenhagen LK, Fieldhouse JK, Seto D, Gray GC. 2019. Are adenoviruses zoonotic? A systematic review of the evidence. Emerg Microbes Infect 8:1679–1687. doi: 10.1080/22221751.2019.1690953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Medkour H, Amona I, Akiana J, Davoust B, Bitam I, Levasseur A, Tall ML, Diatta G, Sokhna C, Hernandez-Aguilar RA, Barciela A, Gorsane S, La Scola B, Raoult D, Fenollar F, Mediannikov O. 2020. Adenovirus infections in African humans and wild non-human primates: great diversity and cross-species transmission. Viruses 12:657. doi: 10.3390/v12060657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han J-W, La T-M, Kim J-H, Choi I-S, Song C-S, Park S-Y, Lee J-B, Lee S-W. 2018. The possible origin of human adenovirus type 3: evidence of natural genetic recombination between human and simian adenovirus. Infect Genet Evol 65:380–384. doi: 10.1016/j.meegid.2018.08.020 [DOI] [PubMed] [Google Scholar]
- 31. Zhou C, Tian H, Wang X, Liu W, Yang S, Shen Q, Wang Y, Ni B, Chen S, Fu X, Fei R, Zhang W. 2014. The genome sequence of a novel simian adenovirus in a chimpanzee reveals a close relationship to human adenoviruses. Arch Virol 159:1765–1770. doi: 10.1007/s00705-013-1967-1 [DOI] [PubMed] [Google Scholar]
- 32. Kremer EJ. 2021. What is the risk of a deadly adenovirus pandemic? PLoS Pathog 17:e1009814. doi: 10.1371/journal.ppat.1009814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen EC, Yagi S, Kelly KR, Mendoza SP, Tarara RP, Canfield DR, Maninger N, Rosenthal A, Spinner A, Bales KL, Schnurr DP, Lerche NW, Chiu CY. 2011. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog 7:e1002155. doi: 10.1371/journal.ppat.1002155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abbink P, Maxfield LF, Ng’ang’a D, Borducchi EN, Iampietro MJ, Bricault CA, Teigler JE, Blackmore S, Parenteau L, Wagh K, Handley SA, Zhao G, Virgin HW, Korber B, Barouch DH. 2015. Construction and evaluation of novel rhesus monkey adenovirus vaccine vectors. J Virol 89:1512–1522. doi: 10.1128/JVI.02950-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, Stanley K, Kramer J, Macri SC, Permar SR, Schmitz JE, Mansfield K, Brenchley JM, Veazey RS, Stappenbeck TS, Wang D, Barouch DH, Virgin HW. 2012. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell 151:253–266. doi: 10.1016/j.cell.2012.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Capone S, Raggioli A, Gentile M, Battella S, Lahm A, Sommella A, Contino AM, Urbanowicz RA, Scala R, Barra F, et al. 2021. Immunogenicity of a new gorilla adenovirus vaccine candidate for COVID-19. Mol Ther 29:2412–2423. doi: 10.1016/j.ymthe.2021.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Colloca S, Barnes E, Folgori A, Ammendola V, Capone S, Cirillo A, Siani L, Naddeo M, Grazioli F, Esposito ML, Ambrosio M, Sparacino A, Bartiromo M, Meola A, Smith K, Kurioka A, O’Hara GA, Ewer KJ, Anagnostou N, Bliss C, Hill AVS, Traboni C, Klenerman P, Cortese R, Nicosia A. 2012. Vaccine vectors derived from a large collection of simian adenoviruses induce potent cellular immunity across multiple species. Sci Transl Med 4:115ra2. doi: 10.1126/scitranslmed.3002925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pantó L, Podgorski II, Jánoska M, Márkó O, Harrach B. 2015. Taxonomy proposal for Old World monkey adenoviruses: characterisation of several non-human, non-ape primate adenovirus lineages. Arch Virol 160:3165–3177. doi: 10.1007/s00705-015-2575-z [DOI] [PubMed] [Google Scholar]
- 39. Roy S, Vandenberghe LH, Kryazhimskiy S, Grant R, Calcedo R, Yuan X, Keough M, Sandhu A, Wang Q, Medina-Jaszek CA, Plotkin JB, Wilson JM. 2009. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog 5:e1000503. doi: 10.1371/journal.ppat.1000503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Podgorski II, Pantó L, Papp T, Harrach B, Benkö M. 2016. Genome analysis of four Old World monkey adenoviruses supports the proposed species classification of primate adenoviruses and reveals signs of possible homologous recombination. J Gen Virol 97:1604–1614. doi: 10.1099/jgv.0.000465 [DOI] [PubMed] [Google Scholar]
- 41. Podgorski II, Pantó L, Földes K, de Winter I, Jánoska M, Sós E, Chenet B, Harrach B, Benkő M. 2018. Adenoviruses of the most ancient primate lineages support the theory on virus-host co-evolution. Acta Vet Hung 66:474–487. doi: 10.1556/004.2018.042 [DOI] [PubMed] [Google Scholar]
- 42. Kosoltanapiwat N, Tongshoob J, Ampawong S, Reamtong O, Prasittichai L, Yindee M, Tongthainan D, Tulayakul P, Boonnak K. 2022. Simian adenoviruses: molecular and serological survey in monkeys and humans in Thailand. One Health 15:100434. doi: 10.1016/j.onehlt.2022.100434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mancuso DM, Gainor K, Dore KM, Gallagher CA, Cruz K, Beierschmitt A, Malik YS, Ghosh S. 2023. Detection and molecular characterization of adenoviruses in captive and free-roaming African green monkeys (Chlorocebus sabaeus): evidence for possible recombination and cross-species transmission. Viruses 15:1605. doi: 10.3390/v15071605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wevers D, Metzger S, Babweteera F, Bieberbach M, Boesch C, Cameron K, Couacy-Hymann E, Cranfield M, Gray M, Harris LA, Head J, Jeffery K, Knauf S, Lankester F, Leendertz SAJ, Lonsdorf E, Mugisha L, Nitsche A, Reed P, Robbins M, Travis DA, Zommers Z, Leendertz FH, Ehlers B. 2011. Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions. J Virol 85:10774–10784. doi: 10.1128/JVI.00810-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chiu CY, Yagi S, Lu X, Yu G, Chen EC, Liu M, Dick EJ, Carey KD, Erdman DD, Leland MM, Patterson JL. 2013. A novel adenovirus species associated with an acute respiratory outbreak in A baboon colony and evidence of coincident human infection. mBio 4:e00084. doi: 10.1128/mBio.00084-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Foytich KR, Deshazer G, Esona MD, Liu A, Wang Y, Tu X, Jiang B. 2014. Identification of new provisional simian adenovirus species from captive monkeys, China. Emerg Infect Dis 20:1758–1759. doi: 10.3201/eid2010.131255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Malouli D, Howell GL, Legasse AW, Kahl C, Axthelm MK, Hansen SG, Früh K. 2014. Full genome sequence analysis of a novel adenovirus of rhesus macaque origin indicates a new simian adenovirus type and species. Virol Rep 3–4:18–29. doi: 10.1016/j.virep.2014.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kosulin K. 2019. Intestinal HAdV infection: tissue specificity, persistence, and implications for antiviral therapy. Viruses 11:804. doi: 10.3390/v11090804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Grand RJ. 2023. Pathogenicity and virulence of human adenovirus F41: possible links to severe hepatitis in children. Virulence 14:2242544. doi: 10.1080/21505594.2023.2242544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Troeger C, Blacker BF, Khalil IA, Rao PC, Cao S, Zimsen SR, Albertson SB, Stanaway JD, Deshpande A, Abebe Z, et al. 2018. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the global burden of disease study 2016. Lancet Infect Dis 18:1211–1228. doi: 10.1016/S1473-3099(18)30362-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jones MS, Harrach B, Ganac RD, Gozum MMA, Dela Cruz WP, Riedel B, Pan C, Delwart EL, Schnurr DP. 2007. New adenovirus species found in a patient presenting with gastroenteritis. J Virol 81:5978–5984. doi: 10.1128/JVI.02650-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bányai K, Martella V, Meleg E, Kisfali P, Péterfi Z, Benkö M, Melegh B, Szucs G. 2009. Searching for HAdV-52, the putative gastroenteritis-associated human adenovirus serotype in Southern Hungary. New Microbiol 32:185–188. [PubMed] [Google Scholar]
- 53. Wong SSY, Yip CCY, Sridhar S, Leung K-H, Cheng AKW, Fung AMY, Lam H-Y, Chan K-H, Chan JFW, Cheng VCC, Tang BSF, Yuen K-Y. 2018. Comparative evaluation of a laboratory-developed real-time PCR assay and RealStar adenovirus PCR Kit for quantitative detection of human adenovirus. Virol J 15:149. doi: 10.1186/s12985-018-1059-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schnurr D, Bollen A, Crawford-Miksza L, Dondero ME, Yagi S. 1995. Adenovirus mixture isolated from the brain of an AIDS patient with encephalitis. J Med Virol 47:168–171. doi: 10.1002/jmv.1890470210 [DOI] [PubMed] [Google Scholar]
- 55. Suzuki HI, Asai T, Okada K, Kazuyama Y, Takahashi T, Kanda Y, Chiba S, Kurokawa M. 2008. Disseminated adenovirus disease by multiple adenovirus serotypes following allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 14:353–355. doi: 10.1016/j.bbmt.2007.12.001 [DOI] [PubMed] [Google Scholar]
- 56. Metzgar D, Osuna M, Yingst S, Rakha M, Earhart K, Elyan D, Esmat H, Saad MD, Kajon A, Wu J, Gray GC, Ryan MAK, Russell KL. 2005. PCR analysis of Egyptian respiratory adenovirus isolates, including identification of species, serotypes, and coinfections. J Clin Microbiol 43:5743–5752. doi: 10.1128/JCM.43.11.5743-5752.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kosulin K, Geiger E, Vécsei A, Huber W-D, Rauch M, Brenner E, Wrba F, Hammer K, Innerhofer A, Pötschger U, Lawitschka A, Matthes-Leodolter S, Fritsch G, Lion T. 2016. Persistence and reactivation of human adenoviruses in the gastrointestinal tract. Clin Microbiol Infect 22:381. doi: 10.1016/j.cmi.2015.12.013 [DOI] [PubMed] [Google Scholar]
- 58. Kosulin K, Berkowitsch B, Matthes S, Pichler H, Lawitschka A, Pötschger U, Fritsch G, Lion T. 2018. Intestinal adenovirus shedding before allogeneic stem cell transplantation is a risk factor for invasive infection post-transplant. EBioMedicine 28:114–119. doi: 10.1016/j.ebiom.2017.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roy S, Calcedo R, Medina-Jaszek A, Keough M, Peng H, Wilson JM. 2011. Adenoviruses in lymphocytes of the human gastro-intestinal tract. PLoS One 6:e24859. doi: 10.1371/journal.pone.0024859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Curlin ME, Huang M-L, Lu X, Celum CL, Sanchez J, Selke S, Baeten JM, Zuckerman RA, Erdman DD, Corey L. 2010. Frequent detection of human adenovirus from the lower gastrointestinal tract in men who have sex with men. PLoS One 5:e11321. doi: 10.1371/journal.pone.0011321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Garnett CT, Erdman D, Xu W, Gooding LR. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J Virol 76:10608–10616. doi: 10.1128/jvi.76.21.10608-10616.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wigand R. 1987. Pitfalls in the identification of adenoviruses. J Virol Methods 16:161–169. doi: 10.1016/0166-0934(87)90001-2 [DOI] [PubMed] [Google Scholar]
- 63. Ganzenmueller T, Heim A. 2012. Adenoviral load diagnostics by quantitative polymerase chain reaction: techniques and application. Rev Med Virol 22:194–208. doi: 10.1002/rmv.724 [DOI] [PubMed] [Google Scholar]
- 64. Heim A, Ebnet C, Harste G, Pring-Akerblom P. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol 70:228–239. doi: 10.1002/jmv.10382 [DOI] [PubMed] [Google Scholar]
- 65. Miura-Ochiai R, Shimada Y, Konno T, Yamazaki S, Aoki K, Ohno S, Suzuki E, Ishiko H. 2007. Quantitative detection and rapid identification of human adenoviruses. J Clin Microbiol 45:958–967. doi: 10.1128/JCM.01603-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Alsaleh AN, Grimwood K, Sloots TP, Whiley DM. 2014. A retrospective performance evaluation of an adenovirus real-time PCR assay. J Med Virol 86:795–801. doi: 10.1002/jmv.23844 [DOI] [PubMed] [Google Scholar]
- 67. Seto D, Chodosh J, Brister JR, Jones MS, Members of the Adenovirus Research Community . 2011. Using the whole-genome sequence to characterize and name human adenoviruses. J Virol 85:5701–5702. doi: 10.1128/JVI.00354-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aoki K, Benkö M, Davison AJ, Echavarria M, Erdman DD, Harrach B, Kajon AE, Schnurr D, Wadell G, Members of the Adenovirus Research Community . 2011. Toward an integrated human adenovirus designation system that utilizes molecular and serological data and serves both clinical and fundamental virology. J Virol 85:5703–5704. doi: 10.1128/JVI.00491-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Handley SA, Desai C, Zhao G, Droit L, Monaco CL, Schroeder AC, Nkolola JP, Norman ME, Miller AD, Wang D, Barouch DH, Virgin HW. 2016. SIV infection-mediated changes in gastrointestinal bacterial microbiome and virome are associated with immunodeficiency and prevented by vaccination. Cell Host Microbe 19:323–335. doi: 10.1016/j.chom.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Persson BD, John L, Rafie K, Strebl M, Frängsmyr L, Ballmann MZ, Mindler K, Havenga M, Lemckert A, Stehle T, Carlson L-A, Arnberg N. 2021. Human species D adenovirus hexon capsid protein mediates cell entry through a direct interaction with CD46. Proc Natl Acad Sci U S A 118:e2020732118. doi: 10.1073/pnas.2020732118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. de Jong JC, Osterhaus ADME, Jones MS, Harrach B. 2008. Human adenovirus type 52: a type 41 in disguise? J Virol 82:3809. doi: 10.1128/JVI.02457-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lion T. 2019. Adenovirus persistence, reactivation, and clinical management. FEBS Lett 593:3571–3582. doi: 10.1002/1873-3468.13576 [DOI] [PubMed] [Google Scholar]
- 73. Lion T. 2014. Adenovirus infections in immunocompetent and immunocompromised patients. Clin Microbiol Rev 27:441–462. doi: 10.1128/CMR.00116-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lion T, Kosulin K, Landlinger C, Rauch M, Preuner S, Jugovic D, Pötschger U, Lawitschka A, Peters C, Fritsch G, Matthes-Martin S. 2010. Monitoring of adenovirus load in stool by real-time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia 24:706–714. doi: 10.1038/leu.2010.4 [DOI] [PubMed] [Google Scholar]
- 75. Adrian T, Schäfer G, Cooney MK, Fox JP, Wigand R. 1988. Persistent enteral infections with adenovirus types 1 and 2 in infants: no evidence of reinfection. Epidemiol Infect 101:503–509. doi: 10.1017/s0950268800029393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Garnett CT, Talekar G, Mahr JA, Huang W, Zhang Y, Ornelles DA, Gooding LR. 2009. Latent species C adenoviruses in human tonsil tissues. J Virol 83:2417–2428. doi: 10.1128/JVI.02392-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kwon HJ, Rhie YJ, Seo WH, Jang G-Y, Choi BM, Lee JH, Lee C-K, Kim YK. 2013. Clinical manifestations of respiratory adenoviral infection among hospitalized children in Korea. Pediatr Int 55:450–454. doi: 10.1111/ped.12108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lefeuvre C, Salmona M, Feghoul L, Ranger N, Mercier-Delarue S, Nizery L, Alimi A, Dalle J-H, LeGoff J. 2021. Deciphering an adenovirus F41 outbreak in pediatric hematopoietic stem cell transplant recipients by whole-genome sequencing. J Clin Microbiol 59:e03148-20. doi: 10.1128/JCM.03148-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tahmasebi R, Luchs A, Tardy K, Hefford PM, Tinker RJ, Eilami O, de Padua Milagres FA, Brustulin R, Teles M da A, Dos Santos Morais V, Moreira CHV, Buccheri R, Araújo ELL, Villanova F, Deng X, Sabino EC, Delwart E, Leal É, Charlys da Costa A. 2020. Viral gastroenteritis in Tocantins, Brazil: characterizing the diversity of human adenovirus F through next-generation sequencing and bioinformatics. J Gen Virol 101:1280–1288. doi: 10.1099/jgv.0.001500 [DOI] [PubMed] [Google Scholar]
- 80. Holly MK, Smith JG. 2018. Adenovirus infection of human enteroids reveals interferon sensitivity and preferential infection of goblet cells. J Virol 92:e00250-18. doi: 10.1128/JVI.00250-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Renzette N, Bhattacharjee B, Jensen JD, Gibson L, Kowalik TF. 2011. Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog 7:e1001344. doi: 10.1371/journal.ppat.1001344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Parsons LR, Tafuri YR, Shreve JT, Bowen CD, Shipley MM, Enquist LW, Szpara ML. 2015. Rapid genome assembly and comparison decode intrastrain variation in human alphaherpesviruses. mBio 6:e02213-14. doi: 10.1128/mBio.02213-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Renner DW, Szpara ML. 2018. Impacts of genome-wide analyses on our understanding of human herpesvirus diversity and evolution. J Virol 92:e00908-17. doi: 10.1128/JVI.00908-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Götting J, Baier C, Panagiota V, Maecker-Kolhoff B, Dhingra A, Heim A. 2022. High genetic stability of co-circulating human adenovirus type 31 lineages over 59 years. Virus Evol 8:veac067. doi: 10.1093/ve/veac067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang D, Urisman A, Liu Y-T, Springer M, Ksiazek TG, Erdman DD, Mardis ER, Hickenbotham M, Magrini V, Eldred J, Latreille JP, Wilson RK, Ganem D, DeRisi JL. 2003. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol 1:E2. doi: 10.1371/journal.pbio.0000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kalantar KL, Carvalho T, de Bourcy CFA, Dimitrov B, Dingle G, Egger R, Han J, Holmes OB, Juan Y-F, King R, et al. 2020. IDseq—an open source cloud-based pipeline and analysis service for metagenomic pathogen detection and monitoring. Gigascience 9:giaa111. doi: 10.1093/gigascience/giaa111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tcherepanov V, Ehlers A, Upton C. 2006. Genome annotation transfer utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics 7:150. doi: 10.1186/1471-2164-7-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. doi: 10.1128/JVI.73.1.152-160.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Koboldt DC. 2020. Best practices for variant calling in clinical sequencing. Genome Med 12:91. doi: 10.1186/s13073-020-00791-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lin S-C, Haga K, Zeng X-L, Estes MK. 2022. Generation of CRISPR–Cas9-mediated genetic knockout human intestinal tissue–derived enteroid lines by lentivirus transduction and single-cell cloning. Nat Protoc 17:1004–1027. doi: 10.1038/s41596-021-00669-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Isolation and separation of rhAdV-68 and rhAdV-69.
Phylogenetic analysis of rhAdV-68 and rhAdV-69 hexon and short fiber genes.
Schematic overview of genomes of coinfecting rhAdVs.
Comparative analysis of rhAdV-70–rhAdV-73.
Genomic characterization of coinfecting rhAdVs.
Assessment of virus isolation using HTS.
Data Availability Statement
Complete genome sequences are available at GenBank for rhAdV-68 to rhAdV-73 (PP134848, PP134849, PP134850, PP134851, PP134852, and PP134853). Raw sequencing data have been deposited to the Sequence Read Archive as PRJNA1061574.