Abstract
Hepatocytes work in highly structured, repetitive hepatic lobules. Blood flow across the radial axis of the lobule generates oxygen, nutrient, and hormone gradients, which result in zoned spatial variability and functional diversity. This large heterogeneity suggests that hepatocytes in different lobule zones may have distinct gene expression profiles, metabolic features, regenerative capacity, and susceptibility to damage. Here, we describe the principles of liver zonation, introduce metabolomic approaches to study the spatial heterogeneity of the liver, and highlight the possibility of exploring the spatial metabolic profile, leading to a deeper understanding of the tissue metabolic organization. Spatial metabolomics can also reveal intercellular heterogeneity and its contribution to liver disease. These approaches facilitate the global characterization of liver metabolic function with high spatial resolution along physiological and pathological time scales. This review summarizes the state of the art for spatially resolved metabolomic analysis and the challenges that hinder the achievement of metabolome coverage at the single-cell level. We also discuss several major contributions to the understanding of liver spatial metabolism and conclude with our opinion on the future developments and applications of these exciting new technologies.
INTRODUCTION
The liver is a highly heterogeneous and unique structure that supports body homeostasis through blood detoxification, production of bile acids, and several metabolic functions. This myriad of roles is orchestrated in hepatocytes, KCs, HSCs, liver sinusoidal endothelial cells, and cholangiocytes organized in a repeated hexagonal manner. These architectural structures are termed lobules and create a gradient of oxygen, nutrients, and hormones that contribute to hepatic zonation.
The human metabolome is composed of a large number of highly diverse molecules, such as amino acids, lipids, organic acids, and carbohydrates.1 These small molecules represent the substrates, intermediates, and products of the enzymatic reactions and metabolic pathways of a cell, organ, or organism. Constituting the building blocks of the physiological processes, metabolites are also energy storage entities and key regulators of homeostasis systems, participating in metabolic regulation, membrane trafficking, signaling, proliferation, or apoptosis. This results in a metabolome that is highly sensitive to alterations. Even subtle changes in daily habits, such as diet, increased stress, physical activity, lifestyle, stimuli, or environmental changes, can significantly alter the metabolite levels. Thus, metabolomics is a powerful tool for the real-time assessment of a metabolic state or a clinical phenotype, as well as to decode the molecular mechanisms of metabolic-related disorders.
Nevertheless, the highly specialized microenvironment in the liver generates different metabolic profiles, where bulk liver tissue metabolomics may compromise the understanding of spatial and cell-specific regulators in each disease phenotype, leading to the current demand for spatial metabolomic approaches enabled by advances in mass spectrometry imaging (MSI) technology. MSI is a powerful approach to performing in situ analysis of the molecular composition of the biological tissue, while retaining the spatial information. No prior knowledge or labeling is required and hundreds to thousands of molecules can be detected simultaneously. This allows molecular mapping for a range of sample types and chemical classes, revealing an unprecedented level of information on molecular processes at the cellular level. Importantly, single-cell metabolomics allows high-throughput metabolic analysis at the cellular level. Metabolites represent precursors, intermediates, and end products crucial to identify the respective phenotype signatures in each zonate.2 Spatial metabolomics can then reflect the intracellular physiological reactions and functional state of the tissue sample,3 and provide the possibility to explore the spatial metabolic profile and tissue histology at the single-cell level, leading to a deeper understanding of tissue metabolic organization. However, spatial metabolomics is challenged by the fact that, within a single cell, metabolites are present in very low amounts (femtomolar range) and can be more transient (millisecond turnover) than proteins or mRNA.4 Accordingly, the methodology and sampling period will determine the outcome.5,6
In the field of liver disease, the application of spatial metabolomics is expected to strongly empower our understanding of the metabolic processes within specific liver zonations and their impact on disease progression and treatment. On one hand, hepatocyte functions are oxygen dependent. On the other hand, periportal hepatocytes are responsible for glucose delivery, gluconeogenesis from lactate, urea synthesis, fatty acid oxidation, sulfation, and cholesterol synthesis, whereas pericentral hepatocytes are involved in glucose uptake, glycolysis from glucose, glutamine synthesis, bile acid synthesis, and lipogenesis and ketogenesis.7
SPATIAL HETEROGENEITY IN THE MAMMALIAN LIVER
Architecture of the mammalian liver
The liver is the central metabolic organ of higher vertebrates and the largest glandular organ. The mammalian liver is a highly complex 3D structured and functionally heterogeneous organ with a unique dual blood supply from the hepatic portal vein (PV), which brings all the blood from the gastrointestinal tract, gallbladder, pancreas, and spleen, and from the hepatic artery that carries oxygenated blood to the liver. Portal venous and arterial blood mix together in hepatic sinusoids before leaving the liver through the hepatic vein. The mammalian liver is divided into functionally distinct lobes, which in turn are formed by honeycomb-like structures made of repetitive functional structural units, the liver lobules, are organized in a highly irregular manner, and present different sizes and axial orientations (Figure 1A).
FIGURE 1.
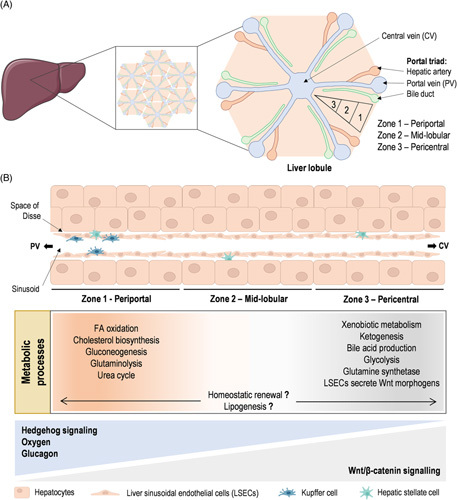
Liver architecture and metabolic zonation. (A) The mammalian liver is a highly complex 3D structure organized in a honeycomb-like arrangement made of liver lobules as repetitive functional structural units. Liver lobules can be further divided into 3 zones: the region surrounding the portal triads, composed of the portal vein (PV), hepatic artery, and the bile duct, is called the periportal (zone 1); hepatocytes adjacent to the central vein (CV) are known as the pericentral (zone 3); and the regions in between are referred to as the midlobular (zone 2). (B) Currently described spatial metabolic zonation of liver metabolic processes and liver immune cells, and the molecular determinants of liver zonation are schematically summarized.
Liver lobules are hexagonal-shaped anatomical functional units, centered on a branch of the hepatic vein called the central vein (CV). The portal triads are located in the hexagonal corners and the junction with the neighboring lobules, and are formed by PV, the hepatic arteriole, and bile ducts. Liver lobules are composed of concentric layers of hepatocytes distributed into hepatic cords and containing two intertwined radially distributed networks, the sinusoids for blood flow and the bile canaliculi for bile flux and secretion.8 In the basolateral domains, hepatocytes face fenestrated liver sinusoidal endothelial cells that form the radial sinusoidal blood vessels. HSCs, the primary cell type involved in fibrosis, reside in the “space of Disse” between the hepatocytes and sinusoids. In contrast, KCs, the liver-resident macrophages, are largely immotile cells in the sinusoids. Liver lobules can be further divided into 3 zones: the region surrounding the portal triads is called periportal (zone 1), hepatocytes adjacent to the CV are known as pericentral (zone 3), and the regions in between are referred to as midlobular (zone 2)9 (Figure 1A). Alternatively, another way to conceptualize the microscopic structure of the liver lobe is by dividing them into portal lobules, centered in the portal triads instead of the CV, or dividing them into acini, the functional units in terms of blood flow. The acinus contains a small portal tract at the center and terminal hepatic venules at the periphery.10 The liver acinus was first divided by Rappaport into zones 1, 2, and 3, wherein zone 1 surrounds the portal tract and zone 3 surrounds the hepatic venule. Acinar zones 1, 2, and 3 correspond to the periportal, middle, and pericentral zones of the lobule.10,11 As blood flows inward from the periportal region to the pericentral region, hepatocytes take up and secrete nutrients and sense hormones. Sequential hepatocyte consumption and production, together with local tissue morphogens, give rise to a graded microenvironment of oxygen concentrations, nutrients, and signaling along the PV-CV axis and create different radial layers of subspecialized hepatocytes, a phenomenon termed zonation.12 Liver metabolic zonation is a highly regulated process critical for optimal liver function by maintaining energy homeostasis, regulating the metabolism of nutrients and xenobiotics, and controlling the production, regulation, and recycling of various proteins.
Molecular determinants of liver zonation
Since liver zonation was first described, a great effort has been dedicated to understanding the molecular determinants of liver zonation. Wnt ligands, generally considered classical morphogens, are insoluble secreted glycoproteins that act through autocrine or paracrine binding to a Frizzled receptor and LDL receptor–related protein 5/6 (LRP5/6) coreceptors. Eventually, the signaling cascade leads to β-catenin accumulation in the cytoplasm, translocation into the nucleus, and interaction with the T-cell factor family of transcription factors to help transcribe target genes. Previous reports have shown that under homeostatic conditions, Wnt/β-catenin activity is restricted to pericentral hepatocytes.13 More importantly, compelling studies have demonstrated that conditional liver-specific β-catenin knockouts,14,15 inducible liver-specific adenomatous polyposis coli knockouts,16 and Lrp5/6 knockouts17 loosen zonation patterns, supporting Wnt/β-catenin as the gatekeeper of liver metabolic zonation. The impact of Wnt/β-catenin–induced metabolic changes on liver carcinogenesis has recently been thoroughly explored.18–20
Other pathways have also been implicated in liver zonation mostly by counteracting Wnt/β-catenin signaling. Hedgehog signaling, although presenting low activity in healthy mature hepatocytes, is higher in the periportal region and hypothetically implicated in the regulation of zone 1 hepatocytes.21 Hedgehog ligands, such as Sonic hedgehog, Indian hedgehog, and Desert hedgehog, bind to Ptch1/2 receptors to relieve patched-mediated suppression of Smoothened. Activated Smoothened leads to the stabilization and nuclear translocation of GLI transcription factors.22 Using conditional Smoothened knockout mice, two key mechanisms of Hedgehog signaling regulation of liver zonation were proposed: (i) Hedgehog signaling affects the Wnt/β-catenin pathway by downregulating its target gene23 and (ii) Hedgehog signaling regulates the insulin-like growth factor axis by means of the GLI3 transcription factor.24 Likewise, blood-borne molecules have been shown to activate Ras signaling, which promotes the expression of periportal genes while suppressing pericentral-associated genes.25 Also, the canonical Wnt signaling has been described to converge on the hepatocyte nuclear factor-4α–driven transcription to modulate liver zonation.26,27 Another well-described antagonist to the Wnt/β-catenin signaling pathway is glucagon, a hormone released from the pancreatic α-cells and distributed to the liver lobules. A gradient drop in glucagon concentration is observed in the sinusoid along the PV-CV axis. Noteworthy, glucagon-deficient mice (Gcg −/−) have perturbed hepatic gene expression and zonation patterns, which can be rescued by glucagon reinfusion, further suggesting that glucagon plays a critical role in shaping liver zonation.28 In addition, the family of hypoxia-induced transcription factors (HIFs) provides another layer of regulation for liver zonation and patterned gene expression profiles.29 HIFs, mainly localized in the perivenous region, are a family of oxygen-sensitive heterodimeric transcription factors that regulate gene expression in response to oxygen availability.30 It has been hypothesized that the oxygen gradient along the PV-CV axis drives transcriptional responses regulated by HIFs controlling the zone-dependent heterogeneity of hepatocytes.31,32 In agreement, ablation of the hif-1α gene in stem cells reduces Wnt/β-catenin gene expression under hypoxic conditions,33 and adenomatous polyposis coli, a negative regulator of β-catenin, directly represses HIF-1α34 (Figure 1B). Finally, the effect of hypophysectomy and subsequent infusion of growth hormone or injections of triiodothyronine were used to show that pituitary-dependent hormones regulate zone-specific xenobiotic metabolism.35 The influence of pituitary hormones on liver zonation was further confirmed using hypopituitary dwarf mice.36
High-throughput methods can identify genome-wide differences in expression between cells enriched for periportal and pericentral hepatocytes. However, until recently, the spatial resolution of these studies was limited to cell populations isolated by separating hepatocytes according to their physical characteristics, such as cell size, cell density, and different binding to lecithins, or more commonly, using digitonin perfusion,37,38 retrograde/anterograde collagenase perfusion,39 laser capture microdissection (LCM),40 or using flow cytometry.41 In recent years, the development of novel technologies such as single-cell RNA sequencing,36 spatial transcriptomics,42 and targeted immunoaffinity-based proteomics,43 among others, have driven the re-examination of earlier discoveries of liver zonation determinants, enlightening some novel regulators of liver zonation. For example, using targeted immunoaffinity-based proteomics, data mining revealed key regulators and preferentially active pathways in either periportal or pericentral hepatocytes. The authors confirmed that β-catenin signaling and nuclear xenosensing receptors are the most prominent pericentral regulators. Several kinase-dependent and G-protein-dependent signaling cascades are active mainly in periportal hepatocytes.43 Also, using single-cell RNA sequencing combined with single-molecule RNA fluorescence in situ hybridization to interrogate hepatocyte zonation in the mouse liver, 50% of the hepatocyte genes were shown to be zonated. The major determinants of liver zonation were Wnt/β-catenin signaling and the oxygen gradient, Ras signaling, which activates periportal genes, and pituitary signals, which inhibit periportal genes. Surprisingly, regulation of the expression of two-thirds of the zonated liver genes remains to be established.36
In summary, although our understanding of the molecular determinants of liver zonation has grown immensely in the last years with the advent of novel technologies, it is evident that many regulators or the interplay among determinants of liver zonation remain to be identified. On the other hand, whereas Wnt/β-catenin signaling has been shown to regulate the expression of one-third of the zonated hepatocyte genes, the factors underlying the control of Wnt/β-catenin signaling remain to be addressed. On this basis, Dicer, an endoribonuclease III type enzyme involved in microRNA biogenesis, has been proven to be essential for the actions of Wnt/β-catenin signaling in liver zonation.44 By applying transcriptomics, microRNA arrays, and mass spectrometry (MS) proteomics to reconstruct spatial atlases of multiple zonated features, protein zonation was demonstrated to largely overlap with microRNA zonation.45 Despite this, the relevance of microRNAs, and other cellular properties, such as DNA methylation patterns, chromosomal conformations and chromatin modifications, proteomes, and metabolomes in the regulation of liver zonation remains to be addressed.
Spatial metabolic zonation of liver metabolic processes
The liver is a central hub regulating carbohydrate, lipid, and amino acid metabolism, ammonia clearance, urea, albumin and bile acid synthesis, and xenobiotic metabolism, among others, and it is also involved in the inflammatory response. The zonal-specific differences in the metabolic capacities of many enzymes of liver hepatocytes allow different and opposing metabolic processes to coexist simultaneously conferring optimal liver function.
The liver regulates systemic glucose and lipid fluxes during feeding and fasting, and relies on these substrates for its own energy needs.46 After a carbohydrate-rich meal, plasma glucose is taken up by the liver and transiently converted into glycogen and used as a substrate for synthesizing fatty acids through de novo lipogenesis. On the contrary, during fasting, the liver becomes a producer of glucose to prevent a dramatic drop in plasma glucose through the glycogenolysis pathways and gluconeogenic synthesis from amino acids, lactate, and glycerol. Early studies using LCM of periportal and pericentral rat liver tissue have shown that the gluconeogenic enzymes fructose-1,6-bisphosphatase, glucose-6-phosphatase, and phosphoenolpyruvate carboxykinase are preferentially expressed in the periportal zone.47,48 In contrast, the glycolytic enzyme glucokinase is mainly expressed in the pericentral zone.47,49,50 In addition, an ultrastructural heterogeneity of glycogen lobular distribution has been described, with periportal hepatocytes showing dense glycogen deposits during fasting while midlobular and pericentral hepatocytes present sparse glycogen particles.51,52 This functional and spatial division of hepatocytes prevents futile cycling when fulfilling both anabolic processes (ATP-consuming, such as gluconeogenesis) in periportal hepatocytes and catabolic requirements (ATP-generating, such as glycolysis) in pericentral cells, avoiding competition for common substrates between pathways. Molecular determinants of zonal glucose homeostasis include (i) glucagon to promote hepatic glucose output in the periportal zone53; (ii) HIF-2α/insulin receptor substrate 2 that preferentially enhances insulin signaling, thereby suppressing gluconeogenesis, and HIF-1α that promotes glycolysis, corroborating the role of HIFs and oxygen supply in maintaining zonal glucose metabolism54; and (iii) Hedgehog signaling by the regulation of insulin-like growth factor-1 homeostasis.24 The fact that glucagon, a hormone secreted in response to fasting, can quickly alter the zonation metabolic profile underlines the liver’s flexibility to adapt to substrate availability.
The list of zonally expressed enzymes can be further extended to metabolic pathways such as lipid metabolism. After a meal, glucose and fructose can be channeled to fatty acid biosynthesis through de novo lipogenesis. On the contrary, during fasting, the liver responds by producing glucose through glycogenolysis and gluconeogenesis. With increased fasting time and a shortage of glycogenic and gluconeogenic substrates, ketone bodies provide an alternative energy source for highly oxidative organs such as the brain. Under these circumstances, hepatic fatty acid β-oxidation is critical by providing the carbon backbone for ketogenesis (acetyl-CoA) and supporting gluconeogenesis energetic demands. Early reports indicate that the rate of fatty acid synthesis and activity of acetyl-CoA carboxylase are markedly enhanced in pericentral hepatocytes.55 However, this seems far more complex, as the staining of fatty acid synthase, the major lipogenic enzyme, has been shown to be diffused from the periportal to the midzone under healthy conditions. In contrast, in transgenic mice with hepatocyte-targeted expression of all HCV proteins and presenting steatosis, fatty acid synthase is concentrated in the midzone and extended toward the centrilobular region.56 Moreover, the oxidative process of fatty acid oxidation is a zonated process occurring preferentially in periportal hepatocytes.55,57 In addition, both cholesterol biosynthesis, localized in the periportal zone, and bile acid production, in the pericentral region, are also localized metabolic processes.58,59 The zonation of complementary tasks is important for maintaining liver lipid homeostasis.
One of the best known examples of liver metabolic processes carried out by zonated enzymes is ammonia detoxification. In fact, the enzyme glutamine synthetase is highly expressed within 1 or 2 layers of pericentral hepatocytes.60 The zonal-specific expression has also been established for enzymes of the urea cycle in periportal cells compared with perivenous hepatocytes.61 Hepatic ammonia detoxification is a clear example of spatial recycling where periportal hepatocytes detoxify ammonia to generate urea. This process requires the breakdown of glutamine into glutamate by the action of the co-localized enzyme glutaminase, and pericentral hepatocytes take up the excess glutamate and recover it to glutamine, by the activity of glutamine synthetase, maintaining the amino acid balance. Nevertheless, under steatotic pathological conditions, this hepatic nitrogen homeostasis is hampered due to loss of periportal glutaminase 2 isoform expression and appearance of glutaminase 1 throughout the liver parenchyma.62 Also, liver-specific knockout of β-catenin leads to a loss of glutamine synthetase expression.14 Moreover, adenomatous polyposis coli hepatic deletion results in the upregulation of Wnt/β-catenin signaling in the periportal region and dysregulated ammonia metabolism,16 indicating that β-catenin regulates the periportal urea cycle and pericentral glutamine synthesis genes. However, Gcg -/- mice exhibit extended glutamine synthase expression.28
Many enzymes of xenobiotic metabolism also exhibit zonal-specific differences in protein or mRNA levels, with a preferential pericentral expression of the main detoxification enzymes, such as the cytochrome P450 monooxygenase isoforms.49 β-catenin is thought responsible for the localization of drug-metabolizing enzymes in pericentral hepatocytes. Indeed, liver-specific β-catenin knockout leads to a complete loss of cytochrome CYP1A2 and CYP2E1 expression and attenuated cytochrome P450 enzyme activity.14 Moreover, gene-set analysis of pericentral hepatocytes residing in hypoxic environments and exposed to xenobiotics and toxic metabolites showed enrichment for pathways regulating protein synthesis, proteasomal activity, and mitophagy, most probably reflecting the high need for recycling damaged mitochondria.63
Finally, hepatocytes suffer a gradual turnover under homeostatic conditions. In the last few years, the concept of hepatic zonation has been coupled with the homeostatic renewal of hepatocytes by controversial mechanisms. Zone-dependent transcriptome analysis of normal human liver was performed using LCM to demonstrate that the focal activation of the canonical Wnt pathway sets in the pericentral zone as the site of homeostatic renewal.40 Alternatively, others identified the midlobular zone as having the highest rate of homeostatic renewal activity. In agreement, lineage tracing using the Wnt-responsive gene Axin2 in mice identified a population of proliferating and self-renewing cells adjacent to the CV in the liver lobule.64 On the opposite, periportal hepatocytes have been shown to ensure hepatocyte renewal,65 whereas others identified the midlobular zone as having the highest rate of homeostatic renewal activity. The latter suggests that midlobular hepatocytes are somehow protected from toxic injuries and therefore gain an advantage for regeneration activity. This midlobular zone repopulation is driven by the insulin growth factor binding protein 2—a mechanistic target of the rapamycin-cyclin D1 axis.66 Of relevance, the gene Hamp encoding for hepcidin, a central regulator of systemic iron homeostasis, has the highest expression in the mid-layers of the lobule, and iron-regulatory gene expression during liver regeneration has been proposed.67
Overall, metabolic zonation is a dynamic phenomenon being compartmentalized spatiotemporally at the sublobular scale.63 Indeed, most hepatic gene expression patterns and enzyme distributions change during the daily feeding/fasting cycles and in response to drugs, hormones, and other blood-borne factors. In addition, the sexual dimorphic expression of a variety of zonated genes and metabolic processes has been reported, at least in the mouse liver.68 Periportal hepatocytes, existing in an oxygen and nutrient-rich environment, carry out most of the liver metabolic functions with high energetic demand, including fatty acid β-oxidation, gluconeogenesis, urea, and protein synthesis, and lipid metabolism. In contrast, pericentral hepatocytes, that exist in a low oxygen environment, are characterized by glycolysis, xenobiotic biotransformation reactions, and glutamine synthesis, overall, less energy-demanding processes (Figure 1B). Topographical distributions of liver metabolic processes have been recently re-examined by LCM approaches coupled with RNA sequencing to provide a comprehensive transcriptome analysis of human and mouse liver zonation.40 In addition, a recent study using MSI has shown hepatocyte heterogeneity in terms of amino acid and lipid composition.69 Also, using MSI, liver metabolome was shown to change with fasting in a localized pattern, including increased levels of fatty acids and tricarboxylic acid cycle intermediates.70
Spatial metabolic zonation of liver immune cells
Most methodologies historically used to study liver zonation select hepatocytes for analysis. Therefore, it is not surprising that our current knowledge of hepatocyte metabolic zonation stands out in contrast to our contemporary understanding of the immune system of the liver. In particular, the identity and precise localization of most hepatic immune cells in healthy and diseased human livers are unknown.
Recently, quantitative multiplex imaging, genetic manipulations, transcriptomics, infection-based assays, and mathematical modeling were used to reassess the relationship between the localization of immune cells in the liver and host protection in mice. These authors revealed that periportal regions in the liver are enriched with myeloid and lymphoid cells upon bacterial infection.71 Another study using spatial transcriptomics demonstrated unaltered proportions of KCs in the periportal cluster, although an enrichment in genes related to immune system processes was found in this zone.42 The localization of proinflammatory responses in periportal regions may help to protect unique cell populations in the pericentral region. In addition, spatial proteogenomics has been used to unravel a population of lipid-associated macrophages that surround the bile ducts in the healthy liver and that during steatosis are preferentially recruited to the steatotic regions of the liver.72
In the quest for a better characterization of immune cells in the liver, doublets of hepatocytes and liver sinusoidal endothelial cells were sequenced and hepatocyte single-cell zonation data were used to infer the zonation of the latter. This analysis showed that LSEC genes are significantly zonated in the pericentral region and enriched with Wnt signaling genes and modulators.36,73 Indeed, much evidence shows that diffusible Wnt morphogens are secreted by endothelial cells surrounding the CV and act upon nearby hepatocytes.36,64,73–75 In addition, liver sinusoidal endothelial cells were demonstrated to sense gut-derived bacteria, triggering a signaling cascade mediated by MYD88 and chemokine secretion, which ultimately orchestrates immune cell localization to periportal regions.71
Altogether, a spatiotemporally complex intracellular crosstalk is crucial for shaping liver zonation. In addition, deregulated immune zonation and/or disrupted intracellular crosstalk leading to altered liver zonation often underlies the liver disease and vice versa, meaning that several types of liver injury may disrupt metabolic features of liver zonation.
DECIPHERING METABOLIC HETEROGENEITY
The diversity of the metabolites is based on chemical structures and biochemical functions,1 and also on the wide range of abundances. A clear example is found among lipids, which highly dominate the diverse composition of the human metabolome. Triglycerides, fatty acids, and phospholipids, such as phosphatidylcholines, are highly abundant, while other lipids that play a role in signaling, such as phosphoinositides76 and ceramide phosphates,77 are at trace level in biological samples.
Bulk metabolomics
Although metabolomics is considered the youngest among the omics sciences, it has evolved into a well-established discipline built on the advances made in the analytical chemistry, biochemistry, and bioinformatics fields. Despite the noteworthy technological developments over the past 2 decades, metabolite diversity poses a technical challenge for analytical science, as there is no single methodology that is able to comprehensively, or even broadly, cover the metabolome of complex biological samples. The extensive array of techniques available in the field, although robust and reproducible, are biased with respect to the coverage of metabolites, being more sensitive to specific classes of metabolite of interest. Thus, cohesive workflows, combining different extraction techniques and complementary analytical platforms, are required to cover the metabolome diversity without compromising the data output quality.
Nuclear magnetic resonance (NMR) spectroscopy and MS are the 2 most common analytical techniques used in metabolomics.78,79 Both techniques can also provide structural information on the molecules detected.80 A summary of the main features of MS versus NMR-based metabolomics is presented in Table 1.
TABLE 1.
Mass spectrometry-based bulk metabolomics and nuclear magnetic resonance spectroscopy: strengths and weaknesses
| Method | Description | Advantages | Limitations |
|---|---|---|---|
| MS | • Based on the analysis of gas-phase ions and measurement of the mass-to-charge ratio of the ions • Different configurations are available depending on: – Ionization technique, used for generation of gaseous ions from the sample/extract – Mass analyzer, used for separation of analytes according to mass-to-charge ratio – Direct injection or coupled to separations-based techniques: gas chromatography, liquid chromatography, supercritical fluid chromatography or capillary electrophoresis |
• High sensitivity and versatility • High throughput • Broad coverage of metabolites or tailored analysis associated with specific sample preparation, separation method, and chosen MS-configuration • Wide dynamic range • Low amount of sample |
• Noninherently quantitative • Not equally sensitive for all the metabolites: the ionization efficiency differs among compounds • Matrix effect on complex samples such as hepatic tissue • High competition for the ionization of the metabolites, generating ion suppression of molecules less prone to be ionized • Destructive technique |
| NMR spectroscopy | • Based on the magnetic properties of atomic nuclei • It can be applied to the analysis of small molecules to large macromolecular complexes (ie, lipoprotein profiling): – 1H NMR: distinctive signal for each proton or group of equivalent protons – 31P NMR: analysis of phosphorylated compounds involved in central carbon and phospholipid metabolism – 13C NMR: flux analysis |
• Quantitative technique • High level of reproducibility and instrument stability • Low experimental variability between laboratories • Nondestructive • Minimal sample preparation of biofluids or analysis of intact tissue using 1H magic angle spinning NMR |
• Low sensibility • Requires higher amount of sample • Lower metabolite coverage |
Abbreviations: MS, mass spectrometry; NMR, nuclear magnetic resonance.
Widely used in the structural characterization of small organic compounds, NMR spectroscopy is a robust and reliable technique for the application of metabolomics. The main analytical characteristics of this technique are its inherently quantitative character, high level of reproducibility, and instrument stability.78,81–83 NMR is a nondestructive technique and the analysis of biofluids requires minimal sample preparation, often limited to dissolving the sample in a buffered solution. However, the analysis of tissue samples requires further sample preparation, to obtain hydrophilic and lipophilic fractions of the tissue extracts. Both fractions can be analyzed by 1H NMR84 and 31P NMR85 spectroscopy, with the latter enabling specific quantification of phosphorylated compounds, such as those involved in central carbon metabolism and phospholipid metabolism.
The number of MS-based metabolomics studies is rapidly increasing, and it is currently the most widely used technique for the analysis of metabolites, mainly due to its sensitivity and versatility. Based on the analysis of gas-phase ions, the metabolite coverage is not only dependent on sample extraction methodology, but also on the choice of ionization technique (for ion formation), mass analyzer (for ion selection), or acquisition mode.78,86 However, as ionization efficiency differs among compounds due to their chemical structure, MS is not equally sensitive for metabolites.87
Mass spectrometers can be coupled to separation techniques or use direct flow injection. Without prior separation, direct infusion (shotgun) MS is valuable for high-throughput screening studies as it reduces the time of analysis and improves inter-sample reproducibility. However, this approach cannot differentiate between isomeric compounds and is subjected to ion suppression effects.87 These technical limitations can be mitigated by interfacing MS with separation techniques. Among them, liquid chromatography (LC) coupled to MS is the gold standard, particularly for lipidomics, with high sensitivity and dynamic range. As for NMR analysis, LC-MS analysis of hepatic tissue first requires prior metabolite extraction of the homogenized tissue. The main characteristics of the most common analytical approaches for MS-based metabolomics are summarized in Table 2.
TABLE 2.
Most common analytical methodologies for mass spectrometry-based bulk metabolomics
| Method | Description | Advantages | Limitations |
|---|---|---|---|
| Direct infusion (shotgun) MS | • Direct infusion of sample extracts to the MS detector • Mainly used as a fingerprinting method • Relative quantification • Used with low-resolution, high-resolution MS, or tandem MS instruments |
• Increased analytical throughput • Increased intersample reproducibility |
• Matrix-dependent signal suppression or enhancement in the analysis of complex samples; ion suppression may lead to decreased sensitivity, especially of less abundant species • Difficulties in resolving isobaric metabolites (compounds with the same nominal mass) in low resolution MS • No resolution of isomeric compounds |
| LC-high resolution MS | • LC interfaced with the MS detector: metabolites are separated before their detection • Nontarget analysis or broad profiling of aqueous and lipid metabolites, typically covering 400–2000 compounds |
• High sensitivity • Broad coverage of metabolites or tailored analysis associated with specific sample preparation, separation method • Ideally suited to lipidomics • Possible combination of relative (semiquantitative) and quantitative analysis of some selected metabolites • Separation of isomeric compounds |
• Usually semiquantitative; full quantification of all detected compounds is not possible • Matrix effect and ion suppression, although reduced compared with shotgun-MS • Generation of large amounts of data |
| Targeted LC-MS | • Typically, LC coupled with a triple quadrupole MS detector for the analysis of preselected metabolites | • High selectivity and sensitivity • Approach of choice for quantitative analysis • Suitable for the analysis of metabolites at low concentration |
• Information of preselected compounds only • Specific sample preparation focused on the selected metabolites |
Abbreviations: LC, liquid chromatography; MS, mass spectrometry.
While the strengths of NMR are its reproducibility and direct quantification, NMR has lower sensitivity than MS, which limits its application to detect lower abundance metabolites. However, despite the high sensitivity of MS, the relative intensity of the MS-measured metabolites does not necessarily correlate to absolute concentrations, being dependent on ionization efficiency and ion suppression effects in complex matrices. Considering their advantages and drawbacks, both techniques are highly complementary. However, NMR and LC-MS metabolomic approaches provide a bulk analysis of homogenized liver tissue or cell cultures, and cannot determine the spatial localization of the metabolites.
Spatial metabolomics and emerging technologies
The first application of MSI on the tissue was carried out in 1997 by Caprioli et al.88 Subsequently, there have been substantial technological advances and an ever-increasing range of applications.6,89–92 MSI can measure endogenous molecules, such as lipids, nucleotides, peptides, and intact proteins, and study drug uptake and distribution.93 Recent improvements in instrument design have enabled single-cell and subcellular imaging,94–97 and 3D molecule imaging.94,98
Technology overview
MSI instrumentation has two main functions: first, the generation of ions from biomolecules in the tissue; and second, the separation and detection of the generated ions (Figure 2). There are several means of generating ions for MSI, with secondary ion mass spectrometry, desorption electrospray ionization (DESI), and matrix-assisted laser desorption ionization (MALDI) being the most frequently used (Figure 2A).
FIGURE 2.
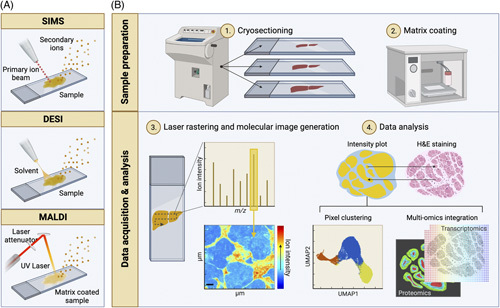
Mass spectrometry imaging (MSI) modalities and typical workflow. (A) Schematic drawing of the main ion sources used in MSI. From left to right; secondary ion mass spectrometry (SIMS), desorption electrospray ionization (DESI), and matrix-assisted laser desorption ionization (MALDI). In SIMS, a primary ion beam is applied to sputter secondary analyte ions off the sample. DESI uses an electrically charged stream of solvent, which is sprayed over the tissue surface to desorb analyte ions. To aid ionization, MALDI requires the coating of the sample with an energy-absorbing matrix. A laser is then rastered over the sample to desorb and release analyte ions. (B) A typical workflow for MALDI-MSI. (1) Frozen tissue specimens are cryosectioned, mounted onto glass slides, and (2) coated with an energy-absorbing matrix such as 2,5-dihydroxybenzoic acid using a robotic sprayer. (3) Once loaded on the instrument, a laser beam is rastered across the tissue on a pixel-by-pixel basis, producing a mass spectrum per (x, y) coordinate or pixel. For any m/z value, a heatmap can be generated by mapping the ion intensity values across the coordinates analyzed. (4) The resulting MSI data set is customarily interrogated in the context of the specimen histology (top). Spatial segmentation can be carried out by clustering pixels based on their spectral similarity (bottom left). MSI can also be integrated with other molecular imaging modalities such as immunofluorescence, imaging mass cytometry, and spatial transcriptomics (bottom right). Created with BioRender.com.
In secondary ion mass spectrometry, a focused primary ion beam is fired at the tissue under ultrahigh vacuum conditions. Gold, gallium, bismuth, and fullerene (C60) cations are used as primary ions. Applying these over the tissue induces the generation and sputtering of secondary ions. Secondary ion mass spectrometry offers the best spatial resolution (nanometer) capabilities that can reach subcellular distributions; however, the high-energy beam may induce molecular fragmentation, making this technique more commonly used to study low molecular weight or inorganic species.
DESI is the so-called ambient ionization technique, meaning that no sample preparation is required, and ions are formed at atmospheric conditions. During DESI, an electrically charged solvent stream is sprayed at the tissue surface while the X-Y stage, holding the sample, moves. Ions are desorbed from the surface and enter the mass spectrometer through the inlet capillary. DESI has the advantage of being a “soft” ionization method; therefore, intact molecules can be readily analyzed. The pixel-to-pixel resolution achieved using DESI is typically 50–200 µm. However, higher spatial resolution is achievable with newer designs and/or over-sampling methodologies.99
MALDI first requires the application of an organic chemical matrix to the tissue, which aids ionization and desorption of the analyte. MALDI can be performed under vacuum or at atmospheric pressure. Typically a laser (eg, Nd:YAG; N2) is rastered across the tissue of interest. The matrix absorbs the laser energy and through localized disintegration and charge transfer, analyte ions are released. Matrix adducts can give rise to interfering peaks in the low mass to charge (m/z) range; in addition, the process of applying the matrix can introduce variability. However, MALDI tends to offer higher spatial resolution than DESI, typically in the range of 5–20 µm, but recent advances have seen a lateral resolution of less than 2 µm.98,100
Following ionization, ions enter the high vacuum region of the mass spectrometer, where they are separated in the mass analyzer before detection. The choice of mass analyzer will depend on the required sensitivity, mass resolution and accuracy, scan speed, and the need for ion fragmentation. The highest mass resolving power can be achieved with a Fourier transform ion cyclotron resonance mass spectrometer, whereas time-of-flight mass spectrometers offer fast scanning speeds but lower mass resolution. Excellent mass accuracy can be achieved using Orbitrap mass spectrometers; when coupled with a quadrupole or linear ion trap mass analyzer, this enables tandem MS (MS/MS) for selected fragmentation of ions to aid metabolite assignment.
Workflow
The typical workflow for MSI (Figure 2B) would first involve tissue collection and preservation. Fresh frozen tissue is preferable over formalin-fixed and paraffin-embedded tissue as the molecular availability is decreased, and deparaffinization may give rise to the loss of specific molecular species such as lipids.101 Next, tissue is cut into slices using a cryomicrotome (10–20 µm thickness). The use of an “optimal cutting temperature” compound for embedding before cryosectioning is to be avoided as this gives rise to ion suppression and background signal on the mass spectrometer. When samples are fragile, alternative MS-compatible embedding agents may be used, such as gelatin or a hydrogel matrix composed of hydroxypropyl methylcellulose and polyvinylpyrrolidone.102 Tissue sections are thaw mounted onto glass microscope slides.
Before MALDI-MSI, it is necessary to apply a matrix to the tissue surface. These are small organic compounds, which aid the ionization and desorption of the analyte. In positive ion mode, 2,5-dihydroxybenzoic acid and α-cyano-4-hydroxycinnamic acid are popular choices, whereas, for negative ion mode, 9-aminoacridine and 1,5-diaminonaphthalene are commonly used.103 The matrix application method is critical as this needs to be applied homogeneously, forming small crystal sizes, and without introducing any delocalization effects in the sample. The main approaches include sublimation and robotic spraying, the latter being the most common method.
Data analysis
During MSI, biomolecular species are ionized across a tissue slice at distinct points in a grid-like manner, generating complex data sets, where each coordinate or pixel is associated with a mass spectrum, a list of m/z values, and their corresponding intensities. These data sets are large and complex due to their high dimensionality. Data are converted into a universal (imzML) format, which can then be interrogated with commercial or open-source software or parsed into a programming language of choice for further analysis. Raw data requires preprocessing steps, such as “binning,” baseline correction, normalization, smoothing, and potentially transformation and scaling.89 Data processing pipelines can plot the intensity for a single m/z value across the area sampled. This results in a heatmap image, which shows the abundance of the selected species across the tissue, with spatial resolution governed by the acquisition of pixel-to-pixel distance and area ablated. This “single ion” image can be compared or overlaid with histological staining of an adjacent tissue slice to match the ion images with tissue architecture. Alternatively, where acquisition has been nondestructive (eg, DESI), the same section can be stained after MSI, which aids image co-registration.104 Analysis of MSI data can also be performed on the entire data set with dimensionality reduction (eg, principal component analysis, t-distributed stochastic neighbor embedding, uniform manifold approximation, and projection) or clustering pixels with similar spectral properties.6 Molecular identification is one of the major bottlenecks in metabolomics; annotation is facilitated by matching experimentally obtained accurate mass with databases such as the LIPID MAPS Structure Database105 and the Human Metabolome Database.1 Information from isotopic patterns and adducts can add confidence in metabolite annotation, which is exploited in Metaspace.106
Multimodal imaging
The combination of multiple MSI methods applied to the same sample can offer complementary information and significantly extended molecular coverage.107 MSI can be integrated with numerous analytical modalities such as microscopy, imaging mass cytometry, and spatial transcriptomics. This is referred to as multimodal imaging.102,108 Classically, MSI data has been interpreted in conjunction with morphological assessment through histological staining such as hematoxylin and eosin and visualization by optical microscopy. In addition, immunohistochemistry and immunofluorescence allow for comparing protein expression and MSI-detected metabolite distribution.109,110 However, the availability of chromogens or fluorophores is a limiting factor for multiplexing. By coupling laser ablation to elemental MS, imaging mass cytometry allows the detection of up to 40 targets labeled with metal-tagged antibodies.111 Interpretation of MSI data in the context of spatially resolved endogenous cell markers can provide a powerful strategy to elucidate metabolic microenvironments and their relationship to distinct cell subpopulations. Furthermore, the integration of MSI (spatial metabolomics) with rapidly developing spatial transcriptomics and proteomics methodologies can facilitate multiomics for a spatial systems understanding of cell and tissue organization in the liver.112
APPLICATIONS IN LIVER DISEASE
With ~2 million deaths per year worldwide, liver disease represents a major economic and societal burden that is expected to increase in the next years. Liver disease includes a broad spectrum of pathologies including alcohol-associated liver disease, NAFLD, viral hepatitis, cholangitis, cirrhosis, and HCC, among others.113 While viral hepatitis constitutes the leading cause of acute liver disease, the generalized adoption of the so-called Western diet is contributing to the swift rise of NAFLD as the leading cause of chronic liver disease, progressing to HCC and becoming the number 1 indication for liver transplantation.114
In recent years, the dawn of metabolomics and lipidomics has brought novel tools to the understanding of liver disease, including the pathophysiology of NAFLD. Changes in the liver lipid composition associated with NAFLD development have been characterized115 and used to establish NAFLD plasma signature.116 Considering that around 40% of cases with NASH progress to liver fibrosis, a significant risk factor for HCC development117 understanding the molecular changes at the metabolic and lipidic levels is paramount for disease diagnosis and management, where metabolomic-based and lipidomic-based approaches have been increasingly used.118–120 By combining genetic and dietary murine models of NAFLD, the role of receptor-interacting protein 3 in NAFLD pathophysiology was demonstrated,121 which impacted necroptosis and hepatic lipid profiles associated with NAFLD.122,123 Animal models have also been used to deepen the knowledge of organ-organ communication and its impact on liver metabolism, where the relevance of the gut microbiota is emerging. Changes in mouse gut microbiota and metabolites can drive NAFLD progression to HCC.124 Moreover, cecum and liver metabolic signatures have been described in obese mice using the high-fat diet (HFD) model.125 Interestingly, circulating microRNAs are known modulators of metabolic profiles45,126,127 and might also influence gut microbiota,128–130 which in turn may alter the systemic metabolome. Therefore, it is crucial to localize the origin of specific metabolites to precisely address the influence of each factor on liver disease onset and progression.
Metabolic liver disease
The application of spatial metabolomic methodologies such as MALDI-MSI aids in the understanding of metabolic zonation in the liver tissue and metabolite spatial distribution changes associated with (patho)physiological processes (Table 3). Many of these efforts have focused on NAFLD and its progression to NASH.131,133,134 Changes in specific phospholipid distributions were linked to the upregulation of enzymes involved in membrane remodeling and eicosanoid formation.133 This resulted in the exacerbation of inflammation contributing to regional oxidative damage in NASH. In another study, alterations in N-linked glycosylation were linked to histopathological features of NASH, with an increase in fucosylated and high mannose glycans in the fibrotic and steatotic regions of the tissue, respectively.143 MALDI-MSI has also been used to differentiate macro and micro steatosis in the human liver by their constituent lipids. Macrosteatosis was found to be enriched in saturated triglyceride species, indicating that the origin of these may be from de novo lipogenesis.135
TABLE 3.
Application of spatial metabolomics in the study of liver pathophysiology and its implication in zonation
| Sample/study description | Technique | Metabolic differences | Year | References |
|---|---|---|---|---|
| Liver tissue from obese normal, simple steatosis and NASH of class III obese women | MALDI-MSI | • Zonal distributions for 32, 34, and 36 carbon PC in controls lost in simple steatosis or NASH | 2013 | Wattacheril et al131 |
| Primary liver cells from ethanol-treated and silymarin-treated mice | MFD-MD | • Hydrogen peroxide, glutathione, and cysteine were detected simultaneously with high specificity and sensitivity • Identification of significant cell diversity or cell heterogeneity among primary liver cells |
2016 | Li et al132 |
| Liver tissue from mice under HFD, WD, and MCD; human liver biopsies from control, simple steatosis, NASH, and patients with cirrhosis | MALDI-MSI | • Zonal location of arachidonic acid-containing phospholipids; loss of lipid zonation in NASH | 2017 | Hall et al133 |
| Fresh frozen human liver biopsies from obese subjects undergoing bariatric surgery with various degrees of NAFLD | MALDI-TOF MSI | • Marked differences between spatially resolved lipid profiles from nonsteatotic and steatotic tissues • PI and arachidonic acid metabolism in nonsteatotic regions • LDL and VLDL metabolism associated with steatotic tissue |
2018 | Ščupáková et al134 |
| Human liver biopsies from control, simple steatosis, NASH, and patients with cirrhosis | Au-CBS-LDI MS | • Differential distribution of TGs: in low steatosis levels, TG accumulated around the pericentral zone • Macro lipid droplets hepatocytes enriched in fully saturated TG • In NASH and cirrhosis biopsies, TG enrichment observed in ballooned areas |
2019 | Alamri et al135 |
| Liver tissue from mice exposed or not to a single dose of B[a]P | MALDI-MSI | • B[a]P exposed mice with an altered abundance of PI, PC, TG, PE, LysoPEs, LysoPC, FFA, and eicosanoids | 2020 | Li et al136 |
| Liver tissue from mice under HFD | (Au)LDI−MS | • The lipogenic activity is shown to be regiospecific and not always associated with the TG pool size in a given region | 2020 | Downes et al137 |
| OCT-embedded and snap-frozen wild-type mouse and human fibrotic liver tissues | TOF-SIMS | • Different ion species associated as metabolic markers for different liver cell types: – hepatocytes: m/z 255, 279, and 281 – endothelial cells: m/z 60, 76, and 77 – KCs: m/z 134, 181, and 91 • Hepatocytes subclassified by C1–C4 each presenting different metabolic fingerprints: • hepatocytes C1 localized around the CV and near fibrotic boundaries, and associated with metabolic markers m/z 69, 55, and 57 |
2021 | Yuan et a.138 |
| Differentiated human hepatocytes cell line; liver tissue from mice under WD | MALDI-MSI | • Steatotic hepatocytes presented aberrant accumulation of lipid droplets and neutral lipids (TG and DG), and glycerophospholipid ER-enrichment • Inflammatory steatotic hepatocytes with increased sphingomyelins |
2021 | Rappez et al139 |
| Liver tissues from mice exposed to third-hand smoke treated or not with antioxidants | LDI-MSI | • Third-hand smoke-exposed mice presented increased TG and decreased PC and SM lipid species, accumulating in larger and more abundant lipid droplets • Third-hand smoke mainly dysregulated glutathione metabolism, D-glutamine and D-glutamate metabolism, and oxidative phosphorylation |
2021 | Torres et al140 |
| Liver tissue from mice under HFD treated with Eurycoma longifolia | DESI-MSI | • E. longifolia downregulated lipid accumulation and FA biosynthesis, and induced changes in amino acids, organic acids, phospholipids, and glycerolipids | 2021 | Zhang et al141 |
| Liver tissue from mice exposed to aristolochic acid I | Atmospheric pressure-MALDI-MSI | • Aristolochic acid I-induced hepatotoxicity associated with changes in taurine, hypotaurine, glycerophospholipid, D-glutamine, D-glutamate, and arachidonic acid metabolisms | 2021 | Guo et al142 |
| Liver tissues from mice under LFD, HFD, and WD; human FFPE liver biopsy samples from NASH patients | MALDI-MSI | • N-glycan structures upregulated in portal triad area • N-glycosylation modifications correlated with fibrosis score and liver histology changes |
2022 | Ochoa-rios et al143 |
| Liver tissue from mice under fasting conditions, ND, and HFD | MALDI-MSI | • Hepatocyte FA enrichment dependent on proximity to vasculature • HFD disrupted tissue metabolic compartmentalization • Lipid droplets accumulated away from the vasculature and glutathione increased in extravascular regions • HFD associated with oxidative stress, PPP, and purine metabolism |
2022 | Stopka et al70 |
Abbreviations: (Au)LDI-MS, gold-laser desorption/ionization mass spectrometry; Au-CBS-LDI MS, sodium-doped gold-assisted laser desorption ionization mass spectrometry; B[a]P, benzo[a]pyrene; CV, central vein; DG, diglyceride; ER, endoplasmic reticulum; FA, fatty acid; FFA, free fatty acid; FFPE, formalin-fixed paraffin-embedded; HFCD, high-fat choline-deficient diet; HFD, high-fat diet; HFHD, high-fat high-calorie diet; LFD, low-fat diet; MALDI, matrix-assisted laser desorption/ionization; MCD, methionine–choline-deficient diet; MFD-MD, multicolor fluorescence detection-based microfluidic device; MSI, mass spectrometry imaging; OCT, optimal cutting temperature compound; PC, phosphatidylcholine; PE, phosphotidylethanolamine; PI, phosphatidylinositol; PPP, pentose phosphate pathway; SIMS, secondary ion mass spectrometry; TG, triglyceride; TOF, time-of-flight; WD, Western diet.
As referred to previously, liver zonation contributes to specific protein, glucose, and lipid metabolism patterns across the tissue. In 2013, using this evidence and the possible role of phosphatidylcholine in NASH pathogenesis, phosphatidylcholine specimens with 32, 34, and 36 carbons were found to be decreased in simple steatosis and NASH patients compared with controls. Using MALD-MSI, the authors determined that the distribution of these phosphatidylcholines was highly zonated and that this feature was lost in simple steatosis and NASH. This work highlighted the importance of phospholipid zonation and its implications for the onset and progression of NAFLD.131 In line with this, zonation of lipid species in human and murine NAFLD was linked to the colocalization of calcium-dependent lysophosphatidylcholine acyltransferase 2 enzyme. Lysophosphatidylcholine acyltransferase 2 enzyme had a higher expression adjacent to pericentrally located lipid droplets and infiltrating macrophages, suggesting an interplay between lipid zonation and macrophage infiltration in liver disease progression.133
An HFD is known to alter the all-body metabolome, which in turn influences interorgan communication. Using an HFD mouse model, metabolic dysregulation was analyzed in a variety of tissues including adipose and liver, before and after HFD. White and brown adipose tissue and liver suffered higher metabolomic alterations when comparing HFD with the chow diet. In liver tissue, carbohydrates were the main metabolites reaching 53% of all metabolites on chow contrary to only 8% on HFD, while on the other hand lipids rose from 11% to 52% on the chow diet and HFD, respectively. Overall, this study contributed to mapping the interorgan communication within selected tissues after HFD and during the mouse circadian circle, which was essential for the chronological reconstruction of the complex metabolome across tissues.144 In another study, spatially resolved lipids were measured by MALDI-MSI in the liver tissue with varying degrees of NAFLD progression. A profound difference was shown between steatotic and nonsteatotic zonated liver tissues. Nonsteatotic regions were predominantly associated with phosphatidylinositols and arachidonic acid metabolism, while steatotic areas were more prone to metabolize low-density and very LDLs.134 Using human liver samples with no alterations, simple steatosis, steatosis, and cirrhosis, saturated triglycerides were found to accumulate in hepatocytes with macrovesicular steatosis. During short-term steatosis, this effect might be beneficial as it may protect cells against high levels of saturated fatty acids. However, if this becomes chronic, macrovesicular steatotic areas enriched with saturated triglycerides will undergo ballooning, suggesting a crucial role for saturated triglycerides in the development of NASH.135
In the same line, to understand how lipid metabolism modulates liver steatosis, sodium adduct ions were used along with gold and stable isotope tracer protocol coupled with laser desorption ionization–mass spectrometry (LDI-MS) to capture the spatial characterization of triglyceride isotopic patterns (lipogenic flux) at 50 μm spatial resolution. Gold LDI−MS showed, in an in vivo model of diet-induced obesity mice, an increase in lipogenesis with the highest average isotope ratio observed in chow-fed mice across 2 triglycerides (TG 52:2 and TG 54:3) and 1 cholesterol ester (CE 18:1). Indeed, lipogenic activity was region specific; however, contrary to what would be expected, this activity was not determined by triglyceride pool size.137
Recently, another important discovery was the spatial distribution of abundances of released N-glycans during NASH. Using both human samples and mice models, a correlation was identified between fibrotic and fatty areas within the tissue and a significant alteration of complex/fucosylated glycans and high mannose glycans. Here, the authors suggested that during early stages of liver disease, significant changes in the core fucosylation might drive liver damage progression to NASH.143
The spatial single-nuclear metabolomics method was the first to allow the segmentation and analysis of a single nuclear metabolic profile directly on tissue sections. This new multiscale spatial resolution platform combines experiments and computational algorithms to quantitatively characterize the metabolic intracellular or intercellular features. In a recent study, spatial single-nuclear metabolomics was applied to mice liver and human fibrotic liver samples.138 In mice, 9 zones were arithmetically determined from the CV to the periportal vein, and 6 metabolic markers showed a gradient decrease from the CV to zone 9. In human fibrotic liver tissue samples, 7 subpopulations were identified (3 hepatocytes, KCs, immune cell, fibroblast, and endothelial cell populations). Among the 3 different populations of hepatocyte subtypes, the differential metabolic states were associated with spatial localization. Interestingly, one subtype is closer to the fibrotic tissue, proving that the proximity to the fibrotic tissue alters the metabolic profile of the neighbor hepatocytes.138
Alexandrov and his team have recently developed a new method for spatially resolved single-cell metabolomics that integrates metabolic profile, morphometric, and fluorescence intensities, named SpaceM.139 By combining light microscopy and MALDI-MSI, they characterized the metabolic states of lipid-stimulated hepatocytes in an inflammatory environment, revealing populations with distinct metabolic states that were constituent with murine models of NASH. This method can be used for a wide range of adherent cell cultures and enables the analysis of thousands of cells, each sampled in situ. The use of SpaceM in human hepatocyte fatty acids allowed the separation of hepatocytes into 2 distinct subpopulations, high and low steatotic levels. Among the high-fat hepatocyte subpopulation, it was possible to distinguish between “benign” and “inflammatory” steatotic hepatocytes, as both presented different metabolic profiles, with ceramide phosphocholines (sphingomyelins) being highly enriched in the “inflammatory” steatotic hepatocytes. Thus, the SpaceM method allows single-cell metabolomics of in vitro cell cultures and contributes to deciphering in vivo results.
Spatial liver metabolomics was recently used in another study to achieve broad coverage of central carbon, nucleotide, and lipid metabolism pathways.70 A workflow was implemented to prepare tissue specimens for MALDI-MSI. This technique allowed the visualization of the liver zonates up to 30 μm pixel resolution and enabled the identification of the disruption of highly organized metabolic liver zonates in mice. This method was validated using 2 mice models, fasted versus nonfasted and regular chow diet versus HFD. In the first model, hepatocyte fatty acid content within the liver microenvironment was specific to proximity to the vasculature under nonfasting conditions. Specifically, fatty acid docosahexaenoic and arachidonic acid were spatially distributed, with docosahexaenoic closer to the blood vessels opposing to the arachidonic acid location. Under fasting conditions, this feature was abolished, with increased docosahexaenoic and decreased glycolytic intermediates, indicating a metabolic switch from glucose to lipid metabolism. During HFD, the pentose phosphate pathway and purine metabolism were increased, which represents a response to increased oxidative stress through NADH production, supported in part by the detection of glutathione in extracellular tissue areas. Reinforcing the unbalance in the redox state is the increase in purine metabolism, which can be triggered to repair DNA damage. This technique revealed new perspectives of looking at spatial liver zonation to better understand how liver cells cope with different nutrient supplies and how it modulates liver disease.70
The alleviation effect of the Asian herb Eurycoma longifolia on HFD-fed mice resulted in weight loss with beneficial effects for the treatment of NAFLD, namely enhanced decomposition and inhibition of accumulation of lipids.141 Using DESI, HFD mice treated with E. longifolia showed an altered metabolic profile, mainly affecting the levels of amino acids, organic acids, phospholipids, and glycerolipids. Combining these results with optical images of hematoxylin and eosin–stained liver sections exhibiting cell-originated imaging signal, extracted heatmaps, and differential metabolites, the authors demonstrated the importance of the metabolomics approach in understanding the effect of a therapeutic approach through metabolite analysis and bioactivity assessment.141
Toxin-induced liver injury
As one of the main organs for blood clearance, the liver is highly impacted by environmental toxins, such as third-hand smoke, that cause liver metabolic dysfunction. To disclose the intracellular impact of third-hand smoke, an untargeted metabolomics multiplatform method, including NMR, LC coupled to high-resolution MS, and LDI-MSI, was applied in mice liver samples.140 In third-hand smoke-exposed mice, this strategy allowed the detection of 88 significant metabolites included in 21 metabolic pathways, among which D-glutamine and D-glutamate metabolism, glycerophospholipid metabolism, oxidative phosphorylation, and glutathione metabolism, being the last 2 related to oxidative stress. These were associated with increased lipid accumulation and choline deficiency, mimicking NAFLD and steatosis. Indeed, the lipid droplet layer in liver tissue of third-hand smoke-exposed mice was formed by phosphatidylethanolamine (36.6) and phosphatidylcholine (36.6), while TG (52:6), TG (54:7), and TG (52:6) were located at the center of the lipid droplet. Antioxidant treatment, in turn, reduced TG droplet content but not phosphatidylethanolamine (36.6) or phosphatidylcholine (36.6) droplet layers.140 This agrees with the fact that both lipid accumulation and the specific lipid content are crucial to aggravate liver disease.
In another study, the effects of benzo[a]pyrene, a polycyclic aromatic hydrocarbon known to have detrimental effects on human health, was evaluated on the liver through the combination of LC-MS-based and MALDI-MSI-based lipidomics platforms.136 Hepatic lipid metabolic disorders induced by intratracheally instilled polycyclic aromatic hydrocarbons were investigated, and distinctly altered glycerophospholipids, glycerolipids, and fatty acid metabolism were found in the mouse liver, with increasing triacylglycerol, phosphatidylinositol, and phosphatidylcholine, and decreasing lysophosphatidylcholines, lysophosphatidylethanolamine, free fatty acids, and eicosanoids, thus concluding that benzo[a]pyrene might induce NAFLD and contribute to hepatocyte membrane injury and inflammation.136
The use of weight-loss pills containing aristolochic acids, a chemical derived from Aristolochia plants, was highly detrimental to human health, having been related to urothelial and renal diseases and cancers. Atmospheric pressure MALDI-MS was used to analyze metabolic alterations in aristolochic acid I-induced liver damage, which found a high abundance of small metabolites combined with lower abundances of lipid species compared with the control.142 Indeed, glycerol phosphate, glyceryl-phophorylethanolamine, and taurocholic acid exhibited significant increases, while phosphatidic acids, phosphatidylethanolamine, and phosphatidylinositols were considerably reduced in the liver after aristolochic acid I exposure. Machine learning metabolomics pathway analysis allowed the authors to observe evidence of altered taurine and hypotaurine metabolism, glycerophospholipid metabolism, D-glutamine and D-glutamate metabolism, and arachidonic acid.142
Multicolor fluorescence detection-based microfluidic device for single-cell metabolomics was another technique that improved the understanding of alcohol-associated liver disease. A device with 2-laser excitation and 3-channel fluorescence has been developed to analyze liver cells from acute ethanol-stimulated mice.132 This technique enabled the study of oxidative stress through single-cell analysis of H2O2, GSH, and Cys, revealing a high metabolic variance within primary liver cells, while accurately improving our view of the intercellular oxidative/antioxidative molecular mechanism in response to external stimuli.132
Liver cancer
The heterogeneity of cancer cells within a specific tissue offers several potential targets for the use of spatial metabolomics. Using HCC patient samples, a recent study evaluated tumor heterogeneity using mass spectrometry-based proteomics and metabolomics, single-cell analysis, whole-exome sequencing, cytometry by time-of-flight, and RNA sequencing.145 Although high heterogeneity was described with the use of genomes, transcriptomes, proteomes, and metabolomes, less variability was found within the HCC microenvironmental immune status. Indeed, 3 novel HCC immunophenotypic subtypes were identified, including immunocompetent (subtype 1), immunodeficient (subtype 2), and immunosuppressive (subtype 3). Further, the metabolome was the best correlator for the immunome subtype contrary to transcriptome or proteome. Subtype 1 was characterized by upregulated urea cycle, subtype 2 showed increased activity of nucleotide biosynthesis, and subtype 3 showed inhibited glycolysis and enhanced mitochondrial respiration.145 Thus, through a multiomics approach, this study dissected tumor heterogeneity, revealing the landscape of tumor cells and the spatial microenvironment.
The studies highlighted above reflect only the beginning of an exciting era in spatial metabolomics and its application to liver biology and disease pathology.
CHALLENGES AND FUTURE DIRECTION
The successful adoption of spatial metabolomics to characterize pathobiological phenomena in situ and, more specifically, in the liver pathology space, depends on the collaboration between (i) technology developers to improve the sensitivity, coverage, and resolution of MSI and analysis of metabolites in specific regions of tissue, at the edges of lesions or at single-cell resolution; (ii) experimentalists knowledgeable of the key biological unknowns, able to design meaningful experiments about specific biological and pathobiological problems to generate the “ground truth data”; and (iii) experts in data science needed to develop artificial intelligence (AI), machine learning, or deep learning algorithms to deal with the vast amount of information generated by the MSI technology.6
MSI is still suboptimal due to the overlap of signals, noise, and variability of spectrum intensities. Also, there are important issues in data handling and signal processing, data storage and export, comparability of data, and the need to optimize for each type of tissue.6,146 However, there is a considerable incentive to solve these problems because of its potential large market in biomedicine and, more specifically, in liver disease with potential relevance for diagnosis, treatment, and prognosis in NAFLD/NASH and liver cancer.
In the context of liver disease, the great opportunity is the convergence with current efforts in molecular digital pathology. Spatial metabolomics is well positioned to contribute to cancer simply because there are high-quality histology data that, if combined with spatial metabolomics, could be used in supervised AI, connecting in situ specific metabolic signatures with already validated clinical, histopathology, and molecular profiling as well as documented responses to treatments with known mechanism of action. This structure of data permits AI-based supervised approaches, as the availability of the outcome allows labeling of the cases required for supervised algorithms to be predictive, according to the metabolic signature of specific cells, with inferences about prognosis and potential treatments. The viability of this approach might be enhanced by the sufficient availability of data to train the machine and by independent data validation.
Spatial metabolomics might generate unique insights on cancer treatments147–150 based on characterizing the vulnerabilities resulting from specific tumor cell reprogramming as well from the adaptation of the ancillary supportive systems (eg, vascularization, immune) supporting cell growth, proliferation, differentiation, and other factors that affect cancer progression. Identifying the cell-specific metabolic signatures in typically heterogeneous tumors might identify cell-specific Achilles heel tumor metabolism, providing the rationale for relatively targeted therapeutic interventions based on potential metabolic vulnerabilities of selected types of cells. Thus, a higher-throughput spatial metabolomics imaging method is a technological challenge but might add enormous value to diagnosis, prognosis, and personalized treatment.
The approach to NAFLD/NASH seems more complex5 at this stage as its diagnosis is usually based on histopathology, and evolution takes a long time with insufficient well-characterized intermediate diagnostic time points and no biomarkers indicating the likelihood of progression toward more severe stages or early responsiveness to treatments. There is an opportunity to address these shortcomings by combining digital pathology with spatial metabolomics to identify heterogeneity behind the current boundaries defined by morphological-based diagnostic stages. The combination of digital pathology and spatial metabolomics aims for better stratification of the patients for diagnostic, prognostic, and therapeutic approaches. This approach is, in principle, technologically feasible, but it requires upscaling the information available, training on the technology for this indication, increasing the technological demand, and hopefully adjusting the prices to make the technology more broadly available. This is required to generate background data required to accelerate the development of the data analysis tools able to make sense of the NAFLD/NASH information. In some way, the experience in the cancer field might contribute to facilitating transfer to NAFLD/NASH.
Spatial metabolomics can also successfully thrive thanks to the current success of other spatial omics technologies. Laser capture microdissection-based, in situ hybridization imaging-based, or spatial barcoding-based transcriptomics have successfully been applied to study liver physiology and disease biology.151 Although the spatially resolved study of the proteome remains limited by the existence and performance of antibodies, great strides have been made toward the improvement of the technology multiplexity. These mainly consist of the utilization of MS to image photocleavable, metal, or peptide-tagged antibodies.152 Nonetheless, such strategies introduce a bias resulting from the choice of the antibody panel and do not suffice for proteome-wide studies. However, the current throughput could potentially be increased to proteome-wide using MALDI-MSI to analyze protease-treated specimens.153 One of the greatest challenges remains the coupling and integration of the technologies hitherto discussed, although multimodal MS has already successfully been applied to define liver metabolic zones and single-cell identity beyond histological annotation (Tian et al. unpublished data, 2022).
The explosion of data at single-cell resolution also entails a key event on the progress of spatial metabolomics. Although cells are part of their niche and do not work in isolation, single-cell studies have confirmed the existence of heterogeneity among specific types of cells in healthy liver, NAFLD, NASH, and in cancer, highly determined by their topographic location in the functional liver structure. Characterization of the functional relevance of cellular heterogeneity would help to understand better the natural history of NAFLD/NASH or liver cancer and more specifically about their diverse trajectories.
Identifying specific subsets of immune cells or SCs in situ with unique transcriptomes, proteomes, and functionality, raises the unresolved questions of which, how, and where the different subtypes of cells might accumulate and exert their unique functional roles influencing the progression of the disease. Associating cellular metabolomic information with their spatial context might be used to deconvolute each subset of cells according to their metabolome as a surrogate of their functional state or capabilities. Moreover, learning about the metabolites in different types of cells, in their specific location in the liver combined with other molecular data at single-cell resolution opens an excellent opportunity to learn about the pathogenesis of the disease, unexpected cellular cross talks, chronological sequence of cellular events and hot spots, triggering the initiation of fibroinflammatory changes in response to hepatocyte injury. From a metabolic perspective, it should be possible to identify “primary movers” triggering irreversible fibroinflammatory responses representing putative targets that, if contained, might limit the progression of NASH, accelerated tumoral growth, or dissemination. Moreover, by coupling spatial cellular information with the metabolic fingerprints of specific types of cells, it might be possible to do targeted nutritional/metabolic substrate-based treatments, cell therapy to selectively enrich some cells, or gene therapy to lockdown the configuration of specific subtypes preventing the progression of the disease. This information might help to identify essential cross talk within the niche cells in each zone and devise novel approaches aiming at restoring their functional homeostasis.
The success of spatial metabolomics depends on the quality of the data and their analysis. Whereas the technological revolution leading to MS-based spatial imaging started 20 years ago, the main innovation of the last 10 years has occurred in the domain of AI-based data analysis. This is easily understood if we consider AI development depends on data availability. The amount of data generated, and their complexity are useless without the parallel development of powerful tools to handle, analyze, visualize, and interpret them. AI offers a unique opportunity, but it departs from available trustable data that can be used to train the AI machine. Whereas the data related to cancer is more complete, the data available on NASH are typically not sufficient nor of standardized quality to allow supervised AI approaches to deliver the whole potential of MSI. Conversely, the scarcity of AI expertise in the context of the excessive demands from many biomedical and nonbiomedical domains creates a bottleneck for a fully developed optimized tissue-specific platform generating high-quality data quickly. For this to succeed, it is essential the connectivity between computing data analysts and experimentalists to ensure that the experimental design enables high-quality data with an appropriate structure for AI analysis. The success in using data also represents an incentive to improve the resolution of the technology by increasing the boundaries of sensitivity, time of resolution, and technology innovation. The current cost of the technology and expertise required is another brake to the broader adoption and upscaling of technology.
CONCLUSIONS
Technology is a primary driver for innovation in biomedicine. It takes years to advance the precompetitive technological development before it reaches the level of maturity required to be derisked enough to enter the clinical application and industrial adoption and upscaling. In the case of spatial metabolomics, the technology has reached enough academic maturity to penetrate the many fields in biomedicine. The liver, due to its well-defined spatial functional organization, is well suited to exploit this technology in cancer and also in highly prevalent NAFLD/NASH, providing a unique opportunity to reach landmarks in diagnosis, prognosis, and treatment. However, its definitive validation requires AI-based data analysis tools and high-quality datasets. We are optimistic concerning its future incorporation in health care, not only because of the quality of the information it generates but also because its excellent market size provides unique incentives for collaboration and open innovation between academics, the pharma industry, private and governmental funders, and technology and data-driven ventures to generate the evidence-based validation required for its global adoption.
ACKNOWLEDGMENTS
Cecilia M.P. Rodrigues is supported by grants from Fundação para a Ciência e Tecnologia (PTDC/MED-FAR/3492/2021) and La Caixa Scientific Foundation (LCF/PR/HR21/52410028). María Luz Martínez-Chantar is supported by grants from Ministerio de Ciencia, Innovación y Universidades (MICINN: PID2020-117116RB-I00 integrado en el Plan Estatal de Investigación Científica y Técnica e Innovación, cofinanciado con Fondos FEDER), Ministerio de Ciencia e Innovación (Programa Retos-Colaboración RTC2019-007125-1), Instituto de Salud Carlos III (Proyectos Investigación en Salud DTS20/00138), La Caixa Scientific Foundation (HR17-00601) and PID2020-117116RB-I00 CEX2021-001136-S CEX2021-001136-S integrado en el Plan Estatal de Investigación Científica y Técnica e Innovación, cofinanciado con Fondos FEDER. Teresa C. Delgado is funded by Ayuda RYC2020-029316-I financiada por MCIN/AEI/10.13039/501100011033 y por El FSE invierte en tu futuro. Zoe Hall is funded by the Medical Research Council, UK (MR/W019132/1) and CAMS-UK Fellowship (Analytical Chemistry Trust Fund).
CONFLICTS OF INTEREST
Cristina Alonso is employed by OWL Metabolomics (One Way Liver, S.L.). The remaining authors have no conflicts to report.
Footnotes
Abbreviations: AI, artificial intelligence; APC, adenomatous polyposis coli; CV, central vein; DESI, desorption electrospray ionization; ESI-MS, electrospray ionization-mass spectrometry; HFD, high-fat diet; HIF, hypoxia-induced transcription factor; LC, liquid chromatography; LCM, laser capture microdissection; LDI-MS, laser desorption ionization–mass spectrometry; MALDI, matrix-assisted laser desorption ionization; MS, mass spectrometry; MSI, mass spectrometry imaging; NMR, nuclear magnetic resonance spectroscopy; PV, portal vein; SIMS, secondary ion mass spectrometry.
Contributor Information
André A. Santos, Email: afasantos@ff.ulisboa.pt.
Teresa C. Delgado, Email: tcardoso@cicbiogune.es.
Vanda Marques, Email: vismsmarques@ff.ulisboa.pt.
Carmen Ramirez-Moncayo, Email: carmen.ramaorez-moncayo18@imperial.ac.uk.
Cristina Alonso, Email: calonso@owlmetabolomics.com.
Antonio Vidal-Puig, Email: ajv22@medschl.cam.ac.uk.
Zoe Hall, Email: zoe.hall@imperial.ac.uk.
María Luz Martínez-Chantar, Email: mlmartinez@cicbiogune.es.
Cecilia M.P. Rodrigues, Email: cmprodrigues@ff.ulisboa.pt.
REFERENCES
- 1.Wishart DS, Guo AC, Oler E, Wang F, Anjum A, Peters H, et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022;50:D622–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanekoff I, Sharma VV, Marques C. Single-cell metabolomics: where are we and where are we going? Curr Opin Biotechnol. 2022;75:102693. [DOI] [PubMed] [Google Scholar]
- 3.Duncan KD, Fyrestam J, Lanekoff I. Advances in mass spectrometry based single-cell metabolomics. Analyst. 2019;144:782–793. [DOI] [PubMed] [Google Scholar]
- 4.Milo R. What is the total number of protein molecules per cell volume? A call to rethink some published values. BioEssays. 2013;35:1050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science. 2013;342:1243259. [DOI] [PubMed] [Google Scholar]
- 6.Alexandrov T. Spatial metabolomics and imaging mass spectrometry in the age of artificial intelligence. Annu Rev Biomed Data Sci. 2020;3:61–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kietzmann T. Metabolic zonation of the liver: the oxygen gradient revisited. Redox Biol. 2017;11:622–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saxena R, Theise ND, Crawford JM. Microanatomy of the human liver—exploring the hidden interfaces. Hepatology. 1999;30:1339–46. [DOI] [PubMed] [Google Scholar]
- 9.Gebhardt R, Matz-Soja M. Liver zonation: novel aspects of its regulation and its impact on homeostasis. World J Gastroenterol. 2014;20:8491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rappaport AM, Wilson WD. The stuctural and functional unit in the human liver (liver acinus). Anat Rec. 1958;130:673–89. [DOI] [PubMed] [Google Scholar]
- 11.Crawford AR, Lin XIZ, Crawford JM. The normal adult human liver biopsy: a quantitative reference standard. Hepatology. 1998;28:323–31. [DOI] [PubMed] [Google Scholar]
- 12.Paris J, Henderson NC. Liver zonation, revisited. Hepatology. 2022;76:1219–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monga SPS. Role of Wnt/β-catenin signaling in liver metabolism and cancer. Int J Biochem Cell Biol. 2011;43:1021–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sekine S, Lan BYA, Bedolli M, Feng S, Hebrok M. Liver-specific loss of β-catenin blocks glutamine synthesis pathway activity and cytochrome P450 expression in mice. Hepatology. 2006;43:817–25. [DOI] [PubMed] [Google Scholar]
- 15.Gougelet A, Torre C, Veber P, Sartor C, Bachelot L, Denechaud PD, et al. T-cell factor 4 and β-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology. 2014;59:2344–57. [DOI] [PubMed] [Google Scholar]
- 16.Benhamouche S, Decaens T, Godard C, Chambrey R, Rickman DS, Moinard C, et al. APC tumor suppressor gene is the “zonation-keeper” of mouse liver. Dev Cell. 2006;10:759–770. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Mowry LE, Nejak-Bowen KN, Okabe H, Diegel CR, Lang RA, et al. Beta-catenin signaling in murine liver zonation and regeneration: a Wnt-Wnt situation. Hepatology. 2014;60:964–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gougelet A, Sartor C, Senni N, Calderaro J, Fartoux L, Lequoy M, et al. Hepatocellular carcinomas with mutational activation of beta-catenin require choline and can be detected by positron emission tomography. Gastroenterology. 2019;157:807–22. [DOI] [PubMed] [Google Scholar]
- 19.Senni N, Savall M, Cabrerizo Granados D, Alves-Guerra MC, Sartor C, Lagoutte I, et al. Β-catenin-activated hepatocellular carcinomas are addicted to fatty acids. Gut. 2019;68:322–34. [DOI] [PubMed] [Google Scholar]
- 20.Dai W, Shen J, Yan J, Bott AJ, Maimouni S, Daguplo HQ, et al. Glutamine synthetase limits b-catenin-mutated liver cancer growth by maintaining nitrogen homeostasis and suppressing mTORC1. J Clin Invest. 2022;132:e161408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matz-Soja M, Hovhannisyan A, Gebhardt R. Hedgehog signalling pathway in adult liver: a major new player in hepatocyte metabolism and zonation. Med Hypotheses. 2013;80:589–94. [DOI] [PubMed] [Google Scholar]
- 22.Salaritabar A, Berindan-Neagoe I, Darvish B, Hadjiakhoondi F, Manayi A, Devi KP, et al. Targeting Hedgehog signaling pathway: paving the road for cancer therapy. Pharmacol Res. 2019;141:466–80. [DOI] [PubMed] [Google Scholar]
- 23.Kolbe E, Aleithe S, Rennert C, Spormann L, Ott F, Meierhofer D, et al. Mutual zonated interactions of Wnt and Hh signaling are orchestrating the metabolism of the adult liver in mice and human. Cell Rep. 2019;29:4553–4567.e7. [DOI] [PubMed] [Google Scholar]
- 24.Matz-Soja M, Aleithe S, Marbach E, Böttger J, Arnold K, Schmidt-Heck W, et al. Hepatic Hedgehog signaling contributes to the regulation of IGF1 and IGFBP1 serum levels. Cell Commun Signal. 2014;12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braeuning A, Menzel M, Kleinschnitz EM, Harada N, Tamai Y, Köhle C, et al. Serum components and activated Ha-ras antagonize expression of perivenous marker genes stimulated by β-catenin signaling in mouse hepatocytes. FEBS J. 2007;274:4766–77. [DOI] [PubMed] [Google Scholar]
- 26.Colletti M, Cicchini C, Conigliaro A, Santangelo L, Alonzi T, Pasquini E, et al. Convergence of Wnt signaling on the HNF4α-driven transcription in controlling liver zonation. Gastroenterology. 2009;137:660–72. [DOI] [PubMed] [Google Scholar]
- 27.Stanulović VS, Kyrmizi I, Kruithof-De Julio M, Hoogenkamp M, Vermeulen JLM, Ruijter JM, et al. Hepatic HNF4α deficiency induces periportal expression of glutamine synthetase and other pericentral enzymes. Hepatology [Internet]. 2007;45:433–44. [DOI] [PubMed] [Google Scholar]
- 28.Cheng X, Kim SY, Okamoto H, Xin Y, Yancopoulos GD, Murphy AJ, et al. Glucagon contributes to liver zonation. Proc Natl Acad Sci U S A. 2018;115:E4111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kietzmann T. Liver zonation in health and disease: hypoxia and hypoxia-inducible transcription factors as concert masters. Int J Mol Sci. 2019;20:2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kietzmann T, Cornesse Y, Brechtel K, Modaressi S, Jungermann K. Perivenous expression of the mRNA of the three hypoxia-inducible factor α-subunits, HIF1α HIF2α and HIF3α, in rat liver. Biochem J. 2001;354:531–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rappaport AM, Borowy ZJ, Lougheed WM, Lotto WN. Subdivision of hexagonal liver lobules into a structural and functional unit. Role in hepatic physiology and pathology. Anat Rec. 1954;119:11–33. [DOI] [PubMed] [Google Scholar]
- 32.Jungermann K, Kietzmann T. Oxygen: modulator of metabolic zonation and disease of the liver. Hepatology [Internet]. 2000;31:255–60. [DOI] [PubMed] [Google Scholar]
- 33.Mazumdar J, O’Brien WT, Johnson RS, Lamanna JC, Chavez JC, Klein PS, et al. O2 regulates stem cells through Wnt/β-catenin signalling. Nat Cell Biol. 2010;12:1007–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newton IP, Kenneth NS, Appleton PL, Näthke I, Rocha S. Adenomatous polyposis coli and hypoxia-inducible factor-1α have an antagonistic connection. Mol Biol Cell. 2010;21:3630–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oinonen T, Mode A, Lobie PE, Lindros KO. Zonation of cytochrome P450 enzyme expression in rat liver: isozyme-specific regulation by pituitary dependent hormones. Biochem Pharmacol. 1996;51:1379–87. [DOI] [PubMed] [Google Scholar]
- 36.Halpern KB, Shenhav R, Matcovitch-Natan O, Tóth B, Lemze D, Golan M, et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindros KO, Penttila KE. Digitonin-collagenase perfusion for efficient separation of periportal or perivenous hepatocytes. Biochem J. 1985;228:757–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tordjmann T. An improved digitonin-collagenase perfusion technique for the isolation of periportal and perivenous hepatocytes from a single rat liver: physiological implications for lobular heterogeneity. Hepatology. 1997;26:1592–9. [DOI] [PubMed] [Google Scholar]
- 39.Gumucio JJ, May M, Dvorak C, Chianale J, Massey V. The isolation of functionally heterogeneous hepatocytes of the proximal and distal half of the liver acinus in the rat. Hepatology. 1986;6:932–44. [DOI] [PubMed] [Google Scholar]
- 40.McEnerney L, Duncan K, Bang BR, Elmasry S, Li M, Miki T, et al. Dual modulation of human hepatic zonation via canonical and non-canonical Wnt pathways. Exp Mol Med [Internet]. 2017;49:e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thalhammer T, Gessl A, Braakman I, Graf J. Separation of hepatocytes of different acinar zones by flow cytometry. Cytometry. 1989;10:772–8. [DOI] [PubMed] [Google Scholar]
- 42.Hildebrandt F, Andersson A, Saarenpää S, Larsson L, Van Hul N, Kanatani S, et al. Spatial transcriptomics to define transcriptional patterns of zonation and structural components in the mouse liver. Nat Commun. 2021;12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kling S, Lang B, Hammer HS, Naboulsi W, Sprenger H, Frenzel F, et al. Characterization of hepatic zonation in mice by mass-spectrometric and antibody-based proteomics approaches. Biol Chem. 2022;403:331–43. [DOI] [PubMed] [Google Scholar]
- 44.Sekine S, Ogawa R, Mcmanus MT, Kanai Y, Hebrok M. Dicer is required for proper liver zonation. J Pathol. 2009;219:365–72. [DOI] [PubMed] [Google Scholar]
- 45.Ben-Moshe S, Shapira Y, Moor AE, Manco R, Veg T, Bahar Halpern K, et al. Spatial sorting enables comprehensive characterization of liver zonation. Nat Metab. 2019;1:899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones JG. Hepatic glucose and lipid metabolism. Diabetologia. 2016;59:1098–1103. [DOI] [PubMed] [Google Scholar]
- 47.Katz N, Teutsch HF, Jungermann K, Sasse D. Heterogeneous reciprocal localization of fructose-1,6-bis-phosphatase and of glucokinase in microdissected periportal and perivenous rat liver tissue. FEBS Lett. 1977;83:272–6. [DOI] [PubMed] [Google Scholar]
- 48.Jungermann K, Kietzmann T. Zonation of parenchymal and nonparenchymal metabolism in liver. Annu Rev Nutr. 1996;16:179–203. [DOI] [PubMed] [Google Scholar]
- 49.Jungermann K. Functional significance of hepatocyte heterogeneity for glycolysis and gluconeogenesis. Pharmacol Biochem Behav. 1983;18:409–414. [DOI] [PubMed] [Google Scholar]
- 50.Quistorff B. Gluconeogenesis in periportal and perivenous hepatocytes of rat liver, isolated by a new high-yield digitonin/collagenase perfusion technique. Biochem J. 1985;229:221–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chamlian A, Benkoel L, Minko D, Njee T, Gulian JM. Ultrastructural heterogeneity of glycogen in human liver. Liver. 1989;9:346–50. [DOI] [PubMed] [Google Scholar]
- 52.Cardell RR, Cardell EL. Heterogeneity of glycogen distribution in hepatocytes. J Electron Microsc Tech. 1990;14:126–39. [DOI] [PubMed] [Google Scholar]
- 53.Ramnanan CJ, Edgerton DS, Kraft G, Cherrington AD. Physiologic action of glucagon on liver glucose metabolism. Diabetes Obes Metab. 2011;13:118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramakrishnan SK, Shah YM. A central role for hypoxia-inducible factor (HIF)-2α in hepatic glucose homeostasis. Nutr Heal Aging. 2017;4:207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guzman M, Castro J. Zonation of fatty acid metabolism in rat liver. Biochem J. 1989;264:107–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moreau M, Rivière B, Vegna S, Aoun M, Gard C, Ramos J, et al. Hepatitis C viral proteins perturb metabolic liver zonation. J Hepatol. 2015;62:278–85. [DOI] [PubMed] [Google Scholar]
- 57.Guzman M, Bijleveld C, Geelen MJH. Flexibility of zonation of fatty acid oxidation in rat liver. Biochem J. 1995;311:853–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng H‐C, Yang C‐M, Shiao M‐S. Zonation of cholesterol and glycerolipid synthesis in regenerating rat livers. Hepatology. 1993;17:280–6. [PubMed] [Google Scholar]
- 59.Twisk J, Hoekman MFM, Mager WH, Moorman AFM, De Boer PAJ, Scheja L, et al. Heterogeneous expression of cholesterol 7α-hydroxylase and sterol 27-hydroxylase genes in the rat liver lobulus. J Clin Invest. 1995;95:1235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gebhardt R, Mecke D. Heterogeneous distribution of glutamine synthetase among rat liver parenchymal cells in situ and in primary culture. EMBO J. 1983;2:567–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poso AR, Penttila KE, Suolinna EM, Lindros KO. Urea synthesis in freshly isolated and in cultured periportal and perivenous hepatocytes. Biochem J. 1986;239:263–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simon J, Nuñez-García M, Fernández-Tussy P, Barbier-Torres L, Fernández-Ramos D, Gómez-Santos B, et al. Targeting hepatic glutaminase 1 ameliorates non-alcoholic steatohepatitis by restoring very-low-density lipoprotein triglyceride assembly. Cell Metab. 2020;31:605–622.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Droin C, Kholtei J El, Bahar Halpern K, Hurni C, Rozenberg M, Muvkadi S, et al. Space-time logic of liver gene expression at sub-lobular scale. Nat Metab. 2021;3:43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2 + cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. [DOI] [PubMed] [Google Scholar]
- 66.Wei Y, Wang YG, Jia Y, Li L, Yoon J, Zhang S, et al. Liver homeostasis is maintained by midlobular zone 2 hepatocytes. Science. 2021;371:Eabb1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mollbrink A, Holmström P, Sjöström M, Hultcrantz R, Eriksson LC, Stål P. Iron-regulatory gene expression during liver regeneration. Scand J Gastroenterol. 2012;47:591–600. [DOI] [PubMed] [Google Scholar]
- 68.Saito K, Negishi M, James, Squires E. Sexual dimorphisms in zonal gene expression in mouse liver. Biochem Biophys Res Commun. 2013;436:730–5. [DOI] [PubMed] [Google Scholar]
- 69.Rodimova SA, Kuznetsova DS, Bobrov NV, Gulin AA, Vasin AA, Gubina MV, et al. Multiphoton microscopy and mass spectrometry for revealing metabolic heterogeneity of hepatocytes in vivo. Sovrem Tehnol Med. 2021;13:18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stopka SA, van der Reest J, Abdelmoula WM, Ruiz DF, Joshi S, Ringel AE, et al. Spatially resolved characterization of tissue metabolic compartments in fasted and high-fat diet livers. PLoS One. 2022;17:e0261803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gola A, Dorrington MG, Speranza E, Sala C, Shih RM, Radtke AJ, et al. Commensal-driven immune zonation of the liver promotes host defence. Nature. 2021;589:131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guilliams M, Bonnardel J, Haest B, Vanderborght B, Wagner C, Remmerie A, et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell. 2022;185:379–396.e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Halpern KB, Shenhav R, Massalha H, Toth B, Egozi A, Massasa EE, et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat Biotechnol. 2018;36:962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rocha AS, Vidal V, Mertz M, Kendall TJ, Charlet A, Okamoto H, et al. The angiocrine factor rspondin3 is a key determinant of liver zonation. Cell Rep. 2015;13:1757–64. [DOI] [PubMed] [Google Scholar]
- 75.Preziosi M, Okabe H, Poddar M, Singh S, Monga SP. Endothelial Wnts regulate β-catenin signaling in murine liver zonation and regeneration: a sequel to the Wnt–Wnt situation. Hepatol Commun. 2018;2:845–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Presa N, Gomez-Larrauri A, Dominguez-Herrera A, Trueba M, Gomez-Muñoz A. Novel signaling aspects of ceramide 1-phosphate. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865:158630. [DOI] [PubMed] [Google Scholar]
- 78.Lenz EM, Wilson ID. Analytical strategies in metabonomics. J Proteome Res. 2007;6:443–58. [DOI] [PubMed] [Google Scholar]
- 79.Dunn WB, Ellis DI. Metabolomics: current analytical platforms and methodologies. Trends Anal Chem. 2005;24:285–294. [Google Scholar]
- 80.Dias DA, Jones OAH, Beale DJ, Boughton BA, Benheim D, Kouremenos KA, et al. Current and future perspectives on the structural identification of small molecules in biological systems. Metabolites. 2016;6:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takis PG, Ghini V, Tenori L, Turano P, Luchinat C. Uniqueness of the NMR approach to metabolomics. Trends Anal Chem. 2019;120:115300. [Google Scholar]
- 82.Beckonert O, Keun HC, Ebbels TMD, Bundy J, Holmes E, Lindon JC, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–703. [DOI] [PubMed] [Google Scholar]
- 83.Amathieu R, Triba MN, Goossens C, Bouchemal N, Nahon P, Savarin P, et al. Nuclear magnetic resonance based metabolomics and liver diseases: recent advances and future clinical applications. World J Gastroenterol. 2016;22:417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vinaixa M, Ángel Rodríguez M, Rull A, Beltrán R, Bladé C, Brezmes J, et al. Metabolomic assessment of the effect of dietary cholesterol in the progressive development of fatty liver disease. J Proteome Res. 2010;9:2527–38. [DOI] [PubMed] [Google Scholar]
- 85.Bernardo-Seisdedos G, Bilbao J, Fernández-Ramos D, Lopitz-Otsoa F, Gutierrez de Juan V, Bizkarguenaga M, et al. Metabolic landscape of the mouse liver by quantitative 31P nuclear magnetic resonance analysis of the phosphorome. Hepatology. 2021;74:148–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ren JL, Zhang AH, Kong L, Wang XJ. Advances in mass spectrometry-based metabolomics for investigation of metabolites. RSC Adv. 2018;8:22335–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alseekh S, Aharoni A, Brotman Y, Contrepois K, D’Auria J, Ewald J, et al. Mass spectrometry-based metabolomics: a guide for annotation, quantification and best reporting practices. Nat Methods. 2021;18:747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–60. [DOI] [PubMed] [Google Scholar]
- 89.Buchberger AR, DeLaney K, Johnson J, Li L. Mass spectrometry imaging: a review of emerging advancements and future insights. Anal Chem. 2018;90:240–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Addie RD, Balluff B, Bovée JVMG, Morreau H, McDonnell LA. Current state and future challenges of mass spectrometry imaging for clinical research. Anal Chem. 2015;87:6426–33. [DOI] [PubMed] [Google Scholar]
- 91.Gilmore IS, Heiles S, Pieterse CL. Metabolic imaging at the single-cell scale: recent advances in mass spectrometry imaging. Annu Rev Anal Chem. 2019;12:201–24. [DOI] [PubMed] [Google Scholar]
- 92.Römpp A, Spengler B. Mass spectrometry imaging with high resolution in mass and space. Histochem Cell Biol. 2013;139:759–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morawietz CM, Peter Ventura AM, Grevelding CG, Haeberlein S, Spengler B. Spatial visualization of drug uptake and distribution in Fasciola hepatica using high-resolution AP-SMALDI mass spectrometry imaging. Parasitol Res. 2022;121:1145–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Passarelli MK, Pirkl A, Moellers R, Grinfeld D, Kollmer F, Havelund R, et al. The 3D OrbiSIMS—label-free metabolic imaging with subcellular lateral resolution and high mass-resolving power. Nat Methods. 2017;14:1175–83. [DOI] [PubMed] [Google Scholar]
- 95.Capolupo L, Khven I, Lederer AR, Mazzeo L, Glousker G, Ho S, et al. Sphingolipids control dermal fibroblast heterogeneity. Science. 2022;376:eabh1623. [DOI] [PubMed] [Google Scholar]
- 96.Suvannapruk W, Edney MK, Kim DH, Scurr DJ, Ghaemmaghami AM, Alexander MR. Single-cell metabolic profiling of macrophages using 3D OrbiSIMS: correlations with phenotype. Anal Chem. 2022;94:9389–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seydel C. Single-cell metabolomics hits its stride. Nat Methods. 2021;18:1452–6. [DOI] [PubMed] [Google Scholar]
- 98.Kompauer M, Heiles S, Spengler B. Autofocusing MALDI mass spectrometry imaging of tissue sections and 3D chemical topography of nonflat surfaces. Nat Methods. 2017;14:1156–8. [DOI] [PubMed] [Google Scholar]
- 99.Tillner J, Wu V, Jones EA, Pringle SD, Karancsi T, Dannhorn A, et al. Faster, more reproducible DESI-MS for biological tissue imaging. J Am Soc Mass Spectrom. 2017;28:2090–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zavalin A, Yang J, Hayden K, Vestal M, Caprioli RM. Tissue protein imaging at 1 µm laser spot diameter for high spatial resolution and high imaging speed using transmission geometry MALDI TOF MS. Anal Bioanal Chem. 2015;407:2337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ly A, Buck A, Balluff B, Sun N, Gorzolka K, Feuchtinger A, et al. High-mass-resolution MALDI mass spectrometry imaging of metabolites from formalin-fixed paraffin-embedded tissue. Nat Protoc. 2016;11:1428–43. [DOI] [PubMed] [Google Scholar]
- 102.Dannhorn A, Kazanc E, Ling S, Nikula C, Karali E, Serra MP, et al. Universal sample preparation unlocking multimodal molecular tissue imaging. Anal Chem. 2020;92:11080–8. [DOI] [PubMed] [Google Scholar]
- 103.Angerer TB, Bour J, Biagi JL, Moskovets E, Frache G. Evaluation of 6 MALDI-matrices for 10 μm lipid imaging and on-tissue MSn with AP-MALDI-Orbitrap. J Am Soc Mass Spectrom. 2022;33:760–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Balluff B, Heeren RMA, Race AM. An overview of image registration for aligning mass spectrometry imaging with clinically relevant imaging modalities. J Mass Spectrom Adv Clin Lab. 2022;23:26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Palmer A, Phapale P, Chernyavsky I, Lavigne R, Fay D, Tarasov A, et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat Methods. 2016;14:57–60. [DOI] [PubMed] [Google Scholar]
- 107.Fincher JA, Korte AR, Yadavilli S, Morris NJ, Vertes A. Multimodal imaging of biological tissues using combined MALDI and NAPA-LDI mass spectrometry for enhanced molecular coverage. Analyst [Internet]. 2020;145:6910–8. [DOI] [PubMed] [Google Scholar]
- 108.Neumann EK, Djambazova KV, Caprioli RM, Spraggins JM. Multimodal imaging mass spectrometry: next generation molecular mapping in biology and medicine. J Am Soc Mass Spectrom. 2020;31:2401–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goossens P, Lu C, Cao J, Gijbels MJ, Karel JMH, Wijnands E, et al. Integrating multiplex immunofluorescent and mass spectrometry imaging to map myeloid heterogeneity in its metabolic and cellular context. Cell Metab. 2022;34:1214–1225.e6. [DOI] [PubMed] [Google Scholar]
- 110.Solé-Domènech S, Sjövall P, Vukojević V, Fernando R, Codita A, Salve S, et al. Localization of cholesterol, amyloid and glia in Alzheimer’s disease transgenic mouse brain tissue using time-of-flight secondary ion mass spectrometry (ToF-SIMS) and immunofluorescence imaging. Acta Neuropathol. 2013;125:145–57. [DOI] [PubMed] [Google Scholar]
- 111.Elaldi R, Hemon P, Petti L, Cosson E, Desrues B, Sudaka A, et al. High dimensional imaging mass cytometry panel to visualize the tumor immune microenvironment contexture. Front Immunol. 2021;12:1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ravi VM, Will P, Kueckelhaus J, Sun N, Joseph K, Salié H, et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell. 2022;40:639–655.e13. [DOI] [PubMed] [Google Scholar]
- 113.Asrani SK, Devarbhavi H, Eaton J, Kamath PS. Burden of liver diseases in the world. J Hepatol. 2019;70:151–71. [DOI] [PubMed] [Google Scholar]
- 114.Younossi Z, Tacke F, Arrese M, Chander Sharma B, Mostafa I, Bugianesi E, et al. Global perspectives on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology. 2019;69:2672–82. [DOI] [PubMed] [Google Scholar]
- 115.Puri P, Baillie RA, Wiest MM, Mirshahi F, Choudhury J, Cheung O, et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46:1081–90. [DOI] [PubMed] [Google Scholar]
- 116.Puri P, Wiest MM, Cheung O, Mirshahi F, Sargeant C, Min HK, et al. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 2009;50:1827–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease—meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. [DOI] [PubMed] [Google Scholar]
- 118.Mayo R, Crespo J, Martínez-Arranz I, Banales JM, Arias M, Mincholé I, et al. Metabolomic-based noninvasive serum test to diagnose nonalcoholic steatohepatitis: results from discovery and validation cohorts. Hepatol Commun. 2018;2:807–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Perakakis N, Polyzos SA, Yazdani A, Sala-Vila A, Kountouras J, Anastasilakis AD, et al. Non-invasive diagnosis of non-alcoholic steatohepatitis and fibrosis with the use of omics and supervised learning: a proof of concept study. Metabolism. 2019;101:154005. [DOI] [PubMed] [Google Scholar]
- 120.Marques V, Afonso MB, Bierig N, Duarte-Ramos F, Santos-Laso Á, Jimenez-Agüero R, et al. Adiponectin, Leptin, and IGF-1 are useful diagnostic and stratification biomarkers of NAFLD. Front Med. 2021;8:683250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Afonso MB, Rodrigues PM, Carvalho T, Caridade M, Borralho P, Cortez-Pinto H, et al. Necroptosis is a key pathogenic event in human and experimental murine models of non-alcoholic steatohepatitis. Clin Sci. 2015;129:721–739. [DOI] [PubMed] [Google Scholar]
- 122.Afonso MB, Rodrigues PM, Mateus-Pinheiro M, Simão AL, Gaspar MM, Majdi A, et al. RIPK3 acts as a lipid metabolism regulator contributing to inflammation and carcinogenesis in non-alcoholic fatty liver disease. Gut. 2021;70:2359–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Afonso MB, Islam T, Magusto J, Amorim R, Lenoir V, F, Simões R, et al. RIPK3 dampens mitochondrial bioenergetics and lipid droplet dynamics in metabolic liver disease. Hepatology. 2022. doi: 10.1002/hep.32756. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang X, Coker OO, Chu ESH, Fu K, Lau HCH, Wang YX, et al. Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut. 2021;70:761–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cai H, Wen Z, Meng K, Yang P. Metabolomic signatures for liver tissue and cecum contents in high-fat diet-induced obese mice based on UHPLC-Q-TOF/MS. Nutr Metab. 2021;18:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li Y, Meng Y, Zhu X, Van Wijnen A, Eirin A, Lerman LO. Metabolic syndrome is associated with altered mRNA and miRNA content in human circulating extracellular vesicles. Front Endocrinol (Lausanne). 2021;12:973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kieran NW, Suresh R, Dorion MF, MacDonald A, Blain M, Wen D, et al. MicroRNA-210 regulates the metabolic and inflammatory status of primary human astrocytes. J Neuroinflammation. 2022;19:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Teng Y, Ren Y, Sayed M, Hu X, Lei C, Kumar A, et al. Plant-derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe. 2018;24:637–652.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu S, Da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe. 2016;19:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Santos AA, Afonso MB, Ramiro RS, Pires D, Pimentel M, Castro RE, et al. Host miRNA-21 promotes liver dysfunction by targeting small intestinal Lactobacillus in mice. Gut Microbes. 2020;12:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wattacheril J, Seeley EH, Angel P, Chen H, Bowen BP, Lanciault C, et al. Differential intrahepatic phospholipid zonation in simple steatosis and nonalcoholic steatohepatitis. PLoS One. 2013;8:e57165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li Q, Chen P, Fan Y, Wang X, Xu K, Li L, et al. Multicolor fluorescence detection-based microfluidic device for single-cell metabolomics: simultaneous quantitation of multiple small molecules in primary liver cells. Anal Chem. 2016;88:8610–6. [DOI] [PubMed] [Google Scholar]
- 133.Hall Z, Bond NJ, Ashmore T, Sanders F, Ament Z, Wang X, et al. Lipid zonation and phospholipid remodeling in nonalcoholic fatty liver disease. Hepatology. 2017;65:1165–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ščupáková K, Soons Z, Ertaylan G, Pierzchalski KA, Eijkel GB, Ellis SR, et al. Spatial systems lipidomics reveals nonalcoholic fatty liver disease heterogeneity. Anal Chem. 2018;90:5130–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Alamri H, Patterson NH, Yang E, Zoroquiain P, Lazaris A, Chaurand P, et al. Mapping the triglyceride distribution in NAFLD human liver by MALDI imaging mass spectrometry reveals molecular differences in micro and macro steatosis. Anal Bioanal Chem. 2019;411:885–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Li F, Xiang B, Jin Y, Li C, Ren S, Wu Y, et al. Hepatotoxic effects of inhalation exposure to polycyclic aromatic hydrocarbons on lipid metabolism of C57BL/6 mice. Environ Int. 2020;134:105000. [DOI] [PubMed] [Google Scholar]
- 137.Downes DP, Zhong W, Zhang J, Chen B, Satapati S, Metzger D, et al. Mapping lipogenic flux: a gold LDI-MS approach for imaging neutral lipid kinetics. J Am Soc Mass Spectrom. 2020;31:2421–5. [DOI] [PubMed] [Google Scholar]
- 138.Yuan Z, Zhou Q, Cai L, Pan L, Sun W, Qumu S, et al. SEAM is a spatial single nuclear metabolomics method for dissecting tissue microenvironment. Nat Methods. 2021;18:1223–32. [DOI] [PubMed] [Google Scholar]
- 139.Rappez L, Stadler M, Triana S, Gathungu RM, Ovchinnikova K, Phapale P, et al. SpaceM reveals metabolic states of single cells. Nat Methods. 2021;18:799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Torres S, Samino S, Ràfols P, Martins-Green M, Correig X, Ramírez N. Unravelling the metabolic alterations of liver damage induced by thirdhand smoke. Environ Int. 2021;146:106242. [DOI] [PubMed] [Google Scholar]
- 141.Zhang D, Zheng W, Li X, Liang G, Ye N, Liu Y, et al. Investigation of obesity-alleviation effect of Eurycoma longifolia on mice fed with a high-fat diet through metabolomics revealed enhanced decomposition and inhibition of accumulation of lipids. J Proteome Res. 2021;20:2714–24. [DOI] [PubMed] [Google Scholar]
- 142.Guo W, Shi Z, Zeng T, He Y, Cai Z, Zhang J. Metabolic study of aristolochic acid I-exposed mice liver by atmospheric pressure matrix-assisted laser desorption/ionization mass spectrometry imaging and machine learning. Talanta. 2022;241:123261. [DOI] [PubMed] [Google Scholar]
- 143.Ochoa-Rios S, O’Connor IP, Kent LN, Clouse JM, Hadjiyannis Y, Koivisto C, et al. Imaging mass spectrometry reveals alterations in N-linked glycosylation that are associated with histopathological changes in nonalcoholic steatohepatitis in mouse and human. Mol Cell Proteomics. 2022;21:100225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dyar KA, Lutter D, Artati A, Ceglia NJ, Liu Y, Armenta D, et al. Atlas of circadian metabolism reveals system-wide coordination and communication between clocks. Cell. 2018;174:1571–1585.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang Q, Lou Y, Yang J, Wang J, Feng J, Zhao Y, et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut. 2019;68:2019–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Petras D, Jarmusch AK, Dorrestein PC. From single cells to our planet—recent advances in using mass spectrometry for spatially resolved metabolomics. Curr Opin Chem Biol. 2017;36:24–31. [DOI] [PubMed] [Google Scholar]
- 147.Wang J, Kunzke T, Prade VM, Shen J, Buck A, Feuchtinger A, et al. Spatial metabolomics identifies distinct tumor-specific subtypes in gastric cancer patients. Clin Cancer Res. 2022;28:2865–77. [DOI] [PubMed] [Google Scholar]
- 148.Wu R, Guo W, Qiu X, Wang S, Sui C, Lian Q, et al. Comprehensive analysis of spatial architecture in primary liver cancer. Sci Adv. 2021;7:3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun C, Li T, Song X, Huang L, Zang Q, Xu J, et al. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc Natl Acad Sci U S A. 2019;116:52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol. 2019;16:411–28. [DOI] [PubMed] [Google Scholar]
- 151.Saviano A, Henderson NC, Baumert TF. Single-cell genomics and spatial transcriptomics: discovery of novel cell states and cellular interactions in liver physiology and disease biology. J Hepatol. 2020;73:1219–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yagnik G, Liu Z, Rothschild KJ, Lim MJ. Highly multiplexed immunohistochemical MALDI-MS imaging of biomarkers in tissues. J Am Soc Mass Spectrom. 2021;32:977–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Darie-Ion L, Whitham D, Jayathirtha M, Rai Y, Neagu AN, Darie CC, et al. Applications of MALDI-MS/MS-based proteomics in biomedical research. Molecules. 2022;27:6196. [DOI] [PMC free article] [PubMed] [Google Scholar]


