Abstract
Background:
This study aimed to develop an automated segmentation system for biliary structures using a deep learning model, based on data from magnetic resonance cholangiopancreatography (MRCP).
Materials and methods:
Living liver donors who underwent MRCP using the gradient and spin echo technique followed by three-dimensional modeling were eligible for this study. A three-dimensional residual U-Net model was implemented for the deep learning process. Data were divided into training and test sets at a 9:1 ratio. Performance was assessed using the dice similarity coefficient to compare the model’s segmentation with the manually labeled ground truth.
Results:
The study incorporated 250 cases. There was no difference in the baseline characteristics between the train set (n=225) and test set (n=25). The overall mean Dice Similarity Coefficient was 0.80±0.20 between the ground truth and inference result. The qualitative assessment of the model showed relatively high accuracy especially for the common bile duct (88%), common hepatic duct (92%), hilum (96%), right hepatic duct (100%), and left hepatic duct (96%), while the third-order branch of the right hepatic duct (18.2%) showed low accuracy.
Conclusion:
The developed automated segmentation model for biliary structures, utilizing MRCP data and deep learning techniques, demonstrated robust performance and holds potential for further advancements in automation.
Keywords: cholangiopancreatography, deep learning, liver, living-donor, magnetic resonance
Introduction
Highlights
Developed an automated segmentation system utilizing a 3D residual U-Net deep learning model to delineate biliary structures from magnetic resonance cholangiopancreatography data, aimed at improving preoperative planning in living liver donors.
Achieved a mean dice similarity coefficient of 0.80, demonstrating a high level of accuracy in the model’s segmentation capability when compared to manually labeled ground truth, particularly in segmenting major biliary structures.
The proposed model was able to connect originally disconnected voxels in certain cases, which shows its potential to emulate expert-driven segmentation based on anatomical knowledge.
Liver transplantation is an essential therapeutic option for acute liver failure and hepatic malignancies1–3, but the limited availability of deceased donor organs has led to the emergence of living-donor liver transplantation, expanding the pool of available organs4–6. Ensuring donor safety during hepatectomy is of utmost importance, and the quality of the donor surgery significantly affects the recipient’s post-transplantation recovery7.
A crucial step in the donor hepatectomy procedure is the meticulous division of the bile duct, which requires precise dissection and resection at an anatomically specific point to optimize outcomes for both the donor and recipient8. Effective preoperative planning, therefore, becomes paramount in navigating the intricate anatomical landscape of the liver. The complexity of biliary anatomy necessitates detailed and accurate visualization of these structures to guide the surgical approach, reduce the risk of complications, and ensure the safety of the donor. To address this challenge, recent clinical guidelines recommend the incorporation of preoperative magnetic resonance cholangiopancreatography (MRCP) for all potential donors9. MRCP allows for the evaluation of biliary structures and the identification of any unexpected anatomical variations. In our institution, the biliary structures obtained by MRCP are manually segmented and reconstructed into three-dimensional images for better visualization during operation. This approach has proved instrumental in facilitating image-guided surgical procedures10,11.
The aim of the study is to leverage the accumulated annotated dataset from the manually segmented biliary structure to train a deep learning model capable of automatically segmenting the bile ducts using MRCP images. We thoroughly evaluated the model’s performance and present our findings.
Materials and methods
This retrospective study was approved by the institutional review board of Samsung Medical Center (SMC 2021-03-056) and the need for informed consent was waived by the IRB due to the retrospective nature of the study. It was carried out in accordance with the principles of the Declaration of Helsinki.
Patients
Living liver donors at Samsung Medical Center, between January 2014 and February 2021 were included in this study. All donors underwent a comprehensive evaluation to determine their suitability for liver donation, including an assessment of residual liver volume using computed tomographic angiography. Additionally, MRCP was performed to accurately evaluate the structure of the biliary tract prior to surgery. Segmentation and 3D modeling was prospectively performed to enhance the understanding of the biliary structure for donor surgery. Demographic data including age, sex, and BMI were retrieved in a de-identified state from the Clinical Data Warehouse DARWIN-C of Samsung Medical Center. The types of bile ducts were classified according to the modified classification system proposed by Huang et al. (Fig. 1).
Figure 1.
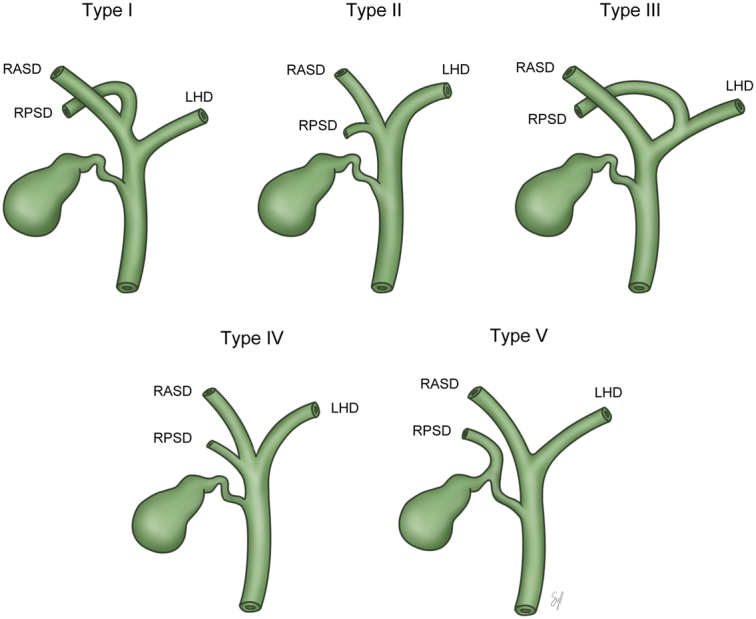
The types of bile ducts were classified according to the modified classification system proposed by Huang et al.: type I, normal type; type II, trifurcation of right anterior, right posterior, and left hepatic duct; type III, right posterior duct draining into left hepatic duct; type IV, early branching of right posterior duct from the common hepatic duct; type V, right posterior duct draining into cystic duct; and other types of variation in bile duct anatomy.
Dataset
The study used three-dimensional (3D) MRCP using a gradient and spin echo (GRASE) technique for visualization of biliary structures. The 3D GRASE MRCP datasets were manually labeled slice by slice for the common bile duct (CBD), common hepatic duct (CHD), gall bladder (GB), cystic duct, and intrahepatic duct (IHD) by two experienced biomedical engineers using an annotation tool called 3D slicer. The results were verified by a board-certified abdominal radiologist and several liver surgeons, and it the annotations were not appropriate, they were reannotated until the supervising physicians were satisfied. Training and test sets were allocated in 9:1 ratio.
3D residual U-Net model and implementation
We utilized a 3D residual U-Net model for this study. The model extracts context features through an encoding process and uses these features to reconstruct the segmented image during decoding. The model is trained to minimize a loss function, optimizing the network parameters for hierarchical feature extraction. During encoding, the convolutional residual operation is employed, while during decoding, multitarget segmented images are reconstructed with deconvolutional feature maps, including skip connections at each resolution level.
The 3D residual U-Net model was implemented in TensorFlow 1.14 and trained on a workstation with four GPUs (NVIDIA TITAN XP 16GB). During preprocessing, images were cropped, resized, and normalized. The training data was augmented through various techniques, such as 3D rotation, scaling, random flipping, and cropping. The dice similarity coefficient (DSC) loss (L) was used as the loss function. The network was trained using the Adam optimizer with a learning rate of 0.0001, with 1000 epochs and a batch size of 4. During the testing phase, preprocessed images of the entire MRI scan were fed into the proposed network (for detailed methodological information, see the Supplementary Document, Supplemental Digital Content 1, http://links.lww.com/JS9/B685).
Evaluation
The performance of the deep learning model to segment biliary structure was compared with manually delineated ground truth. Since the ground truth was masked by connecting the originally disconnected structures, comparison was additionally performed against the initially masked image, which did not connect the disconnected parts from the original image. Both quantitative and qualitative assessment were performed. As a quantitative measure, DSC12 was used to quantify the segmentation performance of the deep learning model (Fig. 2). As a qualitative measure, the biliary structure was subdivided according to the orders. Then the results of auto-segmentation were assessed by two expert radiologists, and classified as follows: ‘complete’, as for segmentation done without disconnection, ‘partial’, as for segmentation but disconnection between proximal and distal parts, and ‘absent’, as for no segmentation. To evaluate the accuracy, proportion of complete segmentation of the deep learning model compared to both the original MRCP image and the ground truth was assessed. Furthermore, a modified assessment of accuracy was performed by allocating score 1 for complete segmentation and score 0.5 for partial segmentation and comparing the score of the deep learning model compared to the original MRCP and the ground truth. For visualization, biliary structures were 3-dimensionally reconstructed with Mimics Medical (Materialise). For assessing the DSC coefficient, 3D slicer was used for calculation.
Figure 2.
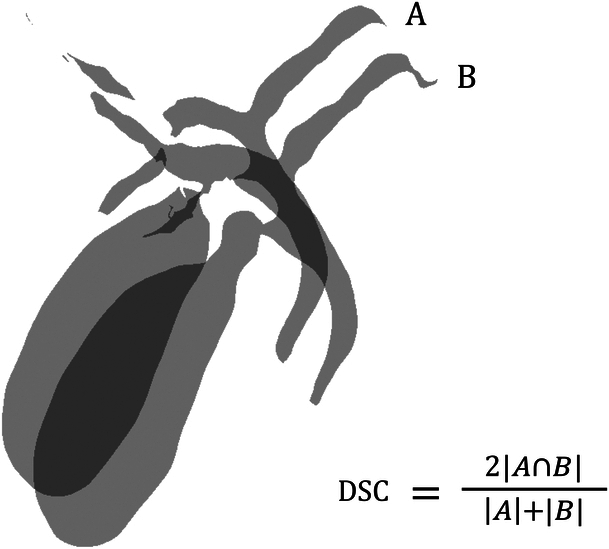
Illustration of Dice similarity coefficient (DSC) for biliary structure segmentation. The shaded areas represent the segmented regions from two different methods: A (e.g. manual segmentation) and B (e.g. automated segmentation by the deep learning model). The intersection of A and B represents the common area correctly identified by both methods. The DSC is calculated by the formula shown, which quantifies the similarity between the two segmentation results. The closer the DSC value is to 1, the higher the degree of agreement between the two methods.
Statistical analysis
Continuous variables were presented as mean±SD and analyzed using the independent t-test or Mann–Whitney test, as appropriate. Categorical data were presented as numbers and percentages and analyzed using the χ2 or Fisher’s exact test. All statistical analyses were performed using R Statistical Software (version 3.6.3; Foundation for Statistical Computing).
Results
Patients
A total of 250 living liver donors were included in the study, whose mean age was 34.4±11.3 years old, with 58% of male (145/250) and type I bile duct as the most common (183/250, 73.2%) anatomical type. There were no statistical differences in demographic and morphological characteristics between training and test sets (Table 1).
Table 1.
Baseline characteristics.
| Overall | Training set | Test set | P | |
|---|---|---|---|---|
| N | 250 | 225 | 25 | |
| Sex (%) | ||||
| Male | 145 (58.0) | 131 (58.2) | 14 (56.0) | 1 |
| Female | 105 (42.0) | 94 (41.8) | 11 (44.0) | |
| Age | 34.4±11.3 | 34.7±11.2 | 31.5±12.6 | 0.231 |
| BMI | 23.3±2.9 | 23.3±2.9 | 23.4±2.7 | 0.769 |
| BD type (%) | ||||
| I | 183 (73.2) | 165 (73.3) | 18 (72.0) | 0.523 |
| II | 14 (5.6) | 12 (5.3) | 2 (8.0) | |
| III | 35 (14.0) | 33 (14.7) | 2 (8.0) | |
| IV | 14 (5.6) | 11 (4.9) | 3 (12.0) | |
| V | 2 (0.8) | 2 (0.9) | 0 (0.0) | |
| others | 2 (0.8) | 2 (0.9) | 0 (0.0) | |
BD type, bile duct type.
Quantitative evaluation
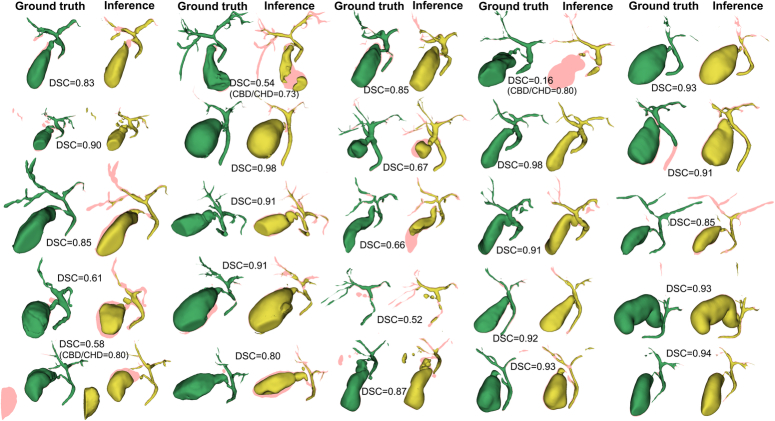
The results of the manual segmentation and automatic segmentation using the 3D residual U-Net model for each case are summarized in Figure 3, showing the 3D reconstructed structures. The mean DSC for the whole biliary structure was 0.79±0.18 against the initially masked image and 0.80±0.20 against the ground truth (Table 2). DSC were also calculated for subdivided structures. For CBD, CHD, GB, and main IHD, DSC against the initially masked model and ground truth were 0.80±0.19 and 0.81±0.20, respectively. For CBD, CHD, and GB, DSC against the initially masked model and ground truth were 0.80±0.21 and 0.81±0.21, respectively. For CBD and CHD, DSC against the initially masked model and ground truth were 0.70±0.11 and 0.74±0.08, respectively.
Figure 3.
Three-dimensionally reconstructed models of ground truth and inference of the auto-segmentation model. The green models represent ground truth while yellow models represent inference of the auto-segmentation model. Dice similarity coefficient is presented case by case. The pink shadow represents the errors between the models.
Table 2.
Dice similarity coefficient (DSC) for the test set against initially masked image and ground truth according to the anatomical orders.
| Against initially masked image | Against ground truth | |
|---|---|---|
| Whole biliary structure | 0.79±0.18 | 0.80±0.20 |
| CBD/CHD/GB/Main IHD | 0.80±0.19 | 0.81±0.20 |
| CBD/CHD/GB | 0.80±0.21 | 0.81±0.21 |
| CBD/CHD | 0.70±0.11 | 0.74±0.08 |
CBD, common bile duct; CHD, common hepatic duct; GB, gall bladder; IHD, intrahepatic duct.
Qualitative evaluation
Table 3 shows the qualitative assessment for auto-segmentation accuracy. When only the complete segmentations were assessed, CBD, the right hepatic duct, and the second order of the left hepatic duct showed 100% reconstruction compared to the original MRCP image. CHD and hilum showed better outcomes compared to the original MRCP image. When the accuracy was assessed against the ground truth, only the right hepatic duct showed 100% reconstruction. While most of the structures showed relatively high accuracy, the third-order of right hepatic duct showed a low reconstruction rate of 18.2% when compared to the ground truth. The modified assessment method, which also included partially segmented structure showed that all the structures except the cystic duct showed high accuracy exceeding 90.0% of reconstruction. Similarly, modified assessment comparing to the ground truth showed that except for the cystic duct (70.2%), right posterior duct (72.0%), and 3rd order of the right hepatic duct (46.7%), all the structures showed accuracy exceeding 80.0%.
Table 3.
Qualitative assessment of performance of the deep learning model compared to initially masked image (original) and the ground truth.
| Initially masked model | Ground truth | Inference of Auto-segmentation model | Assessment for Accuracy | Modified assessment for accuracy | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Complete | Partial | Absent | Complete | Partial | Absent | Complete | Partial | Absent | Against original | Against GT | Against original | Against GT | |
| CBD | 22 (88%) | 2 (8%) | 1 (4%) | 25 (100%) | — | — | 22 (88%) | 2 (8%) | 1 (4%) | 22/22 (100%) | 22/25 (88%) | 23/23 (100%) | 23/25 (92%) |
| CHD | 22 (88%) | 3 (12%) | — | 25 (100%) | — | — | 23 (92%) | 2 (8%) | — | 23/22 (104.5%) | 23/25 (92%) | 24/23.5 (102%) | 24/25 (96%) |
| Cystic | 20 (80%) | 3 (12%) | 2 (8%) | 23 (92%) | 1 (4%) | 1 (4%) | 15 (60%) | 3 (12%) | 7 (28%) | 15/20 (75%) | 15/23 (65%) | 16.5/21.5 (76.7%) | 16.5/23.5 (70.2%) |
| GB | 20 (80%) | 4 (16%) | 1 (4%) | 24 (96%) | 1 (4%) | 19 (76%) | 5 (20%) | 1 (4%) | 19/20 (95%) | 19/24 (79%) | 21.5/22 (97.7%) | 21.5/24 (89.6%) | |
| Hilum | 23 (92%) | 1 (4%) | 1 (4%) | 25 (100%) | — | — | 24 (96%) | 1 (4%) | — | 24/23 (104%) | 24/25 (96%) | 24.5/23.5 (104%) | 24.5/25 (98%) |
| RHD | 25 (100%) | — | — | 25 (100%) | — | — | 25 (100%) | — | — | 25/25 (100%) | 25/25 (100%) | 25/25 (100%) | 25/25 (100%) |
| RAHD | 20 (80%) | 2 (8%) | 3 (12%) | 25 (100%) | — | — | 19 (76%) | 5 (20%) | 1 (4%) | 19/20 (95%) | 19/25 (76%) | 21.5/22 (97.7%) | 21.5/25 (86%) |
| RPHD | 17(68%) | 5 (20%) | 3 (12%) | 25 (100%) | — | — | 14 (56%) | 8 (32%) | 3 (12%) | 14/17 (82.3%) | 14/25 (56%) | 18/19.5 (92.3%) | 18/25 (72%) |
| 3rd order | 6 (24%) | 10 (40%) | 9 (36%) | 22 (88%) | 1 (4%) | 2 (8%) | 4 (16%) | 13 (52%) | 8 (32%) | 4/6 (66.6%) | 4/22 (18.2%) | 10.5/11 (95.5%) | 10.5/22.5 (46.7%) |
| LHD | 25 (100%) | — | — | 25 (100%) | — | — | 24 (96%) | 1 (4%) | — | 24/25 (96%) | 24/25 (96%) | 24.5/15 (98%) | 24.5/25 (98%) |
| 2nd order | 15 (60%) | 5 (20%) | 5 (20%) | 21 (84%) | 1 (4%) | 3 (12%) | 15 (60%) | 5 (20%) | 5 (20%) | 15/15 (100%) | 15/21 (71.4%) | 17.5/17.5 (100%) | 17.5/21.5 (81.4%) |
CBD, common bile duct; CHD, common hepatic duct; GB, gall bladder; LHD, left hepatic duct; RAHD, right anterior hepatic duct; RHD, right hepatic duct; RPHD, right posterior hepatic duct.
Discussion
The present study aimed to develop a deep learning model for the automatic segmentation of biliary structure using MRCP images for preoperative planning of living-donor liver transplantation. The results demonstrated the promising performance of the deep learning model in segmenting bile ducts from MRCP images. The mean DSC for the whole biliary structure was 0.80±0.20. This result indicates a relatively high degree of similarity between the automatic segmentation and human-labeled manual segmentation.
The development of an automated segmentation model holds significant potential in preoperative planning in LDLT. By reducing the time and labor required for 3D reconstruction, this model can streamline the surgical planning process and allow for more rapid decision-making. Since 2019, our transplantation center has initiated the image-guidance program for precise surgical planning using 3D modeling software such as Mimics Medical and 3D slicer9. This approach not only helps young surgeons but also surgeons who already overcome the learning curve for laparoscopic living-donor hepatectomy. The biliary anatomy of the donor is of best importance in laparoscopic living-donor hepatectomy due to its high relationship with donor and recipient morbidity7,8,13. Living-donor liver transplantation is related to high rate of biliary complication compared to deceased donor liver transplantation14,15. The difference derives from the anatomical difference in that while deceased donor liver transplantation is mostly performed with whole liver with large single anastomosis of the CBD, living-donor liver transplantation is usually performed with partial liver grafts with IHD with small diameter. A precise surgical technique is required to achieve the best outcome in living-donor liver transplantation to reduce the bile duct opening of the liver graft.
However, these processes require a significant amount of time and human work to finalize the 3D-models. The anatomical understanding to model a precise structure is not given for free and requires significant experience. Therefore, an automated process for this procedure is deemed to enhance the workflow of image-guidance program16.
The automated segmentation model yielded a reasonable outcome for masking the biliary structure. While including the gall bladder for the evaluation enhanced the performance, the outcome for IHDs showed relatively poor performance. This is partly due to the original image itself since IHDs are usually invisible due to its small caliber. These structures are frequently disconnected in MRCP and the ground truth is modeled by connecting the missing voxels by the biomedical artists. While the disconnected parts are connected by human hands, the deep learning model fails to connect it since there is no high signal intensity in the original DICOM image. While the deep learning model showed reasonable outcomes for visually distinguishable structures, it did not reach the level of connecting the isolated structures.
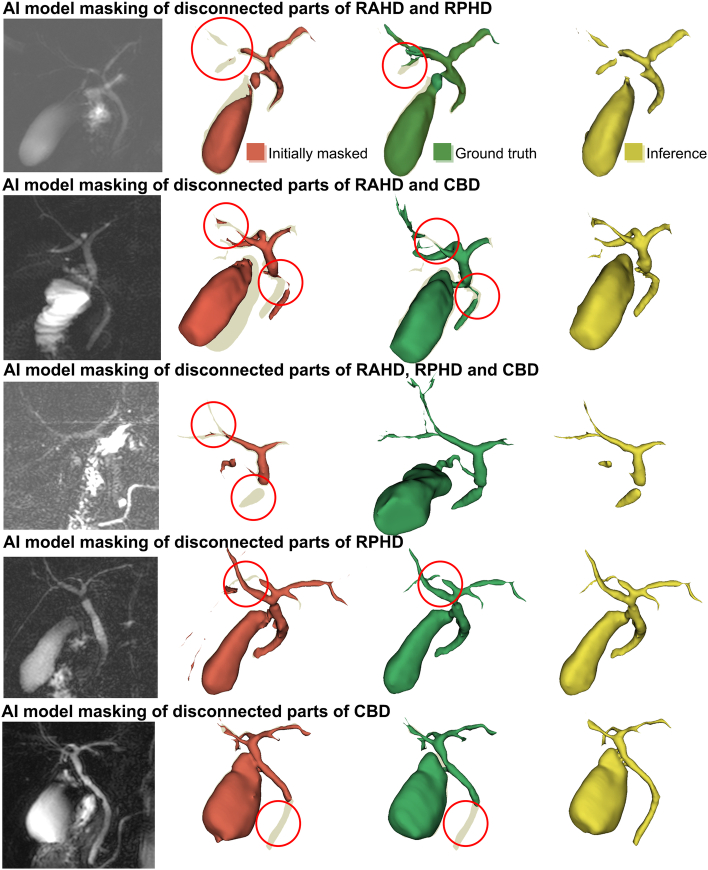
However, there were cases where the model connected the originally disconnected voxels in the original image. Figure 4 summarized those cases where the deep learning model outperformed either the ground truth or the initially masked model in certain anatomical area. These findings show the possibility of the automated model to mimic the human biomedical engineers who knows the anatomical structures and can reconstruct the structure based on their knowledge. The reason for the disconnection in the original image is the unique characteristics of the biliary structure in contrast to the vascular structure. While the hepatic artery, portal vein, or hepatic vein are not disconnected due to its volume and pressure inside, biliary structures can be decompressed by the filling of the GB or the drainage through the duodenum. Therefore, these structures are usually very thin and peripheral structures cannot be observed17,18.
Figure 4.
Cases where the auto-segmentation model showed extra-performance connecting the disconnected structures in the original DICOM images. Images in red represent initially masked 3D-model, whereas 3D-models in green represent ground truth. These five cases show that auto-segmentation model can connect the structures that were disconnected in the original image for reconstructing a biliary structure.
The reason for including qualitative assessment in this study was that the DSC score is considered not to be a perfect method for evaluating the accuracy of the bile duct of MRCP. While the DSC score evaluates the correctness of each voxel objectively, the score itself is highly dependent on the number of total voxels included12. This means that large structures tend to show high scores while small structures show low scores. For biliary structures of MRCP, the DSC score has the disadvantage to underestimate the performance since the biliary structure is a very thin structure in regards of the original DICOM image.
Based on the qualitative method, the deep learning model showed reasonable performance compared to the original MRCP. For CHD and hilum, it even showed a better outcome than the original image. The reason for this seems to originate from the fact that the model was trained based on the ground truth, which was reconstructed by connecting the disconnected structures by the biomedical artist. Since the model was trained with the expert intervening in the disconnected structures, the model also showed promising results.
The dataset used for training and validation was collected from a single institution and predominantly features a homogeneous patient demographic, primarily young individuals with an ideal BMI. This specificity raises concerns about the model’s performance across a more diverse patient population, particularly in older patients and those with a higher BMI, who may present with anatomical and imaging differences that could impact segmentation results. In addition, our study did not explore the clinical impact of the automated segmentation technique nor its integration into the surgical workflow. To bridge this gap, we underscore the importance of conducting further research and validation studies to evaluate the real-world benefits and challenges associated with the implementation of this technology in clinical practice.
Another issue that should be discussed is that the evaluation process for this type of study has innate limitations. The biliary structure is a relatively small and thin structure compared to the liver itself. While the DSC or IOU (intersection of union) score usually shows high performance for relatively large structures, it is relatively poor for small structures. Error of single voxel has significant impact on outcome for structures such as biliary structures. Therefore, readers should consider for this innate limitation of DSC score as the evaluation method.
In conclusion, the developed deep learning model demonstrated promising performance in automatically segmenting bile ducts from MRCP images. The application of this automated segmentation technique holds significant promise in enhancing the preoperative understanding of bile duct structures and augmenting surgical guidance during living-donor liver transplantation procedures. Further research and validation are necessary to establish the clinical utility and wider applicability of this approach.
Ethical approval
This retrospective study was approved by the institutional review board of Samsung Medical Center (SMC 2021-03-056).
Consent
The need for informed consent was waived by the IRB due to the retrospective nature of the study.
Sources of funding
This study was supported by Future Medicine 2030 Project of the Samsung Medical Center [SMX1230771, SMX 1230031], and the Korea Health Technology R&D project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare (HI23C038700).
Author contribution
J.R. and W.K.J.: study design; N.O., J.R., G.-S.C., J.M.K., and J.-W.J.: data collection; N.O. and J.-H.K.: data analysis; N.O., J.-H.K., J.R., and W.K.J.: writing the paper.
Conflicts of interest disclosure
All authors have no conflicts of interest.
Research registration unique identifying number (UIN)
Name of the registry: not applicable.
Unique identifying number or registration ID: not applicable.
Hyperlink to your specific registration (must be publicly accessible and will be checked): not applicable.
Guarantor
Jinsoo Rhu and Woo Kyoung Jeong.
Data availability statement
Data analyzed during the study are available from the corresponding author by request.
Provenance and peer review
Not applicable.
Supplementary Material
Footnotes
Namkee Oh and Jae-Hun Kim contributed equally as first authors.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, www.lww.com/international-journal-of-surgery.
Published online 11 January 2024
Contributor Information
Namkee Oh, Email: ngnyou@gmail.com.
Jae-Hun Kim, Email: jaehun1115.kim@samsung.com.
Jinsoo Rhu, Email: jsrrules@gmail.com.
Woo Kyoung Jeong, Email: jeongwk@gmail.com.
Gyu-Seong Choi, Email: med9370@gmail.com.
Jong Man Kim, Email: yjongman21@gmail.com.
Jae-Won Joh, Email: jwjoh@naver.com.
References
- 1.Merion RM, Schaubel DE, Dykstra DM, et al. The survival benefit of liver transplantation. Am J Transplant 2005;5:307–313. [DOI] [PubMed] [Google Scholar]
- 2.Berg CL, Merion RM, Shearon TH, et al. Liver transplant recipient survival benefit with living donation in the model for endstage liver disease allocation era. Hepatology 2011;54:1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwong AJ, Lai JC, Dodge JL, et al. Outcomes for liver transplant candidates listed with low model for end‐stage liver disease score. Liver Transpl 2015;21:1403–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rela M, Rammohan A. Why are there so many liver transplants from living donors in Asia and so few in Europe and the US? J Hepatol 2021;75:975–980. [DOI] [PubMed] [Google Scholar]
- 5.Jackson WE, Malamon JS, Kaplan B, et al. Survival benefit of living-donor liver transplant. JAMA Surg 2022;157:926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh N, Kim JM, Han S, et al. Survival after living donor liver transplantation versus best supportive care in patients with end-stage liver disease with various MELD-Na scores: retrospective cohort study. BJS Open 2023;7:zrad127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhu J, Choi G-S, Kim JM, et al. Risk factors associated with surgical morbidities of laparoscopic living liver donors. Ann Surg 2023;278:96–102. [DOI] [PubMed] [Google Scholar]
- 8.Rhu J, Kim MS, Choi G-S, et al. A novel technique for bile duct division during laparoscopic living donor hepatectomy to overcome biliary complications in liver transplantation recipients:“cut and clip” rather than “clip and cut”. Transplantation 2021;105:1791–1799. [DOI] [PubMed] [Google Scholar]
- 9.Cherqui D, Ciria R, Kwon CHD, et al. Expert consensus guidelines on minimally invasive donor hepatectomy for living donor liver transplantation from innovation to implementation: a joint initiative from the International Laparoscopic Liver Society (ILLS) and the Asian-Pacific Hepato-Pancreato-Biliary Association (A-PHPBA). Ann Surg 2021;273:96–108. [DOI] [PubMed] [Google Scholar]
- 10.Rhu J, Choi GS, Kim MS, et al. Image guidance using two‐dimensional illustrations and three‐dimensional modeling of donor anatomy during living donor hepatectomy. Clin Transplant 2021;35:e14164. [DOI] [PubMed] [Google Scholar]
- 11.Oh N, Kim J-H, Rhu J, et al. Automated 3D liver segmentation from hepatobiliary phase MRI for enhanced preoperative planning. Sci Rep 2023;13:17605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 2015;15:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong SK, Kim J-Y, Lee J, et al. Pure laparoscopic donor hepatectomy: experience of 556 cases at Seoul National University Hospital. Am J Transplant 2023. 10.1016/j.ajt.2023.06.007 [DOI] [PubMed] [Google Scholar]
- 14.Zimmerman MA, Baker T, Goodrich NP, et al. Development, management, and resolution of biliary complications after living and deceased donor liver transplantation: a report from the adult‐to‐adult living donor liver transplantation cohort study consortium. Liver Transpl 2013;19:259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan P, Yu X, Xia Q. Operative outcomes of adult living donor liver transplantation and deceased donor liver transplantation: a systematic review and meta‐analysis. Liver Transpl 2014;20:425–436. [DOI] [PubMed] [Google Scholar]
- 16.Jones DB, Sung R, Weinberg C, et al. Three-dimensional modeling may improve surgical education and clinical practice. Surg Innov 2016;23:189–195. [DOI] [PubMed] [Google Scholar]
- 17.Nandalur KR, Hussain HK, Weadock WJ, et al. Possible biliary disease: diagnostic performance of high-spatial-resolution isotropic 3D T2-weighted MRCP. Radiology 2008;249:883–890. [DOI] [PubMed] [Google Scholar]
- 18.Nam JG, Lee JM, Kang H-J, et al. GRASE Revisited: breath-hold three-dimensional (3D) magnetic resonance cholangiopancreatography using a Gradient and Spin Echo (GRASE) technique at 3T. Eur Radiol 2018;28:3721–3728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data analyzed during the study are available from the corresponding author by request.