Abstract
During EBV infection, lytic DNA replication activates late gene expression in trans via an uncharacterized pathway. In this study, we mapped the target of this regulatory cascade to a variant TATA box (TATTAAA) and the 3′ flanking region within the core promoter of the BcLF1 gene. The inherent late activity of this core promoter is, surprisingly, disrupted by a heterologous enhancer, suggesting that late gene expression is regulated through core promoter sequences located in a transcriptionally inert environment.
Lytic infections of DNA viruses are characterized by cascades of temporally regulated gene expression ultimately leading to the release of infectious virus. Progression through the viral life cycle is monitored at specific checkpoints; the basic organization of these checkpoints is strikingly similar from bacteriophage to mammalian viruses (33). In the case of Epstein-Barr virus (EBV), entry into the lytic phase of infection is regulated at the level of expression of the immediate-early genes for ZEBRA and Rta (4, 10, 31, 45). Transcription of these products proceeds in the presence of translational inhibitors (2, 44). Expression of the early genes is directed by the immediate-early factors (24), and together, these two groups stimulate replication of the viral genome (2, 7, 8, 44). Following this checkpoint, the structural proteins are synthesized (5, 21, 32, 35, 43) and assembled into mature virions during the late phase of infection. Virion particles encapsidate the newly replicated viral genomes and are then released to initiate a new round of infection.
While lytic replication of the viral genome serves as a key checkpoint in the production of infectious virus, the mechanism by which this event releases the expression of late products has not been elucidated in any viral system. The available information, however, suggests a complex relationship between these two processes. In EBV infection, for example, the viral genome must be amplified for infected cells to support expression of late products (24); however, the transcriptional template itself need not be replicated (36). EBV DNA replication acts in trans, likely initiating a cascade of events which culminates at specific late promoter sequences. By using a reporter assay with EBV-infected human B cells as a model system, we have identified the cis target of this regulatory pathway as the first step in understanding the link between replication and late gene expression.
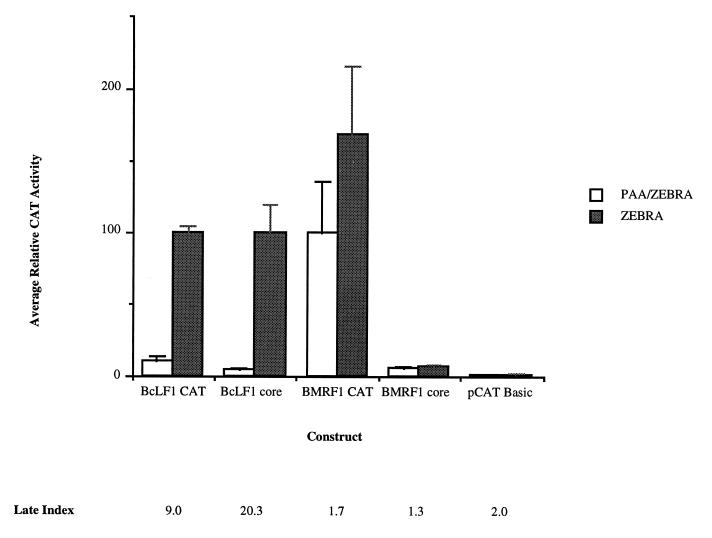
We have previously demonstrated in EBV-infected human B cells that cloned viral promoters of different temporal classes retain the ability to direct viral stage-specific expression of a reporter gene (36). Expression from the BcLF1 late promoter, BcLF1 chloramphenicol acetyltransferase (CAT), exhibits replication-dependent activity: reporter gene expression is markedly decreased by treatment with an inhibitor of the viral DNA polymerase, phosphonoacetic acid (32, 43) (Fig. 1). In contrast, amplification of the viral genome does not alter the level of reporter gene expression from an early viral promoter, BMRF1 CAT, in comparison with the vector control, pCAT Basic (Fig. 1). The faithful recapitulation of viral gene expression by these cloned promoters in infected cells has provided us with an experimentally tractable model to study the regulation of late gene expression in EBV. We have employed a combination of deletional mutagenesis and hybrid promoter construction between BcLF1 and BMRF1 sequences (Fig. 2) to elucidate the cis modulator of late gene expression reported here.
FIG. 1.
Activities of lytic promoters and their corresponding core fragments in EBV-infected cell line HH514-16. The activities of the promoter fragments are expressed as average CAT activity relative to BcLF1 CAT activity in HH514-16 cells (30) cotransfected with ZEBRA as previously described (36). CAT activities are standardized to luciferase activities expressed from the cotransfected pGL2 promoter plasmid (36). The data represents the average of at least two separate transfections, and the error bars indicate the standard error of the mean. The white bars represent the activity of the promoter fragments during the early lytic cycle (PAA [phosphonoacetic acid]/ZEBRA), while promoter activity during the late lytic cycle (ZEBRA) is shown in gray. The ratio of late activity to early activity (late index) for each promoter fragment is shown below the graph.
FIG. 2.
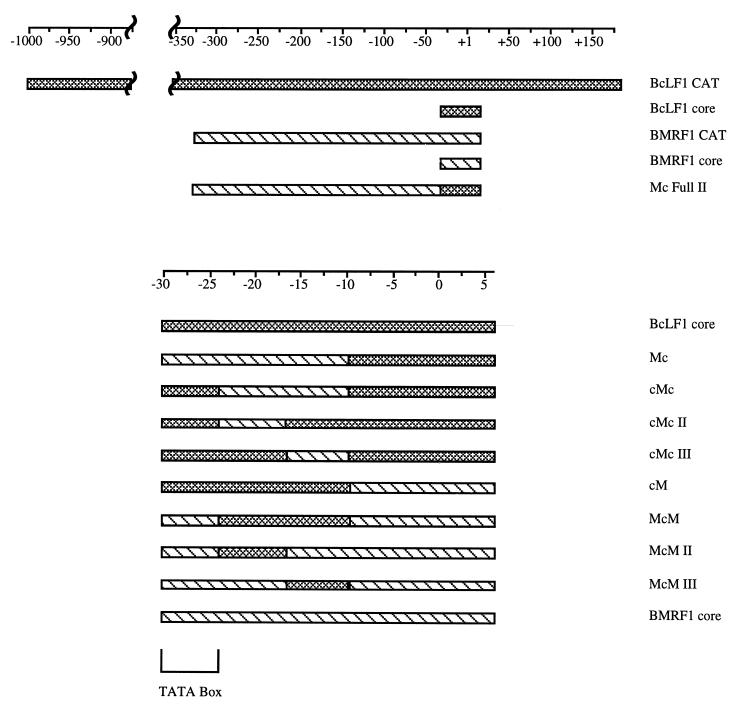
Schematic diagram of promoter fragments. Checked bars represent sequences from the BcLF1 late promoter, while striped bars correspond to sequences from the BMRF1 early promoter. Numbering relative to the transcriptional start site is shown above the schematics, and an expanded view of the core promoters is shown in the lower portion of the diagram.
Deletional analysis of the BMRF1 and BcLF1 promoters suggested that their distinct expression patterns were reflected in different structural organizations. Promoters transcribed by RNA polymerase II are composed of multiple cis-active components residing in two regions, the core promoter and the enhancer (for a review, see reference 49). The core promoter is defined by the presence or absence of two basal DNA elements: the TATA box and the initiator (41). These elements are thought to act in concert to position the basal transcriptional machinery correctly for accurate initiation of transcription. Enhancers of transcription are believed to increase the occupancy of these sites from a distance, thus stimulating gene expression (for a review, see reference 49).
The BMRF1 early promoter exhibited classic promoter construction (49) with strong enhancer sequences 5′ to the basal core elements (Fig. 1, compare BMRF1 CAT to BMRF1 core) (13, 14, 22, 46). In contrast, the BcLF1 late promoter represented a novel promoter class in which regulation occurred exclusively through a core promoter with a unique TATA box, independently of upstream or downstream sequences. Progressive removal of sequences outside of the late BcLF1 core promoter (i.e., upstream of the TATA box and downstream of +6 relative to the transcriptional start site) had no consequence for either the replication dependence or amplitude of reporter gene expression in this system (Fig. 1). Indeed, expression from a 36-bp promoter spanning −30 to +6 of the BcLF1 late promoter, BcLF1 core (Fig. 2), exhibited an even greater replication dependence than did the full-length promoter (Fig. 1, compare BcLF1 CAT to BcLF1 core). This ability to direct strong late temporal expression exclusively through the core region was an intrinsic property of the BcLF1 promoter, as the analogous region of an early promoter, BMRF1 core, did not support this pattern (Fig. 1, compare BcLF1 core to BMRF1 core). This functional difference between early and late core promoters, furthermore, suggested that DNA replication stimulates gene expression by a sequence-dependent mechanism rather than by evoking a globally acting change in the basal transcriptional machinery.
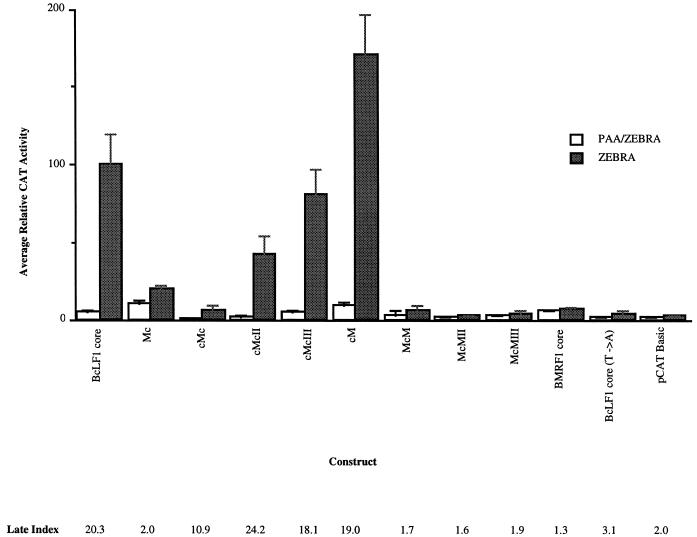
Exploiting the functional differences between late and early promoter core sequences to characterize cis-active regulatory elements further, we constructed a series of hybrid early and late core promoters (Table 1 and Fig. 2, bottom) and analyzed their ability to direct replication-dependent expression of CAT in EBV-infected human B cells. Bisection of the core promoters at −9 relative to the transcriptional start site localized the replication-dependent cis-active site to the 5′ end of the late core promoter, between −30 and −10, suggesting that the initiator element was neither necessary nor sufficient for late gene expression (Fig. 2 and 3, compare cM to Mc). Substitution of the BMRF1 early gene TATA box into cM (Fig. 2, bottom) abolished replication-dependent activity from that promoter (Fig. 3, compare cM to McM). Reciprocally, introduction of the BcLF1 late gene TATA box into Mc (Fig. 2, bottom) imparted increased replication dependence to this promoter, albeit at a reduced level (Fig. 3, compare Mc to cMc). Sequences between −25 and −10 of the BcLF1 promoter restored full-level activity to this hybrid promoter (Fig. 3, compare cMcII and cMcIII to cMc), but this region did not alter expression in the presence of an early gene TATA box (Fig. 3, compare BMRF1 core to McM, McMII, and McMIII).
TABLE 1.
Oligonucleotides used to construct promoters
| Construct | Sequence(s)a |
|---|---|
| cmin | AACTGCAGTATTAAACCGGGTGGCAGCTCCT GGCAGTCATTCATCTAGAGC |
| Mmin | AACTGCAGCATAAATTCTCCTGCCTGCCTCTGCTCTGGTACGTTGTCTAGAC |
| Mc | AACTGCAGCATAAATTCTCCTGCCTGCCTCCTGGCAGTCATTCATCTAGAGC |
| cMc | AACTGCAGTATTAAATC TCCTGCCTGCCCCTGGCAGTCATTCATTCTAGAGC |
| cMcII | AACTGCAGTATTAAATCTCCTGGCAGCTCCTGGCAGTCATTCATTCTAGAGC |
| cMcIII | AACTGCAGTATTAAACCGGGTGCCTGCCCCTGGCAGTCATTCATTCTAGAGC |
| cM | AACTGCAGTATTAAACCGGGTGGCAGCTCTGCTCTGGTACGTTCTCTAGAGC |
| McM | AACTGCAGCATAAATCCGGGTGGCAGCTCTGCTCTGGTACGTTGTCTAGAGC |
| McMII | AACTGCAGCATAAATCCGGGTGCCTGCCTCTGCTCTGGTACGTTGCTAGAGC |
| McMIII | AACTGCAGCATAAATTCTCCTGGCAGCTTCTGCTCTGGTACGTTGTCTAGAC |
| cmin (T→A) | AACTGCAGTATAAAACCGGGTGGCAGCTCCTGGCAGTCATTCATCTAGAGC |
| McFull IIb | AACTGCAGGGATCCAGGAGCTGTT |
| GAGGCGGCAGGAGATTTAATAATTCT | |
| TCTCCTGCCTGCCTCTGCTCTGGTATTCATTCGGATAAC | |
| GCTCTAGATGAATGACTGCCAGGAGCTGCCACCCGGTTTAA TACCCAGAAA |
TATA sequences are italicized, and the transcriptional start site is underlined for the minimal promoters. Core promoters were cloned between the PstI and XbaI sites of pCAT Basic (Promega).
Mc Full II fuses BMRF1 promoter sequences between −335 and −31 relative to the start of transcription to BcLF1 promoter sequences between −30 and +6. The oligonucleotides listed were used in a two-step PCR.
FIG. 3.
Activities of hybrid core promoter fragments in EBV-infected cell line HH514-16. Shading is described in the legend to Fig. 1. The data represents the average of at least two separate transfections. PAA, phosphonoacetic acid.
Despite the functional differences between the early and late promoter TATA sequences, both were capable of binding to purified TATA binding protein (TBP) in vitro (data not shown). Surprisingly, core promoters with late activity produced identical electrophoretic mobility shift patterns when incubated with nuclear extracts from virally infected cells during the early or late phase (data not shown). Therefore, late gene expression in EBV infection was mediated through a basal transcriptional element, the TATA box, without obvious recruitment of additional DNA binding factors, a mechanism proposed in other viral systems (27).
Comparison of the early BMRF1 TATA box sequence (CATAAAT) to that of the late BcLF1 TATA box sequence (TATTAAA) (1) implicated three potential positions for regulation. A total of 13 (72%) of 18 TATA box sequences from known EBV late promoters contained a T at the fourth position, while this variation was not observed in any latent or early EBV TATA boxes (1). By comparison, the promoters of several homologs of EBV late genes in Kaposi sarcoma-associated herpesvirus, a related gammaherpesvirus, contained a T in the fourth position of the TATA box (34), and this variant also occurred in 30% of the promoters in the eukaryotic database (3). Mutation of this T to an A, restoring the consensus TATA box sequence (20, 29, 48) to the late BcLF1 core promoter, abolished replication-dependent activity (Fig. 3, compare BcLF1 core to BcLF1 core [T→A]). While the activity of this promoter is extremely low, expression was reproducibly higher than that obtained with both the vector control and an A-to-G mutation in the second position of the TATA box (TGTTAA) which disrupts TBP binding (48 and data not shown). These data strongly implicate the alternative TATA box sequence (TATTAAA) in late temporal regulation of viral gene expression in EBV-infected human B cells.
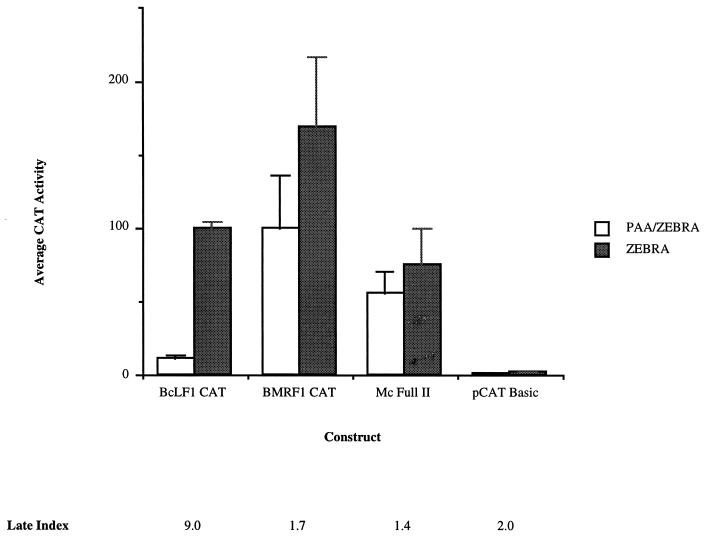
An obvious explanation for these observations was active repression of late promoter activity through the unique TATA box prior to lytic viral DNA replication. To address this possibility, we constructed a hybrid promoter linking the BMRF1 early enhancer region (13, 14, 22, 46) to the BcLF1 late core promoter (Mc Full II, Fig. 2). As is characteristic of early promoters, this promoter directed reporter gene expression to nearly identical levels before and after lytic replication of the viral genome (Fig. 4). The ratio of these activities was nearly identical to that of the wild-type BMRF1 early promoter (Fig. 4, compare BMRF1 CAT to Mc Full II). The late TATA box sequence was, therefore, able to support expression from an upstream enhancer element, fulfilling its well-documented role as a basal transcriptional element (49). In the absence of an upstream enhancer, however, the late TATA box sequence functioned in temporal regulation, directing replication-dependent gene expression (Fig. 3). These observations were consistent with the activity of the BcLF1 core promoter within its normal context: the region 5′ to the BcLF1 core promoter in the viral genome did not confer activity when transferred to a heterologous core promoter, and removal of this region did not alter expression from its own promoter (Fig. 1).
FIG. 4.
Effect of fusion of the BcLF1 core promoter to the BMRF1 enhancer. See Fig. 1 for a description of the constructs. Shading is described in the legend to Fig. 1. The data represents the average of at least two separate transfections. PAA, phosphonoacetic acid.
The TATA box has classically been regarded as a static component in the temporal regulation of gene expression. While this element is absolutely required for activity from most promoters, the expression pattern is generally dictated by specific regulators acting outside of the core promoter (19, 28). Recently, however, general transcription factors, such as alternate TBPs or TBP-associated factors, have been directly implicated in temporal or cell type-specific expression (6, 12, 37), although the mechanisms by which these factors target a specific promoter have yet to be elucidated. Other studies have hinted at the importance of enhancer-TATA sequence cross talk (25, 39, 40, 47, 48), but the dominant regulation in these cases is always the promoter-specific enhancer rather than the basal element. Here, we have characterized a system in which an alternate TATA sequence itself directs temporal expression which is enhanced by nearby cis-active sequences.
Our findings indicate the existence of at least two distinct promoter architectures in the EBV genome. The BMRF1 early promoter directs expression through sequences outside of the core promoter (Fig. 1) (13, 14, 22, 46). In contrast, temporal expression from the BcLF1 late promoter is solely dependent upon a variant TATA element and its interaction with flanking sequences within the core promoter (Fig. 3). While both the early enhancer and late core promoter are autonomous regulatory units, their combination into a heterologous system (Fig. 4) leads to an observed dominance of the early enhancer, rather than a hybrid expression pattern. This complex interaction suggests that regulatory mechanisms active before and after amplification of the EBV genome are unique and highlights the importance of studying both basal expression and enhancer contributions to observed expression patterns.
Comparison of these results with the extensive work completed on herpes simplex virus (HSV) late gene expression reveals both striking similarities and unique adaptations by EBV. For example, late gene expression in both EBV and HSV (γ2) is dependent upon lytic DNA replication, but the requirement is in cis for HSV (18, 26, 38, 48) and in trans for EBV (36). A cis regulator of late gene expression in HSV, as in EBV, occurs near the core promoter (9, 11, 15, 16, 18, 23). In both HSV and EBV, early minimal promoters cannot functionally substitute for late minimal promoters (Fig. 3) (15), but late minimal promoters can support replication-independent expression from early enhancers (Fig. 4) (15). While both viruses have targeted regulatory elements near late core promoters, the specific targets are unique. HSV late gene expression requires specific sequences within the vicinity of the transcriptional start site (9, 11, 15, 16, 18, 23), but in contrast to EBV, the TATA boxes of early and late genes are interchangeable in HSV (17, 42). Identification of the cis regulators of late gene expression in both HSV and EBV are important initial steps in ultimately understanding the complex link to lytic DNA replication, but a complete understanding requires identification of trans factors, basal or promoter specific, interacting with these elements. The work presented here provides a foundation for a detailed mechanistic understanding of late gene expression regulation in EBV.
Acknowledgments
This work was supported by NIH grants (CA 12055 and CA 16038) to G.M. and an HHMI predoctoral fellowship to T.R.S.
We thank Sean Juo for supplying purified TBP and Bernard Roizman and Lucia Rothman-Denes for helpful discussions and comments on the manuscript.
REFERENCES
- 1.Baer R, Bankier A T, Biggin M D, Deininger P, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 2.Biggin M, Bodescot M, Pericaudet M, Farrell P. Epstein-Barr virus gene expression in P3HR-1-superinfected Raji cells. J Virol. 1987;61:3120–3132. doi: 10.1128/jvi.61.10.3120-3132.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 4.Countryman J, Miller G. Activation of expression of latent Epstein-Barr virus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datta A K, Hood R E. Mechanism of inhibition of Epstein-Barr virus replication by phosphonoformic acid. Virology. 1981;114:52–59. doi: 10.1016/0042-6822(81)90251-8. [DOI] [PubMed] [Google Scholar]
- 6.Dikstein R, Zhou S, Tjian R. Human TAFII105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- 7.Fixman E D, Hayward G S, Hayward S D. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol. 1995;69:2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fixman E D, Hayward G S, Hayward S D. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flanagan W M, Papavassiliou A G, Rice M, Hecht L B, Silverstein S, Wagner E K. Analysis of the herpes simplex virus type 1 promoter controlling the expression of UL38, a true late gene involved in capsid assembly. J Virol. 1991;65:769–786. doi: 10.1128/jvi.65.2.769-786.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grogan E, Jenson H, Countryman J, Heston L, Gradoville L, Miller G. Transfection of a rearranged viral DNA fragment, WZhet, stably converts latent Epstein-Barr viral infection to productive infection in lymphoid cells. Proc Natl Acad Sci USA. 1987;84:1332–1336. doi: 10.1073/pnas.84.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzowski J F, Wagner E K. Mutational analysis of the herpes simplex virus type 1 strict late UL38 promoter/leader reveals two regions critical in transcriptional regulation. J Virol. 1993;67:5098–5108. doi: 10.1128/jvi.67.9.5098-5108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen S, Takada S, Jacobson R, Lis J, Tjian R. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell. 1997;91:71–83. doi: 10.1016/s0092-8674(01)80010-6. [DOI] [PubMed] [Google Scholar]
- 13.Hardwick J M, Lieberman P M, Hayward S D. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol. 1988;62:2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holley-Guthrie E A, Quinlivan E B, Mar E-C, Kenney S. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J Virol. 1990;64:3753–3759. doi: 10.1128/jvi.64.8.3753-3759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Homa F L, Glorioso J C, Levine M. A specific 15-bp TATA box promoter element is required for expression of a herpes simplex virus type 1 late gene. Genes Dev. 1988;2:40–53. doi: 10.1101/gad.2.1.40. [DOI] [PubMed] [Google Scholar]
- 16.Homa F L, Otal T M, Glorioso J C, Levine M. Transcriptional control signals of a herpes simplex virus type 1 late (γ2) gene lie within bases −34 to +124 relative to the 5′ terminus of the mRNA. Mol Cell Biol. 1986;6:3652–3666. doi: 10.1128/mcb.6.11.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imbalzano A N, DeLuca N A. Substitution of a TATA box from a herpes simplex virus late gene in the viral thymidine kinase promoter alters ICP4 inducibility but not temporal expression. J Virol. 1992;66:5453–5463. doi: 10.1128/jvi.66.9.5453-5463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson P A, Everett R D. The control of herpes simplex virus type-1 late gene transcription: a ‘TATA-box’/cap site region is sufficient for fully efficient regulated activity. Nucleic Acids Res. 1986;14:8247–8264. doi: 10.1093/nar/14.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson P F, McKnight S L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- 20.Juo Z S, Chiu T K, Leiberman P M, Baikalov I, Berk A, Dickerson R. How proteins recognize the TATA box. J Mol Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 21.Katz B Z, Raab-Traub N, Miller G. Latent and replicating forms of Epstein-Barr virus DNA in lymphomas and lymphoproliferative diseases. J Infect Dis. 1989;160:589–598. doi: 10.1093/infdis/160.4.589. [DOI] [PubMed] [Google Scholar]
- 22.Kenney S, Kamine J, Holley-Guthrie E, Lin J-C, Mar E C, Pagano J. The Epstein-Barr virus (EBV) BZLF1 immediate-early gene product differentially affects latent versus productive EBV promoters. J Virol. 1989;63:1729–1736. doi: 10.1128/jvi.63.4.1729-1736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kibler P K, Duncan J, Keith B D, Hupel T, Smiley J R. Regulation of herpes simplex virus true late gene expression: sequences downstream from the US11 TATA box inhibit expression from an unreplicated template. J Virol. 1991;65:6749–6760. doi: 10.1128/jvi.65.12.6749-6760.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieff E. Epstein-Barr virus and its replication. In: Fields B, Knipe D, Howley P, editors. Fields virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 25.Leitman D C, Mackow E R, Williams T, Baxter J D, West B L. The core promoter region of the tumor necrosis factor alpha gene confers phorbol ester responsiveness to upstream transcriptional activators. Mol Cell Biol. 1992;12:1352–1356. doi: 10.1128/mcb.12.3.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mavromara-Nazos P, Roizman B. Activation of herpes simplex virus 1 gamma 2 genes by viral DNA replication. Virology. 1987;161:593–598. doi: 10.1016/0042-6822(87)90156-5. [DOI] [PubMed] [Google Scholar]
- 27.Miller A, Wood D, Ebright R, Rothman-Denes L. RNA polymerase b′ subunit: a target of DNA binding-independent activation. Science. 1997;275:1655–1657. doi: 10.1126/science.275.5306.1655. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell P J, Tijan R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 29.Nikolov D B, Chen H, Halay E D, Hoffmann A, Roeder R G, Burley S K. Crystal structure of a human TATA box-binding protein/TATA element complex. Proc Natl Acad Sci USA. 1996;93:4862–4867. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabson M, Heston L, Miller G. Identification of a rare Epstein-Barr virus variant that enhances early antigen expression in Raji cells. Proc Natl Acad Sci USA. 1983;80:2762–2766. doi: 10.1073/pnas.80.9.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ragoczy T, Heston L, Miller G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72:7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rickinson A B, Finerty S, Epstein M A. Inhibition by phosphonoacetate of the in vitro outgrowth of Epstein-Barr virus genome-containing cell lines from the blood of infectious mononucleosis patients. IARC Sci Publ. 1978;1978:721–728. [PubMed] [Google Scholar]
- 33.Roizman B, Palese P. Multiplication of viruses: an overview. In: Fields B, Knipe D, Howley P, editors. Fields virology. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 101–112. [Google Scholar]
- 34.Russo J, Bohenzky R, Chien M, Chen J, Yan M, Maddalena D, Parry J, Peruzzi D, Edelman I, Chang Y, Moore P. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seibel R, Wolf H. Mapping of Epstein-Barr virus proteins on the genome by translation of hybrid-selected RNA from induced P3HR1 cells and induced Raji cells. Virology. 1985;141:1–13. doi: 10.1016/0042-6822(85)90177-1. [DOI] [PubMed] [Google Scholar]
- 36.Serio T R, Kolman J L, Miller G. Late gene expression from the Epstein-Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J Virol. 1997;71:8726–8734. doi: 10.1128/jvi.71.11.8726-8734.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen W, Green M. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 38.Silver S, Roizman B. γ2-Thymidine kinase chimeras are identically transcribed but regulated as γ2 genes in herpes simplex virus genomes and as β genes in cell genomes. Mol Cell Biol. 1985;5:518–528. doi: 10.1128/mcb.5.3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon M C, Fisch T M, Benecke B J, Nevins J R, Heintz H. Definition of multiple, functionally distinct TATA elements, one of which is a target in the hsp70 promoter for E1A regulation. Cell. 1988;52:723–729. doi: 10.1016/0092-8674(88)90410-2. [DOI] [PubMed] [Google Scholar]
- 40.Simon M C, Rooney R J, Fisch T M, Heintz N, Nevins J R. E1A-dependent trans-activation of the c-fos promoter requires the TATAA sequence. Proc Natl Acad Sci USA. 1990;87:513–517. doi: 10.1073/pnas.87.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smale S, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 42.Steffy K R, Weir J P. Mutational analysis of two herpes simplex virus type 1 late promoters. J Virol. 1991;65:6454–6460. doi: 10.1128/jvi.65.12.6454-6460.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Summers W C, Klein G. Inhibition of Epstein-Barr virus DNA synthesis and late gene expression by phosphonoacetic acid. J Virol. 1976;18:151–155. doi: 10.1128/jvi.18.1.151-155.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takada K, Shimizu N, Sakuma S, Ono Y. trans activation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J Virol. 1986;57:1016–1022. doi: 10.1128/jvi.57.3.1016-1022.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urier G, Buisson M, Chambard P, Sergeant A. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 1989;8:1447–1453. doi: 10.1002/j.1460-2075.1989.tb03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wefald F, Devlin B, Williams R. Functional heterogeneity of mammalian TATA-box sequences revealed by interaction with a cell-specific enhancer. Nature. 1990;344:260–262. doi: 10.1038/344260a0. [DOI] [PubMed] [Google Scholar]
- 48.Wobbe C R, Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol Cell Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zawel L, Reinberg D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog Nucleic Acid Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]