Abstract
The Epstein-Barr virus (EBV) homolog of the conserved herpesvirus glycoprotein gN is predicted to be encoded by the BLRF1 open reading frame (ORF). Antipeptide antibody to a sequence corresponding to residues in the predicted BLRF1 ORF immunoprecipitated a doublet of approximately 8 kDa from cells expressing the BLRF1 ORF as a recombinant protein. In addition, four glycosylated proteins of 113, 84, 48, and 15 kDa could be immunoprecipitated from virus-producing cells by the same antibody. The 15-kDa species was the mature form of gN, which carried α2,6-sialic acid residues. The remaining glycoproteins which associated with gN were products of the BBRF3 ORF of EBV, which encodes the EBV gM homolog. The 8-kDa doublet seen in cells expressing recombinant gN comprised precursors of the mature 15-kDa gN. Coexpression of EBV gM with EBV gN was required for authentic processing of the 8-kDa forms to the 15-kDa form.
The envelopes of herpesviruses include multiple glycoproteins which are known or inferred to play various roles in the attachment, entry, egress, and spread of virus. Some of these glycoproteins appear to be unique to individual members or subfamilies of herpesviruses, presumably reflecting adaptations to specialized biological niches; others are more obviously conserved in either sequence or genomic organization, reflecting a common evolutionary origin, if not a current commonality of function. By general consensus, five glycoproteins, gB, gH, gL, gN, and gM, fall under the heading of conserved molecules, although not all have been well characterized, even in some of the most extensively studied of herpesviruses.
The five Epstein-Barr virus (EBV) open reading frames (ORF) that are predicted to encode members of the conserved group of glycoproteins are BXLF2 (encoding gH), BKRF2 (encoding gL), BALF4 (encoding gB), BLRF1 (encoding gN), and BBRF3 (encoding gM) (1). Glycoprotein gp85, the EBV gH (8, 22), glycoprotein gp25, the EBV gL (31), and glycoprotein gp110, the EBV gB (6), have all been characterized biochemically and functionally, and a mixed picture is emerging when their behavior is compared to that of their counterparts in other herpesviruses. The EBV gB is not a major virion component as it is in many herpesviruses but, rather, is localized primarily to the nuclear membrane of virion-producing cells (5), and although in many herpesviruses gB is critical to virus entry, in EBV it plays a major role in virus assembly (9, 14). In contrast, although the EBV gH-gL complex includes a third poorly conserved but functionally important glycoprotein gp42 (18), in general it behaves like that of other herpesviruses. Glycoprotein gH is retained in the endoplasmic reticulum in the absence of its partner gL (25, 31), and the virion-associated complex plays a critical role in virus entry (7, 17, 18, 30).
Neither the gN nor the gM homologs of EBV have yet been described. All gN homologs are predicted to be small type 1 membrane proteins. The EBV BLRF1 ORF is predicted to encode a type 1 membrane protein of 102 amino acids with an Mr of 10,944 without posttranslational modification (1). The putative sequence includes no potential N-linked glycosylation sites, but 12 of the 102 amino acids are serine or threonine residues that are predicted to lie outside the signal sequence or carboxyl-terminal transmembrane domain and might be targets for O-linked glycosylation. Descriptions of gN homologs in the alphaherpesviruses, however, indicate a variety of processing differences. The pseudorabies virus (PRV) gN homolog is a 14-kDa O-glycosylated protein which is associated with the virion (11). The herpes simplex virus type 1 (HSV-1) homolog is a 12-kDa protein (3) that does not appear to carry posttranslational modifications (26). The bovine herpesvirus 1 homolog is a 9-kDa nonglycosylated membrane protein found on the surface of the virion (19). It is reported to be disulfide linked to a second virion component and to be more tightly associated with the tegument than is the envelope glycoprotein gD. The varicella-zoster virus homolog is a 7-kDa protein which is very close to the predicted size of the molecule after cleavage of the signal peptide and without further posttranslational modifications (27). Most recently, the PRV gN homolog has been shown to exist in a disulfide-linked complex with glycoprotein gM and to be dependent on gM for localization in the virion (10).
To examine the structure and processing of the EBV gN homolog, we expressed the protein as a recombinant molecule and compared its biosynthesis with that of the native protein in virus. We report here that the BLRF1 ORF of EBV encodes a 15-kDa glycoprotein which carries O-linked sugars, whose processing differs for the recombinant and the native proteins. Like all the glycoprotein gH homologs, the EBV gN associates with and requires the presence of a second glycoprotein for authentic processing. As in PRV, the EBV glycoprotein gN associates with the gM homolog, the product of the BBRF3 ORF.
MATERIALS AND METHODS
Cells.
Akata, a Burkitt lymphoma-derived cell line that carries and can be induced to make EBV (28) (a gift of John Sixbey, St. Jude Children’s Research Hospital, Memphis, Tenn.), was grown in RPMI 1640 (Sigma Chemical Co., St. Louis, Mo.) supplemented with 10% heat-inactivated fetal bovine serum (Gibco/BRL Life Technologies, Grand Island, N.Y.). CV-1 cells were grown in Dulbecco’s modified Eagle’s medium containing 10% heat-inactivated fetal bovine serum.
Virus production.
EBV was obtained from Akata cells which were resuspended at 2 × 106 per ml and induced with 100 μg of anti-human immunoglobulin G per ml for 5 days. Stocks of vaccinia virus expressing the T7 RNA polymerase (vvT7) (21) were grown in CV-1 cells infected at a multiplicity of infection of 0.01 and harvested by freeze-thawing and sonication of cells.
Expression in the pTM1 vector.
The BLRF1 ORF was amplified by a PCR method from the EcoRI G fragment of Akata virus DNA. The 5′ primer (GGC GC↓C ATG GGG AAG GTC CT) included the first ATG of the ORF and an NcoI site. The 3′ primer (GGC GC↓T CGA GCA TCT AAT CCG) included the BLRF1 stop codon and an XhoI site. The DNA amplified with these primers was cut with NcoI and XhoI and inserted into pTM1 (21) that had previously been cut with the same enzymes to make plasmid pTM1-gN. The BBRF3 ORF was amplified from Akata virus DNA. The 5′ primer (GGC GT↓C ATG AAG TCC AAG A) included the first ATG of the ORF and a BspHI site. The 3′ primer (GGC GGA GCT↓ CTT AGG GGA AGA T) included the BBRF3 stop codon and a SacI site. The DNA amplified with these primers was cut with BspHI and SacI and inserted into pTM1 that had been cut with SacI and NcoI (which leaves ends compatible with those produced by BspHI) to make plasmid pTM1-gM. Plasmids were transfected into CV-1 cells 30 min after the cells had been infected with vvT7. For transfections of a single plasmid, 5 μg of DNA was mixed with 50 μl of Lipofectin (Gibco/BRL) made up to a total volume of 800 μl with serum-free medium. For transfections of two plasmids, the total amount of DNA used was 5 μg. Each mixture was incubated for 45 min at room temperature before being added to cells.
Antibodies.
Three antipeptide antibodies were made, as described previously (22), to synthetic peptides corresponding respectively to residues 125 to 137 of the BKRF2 ORF (31) (anti-gL), to residues 44 to 55 and 55 to 69 of the predicted BLRF1 ORF (anti-gN), and to residues 346 to 364 of the BBRF3 ORF (anti-gM). A cysteine residue not present in the virus sequence was added to the amino terminus of the BBRF3 and BKRF2 peptides for ease of coupling to keyhole limpet hemocyanin. All antibodies were purified by chromatography on protein A (Sigma) coupled to Affi-Gel-15 (Bio-Rad, Richmond, Calif.).
Radiolabeling and immunoprecipitation.
EBV was labeled extrinsically with 125I (Amersham Corp., Arlington Heights, Ill.) after concentration by centrifugation from 4 liters of spent culture medium as previously described (31). EBV proteins were labeled biosynthetically with [3H]leucine or [3H] glucosamine (20 Ci/mmol; Amersham) for 20 h at 6 h after induction with anti-human immunoglobulin G as previously described (31). CV-1 cells that had been infected with vvT7 and transfected with pTM1 DNA, pTM1-gN DNA, pTM1-gM DNA, or a mixture of plasmids were labeled with [3H]leucine or [3H] glucosamine as previously described (16). Labeled cells were solubilized in radioimmunoprecipitation buffer (50 mM Tris-HCl [pH 7.2], 0.15 M NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 0.1 mM phenylmethylsulfonyl fluoride, 100 U of aprotinin per ml) and immunoprecipitated with antibody and protein A-Sepharose CL4B (Sigma). Immunoprecipitated proteins were washed, dissociated either by boiling for 2 min or by heating to 37°C for 30 min in sample buffer with or without 2-mercaptoethanol, and analyzed by SDS-polyacrylamide gel electrophoresis in 18% acrylamide cross-linked with 0.09% bisacrylamide or in 9 to 18% polyacrylamide cross-linked with 0.28% N,N′-diallyltartardiamide followed by fluorography.
Oligosaccharide digestion.
For removal of sugars, immunoprecipitated proteins were washed in radioimmunoprecipitation buffer and resuspended in buffer (0.1% SDS, 50 mM EDTA, 1% 2-mercaptoethanol, 100 mM sodium phosphate, 0.5% N-octylglucoside [pH 7.2 or pH 5.0]), boiled for 2 min, cooled, and incubated at 37°C for 20 h with the addition of either 5 mU of neuraminidase from Newcastle disease virus or 5.0 mU of neuraminidase from Arthrobacter, or neuraminidase and 2.5 mU of O-glycosidase. All enzymes were from Boehringer Mannheim. Digested samples were analyzed by SDS-polyacrylamide gel electrophoresis and fluorography.
RESULTS
Expression of gN as a recombinant protein.
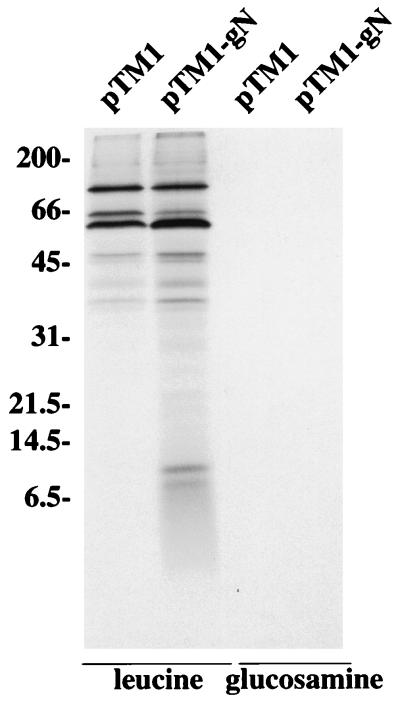
The BLRF1 ORF is predicted to encode the gN homolog of EBV, a molecule which is expected, without posttranslational modification, to be a protein of approximately 10 kDa. Structural glycoproteins of EBV can be expressed only by inducing virus in a proportion of latently infected B cells, usually fewer than 50%, to switch from the latent to the lytic cycle. Hence, virus proteins which are not particularly abundant are very difficult to visualize against a background of cellular protein synthesis. To make a preliminary identification of the putative EBV gN, we therefore cloned the BLRF1 ORF into the pTM1 vector for expression under the control of the strong bacteriophage T7 promoter. A rabbit antibody, anti-gN, was made to two peptides corresponding to residues 44 through 55 and 55 through 69 of the predicted BLRF1 sequence and used to examine the expression of pTM1-gN in CV-1 cells that had been concurrently infected with a recombinant vaccinia virus vvT7 that expresses the T7 polymerase. Anti-gN immunoprecipitated a unique doublet of approximately 8 kDa from cells that had been labeled with [3H] leucine (Fig. 1). Longer exposures of the gel revealed the two faint bands at just above the 21.5-kDa marker and just below the 31-kDa marker to be present in cells transfected with vector alone. In contrast, no protein was immunoprecipitated from cells that had been labeled with [3H] glucosamine.
FIG. 1.
Electrophoretic analysis (in 18% polyacrylamide gels) of proteins immunoprecipitated with anti-gN from CV-1 cells transfected with pTM1 vector or pTM1-gN, infected with vvT7 and labeled with [3H] leucine or [3H] glucosamine. Sizes are indicated on the left in kilodaltons.
Expression of gN in Akata cells.
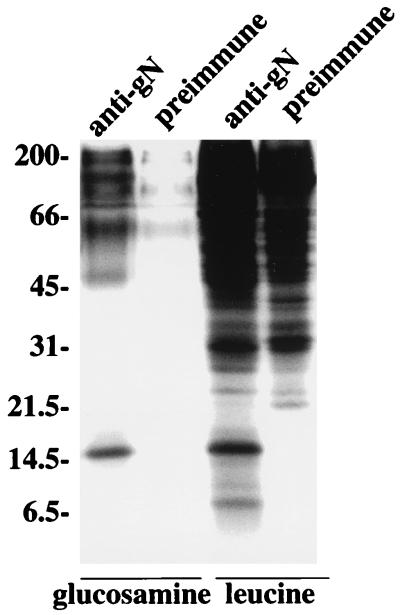
With the preliminary identification of EBV gN as a protein slightly smaller than predicted, perhaps as the result of posttranslational cleavage of the putative signal peptide, the anti-gN antibody was used to look for expression in Akata cells that had been induced with anti-immunoglobulin. The antibody reacted with both fixed and unfixed cells in indirect-immunofluorescence assays (18), suggesting that gN was expressed on the cell surface (data not shown). Anti-gN but not preimmune antibody from the same animal immunoprecipitated a doublet of approximately 8 kDa from Akata cells that had been labeled with [3H] leucine which was similar to that seen in CV-1 cells transfected with pTM1- gN (Fig. 2). In addition, however, a larger, more prominent 15- kDa species was seen. Both anti-gN and preimmune antibody nonspecifically immunoprecipitated a large number of [3H] leucine-labeled proteins with masses greater than approximately 30 kDa. The autoradiographs were clearer when proteins were instead labeled with [3H] glucosamine. The 8- kDa doublet carried no detectable sugar label. This had also been true for the recombinant protein expressed in pTM1. However, the 15- kDa species labeled well with [3H] glucosamine. A very long exposure of the autoradiograph gave evidence of faint sugar labeling of the upper of the two 8- kDa species (data not shown). Additional diffuse bands only faintly visible with preimmune antibody were evident just above the 45- and 66- kDa markers and at the top of the gel, although their apparent masses were uncertain in the high percentage polyacrylamide (18%) that had been used for optimum visualization of low-molecular-weight proteins.
FIG. 2.
Electrophoretic analysis in 18% polyacrylamide of proteins immunoprecipitated from Akata cells that had been induced with anti-immunoglobulin and labeled with [3H] glucosamine or [3H] leucine. Proteins were immunoprecipitated with anti-gN or with preimmune antibody from the same rabbit. Sizes are indicated on the left in kilodaltons.
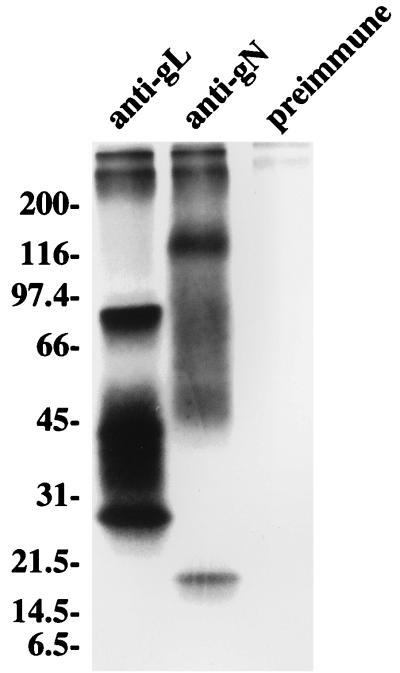
To obtain better resolution of the higher-molecular-weight species, proteins immunoprecipitated from Akata cells labeled with [3H] glucosamine were heated to 37°C rather than 100°C to dissociate them from protein-A agarose and were electrophoresed in 9 to 18% polyacrylamide. Although the higher-molecular-weight species still ran very diffusely, producing a smear of radioactivity throughout the gel, they were better resolved, since at least three glycoproteins of approximately 48, 84, and 113 kDa were visible (Fig. 3). For comparative purposes, the EBV gH-gL complex, which comprises three proteins of 85, 42/38, and 25 kDa, was included in the gel.
FIG. 3.
Electrophoretic analysis in 9 to 18% polyacrylamide of proteins immunoprecipitated from Akata cells that had been induced with anti-immunoglobulin and labeled with [3H] glucosamine. Proteins were immunoprecipitated with a rabbit anti-peptide antibody to the EBV gL (gp25), which immunoprecipitates the EBV gH-gL complex of gp85, gp42/38, and gp25, with rabbit anti-peptide antibody anti-gN, or with preimmune rabbit antibody. Immunoprecipitated proteins were eluted from protein A-agarose beads by heating at 37°C. Sizes are indicated on the left in kilodaltons.
Association of gN with gM.
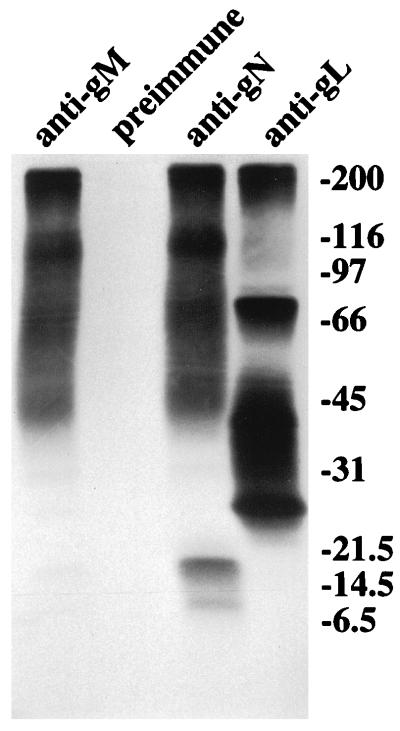
The gN homologs of all other herpesviruses studied to date are relatively small proteins. The immunoprecipitation of such high-molecular-weight species from Akata cells by the anti-gN antibody was thus a surprise. The very recent observation that the PRV gN homolog associates with gM (10) then suggested that the higher-molecular-weight species seen in Akata cells, but not in CV-1 cells transfected with pTM1-gN alone, might be products of the EBV gM homolog. To test the hypothesis, an anti-peptide antibody, anti-gM, was made to a peptide corresponding to residues 346 to 364 encoded by the BBRF3 ORF, which is predicted to encode EBV gM. The anti-gM antibody immunoprecipitated proteins from induced Akata cells labeled with [3H] glucosamine that comigrated with the higher-molecular-weight proteins immunoprecipitated by anti-gN (Fig. 4). A very faint band at 15 kDa comigrated with the prominent 15- kDa species immunoprecipitated by anti-gN. In this experiment, the larger of the doublet of proteins at 8 kDa labeled more convincingly with [3H] glucosamine.
FIG. 4.
Electrophoretic analysis in 9 to 18% polyacrylamide of proteins immunoprecipitated from Akata cells that had been induced with anti-immunoglobulin and labeled with [3H] glucosamine. Proteins were immunoprecipitated with anti-peptide antibody anti-gN, anti-peptide antibody anti-gM, anti-peptide antibody to EBV gL, or pre-immune antibody. Immunoprecipitated proteins were eluted from protein A-agarose beads by heating at 37°C. Sizes are indicated on the right in kilodaltons.
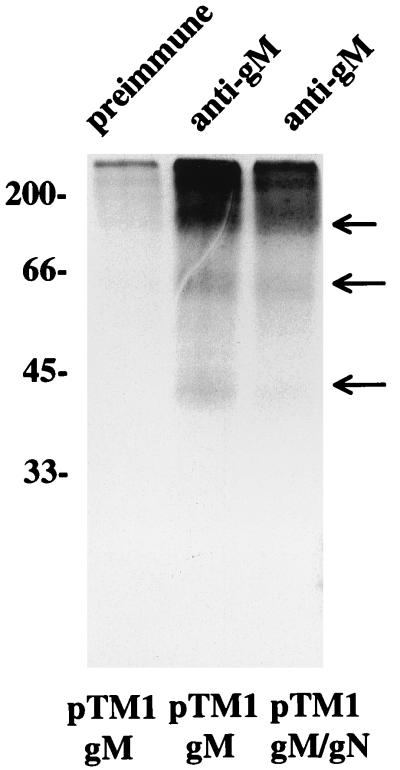
The results of this experiment suggested that the higher-molecular-weight species were not products of the BLRF1 ORF but, rather, represented those of the gene encoding the EBV gM homolog, the BBRF3 ORF. To confirm this hypothesis, the BBRF3 ORF was cloned into the pTM1 vector and expressed in CV-1 cells that were infected with vvT7. Anti-gM immunoprecipitated the three-higher-molecular weight glycoproteins from these cells (Fig. 5). Their mobilities did not change appreciably if pTM1-gM were cotransfected with pTM1-gN.
FIG. 5.
Electrophoresis in 18% polyacrylamide of proteins immunoprecipitated with anti-gM or preimmune rabbit antibody from CV-1 cells labeled with [3H] glucosamine, infected with vvT7, and transfected with pTM1-gM or cotransfected with pTM1-gM and pTM1-gN. Immunoprecipitated proteins were eluted from protein A-agarose beads by heating at 37°C. Arrows indicate three glycoproteins specifically immunoprecipitated by anti-gM antibody.
Glycosylation of gN.
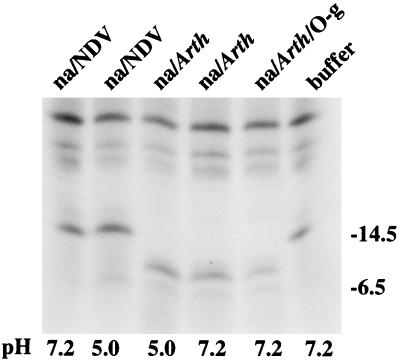
The observation that the 15- kDa glycoprotein (gp15) was immunoprecipitated well by the anti-gN antibody and very poorly if at all by the anti-gM antibody suggested that although it was not apparent in CV-1 cells transfected with pTM1-gN, it was a product of the BLRF1 ORF. Since the product of the BLRF1 ORF is predicted to include no potential N-linked glycosylation sites, the possible relationship of gp15 to the 8- kDa doublet was examined by immunoprecipitating proteins with anti-gN from induced Akata cells that had been labeled with [3H] leucine and digesting them with neuraminidase or neuraminidase plus O-glycanase. Digestions were done at pH 7.2 or 5.0, which is the optimum pH of the neuraminidases used. Neuraminidase derived from Arthrobacter, which hydrolyzes terminal α2,3-, α2,6-, and α2,8- ketosidic bonds that join sialic acid to oligosaccharides, reduced the mass of the gp15 to that of the 8- kDa doublet found in both Akata cells and cells transfected with pTM1-gN (Fig. 6). No further digestion was achieved by the addition of O-glycosidase. The neuraminidase derived from Newcastle disease virus cleaves α2,3- and α2,8- but not α2,6- ketosidic bonds and was unable to alter the mobility of gp15, indicating that the majority of the sugars on the molecule were α2,6-linked sialic acid residues.
FIG. 6.
Electrophoretic analysis in 18% polyacrylamide of proteins immunoprecipitated with anti-gN from induced Akata cells and labeled with [3H] leucine. After immunoprecipitation and before electrophoresis, the proteins were incubated overnight in buffer at pH 7.2 or 5.0 or buffer at either pH containing neuraminidase from Arthrobacter (na/Arth), neuraminidase and O-glycanase (na/Arth/O-g), or neuraminidase from Newcastle disease virus (na/NDV). Sizes are indicated on the right in kilodaltons.
Coexpression of gN and gM facilitates authentic processing of gp15.
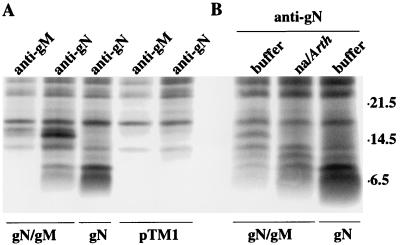
The failure of the pTM1-gN product to be processed to a 15- kDa or higher-molecular-weight species in CV-1 cells and the apparent association of gN and gM in virus-producing cells suggested that just as gL serves as a chaperone-like protein for gH, glycoprotein gM might be required for authentic processing of gN. To test this hypothesis, CV-1 cells were cotransfected with pTM1-gN and pTM1-gM. Cotransfection resulted in the appearance of the 15- kDa form of gN, which could be immunoprecipitated by anti-gN and, less efficiently, by anti-gM (Fig. 7A). Very long exposure of the gel (data not shown) revealed that a small amount of the incompletely processed form of gN could also be immunoprecipitated by anti-gM. Neuraminidase digestion of the 15- kDa species produced in cotransfected cells reduced its mass to that of the 8- kDa doublet (Fig. 7B).
FIG. 7.
(A) Electrophoretic analysis in 18% polyacrylamide of proteins immunoprecipitated with anti-gN or anti-gM from CV-1 cells labeled with [3H] leucine, infected with vvT7, and transfected with pTM1 vector alone, pTM1-gN (gN), or pTM1-gN and pTM1-gM (gN/gM). (B) All proteins were immunoprecipitated with anti-gN and were incubated overnight in buffer or buffer containing neuraminidase from Arthrobacter (na/Arth) before being subjected to electrophoresis. Sizes are indicated on the right in kilodaltons.
Linkage of gN and gM.
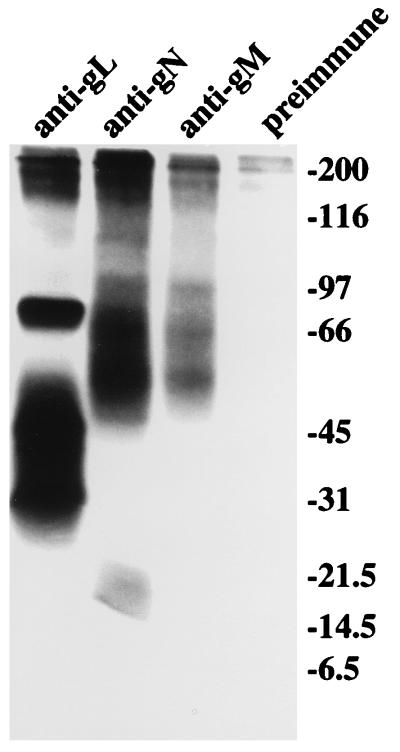
The PRV gN is reported to be disulfide linked to gM, and the BHV-1 gN is reported to be disulfide linked to an as yet unidentified virion component. To determine the nature of the interaction between gN and gM, virus-producing cells that had been radiolabeled with [3H] glucosamine were immunoprecipitated with antibodies to gN and gM and analyzed by electrophoresis under nonreducing conditions (Fig. 8). Although the proteins ran a little differently under nonreducing conditions, there was no indication that higher- molecular-weight complexes of the individual gM and gN species were formed. The EBV gH-gL complex was included in the gel for comparative purposes. This complex is not disulfide linked. The positions of the gN and gM species relative to the proteins in the gH-gL complex were similar to those seen under reducing conditions in Fig. 3, in which samples of the same immunoprecipitates were electrophoresed, or in Fig. 4.
FIG. 8.
Electrophoretic analysis in 9 to 18% polyacrylamide under nonreducing conditions of proteins immunoprecipitated by preimmune antibody, anti-peptide antibody to EBV gL, anti-gN, or anti-gM from Akata cells that had been induced with anti-immunoglobulin and labeled with [3H] glucosamine. Immunoprecipitated proteins were eluted from protein A-agarose beads by heating at 37°C. Positions of molecular size markers (in kilodaltons) electrophoresed under reducing conditions several lanes away from the samples are indicated on the right.
DISCUSSION
The association of glycoproteins gH and gL and the dependence of gH on coexpression with gL for folding and intracellular transport are well- known themes in herpesvirology. The essential nature of these two proteins and their apparently conserved roles in virus penetration provide a framework within which to accommodate such a conserved structural paradigm. It now appears, somewhat surprisingly, that two additional conserved glycoproteins, gN and gM, which have not so far proven to be essential for replication of any herpesvirus, may follow a similar pattern.
The expression of the EBV gN homolog as a recombinant protein first suggested that the gene product was a nonglycosylated protein that migrated in polyacrylamide as a doublet of approximately 8 kDa. The implication that was most consistent with predictions from the sequence of the gene was that gN was a type 1 membrane protein from which the signal peptide had been cleaved. Examination of the molecule in virus-producing cells, however, suggested a somewhat different processing pathway. The mature 15- kDa protein was glycosylated, carrying sugars that could be removed only by a neuraminidase that cleaves α2,6-linked sialic acid residues, and on long exposure of autoradiographs the upper of the proteins in the doublet also appeared to carry some sugar. This suggested that after cleavage of the signal peptide to create the nonglycosylated smaller protein in the doublet, the larger of the doublet proteins was made by posttranslational addition of O-linked N-acetylgalactosamine in the endoplasmic reticulum or cis Golgi. This, in turn, would allow for the addition of α2,6-linked sialic acid, possibly in the trans Golgi apparatus (29). It further implied that gN expressed in the pTM1 vector in the absence of other virus proteins was not completely processed. Whether or not this represents a defect in transportation as well is uncertain. CV-1 cells that had been transfected with pTM1-gN alone and fixed with 4% paraformaldehyde to inactivate vaccinia virus did react with anti-gN in indirect-immunofluorescence assays (data not shown), but since the transfected cells were significantly damaged by the vaccinia virus infection, some or all of this reactivity may have been due to penetration of the cell by antibody.
The recent observation that the PRV gN associates with the PRV gM (10) and some preliminary data with an anti-peptide antibody made to residues corresponding to the predicted sequence of the EBV gM homolog fortunately provided some clues to the identity of the virus protein that might be required to facilitate processing of gN. Not only were products of the BBRF3 ORF associated with gN, but also coexpression of gM with gN in the absence of other virus proteins was sufficient to restore authentic gN processing.
All gM homologs are predicted to be type 3 membrane proteins with multiple transmembrane domains (2, 23, 24). In all viruses in which they have been studied, i.e., PRV (4), HSV- 1 (2, 20), human cytomegalovirus (12, 15) and equine herpes virus 1 (EHV-1) (23), the proteins are virion components that carry N-linked sugars. The EBV BBRF3 ORF is predicted to encode a protein of approximately 46 kDa without posttranslational modification with two potential N-linked glycosylation sites and eight putative transmembrane domains (1). The three glycosylated species of 48, 84, and 113 kDa probably represent differential processing of polysaccharide chains, as is the case for the HSV-1 gM (2) and EHV-1 gM (23), although as yet the apparent low abundance of the protein and the availability of only a peptide antibody for immunoprecipitation has made it impossible to confirm the assumption. Both the EHV-1 gM (23) and the human cytomegalovirus gM (12) are thought to aggregate because of the very hydrophobic nature of the sequence. There was no appreciable difference in the relative mobility of any of the EBV gM species if they were dissociated from immunoprecipitating agarose beads at 37°C instead of by boiling, which suggests that this may not be the case for EBV. However, another possible explanation for the apparent low abundance of the EBV gM, even when expressed as a recombinant molecule under control of the strong T7 promoter, is that it is not being completely solubilized by the protocols normally used to extract virus membrane proteins.
There are both similarities and differences between the association of the EBV and PRV gN and gM homologs. The PRV gN-gM complex is disulfide linked (10), whereas there is little evidence for covalent linkage of the EBV gN-gM complex. On the other hand, transport of both PRV and EBV gN is dependent on coexpression of gM. Although the sugars on PRV gN can be processed in the absence of gM, it is much less efficient and the protein is not found in the virion in a virus with expression of gM deleted. At the same time, in neither virus does it appear that the maturation of gM is influenced by gN. Thus, gM is found in the PRV virion in a virus with expression of gN deleted, and no difference could be seen in the processing of recombinant EBV gM in the presence of recombinant gN. It is not currently possible to determine whether either gN or gM is a virion protein in EBV. It is difficult to see even relatively abundant proteins in purified virions harvested from spent culture media of induced cells. Indirect immunofluorescence with the anti-gN antibody, which, unlike anti-gM, was made against a peptide that is predicted to be on the luminal or extracellular side of the membrane, does, however, suggest that gN at least is present in the plasma membrane of the virus-producing cell.
As pointed out previously (10), the report that the BHV-1 gN homolog is disulfide linked to a 39- kDa protein that is more tightly associated with the tegument than membrane proteins known to span the membrane only once (19) strongly suggests that there is a gN-gM complex in this virus as well, and one in which gM is most difficult to solubilize. To date, no essential functions have been ascribed to either molecule in any virus, but defects in penetration have been reported for PRV lacking gN (10) and deletion of gN in varicella-zoster virus results in a modest reduction in syncytium formation. Preliminary results with a recombinant EBV with expression of gN deleted indicate that it has significantly reduced infectivity for B lymphocytes (13). Although these effects are subtle, they are consistent with a role for the protein at or close to the point of membrane penetration and suggest that like gH-gL, gN-gM may represent a second glycoprotein complex that is conserved in both structure and function across more than one herpesvirus subfamily.
ACKNOWLEDGMENT
This research was supported by Public Health Service grant AI20662 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tuffnell P S, Barrell B G. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 2.Baines J D, Roizman B. The UL10 gene of herpes simplex virus 1 encodes a novel viral glycoprotein, gM, which is present in the virion and in the plasma membrane of infected cells. J Virol. 1993;67:1441–1452. doi: 10.1128/jvi.67.3.1441-1452.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker D E, Roizman B. The unique sequence of herpes simplex virus 1 L component contains an additional translated open reading frame designated UL49.5. J Virol. 1992;66:562–566. doi: 10.1128/jvi.66.1.562-566.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dijkstra J M, Visser N, Mettenleiter T C, Klupp B G. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J Virol. 1996;70:5684–5688. doi: 10.1128/jvi.70.8.5684-5688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong M, Kieff E. Intracellular trafficking of two major Epstein-Barr virus glycoproteins, gp350/220 and gp110. J Virol. 1990;64:1507–1516. doi: 10.1128/jvi.64.4.1507-1516.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong M, Ooka T, Matsuo T, Kieff E. Epstein-Barr virus glycoprotein homologous to herpes simplex virus gB. J Virol. 1987;61:499–508. doi: 10.1128/jvi.61.2.499-508.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haddad R S, Hutt-Fletcher L M. Depletion of virosomes made with Epstein-Barr virus proteins abolishes their ability to fuse with virus receptor-bearing cells. J Virol. 1989;63:4998–5005. doi: 10.1128/jvi.63.12.4998-5005.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heineman T, Gong M, Sample J, Kieff E. Identification of the Epstein-Barr virus gp85 gene. J Virol. 1988;62:1101–1107. doi: 10.1128/jvi.62.4.1101-1107.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrold R E, Marchini A, Frueling S, Longnecker R. Glycoprotein 110, the Epstein-Barr virus homolog of herpes simplex virus glycoprotein B, is essential for Epstein-Barr virus replication in vivo. J Virol. 1995;70:2049–2054. doi: 10.1128/jvi.70.3.2049-2054.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jöns A, Dijkstra J M, Mettenleiter T C. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J Virol. 1998;72:550–557. doi: 10.1128/jvi.72.1.550-557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jöns A, Granzlow H, Kuchling R, Mettenleiter T C. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the virus envelope. J Virol. 1996;70:1237–1241. doi: 10.1128/jvi.70.2.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kari B, Li W, Cooper R, Goertz R, Radeke B. The human cytomegalovirus UL100 gene encodes the gC-II glycoproteins recognized by group 2 monoclonal antibodies. J Gen Virol. 1994;75:3081–3086. doi: 10.1099/0022-1317-75-11-3081. [DOI] [PubMed] [Google Scholar]
- 13.Lake, C. M., and L. M. Hutt-Fletcher. Unpublished data.
- 14.Lee S K, Longnecker R. The Epstein-Barr virus glycoprotein 110 carboxy-terminal tail domain is essential for lytic virus replication. J Virol. 1997;71:4092–4097. doi: 10.1128/jvi.71.5.4092-4097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehner R, Meyer H, Mach M. Identification and characterization of a human cytomegalovirus gene coding for a membrane protein that is conserved among human herpesviruses. J Virol. 1989;63:3792–3800. doi: 10.1128/jvi.63.9.3792-3800.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q X, Buranathai C, Grose C, Hutt-Fletcher L M. Chaperone functions common to nonhomologous Epstein-Barr virus gL and varicella-zoster virus gL proteins. J Virol. 1997;71:1667–1670. doi: 10.1128/jvi.71.2.1667-1670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q X, Spriggs M K, Kovats S, Turk S M, Comeau M R, Nepom B, Hutt-Fletcher L M. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J Virol. 1997;71:4657–4662. doi: 10.1128/jvi.71.6.4657-4662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q X, Turk S M, Hutt-Fletcher L M. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol. 1995;69:3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang X, Chow B, Raggo C, Babiuk L A. Bovine herpesvirus 1 UL49.5 homolog gene encodes a novel viral envelope protein that forms a disulfide-linked complex with a second virion structural protein. J Virol. 1996;70:1448–1454. doi: 10.1128/jvi.70.3.1448-1454.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacLean C A, Robertson L M, Jamieson F E. Characterization of the UL10 gene product of herpes simplex virus type 1 and investigations of its role in vivo. J Gen Virol. 1991;74:975–983. doi: 10.1099/0022-1317-74-6-975. [DOI] [PubMed] [Google Scholar]
- 21.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 22.Oba D E, Hutt-Fletcher L M. Induction of antibodies to the Epstein-Barr virus glycoprotein gp85 with a synthetic peptide corresponding to a sequence in the BXLF2 open reading frame. J Virol. 1988;62:1108–1114. doi: 10.1128/jvi.62.4.1108-1114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osterreider N, Neubauer A, Brandmuller C, Braun B, Kaaddden O-R, Baines J. The equine herpesvirus type 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell-to-cell spread of virions. J Virol. 1996;70:4110–4115. doi: 10.1128/jvi.70.6.4110-4115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilling A, Davison A J, Telford E, Meredith D. The equine herpesvirus type 1 glycoprotein homologous to herpes simplex virus type 1 glycoprotein M is a major constituent of the virus particle. J Gen Virol. 1994;75:439–442. doi: 10.1099/0022-1317-75-2-439. [DOI] [PubMed] [Google Scholar]
- 25.Pulford D J, Lowrey P, Morgan A J. Co-expression of the Epstein-Barr virus BXLF2 and BKRF2 genes with a recombinant baculovirus produces gp85 on the cell surface with antigenic similarity to the native protein. J Gen Virol. 1995;76:3145–3152. doi: 10.1099/0022-1317-76-12-3145. [DOI] [PubMed] [Google Scholar]
- 26.Pyles R B, Sawtell N M, Thompson R L. Herpes simplex virus type 1 dUTPase mutants are attenuated for neurovirulence, neuroinvasiveness, and reactivation from latency. J Virol. 1992;66:6706–6713. doi: 10.1128/jvi.66.11.6706-6713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross J, Williams M, Cohen J I. Disruption of the varicella-zoster virus dUTPase and the adjacent ORF9A gene results in impaired growth and reduced syncytia formation in vitro. Virology. 1997;234:186–195. doi: 10.1006/viro.1997.8652. [DOI] [PubMed] [Google Scholar]
- 28.Takada K. Cross-linking of cell surface immunoglobulin induces Epstein-Barr virus in Burkitt lymphoma lines. Int J Cancer. 1984;33:27–32. doi: 10.1002/ijc.2910330106. [DOI] [PubMed] [Google Scholar]
- 29.Varki A. Factors controlling the glycosylation potential of the Golgi apparatus. Trends Cell Biol. 1998;8:34–40. doi: 10.1016/s0962-8924(97)01198-7. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Hutt-Fletcher L M. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J Virol. 1998;72:158–163. doi: 10.1128/jvi.72.1.158-163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaswen L R, Stephens E B, Davenport L C, Hutt-Fletcher L M. Epstein-Barr virus glycoprotein gp85 associates with the BKRF2 gene product and is incompletely processed as a recombinant protein. Virology. 1993;195:387–396. doi: 10.1006/viro.1993.1388. [DOI] [PubMed] [Google Scholar]