Abstract
Background
Out-of-hospital cardiac arrest (OHCA) represents a major burden for society and health care, with an average incidence in adults of 67 to 170 cases per 100,000 person-years in Europe and in-hospital survival rates of less than 10%. Patients and practitioners would benefit from a prognostication tool for long-term good neurological outcomes.
Objective
We aim to develop a machine learning (ML) pipeline on a local database to classify patients according to their neurological outcomes and identify prognostic features.
Methods
We collected clinical and biological data consecutively from 595 patients who presented OHCA and were routed to a single regional cardiac arrest centre in the south of France. We applied recursive feature elimination and ML analyses to identify the main features associated with a good neurological outcome, defined as a Cerebral Performance Category score less than or equal to 2 at six months post-OHCA.
Results
We identified 12 variables 24 h after admission, capable of predicting a six-month good neurological outcome. The best model (extreme gradient boosting) achieved an AUC of 0.96 and an accuracy of 0.92 in the test cohort.
Conclusion
We demonstrated that it is possible to build accurate, locally optimised prediction and prognostication scores using datasets of limited size and breadth. We proposed and shared a generic machine-learning pipeline which allows external teams to replicate the approach locally.
Keywords: Machine learning, neurological outcome, OHCA, prognostication
Introduction
Out-of-hospital cardiac arrest (OHCA) represents a major healthcare burden and one of the leading causes of death. 1 In Europe, the incidence of OHCA with attempted resuscitation is estimated between 67 and 170 per 100,000 person-years. 2 Nowadays, thanks to communication campaigns, improved early recognition and ‘chain of survival’, more and more patients benefit from specialised care.3–5 Over the years, specialised care improved outcomes thanks to intensive resuscitation and invasive organ support, from mechanical ventilation to extracorporeal membrane oxygenation (ECMO).6,7 However, the current survival rate at hospital discharge is estimated at only 8.8%, 8 and many survivors suffer from severe neurological damage secondary to cerebral anoxia. 5 Return to normal cerebral function (cerebral performance category (CPC) score = 1) or moderate cerebral disability (CPC score = 2) varies between 2.8% and 18.2%. 9
Prognostication post-OHCA is central to clinical decision-making, since it can inform family discussions, level of care and the initiation or not of invasive therapies, in the complex medical, ethical and economic landscape of post-cardiac arrest care. While many intensive care unit (ICU) prediction scores exist (OHCA, CAST, MIRACLE2, etc.), 10 their performance fluctuates and invariably degrades when applied to populations different from those used to derive the scores. Machine learning (ML) holds the promise of generating new insight into a vast number of medical topics,11–13 including for the prediction of sepsis or renal failure.14,15 Data-driven analyses with built-in explainability might become the new go-to approach for building prediction scores. The purpose of this study was to build an ML model to predict good neurological outcomes after OHCA and identify key clinical predictors.
Methods
Study design
Data was collected retrospectively from patients admitted between the 21st of January 2014 and the 22nd of December 2021 to Toulouse University Hospital, a tertiary centre located in the south of France. Toulouse University Hospital provides 24/7 access to a large technical platform including a cardiac arrest centre, and cardiovascular surgery. We included patients older than 18 years old, admitted for OHCA regardless of the aetiology. We excluded patients who died before ICU admission, and patients who did not remain comatose after return to spontaneous circulation (ROSC), who were directly routed to another specialised cardiology care centre. Upon arrival, after the patient's assessment and initial blood tests, the decision was made by clinicians to perform coronary angiography and/or to start ECMO, based on available local guidelines. According to the French ethics and regulatory law (public health code), this study was registered in the register of retrospective studies of the Toulouse University Hospital (number RnIPH 2023-02) and covered by reference methodology of the French National Commission for Informatics and Liberties (CNIL). Due to the retrospective nature of the study, passing through an ethics committee was not mandatory.
Outcome
The primary outcome was good neurological outcome, defined as a CPC score of 1 or 2 at six months post-OHCA. Non-survivors were classified as CPC 5.
Data collection
Data were collected from patients’ electronic medical records. The physiological variables, specifically arterial pressure, heart rate, body temperature, and blood results were collected at the time of ICU admission. Pre-hospital data including time of no-flow and low-flow, location of the cardiac arrest (home, street, or workplace) and bystander CPR, were extracted from the emergency department records. Simplified Acute Physiology Score II (SAPS II) was collected at 24 h after ICU admission. Patient medical history and outcomes of interest were also extracted. In total, we included 30 features for each patient.
Data preprocessing
Two features were removed due to high missingness: Neuron-specific enolase (NSE 16 ) at day 3, and duration before ECMO. Categorical variables were transformed into binary variables using the one hot encoding method. We split the dataset into training and test sets, with respectively 80% and 20% of the cohort. Next, we used a feature selection algorithm in the training dataset called recursive feature elimination (RFE) to select only the relevant features for predicting good neurological outcomes. RFE consists of evaluating a model over the recursive deletion of the least contributive input feature, in order to keep the minimal set of features that preserve a high model performance. To perform RFE, we follow the following steps. First, we randomly split the training dataset into training and validation sets (respectively 80% and 20% of the training data), and we fit a model (we selected XGBoost) on the data using grid search cross-validation. It is crucial to use grid search cross-validation at this step because the best hyperparameters for the model may change depending on the number of features. 17 Then, we assess the feature's contributions using SHapeley Additive exPlanations (SHAP), and we remove the least contributive feature from the dataset. We repeat these two steps (fitting the model and removing the least contributive feature) until there is no feature left and report the model's performances in train and validation each time. We repeated the whole RFE procedure 100 times to ensure the robustness of our method.
Model training and performance
Once the final set of features were selected, we trained a model to predict good neurological outcome using the training data (80% of the entire cohort). We built and compared four different models: random forest (RF), gradient boosting model (XGBoost, XGB), logistic regression, and neural network (multilayer perceptron). We then tested each model on the test dataset (representing 20% of the cohort) and reported train and test set confusion matrices, accuracy (ratio of number of correct predictions divided by the total number of predictions), and the area under the receiver operating characteristic (ROC) curves. We selected our best final model based on model accuracy.
Statistical analysis
Analyses were performed using GraphPad Prism 9.0 (GraphPad Software, Inc., San Diego, CA). Student's t-test or Mann–Whitney tests were used for continuous variables, while the Chi-squared test or Fisher's exact test was used for binary features. Variables are presented as mean and standard derivation or percentage. A two-sided p-value of less than .05 was considered to be statistically significant. The code of the analysis is available for reference. 18
Explainability
We used SHAP, which is a post-hoc explainability method where the contribution of each variable to the prediction is estimated for each individual using game theory Shapeley values.19,20
Results
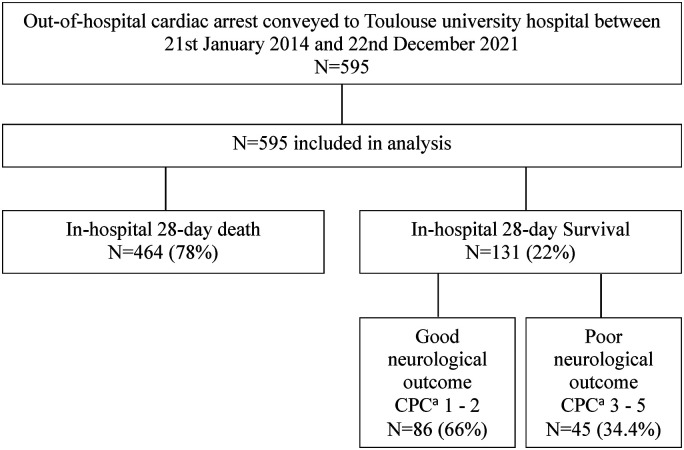
Figure 1 presents a flowchart of the patients included in the study. All patients admitted for OHCA included in the study received CPR and presented ROSC upon arrival. They were all attended to by pre-hospital specialised medical teams which included a medical doctor (the French ‘SAMU’). All the patients could receive advanced medical care during transportation including intubation and mechanical ventilation, mechanical CPR (LUCAS® chest compression system), epinephrine, and other advanced cardiac arrest treatment (amiodarone, lignocaine) as indicated in recommendations at the time of the event.21,22 Patients’ characteristics, in particular severity index, the suspected cause of cardiac arrest, and treatments administered are detailed in Table 1. The median age in the cohort was 56 years old (±17), men were overrepresented (68.2%) and the median SAPS II score was 68.9 (±19.3). Most of the patients received bystander CPR (76.5%), and 62.5% had a no-flow time of less than 5 min. The aetiology remained unknown in 31.6% of cases, and almost a quarter of patients presented ST-elevated myocardial infarction (24.5%).
Figure 1.
Flow chart of the patient included in the study.a
aCPC: cerebral performance category score.
Table 1.
Development cohort: descriptive data of patients included in the study.
| Parameters | Total | CPC ≤ 2 | CPC > 2 | p |
|---|---|---|---|---|
| N | 595 | 89 | 506 | / |
| Age, years (SD) | 56 (17) | 50 (18) | 57 (17) | .0002 |
| Male, N (%) | 403 (68.2) | 61 (68.5) | 342 (68.1) | .9920 |
| Weights, KG (SD) | 78.2 (17.2) | 79.6 (15.1) | 77.9 (17.6) | .4948 |
| Height, CM (SD) | 172.1 (9.1) | 173.3 (8.6) | 171.8 (9.2) | .2085 |
| BMIa (SD) | 26.3 (5.8) | 26.4 (4.4) | 26.3 (6.1) | .9457 |
| SAPS IIb (SD) | 68.9 (19.3) | 56.8 (19.2) | 71 (18.5) | <.0001 |
| Admission temperature, °C (SD) | 35.1 (1,76) | 35.8 (1.1) | 35 (1.9) | .0002 |
| Place of residence, N (%) | 361 (64.4) | 45 (53.6) | 316 (66.3) | .0380 |
| Public place, N (%) | 182 (32.4) | 36 (42.9) | 146 (30.6) | .0416 |
| Working place, N (%) | 17 (3) | 2 (2.4) | 15 (3.1) | .7187 |
| Bystander CPRc, N (%) | 446 (76.5) | 78 (87.6) | 368 (79.5) | .0081 |
| No-flow time, Min (SD) | 4.2 (6.0) | 2.1 (3.1) | 4.6 (6.4) | .0006 |
| No flow time under 5 Min, N (%) | 362 (62.5) | 74 (85.1) | 288 (58.5) | <.0001 |
| Low-flow time, Min (SD) | 38.6 (29.4) | 18.2 (14.5) | 42.2 (29.8) | <.0001 |
| Shockable initial rhythm, N (%) | 276 (46.5) | 67 (75.3) | 209 (41.4) | <.0001 |
| TTMd, N (%) | 241 (44.5) | 47 (54.7) | 194 (42.5) | .0398 |
| Epinephrin administration, N (%) | 391 (67.7) | 23 (27.1) | 368 (74.7) | <.0001 |
| Use of chest compression system, N (%) | 166 (28.4) | 8 (9.2) | 158 (31.8) | <.0001 |
| Veno-Arterial Ecmoe AT Admission, n (%) | 83 (14) | 10 (11.2) | 73 (14.4) | .4328 |
| Emergency coronarographyf, N (%) | 200 (33.8) | 64 (71.9) | 136 (27.0) | <.0001 |
| PCIg, N (%) | 104 (17.2) | 33 (37.5) | 71 (14.2) | <.0001 |
| Admission PH (SD) | 7.1 (0.3) | 7.2 (0.2) | 7.0 (0.3) | <.0001 |
| Blood lactate, MMOL/L (SD) | 10.5 (6.6) | 5.4 (5.3) | 11.4 (6.4) | <.0001 |
| Serum troponin T HSh, NG/L (SD) | 1162.2 (3987.4) | 1562.2 (3998.1) | 1064.3 (3984.7) | .2977 |
| STEMIi, N (%) | 146 (24.5) | 37 (41.6) | 109 (21.5) | <.0001 |
| Hypoxia, N (%) | 122 (20.5) | 15 (16.9) | 107 (21.2) | .3795 |
| Unkown etiology, N (%) | 188 (31.6) | 14 (15.7) | 174 (34.4) | .0006 |
| Intoxication, N (%) | 22 (3.7) | 3 (3.4) | 19 (3.8) | .8740 |
| 28-day death, N (%) | 464 (78) | / | 463 (94.5) | / |
All values are reported as mean and standard derivation (SD) for continuous variables, and percentage for categorical variables.
BMI: body mass index.
SAPS II: simplified acute physiology score version II.
CPR: cardio-pulmonary resuscitation.
TTM: targeted temperature management.
ECMO: extracorporeal membrane oxygenation.
Coronarography within 2 h after admission.
PCI: percutaneous coronary intervention.
HS: high-sensitive.
STEMI: ST-elevated myocardial Infarction
Table 2.
Predictive performance comparison of the four machine learning models in the test cohort.
| Parameters | XGBa | RFb | LRc | MLPd |
|---|---|---|---|---|
| Sensitivity | 0.89 | 0.5 | 0.72 | 0.67 |
| specificity | 0.92 | 1 | 0.96 | 0.95 |
| Positive predictive value (%) | 66.43 | 100 | 76.20 | 70.44 |
| Negative predictive value (%) | 97.92 | 91.93 | 95.07 | 94.18 |
| Accuracy | 0.92 | 0.92 | 0.92 | 0.91 |
| Precision | 0.67 | 1 | 0.76 | 0.71 |
| F1 score | 0.76 | 0.67 | 0.74 | 0.68 |
| AUC | 0.96 | 0.96 | 0.95 | 0.96 |
| Average precision | 0.84 | 0.85 | 0.77 | 0.85 |
XGB: XGBoost.
RF: Random forest.
LR: Logistic regression.
MLP: Multilayer perceptron.
Variables of interest
The results of the RFE to predict good neurological outcome is presented in Figure 2. We identified that a minimum of 12 features were necessary before model accuracy started to deteriorate. We showed that removing features was not associated with any accuracy improvement as it could have reduced background noise.
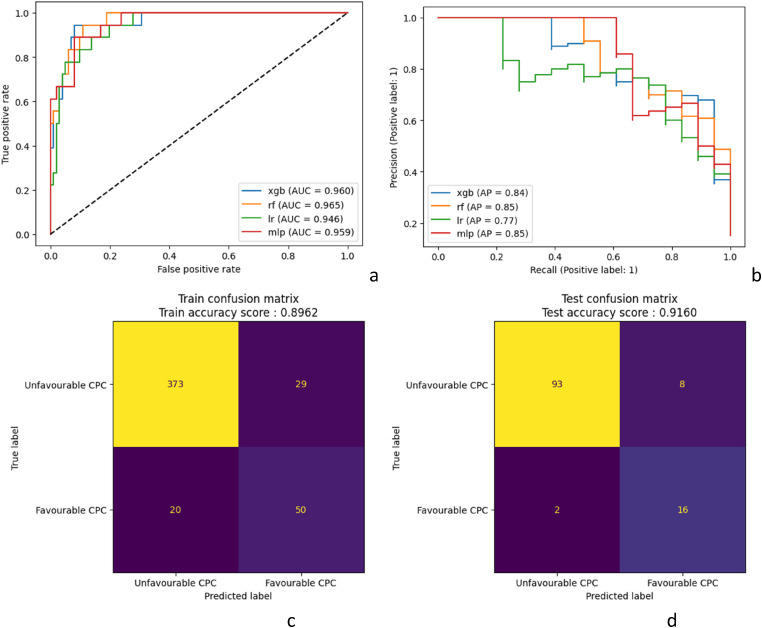
Figure 2.
Receiver operating curves and precision-recall curves for the four models in the test cohorta,b; confusion matrix for neurological outcome prediction in (b) training cohort and (c) test cohort for the best model (xGBoost).
XGB: xGBoost; rf: Random Forest; lr: Logistic Regression; mlp: Multilayer Perceptron. CPC: cerebral performance category.
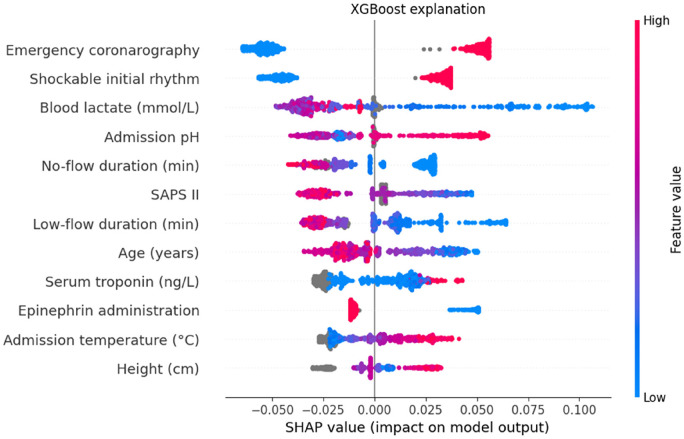
Figure 3.
Mean SHAP value for the top 12 features of the XGBoost model.
Variables are shown from top to bottom in order of importance (average absolute SHAP values). The contribution of each feature on each prediction is shown on the x-axis. Each dot represents one patient in the test set, and their colour encodes the value of the associated variable for each individual. Overlapping points are vertically separated for clarity. Grey points represent missing values, estimated by the model based on the values of other features. A positive SHAP value means that the variable, for this individual, contributes positively to the final outcome prediction. For example, a low lactate (blue colour) is positively associated (positive SHAP value) with good neurological outcome, whilst long low-flow duration (red colour) is negatively associated (negative SHAP value) with good neurological outcome.
SHAP: SHapeley Additive exPlanations.
Discrimination performance was determined using ROC curves and precision-recall curves analysis (Figure 2), while calibration was assessed with confusion matrices (Figure 2(b) and (c)). In the train cohort, the different models achieved AUC between 0.91 and 0.95 and average precision between 0.68 and 0.82. The results were higher in the test cohort with AUC between 0.95 and 0.96 and average precision between 0.77 and 0.85. The best F1-score (0.76) was obtained by the XGBoost model (Table 2). The confusion matrix in the test cohort reveals that the XGBoost model misclassified 20 patients out of 119. Out of the misclassified patients, two were false negatives. The false negative rate was 8% in that model. The O:E ratio was 0.75 in the validation cohort and 0.89 in the development cohort, showing an overestimation of good neurological outcomes.
Explainability
Mean SHAP value extracted from the XGBoost model identified 12 features associated with a good neurological outcome (Figure 3 and Figure 4): emergency coronary angiography, shockable initial rhythm, blood lactate, pH, no-flow duration, SAPS II, low-flow duration, age, serum troponin, epinephrine administration, admission temperature, patient height. The aetiology of OHCA was not identified by the model as a significant contributing feature.
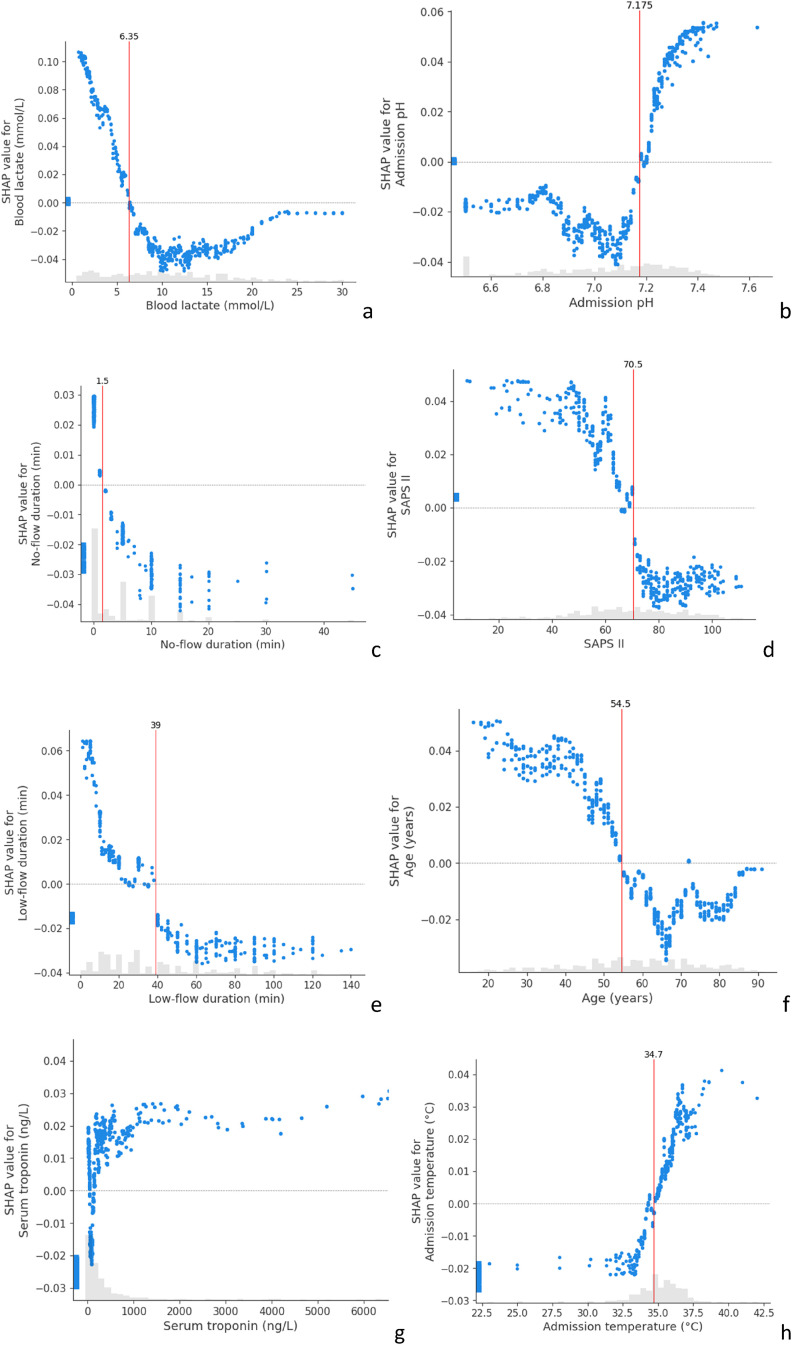
Figure 4.
SHAP dependence contribution plots, illustrating the link between individual feature values and SHAP values for the prediction of a good neurological outcome. For each feature, we identified the thresholds which differentiate between positive and negative SHAP values (vertical red lines). The grey histograms represent the distribution of patients for each feature value.
(a) SHAP value for blood lactate; (b) for admission pH; (c) no-flow duration (min); (d) for SAPS II; (e) for low-flow duration (min); (f) for Age (years); (g) for admission serum troponine; (h) for admission temperature.
SHAP: SHapeley Additive exPlanations; SAPS II: Simplified Acute Physiology Score II.
The SHAP analysis also allows us to build dependence contribution plots for each feature, which illustrates further the link between individual feature values and SHAP values. We identified the thresholds which differentiate between positive and negative SHAP values. Figure 4 shows that the SAPS-II score higher than 70.5 at 24 h after admission, no-flow longer than 1.5 min, low-flow longer than 39 min, lactate level higher than 6.35 mmol/l, age under 54.5 years and an admission temperature under 34.7°C were associated with a worse outcome.
Discussion
We deployed ML methods on a cohort of patients following OHCA and constructed a sparse model that accurately predicts a six-months neurological outcome. The main contribution of this study is related to the establishment of a locally tailored model development pipeline, identifying recognised predictive variables, and capable of achieving satisfactory performance. XGBoost-based predictive model achieved the highest prediction score in terms of Accuracy, AUC, F1 score and average precision (respectively 0.92, 0.96, 0.76, 0.84). The aim of this study was to demonstrate the value of modern prediction processes with a new level of dynamism and adaptability. While recent publications focus on utilising vast cohorts 23 that are inaccessible to many clinicians, we aimed to highlight the significant extrapolation capabilities provided by ML-based methods today.
The analyses highlighted 12 features that were considered critical to determine prognosis. Blood lactate was the main predictive feature identified. A low value has the most important impact on the prognostication as the SHAP value might reach a maximum of 0.10. The role of blood lactate as a mortality prognostication marker has been known for a long time. Its metabolism in circulation failure or ischaemia is also well known. This ML approach emphasised its role respecting the known physiological model concerning cardiac arrest. On the contrary, some other features have a more unclear impact. Serum troponin seemed to have either a positive or negative impact. The model interpreted missing serum troponin value as an unfavourable prognostication marker. In fact, some patients were routed to our ICU with an unclear history of the disease and without return on spontaneous circulation. Resuscitation was then stopped at arrival for obvious ethical reasons (such as excessive no-flow duration, known palliative disease, advances directives, etc.). These data, however, were not collected in the study dataset. Height was the least contributive feature in the model. There is no literature comforting its role in OHCA outcomes prognostication. As the serum troponin, it's the missing values that are predictive. We think that it emphasises that our ML model extrapolates from data regardless of the known prevailing physiological model and extracts data even from less obvious features. It emphases also the main limitation of the model prediction as it is strongly affected by initial features selection. This limitation concerns all the prognostication models with or without ML. 24
The thresholds identified by the algorithm were close to those already described in the previous study. 25 Some of the variables we identified were already used in previous prognostication models for neurological outcomes post cardiac arrest at six months, such as SALTED and MIRACLE2 score.26,27 Both these scores were built with the intent of being as parsimonious as possible to simplify prognostication at the time of admission and included only a limited set of features. As a consequence, they likely suffer from low temporal (across time) and external (in new settings) validity. 28 We obtained higher AUC values in the development and validation cohort than SALTED (respectively 0.90 and 0.82), and MIRACLE2 (respectively 0.90 and 0.92). C-GRApH and CAST scores predict neurological outcomes at discharge and reach AUC values of respectively 0.81 and 0.86 (in validation cohorts).29,30
We created an adaptive tool allowing optimised prediction in a dataset of relatively limited size (number of patients) and breadth (number of features). As our study shows, a 600-patient dataset with only 30 features appears large enough to generate a locally robust and highly accurate ML prognostication system. The danger of building prediction models from small datasets is the risk of over-fitting, when the model gives (highly) accurate predictions for the training data but not for new data. We showed that this obstacle could be overcome by using cross-validation, and RFE. 31 Generalisability (the ability of an ML model to perform well across time and/or settings) has been put into question. 32 Instead, ML experts have called for “locally optimised” models that are more likely to find clinical utility at the bedside. The pipeline we propose (data pre-processing, RFE, then model selection) could be replicated by external teams in other institutions to enable them to build local prediction models and prognostication tools. We argue that external validation of the current model in a different centre would be of limited value. Instead, we call for the assessment of the performance of a new model, built implementing our strategy (RFE, model selection and then algorithm testing) with the features and data available at that centre.
The main limitation of our study is the absence of prospective validation in the clinical setting, which represents the next step in the assessment of its clinical utility. However, the model's performance showed great stability between the test set and the development set, as well as during cross-validation. These are positive indicators that mitigate the risk of overfitting. Another point of discussion is that we did not estimate how active withdrawal of care changed the results of neurological prognostication. Neurological outcome evaluation in OHCA patients was based at the beginning of this study (2014), on the review from Sandroni and al. 33 This evaluation considers mainly day-3 features (as blood NSE, EEG results and cerebral scan results) to determine end-of-life support. More recent studies did not bring substantial changes.33,34 We believe that based on that strategy, the patients angled towards end-of-life support had worse feature values and would mainly have been classified in the poor neurological outcome prediction group (CPC > 2).
A central concern with the use of predictive scores such as this one is the rate of false negatives, where patients are erroneously predicted to have a poor neurological outcome, when in reality they evolved favourably. This could have serious consequences, as it might lead to withdrawing assistance from a patient who could have benefited from it. The rate of false negatives in our study in the test cohort was 8%, which is higher than the recommended value of 5%. 34 Modulating the hyperparameters of the models did not allow us to achieve better performance on this element. This limitation probably stems from deficiencies in the dataset.
The ability of models to extrapolate data, to account for feature correlation, and to reduce noise enable its use in many diverse settings. Adaptive prediction models could help evaluate the specificity of predictive features for example: population base, cohort's derivation with time, and calibration drift. 35 As computational power grows and analytical skills in healthcare institutions become more ubiquitous, developing algorithm strategies for outcome prognostication based on ML will become the new standard. This study shows that the future of ICU patient prognostication may include evolving ML models, and should be ICU-specific (regarding the level of care and the population recruitment).
Conclusion
ML models managed to accurately identify the predictive factors of good neurological outcomes following OHCA in a single centre cohort of 595 patients. We proposed and shared a pipeline that can be replicated by external teams in different centres to generate locally optimised prediction and prognostication tools.
Acknowledgements
The authors thank Anais Chekroun and Louise Richez for their participation in collecting data and revising the manuscript.
Footnotes
Contributorship: Vincent Pey and Emmanuel Doumard wrote the article, and take responsibility for the content of the manuscript. Matthieu Komorowski, Antoine Rouget, Clément Delmas, Fanny Vardon-Bounes, Michaël Poette, Valentin Ratineau, Cédric Dray and Isabelle Ader helped revise the manuscript and Vincent Minville helped draft and revise the manuscript.
Consent statement: According to French law on ethics, patients were informed that their codified data will be used for the study. According to the French ethics and regulatory law (public health code) retrospective studies based on the exploitation of usual care data should be submitted at an ethic committee but they have to be declared or covered by reference methodology of the French CNIL. A collection and computer processing of personal and medical data was implemented to analyze the results of the research. Toulouse University Hospital signed a commitment of compliance to the reference methodology MR-004 of the French CNIL. After evaluation and validation by the data protection officer and according to the General Data Protection Regulation*, this study completes all the criteria, and it is registered in the register of a retrospective study of the Toulouse University Hospital (number's register: RnIPH 2023-02) and cover by the MR-004 (CNIL number: 2206723 v 0). This study was approved by Toulouse University Hospital and confirmed that ethic requirements were totally respected in the above report.
*Regulation (EU) 2016/679 of the European Parliament and of the Council of 27 April 2016.
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article
Guarantor: Vincent Pey: This author wrote the article and takes responsibility for the content of the manuscript.
ORCID iD: Vincent Pey https://orcid.org/0000-0001-6660-0576
References
- 1.Ong MEH, Perkins GD, Cariou A. Out-of-hospital cardiac arrest: prehospital management. The Lancet 2018; 391: 980–988. [DOI] [PubMed] [Google Scholar]
- 2.Gräsner JT, Wnent J, Herlitz J, et al. Survival after out-of-hospital cardiac arrest in Europe - results of the EuReCa TWO study. Resuscitation 2020; 148: 218–226. [DOI] [PubMed] [Google Scholar]
- 3.Cummins RO, Ornato JP, Thies WHet al. et al. Improving survival from sudden cardiac arrest: the “chain of survival” concept. A statement for health professionals from the Advanced Cardiac Life Support Subcommittee and the Emergency Cardiac Care Committee, American Heart Association. Circulation 1991; 83: 1832–1847. [DOI] [PubMed] [Google Scholar]
- 4.Chan PS, McNally B, Tang Fet al. et al. Recent trends in survival from out-of-hospital cardiac arrest in the United States. Circulation 2014; 130: 1876–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sondergaard KB, Wissenberg M, Gerds TA, et al. Bystander cardiopulmonary resuscitation and long-term outcomes in out-of-hospital cardiac arrest according to location of arrest. Eur Heart J 2019; 40: 309–318. [DOI] [PubMed] [Google Scholar]
- 6.Goldsweig AM, Tak HJ, Alraies MC, et al. Mechanical circulatory support following out-of-hospital cardiac arrest: insights from the national cardiogenic shock initiative. Cardiovasc Revasc Med 2021; 32: 58–62. [DOI] [PubMed] [Google Scholar]
- 7.Yannopoulos D, Bartos J, Raveendran G, et al. Advanced reperfusion strategies for patients with out-of-hospital cardiac arrest and refractory ventricular fibrillation (ARREST): a phase 2, single centre, open-label, randomised controlled trial. The Lancet 2020; 396: 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan S, Gan Y, Jiang N, et al. The global survival rate among adult out-of-hospital cardiac arrest patients who received cardiopulmonary resuscitation: a systematic review and meta-analysis. Crit Care 2020; 24: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiguchi T, Okubo M, Nishiyama C, et al. Out-of-hospital cardiac arrest across the world: first report from the International Liaison Committee on Resuscitation (ILCOR). Resuscitation 2020; 152: 39–49. [DOI] [PubMed] [Google Scholar]
- 10.Naik R, Mandal I, Gorog DA. Scoring systems to predict survival or neurological recovery after out-of-hospital cardiac arrest. Eur Cardiol 2022; 17: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magunia H, Lederer S, Verbuecheln R, et al. Machine learning identifies ICU outcome predictors in a multicenter COVID-19 cohort. Crit Care 2021; 25: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moghadam MC, Masoumi E, Bagherzadeh Net al. et al. Supervised machine-learning algorithms in real-time prediction of hypotensive events. In: 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC) [Internet], 2020. [cited 2022 May 16], pp. 5468–5471. Montreal, QC, Canada: IEEE. Available from: https://ieeexplore.ieee.org/document/9175451/ [DOI] [PubMed] [Google Scholar]
- 13.Quer G, Arnaout R, Henne Met al. et al. Machine learning and the future of cardiovascular care. J Am Coll Cardiol 2021; 77: 300–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyland SL, Faltys M, Hüser M, et al. Early prediction of circulatory failure in the intensive care unit using machine learning. Nat Med 2020; 26: 364–373. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Ho KM, Hong Y. Machine learning for the prediction of volume responsiveness in patients with oliguric acute kidney injury in critical care. Crit Care 2019; 23: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogel W, Krieger D, Veith M, et al. Serum neuron-specific enolase as early predictor of outcome after cardiac arrest. Crit Care Med 1997; 25: 1133–1138. [DOI] [PubMed] [Google Scholar]
- 17.Guyon I, Weston J, Barnhill S. Gene Selection for Cancer Classification using Support Vector Machines.
- 18.Doumard E. E. Doumard, CPC_prediction (2023), GitHub repository, https://github.com/EmmanuelDoumard/CPC_prediction. 2023.
- 19.A Value for N-Person Games [Internet]. RAND Corporation; 1952 [cited 2023 Jan 11]. Available from: https://www.rand.org/pubs/papers/P295.html
- 20.Lundberg S, Lee SI. A Unified Approach to Interpreting Model Predictions [Internet]. arXiv; 2017 [cited 2023 Jan 11]. Available from: http://arxiv.org/abs/1705.07874
- 21.Soar J, Böttiger BW, Carli P, et al. European Resuscitation Council guidelines 2021: adult advanced life support. Resuscitation 2021; 161: 115–151. [DOI] [PubMed] [Google Scholar]
- 22.Soar J, Nolan JP, Böttiger BW, et al. European Resuscitation Council guidelines for resuscitation 2015. Resuscitation 2015; 95: 100–147. [DOI] [PubMed] [Google Scholar]
- 23.Lou SS, Liu H, Lu Cet al. et al. Personalized surgical transfusion risk prediction using machine learning to guide preoperative type and screen orders. Anesthesiology 2022; 137: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinze G, Wallisch C, Dunkler D. Variable selection - A review and recommendations for the practicing statistician. Biom J 2018; 60: 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinell L, Nielsen N, Herlitz J, et al. Early predictors of poor outcome after out-of-hospital cardiac arrest. Crit Care 2017; 21: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pareek N, Kordis P, Beckley-Hoelscher N, et al. A practical risk score for early prediction of neurological outcome after out-of-hospital cardiac arrest: MIRACLE2. Eur Heart J 2020 Dec 14; 41(47): 4508–4517. [DOI] [PubMed] [Google Scholar]
- 27.Pérez-Castellanos A, Martínez-Sellés M, Uribarri A, et al. Development and external validation of an early prognostic model for survivors of out-of-hospital cardiac arrest. Revista Española de Cardiología (English Edition) 2019; 72: 535–542. [DOI] [PubMed] [Google Scholar]
- 28.Ridley SA. Uncertainty and scoring systems: uncertainty and scoring systems. Anaesthesia 2002; 57: 761–767. [DOI] [PubMed] [Google Scholar]
- 29.Nishikimi M, Matsuda N, Matsui K, et al. A novel scoring system for predicting the neurologic prognosis prior to the initiation of induced hypothermia in cases of post-cardiac arrest syndrome: the CAST score. Scand J Trauma Resusc Emerg Med 2017; 25: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiehl EL, Parker AM, Matar RM, et al. C-GRApH : a validated scoring system for early stratification of neurologic outcome after out-of-hospital cardiac arrest treated with targeted temperature management. JAHA 2017; 6: e003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kernbach JM, Staartjes VE. Foundations of machine learning-based clinical prediction modeling: part II—generalization and overfitting. In: Staartjes VE, Regli L, Serra C. (ed.) Machine learning in clinical neuroscience [internet]. Cham: Springer International Publishing, 2022. [cited 2023 Aug 8], pp. 15–21. (Acta Neurochirurgica Supplement; vol. 134). Available from: https://link.springer.com/10.1007/978-3-030-85292-4_3 [DOI] [PubMed] [Google Scholar]
- 32.Futoma J, Simons M, Panch Tet al. et al. The myth of generalisability in clinical research and machine learning in health care. The Lancet Digital Health 2020; 2: e489–e492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandroni C, Cariou A, Cavallaro F, et al. Prognostication in comatose survivors of cardiac arrest: an advisory statement from the European Resuscitation Council and the European Society of Intensive Care Medicine. Intensive Care Med 2014; 40: 1816–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nolan JP, Sandroni C, Böttiger BW, et al. European Resuscitation Council and European society of intensive care medicine guidelines 2021: post-resuscitation care. Resuscitation 2021; 161: 220–269. [DOI] [PubMed] [Google Scholar]
- 35.Davis SE, Greevy RA, Lasko TAet al. et al. Detection of calibration drift in clinical prediction models to inform model updating. J Biomed Inform 2020; 112: 103611. [DOI] [PMC free article] [PubMed] [Google Scholar]