Abstract
In this study, we used a dicistronic vesicular stomatitis virus (VSV) minigenome to investigate the effects of either single or multiple nucleotide insertions placed immediately after the nontranscribed intergenic dinucleotide of the M gene on VSV transcription. Both Northern blot and primer extension analysis showed that the polymerase responded to the inserted nucleotides in a sequence-specific manner such that some insertions had no effect on mRNA synthesis from the downstream G gene, nor on the site of transcript initiation, whereas other insertions resulted in dramatic reductions in transcript accumulation. Some of these transcripts were initiated at the wild-type site, while others initiated within the inserted sequence. We also examined the transcriptional events that occurred when a natural, 21-nucleotide intergenic region located between the G and L genes from the New Jersey (NJ) serotype of VSV was inserted into the minigenome gene junction. In contrast to the normal 25 to 30% attenuation observed for downstream transcription at gene junctions containing the typical dinucleotide (3′-GA-5′) intergenic region, the NJ variant showed greater than 75% attenuation at the gene junction. In addition, the polymerase initiated transcription at two major start sites, one of which was located within the intergenic sequence. Collectively, these data suggest that the polymerase “samples” the intergenic sequences following polyadenylation and termination of the upstream transcript by scanning until an appropriate start site is found. One implication of a scanning polymerase is that the polymerase presumably switches states from a processive elongation mode to a stuttering mode for polyadenylation to one in which no transcription occurs, before it reinitiates at the downstream gene. Our data support the hypothesis that sequences surrounding the intergenic region modulate these events such that appropriate amounts of each mRNA are synthesized.
Vesicular stomatitis virus (VSV) is a nonsegmented negative-strand RNA virus from the Rhabdoviridae family. Although VSV has served as a useful model to study virus entry, replication, and assembly, the mechanism by which the VSV polymerase transcribes and processes its transcripts has been a topic of debate for many years. Moreover, progress in this area has been hampered by the inability to introduce site-specific mutations into the VSV genome. The recent development of systems that allow directed genetic manipulation (reverse genetics) of VSV and the ability to recover full-length virus (22, 39) or minivirus particles (26, 35) entirely from cDNAs have now provided additional tools to dissect and understand the events that occur during VSV transcription.
The replication cycle of VSV begins when an endocytosed virion fuses with the endosomal membrane and the ribonucleocapsid core particle (RNP) is uncoated and released into the host cell cytoplasm. Uncoating occurs either during or immediately following fusion of the viral envelope with the endosomal membrane and results in the release of the matrix (M) protein from the RNP. The uncoated RNP, which consists of the genomic RNA tightly encapsidated by ∼1,200 molecules of the nucleocapsid (N) protein, serves as a template for both transcription of mRNAs and replication of the genome by the viral polymerase, which is also brought into the cell by the incoming virion.
One of the characteristic features of VSV transcription, as well as that of other nonsegmented, negative-strand RNA viruses, is that mRNA synthesis is both sequential and polar (19, 20). For VSV, the polymerase first transcribes a small 47-nucleotide RNA called the leader from the extreme 3′ end of the genome, and then each of the five mRNAs encoding the VSV proteins are synthesized in the order in which they appear from the 3′ end of the genome. For the gene junctions that have been studied, the downstream gene is transcribed approximately 30% less than that of the upstream gene (19), and as a consequence, the abundance of the five mRNAs also follows the order of genes on the genome (e.g., N>P>M>G>L). In addition to transcription and replication, the VSV polymerase is responsible for capping and methylating (17, 25, 27, 28) as well as polyadenylating (32) VSV transcripts.
The recent demonstration that a sequence of 23 conserved nucleotides, which is found at the 3′ and 5′ junction of all VSV genes, can direct the expression of foreign genes within recombinant VSV genomes provided the first evidence that this sequence contains all the cis-acting signals necessary to polyadenylate and terminate transcripts from the upstream gene and then to reinitiate transcription at the adjacent downstream gene (31). The conserved sequence 3′-AUAC(U)7-5′ which is found at the end of each gene is critical for both polyadenylation and termination of VSV transcripts (3, 18). When this sequence is encountered, the polymerase reiteratively copies, or stutters, over the seven U’s to produce a poly(A) tail approximately 150 nucleotides in length (32). Immediately following the polyadenylation signal, there are two nontranscribed intergenic nucleotides, which are usually 3′-GA-5′ (29). Recently, it was suggested that this dinucleotide is important for efficient termination of the upstream transcript because certain nucleotide substitutions resulted in higher levels of readthrough transcripts at the mutated gene junction (4, 36). Substitutions at the first nucleotide were found to have the greatest effect, while single substitutions at the second position resulted in only slightly higher levels of readthrough transcripts (4). Based on those results, it was proposed that the minimum polyadenylation and termination signal is 3′-AUAC(U)7G-5′ (3). However, it appears that the second nucleotide does play a small role in upstream mRNA termination, since nucleotide changes at both positions resulted in higher levels of readthrough transcription than single nucleotide changes at the first position (36). Still others question the interpretation that the dinucleotide is important for efficient termination, because only minimal effects on the upstream transcript levels were observed when this sequence was deleted (18). Following the intergenic dinucleotide is the sequence 3′-UUGUCnnUAC-5′ (with n being any nucleotide). The first three nucleotides of this sequence are most important for efficient gene expression because mutations at these positions severely reduced the amount of mRNA produced from the mutated gene (36). Presumably, this sequence is important for reinitiation following polyadenylation and release of the upstream mRNA, although it is possible that the reduced mRNA levels from the mutated gene may be the result of mRNA instability due to the lack of a 5′ cap or prevention of some other postinitiation step by the polymerase.
Although the precise mechanisms responsible for transcript initiation and termination have not been defined, several different models have been proposed over the last two decades to explain certain features of VSV transcription (2). There is some evidence that the viral polymerase may initiate at internal sites on the genome (7, 37), and it has been suggested that the polarity observed during VSV transcription is the result of elongation of internally initiated polymerase complexes being dependent on transcription of the upstream gene (37). However, the most widely accepted model is that the viral polymerase initiates transcription de novo from the extreme 3′ terminus of the genomic RNA (12) and then genes are sequentially transcribed by a start-stop mechanism (12, 20). Based on our analysis of transcription start site mutants, we proposed an additional feature to the start-stop model which suggested that after polyadenylation and termination of the upstream message, the polymerase scans the intergenic junction and initiates transcription at the first U it encounters, even if the U is not in the context of an optimal start sequence (36). Recently, an alternative model which suggests that the 5′ end of a transcript is generated following an RNA cleavage event was revived and modified (33). The chemistry of an RNA cleavage event would explain the atypical 5′ cap found on VSV mRNAs in which the α and β phosphates of the cap are provided by the cap donor (GDP) and only the α phosphate of the 5′ RNA acceptor is used in the 5′-5′ triphosphate bridge (1). In this current model, the last two nucleotides of the poly(A) tail of the upstream message serve as the primer for transcript initiation at the adjacent downstream gene. The priming event occurs by a forward slippage of the nascent poly(A) tail over the intergenic dinucleotide and subsequent base pair formation between the last two nucleotides of the poly(A) tail and the first two nucleotides (UUGUC…) of the downstream start sequence. Addition of the 5′ cap results from a GDP-dependent, polymerase-mediated cleavage event.
In this study, we used a transcriptionally active, dicistronic VSV minivirus (G and M gene minigenome [GMMG]) to test several aspects of the cleavage-capping model (33) and the U-dependent start model (36) for initiation of VSV transcription. Through insertional mutagenesis at the VSV intergenic dinucleotide within GMMG, we determined that the polymerase can efficiently perform all of the transcriptional events at gene junctions containing at least a 6-nucleotide intergenic region. However, not all sequences were tolerated by the VSV polymerase, suggesting that, in contrast to the proposed cleavage-capping model, the polymerase appears to scan the region and is affected in a sequence-specific manner. We also show that the first U following the polyadenylation signal of the upstream gene is not the sole determinant for transcript initiation. These data support a model for VSV transcription which includes a scanning polymerase that samples sequences downstream of the polyadenylation signal until an appropriate start site is encountered.
MATERIALS AND METHODS
Expression plasmids and minigenome constructs.
Plasmids encoding the wild-type (wt) GMMG as well as the VSV Indiana N, P, G, and L proteins have been described previously (14, 35). To generate minigenome mutants with insertions in the G and M intergenic region, we performed region-specific mutagenesis by using a PCR-based strategy. Genomic-sense, degenerate oligonucleotides spanning the M-G gene junction [5′-CATAGTGACGCGTAAACAGATCGATCTCTGTT(N/V4)AGTTTTTTTCATAGGG-3′; N = G, A, T, or C; V = G, A, or C] were used with an antigenomic-sense oligonucleotide (MW-28, 5′-TATAGGGCCCTCGCGAAGACAAACAAACCATTATTATC-3′) complementary to the leader region (in boldface type) to generate PCR products from a wt pBS-GMMG template. The primers used to generate the intergenic mutants were synthesized by delivering equimolar amounts of the indicated phosphoramidites at each target position. For the VSV New Jersey G-L intergenic mutants, primers that included the sequences shown in Fig. 1 were used in the PCR. Following PCR amplification using standard conditions (14), the PCR products were digested with MluI and StuI. The resulting 210- to 229-bp fragments were used with a StuI-to-BglII fragment (391 bp) from the M gene in a three-way ligation to replace the corresponding regions in the wild-type pBS-GMMG plasmid. Direct sequence analysis (thermal cycle dideoxy sequencing method; New England Biolabs) was used to identify plasmids containing the site-specific mutations and to confirm that deleterious mutations were not introduced during PCR amplification.
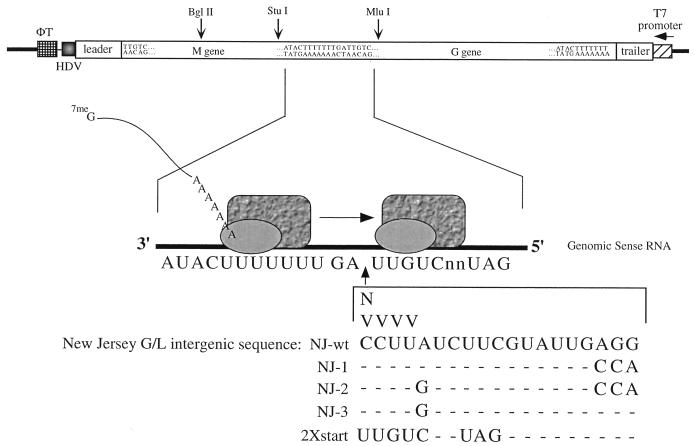
FIG. 1.
Sequences of GMMG intergenic insertion mutants. A diagram of pBS-GMMG is shown. Expression from the T7 promoter results in the synthesis of a genome-sense RNA which becomes encapsidated by the N protein and serves as a template for the synthesis of the M and G mRNAs by the VSV polymerase (35). The sequence of the M-G gene junction is enlarged, and the inserted sequences are shown below it. All insertions were between the wt nontranscribed intergenic dinucleotide (3′-GA-5′) and the G gene start sequence. The VSV polymerase is represented by the shaded rectangles and ovals. HDV, hepatitis delta virus ribozyme sequence; φT, the T7 terminator. N corresponds to G, A, T, or C; V corresponds to G, A, or C. Dashes indicate nucleotides that are identical to the nucleotides found in the NJ-wt sequence.
Minigenome recovery and passaging.
Recoveries were performed essentially as described previously (36). Baby hamster kidney (BHK-21) cells in 35-mm-diameter dishes were infected with vTF7-3 (15) and then transfected with 10 μg of plasmid DNA encoding either wt or mutant VSV minigenomes, together with 5, 4, 3, and 1 μg of plasmids encoding the N, P, G, and L proteins, respectively, by using TransfectACE (30). Culture supernatants were harvested 18 to 24 h posttransfection.
Minigenomes were passaged by transfecting cells with 5, 4, 3, and 1 μg of plasmids encoding the N, P, G, and L proteins, respectively, prior to addition of the harvested supernatant. The plasmid encoding G protein was included to obtain high-titer supernatants of minigenome mutants that could not passage efficiently due to insufficient G protein expression. Minigenome expression and passaging were monitored by detecting M protein expression in GMMG-infected cells by indirect immunofluorescence microscopy as described previously (36).
RNA analysis (primer extension and Northern blot assays).
All RNA analyses were performed exactly as described before (36). For primer extensions, we used an oligonucleotide (3188; 5′-GTCATTATGCCAATTTAAATC-3′) complementary to a sequence approximately 180 nucleotides from the 5′ end of the G mRNA. The sequencing ladder was generated by using the same end-labeled primer and wt pBS-GMMG plasmid, unless otherwise indicated. For Northern blot and primer extension analysis, total RNA was isolated at 5 to 6 h postinfection after infection with third- or fourth-passaged (P3 or P4) supernatants. The RNAs used for both Northern blot analysis and primer extension assays were isolated from the same minigenome infections except for the 3′-GAGGCA-5′ and 3′-GAGGUU-5′ mutants in Fig. 4, in which the RNA for each assay was isolated from separate minigenome infections. The templates used to generate the G mRNA-specific and the M mRNA-specific probes were described previously (35).
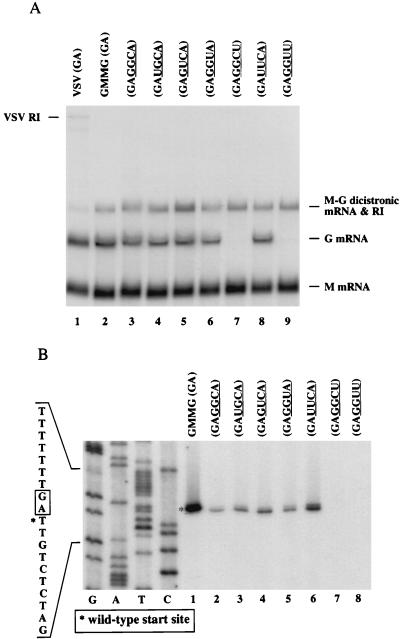
FIG. 4.
Analysis of U-dependent transcription initiation. (A) Northern blot analysis. Approximately 4 μg of total RNA was probed essentially as described in the legend to Fig. 2. Inserted nucleotides are underlined. The percentage of G mRNA expression for each mutant that contained a U ranged from 83 to 106% relative to that of the parental 3′-GAGGCA-5′ mutant and was determined by densitometry of digitized fluorograms. Inserted nucleotides are underlined. RI, replicative intermediate. (B) Primer extension analysis. The initiating nucleotide from the G mRNA was determined essentially as described in the legend to Fig. 3. One hundred times more RNA was used for the mutants shown in lanes 7 and 8. Since different amounts of RNA were used for this assay, the differences in signal intensity for the mutants compared to that of wt GMMG are not quantitative.
RESULTS
Previous data from saturation mutagenesis of the nontranscribed intergenic dinucleotide and the G gene start sequence suggested that the intergenic dinucleotide is important for efficient termination of the upstream gene mRNA whereas the first 3 nucleotides of the start sequence are most critical for efficient expression of the downstream gene. From these data, we suggested that the polymerase scans the intergenic region and attempts to initiate transcription at the first U encountered (36). Recently, Shuman proposed a novel cleavage-capping mechanism to explain the unique capping reaction chemistry and mRNA synthesis that occur during VSV transcription (33). In order to (i) test our U-dependent initiation hypothesis, (ii) test one aspect of the cleavage-capping model, and (iii) further examine the role that the conserved intergenic sequences play during polyadenylation and termination of the upstream mRNA and reinitiation at the next downstream gene, we utilized insertional mutagenesis at the M-G gene junction in the context of the dicistronic minigenome, GMMG. Either 1 or 4 nucleotides was inserted after the wild-type nontranscribed intergenic dinucleotide and before the G start sequence (Fig. 1). We also constructed a GMMG variant containing the 21-nucleotide intergenic region found between the G and L genes of VSV New Jersey (Ogden strain). Nucleotide substitutions were then made within this intergenic sequence either to create a minimal start sequence (5′-UUGUC…3′) (Fig. 1, NJ-2 and NJ-3) or to disrupt the potential base-pairing between the margins of this intergenic region (Fig. 1, NJ-1 and NJ-2). Base-pairing between the intergenic margins was suggested in the cleavage-capping model to be necessary to loop out the extra intergenic sequence (33). A total of 22 GMMG mutants were constructed and analyzed for M and G mRNA expression.
Differential effects of intergenic insertions on transcript initiation and accumulation.
To examine the effects of 1- or 4-nucleotide insertions on M and G mRNA expression, total RNA from cells replicating either wild-type or mutant minigenomes were analyzed by Northern blot analysis. G and M antisense probes were used to detect G mRNA, M mRNA, and two types of M-G RNA species, which were the positive-sense replicative intermediate and an M-G dicistronic mRNA. The replicative intermediate and M-G mRNA from GMMG-infected cells are approximately the same size because the latter is polyadenylated.
Figure 2 shows that insertions of a single G, A, or C immediately following the wt 3′-GA-5′ dinucleotide had no effect on either the M or G mRNA level, relative to that found in wt GMMG-infected cells (lanes 2 to 4 and 6). These data indicate that the VSV polymerase can tolerate a 1-nt insertion and confirmed previous results which showed that a single A insertion is tolerated (4). However, a U insertion almost completely eliminated G mRNA expression (lane 5). Primer extension analysis of these mutants indicated that the G mRNA initiated at the correct 5′ start site when G, A, or C was inserted (data not shown). In contrast, the low levels of G mRNA expressed from the U insertion mutant initiated at the inserted U. These results were consistent with our previous observations (36) which indicated that the polymerase attempts to initiate transcription at the first U it encounters. Based on our mutational analysis of the transcription start site, it is likely that the reduction in G mRNA level resulted from changing the context of the start sequence by 1 nucleotide from 3′-UUGUC-5′ to 3′-UUUGUC-5′. In this context, a U is in the third position of the start sequence (shown in boldface type), which we had shown results in almost undetectable amounts of mRNA (36).
FIG. 2.
Northern blot analysis of single-nucleotide insertion mutants. Approximately 4 μg of total RNA was fractionated on a 1% agarose–formaldehyde gel, transferred to a nylon membrane, and hybridized with a mixture of two antisense RNA probes specific for either G or M mRNA sequence. The inserted nucleotide for each mutant is underlined.
Previously, we and others (4, 36) had reported that some mutations in the conserved 3′-GA-5′ nontranscribed intergenic dinucleotide increased the levels of readthrough transcription at that gene junction. In contrast to the intergenic substitution mutants, none of the four insertion mutants had increased levels of readthrough transcription as determined by calculating the ratio of M-G dicistronic species to M mRNA. These data show that the normal (GA) dinucleotide intergenic sequence in conjunction with the upstream 3′-AUAC(U)7-5′ sequence is sufficient for M protein mRNA termination, even when the downstream G mRNA expression is severely reduced by an insertion of a U (Fig. 2, lane 5). This observation supports previous studies showing that termination of an upstream transcript is not dependent on initiation of the downstream transcript (3, 18, 36).
We next examined the effect of 4-nucleotide insertions placed immediately after the wild-type intergenic dinucleotide on transcription. Because our data suggested that the polymerase may attempt to initiate transcription at a U in the nontranscribed intergenic region, we generated GMMG mutants that contained an additional 4 nucleotides of random G, A, and C sequences after the normal 3′-GA-5′ dinucleotide. Seven different mutants were randomly selected, and the RNA species expressed were analyzed (Fig. 3A). Interestingly, the amount of G mRNA expressed from the mutants ranged from wild-type levels to nearly undetectable amounts. These data suggest that the polymerase can tolerate larger intergenic regions, although there is some specificity that determines what sequences are allowable. In fact, this has been observed by others for a single mutant containing a 4-nucleotide insertion, 3′-GAGCUC-5′ (4). For the seven insertion mutants examined here, there was no obvious pattern which could be used to predict the sequences that can be tolerated from those that cannot.
FIG. 3.
Analysis of “random” 4-nucleotide insertion mutants. (A) Northern blot analysis. Approximately 4 μg of total RNA was probed essentially as described in the legend to Fig. 2. Lane 1, wt VSV; lane 2, wt GMMG; lanes 3 to 9, 4-nt intergenic insertion mutants. The percentage of G mRNA expression for each mutant compared to that of wt GMMG is shown below each lane and was determined by densitometry of digitized fluorograms. Inserted nucleotides are underlined. (B) Primer extension. The total RNA was hybridized to an end-labeled oligonucleotide complementary to sequences in the G mRNA. Following extension, the products were resolved on a denaturing 6% polyacrylamide gel. A sequencing ladder generated from pBS-GMMG and the end-labeled oligonucleotide is also shown. The conserved intergenic dinucleotide is boxed, and the position of the wt initiating nucleotide is marked by an asterisk. The amount of total RNA used for mutant GMMG extension reactions ranged between 2 and 100 times more than that used for wt GMMG. The nucleotides where aberrant initiation occurred are indicated (lanes 4 to 8, outline font). A sixfold-longer exposure of lanes 5 to 8 is shown in the panel on the right.
To determine if any of the insertions altered the site of transcript initiation, we mapped the position of the 5′ end by primer extension. As much as 100 times more total RNA was used for the mutants that expressed very low levels of G mRNA. The results (Fig. 3B) show that for the two mutants which expressed near-wt levels of G mRNA (GAGGCA and GACCAG) (lanes 3 and 4, respectively), and the two mutants which had reduced amounts of mRNA (GAGGGC and GAGCCC) (lanes 2 and 5, respectively), the major transcription product initiated at the correct (wild-type) start site. However, transcripts from many of the mutants (lanes 4 to 8) initiated at multiple sites within the inserted region as well as the normal start site at various ratios. Importantly, all of the internal initiation sites mapped to C residues. It is important to note that for the more abundant extension products, there is a minor product which was 1 nucleotide longer (e.g., wt GMMG) (lane 1). This likely resulted from extension through the cap on the 5′ end of the message (11).
The VSV polymerase does not attempt to initiate transcription at the first U encountered.
To determine if the polymerase attempts to initiate transcription at the first U encountered following polyadenylation of the upstream mRNA, we substituted one or two U’s at nucleotides 3 through 6 in the 3′-GAGGCA-5′ mutant (nucleotides 3 to 6 are underlined), which expressed wild-type levels of M and G mRNAs, and then examined the level of G mRNA expressed from each mutant minigenome. Northern blot analysis (Fig. 4A) revealed that a U at position 3, 4, or 5 did not drastically alter downstream G expression relative to that seen for the 3′-GAGGCA-5 mutant (lanes 4 to 6 and 3, respectively). In addition, U residues at both positions 3 and 4 did not drastically alter G mRNA expression (lane 8). However, a U at position 6 or two U’s at positions 5 and 6 almost completely eliminated G mRNA expression (lanes 7 and 9). These data indicate that the presence of a U directly preceding the downstream start sequence is deleterious for gene expression.
Primer extension analysis showed that for the mutants which had near-wt levels of G mRNA expression, the transcripts initiated at the wt start site (Fig. 4B). Due to the extremely low levels of G mRNA present, it was difficult to determine at which nucleotide the 3′-GAGGUU-5′ and 3′-GAGGCU-5′ mutants initiated transcription. A longer exposure revealed that transcripts from the 3′-GAGGCU-5′ mutant were 1 nucleotide longer than the extension product for wt GMMG (data not shown). These data indicated that the polymerase did not attempt to initiate transcription at the first U it encountered following the wt intergenic dinucleotide (3′-GA-5′) unless the U directly preceded the wt start site.
Analysis of an intergenic variant.
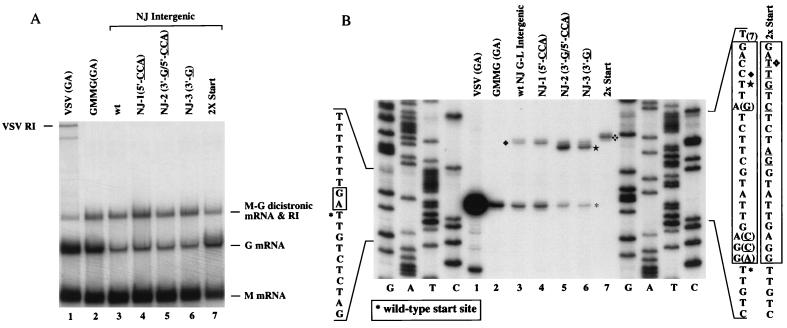
Whereas all intergenic junctions of VSV Indiana are 2 nucleotides (usually 3′-GA-5′), the G-L intergenic junction from VSV New Jersey contains an extra 19 nucleotides between the wt (3′-GA-5′) dinucleotide and the L gene start sequence. To examine this G-L gene junction, we generated a GMMG mutant (Fig. 1, NJ-wt) which contained the intergenic sequence from the Ogden strain of VSV New Jersey. Northern blot analysis (Fig. 5A) showed that the G mRNA level was reduced to approximately 25% of wt (VSV Indiana) levels (Fig. 5A, compare lanes 2 and 3), and primer extension analysis showed that the NJ-wt G gene transcripts were initiated at two sites (Fig. 5B). The majority of the transcripts initiated at the consensus start sequence located 21 nucleotides downstream from the M gene poly(A) signal. A less abundant transcript which was 18 nucleotides larger was also detected (Fig. 5B). This transcript would have initiated at the fourth nucleotide of the intergenic sequence. Interestingly, no transcripts were initiated at any of the U’s within the sequence, further confirming that the polymerase does not attempt to initiate transcription at the first U following the upstream polyadenylation sequence.
FIG. 5.
Mutagenesis of the 21-nucleotide G-L intergenic region from VSV New Jersey, Ogden strain. (A) Northern blot analysis. Approximately 4 μg of total RNA was probed essentially as described in the legend to Fig. 2. Lane 1, wt VSV Indiana; lane 2, wt GMMG; lane 3, NJ-wt G-L intergenic variant; lane 4, mutant NJ-1 that eliminates the potential base pairing at margins; lane 5, mutant NJ-2 that eliminates potential base pairing at margins and that contains an internal minimal start sequence; lane 6, mutant NJ-3 that contains an internal minimal start sequence; lane 7, VSV New Jersey intergenic region that contains two optimal start sequences (2X Start). RI, replicative intermediate. (B) Primer extension analysis. The initiating nucleotide of the G mRNA was determined essentially as described in the legend to Fig. 3. To obtain equivalent band intensities, four times more total RNA was used for the primer extension reactions of mutant GMMGs that had reduced G mRNA levels. Sequences for the various intergenic mutants with the corresponding start sites are shown. The Indiana wt sequence is shown on the left; the sequences of the NJ-wt and the 2xStart mutant are shown on the right. Intergenic sequences are boxed. Nucleotides that are in parentheses and/or are underlined indicate changes from the NJ-wt sequence. Alternative start sites are indicated by ⧫, ★, and ❖.
It was suggested that base pairing of nucleotides at the margins of the VSV New Jersey G-L intergenic region may be critical for looping out the extra sequence to allow the polymerase to transverse this intergenic junction during transcription (33). To examine this hypothesis, we generated mutants in which the potential base pairing was eliminated (mutants NJ-1 and NJ-2) (Fig. 1). We also produced mutants that resulted in the creation of an internal minimal start sequence by altering the A at position 7 to a G (mutants NJ-2 and NJ-3) (Fig. 1). Northern blot analysis indicated that elimination of the potential base pairing or the presence of an internal start sequence did not alter the levels of G mRNA expressed from these mutants as compared to the parental NJ-wt minigenome (Fig. 5A). All of the mutants expressed G mRNA at about 25% the level expressed from wt GMMG. In addition, primer extension analysis showed that the relative abundances of the two transcripts expressed from the variant in which the potential base pairing was eliminated (mutant NJ-1) were similar to those expressed from NJ-wt (Fig. 5B, lanes 3 and 4). Together, these results suggest that the extra sequence is not looped-out and that the polymerase can utilize multiple signals within the sequence for transcript initiation. Importantly, transcripts from the mutants which contained an internal minimal start sequence, either with or without complementary ends (mutants NJ-2 and NJ-3, respectively), were initiated approximately 80% of the time at the internal start sequence (Fig. 5B, lanes 5 and 6) and only 20% of the time at the downstream wt start sequence. These data are analogous to our findings with the four random nucleotide insertion mutants which suggest that the polymerase does not simply bypass the intergenic sequence but that it scans the intergenic region and responds to signals within this extra sequence.
To examine the effect of two consecutive optimal start sequences on G mRNA levels, we constructed an additional variant in which nucleotides 3 to 12 were altered to generate a consensus start sequence following the wt 3′-GA-5′ intergenic dinucleotide (2X Start) (Fig. 1). Northern blot analysis indicated that G mRNA levels for this variant were now at wt levels (Fig. 5A, lane 7). Moreover, primer extension showed that only one major transcript was produced (Fig. 5B, lane 7) which mapped to the first consensus start sequence immediately following the 3′-GA-5′ intergenic dinucleotide. Therefore, downstream start sites are not used if the polymerase encounters an optimal start sequence first.
DISCUSSION
In this study, we used insertional mutagenesis to test several aspects of two models that have been proposed for initiation of VSV transcription (33, 36). One model proposed that the 5′ end of transcripts are generated following a polymerase-mediated cleavage-capping event in which the 3′ end of the upstream polyadenylated mRNA is used as a primer while the intergenic region is simultaneously looped out (33). Another model suggested that after polyadenylation and termination of the upstream message, the polymerase scans the intergenic junction and initiates transcription de novo at the first U it encounters, even if the U is not in the context of an optimal start sequence (36). The data presented here demonstrate unequivocally that this later model is not completely correct and that other factors are most certainly involved in determining at which nucleotide the VSV polymerase initiates transcription. However, the data also show that the polymerase does sample the sequence of the intergenic region, presumably by a nontranscriptive scanning mechanism, and can respond by initiating transcripts at suboptimal, nonconsensus start sites within the intergenic region. Therefore, longer intergenic regions are not simply bypassed or looped out, as suggested in the cleavage-capping model.
No sequence patterns emerged to indicate why some of the inserted sequences were bypassed (e.g., 3′-GAGGCA-5′ and 3′-GACCAG-5′) while others were not (e.g., 3′-GACGGG-5′) nor why some of the sequences influenced the polymerase to initiate at a suboptimal start sequence. Perhaps the sequence itself or the events resulting from initiation at a suboptimal start sequence cause a portion of the polymerase population to disengage from the template. Alternatively, since suboptimal start sequences can be utilized and the resulting transcripts do not accumulate to high levels, the conserved sequence found at the beginning of each VSV gene may not be needed only for transcript initiation. Perhaps this sequence is also crucial for a later step, such as capping, methylation, and/or elongation of the nascent mRNA, and therefore is critical for transcript stability. We are currently screening a larger population of these insertion mutants to ascertain if there is a predictive rule regarding which sequences will be tolerated by the polymerase. One rule did emerge in that all initiations at suboptimal start sequences which resulted in detectable amounts of mRNA occurred at a C. This observation supports our previous conclusion (36) that there is a strong bias for the VSV polymerase to initiate transcripts with a purine.
Several nonsegmented negative-strand RNA viruses contain nontranscribed intergenic regions with a consensus length and sequence (3′-GA-5′ for VSV and 3′-GAA-5′ for Sendai virus, parainfluenza virus type 3, and measles virus [6, 16, 29, 34]), while others contain intergenic regions with both heterogeneous length and sequence (respiratory syncytial virus, rabies virus, Ebola virus, and Marburg virus [8, 13, 38]). Recently, it was shown, by using a similar type of minigenome system for respiratory syncytial virus, that the heterogeneous intergenic regions of respiratory syncytial virus do not appear to increase or decrease the attenuation that occurs at each gene junction (21). Therefore, while our results suggest that the VSV polymerase responds to the nontranscribed intergenic region in a sequence-specific manner, it appears that not all of the polymerases from the order Mononegavirales are affected by the intergenic region in the same manner.
From previous data (4, 36) and the data presented here, it is clear that transcript levels are severely decreased when a U directly precedes the start sequence. Primer extension analysis showed that a U at this position resulted in the accumulation of very low levels of transcripts which were initiated at this U. Presumably, the decreased levels of transcript are a result of altering the context of the start sequence. If the mechanism of transcription proceeds through some variation of the cleavage-capping model, then perhaps when the start sequence contains three U’s (e.g., 3′-UUUGUC-5′), the poly(A) sequence of the upstream transcript forms a more stable hybrid through base pairing with three U’s instead of the normal two. The increased stability of this hybrid may alter the strained complex proposed to be required for the polymerase-mediated attack by GDP across the ApA dinucleotide and inhibit the cleavage event. However, our previous observations (36) that nucleotide substitutions at position 3 of the transcription start site (3′-UUGUC-5′) can also severely reduce the amount of transcript produced and the fact that the polymerase can utilize suboptimal start sequences indicate that there are additional sequence-specific requirements for transcript initiation that cannot be explained by the cleavage-capping model.
One of the early models for VSV transcript initiation proposed that some fraction of polymerase molecules can initiate transcription at internal start sequences (37). More recent studies with the polR mutant of VSV have provided compelling evidence that internal initiation can occur at the leader-N gene junction in vitro (7); however, the authors cautioned that internal initiation may be specific for the leader-N gene junction and suggested that sequential transcription likely occurs once the N gene is transcribed. Although we did not examine the leader junction, our results with a GMMG mutant which contained two optimal start sequences after the M-G gene junction support this conclusion because we detected only transcripts that were initiated at the first consensus start sequence. If the polymerase had been able to initiate internally, we would have expected to see transcripts which initiated at both sites. The data obtained with this mutant also eliminated a potential mechanism for attenuation, namely, that some fraction of polymerase molecules bypass the start sequence and then disengage from the template at some point downstream. Had this been the mechanism, we again would have expected to see transcripts which had initiated at both sites, leading to an increase in the levels of the G gene transcript compared to wt levels.
By using a GMMG variant that contained the intergenic region from the G-L gene junction of the New Jersey serotype (Ogden strain), we found that expression from the downstream G gene was reduced 75% relative to that of wt (VSV Indiana) GMMG. Although it has not been determined if there is a similar increase in attenuation at the G-L gene junction of VSV New Jersey, our data suggest that this extra sequence may have been maintained to down-regulate polymerase expression. It is known that some negative-strand RNA viruses utilize a variety of strategies to down-regulate expression of not only their polymerase (9, 10, 24) but of other genes as well (5). We also found that transcripts were initiated from two sites. Approximately 22% initiated at the fourth nucleotide of the intergenic region, while the majority initiated at the consensus start site. These results contrast those of Luk et al., who used primer extension to map the start site of the L protein transcript in VSV New Jersey-infected cells as well as following in vitro transcription (23). Analysis of in vivo-generated transcripts indicated that only the consensus start sequence was used, whereas it appeared that in vitro-generated transcripts were initiated at two distinct sites, one following the intergenic dinucleotide and one at the consensus start sequence. The differences between the initiation sites that we observed and those previously reported (23) could be due to inherent differences between the VSV Indiana and VSV New Jersey polymerases and their ability to navigate this intergenic sequence. However, we cannot exclude the possibility that the differences are due to other factors such as the ratios of the N, P, and L proteins following transfection compared to that found in VSV New Jersey-infected cells.
Collectively, our data suggest that the nontranscribed intergenic region is not simply bypassed or looped out during transcription. While other aspects of the cleavage-capping model were not tested (i.e., that initiation of transcription occurs via a concomitant cleavage and capping event), this model is worthy of further investigation because of its potential to explain the unusual chemistry of the 5′ cap present on VSV mRNAs and the possible identification of a primer for transcript initiation. To further test this model, we are currently sequencing the 5′ ends of mRNAs generated from mutants in which the first two nucleotides of the start sequence have been altered. Should the poly(A) tail of the upstream message serve as the primer for the downstream transcript, one might expect to see A’s as the initiating nucleotides as opposed to the corresponding nucleotide change that was made in the template. Defining the mechanism responsible for VSV transcript initiation and capping will likely provide important clues as to how other viruses in the order Mononegavirales transcribe their genes. Although the fundamental mechanisms may be similar, sequence differences at the transcription start and stop polyadenylation sites and the potential utilization of different cellular factors important for polymerase activity may provide avenues to develop specific antiviral agents capable of selectively inhibiting the replication of members of this group of medically important pathogens.
ACKNOWLEDGMENTS
We thank Tim Higgins for figure preparation. We also thank Himangi Jayakar for comments after reading the manuscript. Oligonucleotides were synthesized by the Molecular Resource Center at the University of Tennessee, Memphis.
This work was supported by NIH grant GM-53726 to M.A.W.
REFERENCES
- 1.Abraham G, Rhodes D P, Banerjee A K. The 5′ terminal structure of the methylated mRNA synthesized in vitro by vesicular stomatitis virus. Cell. 1975;5:51–58. doi: 10.1016/0092-8674(75)90091-4. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee A K. Transcription and replication of rhabdoviruses. Microbiol Rev. 1987;51:66–87. doi: 10.1128/mr.51.1.66-87.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr J N, Whelan S P J, Wertz G W. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr J N, Whelan S P J, Wertz G W. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J Virol. 1997;71:1794–1801. doi: 10.1128/jvi.71.3.1794-1801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bousse T, Takimoto T, Murti K G, Portner A. Elevated expression of the human parainfluenza virus type 1 F gene downregulates HN expression. Virology. 1997;232:44–52. doi: 10.1006/viro.1997.8524. [DOI] [PubMed] [Google Scholar]
- 6.Cattaneo R, Rebmann G, Schmid A, Baczko K, ter Meulen V, Billeter M A. Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 1987;6:681–688. doi: 10.1002/j.1460-2075.1987.tb04808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuang J L, Perrault J. Initiation of vesicular stomatitis virus mutant polR1 transcription internally at the N gene in vitro. J Virol. 1997;71:1466–1475. doi: 10.1128/jvi.71.2.1466-1475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins P L, Dickens L E, Buckler-White A, Olmsted R A, Spriggs M K, Camargo E, Coelingh K V W. Nucleotide sequences for the gene junctions of human respiratory syncytial virus reveal distinctive features of intergenic structure and gene order. Proc Natl Acad Sci USA. 1986;83:4594–4598. doi: 10.1073/pnas.83.13.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins P L, Olmsted R A, Spriggs M K, Johnson P R, Buckler-White A J. Gene overlap and site-specific attenuation of transcription of the viral polymerase L gene of human respiratory syncytial virus. Proc Natl Acad Sci USA. 1987;84:5134–5138. doi: 10.1073/pnas.84.15.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conzelmann K K, Cox J H, Schneider L G, Thiel H J. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology. 1990;175:485–499. doi: 10.1016/0042-6822(90)90433-r. [DOI] [PubMed] [Google Scholar]
- 11.Davison A J, Moss B. Structure of vaccinia virus early promoters. J Mol Biol. 1989;210:749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 12.Emerson S U. Reconstitution studies detect a single RNA polymerase entry site on the vesicular stomatitis virus genome. Cell. 1982;31:635–642. doi: 10.1016/0092-8674(82)90319-1. [DOI] [PubMed] [Google Scholar]
- 13.Feldmann H, Klenk H D, Sanchez A. Molecular biology and evolution of filoviruses. Arch Virol Suppl. 1993;7:81–100. doi: 10.1007/978-3-7091-9300-6_8. [DOI] [PubMed] [Google Scholar]
- 14.Fredericksen B L, Whitt M A. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J Virol. 1995;69:1435–1443. doi: 10.1128/jvi.69.3.1435-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuerst T R, Niles E G, Studier R W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta K C, Kingsbury D W. Complete sequences of the intergenic and mRNA start signals in the Sendai virus genome: homologies with the genome of vesicular stomatitis virus. Nucleic Acids Res. 1984;12:3829–3841. doi: 10.1093/nar/12.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hercyk N, Horikami S M, Moyer S A. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology. 1988;163:222–225. doi: 10.1016/0042-6822(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 18.Hwang L N, Englund N, Pattnaik A K. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J Virol. 1998;72:1805–1813. doi: 10.1128/jvi.72.3.1805-1813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iverson L E, Rose J K. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 20.Iverson L E, Rose J K. Sequential synthesis of 5′-proximal vesicular stomatitis virus mRNA sequences. J Virol. 1982;44:356–365. doi: 10.1128/jvi.44.1.356-365.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo L, Fearns R, Collins P L. The structurally diverse intergenic regions of respiratory syncytial virus do not modulate sequential transcription by a dicistronic minigenome. J Virol. 1996;70:6143–6150. doi: 10.1128/jvi.70.9.6143-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawson N, Stillman E, Whitt M A, Rose J K. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luk D, Masters P S, Gill D S, Banerjee A K. Intergenic sequences of the vesicular stomatitis virus genome (New Jersey serotype): evidence for two transcription initiation sites within the L gene. Virology. 1987;160:88–94. doi: 10.1016/0042-6822(87)90048-1. [DOI] [PubMed] [Google Scholar]
- 24.Morimoto K, Ohkubo A, Kawai A. Structure and transcription of the glycoprotein gene of attenuated HEP-Flury strain of rabies virus. Virology. 1989;173:465–477. doi: 10.1016/0042-6822(89)90559-x. [DOI] [PubMed] [Google Scholar]
- 25.Moyer S A, Banerjee A K. In vivo methylation of vesicular stomatitis virus and its host cell messenger RNA species. Virology. 1976;70:339–351. doi: 10.1016/0042-6822(76)90276-2. [DOI] [PubMed] [Google Scholar]
- 26.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 27.Rhodes D, Banerjee A K. 5′-Terminal sequence of vesicular stomatitis virus genome RNA. J Virol. 1976;17:33–42. doi: 10.1128/jvi.17.1.33-42.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose J K. Heterogeneous 5′-terminal structures occur on vesicular stomatitis virus mRNAs. J Biol Chem. 1975;250:8098–8104. [PubMed] [Google Scholar]
- 29.Rose J K. Complete intergenic and flanking gene sequences from the genome of vesicular stomatitis virus. Cell. 1980;19:415–421. doi: 10.1016/0092-8674(80)90515-2. [DOI] [PubMed] [Google Scholar]
- 30.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 31.Schnell M J, Buonocore L, Whitt M A, Rose J K. The minimal conserved transcription stop/start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J Virol. 1996;70:2318–2323. doi: 10.1128/jvi.70.4.2318-2323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schubert M, Keene J D, Herman R C, Lazzarini R A. Site of the vesicular stomatitis virus genome specifying polyadenylation and the end of the L gene mRNA. J Virol. 1980;34:550–559. doi: 10.1128/jvi.34.2.550-559.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuman S. A proposed mechanism of mRNA synthesis and capping by vesicular stomatitis virus. Virology. 1997;227:1–6. doi: 10.1006/viro.1996.8305. [DOI] [PubMed] [Google Scholar]
- 34.Spriggs M K, Collins P L. Human parainfluenza virus type 3: messenger RNAs, polypeptide coding assignments, intergenic sequences, and genetic map. J Virol. 1986;59:646–654. doi: 10.1128/jvi.59.3.646-654.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stillman E A, Rose J K, Whitt M A. Replication and amplification of novel vesicular stomatitis virus minigenomes encoding viral structural proteins. J Virol. 1995;69:2946–2953. doi: 10.1128/jvi.69.5.2946-2953.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stillman E A, Whitt M A. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J Virol. 1997;71:2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Testa D, Chanda P K, Banerjee A K. Unique mode of transcription in vitro by vesicular stomatitis virus. Cell. 1980;21:267–275. doi: 10.1016/0092-8674(80)90134-8. [DOI] [PubMed] [Google Scholar]
- 38.Tordo N, Poch O, Ermine A, Keith G, Rougeon F. Walking along the rabies genome: is the large G-L intergenic region a remnant gene? Proc Natl Acad Sci USA. 1986;83:3914–3918. doi: 10.1073/pnas.83.11.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whelan S P J, Ball L A, Barr J N, Wertz G T W. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]