Abstract
Among the three viral proteins present in the hepatitis B virus (HBV) envelope, both the small and large polypeptides, but not the middle polypeptide, are necessary for the production of complete viral particles. Whereas it has been established that the C-terminal extremity of the pre-S1 region is required for HBV morphogenesis, whether the pre-S2 region of the large surface protein plays a critical role remains questionable. In the present study, we have analyzed the role of the large-polypeptide pre-S2 region in viral maturation and infectivity. For this purpose, mutants bearing contiguous deletions covering the entire pre-S2 domain were generated. First, the efficient expression of all the mutant large envelope proteins was verified and their ability to substitute for the wild-type form in virion secretion was tested. We found that distinct deletions covering the domain between amino acids 114 and 163 still allowed virion production. In contrast, the polypeptide lacking the first 5 amino acids of pre-S2 (amino acids 109 to 113) was unable to support viral secretion. This result shows that the domain of the large surface protein, required for this process, must be extended to the N-terminal extremity of pre-S2. We then demonstrated that all the mutants competent for virion release were able to infect normal human hepatocytes in primary culture. Taken together, these results indicate that only 10% of the large-protein pre-S2 region at its N-terminal extremity is essential for virion export and that the remaining part, dispensable for viral secretion, is also dispensable for infectivity.
The hepatitis B virus (HBV) envelope contains three transmembrane proteins known as hepatitis B surface (HBs) proteins: the small (S), the middle (M), and the large (L) polypeptides. All these proteins have a common hydrophobic S region with additional N-terminal pre-S extensions for the M and L proteins. The S protein, a 226-amino-acid polypeptide, is the most abundant and is encoded by the S gene. The M protein is formed by the S peptide extended by 55 amino acids at its amino terminus, corresponding to the pre-S2 region. The L protein contains all the amino acids found in the M protein and has an additional N-terminal sequence of 108 amino acids (ayw subtype), referred to as the pre-S1 region. The three proteins are posttranslationally modified: each of them exhibits a partially glycosylated site in the S domain, an additional glycosylation occurs in the pre-S2 region of the M protein, and the L protein differs from the other envelope proteins by a N-terminal myristylation (22).
The S and the M proteins undergo their final transmembrane folding in the endoplasmic reticulum (ER) membrane during their synthesis. Their S domains span the lipid bilayer at two topogenic signals (I and II) and probably at their hydrophobic C terminus (7). Their N-terminal extremities, and particularly the pre-S2 region of the M protein, are located into the ER lumen and so appear on the surface of secreted viral particles. The early luminal disposition of the pre-S2 domain is confirmed by its carbohydrate modification. Conversely, in the L protein, no glycan is linked at the two potential sites located in the pre-S1 and pre-S2 regions. This is explained by the fact that in the primary translation product, the type I signal does not cross the ER membrane and all the pre-S sequence remains in the cytosol. During viral morphogenesis, half of the L protein population keep this topology, with the pre-S region inside the virion (i-preS form) (4, 21, 24). A posttranslational reorganization occurs for the other half since this protein displays a topology similar to the M protein in complete viral particles, with an external pre-S domain (e-preS form).
Several studies have investigated the role of these surface proteins in the viral cycle. Expression of the S protein appears sufficient for the secretion of empty envelope particles, whereas the expression of both the S and L peptides is required for the production of mature virions (2). Experiments based on truncations of the L protein have shown that the N-terminal five-sixths of the pre-S1 sequence is dispensable for this process (5). Inhibition of the M protein expression has no effect on viral morphogenesis (2). Concerning the infection step, only a few data on the contribution of viral envelope proteins have been reported. Thus, the in vitro infectious ability of HBV requires the presence of the myristate moiety of the L protein (3, 11). By contrast, the M protein is not likely to be involved in viral infectivity (8, 20).
Finally, although no precise function is obviously attributed to the pre-S2 region of the M protein, it remains to be determined whether the pre-S2 domain of the L protein could be involved in the viral life cycle. To investigate this point, a set of deletions extending into this domain was created and the ability of the modified proteins to substitute for the wild-type (WT) form in virion secretion and infectivity was evaluated.
MATERIALS AND METHODS
Plasmid engineering.
Plasmid pHBV-ΔEcoRI contains an HBV DNA insert of more than one genome length, starting at position 1232 and ending at position 1984 (11). This viral sequence contains only one copy of the pre-S1-pre-S2-S open reading frame (ORF) and is able to support viral replication. To introduce a point mutation into the viral sequence, we used the U-DNA mutagenesis kit from Boehringer Mannheim as specified by Kunkel et al. (18). The mutagenic oligonucleotide used to introduce an L protein-defective mutation was 5′ CTGCCTTCCTCACTGGCGATT 3′. Mutant viral genomes were recovered, and a 361-bp pre-S1 fragment, flanked by BstXI and EcoRI sites, was transferred back into pHBV-ΔEcoRI to form the pHBV L− plasmid. The whole sequence of the subcloned fragment was sequenced to verify that no fortuitous mutation had been introduced during mutagenesis.
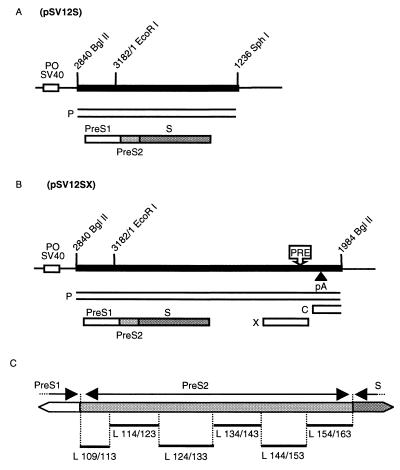
Plasmids have been constructed to express the L surface protein in WT or mutated forms. Plasmid pSV12S WT (Fig. 1A) contains the 1,578-bp BglII-SphI fragment of WT HBV DNA, bearing the entire pre-S-S coding regions, cloned downstream of the simian virus 40 early promoter-origin region in plasmid pSV-SPORT 1 (Life Technologies). Deletions were introduced into pSV12S by PCR. This method is based on the amplification of a first fragment located upstream of the deletion with primers DEL A (5′ ATTTATGCAGAGGCCGAGGC 3′) and DEL B. A second fragment was amplified downstream of the deletion with primers DEL C and DEL D (5′ AGGTTGGGGACTGCGAATTT 3′). Table 1 describes primers DEL B and C according to the expected deletion. After purification, these two products were mixed and used in a second round of amplification with primers DEL A and DEL D. Amplified fragments were digested with EcoRI and XbaI and inserted into EcoRI-XbaI-digested pSV12S WT. In contrast, the first N-terminal deletion eliminates the EcoRI site. In this case, the KpnI site was used instead of the EcoRI site. In new plasmids, inserts were sequenced to confirm the expected deletion (L x/y, where x is the position of the N-terminal amino acid of the deletion and y is the position of the C-terminal amino acid of the deletion) without other mutation (Fig. 1C).
FIG. 1.
(A and B) Expression vectors of the L protein. The heavy line indicates the HBV sequence; the thin line indicates plasmid pSV-SPORT 1 sequences with its simian virus 40 early promoter-origin region (PO SV40); boxes indicate ORFs for viral X, P, C, and S proteins. The envelope ORF is divided into pre-S1, pre-S2, and S domains. The approximate locations of the posttranscriptional regulatory element (PRE) and the polyadenylation site (pA) in the HBV sequence are shown. (C) Amino acid deletions in the pre-S2 region of the S gene cloned in different L protein expression plasmids. Deletions (L x/y; x is the wild-type position of the N-terminal amino acid flanking the deletion; y is the wild-type position of the C-terminal amino acid flanking the deletion) are indicated below as thin lines.
TABLE 1.
Primers used for deletion
| Mutanta | Sequence of primer DEL B and DEL Cb | Deleted amino acid sequencesc |
|---|---|---|
| L 109/113 | 5′ TTGGTGGAAGGTTGTGGCCTGAGGATGAGTGTTTC 3′ | MQWNS (5) |
| 5′ ACTCATCCTCAGGCCACAACCTTCCACCAAACTCT 3′ | ||
| L 114/123 | 5′ CAGGCCTCTCACTCTGGAATTCCACTGCATGGCCT 3′ | TTFHQTLQDP (10) |
| 5′ ATGCAGTGGAATTCCAGAGTGAGAGGCCTGTATTT 3′ | ||
| L 124/133 | 5′ TCCTGAACTGGAGCCGGGATCTTGCAGAGTTTGGT 3′ | RVRGLYFPAG (10) |
| 5′ ACTCTGCAAGATCCCGGCTCCAGTTCAGGAACAGT 3′ | ||
| L 134/143 | 5′ AGAGGCAGTAGTCAGACCAGCAGGGAAATACAGGC 3′ | GSSSGTVNPV (10) |
| 5′ TATTTCCCTGCTGGTCTGACTACTGCCTCTCCCTT 3′ | ||
| L 144/153 | 5′ CCCAATCCTCGAGAAAACAGGGTTTACTGTTCCTG 3′ | LTTASPLSSI (10) |
| 5′ ACAGTAAACCCTGTTTTCTCGAGGATTGGGGACCC 3′ | ||
| L 154/163 | 5′ TGTGATGTTCTCCATGATTGACGATAAGGGAGAGG 3′ | FSRIGDPALN (10) |
| 5′ CCCTTATCGTCAATCATGGAGAACATCACATCAGG 3′ |
Numbers indicate the wild-type positions of amino acids flanking the deleted sequences.
The sequence of DEL C is given below the sequence of DEL B for each mutant.
Numbers in parentheses indicate the length of the deletion in amino acids.
Other expression vectors, WT and mutants pSV12SX (Fig. 1B), were constructed. They are derived from WT and mutants pSV12S, respectively. These plasmids therefore contain a longer HBV DNA insert, starting at position 2840 and ending at position 1986.
Cell line and transfection.
To produce viral proteins or virions, the permissive HepG2 human hepatoma cell line (1, 28) was transfected with HBV DNA by electroporation. HepG2 cells were cultured in H medium (75% minimum essential medium, 25% medium 199, 5 mg of insulin per liter, 1 g of bovine serum albumin per liter, 4.5 mg of penicillin per liter, 50 mg of streptomycin per liter) supplemented with 3.5 × 10−7 M hydrocortisone hemisuccinate and 10% fetal calf serum (FCS).
Virus purification.
HBV particles were isolated from the culture medium of transfected HepG2 cells by precipitation with 6% polyethylene glycol (PEG 8000; Sigma) for 12 h at 4°C. The precipitates were recovered by centrifugation (10,000 × g for 45 min at 4°C) and concentrated 200-fold in phosphate-buffered saline (PBS) with 25% FCS.
Primary cell culture and infection.
Normal adult human liver fragments were obtained from patients undergoing hepatic resection for liver metastases (the fragments were taken at a distance from the metastasis in macroscopically normal liver). Access to this biopsy material was in agreement with French laws and satisfied the requirements of the Ethics Committee of the institution. Hepatocytes were isolated by the procedure of Guguen-Guillouzo and Guillouzo (12) and cultured in H medium supplemented with 3.5 × 10−6 M hydrocortisone hemisuccinate, 2% dimethyl sulfoxide, 5% adult human serum, and 5% FCS. The combination of these two sera favors long-term culture compared with the only use of 10% FCS (10) or 10% porcine serum (11, 25). At 3 days after seeding, the cells were infected as described previously (9). Hepatocytes (1.5 × 106 per 10-cm2 petri dish) were covered with 1 ml of serum-free culture medium containing 5% PEG 8000 and 100 μl of inoculum. Infection was performed for 12 h at 37°C. The cells were then washed three times with culture medium and further cultured. Virus obtained from the culture supernatant of the clonal cell line designated 2.2.15 was used as a positive control for the infection assays (26).
Cellular protein analysis.
Transfected cells were washed with PBS and scraped in PBS containing 10% FCS. Cells were recovered by centrifugation for 3 min at 200 × g, lysed at 0°C for 20 min in a lysis buffer (25 mM Tris-HCl [pH 7.4], 250 mM NaCl, 5 mM EDTA, 1% Nonidet P-40), and then centrifuged at 12,000 × g for 15 min at 4°C to remove nuclei. Sample buffer (3% sodium dodecyl sulfate [SDS], 2% 2-mercaptoethanol, 10% glycerol, 0.1% bromophenol blue, 5 mM EDTA, 200 mM Tris-HCl [pH 6.8]) was added to the supernatants, and the mixture was boiled for 5 min before being loaded into gels. Proteins were separated by SDS-polyacrylamide gel electrophoresis (10% polyacrylamide) and transferred onto a nitrocellulose filter (Amersham). Nonspecific binding sites were blocked for 1 h in Tris-buffered saline (TBS) (10 mM Tris-HCl, [pH 7.6], 150 mM NaCl)–0.1% Tween 20–5% dried milk. For the detection of the L protein, the filters were incubated for 2 h at room temperature with a polyclonal antibody, raised against a pre-S1 peptide (a generous gift of H. J. Hong), at a dilution of 1:500 in block solution. After three washes in TBS–0.1% Tween 20, the filters were incubated for 30 min in block solution. The antibody-antigen complex was visualized by using goat anti-mouse immunoglobulin G coupled to horseradish peroxidase conjugate at a dilution of 1:25,000 (Jackson Immuno Research Laboratories, Inc.) in TBS–0.1% Tween 20 and the enhanced-chemiluminescence Western blotting analysis system (Pierce, Rockford, Ill.).
Assays for HBV-specific proteins.
HBs antigen was detected with a radioimmunoassay kit (Abbott Laboratories, Abbott Park, Ill.) under conditions recommended by the manufacturer. A signal/noise ratio (P/N ratio) of >2.1 was considered positive.
DNA extraction and analysis.
Intracellular nucleocapsids were isolated from the cytoplasmic fraction of transfected HepG2 cells. Cells were recovered as described above for cellular protein analysis and lysed at 0°C for 20 min in a lysis buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 1% Nonidet P-40). Nuclei were removed by centrifugation at 12,000 × g for 20 s at 4°C. The core particles were then immunoprecipitated with an anti-hepatitis B core (HBc) antibody (Dako), and viral DNA was extracted as described below.
Complete viral particles were isolated from the supernatant of transfected HepG2 cells. Viruses were immunoprecipitated with a polyclonal anti-HBs antibody (Dako) or with a monoclonal anti-pre-S1 antibody, 5a-19 (6). Nucleic acids were extracted as described below, after overnight lysis at 37°C in a buffer consisting of 0.5% SDS, 10 mM Tris-HCl (pH 8), 10 mM EDTA, 10 mM NaCl, 40 μg of tRNA per ml, and 200 μg of proteinase K per ml.
The covalently closed circular form of HBV DNA (cccDNA) was extracted from human adult hepatocyte cultures. To remove virions adsorbed onto the cell monolayer, the cells were incubated with 0.5 mg of trypsin per ml–0.5 mM EDTA in PBS for 10 min at 37°C. The reaction was stopped by adding 20% FCS. The cells were then recovered by centrifugation for 3 min at 200 × g. The pellet was washed with PBS, and the cells were lysed at 20°C for 2 h with 0.5% SDS–10 mM Tris-HCl (pH 8)–10 mM EDTA–10 mM NaCl. Cellular DNA was precipitated overnight at 4°C in lysis buffer supplemented with 1 M NaCl (14). Chromosomal DNA was pelleted at 12,000 × g for 15 min at 4°C.
For nucleic acid extraction, the proteins were removed by two phenol-chloroform-isoamyl alcohol extractions followed by one chloroform-isoamyl alcohol extraction. Then, in the absence of salt, 0.3 M sodium acetate (pH 5.5) was added to the aqueous phase and the nucleic acids were precipitated with 2 volumes of ethanol.
DNA was analyzed on 1.5% agarose gels. These gels were soaked in 0.25 N HCl for 15 min, and DNA was denatured in situ in 0.4 M NaOH and transferred onto positively charged nylon membranes (Amersham) by the Southern method (27). Hybridization was performed at 65°C with linearized HBV genomic [α-32P]DNA as a probe.
RESULTS
Experimental strategy.
Plasmid pHBV L−, containing an HBV DNA insert of more than one genome length, allowed the transcription of all known viral RNAs under the control of their own promoters. Only the L-protein expression was suppressed by introducing an opal mutation into codon 90 of the pre-S1 frame. This point mutation remained silent in the overlapping polymerase gene. The defect can be complemented in trans by cotransfecting pHBV L− with an expression vector carrying the missing WT protein (Fig. 1A) (2). To evaluate the role of the pre-S2 region of the L protein, constructs carrying a mutation were used as cotransfecting plasmids and the ability of the mutant proteins to replace the WT form in viral cycle was investigated. Six contiguous deletions, covering the entire pre-S2 region, were introduced in six distinct plasmids expressing the L protein (Fig. 1C).
Expression of the mutant proteins.
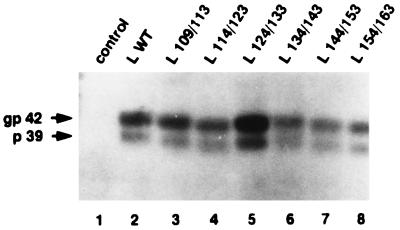
One trivial condition for the ability of a mutant protein to complement the HBV genome defective for the L protein expression is that it must be expressed efficiently. To investigate this point, plasmids expressing the WT or mutant L protein were transiently transfected into HepG2 cells and intracellular viral proteins were analyzed by Western blotting with a polyclonal anti-pre-S1 antibody. A first set of plasmids, pSV12S (Fig. 1A), was used. They carried only the WT or the mutant pre-S1-pre-S2-S genes. The amount of viral surface proteins synthesized in transfected cells was too weak to be detectable under our Western blot conditions (data not shown). A second set of plasmids, pSV12SX (Fig. 1B), was constructed and transfected into HepG2 cells. They contained the genes coding for the envelope proteins and also had the downstream HBV sequence beyond to the polyadenylation signal. This additional viral sequence contained the X gene/enhancer II region, which is known to increase the level of surface gene transcripts. In particular, the presence of the posttranscriptional regulatory element facilitates cytoplasmic accumulation of transcripts (15). Indeed, the WT and modified L proteins became detectable (Fig. 2). The 39-kDa unglycosylated (p39) and the 42-kDa glycosylated (gp42) forms of the L protein were found in the lysate of cells transfected with WT protein expression vector (13) (Fig. 2, lane 2). As expected, proteins from all mutant forms migrated faster than the WT protein but their expression level was similar to that of the WT protein (lanes 3 to 8), with the exception of the L 124/133 mutant protein, for which the amount of the intracellular protein was substantially increased (approximately twofold) for unknown reasons. In all cases, two bands with a 3-kDa difference in their molecular masses, probably corresponding to the unglycosylated and glycosylated forms, respectively, were detected.
FIG. 2.
Western blot analysis of the L protein expressed in transfected cells. HepG2 cells were transfected with 20 μg of different L expression vectors: control plasmid without HBV insert (lane 1), L WT expression plasmid driving the synthesis of the WT L protein (lane 2), and L x/y expression plasmids driving the synthesis of different mutant L proteins (lanes 3 to 8). Proteins were extracted from cells 7 days after transfection and analyzed by electrophoresis through a 10% polyacrylamide–SDS gel. The primary polyclonal antibody was directed against the pre-S1 region. gp 42 and p 39 indicated the migration positions of the glycosylated and unglycosylated L proteins, respectively.
Analysis of nucleocapsid assembly.
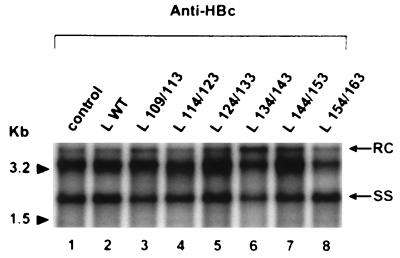
To estimate a possible trans influence of the mutant L proteins on intracellular core particle assembly, HepG2 cells were cotransfected with the replication-competent L-defective genome and the different L protein expression plasmids. DNA of cytoplasmic nucleocapsids was selectively extracted 10 days after cotransfection and analyzed by the Southern blot procedure (Fig. 3). As expected from the results of Bruss et al. (2), the DNA patterns were identical whether the WT L protein was present or absent. Furthermore, the viral DNA patterns observed with all mutant forms of the L protein appeared unchanged. Comparison of different samples showed identical amounts of encapsidated DNA resulting from similar transfection efficiency.
FIG. 3.
Southern blot analysis of HBV DNA from intracellular core particles. An anti-HBc antibody was used to immunoprecipitate core particles from cells transfected with the L-defective genome complemented with different L expression vectors: control plasmid without HBV insert (lane 1), L WT expression plasmid driving the synthesis of the WT L protein (lane 2), and L x/y expression plasmids driving the synthesis of different mutant L proteins (lanes 3 to 8). DNA was extracted and analyzed on a 1.5% agarose gel. Molecular size markers are indicated in kilobases; the positions of relaxed-circular DNA (RC) and single-stranded DNA (SS) are shown.
Effects of pre-S2 deletions in the L protein on HBV particle production.
The ability of the mutant peptides to complement a replication-competent L-defective genome for viral particle secretion was tested in comparison with a WT L protein. Secreted HBs antigen (HBsAg) was measured from the pooled culture supernatants collected between days 3 and 6 posttransfection (data not shown). HBsAg was found to be actively produced by cells transfected with the L-defective genome either uncomplemented or complemented with the WT or mutant L protein expression plasmids. In all cases, slightly smaller amounts were observed compared to those obtained for the control without L protein, in agreement with previous data showing that expression of the L polypeptide is responsible for retention of viral envelope proteins (19).
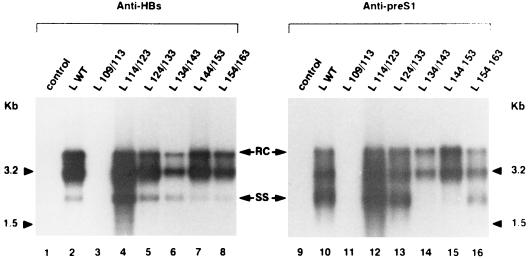
To evaluate the production of complete virions by transfected HepG2 cells, immunoprecipitation with anti-HBs antibodies was performed on the culture supernatants. Viral DNA was extracted from the immunoprecipitates and analyzed by Southern blotting (Fig. 4). As previously shown (2), the mutation preventing L protein expression in the viral genome abolished virion release, but its complementation with a WT L protein expression vector restored viral secretion (Fig. 4, lanes 1 and 2, respectively). Cotransfection with constructs producing deleted proteins clearly showed that mutants with a 10-amino-acid deletion between amino acids 114 and 163 were able to complement the L defect for virion export (lanes 4 to 8). Depending on the deletion, the amount of secreted virions was slightly different compared to that for the WT L protein. Thus, while complete viral particles were more efficiently released when amino acids 114 to 123 (L 114/123) were deleted (lane 4), the level of viral secretion was slightly inhibited with L 134/143 (lane 6). Conversely, no complete viral particle was secreted in the presence of the mutant L 109/113 protein (lane 3). These observations provide evidence that the first 5 amino acids of the pre-S2 region in the L protein are required for nucleocapsid budding.
FIG. 4.
Southern blot analysis of HBV DNA in particles secreted by transfected HepG2 cells. Cells were transfected with the L-defective genome complemented with different L expression vectors: control plasmid without HBV insert (lanes 1 and 9), L WT expression plasmid driving the synthesis of the WT L protein (lanes 2 and 10), and L x/y expression plasmids driving the synthesis of different mutant L proteins (lanes 3 to 8 and 11 to 16). Complete viral particles were immunoprecipitated from HepG2 supernatants, collected between days 3 and 6 posttransfection, with a polyclonal anti-HBs antibody (left panel) or with a monoclonal anti-pre-S1 antibody (right panel). Then DNA was extracted from the immunoprecipitates and analyzed on a 1.5% agarose gel. Molecular size markers are indicated in kilobases; the positions of relaxed-circular DNA (RC) and single-stranded DNA (SS) are shown.
To verify that the deletions did not interfere with the presence of the pre-S1 region on the surface of virions, another immunoprecipitation was performed with a monoclonal antibody directed against a pre-S1 epitope (Fig. 4). All the assembled viruses were successfully immunoprecipitated with this monoclonal antibody (Fig. 4, lanes 10 and 12 to 16). Thus, despite a deletion in the pre-S2 region of the L protein, the pre-S1 epitope remained present at the surface of extracellular virions.
Infectivity of the mutant virions.
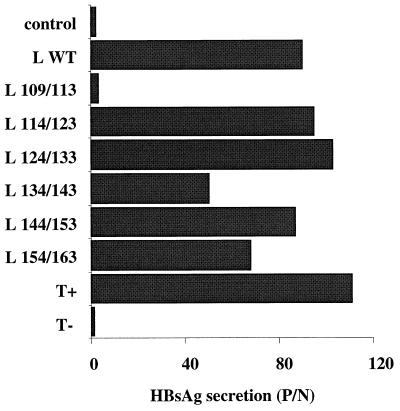
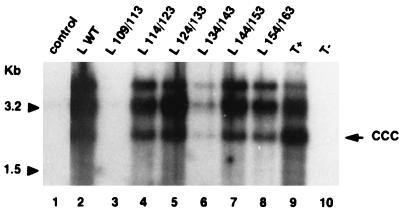
To determine whether the pre-S2 region of the L protein was involved in viral infectivity, in vitro infections of normal adult human hepatocytes were performed with both mutant and WT virions. Cells were incubated for 12 h with viral particles prepared by concentrating the supernatant from cotransfected HepG2 cells. As shown in Fig. 5, long-term HBsAg production was demonstrated in experiments performed with viruses containing the WT L protein and with all mutant viruses, which were secreted. These observations strongly suggest that the mutant viruses were infectious. To further sustain this assertion, we looked for the presence of supercoiled HBV DNA (cccDNA) in infected hepatocytes (Fig. 6). As in positive controls (Fig. 6, lanes 2 and 9), cccDNA was detected in hepatocytes infected by all mutant viral particles (lanes 4 to 8). The results of infectivity for mutant L 134/143 were positive but partially impaired by the limited amount of secreted virions. As expected, both HBsAg secretion and cccDNA detection in cultures infected with the supernatants of HepG2 cells cotransfected with L 109/113 expression plasmid were negative, since no virus was secreted. These results demonstrate that the C-terminal ten-elevenths of the pre-S2 region, dispensable for viral production, is also dispensable for infectivity.
FIG. 5.
HBsAg secretion by human hepatocytes following in vitro infection assays. Hepatocytes were incubated with PBS (T−) or with concentrated supernatants obtained from either 2.2.15 cell cultures (T+) or HepG2 cells transfected with the L-defective genome complemented with different L expression vectors: control plasmid without HBV insert, L WT expression plasmid driving the synthesis of the WT L protein, or L x/y expression plasmids driving the synthesis of different mutant L proteins. HBsAg was measured by a conventional radioimmunoassay in primary hepatocyte culture supernatants collected 10 days postinfection.
FIG. 6.
Southern blot analysis of HBV cccDNA in human hepatocytes following in vitro infection assays. Hepatocytes were incubated with PBS (T−) or with concentrated supernatants obtained from either 2.2.15 cell cultures (T+) or HepG2 cells transfected with the L-defective genome complemented with different L expression vectors: control plasmid without HBV insert, L WT expression plasmid driving the synthesis of the WT L protein, or L x/y expression plasmids driving the synthesis of different mutant L proteins. Supercoiled viral DNA was selectively extracted from hepatocytes collected 10 days postinfection and analyzed on a 1.5% agarose gel. Molecular size markers are indicated in kilobases to the left; the position of cccDNA is shown to the right (ccc).
DISCUSSION
Although the assembly of HBV requires the L protein, most of the pre-S1 region is dispensable for this process, as demonstrated by experimental mutagenesis (2, 5). Indeed, N-terminal truncation up to amino acid 102 of the subtype adw L protein, which corresponds to residue 91 of subtype ayw used in the present work, interferes neither with the transmembrane topology of the protein nor with the production of virus. When the deletion was located further along the protein, the virion release was totally inhibited. This leads us to postulate that the crucial domain involved in the formation of complete viral particles and their secretion might be located beyond amino acid 91 (subtype ayw).
Different contiguous deletions were selectively introduced in the pre-S2 region of the L protein to identify the domain involved in the assembly process. By Western blot analysis, we found that synthesis of deleted proteins was not impaired by the different mutations. In addition, the deletions beyond the fifth of the 55 amino acids of the pre-S2 domain still allowed viral secretion. These results suggest that amino acids 114 to 163 of the L protein are not involved in virion production. By contrast, the deletion of the N-terminal five residues of the pre-S2 domain prevents the release of complete viral particles. Different hypotheses concerning the involvement of this sequence can be advanced; all are based on direct or indirect alteration of an interaction site with cytoplasmic nucleocapsids. First, a possible indirect mechanism, which involves the translocation of the pre-S domain, may be suggested. Indeed, because of the mutation, an early translocation of the pre-S region through the ER membrane could prevent interaction with the nucleocapsid. If this really occurred, the pre-S1 glycosylation site should be glycosylated and we should observe the diglycosylated form showing a reduced migration on gels. It is obvious from Western blot experiments that this did not occur. Second, another indirect mechanism could be involved. Inhibition of interaction with cytoplasmic nucleocapsids, despite a normal cytosolic position of the pre-S domain, could be explained by a conformational modification of the cytosolic sequence, which might reduce the accessibility of the interaction site. Third, the lack of virion formation could result from a direct alteration of the putative interaction site. This conclusion is supported by the observations of Poisson et al. (23), who have tested the ability of peptides corresponding to different regions of the envelope proteins to bind core particles. The peptide found to have the greatest binding affinity to the nucleocapsid was a peptide equivalent to positions 96 to 116, corresponding to the 13 C-terminal amino acids of pre-S1 plus the 8 N-terminal amino acids of pre-S2. This observation, together with our data, strongly argues for the involvement of the pre-S2 amino-terminal extremity of the L protein in the envelope-nucleocapsid interaction.
Another important feature of the virion biology is the infection process. Experiments with the avian Hepadnaviridae model suggest that the pre-S domain of the L protein must be involved in contacts between the virus and the host cell (16, 17). Therefore, the presence of the e-preS topology at the surface of virions is a necessary condition to preserve the infectious ability of the virus. Accordingly, we first verified that deletions did not affect the presence of the e-preS form in virions. A monoclonal antibody directed against a pre-S1 epitope was able to immunoprecipitate all mutant virions. Accessibility of the pre-S1 sequence at the surface of mutant virions indicated that the e-preS form of the mutant L protein was also preserved and could potentially ensure contact(s) with the host cell surface, provided that the site(s) of interaction with a putative cell receptor remained intact. All our mutant viruses conserved their infectious ability. The deleted sequences are consequently not involved during the infection process. Thus, our experiments show that 90% of the pre-S2 region of the L protein is not involved in the infection step. By contrast, we cannot exclude that the N-terminal 5 amino acids of pre-S2 could potentially contribute to attachment to and entry into target cells. This region would ensure two distinct properties: interactions with the nucleocapsid in the i-preS form and interactions with the putative cell receptor in the e-preS form. However, it is most likely that a distinct region is responsible for cell recognition.
ACKNOWLEDGMENTS
This work was supported by INSERM, the Association pour la Recherche contre le Cancer, and the Ligue Nationale contre le Cancer (comité d’Ille et Vilaine). Jacques Le Seyec and Philippe Chouteau were recipients of fellowships from the Ministère de l’Education nationale de la Recherche et de la Technologie and from the Ligue Nationale contre le Cancer (comité des Côtes d’Armor), respectively.
We are indebted to Pascal Loyer, Olivier Loreal and André Guillouzo for helpful criticism of the manuscript. We gratefully acknowledge Agatha Budkowska for the gift of the 5a-19 antibody and Hyo Jeong Hong for the gift of the pre-S1 peptide.
REFERENCES
- 1.Aden D P, Fogel A, Plotkin S, Damjanov I, Knowles B B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature (London) 1979;282:615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- 2.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruss V, Hagelsten J, Gerhardt E, Galle P R. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology. 1996;218:396–399. doi: 10.1006/viro.1996.0209. [DOI] [PubMed] [Google Scholar]
- 4.Bruss V, Lu X Y, Thomssen R, Gerlich W H. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13:2273–2279. doi: 10.1002/j.1460-2075.1994.tb06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruss V, Thomssen R. Mapping a region of the large envelope protein required for hepatitis B virion maturation. J Virol. 1994;68:1643–1650. doi: 10.1128/jvi.68.3.1643-1650.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budkowska A, Quan C, Groh F, Bedossa P, Dubreuil P, Bouvet J P, Pillot J. Hepatitis B virus (HBV)-binding factor in human serum—candidate for a soluble form of hepatocyte HBV receptor. J Virol. 1993;67:4316–4322. doi: 10.1128/jvi.67.7.4316-4322.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eble B E, Lingappa V R, Ganem D. The N-terminal (pre-S2) domain of a hepatitis B virus surface glycoprotein is translocated across membranes by downstream signal sequences. J Virol. 1990;64:1414–1419. doi: 10.1128/jvi.64.3.1414-1419.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernholz D, Galle P R, Stemler M, Brunetto M, Bonino F, Will H. Infectious hepatitis-B virus variant defective in pre-S2 protein expression in a chronic carrier. Virology. 1993;194:137–148. doi: 10.1006/viro.1993.1243. [DOI] [PubMed] [Google Scholar]
- 9.Gripon P, Diot C, Guguen-Guillouzo C. Reproducible high level infection of cultured adult human hepatocytes by hepatitis-B virus—effect of polyethylene glycol on adsorption and penetration. Virology. 1993;192:534–540. doi: 10.1006/viro.1993.1069. [DOI] [PubMed] [Google Scholar]
- 10.Gripon P, Diot C, Theze N, Fourel I, Loreal O, Brechot C, Guguen-Guillouzo C. Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J Virol. 1988;62:4136–4143. doi: 10.1128/jvi.62.11.4136-4143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gripon P, Le Seyec J, Rumin S, Guguen-Guillouzo C. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology. 1995;213:292–299. doi: 10.1006/viro.1995.0002. [DOI] [PubMed] [Google Scholar]
- 12.Guguen-Guillouzo C, Guillouzo A. Methods for preparation of adult and fetal hepatocytes. In: Guillouzo A, Guguen-Guillouzo C, editors. Isolated and cultured hepatocytes. London, United Kingdom: Les Éditions INSERM Paris. John Libbey and Co, Ltd.; 1986. pp. 1–12. [Google Scholar]
- 13.Heermann K H, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich W H. Large surface proteins of hepatitis B virus containing the pre-S sequence. J Virol. 1984;52:396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 15.Huang Z M, Yen T S. Hepatitis B virus RNA element that facilitates accumulation of surface gene transcripts in the cytoplasm. J Virol. 1994;68:3193–3199. doi: 10.1128/jvi.68.5.3193-3199.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishikawa T, Ganem D. The pre-S domain of the large viral envelope protein determines host range in avian hepatitis B viruses. Proc Natl Acad Sci USA. 1995;92:6259–6263. doi: 10.1073/pnas.92.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klingmuller U, Schaller H. Hepadnavirus infection requires interaction between the viral pre-S domain and a specific hepatocellular receptor. J Virol. 1993;67:7414–7422. doi: 10.1128/jvi.67.12.7414-7422.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkel T A, Roberts J D, Zabour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 19.Kuroki K, Russnak R, Ganem D. Novel N-terminal amino acid sequence required for retention of a hepatitis B virus glycoprotein in the endoplasmic reticulum. Mol Cell Biol. 1989;9:4459–4466. doi: 10.1128/mcb.9.10.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Seyec, J., C. Guguen-Guillouzo, and P. Gripon. Unpublished data.
- 21.Ostapchuk P, Hearing P, Ganem D. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 1994;13:1048–1057. doi: 10.1002/j.1460-2075.1994.tb06353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Persing D H, Varmus H E, Ganem D. The pre-S1 protein of hepatitis B virus is acylated at its amino terminus with myristic acid. J Virol. 1987;61:1672–1677. doi: 10.1128/jvi.61.5.1672-1677.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poisson F, Severac A, Hourioux C, Goudeau A, Roingeard P. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology. 1997;228:115–120. doi: 10.1006/viro.1996.8367. [DOI] [PubMed] [Google Scholar]
- 24.Prange R, Streeck R E. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 1995;14:247–256. doi: 10.1002/j.1460-2075.1995.tb06998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rumin S, Gripon P, Le Seyec J, Corral-Debrinski M, Guguen-Guillouzo C. Long-term productive episomal hepatitis B virus replication in primary cultures of adult human hepatocytes infected in vitro. J Viral Hepatol. 1996;3:227–238. doi: 10.1111/j.1365-2893.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 26.Sells M A, Chen M L, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 28.Sureau C, Romet-Lemonne J L, Mullins J I, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1986;47:37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]