Abstract
The reovirus M1, L1, and L2 genes encode proteins found at each vertex of the viral core and are likely to form a structural unit involved in RNA synthesis. Genetic analyses have implicated the M1 gene in viral RNA synthesis and core nucleoside triphosphatase activity, but there have been no direct biochemical studies of μ2 function. Here, we expressed μ2 in vitro and assessed its RNA-binding activity. The expressed μ2 binds both poly(I-C)- and poly(U)-Sepharose, and binding activity is greater in Mn2+ than in Mg2+. Heterologous RNA competes for μ2 binding to reovirus RNA transcripts as effectively as homologous reovirus RNA does, providing no evidence for sequence-specific RNA binding by μ2. Protein μ2 is now the sixth reovirus protein demonstrated to have RNA-binding activity.
Genetic analyses have demonstrated that the reovirus M1, L1, and L2 genes are determinants of acute myocarditis in mice (19, 20). These three genes encode core proteins forming a structural unit at each vertex of the viral core (7, 9, 14). The L2-encoded λ2 protein is a guanylyltransferase (5, 13), and the L1-encoded λ3 protein has RNA polymerase activity (8, 23). While genetic analyses have identified the M1 gene as a determinant of reovirus RNA synthesis (6, 18, 28) and as a determinant of nucleoside triphosphatase (NTPase) activity in the viral core (15), there have been no direct biochemical studies of the function of the M1-encoded protein μ2.
The 736-amino-acid sequence of protein μ2 is well conserved between reovirus serotypes 1 and 3 (27, 30). Upon examination of the amino acid sequence, we noticed several regions that contained an unusually high number of arginine and lysine residues. Such basic regions are frequently associated with RNA-binding activity (3, 4, 24); therefore, we cloned and expressed the M1 gene and examined its capacity to bind synthetic RNA analogs as well as single-stranded RNA (ssRNA).
Reovirus protein μ2 binds both dsRNA and ssRNA analogs.
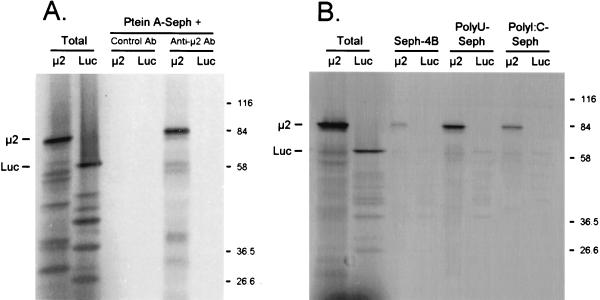
High-fidelity reverse transcription-PCR (Vent polymerase; Promega, Madison, Wis.) was used to insert a copy of the M1 gene (from reovirus strain 8B [21]) into pBluescript II (Stratagene, La Jolla, Calif.), and in vitro transcripts were synthesized with T7 RNA polymerase. M1 transcripts and control luciferase mRNA were translated in rabbit reticulocyte lysates, and the [35S]methionine-labeled proteins were precipitated with anti-μ2 antiserum, poly(U)-Sepharose, or poly(I-C)–Sepharose and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1). In vitro translation of M1 transcripts generated an 83-kDa protein, as expected for μ2 (Fig. 1), that was immunoprecipitated by anti-μ2 antiserum but not by control antiserum (Fig. 1A). The lower-molecular-weight products are routinely obtained in M1 translations (17, 31, 32). The translated luciferase mRNA generated the expected 61-kDa protein (Fig. 1), which was not precipitated by anti-μ2 antiserum or control antiserum (Fig. 1A). μ2 and luciferase bound control Sepharose CL4B insignificantly (4% binding for each [Fig. 1B]). μ2, however, bound both poly(U)-Sepharose and poly(I-C)–Sepharose (increased sevenfold and threefold, respectively, relative to Sepharose CL4B binding), while luciferase did not (no measurable increase). Thus, μ2 binds both ssRNA and double-stranded RNA (dsRNA) analogs.
FIG. 1.
μ2 binds poly(U)-Sepharose and poly(I-C)–Sepharose. (A) Immunoprecipitations. T7-generated M1 transcripts and control luciferase mRNA (Promega) were translated in rabbit reticulocyte lysates (Promega) containing [35S]Met, and the products were precleared with protein A-Sepharose CL4B (Ptein A-Seph) beads (Pharmacia) that had been washed in TNET buffer (50 mM Tris [pH 8.0], 100 mM NaCl, 5 mM EDTA, 1% Triton X-100). Hyperimmune rabbit antiserum prepared against a serotype 3 Dearing (T3D) μ2-Trp-E fusion protein (32), cross-reactive with T1L-(or 8B)-μ2, was incubated with washed protein A-Sepharose CL4B beads and then resuspended in TNET buffer. Precleared supernatants were then incubated with complexed beads. After extensive washing in radioimmunoprecipitation assay buffer (50 mM Tris [pH 8.0], 100 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS), bound protein was eluted by boiling in Laemmli sample buffer (11). Eluted protein and total translated protein were resolved by electrophoresis on a 10% Laemmli SDS-polyacrylamide gel (11), fixed in 5% trichloroacetic acid, dried, and exposed to film. Luc, luciferase; Ab, antibody. (B) RNA binding assays. Sepharose CL4B, poly(I-C)–Sepharose type 6, and poly(U)-Sepharose type 6 (Pharmacia, Piscataway, N.J.) were washed in RNA binding buffer (70 mM NaCl, 10 mM Tris [pH 7.4], 5 mM MnCl2, 1 mM dithiothreitol) and then resuspended in the same buffer. Translated proteins were precleared in Sepharose CL4B, incubated with the indicated beads, and washed in RNA binding buffer. The same gel was used as for panel A.
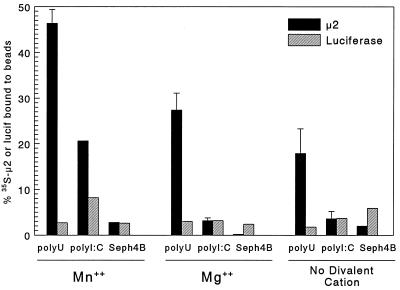
Next, the effects of divalent cations on μ2 binding to poly(U)-Sepharose and poly(I-C)-Sepharose were examined (Fig. 2). In 5 mM Mn2+, μ2 bound significantly to both poly(U) and poly(I-C)-Sepharose, while luciferase did not, and neither bound the control Sepharose CL4B. However, in 5 mM Mg2+, μ2 binding to poly(U)-Sepharose was reduced and binding to poly(I-C)–Sepharose was eliminated, and similar results were seen in the absence of divalent cations. Thus, μ2 binding to RNA analogs was optimal in Mn2+. Similar results were observed over a pH range of 6.8 to 8.0 (data not shown). The observation that μ2 bound RNA better in Mn2+ than in Mg2+ is consistent with evidence that in vitro poly(C)-dependent poly(G) polymerase activity of purified reovirus protein λ3 is higher in Mn2+ than in Mg2+ (23) and that μ2 and λ3 likely form a complex in the viral core (9) for viral RNA synthesis.
FIG. 2.
Effects of divalent cations on μ2 binding to poly(U)-Sepharose and poly(I-C)–Sepharose. Sepharose CL4B (Seph4B), poly(U)-Sepharose, and poly(I-C)–Sepharose were washed as for Fig. 1 RNA binding assays, except that 5 mM Mn2+ was substituted for with 5 mM Mg2+ or no divalent cation where indicated. Translated products (as for Fig. 1) were precleared with Sepharose CL4B and then incubated with the indicated beads as for Fig. 1 RNA binding assays, except that all incubations and washes contained the indicated divalent cation. Total translated product and triplicate samples (μ2) or single samples (luciferase [lucif]) bound to the indicated beads were resolved by SDS-PAGE and scanned with a Packard instant imager. The manufacturer’s software was used to select bands of the appropriate molecular weight for quantitation, and the percent of protein bound was calculated relative to total translated μ2 or luciferase (mean ± standard deviation).
Baculovirus-expressed μ2 binds nucleic acid, with no evidence for sequence-specific binding.
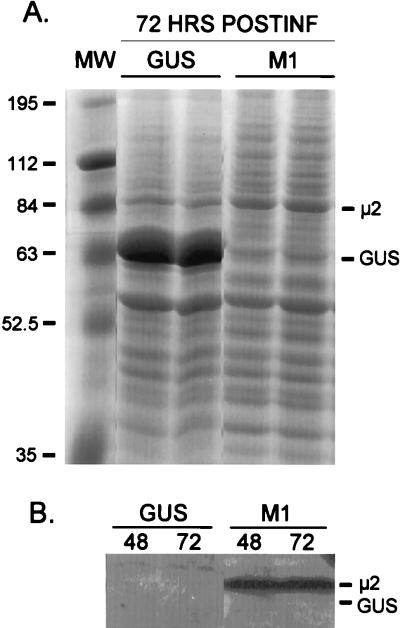
To date, there has been no evidence for sequence specificity in reovirus RNA-binding proteins. To investigate this, μ2 and control GUS protein were expressed from recombinant baculoviruses as follows. Trichoplasma ni (insect) cells were infected with a recombinant baculovirus containing the reovirus M1 gene or control GUS gene, and cell lysates were resolved by SDS-PAGE (Fig. 3A). Coomassie blue staining revealed unique bands at the expected molecular weight for GUS (duplicates in lanes 2 and 3) and μ2 (duplicates in lanes 4 and 5). In Western blots, anti-μ2 antiserum bound exclusively to the 83-kDa protein generated in cells infected with the M1-containing baculovirus (Fig. 3B, lanes 3 and 4).
FIG. 3.
Protein μ2 is expressed from a recombinant baculovirus. The 8B M1 gene was subcloned into recombinant baculovirus by using the Bac-to-Bac baculovirus expression system (GIBCO BRL, Grand Island, N.Y.). Control GUS-expressing recombinant baculovirus was provided by the manufacturer. T. ni insect cells were infected with virus stock that had been passaged in Sf9 insect cells, and cell cultures were harvested at 48 h (B) or 72 h (A and B) postinfection (POSTINF), washed with phosphate-buffered saline supplemented with 1 mM phenylmethylsulfonyl fluoride, and lysed in radioimmunoprecipitation assay buffer. Lysate supernatants were resolved by electrophoresis on 10% Laemmli SDS–polyacrylamide gels. (A) Coomassie blue staining, duplicate samples. MW, molecular mass (kilodaltons) markers. (B) Western Blot analysis. For Western blot analysis, protein was transferred to Immobilon-P membrane with a semidry blotting system (Millipore, Bedford, Mass.). Detection by the ECL (enhanced chemiluminescence) system (Amersham Life Sciences, Arlington Heights, Ill.) was done according to the manufacturer’s protocol with hyperimmune rabbit antiserum as for Fig. 1 immunoprecipitations.
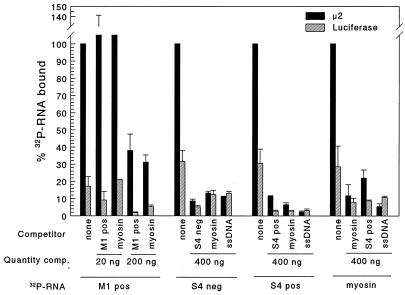
Binding of μ2 to specific RNA sequences was investigated as follows. Baculovirus-expressed μ2 and GUS were incubated with anti-μ2 antiserum and protein A-Sepharose. The antibody-complexed protein was then incubated in 200 mM NaCl with no further addition, with ssDNA (M13mp18; U.S. Biochemical Corp., Cleveland, Ohio), or with the indicated quantity of unlabeled (competitor) T7-generated ssRNA transcripts: reovirus positive- or negative-strand S4 (18), positive-strand M1, or control feline β-myosin. Triplicate samples were then incubated with T7-generated 32P-labeled ssRNA transcripts as indicated. Bound RNA was eluted, resolved by SDS-PAGE, quantitated, and expressed as percent bound in the absence of competitor (Fig. 4). Protein μ2 bound reovirus ssRNA, and while binding was inhibited by homologous ssRNA in a dose-dependent manner, binding was also inhibited by heterologous ssRNA (Fig. 4) (experiments with M1 32P-RNA). Furthermore, μ2 bound myosin ssRNA, and binding to both reovirus and myosin ssRNA was inhibited by up to 90% when excess homologous ssRNA, heterologous ssRNA, or heterologous ssDNA was added (Fig. 4). Thus, while μ2 binds single-stranded nucleic acid, the data provide no evidence for sequence-specific binding. It remains possible that μ2 in a complex with other reovirus proteins recognizes specific reovirus sequences for binding or that ssRNA with authentic termini are required for sequence-specific binding (authentic reovirus transcripts could not be synthesized at a high enough specific activity to be tested). Sequence specificity for dsRNA binding was not examined (again, RNA could not be radiolabeled to a high enough specific activity for testing).
FIG. 4.
Baculovirus-expressed μ2 binds nucleic acid, and binding is not sequence specific. Lysate supernatants from recombinant baculovirus-infected T. ni cells were incubated with rabbit anti-μ2 antisera and then incubated with protein A-Sepharose CL4B. After extensive washing with radioimmunoprecipitation assay buffer, the Sepharose-protein A-immunocomplexed μ2 or control GUS protein was resuspended in high-salt RNA binding buffer (200 mM NaCl, 30 mM Tris [pH 7.4], 5 mM MnCl2, 0.5 mM dithiothreitol) and incubated with no further addition, with unlabeled (competitor [comp.]) ssDNA (M13mp18; U.S. Biochemical Corp.) or with the indicated quantity of unlabeled (competitor) T7-generated ssRNA transcripts: reovirus positive (pos)- or negative (neg)-strand S4 (18), positive-strand M1, or control feline β-myosin. Triplicate samples were then incubated with T7-generated 32P-labeled ssRNA transcripts as indicated (1 ng per reaction, by extrapolation from the predicted specific activity) and then washed extensively with high-salt RNA binding buffer. Bound RNA was eluted with 1 M NaCl–30 mM Tris (pH 7.4), with 5 mM MnCl2, and electrophoresed on a 1% agarose gel. Gels were acid fixed, dried, and scanned with a Packard instant imager. The manufacturer’s software was used to quantitate bands of the appropriate molecular weight. The percent of 32P-RNA bound was calculated relative to 32P-RNA bound in the absence of competing unlabeled nucleic acid (triplicate samples, mean ± standard error of the mean).
The observation that μ2 bound ssRNA at 200 mM NaCl (Fig. 4), with no benefit when ionic strength was reduced (data not shown), indicates the relative stability of μ2 binding to ssRNA. Like μ2, several plant virus movement proteins bind to ssRNAs at concentrations ranging from 100 to 200 mM NaCl (2, 22). In contrast, the binding of bluetongue virus protein NS2 to bluetongue virus ssRNA is severely reduced between 100 and 200 mM NaCl (25). Similarly, the NP protein of influenza virus, although critical for viral transcription and replication, binds ssRNA nonspecifically, and binding can be disrupted at NaCl concentrations greater than 200 mM (26).
Protein μ2 is now the 6th of 11 reovirus proteins to exhibit RNA-binding activity. The five other reovirus proteins (ς2, ς3, λ1, ςNS, and μNS) bind ssRNA, dsRNA, or both; however, no studies have provided evidence for sequence-specific binding (reviewed in reference 14). While μ2 bound ssRNA, this binding was competed by ssDNA (Fig. 4). Replication of reovirus and rotavirus (reviewed in references 10 and 16) occurs outside of the nucleus (although reovirus protein ς3, for unknown reasons, has been found in the nucleus [29]), providing no evolutionary selective pressures for distinguishing between RNA and DNA. Indeed, the reovirus protein λ1 binds dsDNA in addition to dsRNA (12), and the rotavirus protein VP2 binds dsDNA in addition to RNA (1). Most reovirus and rotavirus RNA-binding studies have not included DNA as a control, and therefore it is unclear whether DNA binding is a common property of reovirus and rotavirus RNA-binding proteins.
Role of μ2 RNA-binding activity.
Reovirus cores synthesize positive-sense ssRNA from the enclosed dsRNA template (14), and cryoelectron microscopy suggests that μ2 lies adjacent to the viral polymerase λ3 and the guanylyltransferase λ2 in these cores (9). In addition, genetic evidence has implicated the M1 gene, which encodes μ2, in both positive- and negative-strand RNA synthesis (6, 18, 28). Finally, recent genetic evidence associates μ2 with viral core NTPase activity (15). Together, the data suggest μ2 is part of a heteromeric complex involved in both positive- and negative-strand RNA synthesis, and future studies will address possible enzymatic roles for μ2 in this process.
Acknowledgments
We thank Bill Clay for invaluable help in working with recombinant baculoviruses, Mary Ann Blum for technical assistance, and Max Nibert, Kevin Coombs, and Jon Horowitz for thoughtful review of the manuscript.
This research was supported by Public Health Service grant AI-31250 from the NIAID D.L.N. received fellowship support from the U.S. Department of Education Graduate Assistance in Areas of National Need (GAANN) Program.
REFERENCES
- 1.Boyle J F, Holmes K V. RNA-binding proteins of bovine rotavirus. J Virol. 1986;58:561–568. doi: 10.1128/jvi.58.2.561-568.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brantley J D, Hunt A G. The N-terminal protein of the polyprotein encoded by potyvirus tobacco vein mottling virus is an RNA-binding protein. J Gen Virol. 1993;74:1157–1162. doi: 10.1099/0022-1317-74-6-1157. [DOI] [PubMed] [Google Scholar]
- 3.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 4.Calnan B J, Tidor B, Biancalana S, Hudson D, Frankel A D. Arginine-mediated RNA recognition: the arginine fork. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- 5.Cleveland D R, Zarbl H, Millward S. Reovirus guanylyltransferase is L2 gene product lambda 2 protein. J Virol. 1986;60:307–311. doi: 10.1128/jvi.60.1.307-311.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coombs K M. Identification and characterization of a double-stranded RNA− reovirus temperature-sensitive mutant defective in minor core protein μ2. J Virol. 1996;70:4237–4245. doi: 10.1128/jvi.70.7.4237-4245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coombs K M. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology. 1998;243:218–228. doi: 10.1006/viro.1998.9061. [DOI] [PubMed] [Google Scholar]
- 8.Drayna D, Fields B N. Activation and characterization of the reovirus transcriptase: genetic analysis. J Virol. 1982;41:110–118. doi: 10.1128/jvi.41.1.110-118.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dryden K A, Farsetta D L, Wang G, Keegan J M, Fields B N, Baker T S, Nibert M L. Internal structures containing transcriptase-related proteins in top component particles of mammalian orthoreovirus. Virology. 1998;245:33–46. doi: 10.1006/viro.1998.9146. [DOI] [PubMed] [Google Scholar]
- 10.Estes M K. Rotaviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1625–1656. [Google Scholar]
- 11.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Lemay G, Danis C. Reovirus lambda 1 protein: affinity for double-stranded nucleic acid by a small amino-terminal region of the protein independent from the zinc finger motif. J Gen Virol. 1994;75:3261–3266. doi: 10.1099/0022-1317-75-11-3261. [DOI] [PubMed] [Google Scholar]
- 13.Mao Z X, Joklik W K. Isolation and enzymatic characterization of protein lambda 2, the reovirus guanylyltransferase. Virology. 1991;185:377–386. doi: 10.1016/0042-6822(91)90785-a. [DOI] [PubMed] [Google Scholar]
- 14.Nibert M L, Schiff L A, Fields B N. Reoviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1557–1596. [Google Scholar]
- 15.Noble S, Nibert M L. Core protein μ2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J Virol. 1997;71:7728–7735. doi: 10.1128/jvi.71.10.7728-7735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patton J T. Structure and function of the rotavirus RNA-binding proteins. J Gen Virol. 1995;76:2633–2644. doi: 10.1099/0022-1317-76-11-2633. [DOI] [PubMed] [Google Scholar]
- 17.Roner M R, Roner L A, Joklik W K. Translation of reovirus RNA species m1 can initiate at either of the first two in-frame initiation codons. Proc Natl Acad Sci USA. 1993;90:8947–8951. doi: 10.1073/pnas.90.19.8947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sherry B, Baty C J, Blum M A. Reovirus-induced acute myocarditis in mice correlates with viral RNA synthesis rather than generation of infectious virus in cardiac myocytes. J Virol. 1996;70:6709–6715. doi: 10.1128/jvi.70.10.6709-6715.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherry B, Blum M A. Multiple viral core proteins are determinants of reovirus-induced acute myocarditis. J Virol. 1994;68:8461–8465. doi: 10.1128/jvi.68.12.8461-8465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherry B, Fields B N. The reovirus M1 gene, encoding a viral core protein, is associated with the myocarditic phenotype of a reovirus variant. J Virol. 1989;63:4850–4856. doi: 10.1128/jvi.63.11.4850-4856.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherry B, Schoen F J, Wenske E, Fields B N. Derivation and characterization of an efficiently myocarditic reovirus variant. J Virol. 1989;63:4840–4849. doi: 10.1128/jvi.63.11.4840-4849.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soumounou Y, Laliberte J F. Nucleic acid binding properties of the P1 protein of turnip mosaic potyvirus in Escherichia coli. J Gen Virol. 1994;75:2567–2573. doi: 10.1099/0022-1317-75-10-2567. [DOI] [PubMed] [Google Scholar]
- 23.Starnes M C, Joklik W K. Reovirus protein lambda 3 is a poly(C)-dependent poly(G) polymerase. Virology. 1993;193:356–366. doi: 10.1006/viro.1993.1132. [DOI] [PubMed] [Google Scholar]
- 24.Tan R, Frankel A D. Structural variety of arginine-rich RNA-binding peptides. Proc Natl Acad Sci USA. 1995;92:5282–5286. doi: 10.1073/pnas.92.12.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uitenweerde J M, Theron J, Stoltz M A, Huismans H. The multimeric nonstructural NS2 proteins of bluetongue virus, African horsesickness virus, and epizootic hemorrhagic disease virus differ in their single-stranded RNA-binding ability. Virology. 1995;209:624–632. doi: 10.1006/viro.1995.1294. [DOI] [PubMed] [Google Scholar]
- 26.Wakefield L, Brownlee G G. RNA-binding properties of influenza A virus matrix protein. Nucleic Acids Res. 1989;17:8569–8580. doi: 10.1093/nar/17.21.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiner J R, Bartlett J A, Joklik W K. The sequences of the reovirus serotype 3 genome segments M1 and M3 encoding the minor protein mu2 and the major nonstructural protein muNS, respectively. Virology. 1989;169:293–304. doi: 10.1016/0042-6822(89)90154-2. [DOI] [PubMed] [Google Scholar]
- 28.Yin P, Cheang M, Coombs K M. The M1 gene is associated with differences in the temperature optimum of the transcriptase activity in reovirus core particles. J Virol. 1996;70:1223–1227. doi: 10.1128/jvi.70.2.1223-1227.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yue Z, Shatkin A J. Regulated, stable expression and nuclear presence of reovirus double-stranded RNA-binding protein ς3 in HeLa cells. J Virol. 1996;70:3497–3501. doi: 10.1128/jvi.70.6.3497-3501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou S, Brown E G. Nucleotide sequence comparison of the M1 genome segment of reovirus type 1 Lang and type 3 Dearing. Virus Res. 1992;22:159–164. doi: 10.1016/0168-1702(92)90042-8. [DOI] [PubMed] [Google Scholar]
- 31.Zou S, Brown E G. Stable expression of the reovirus mu2 protein in mouse L cells complements the growth of a reovirus ts mutant with a defect in its M1 gene. Virology. 1996;217:42–48. doi: 10.1006/viro.1996.0091. [DOI] [PubMed] [Google Scholar]
- 32.Zou S, Brown E G. Translation of the reovirus M1 gene initiates from the first AUG codon in both infected and transfected cells. Virus Res. 1996;40:75–89. doi: 10.1016/0168-1702(95)01261-3. [DOI] [PubMed] [Google Scholar]