Abstract
Telomeres are located at the ends of chromosomes and have specific sequences with a distinctive structure that safeguards genes. They possess capping structures that protect chromosome ends from fusion events and ensure chromosome stability. Telomeres shorten in length during each cycle of cell division. When this length reaches a certain threshold, it can lead to genomic instability, thus being implicated in various diseases, including cancer and neurodegenerative disorders. The possibility of telomeres serving as a biomarker for aging and age-related disease is being explored, and their significance is still under study. This is because post-mitotic cells, which are mature cells that do not undergo mitosis, do not experience telomere shortening due to age. Instead, other causes, for example, exposure to oxidative stress, can directly damage the telomeres, causing genomic instability. Nonetheless, a general agreement has been established that measuring telomere length offers valuable insights and forms a crucial foundation for analyzing gene expression and epigenetic data. Numerous approaches have been developed to accurately measure telomere lengths. In this review, we summarize various methods and their advantages and limitations for assessing telomere length.
Keywords: Telomere Lengths, Southern Blotting, Genetic Techniques, Sequence Analysis
1. Introduction
Telomeres are repetitive deoxyribonucleic acid (DNA) sequences located at the ends of chromosomes and act as essential guardians of genomic stability. Telomere sequences in human consist of ‘TTAGGG’ repeats, spanning about 10–15 kilobase pairs (kbp). Structurally, telomeres feature a single-stranded overhang of approximately 300 base pairs (bp) in length at the 3' terminus.
Through interaction with shelterin proteins, telomeres fold into T-loops, playing a pivotal role in maintaining chromosome integrity by preventing end-to-end fusion and DNA loss [1], [2], [3]. With each cell division, telomeres undergo gradual shortening due to the inability of the DNA replication machinery to fully replicate the 3' end, termed the “end replication problem” [4], [5]. Telomerase is a ribonucleoprotein enzyme responsible for maintaining the length of these telomeres [6]. It is composed of the telomerase RNA component (TERC) and telomerase reverse transcriptase (TERT). TERC recognizes the telomeric sequence and serves as the template for the synthesis of telomeric DNA, adding telomeric repeats (TTAGGG) to the single-stranded 3’ overhang, while TERT provides the necessary catalytic activity. Telomerase activity, in particular, tends to be high in germ cells and stem cells, while in most human somatic cells, it tends to be low or even non-existent, leading to a continuous telomere reduction with divisions [7], [8].
When telomeres reach critically short lengths, their protective function diminishes, triggering cellular stress. In post-mitotic cells, there is no telomere shortening, as these cells are mature and do not undergo mitosis anymore. Instead, telomeres can be directly damaged through other causes, such as oxidative stress [9]. Whether through telomere shortening or telomere damage, cells with critically short telomeres can enter a state known as "cellular senescence," where they cease to divide and lose optimal functionality. This phenomenon is recognized to be related to aging and age-related diseases [10], [11]. In addition, shortened telomeres induce chromosomal instability, which may lead to fusion and other types of damage to the unprotected chromosome ends.[12].
The measurement of telomere lengths provides profound insights into diverse biological phenomena, including cellular aging, disease susceptibility, and overall well-being [10]. The link between shorter telomeres and increased risks of age-related conditions such as cardiovascular disease, cancer, and neurodegenerative disorders suggests the role of telomere lengths as a possible biomarker in aging research [11]. Shorter telomeres have been associated with a higher risk of developing cardiovascular complications, such as atherosclerosis and coronary artery disease [13]. Similarly, in cancer, telomere length has been implicated in the development and progression of various malignancies [14]. Neurodegenerative disorders, such as Alzheimer's and Parkinson's disease, have also been linked to telomere shortening, indicating a potential connection between telomere length and cognitive decline [15].
Various techniques are employed to quantify telomere lengths, and these techniques have evolved significantly over time. They range from traditional methods such as polymerase chain reaction (PCR) [16] and Terminal Restriction Fragment (TRF) analysis [7], to more advanced techniques like Next-Generation Sequencing (NGS) [17], which have been developed since the introduction of Single Telomere Length Analysis (STELA) [18]. The data on telomere lengths acquired from these methods offer researchers insights into the dynamics of cellular aging, trends in disease progression, and genetic damage [19]. Our review paper presents a thorough overview of the different methods used to measure telomere lengths (Table 1). Understanding the characteristics of each method and selecting them appropriately can enhance the reliability and significance of research outcomes.
Table 1.
Summary of methods used to measure telomere lengths and their advantages and limitations.
| Methods | Required DNA volume/ cells |
Throughput |
Results | Experiment time | Advantages | Limitations | Reference | |
|---|---|---|---|---|---|---|---|---|
| (Appropriate number of samples) | ||||||||
| TRF | > 1 ug | 30 samples (In 1 gel) | - Average TL | > 48 hr | - Gold standard | - Requires substantial amount of DNA | [7] | |
| - Absolute TL | - Widely available commercialized kit | - Differences in results depending on X region | ||||||
| - Labor intensive | ||||||||
| Q-PCR | 20 ng | 96 - 384 samples (using 96-well or 384-well plates) | - Average TL | < 2 hr | - Requires small amount of DNA | - Need to check the single copy gene of the sample. | [16] | |
| - Relative TL | - Can be performed in high-throughput format | - Does not provide absolute TL values | ||||||
| Q-FISH | 10–15 cells | 10–15 cells | - Shortest TL | > 72 hr | - Can be applied to formalin fixed paraffin-embedded tissue sections | - Adequate number of metaphase cells required | [31] | |
| - Relative TL | - Able to meausure TL in each chromosome | - High-resolution microscopes and equipment required | ||||||
| chromosome specific TL (in metaphase) | - Able to measure shortest TL | - Time consuming and expensive | ||||||
| Flow-FISH | 1 × 10^5 cells | 1 × 10^5cells | - Average TL | > 72 hr | - Able to measure TL in each cell population | - Interpretation challenges | [37] | |
| - Relative TL | - Can be performed in high-throughput format | - Proper sample preparation is crucial | ||||||
| - Cell population specific TL | - Compatibility with other flow cytometry assays | - Time consuming and expensive | ||||||
| STELA | 10–50 ng | 30 samples (In 1 gel) | - Shortest TL | > 72 hr | - Able to meausure TL in each chromosome | - Can only analyze chromosomes with known sub telomere sequences | [18] | |
| (with known sequences) | ||||||||
| - Absolute TL | - Only small amount of DNA required | - Cannot be used in samples with damaged subtelomeric regions | ||||||
| chromosome specific TL (with known subtelomeric regions only) | - Able to measure shortest TL | - Labor intensive | ||||||
| U- STELA | 10–50 ng | 30 samples (In 1 gel) | - Shortest TL | > 72 hr | - Able to measure TL on all chromosomes | - Chromosomes cannot be distinguished | [43] | |
| (chromosomes cannot be distinguished) | - Labor intensive | |||||||
| - Absolute TL | - Only small amount of DNA required | Difficulties in the measurement of long TL | ||||||
| - Able to measure shortest TL | ||||||||
| HT-STELA | 10–50 ng | - | - Shortest TL | > 72 hr | - -Able to meausure TL in each chromosome | - Can only analyze chromosomes with known sub telomere sequences | [44] | |
| (with known sequences) | -Essential equipment required | |||||||
| - Absolute TL | - Only small amount of DNA required | |||||||
| chromosome specific TL (with known subtelomeric regions only) | - Able to measure shortest TL | |||||||
| - High Throughput | ||||||||
| TeSLA | 10–50 ng | 30 samples (In 1 gel) | - Shortest TL | > 72 hr | - Able to measure TL on all chromosomes | - Cannot be used in samples with high heterogeneity | [34] | |
| - Absolute TL | - High accuracy and precision | - Chromosome cannot be distinguished | ||||||
| - Only small amount of DNA required | - High cost and complexity | |||||||
| - Able to measure shortest TL | ||||||||
| SCT-pqPCR | 10 pg(single cell) | 96 samples | - Average TL | > 2 hr | -Measure the length of a single cell | -Accuracy in single cell separation | [45] | |
| - Relative TL | -Utilize from hard-to-get samples | - Need to check the single copy gene of the sample. | ||||||
| - single cell TL | -High Throughput | -process of prePCR | ||||||
| STAR Assay | < 1 ng | 48 samples | - shortes TL | < 3 hr | - Able to profile telomere maintenance mechanisms | - Limited historical data | [47] | |
| - Absolute TL | - Requires small amount of DNA | - High cost and complexity | ||||||
| - Can be performed in high-throughput format | ||||||||
| Optical mapping | 20 ug | - | - shortest TL | > 24 hr | - Able to meausure TL in each chromosome | -Off target issue to CRISPR | [52] | |
| - Absolute TL | (with known sequences) | - High cost and complexity | ||||||
| - Chromosome specific TL | - Able to measure shortest TL | -Requires substantial amount of DNA | ||||||
| TCA | 1–2 × 10^6 cells | 1–2 × 10^6 cells | - Shortest TL | > 72 hr | - Able to directly visualize and quantify telomeres | - Difficult to perform on large samples | [56] | |
| - Absolute TL | - High accuracy and precision | - Limited historical data | ||||||
| - Can be performed in high-throughput format | - High cost and complexity | |||||||
| Short Read | > 2 ug | - | - Average TL | > 72 hr* | - Able to perform telomere length measurement and NGS analysis simultaneously | - Difference in accuracy between WES and WGS | Computel[17] | |
| - Requires substantial amount of DNA | Tel-seq[61] | |||||||
| - High accuracy and precision | - Complex data processing | Telomerecat[62] | ||||||
| - Various analysis tools developed | - High cost and complexity | |||||||
| Long Read | Nanopore ( > 2 ug) | - | - Chromosome specific TL | > 72 hr* | - Able to meausure TL in each chromosome | - Lack of well-established analysis tools | Nanopore[66] | |
| Hifiseq ( > 20 ug) | (all chromosomes) | (all chromosomes) | - Requires substantial amount of DNA | |||||
| -Hifi: Average TL | - Able to perform telomere length measurement and NGS analysis simultaneously | - Complex data processing | Hifiseq-Telomap[68] | |||||
| - High accuracy and precision | - High cost and complexity | |||||||
2. Terminal restriction fragment (TRF)
The process begins with treating a DNA sample with specific restriction enzymes. These enzymes selectively digest DNA, cutting non-TTAGGG portions of the telomeric DNA sequences [7]. Different sets of restriction enzymes can influence the measurement outcome by varying the segments they remove; therefore, the choice of restriction enzymes is critical. It is recommended to consult established protocols [20] to select the most effective enzyme combination for the experiment, ensuring the sub-telomeric regions are excluded as much as possible for the analysis.
The digested DNA samples are then separated based on size using agarose gel electrophoresis. Longer telomere fragments migrate more slowly through the gel, while shorter fragments migrate more quickly. After gel electrophoresis, the DNA fragments are transferred from the gel to a membrane using a technique known as Southern blotting (Fig. 1A). The blotted membrane is then probed with labeled probes complementary to the telomeric DNA sequences [21]. Following hybridization, the probes that have bound to the telomeric sequences are detected using either X-ray or a chemiluminescence imager for chemiluminescently labeled probes for quantification [21]. The resulting pattern observed on the membrane indicates the distribution of telomere lengths within the sample. For quantitative analysis, software tools such as ImageQuant™, TeloTool, and WALTER Tool are utilized to accurately calculate the results [22], [23], [24].
Fig. 1.
Schematic illustrations and examples of telomere lengths measurement methodsA. TRF: DNA is digested using restriction enzymes that do not recognize telomere sequences. Gel electrophoresis is then performed, followed by blotting onto a membrane. Abbreviations: kb: kilobase pairs; MWM: Molecular weight markersB. Q-PCR: The Q-PCR method compares the telomere length (T) to a single-copy gene (S) and represents it relative to each other, with the results expressed as a T/S ratio. We illustrate Q-PCR results from both long and short telomeres. In samples with long telomeres, the peak appears first, whereas in samples with short telomeres, the peak appears later.Abbreviations: S: Single-copy gene; T: Telomere lengthC. Q-FISH: PNA probe with a fluorescent dye is used to hybridize to the telomeric DNA repeats in cells. The PNA bound to telomeres can be observed through fluorescence microscopy. The fluorescence intensity is proportional to the telomere length. By quantifying the fluorescence signals, researchers can determine the relative telomere length of individual chromosomes.Abbreviations: PNA: Peptide nucleic acidD. Flow-FISH: Fluorescently labeled PNA probes specific to telomere repeats are hybridized to the telomeric DNA in cells. These cells are then passed through a flow cytometer, where the fluorescence intensity of each cell is measured. This intensity corresponds to the telomere length, allowing researchers to assess the distribution of telomere lengths in a cell population.Abbreviations: PNA: Peptide nucleic acidE. Southern and PCR base methods.STELA: Tagging telorette at the telomere end, and then the restriction enzyme action proceeds. PCR is performed on chromosomes known in the sub-telomere sequence to perform gel loading and Southern blotting.U-STELA: Tagging telorette at the telomere end, and then the restriction enzyme action proceeds. Attach binding adapters to the cleaved portion with restriction enzymes. PCR is advanced using restriction enzyme site adapter and Telorette.TeSLA: Tagging is performed at the end of the telomere using TeSLA-T. Enzyme digestion and dephosphorylation are performed. A TeSLA adapter that can be complementary to the dephosphorylation part as a white circle is attached. PCR was performed using TeSLA-T and TeSLA adapters.Abbreviations: kb: kilobase pairs; MWM: Molecular weight markers.
TRF is considered the gold standard for measuring the absolute telomere length. The wide availability of commercial kits for experimentation allows for the measurement of telomere length in various tissues and cells [25]. However, TRF analysis provides an average measure of telomere length and does not capture the length of the shortest telomeres [22]. The method requires a substantial amount of DNA (>1 ug), which can be challenging when dealing with limited sample quantities. Additionally, especially due to the Southern blotting step, TRF is time-consuming and requires meticulous attention from the experimenter. The choice of restriction enzymes can influence the results obtained from the same DNA sample, potentially leading to variations in results [20].
3. Quantitative-polymerase chain reaction (Q-PCR)
Telomere length measurement using Q-PCR involves quantifying the abundance of telomeric DNA repeats [16]. This method utilizes a reference DNA sample with a known telomere length to establish a standard curve by measuring the telomere signal (T) and comparing it to a signal from a reference single-copy gene (S), researchers can determine the T/S ratio, which correlates with the average telomere length (Fig. 1B). relative telomere length, and the values obtained may differ between and within samples. This variability is due to factors such as DNA quality, experimental process, and individual differences. [26]. However, the absolute telomere length can be determined by including a standard sample with a known telomere length in the analysis due to advancements in the experimental method [27].
Q-PCR-based telomere length measurement is a valuable tool for exploring telomere dynamics and their potential implications, such as oxidative stress and inflammation [13]. When performing qPCR, it is crucial to recognize the differences between sample types, particularly in the selection of reference genes. While telomere sequences are highly conserved across mammalian species, the expression of single copy reference genes may vary between normal and malignant cells [28]. This variation in reference gene expression can have a significant impact on the accuracy of telomere length measurements. For instance, GAPDH, a commonly used reference gene, can be used as a single copy gene in HEK293T kidney cells, but its expression may be reduced in BAL fluid cells and biopsy tissues, making it unsuitable for these sample types [29]. Therefore, careful evaluation and verification of candidate reference genes for each sample condition are necessary to ensure reliable and comparable results across different studies. Q-PCR captures a snapshot of telomere length at a specific moment, but it can be used to monitor changes in telomere length over time through longitudinal studies. While qPCR is suitable for large-scale epidemiological studies, it is important to match and confirm the appropriate single-copy genes for comparison across different studies[30]. Although qPCR equipment is relatively common in molecular biology laboratories, the cost of reagents and consumables may be a limiting factor for large-scale research.
4. FISH (Fluorescence In Situ Hybridization) based techniques
4.1. Quantitative-FISH (Q-FISH)
Q-FISH (Quantitative Fluorescence In Situ Hybridization) is an advanced technology designed to visualize and quantitatively measure telomeres within cells. Q-FISH employs fluorescently labeled peptide nucleic acid (PNA) probes specific to the telomeric 'TTAGGG' repeat sequence, enabling the probes to bind to telomeres. This method allows for the visualization of all 46 chromosomes in metaphase, including chromosomes 1 to 22 and the sex chromosomes, thereby providing chromosomal-level telomere length information [31] (Fig. 1C).
Q-FISH is also applicable for measuring telomere lengths in interphase cells [31] and can be applied to formalin-fixed paraffin-embedded (FFPE) tissue sections[32], making it suitable for studying telomere length in specific tissues or organs. Q-FISH is particularly useful for investigating the shortest telomeres, which are crucial for genomic stability and cell viability [33], [34]. A fluorescent microscope is essential in Q-FISH for visualizing and analyzing fluorescence signals to accurately measure telomere length from Q-FISH images [35]. However, high-quality microscopes, particularly confocal microscopes, equipped with appropriate filters, are costly. Additionally, issues related to poor chromosome morphology can impact the quality of telomere length measurements [20], [31], [36].
4.2. Flow-FISH
Flow-FISH combines PNA probes with flow cytometry to measure the average telomere length and gather multi-parameter information from thousands of cells simultaneously. Fluorescently labeled PNA probes specific to telomeric repeats are introduced into the cells and bind to the telomere sequences [31]. The cells, now labeled with fluorescent probes, are passed through a flow cytometer [37]. As each cell flows past a laser, the emitted fluorescence is detected. The intensity of fluorescence corresponds to the telomere length in individual cells (Fig. 1D). Flow-FISH can also be enhanced using multicolor antibody staining in conjunction with the PNA probes. Different fluorochrome-labeled antibodies can be used to identify various cellular features in conjunction with telomere length measurement [38], [39]. Flow-FISH allows for the measurement of telomere lengths in each cell population and has been validated for clinical purposes.
However, it is important to note that Flow-FISH analysis is applicable only to cultured or lymphoid cells that exist as single entities. Additionally, the requirement for a flow cytometer, may pose a significant constraint for laboratories operating under budgetary limitations [40]. While Flow-FISH may not be suitable for high-throughput analysis of consecutive samples compared to other methods like TRF and qFISH, it allows for the measurement of telomere lengths in a large number of cells simultaneously. This capability makes Flow-FISH a powerful tool for analyzing telomere lengths in a cell population, providing a relative high-throughput approach compared to other techniques that measure telomere lengths cell by cell. However, it is important to note that Flow-FISH may face challenges in continuously measuring telomere lengths of various cell types using only PNA probes, and it may not be the most optimal technique for single-cell analysis. Despite these limitations, Flow-FISH remains a valuable method for assessing telomere lengths in a large number of cells due to its ability to process cells simultaneously.[41]. Therefore, optimizing the parameters of flow cytometry and exercising meticulous control over all steps within the protocol are crucial to obtaining accurate telomere length measurement results.
5. Single telomere length analysis (STELA) methods
5.1. STELA
STELA is a method that combines ligation-PCR and Southern blotting to accurately determine the lengths of individual chromosome telomeres. A linker is ligated to the 5' end of the telomere, enabling PCR amplification based on unique subtelomeric sequences (Figure1 E). This process is followed by Southern blotting to measure each chromosome's telomere length individually [18]. This technique excels at detecting the shortest telomeres and provides high-resolution analysis of telomere length, thereby offering a precise understanding of telomere heterogeneity within cell populations.
However, the method's applicability is limited for some chromosomes that lack the unique sequences necessary for designing the complementary primer [20]. Challenges may also arise when analyzing samples with DNA damage or regions of chromosome arm deletion, leading to ineffective primer binding. STELA's utility is primarily in cell lines and blood samples, and it is constrained by its labor-intensive nature and the requirements for Southern blotting and long-range PCR[18]. These factors can restrict its practicality for large-scale studies or in resource-constrained settings.
5.2. Universal – STELA (U-STELA)
U-STELA is a method for measuring telomere length that builds upon STELA [42]. Unlike traditional STELA, which targets chromosomes with known specific sequences, U-STELA enables the measurement of telomere lengths across all chromosomes, thus earning the name ‘Universal’. U-STELA incorporates two ligation steps following treatment with restriction enzymes. First, DNA is digested by restriction enzymes, leaving a sticky overhang. Then, a complementary double-stranded oligonucleotide is annealed and ligated to this overhang, tagging the DNA fragments. Telomeric fragments are tagged with a distal primer sequence by using a linker called a telorette on the 3′-overhang of telomeres, enabling PCR amplification. This process allows the tagged DNA fragments to undergo PCR amplification, generating sufficient material for analysis (Fig. 1E).
Like STELA, U-STELA is useful for identifying critically short telomeres in samples of low concentrations. However, one significant limitation is its difficulty in measuring long telomeres (10 kbp or higher) [34]. Furthermore, the method is sensitive to the amount of template DNA added [42], where an excessive amount of template DNA can cause a smear on the gel due to technical artifacts caused by incomplete amplicons acting as primers.
5.3. High throughput STELA (HT-STELA)
High Throughput (HT) STELA employs PCR amplification to amplify telomeric regions for subsequent high-throughput analysis using equipment like a bioanalyzer [43]. This method streamlines telomere length analysis by automating the workflow, significantly enhancing efficiency, and reducing manual labor by eliminating traditional gel electrophoresis and hybridization steps. Specific primers designed to target telomeric regions ensure amplification specificity [18]. This reduces nonspecific amplification of non-telomeric sequences during PCR, and thus improving the accuracy and reliability of this method.
Unlike traditional STELA, which utilizes a complementary TTAGGG probe, HT-STELA's TL measurement is based solely on analyzing the DNA fragment size, as PCR is performed using specific primers that complement known Telorette sequences. This approach enables rapid DNA fragment length measurement in a high-throughput manner, making this technique suitable for large-scale studies. However, this specificity limits TL measurement to only the targeted chromosomes. Furthermore, specialized equipment such as the bioanalyzer contributes to an increase in the overall cost and complexity of the method.
6. Telomere shortest length assay (TeSLA)
TeSLA is designed to measure the length of the shortest telomere in a cell, which plays a significant role in regulating cell cycle arrest and in age-related diseases [7], [11], [33], [34]. This is an advancement over previous methods that focused on measuring average telomere length.
TeSLA is a considered a refined form of telomere length measurement which has evolved from STELA. TeSLA can measure telomeres of all chromosomes simultaneously, while STELA typically focuses on analyzing telomeres of specific chromosomes. Another distinction between TeSLA and STELA is the method employed to generate an artificial sequence for PCR in the sub-telomere region. TeSLA involves dephosphorylating the 5' end using a phosphatase after subjecting the DNA to restriction enzyme treatment, thereby enabling the attachment of TeSLA's adapter. The PCR product, through the artificial sequence thus produced, can measure the length of telomeres through the process of Southern blotting. This method enables specific and accurate measurement of the shortest telomere without depending on chromosome-specific unique sequences (Fig. 1E).
However, one of the drawbacks of TeSLA is that although it provides results for the shortest telomere length across all chromosomes, it lacks the capability to accurately calculate the shortest telomere on a specific chromosome [44]. Furthermore, there may be limitations in measuring telomere lengths in cases where telomeres display pronounced heterogeneity due to the alternative lengthening of telomeres (ALT), which is one of the mechanisms some cancer cells adopt for telomere maintenance. The telomere length measurement capability of TeSLA is capped at a maximum length of approximately 18 kbp in cases of high heterogeneity. This limitation is especially relevant in cancer research, where ALT is frequently observed [34]. Additionally, the TeSLA method involves Southern blotting and long-range PCR techniques, both of which are time-consuming and labor-intensive processes [20].
7. Single-cell telomere length measurement pqPCR (SCT-pqPCR)
The measurement of telomere length in individual cells has traditionally been accomplished using FISH. However, this approach is limited by a slow processing rate as there is a limited number of samples that can be processed at a time. Traditional qPCR analysis also poses challenges in measuring telomere lengths due to the limited amount of DNA found in a single cell, typically around 10 picograms (pg).
To address these challenges, novel techniques named single-cell telomere quantitative PCR (SCT-pqPCR) have been developed [45]. In SCT-pqPCR, DNA is extracted from each isolated single cell using standard procedures. Given the minimal DNA content in a single cell (typically around 10 pg), pre-PCR amplification steps are performed to increase the DNA available for analysis. Subsequently, qPCR is conducted to measure telomere length. This involves the design of specific primer sets for amplifying telomere regions and reference genes (such as Alu or B1) simultaneously. The ratio of telomere signal to reference gene signal is then utilized to calculate the relative telomere length. SCT-pqPCR provides a direct, high-quality approach for analyzing telomere length in individual cells. It establishes a direct correlation between average T/R values obtained through SCT-pqPCR and the absolute telomere length. Moreover, SCT-pqPCR enables the measurement of telomere length in individual cells, regardless of their ability to divide. It also allows for the assessment of telomere length variation within populations of cells [46].
A limitation of SCT-pqPCR is the requirement for specialized equipment and expertise to perform the pre-PCR amplification steps and subsequent qPCR analysis. This may render the method inaccessible to researchers who lack the necessary resources or training. Therefore, careful optimization and validation of the pre-PCR protocol are crucial to ensure accurate results. Another potential limitation is the reliance on multicopy genes, such as Alu and B1, for normalization of telomere measurements. Although these genes are commonly used as reference loci, they may not always accurately represent the true DNA content or amplification efficiency in all cell types or experimental conditions. Furthermore, like any PCR-based method, SCT-pqPCR is vulnerable to potential contamination issues, which could result in false-positive results if not adequately controlled during the experimental procedure.
8. Single telomere absolute-length rapid (STAR) assay
The STAR assay quantifies the abundance of a telomere sequence by employing digital PCR [47]. Digital PCR involves dividing the DNA sample into thousands of individual reactions, with each potentially containing a single DNA molecule. Subsequently, it amplifies and detects the target sequence in each reaction, leading to quantification of the target sequence in samples, even at very low concentrations [48]. During digital PCR, different amounts of synthetic telomere repeats are used. This generates cycle threshold (Ct) values, which are plotted against the total repeat length, forming a precise standard curve. This curve indicates high reproducibility and accuracy in telomere length measurements.
The broad dynamic range of the STAR assay, from 0.2 kb to 320 kb, is demonstrated by its ability to accurately measure telomere lengths across this extensive spectrum. In the no-template controls, negligible amplification is observed up to a Ct value of 27, corresponding to ∼0.2 kb on the standard curve. However, the broad dynamic range is showcased by the assay's capability to reliably quantify individual telomere lengths all the way up to 320 kb, with higher Ct values corresponding to longer telomere fragments. Its ability to accurately profile telomere maintenance mechanisms, including ALT, marks a significant advancement in cancer research. The STAR assay provides insights into cancer cell behavior and deepens our understanding of telomere dynamics during cancer progression [47].
9. Optical mapping in nano-channel array
Optical mapping is a technology that directly visualizes and analyzes the structure and sequence of long biopolymer strands, such as DNA [49]. This technique involves aligning DNA molecules in nanometer-sized channels, labeling them with fluorescent dyes, and observing them under high-resolution microscopy. The nano-channels are microfabricated, typically ranging from tens to hundreds of nanometers in both width and depth, and are etched or manufactured onto a solid substrate, such as silicon or glass. These nano-scale channels serve as platforms for elongating and fixing individual DNA molecules [50]. To mark DNA molecules for visualization and detection within nanochannels, nicks are introduced at specific locations along the DNA strands using nickases. Labeling is achieved by incorporating modified nucleotides or fluorescent markers at these nicked sites [51]. Selectively introducing nicks at predetermined positions allows for the creation of a distinct barcode or signature, facilitating DNA mapping and sequencing.
When optical mapping in a nano-channel array is used for the measurement of telomere lengths, the Cas9n enzyme, a variant of the Cas9 nuclease is used. The Cas9n enzyme is guided by a specific RNA molecule (sgRNA) to target the repetitive sequences in telomeres. It cleaves one strand of the telomeric DNA at the target location, leaving the complementary strand intact. A specifically designed fluorescently labeled probe for the sequence adjacent to the cleavage site (UUAGGGUUAGGGUUAGGGUU) is then introduced [51]. This probe binds to the cleaved DNA, marking the telomeric region with fluorescence. After marking, the telomeric DNA is elongated and fixed within the nanochannels, enabling high-resolution observation and examination. Techniques such as fluorescence microscopy or other imaging methods are used to visualize the labeled telomeres and accurately measure their length [52], [53].
Operating nano-channel systems is complex. There is potential for technical errors as samples are handled at a nano-scale, such as incomplete elongation, twisting, and bending, especially during DNA alignment in nano-channels [54]. Furthermore, previous studies have reported that DNA cleavage patterns may occur unevenly due to differences in the targeting efficiency or specificity of Cas9n [55]. These challenges should be considered in research involving nano-channel technology for measuring telomere lengths.
10. Telomere length combing assay (TCA)
TCA is an advanced technique designed to directly visualize and quantify individual telomere molecules [56]. DNA fibers are stretched onto glass plates, creating an extended conformation that resembles molecular combing. PNA probes, labeled with fluorescent materials, are used for the visualization of telomeres. Fluorescence microscopy quantifies and measures the absolute telomere lengths at a rate of approximately 2 kilobases per micrometer (2kbp/μm). Through this process, TCA can provide a distribution profile of telomere lengths for the examined cell population. It is also expected that TCA may further be improved by using chromosome-specific subtelomere probes enabling measurement of chromosome specific telomere lengths [56]. This method's ability to directly visualize telomeres offers insights into the heterogeneity of telomere lengths, enriching our understanding of the mechanisms behind aging and disease processes [57], [58].
However, the specialized equipment and setups required for operating with long DNA strands on glass plates are not available in all labs. It also incurs significant costs for equipment and reagents, which can increase research expenses. Moreover, TCA involves lengthy sample preparation and may face potential limitations in measuring telomere lengths in multiple samples simultaneously.
11. Sequencing based estimation techniques
11.1. Short-read
The advent of Next-Generation Sequencing (NGS) has significantly advanced DNA sequencing technology, revolutionizing genetic and medical research. Compared to traditional Sanger sequencing methods, NGS can swiftly sequence complete genomes, including both coding and non-coding regions such as telomeres [59], [60]. Computational tools like Telocat, Computel, and Tel-seq estimate telomere length from genomic data, eliminating the need for laboratory experiments [17], [61], [62].
Specifically, TelSeq identifies telomeric reads in Binary Alignment Map (BAM) format by searching for sequences with seven or more TTAGGG repeats. It normalizes telomere counts based on the GC composition of total reads (48–52%), ensuring accurate length estimation by adjusting for GC bias. TelSeq calculates the mean telomere length, assuming a haploid cell contains 23 chromosomes [61]. This tool is compatible with both whole-genome sequencing (WGS) and whole-exome sequencing (WES) data. However, WGS is preferred for more accurate telomere length calculations due to its comprehensive genomic coverage. Although WES primarily targets protein-coding regions, it may still capture non-coding DNA regions, including telomeres. Therefore, while WES may not offer the coverage or precision of WGS, it can still provide valuable data on specific non-coding regions [63].
11.2. Long-read
Long-read sequencing provides the opportunity to generate longer sequences, ranging from hundreds to tens of thousands of base pairs, which allow for the examination of broader regions of the genome in a single sequence. The third generation of NGS technology can theoretically measure telomere length on a chromosome-by-chromosome basis and read the entire human genome [64]. However, its practical application faces several technical challenges. The vast and complex nature of the human genome demands substantial time and resources for comprehensive reading and processing. Additionally, the intricacy of data processing and analysis, coupled with the challenges posed by repetitive sequences in telomeres, complicates accurate sequencing and telomere length measurement [65].
Specialized algorithms are essential for managing these sequences, making the creation of standardized methodologies for precise telomere length measurement crucial. Recent advancements have enabled the measurement of chromosome-specific telomere lengths, uncovering telomere dynamics across different chromosomes. In nanopore sequencing, an adapter-based method has been developed to measure telomere length. This process involves adding poly(A) sequences to the telomeres' 3' ends using terminal transferase, followed by annealing an oligo(dT) primer with a unique adapter sequence, known as TeloTag, to the poly(A) sequence. Analysis of the sequencing data processed in this manner allows for the measurement of telomere length [66].
Additionally, HiFi (High-Fidelity) sequencing technology allows for telomere length measurement in human cells. The PacBio HiFi platform uses circular consensus sequencing (CCS) to achieve high accuracy (>99.5%) for single-molecule real-time (SMRT) sequencing of long reads that have an average length of 10–25 kb [67]. Genomic DNA fragments containing telomeres are enriched using telobaits, designed to bind to telomeric single-stranded 3' overhangs. These telobaits, featuring a single telomere repeat, a unique EcoRI site, and 3' biotin labeling, allow for purification through magnetic beads, enabling precise telomere length measurement on the PacBio HiFi platform [68]. Tools like Telogator are also being used for measuring chromosome specific telomere lengths allowing for empirical investigation into how telomere length is associated with various biological processes such as cellular aging and cancer development. Tools like Telogator have emerged as valuable assets for reporting chromosome-specific telomere lengths, allowing for accurate measurement and enhancing our comprehension of telomere biology [69]. New tools for analyzing long-read data are actively being developed and more advanced analysis pipelines are expected to enhance the value of long-read sequencing data. Compared to the aforementioned short-read approaches, both short-read and long-read telomere length measurement require a large amount of DNA, but long-read sequencing generally requires an even larger quantity of DNA(>2–20 μg).
12. Summary and outlook
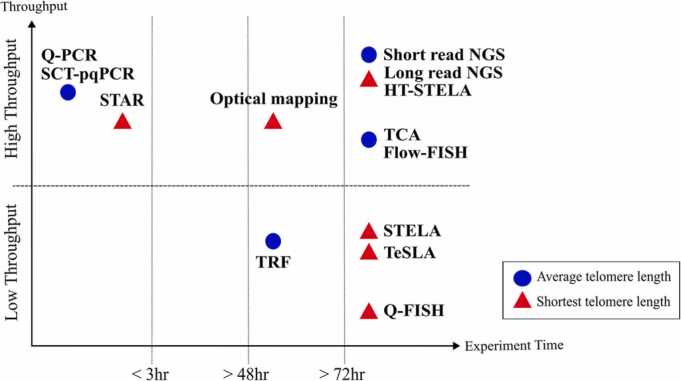
This review has shown the development of telomere length measurement techniques, from TRF, a traditional telomere length measure, to the development of telomere length measurement using genetic analysis (Fig. 2). Each approach has its own merits and suitability for different objectives, such as assessing average telomere length or the shortest telomere length. The choice of an appropriate method depends on the specific research goals. For instance, TRF, qPCR, Flow FISH, and TCA are suitable for measuring mean telomere length, whereas techniques like qFISH, STELA, and TeSLA are better suited for quantifying the shortest telomeres. The field of NGS is also divided into short-read and long-read, which are suitable for different telomere length measurement methods (Fig. 3). While STELA can indeed be used to measure chromosome-specific telomere lengths, further development is needed to achieve the precise differentiation of chromosome-specific telomere lengths, similar to the precision achieved in metaphase cell analysis of qFISH experiments.
Fig. 2.
Timeline of telomere lengths measurement methods.
Fig. 3.
Classification of telomere length measurement methods based on telomere lengths, experiment time, and low/high throughput.Shapes and colors indicate the types of telomere length: Blue circles represent methods that measure average telomere length. Red triangles represent methods that can measure the shortest telomere.
Advanced measurement techniques enable researchers not only to assess average telomere length but also to examine the distribution and potential interactions between telomeres on different chromosomes. As telomere length decreases, interactions among chromosomes within the nucleus may change, affecting chromatin organization and gene expression patterns[70]. Telomeres play a crucial role in maintaining chromosome stability and integrity throughout the cell cycle, which involves dynamic changes in chromatin structure to facilitate DNA replication, repair, and chromosome segregation[71]. This comprehensive approach provides deeper insights into telomere biology, elucidating its significance for genomic stability and cellular function. Precise measurement of telomere length at the chromosome level offers valuable information for investigating gene expression regulation and epigenetic modifications. Epigenetic regulation of telomere maintenance has been shown to play a role in cell cycle control, and future developments in measuring chromosome-specific telomere lengths may shed light on these interactions.
These advancements underscore the critical importance of accurate telomere length measurements in understanding the complexities of cellular biology and disease pathogenesis. They highlight the significance of selecting the appropriate method for measuring telomere length based on research objectives, thereby emphasizing the value of technological advancements in advancing our understanding of telomere biology.
Ethics statement
The IRB of Seoul National University reviewed the study protocol and granted an exemption from full review (IRB no. 2310–046-1473) as the current study posed minimal risk to the subject and did not involve any identifiable personal data. To maintain the confidentiality of personal information, we anonymously processed all data collected from the individuals.
Funding
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI21C0239).
CRediT authorship contribution statement
Chul-Kee Park: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. Hyeon Jong Yu: Visualization, Writing – original draft, Writing – review & editing. Yoon Hwan Byun: Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors have no potential conflicts of interest to disclose. The funder played no role in the study’s design, data collection, analysis, interpretation of the data, writing of the report, or the decision to submit the report for publication.
Data availability
The data supporting the findings of this investigation are available upon reasonable request from the corresponding author.
References
- 1.Greider C.W. Telomeres. Curr Opin Cell Biol. 1991;3(3):444–451. doi: 10.1016/0955-0674(91)90072-7. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E.H. Structure and function of telomeres. Nature. 1991;350(6319):569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 3.Srinivas N., Rachakonda S., Kumar R. Telomeres and telomere length: a general overview. Cancers. 2020;12(3) doi: 10.3390/cancers12030558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sfeir A.J., et al. Telomere-end processing the terminal nucleotides of human chromosomes. Mol Cell. 2005;18(1):131–138. doi: 10.1016/j.molcel.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 5.Olovnikov A.M. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41(1):181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 6.Jafri M.A., et al. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016;8(1):69. doi: 10.1186/s13073-016-0324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harley C.B., Futcher A.B., Greider C.W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345(6274):458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 8.Cong Y.S., Wright W.E., Shay J.W. Human telomerase and its regulation. Microbiol Mol Biol Rev. 2002;66(3):407–425. doi: 10.1128/MMBR.66.3.407-425.2002. (table of contents) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem Sci. 2002;27(7):339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 10.Trybek T., et al. Telomeres and telomerase in oncogenesis. Oncol Lett. 2020;20(2):1015–1027. doi: 10.3892/ol.2020.11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruber H.J., et al. Telomeres and age-related diseases. Biomedicines. 2021;9(10) doi: 10.3390/biomedicines9101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Otin C., et al. Hallmarks of aging: an expanding universe. Cell. 2023;186(2):243–278. doi: 10.1016/j.cell.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Tian R., et al. Association between oxidative stress and peripheral leukocyte telomere length in patients with premature coronary artery disease. Med Sci Monit. 2017;23:4382–4390. doi: 10.12659/MSM.902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C., et al. The association between telomere length and cancer prognosis: evidence from a meta-analysis. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0133174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forero D.A., et al. Meta-analysis of telomere length in Alzheimer's disease. J Gerontol A Biol Sci Med Sci. 2016;71(8):1069–1073. doi: 10.1093/gerona/glw053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cawthon R.M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30(10) doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nersisyan L., Arakelyan A. Computel: computation of mean telomere length from whole-genome next-generation sequencing data. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0125201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baird D.M., et al. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet. 2003;33(2):203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- 19.Blackburn E.H., Epel E.S., Lin J. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science. 2015;350(6265):1193–1198. doi: 10.1126/science.aab3389. [DOI] [PubMed] [Google Scholar]
- 20.Lai T.P., Wright W.E., Shay J.W. Comparison of telomere length measurement methods. Philos Trans R Soc Lond B Biol Sci. 2018;373(1741) doi: 10.1098/rstb.2016.0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura M., et al. Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat Protoc. 2010;5(9):1596–1607. doi: 10.1038/nprot.2010.124. [DOI] [PubMed] [Google Scholar]
- 22.Gohring J., et al. TeloTool: a new tool for telomere length measurement from terminal restriction fragment analysis with improved probe intensity correction. Nucleic Acids Res. 2014;42(3) doi: 10.1093/nar/gkt1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lycka M., et al. WALTER: an easy way to online evaluate telomere lengths from terminal restriction fragment analysis. BMC Bioinforma. 2021;22(1):145. doi: 10.1186/s12859-021-04064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mender I., Shay J.W. Telomere restriction fragment (TRF) analysis. Bio Protoc. 2015;5(22) doi: 10.21769/bioprotoc.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S., et al. The telomere maintenance mechanism spectrum and its dynamics in gliomas. Genome Med. 2022;14(1):88. doi: 10.1186/s13073-022-01095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindrose A.R., et al. Method comparison studies of telomere length measurement using qPCR approaches: a critical appraisal of the literature. PLoS One. 2021;16(1) doi: 10.1371/journal.pone.0245582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Callaghan N.J., Fenech M. A quantitative PCR method for measuring absolute telomere length. Biol Proced Online. 2011;13:3. doi: 10.1186/1480-9222-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krasnov G.S., et al. Pan-cancer analysis of TCGA data revealed promising reference genes for qPCR normalization. Front Genet. 2019;10:97. doi: 10.3389/fgene.2019.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glare E.M., et al. beta-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax. 2002;57(9):765–770. doi: 10.1136/thorax.57.9.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J., et al. Telomere length measurement by qPCR - summary of critical factors and recommendations for assay design. Psychoneuroendocrinology. 2019;99:271–278. doi: 10.1016/j.psyneuen.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poon S.S.S., Lansdorp P.M. Quantitative fluorescence in situ hybridization (Q-FISH) Curr Protoc Cell Biol. 2001 doi: 10.1002/0471143030.cb1804s12. Chapter 18: p. 18 4 1-18 4 21. [DOI] [PubMed] [Google Scholar]
- 32.Aida J., et al. Telomere length variations in 6 mucosal cell types of gastric tissue observed using a novel quantitative fluorescence in situ hybridization method. Hum Pathol. 2007;38(8):1192–1200. doi: 10.1016/j.humpath.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 33.Hemann M.T., et al. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107(1):67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 34.Lai T.P., et al. A method for measuring the distribution of the shortest telomeres in cells and tissues. Nat Commun. 2017;8(1):1356. doi: 10.1038/s41467-017-01291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canela A., et al. High-throughput telomere length quantification by FISH and its application to human population studies. Proc Natl Acad Sci USA. 2007;104(13):5300–5305. doi: 10.1073/pnas.0609367104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slijepcevic P. Telomere length measurement by Q-FISH. Methods Cell Sci. 2001;23(1-3):17–22. [PubMed] [Google Scholar]
- 37.Hultdin M., et al. Telomere analysis by fluorescence in situ hybridization and flow cytometry. Nucleic Acids Res. 1998;26(16):3651–3656. doi: 10.1093/nar/26.16.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Martin I., et al. Telomere length heterogeneity in placenta revealed with high-resolution telomere length analysis. Placenta. 2017;59:61–68. doi: 10.1016/j.placenta.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rufer N., et al. Telomere length dynamics in human lymphocyte subpopulations measured by flow cytometry. Nat Biotechnol. 1998;16(8):743–747. doi: 10.1038/nbt0898-743. [DOI] [PubMed] [Google Scholar]
- 40.Gutierrez-Rodrigues F., et al. Direct comparison of flow-FISH and qPCR as diagnostic tests for telomere length measurement in humans. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0113747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baerlocher G.M., et al. Flow cytometry and FISH to measure the average length of telomeres (flow FISH) Nat Protoc. 2006;1(5):2365–2376. doi: 10.1038/nprot.2006.263. [DOI] [PubMed] [Google Scholar]
- 42.Bendix L., et al. The load of short telomeres, estimated by a new method, Universal STELA, correlates with number of senescent cells. Aging Cell. 2010;9(3):383–397. doi: 10.1111/j.1474-9726.2010.00568.x. [DOI] [PubMed] [Google Scholar]
- 43.Norris K., et al. High-throughput STELA provides a rapid test for the diagnosis of telomere biology disorders. Hum Genet. 2021;140(6):945–955. doi: 10.1007/s00439-021-02257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shay J.W., Wright W.E. Telomeres and telomerase: three decades of progress. Nat Rev Genet. 2019;20(5):299–309. doi: 10.1038/s41576-019-0099-1. [DOI] [PubMed] [Google Scholar]
- 45.Wang F., et al. Robust measurement of telomere length in single cells. Proc Natl Acad Sci USA. 2013;110(21) doi: 10.1073/pnas.1306639110. E1906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antunes D.M., et al. A single-cell assay for telomere DNA content shows increasing telomere length heterogeneity, as well as increasing mean telomere length in human spermatozoa with advancing age. J Assist Reprod Genet. 2015;32(11):1685–1690. doi: 10.1007/s10815-015-0574-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo Y., et al. Massively parallel single-molecule telomere length measurement with digital real-time PCR. Sci Adv. 2020;6(34) doi: 10.1126/sciadv.abb7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanagal-Shamanna R. Digital PCR: principles and applications. Methods Mol Biol. 2016;1392:43–50. doi: 10.1007/978-1-4939-3360-0_5. [DOI] [PubMed] [Google Scholar]
- 49.Yuan Y., Chung C.Y., Chan T.F. Advances in optical mapping for genomic research. Comput Struct Biotechnol J. 2020;18:2051–2062. doi: 10.1016/j.csbj.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang D., Zhang Y., Liu D. DNA nanochannels. F1000Res. 2017;6:503. doi: 10.12688/f1000research.10464.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeffet J., et al. Single-molecule optical genome mapping in nanochannels: multidisciplinarity at the nanoscale. Essays Biochem. 2021;65(1):51–66. doi: 10.1042/EBC20200021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCaffrey J., et al. High-throughput single-molecule telomere characterization. Genome Res. 2017;27(11):1904–1915. doi: 10.1101/gr.222422.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uppuluri L., et al. Single-molecule telomere length characterization by optical mapping in nano-channel array: Perspective and review on telomere length measurement. Environ Toxicol Pharm. 2021;82 doi: 10.1016/j.etap.2020.103562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dai L., Renner C.B., Doyle P.S. The polymer physics of single DNA confined in nanochannels. Adv Colloid Interface Sci. 2016;232:80–100. doi: 10.1016/j.cis.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X.H., et al. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids. 2015;4(11)) doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kahl V.F.S., et al. Telomere length measurement by molecular combing. Front Cell Dev Biol. 2020;8:493. doi: 10.3389/fcell.2020.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu R., et al. A DNA-fiber protocol for single molecule analysis of telomere (SMAT) length and extension events in cancer cells. STAR Protoc. 2022;3(1) doi: 10.1016/j.xpro.2022.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moore G., Jimenez Sainz J., Jensen R.B. DNA fiber combing protocol using in-house reagents and coverslips to analyze replication fork dynamics in mammalian cells. STAR Protoc. 2022;3(2) doi: 10.1016/j.xpro.2022.101371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shendure J., Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 60.Castle J.C., et al. DNA copy number, including telomeres and mitochondria, assayed using next-generation sequencing. BMC Genom. 2010;11:244. doi: 10.1186/1471-2164-11-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding Z., et al. Estimating telomere length from whole genome sequence data. Nucleic Acids Res. 2014;42(9) doi: 10.1093/nar/gku181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farmery J.H.R., et al. Telomerecat: A ploidy-agnostic method for estimating telomere length from whole genome sequencing data. Sci Rep. 2018;8(1):1300. doi: 10.1038/s41598-017-14403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu K., Ghandi M., Huang F.W. Integrated evaluation of telomerase activation and telomere maintenance across cancer cell lines. Elife. 2021;10 doi: 10.7554/eLife.66198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nurk S., et al. The complete sequence of a human genome. Science. 2022;376(6588):44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan K.T., et al. Identifying and correcting repeat-calling errors in nanopore sequencing of telomeres. Genome Biol. 2022;23(1):180. doi: 10.1186/s13059-022-02751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sholes S.L., et al. Chromosome-specific telomere lengths and the minimal functional telomere revealed by nanopore sequencing. Genome Res. 2022;32(4):616–628. doi: 10.1101/gr.275868.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hon T., et al. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci Data. 2020;7(1):399. doi: 10.1038/s41597-020-00743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tham C.Y., et al. High-throughput telomere length measurement at nucleotide resolution using the PacBio high fidelity sequencing platform. Nat Commun. 2023;14(1):281. doi: 10.1038/s41467-023-35823-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stephens Z., et al. Telogator: a method for reporting chromosome-specific telomere lengths from long reads. Bioinformatics. 2022;38(7):1788–1793. doi: 10.1093/bioinformatics/btac005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robin J.D., et al. Telomere position effect: regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 2014;28(22):2464–2476. doi: 10.1101/gad.251041.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galati A., Micheli E., Cacchione S. Chromatin structure in telomere dynamics. Front Oncol. 2013;3:46. doi: 10.3389/fonc.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of this investigation are available upon reasonable request from the corresponding author.