Abstract
Glioblastoma (GBM) is one of the most malignant tumors of the central nervous system. The pattern of immune checkpoint expression in GBM remains largely unknown. We performed snRNA-Seq and spatial transcriptomic (ST) analyses on untreated GBM samples. 8 major cell types were found in both tumor and adjacent normal tissues, with variations in infiltration grade. Neoplastic cells_6 was identified in malignant cells with high expression of invasion and proliferator-related genes, and analyzed its interactions with microglia, MDM cells and T cells. Significant alterations in ligand-receptor interactions were observed, particularly between Neoplastic cells_6 and microglia, and found prominent expression of VISTA/VSIG3, suggesting a potential mechanism for evading immune system attacks. High expression of TIM-3, VISTA, PSGL-1 and VSIG-3 with similar expression patterns in GBM, may have potential as therapeutic targets. The prognostic value of VISTA expression was cross-validated in 180 glioma patients, and it was observed that patients with high VISTA expression had a poorer prognosis. In addition, multimodal cross analysis integrated SnRNA-seq and ST, revealing complex intracellular communication and mapping the GBM tumor microenvironment. This study reveals novel molecular characteristics of GBM, co-expression of immune checkpoints, and potential therapeutic targets, contributing to improving the understanding and treatment of GBM.
Keywords: Glioblastoma, Single-nucleus RNA sequencing, Spatial transcriptomics, Immune checkpoints, Cell-cell communication
Graphical Abstract
1. Introduction
Glioblastoma (GBM) is a typical heterogeneous tumor and the most fatal primary malignant tumor of the central nervous system (CNS)[1]. The conventional treatment for GBM is surgical resection followed by radiotherapy, combined with temozolomide chemotherapy; however the treatment effect is still not ideal[2]. Intratumoral heterogeneity and redundancy of signaling pathways may explain why traditional and targeted therapies cannot achieve long-term remission. Therefore, new therapies are urgently needed to improve the prognosis of glioma patients[3].
At present, cancer immunotherapy is a great breakthrough, with immune checkpoint inhibitors leading the way. It has been shown to be effective in melanoma and non-small cell lung cancer, providing a new approach for the treatment of malignant glioma[4], [5], [6]. Different immune checkpoints have different mechanisms in cancer immunotherapy. Unlike the cytotoxic effects of chemotherapy, immune checkpoints regulate the immune system and induce cancer cell death[7]. However, depleted T cells in the tumor microenvironment (TME) usually express multiple immune checkpoints, and the blockade of a single checkpoint is not sufficient to activate a suppressed immune response[8]. The low mutational burden of GBM suggests that fewer neoantigens trigger an antitumor immune response. Therefore, exploring alternative immune checkpoints may provide new strategies for improving the clinical efficacy of checkpoint blockades in GBM.
With the development of single-nucleus RNA sequencing (snRNA-seq), our understanding of diseases at the genetic level has increased to the single cell levels, which provides an important tool in the study of tumor heterogeneity[9], [10]. Recently, snRNA-seq has revealed the subtype heterogeneity of GBM[11], [12], differences between blood-derived tumor-associated macrophages (TAMs) and microglial TAMs[13], cellular state of malignant cells, and their plasticity and gene-driven regulation in GBM[14]. Nevertheless, snRNA-seq datasets lack spatial location information and cannot reveal cell states. Therefore, we introduced spatial transcriptomics (ST), which combines histological imaging and snRNA-Seq by retaining the positional information for each transcript through spatially immobilized and barcoded cDNA synthesis primers[15], [16]. It can provide quantitative gene expression data and visualization of mRNA distribution in tissue sections, making new types of bioinformatics analysis possible, and has crucial value in clinical research and diagnosis. ST technology has been applied in studies on melanoma[17], prostate cancer[18], pancreatic ductal adenocarcinomas[18], squamous cell carcinoma[19], breast cancer[20], [21] and liver cancer[22], [23].
Here, we revealed the expression of immune checkpoints in GBM and intratumoral heterogeneity via snRNA-seq and ST. We profiled the transcriptome of 32,328 single cells from GBM and paracancerous samples, and produced an atlas of the spatial tumor microenvironment inside GBM tissues. By integrating snRNA-seq and ST via multimodal intersection analysis (MIA), we also generated a potential cellular interaction network of cell populations in the TME of GBM. Our study will improve our understanding of the mechanism of GBM progression, and may be potentially valuable in identifying novel targets for GBM.
2. Results
2.1. Single cell expression and cell typing in GBM and control samples
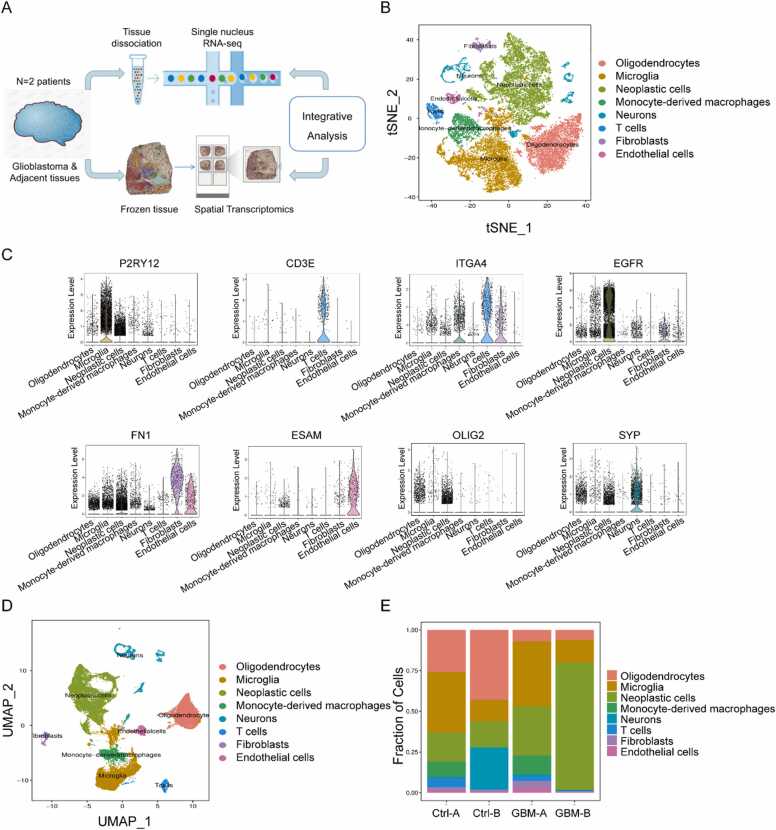
We performed parallel snRNA-seq and ST using the 10X Genomics platform on two untreated GBM patients (GBM-A and GBM-B) (Fig. 1A). Owing to the small number of adjacent tissues in patient A, only patient B was subjected to spatial transcriptome analysis. We performed a second filtering of the abnormal cells to remove low-quality cells. The snRNA-seq data consisted of cells with less than 20,000 unique molecular identifiers (UMIs), approximately 300–7000 uniquely expressed genes, and the proportion of mitochondrial gene expression per cell (less than 5%). The data volume statistics of the cells in each sample after filtration were obtained (Supplementary Figure 1). The single-cell transcriptomic profiles of 18,449 cells from patient A and 13,879 cells from patient B were selected for analysis (Supplementary Table 1).
Fig. 1.
Single cell expression and cell typing in GBM and control samples. (A) Schematic of the experimental design and analysis. ‘GBM’ and ‘Ctrl’ represent glioblastoma tissue and adjacent tissue, respectively. (B) Profiles of the t-SNE plots of 32,328 cells extracted from GBM-A (10,695 cells), Ctrl-A (7754 cells), GBM-B (6115 cells) and Ctrl-B (7764 cells). (C) Expression levels of selected known marker genes across 32,328 unsorted cells illustrated from both normal and tumor tissue in CRC patients. (D) Profiles of the UMAP plots of 32,328 cells extracted from GBM-A (10,695 cells), Ctrl-A (7754 cells), GBM-B (6115 cells) and Ctrl-B (7764 cells). (E) The proportions of the 8 main cell types in different donors.
The t-distributed stochastic neighbor embedding (t-SNE) nonlinear clustering method and unified manifold approximation and projection (UMAP) method were used to further visualize the classification results of a single cell subpopulation. Finally, we identified eight major cell clusters (Fig. 1B&1D, Supplementary Figure 1E–1F). Cluster-specific marker genes were generated by performing differential gene expression analysis to define the identity of each cell cluster, such as P2RY12 for microglia, ITGA4 for monocyte-derived macrophage (MDM), EGFR for neoplastic cells, OLIG2 for oligodendrocytes, ESAM for endothelial cells, CD3E for T cells, FN1 for fibroblasts, SYP for neurons and GFAP for astrocytes[24] (Fig. 1C and Supplementary Figure 1 G). Although all eight major cell types were presented in both tumor and adjacent normal tissues from the two patients (Fig. 1E), the grade of infiltration for each of these major cell types was different, possibly reflecting differences in the stage of GBM progression.
2.2. Distinct functional composition of malignant cells in GBM
Malignant cells extracted from GBM-A, GBM-B, Ctrl-A and Ctrl-B were further divided into sixteen subgroups by UMAP analysis and t-SNE analysis (Fig. 2A-2B). By comparing the gene expression levels, each subgroup expressed a specific set of genes that could be used to distinguish these subgroups (Supplementary Figure 3). Different subgroups of tumor cells accounted for different proportions in the sample. The proportion of the sixth subgroup cells in GBM samples was significantly higher than that in Ctrl samples, and they had the most differentially expressed genes (DEGs) by comparing the gene expression of each other subgroup (Fig. 2C-2D, Table 3). In addition, Neoplastic cells_6 widely expressed the highly invasive gene PTTG1, the cell cycle-related genes CCNB1 and CENPF, and the proliferation-related marker genes MKi67 and TOP2A (Fig. 2E). Infinite proliferation, migration, and invasion are unique malignant biological behaviors of tumor cells. PTTG1 was highly expressed in all glioma cell lines. Down-regulation of PTTG1 expression can significantly reduce cell proliferation, migration, and invasion; increase cell apoptosis; and reduce the deterioration degree of glioma, which is expected to be a new strategy for the clinical treatment of glioma[25], [26]. Next, we conducted cell-to-cell interaction using CellphoneDB to evaluate the ligand-receptor interactions of Neoplastic cells_6, microglia, MDM cells and T cells in the GBM microenvironment[27]. We selected ligand-receptor pairs that were significantly altered between cells from the cellular interaction network, and Neoplastic cells_6 showed markedly altered interactions with other cell types, especially microglia, which may be one of the methods of the cell population to escape the attack of the immune system (Fig. 2F-2 G). The 30 most significant intercellular ligand-receptor pairs were further screened and we found that malignant cells expressed high levels of EGFR, whereas endothelial cells, T cells, and microglia expressed corresponding ligands, such as TGFB1, COPA, and GRN (Fig. 2H&Supplementary Figure 2). It has been reported that EGFR is overexpressed in cancer cells and activates fibroblasts and myeloid cells through molecules such as AREG[28]. EGFR-associated feedback loops promote the development of PASC from ductal cells to cancer cells[29]. In addition, we analyzed the effects of immune regulators on cellular communication. VSIR_IGSF11 was significantly expressed between malignant cells and other cells, whereas HAVCR2_LGALS9 was not significantly expressed, suggesting that the VISTA/VSIG3 signaling pathway may play a crucial role in GBM (Fig. 2H).
Fig. 2.
Distinct functional composition of malignant cells in GBM. (A) Profiles of the UMAP plots of malignant cells extracted from GBM-A, GBM-B, Ctrl-A and Ctrl-B, 16 cell clusters were successfully classified. (B) Profiles of the t-SNE plots of malignant cells extracted from GBM-A, GBM-B, Ctrl-A and Ctrl-B, 16 cell clusters were successfully classified. (C) Proportion of 16 major cell types showing in bar plots in different donors. (D) Total cell number of each cell type. (E) The expression level of representative novel identified markers across the main malignant cell types. (F) Heatmap show number of potential ligand-receptor pairs between cell groups predicted by CellphoneDB in GBM (left) and Ctrl (right). (G) Circle plots displaying the number of ligand-receptor interactions between distinct cellular components, the circle size is proportional to the number of cells in each cluster, the edge width represents communication probability in GBM (up) and Ctrl (down). (H) Bubble plots show ligand-receptor pairs between malignant cells and other cell groups.
Together, our results predicted that the interaction of malignant cells with microglia, MDM cells, and T cells through multiple receptor-ligand signals may have a profound impact on the development of GBM.
2.3. The expression of TIM-3, VISTA, PSGL-1 and VSIG-3 in GBM
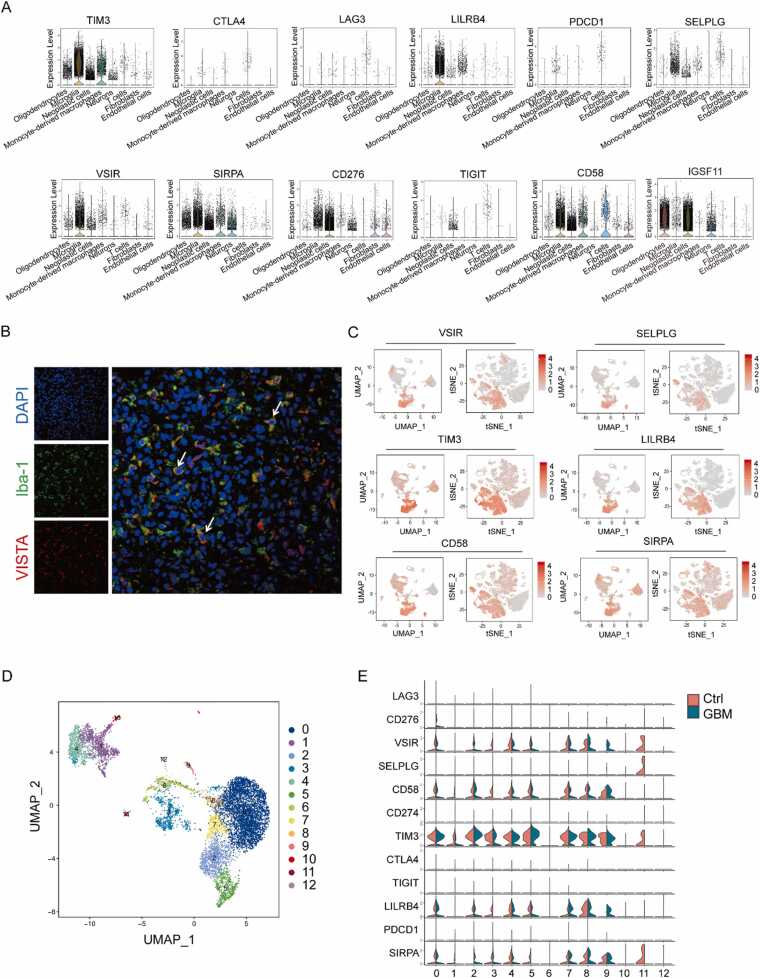
Despite the promise of immunotherapy in the field of cancer treatment, most glioma patients do not respond significantly to the blockage of routine immune checkpoint pathways[30], [31]. To explore the combined strategy for GBM, we evaluated the expression of the co-inhibitory molecules in all cell populations. TIM-3, VISTA, and PSGL-1 (an acidic pH-selective receptor of VISTA) were mainly expressed in microglia and MDM, whereas VSIG-3 (the ligand of VISTA) was mainly expressed in neoplastic cells and oligodendrocytes (Fig. 3A&3 C). Although clinical trials of immune checkpoint inhibitors in GBM mainly focused on monoclonal antibodies against PD-1, PD-L1, and CTLA-4, conventional immune checkpoints (PD-1, PD-L1, CTLA-4, LAG-3 and TIGIT) are not significantly expressed in GBM (Fig. 3A). Furthermore, snRNA-seq and immunofluorescence also demonstrated that VISTA was expressed in microglia (Fig. 3B) and it was highly expressed in GBM tissues compared to adjacent tissues (Supplementary Figure 4). By subdividing microglia into clusters, microglia cells were subdivided into 13 subgroups (Fig. 3D). Due to the difference of communication between Neoplastic cells_6 and microglia between Ctrl and GBM, the expression pattern of common immune checkpoints in microglia cells was analyzed, and the results showed that VISTA was expressed in most microglia cell subgroups, CD58, TIM3, LILRB4 and SIRPA were similar to VISTA expression pattern. TIM3 was highly expressed in microglia (Fig. 3E).
Fig. 3.
The expression distribution of immune checkpoints in GBM. (A) Expression of immune checkpoint genes in all cells from patient A and B. (B) Immunofluorescence staining (n = 3) of IBA1 (FITC, green) and VISTA (CY3, red) in GBM. Scale bar, 25 µm. (C) Expression of immune checkpoint genes with high expression in microglia in all cells from patient A and B. (D) Profiles of the UMAP plots of malignant cells extracted from GBM-A, GBM-B, Ctrl-A and Ctrl-B, 13 cell clusters were successfully classified. (E) Violin plot show expression of immune checkpoint genes in the microglia cell types from patient A and B.
In conclusion, the results suggest that VISTA is highly expressed in microglia, with similar expression profiles to TIM-3 and PSGL-1, and that its ligand VSIG-3 is highly expressed in tumor cells, which may serve as a potential chemotherapy target for GBM.
2.4. MIA of snRNA-seq and ST display the expression of TIM-3, VISTA, PSGL-1 and VSIG-3
ST was applied to analyze patient B samples. A total of 4813 spots of the transcriptome were obtained from the two sections, with an average depth of 5270 UMIs/spots and 2604 genes/spots. Each ST spot captured approximately 1–10 cells, we first normalized the data of expression, then performed principal component analysis (PCA) analysis and grouped the spots to obtain 7 types of subgroups, and finally mapped the spots back to hematoxylin and eosin (H&E) staining tissues, checked the distribution of each subgroup in the tissues[8] (Supplementary Figure 6E). After clustering the points of each ST array according to the principal component scores, we found that the clustering results were consistent with independent histological annotations, supporting the ability to individually identify different spatial regions within the fragment based on ST gene expression (Fig. 4A& Supplementary Figure 6 A). Furthermore, we defined four main areas in tumor and adjacent tissue sections of patient B: cancer, necrosis, stroma and normal brain tissues. In the two sample datasets, we observed that the spatial expression of many variable-expression genes matched the annotated histological regions (Supplementary Figure 7).
Fig. 4.
MIA of snRNA-seq and ST showing VISTA expression patterns in GBM. (A) Annotated tumor cryosection on the ST slide and clustering of the ST spots and color indicates the clustering assignments for patient B(Ctrl-B). (B) Spatial gene expression of immune checkpoint genes from patient B. (C) Standardized expression levels of marker genes in the patient B. (D) Correlation heatmap analysis of immune checkpoint genes in patient B. (E) H&E staining of tissue sections and clustering of ST spots in patient B(Ctrl-B). (F) Immunofluorescence staining (n = 3) of TIM-3 (FITC, green), PSGL-1 (CY5, gold) and VISTA (CY3, red) in GBM. Scale bar, 25 µm. (G-H) Correlation analysis of VISTA and TIM-3, PSGL-1 and VSIG-3 in GEPIA2 and CGGA.
MIA was used to integrate the snRNA-seq and ST[20]. We suggested that neoplastic cells were enriched in all regions except the normal brain tissue regions, showing extensive tumor infiltration of GBM. Normal brain tissue areas were significantly enriched in neurons, astrocytes, and oligodendrocytes. Interestingly, we found that endothelial cells were significantly enriched in stroma. These results supported the widespread application of MIA, providing spatial and functional annotations for the cell population defined by snRNA-seq (Supplementary Figure 5B–5C). In addition, we scored the cell type for each spot. The cancerous region was enriched in neoplastic cells, while the normal brain tissue was enriched in neuron, which is in line with our expectations (Fig. 4C). ST analysis revealed that TIM-3, VISTA, PSGL-1 and VSIG-3 were highly expressed in GBM, which was consistent with the results of the snRNA-seq datasets (Fig. 4B). TIM-3, VISTA, PSGL-1 and VSIG-3 expression in tumor area was higher than that in normal brain tissue (Fig. 4E). Correlation analysis between VISTA and other immune checkpoints showed that VISTA was positively correlated with TIM-3 or PSGL-1 but negatively correlated with VSIG-3, which means the expression pattern of VISTA was similar to those of TIM-3 and PSGL-1 (Fig. 4D). At the protein level, VISTA expression was significantly associated with TIM-3 and PSGL-1 by multi-immunofluorescence (Fig. 4F). Gene correlation analysis of GBM in TCGA database was performed by GEPIA2 (Interactive gene expression profile analysis, http://gepia2.cancer-pku.cn). The results showed that VISTA (C10ORF54) was positively correlated with TIM-3 (HAVCR2) and PSGL-1 (SELPLG) (Fig. 4G). Analysis of the CGGA dataset (http://www.cgga.org.cn) also showed that VISTA was positively correlated with TIM-3 and PSGL-1 and negatively correlated with IGSF11 (Fig. 4H). This result is consistent with our sequencing results.
Taken together, the results elucidate that VISTA, TIM-3, and PSGL-1 may have synergistic effects on GBM through the combined analysis of single cell sequencing and spatial transcriptomics, and were validated in clinical samples, providing reasonable reference for drug combination.
2.5. Subgroup analyses to compare the protein expression and assess the prognostic value of VISTA
VISTA is a novel immune checkpoint that was discovered in 2011, and belongs to the B7 family. The structure of the variable region of extracellular immunoglobulin is similar to that of PD-1 and is also called PD-1 homologue (PD-1 H)[32], [33]. A recent meta-analysis evaluating the prognostic value of VISTA in ten types of solid tumors found that high VISTA expression was associated with prolonged overall survival and CD8+ tumor-infiltrating lymphocytes, suggesting that VISTA is a potential biomarker for solid tumor prognosis[34]. VISTA knockout mice were more resistant to tumor growth than wild-type mice in a radiotherapy mouse glioma model, suggesting that VISTA has the potential to act as an immunomodulatory target in the treatment of glioma[35]. Therefore, we focused on analyzing VISTA to determine the relationship between VISTA and GBM.
We analyzed 180 glioma patients from a tissue microarray (TMA) and 37 patients from Nanjing Drum Tower Hospital, including 4 patients with epilepsy as control samples. Supplementary material showed the clinicopathological characteristics of these patients (Supplementary Table 2). Performed immunohistochemical (IHC) staining of all tissues to determine the correlation between VISTA expression and clinicopathological characteristics. The results showed that the expression profiles of VISTA were associated with age (p < 0.0001), which means VISTA expression is higher in older patients; there was no significant correlation with sex (p = 0.188) or WHO classification (p = 0.584) (Table 1, Fig. 5A,C&5E). Univariate analysis revealed a significant correlation between overall survival (OS) and age (p = 0.009), WHO stage (p < 0.0001), and VISTA expression (p = 0.006). WHO stage (p < 0.0001) and VISTA expression (p = 0.007) were identified as independent prognostic factors in the multifactorial analysis (Table 2). We also compared the effects of different WHO stages, gender, and age groups on VISTA expression and OS, the results showed that patients with low VISTA expression had a better prognosis than those with high VISTA expression in all groups (Figs. 5B,5D&5 F).
Table 1.
Correlation between the expression of VISTA and clinicopathological characteristics.
| Variables |
VISTA expression |
Total |
χ2 |
p value |
||
|---|---|---|---|---|---|---|
| Low | High | |||||
| Age (year) | 17.057 | < 0.0001 | ||||
| ≤ 41 | 56 | 32 | 88 | |||
| > 41 | 25 | 54 | 79 | |||
| Sex | 1.735 | 0.188 | ||||
| Male | 46 | 58 | 104 | |||
| Female | 35 | 29 | 64 | |||
| Grade | 0.3 | 0.584 | ||||
| I/II | 49 | 49 | 98 | |||
| III/IV | 32 | 38 | 70 | |||
Fig. 5.
Subgroup analyses to compare the protein expression and assess the prognostic value of VISTA. (A-B) Comparison of VISTA protein level in glioma patients with different age and Kapla-Meier analyses of OS in patients with age< 41 (left) and age> 41 (right). (C-D) Comparison of VISTA protein level in glioma patients with different gender and Kapla-Meier analyses of OS in patients with male and female. (E-F) Comparison of VISTA protein level in glioma patients with different WHO grade and Kapla-Meier analyses of OS in patients with I&II and III&IV.
Table 2.
Univariate and multivariate analyses of the factors correlated with Overall survival of glioma patients.
| Variables |
Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| p value | HR | 95%CI | p value | |||
| Lower | Upper | |||||
| expression | 0.006 | 2.188 | 1.245 | 3.844 | 0.007 | |
| Age | 0.009 | 2.063 | 1.197 | 3.555 | 0.078 | |
| sex | 0.168 | 0.669 | 0.377 | 1.184 | ||
| Grade stage | < 0.0001 | 13.502 | 6.35 | 28.711 | < 0.0001 | |
In conclusion, the expression of VISTA is likely to be a relatively independent indicator used to evaluate the prognosis of patients in various stages of GBM, providing sufficient evidence for VISTA as a therapeutic target for GBM.
2.6. VISTA was a potential therapeutic target for GBM
Recently, Ghouzlani et al. found that VISTA mRNA expression was significantly associated with glioma grade, histological type, and molecular subtype in TCGA[36]. Kaplan-Meier analysis showed that patients with low VISTA expression had longer survival than those with high VISTA expression. Li-Chong Wang et al. also obtained this conclusion from CGGA and its self-built clinical GBM sample bank[37]. Consistently, our results showed that VISTA was highly expressed in the clinical samples of GBM and positively correlated with glioma grading by analyzing the location of VISTA expression in clinical IHC samples, Pathological staining showed that VISTA was expressed in the membrane and cytoplasm of microglia, macrophages, and lymphocytes but not in tumor cells. Positive staining was also observed in the blood vessels, which were not counted in the pathology score (Fig. 6A-6D). By analyzing the relationship between VISTA OS or disease-free survival (DFS) in glioma patients, we observed that patients with low VISTA expression had a better prognosis than those with high VISTA expression (Fig. 6E). Using bivariate correlation analysis, we observed that the expression of VISTA and EGFR (p < 0.0001, r = 0.309) or PD-L1 (p < 0.05, r = 0.197) was positively correlated, while the expression of VISTA was not correlated with Ki67 (p > 0.05, r = 0.08) (Fig. 6F).
Fig. 6.
VISTA was a potential therapeutic target for GBM. (A) IHC staining sections for VISTA in clinical samples. (B) Comparison of IHC scores of VISTA in 37 clinical samples. (C) Immunohistochemical section staining map of representative grade I-IV glioma tissue. (D) Statistical plot of VISTA positive staining in 180 glioma tissue microarrays. (E) Kaplan–Meier analysis of OS (left) and DFS (right) in patients with low and high VISTA level in TMA according to IHC scores. (F) Correlation analysis between VISTA and EGFR, PDL1, Ki67 in TMA. ns indicates no significant difference and * indicates P < 0.05.
In summary, our results suggest that VISTA expression was closely related to the molecular pathology of glioma. Therefore, VISTA is a promising target for immunotherapy in GBM. Blocking VISTA may provide an alternative therapy for gliomas that are resistant to PD-1/PD-L1 antibody treatment.
3. Discussion
It is traditionally believed that the blood-brain barrier (BBB) prevents normal immune function in the CNS. However, recent studies have shown that astrocytes, microglia, and macrophages are potential antigen-presenting cells. The BBB increases microvascular permeability under inflammatory conditions, which can promote the entry of immune cells into the CNS[38], [39]. In view of the immune characteristics of glioma, immunotherapy is expected to become a new treatment option for gliomas. We generated a single-cell spatial transcriptome map using snRNA-seq and ST to reveal the landscape of the microenvironment in GBM, summarized the expression profiles of common immune checkpoints in GBM and observed that TIM-3, VISTA, PSGL-1, and VSIG-3 were the most highly expressed in GBM at single-cell and spatial levels. We demonstrated that the expression patterns of TIM-3, VISTA, and PSGL-1 were almost the same, showing a significant positive correlation, mainly expressed in microglia, whereas VSIG-3 was highly expressed in neoplastic cells. TIM-3 was found to be mainly expressed in GBM, and its expression was significantly up-regulated in mesenchymal gliomas[24]. Kaplan-Meier analysis was used to analyze the prognosis of 1024 glioma patients, and the expression of TIM-3 was negatively correlated with the patient prognosis. Qing guo et al. also found that TIM-3 was positively correlated with GBM grading and negatively correlated with patient prognosis[40]. Jennifer E. Kim et al. used TIM-3 monoclonal antibody in combination with PD-1 monoclonal antibody and radiotherapy to completely cure an in situ glioma model mouse[41]. The expression of the IGSF11 (also called VSIG-3) gene in high-grade glioma was found to be significantly up-regulated compared with that in low-grade glioma[42]. Increased IGSF11 expression in advanced human gliomas is associated with poor overall survival.
Currently, clinical trials of glioma immune checkpoint inhibitors (mainly monoclonal antibody studies against PD-1/PD-L1 and CTLA-4) are in the early stages, and most trials are still recruiting or in progress. A few trials have reported preliminary results[43], [44]. Scientists are dedicated to finding new immune checkpoints or biomarkers, and considering their combination with radiotherapy, temozolomide or bevacizumab may be useful for the treatment of glioma. Youngmi Kim et al. combined with NanoString GeoMx digital spatial profile (DSP) analysis and single-cell RNA-seq (scRNA-seq) technology characterization archive formalin-fixed paraffin-embedding (FFPE) Glioblastoma specimens identified VSIR as a potential target for high-grade gliomas (n = 3)[45]. Stine Asferg Petterson et al. found that VISTA is down-regulate in the presence of reduced T cells through digital spatial analysis[46]. In this study, we demonstrated that VSIR_IGSF11 was extensively involved in intercellular communication in GBM (Fig. 6), Wang et al. showed that the interaction between VSIG-3 and VISTA expressed on activated human T cells is a poorly regulated pathway that leads to reduced T cell proliferation, decreased production of pro-inflammatory cytokines (IFN-γ, IL-2, and IL-17) and chemokines (CCL3, CCL5, and CXCL11), and decreased production of proinflammatory cytokines (IFN-γ, IL-2, and IL-17). The infiltration of immune cells (namely monocytes, DC, and TAM) into TME was reduced[47]. VSIG3 protein is mainly expressed in the brain and testis, and VISTA protein is expressed in many cancers. Ghouzlani et al. reported that increased expression of VSIG-3 in glioma tissue is associated with high infiltration of immune cells (especially CD4+ and CD8+T cells), in addition, the expression of IL-10 and TGF-β increases with increased VISTA expression[42]. Xi Yang Tang et al. have speculated that VISTA may interact with IGSF11 for immune regulation in other tumors, including non-small cell lung cancer, ovarian cancer, gastric cancer, colorectal cancer, soft tissue sarcoma, and oral squamous cell carcinoma[48]. The Vista-targeting antibody HMBD-002 can effectively block the interaction between VISTA and IGSF11, further inhibiting IGSF11-mediated T cell production of IFN-γ[49]. Our group has reported that M351–0056, a novel compound modulating VISTA, can ameliorated imiquimod-induced psoriasis-like dermatitis in mice, and the crystal structure of VSIG3 protein and its regulator K284–3046[50], [51]. This evidence shows this pathway is expected to become an important therapeutic target for GBM.
Taken together, our GBM transcription map provided a framework for understanding the TME and characterizes tumor-associated subsets. Moreover, we described the immunosuppressive state of GBM in several aspects, which are potential targets for the development of immunotherapy for GBM and other cancers. The combination of single-cell data with spatial information allowed us to summarize spatial and cellular information in GBM. Finally, although we have obtained key interaction networks in the GBM tumor microenvironment through bioinformatics analysis, further functional experiments are needed to explore the biological consequences and potential mechanisms.
4. Material and methods
4.1. Tissue sample source and ethics statement
GBM tissue and patient-matched adjacent para-cancer tissue samples for single-cell sequencing and spatial transcriptome were obtained from Nanjing Gulou Hospital. This study obtained the informed consent of 2 patients and was approved by the Ethics Committee of Nanjing Gulou Hospital. The tissue chips used in this study were purchased from Shanghai Xinchao Biotechnology Co., LTD., and contained samples from 180 patients with grade I-IV brain gliomas. All patients underwent surgery between 2008 and 2011 and were followed up until July 2017. This study was carried out in accordance with the Declaration of Helsinki and was approved by the Medical Ethics Committee of Shanghai Core Biotech Co., LTD., in accordance with institutional guidelines (No.YB M-05–02). In addition, we collected tissues from 37 fresh brain gliomas and control epileptic patients from Nanjing Gulou Hospital. Our study was approved by the Ethics Committee of Nanjing Gulou Hospital, and all included patients received full informed consent.
4.2. snRNA-seq
Cellular suspensions were loaded on a 10X Genomics GemCode Single-cell instrument that generates single-cell gel bead-in-emlusion (GEMs). Libraries were generated and sequenced from the cDNAs with Chromium Next GEM Single Cell 3′ Reagent Kits v3.1. Silane magnetic beads were used to remove leftover biochemical reagents and primers from the post GEM reaction mixture. Full-length, barcoded cDNAs were then amplified by Polymerase Chain Reaction (PCR) to generate sufficient mass for library construction. First, DNA fragments were broken into 200–300 BP fragments by Biorupter Ultrasound Fragmentation Instrument. Next, DNA library was amplified by PCR with sequencing connector P5 and sequencing primer R1. Finally, prepared samples were subjected to the 10Х single-cell sequencing analysis platform.
4.3. ST
The Visium Spatial Tissue Optimization Slide kit is used to fit the time for permeabilization by generating fluorescently labeled cDNA tissue prints. A timer gradient is set for each capture area. Fluorescent cDNA synthesis is performed and fluorescent print of spatial positions where the cDNA reaction took place. The fluorescent print is imaged using fluorescence microscope with tissue removed. The section with the strongest fluorescence signal, minimum diffusion and longest time for permeabilization will be chosen as the fit time for permeabilization.
Sample fixing and imaging have been done in sample preparing and section permeabilization will be performed as follow. Permeabilization processes for the time determined by tissue optimization. The first strand of cDNA is synthesized via reverse transcription and the second strand of cDNA is synthesized via PCR. Then the cDNA is denaturation, making the second strand of cDNA dissociated from slide. The spatially barcoded, full-length cDNA is amplified via PCR to generate sufficient mass for library construction. Enzymatic fragmentation and size selection are used to optimize the cDNA amplicon size. P5, P7, i7 and i5 sample indexes, and TruSeq Read 2 (read 2 primer sequence) are added via End Repair, A-tailing, Adaptor Ligation, and PCR. The final libraries contain the P5 and P7 primers used in Illumina amplification. The Visium Spatial protocol produces Illumina-ready sequencing libraries. A Visium Spatial library comprises standard Illumina paired-end constructs which begin and end with P5 and P7. The Visium Spatial 16 bp spatial barcode and 10 bp UMI are encoded in Read 1, while Read 2 is used to sequence the cDNA fragment. Sample index sequences are incorporated as the i7 index read. Read 1 and Read 2 are standard Illumina® sequencing primer sites used in paired-end sequencing.
4.4. CNV analysis
InferCNV calculated a baseline expression level from the level of gene expression in a normal sample, then subtracted the level of each gene in the cell from the baseline to obtain the relative expression level. Subsequently, a window of 100 genes was set up on the chromosome, and CNV events in single cell chromosome region were predicted by relative gene expression levels. Heatmaps are drawn by relative gene expression levels, but ultimately reflect CNV events in large gene segments. In tumor samples, the red squares in the heat map represent increased gene expression relative to baseline, and the corresponding chromosome segment may have increased copy number. The blue squares represent genes with reduced expression relative to baseline, corresponding to chromosome segments with possible copy number loss. In normal samples, there are red and blue squares, but they are caused by abnormal expression of the gene. A subset of 500 randomly selected neuron cells was removed as a negative control for the analysis.
4.5. TMA
TMA of 180 glioma patient (HBraG180Su01, Shanghai Outdo Biotech Co.Ltd) was applied to explore the expression of VISTA and its impact on the survival of glioma patients. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Medical Ethics Committee of the Shanghai Outdo Biotech Company and performed according to institutional guidelines (No. YB M-05–02). All patients underwent surgery from 2008 to 2011 and were followed up until July 2017.
4.6. IHC
Tris-EDTA buffer (pH 9.0) was used to recover antigen from tissue sections under high temperature and high pressure. Then, TMA were incubated with anti-VISTA rabbit monoclonal antibody (Cell Signaling Technology, Cat#64953) overnight. Perform immunohistochemical staining on TMA samples according to the manufacturer's instructions, and observe and analyze under a microscope (Leica Biosystems, Aperio ImageScope). The level of VISTA protein in the TMA was semi-quantitatively analyzed by two experienced pathologists using a scoring system based on staining intensity and and degree of staining. According to the positive staining ratio of immune cells, the percentage of immunohistochemical positive staining cells is 0–100%. The staining intensity scores are as follows: “Negative” received a score of 0, “0.5 + ” a score of 0.5, “1 + ” a score of 1, “2 + ” a score of 2 and “3 + ” a score of 3. Multiply the positive staining rate score and the staining intensity score to get the total score (0–300%). Samples with scores less than 6% are considered to be the low expression group, while samples with scores greater than 6% are considered to be the high expression group.
4.7. Statistical Analysis
To analyze the association between VISTA expression and clinicopathological features, chi-square test was adopted. Kaplan–Meier survival analysis along with log-rank test was applied to evaluate the impact of VISTA expression on glioma patients’ OS and DFS. Mann–Whitney U-test was used to compare VISTA expression in different groups. Univariate and multivariate Cox regression analyses were employed to identify independent prognostic factors for glioma OS from variables including VISTA expression, age, gender. All statistical analyses were conducted using Prism GraphPad 7 software and R software. Student’s t-test was applied in analyzing our fresh tissue samples. A two-sided P < 0.05 was regarded as statistically significant.
4.8. Immunofluorescence staining of FFPE tissue
FFPE tissues were cut at 4 µm thickness and dried overnight. After deparaffinizing the slides, antigen retrieval was carried out by boiling the samples for 10–20 min in Tris-EDTA (pH 9.0) in a microwave oven. Primary antibodies (VISTA, Cell Signaling Technology, Cat#64953; TIM, abcam, Cat#ab241332; PSGL1, abcam, Cat#ab227836; IBA-1, invitrogen, Cat#MA5–29012) were diluted in goat serum according to the manufacturer's instructions and incubated with slides overnight in a wet chamber kept at 4 °C. Secondary antibodies (PanoPANEL mIHC Kits, Cat#0079100020) were diluted 1:100 in amplification buffer and incubated with slides for 10 min at room temperature before mounting and imaging. After mounting the slide with anti-fluorescence quenching mounting solution (Beyotime, Cat#P0126), take pictures under a laser scanning confocal microscope (Olympus, Cat#FV3000).
4.9. Integrated snRNA-seq and ST by MIA
We integrated our ST data and snRNA-seq data by introducing MIA as previously reported[20]. This analysis proceeds by first delineating sets of cell type-specific and tissue region-specific genes and then determining whether their overlap is higher (enrich ment) or lower (depletion) than expected by chance. In the snRNA-seq data, we defined the gene sets by identifying for each cell type those genes whose expression is statistically higher in the cells annotated to that cell type in comparison with expression in the remaining cells (P < 10−5, two-tailed Student’s t-test; see Methods). For the ST data, we then identified genes with significantly higher expression in each spatial region relative to the others (P < 0.01, two-tailed Student’s t-test). With the gene sets extracted across the snRNA-seq and ST modalities, MIA next computes the overlap between each pair of cell type-specific and region-specific gene sets and performs a hypergeometric test to assess significant enrichment or depletion. Extending this analysis to all pairs of cell types and tumor regions produces an ‘MIA map’.
4.10. Enrichment analysis
GO enrichment analysis provides all GO terms that significantly enriched in differentially expressed genes comparing to the genome background and filter the differentially expressed genes that correspond to biological functions. Firstly, all peak related genes were mapped to GO terms in the Gene Ontology database (http://www.geneontology.org/), gene numbers were calculated for every term, significantly enriched GO terms in differentially expressed genes comparing to the genome background were defined by hypergeometric test.
Genes usually interact with each other to play roles in certain biological functions. Pathway- based analysis helps to further understand genes biological functions. KEGG is the major public pathway related database. Pathway enrichment analysis identified significantly enriched metabolic pathways or signal transduction pathways in differentially expressed genes comparing with the whole genome background. The calculating formula is the same as that in GO analysis.
4.11. SCENIC analysis
To carry out transcription factor network inference, analysis was performed on the SCENIC R package. In brief, log-normalized expression matrix generated using Seurat was used as input, and the pipeline was implanted in three steps. First, we identified gene co-expression network via GENIE3. Second, we pruned each module based on a regulatory motif near a transcription start site via RcisTarget. Precisely, networks were retained if the TF-binding motif was enriched among its targets, while target genes without direct TF-binding motifs were removed. The retained networks were called regulons. Third, we scored the activity of each regulon for each single cell via the AUC scores using AUCell R package. Gene regulatory network (GRN) plots of all regulons were done using the cytoscape software.
4.12. Cell-cell communication analysis
We used cellphoneDB, which contains ligand-receptor information, to analyze expression abundance of ligand-receptor interactions between two cell types on the basis of expression of a receptor by one cell type and a ligand by another cell type. Among them, only receptors and ligands expressed in more than a user-specified threshold percentage of the cells in the specific cluster were considered for the analysis (default is 10%). Based on the above analysis of the expression abundance of ligand-receptor, we obtained the number of ligand-receptor interactions between two cell types, which can make a preliminary assessment of the communication relationship between cells. To identify biological relevance, we used cellphoneDB software to perform pairwise comparisons between all cell types in the dataset, and analyze the number of significantly enriched ligand-receptor interactions between two cell types. First, we randomly permuted the cluster labels of all cells (1000 times by default) and determined the mean of the average ligand expression level in a cluster and the average receptor expression level in the interacting cluster. In this way we generated a null distribution for each ligand-receptor pair in each pairwise comparison between two cell types. We obtained a P value for the likelihood of cell-type enrichment of each ligand-receptor complex by calculating the proportion of the means that are as high as or higher than the actual mean. We defined P value less than 0.05 as a significant enrichment. Based on the analysis of the number of significant enriched ligand-receptor pairs between cell types, we constructed a cell interaction network diagram, which can show the regulation relationship between cells more intuitively.
CRediT authorship contribution statement
Dingyi Yuan: Formal analysis, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Jun Liu: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. Wenting Chen: Data curation, Formal analysis, Project administration, Writing – original draft. Wei Li: Data curation, Resources. Shasha Jin: Formal analysis, Validation, Writing – review & editing. Wanmei Liu: Validation, Visualization. Liu Liu: Formal analysis, Visualization. Yinhao Wu: Validation. Yuxin Zhang: Formal analysis. Xiaoyu He: Formal analysis. Jingwei Jiang: Software. Hongbin Sun: Conceptualization. Xiangyu Liu: Conceptualization, Resources.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgements
This research was funded by the National Natural Science Foundation of China (No. 81973361) and the Natural Science Foundation of Jiangsu Province (BK20202009). We are grateful to Guangzhou Genedenovo Biotechnology Co., Ltd for assisting in sequencing and/or bioinformatics analysis.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2024.04.014.
Contributor Information
Hongbin Sun, Email: 10200307030@cpu.edu.cn.
Xiangyu Liu, Email: liuxiangyumail@163.com.
Jun Liu, Email: junliu@cpu.edu.cn.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
References
- 1.Xiong Z., Raphael I., Olin M., Okada H., Li X., Kohanbash G. Glioblastoma vaccines: past, present, and opportunities. EBioMedicine. 2024;100 doi: 10.1016/j.ebiom.2023.104963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim M., Xia Y., Bettegowda C., Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15:422–442. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 3.Nathanson D.A., Gini B., Mottahedeh J., Visnyei K., Koga T., Gomez G., et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–76. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin X., Xie M., Yao J., Ma X., Qin L., Zhang X.M., et al. Immune-related adverse events in non-small cell lung cancer: Occurrence, mechanisms and therapeutic strategies. Clin Transl Med. 2024;14 doi: 10.1002/ctm2.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Splendiani E., Besharat Z.M., Covre A., Maio M., Di Giacomo A.M., Ferretti E. Immunotherapy in melanoma: Can we predict response to treatment with circulating biomarkers? Pharmacol Ther. 2024;256 doi: 10.1016/j.pharmthera.2024.108613. [DOI] [PubMed] [Google Scholar]
- 6.Regmi M., Wang Y., Liu W., Dai Y., Liu S., Ma K., et al. From glioma gloom to immune bloom: unveiling novel immunotherapeutic paradigms-a review. J Exp Clin Cancer Res. 2024;43:47. doi: 10.1186/s13046-024-02973-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romani M., Pistillo M.P., Carosio R., Morabito A., Banelli B. Immune Checkpoints and Innovative Therapies in Glioblastoma. Front Oncol. 2018;8:464. doi: 10.3389/fonc.2018.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torphy R.J., Schulick R.D., Zhu Y. Newly Emerging Immune Checkpoints: Promises for Future Cancer Therapy. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18122642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papalexi E., Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18:35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 10.Lin S., Sun Y., Cao C., Zhu Z., Xu Y., Liu B., et al. Single-nucleus RNA sequencing reveals heterogenous microenvironments and specific drug response between cervical squamous cell carcinoma and adenocarcinoma. EBioMedicine. 2023;97 doi: 10.1016/j.ebiom.2023.104846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anoop P., Patel I.T., John J., Trombetta Alex, Shalek K., Shawn M., et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Cancer Genom. 2018;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanco-Carmona E., Narayanan A., Hernandez I., Nieto J.C., Elosua-Bayes M., Sun X., et al. Tumor heterogeneity and tumor-microglia interactions in primary and recurrent IDH1-mutant gliomas. Cell Rep Med. 2023;4 doi: 10.1016/j.xcrm.2023.101249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller S., Kohanbash G., Liu S.J., Alvarado B., Carrera D., Bhaduri A., et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol. 2017;18:234. doi: 10.1186/s13059-017-1362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neftel C., Laffy J., Filbin M.G., Hara T., Shore M.E., Rahme G.J., et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell. 2019;178:835–849. doi: 10.1016/j.cell.2019.06.024. e821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S.Q., Gao Z.J., Wu J., Zheng H.M., Li B., Sun S., et al. Single-cell and spatially resolved analysis uncovers cell heterogeneity of breast cancer. J Hematol Oncol. 2022;15:19. doi: 10.1186/s13045-022-01236-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan C.U., Quah F.X., Hemberg M. Single-cell and spatial transcriptomics: Bridging current technologies with long-read sequencing. Mol Asp Med. 2024;96 doi: 10.1016/j.mam.2024.101255. [DOI] [PubMed] [Google Scholar]
- 17.Thrane K., Eriksson H., Maaskola J., Hansson J., Lundeberg J. Spatially Resolved Transcriptomics Enables Dissection of Genetic Heterogeneity in Stage III Cutaneous Malignant Melanoma. Cancer Res. 2018;78:5970–5979. doi: 10.1158/0008-5472.CAN-18-0747. [DOI] [PubMed] [Google Scholar]
- 18.Moncada R., Barkley D., Wagner F., Chiodin M., Devlin J.C., Baron M., et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol. 2020;38:333–342. doi: 10.1038/s41587-019-0392-8. [DOI] [PubMed] [Google Scholar]
- 19.Ji A.L., Rubin A.J., Thrane K., Jiang S., Reynolds D.L., Meyers R.M., et al. Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma. Cell. 2020;182:497–514. doi: 10.1016/j.cell.2020.05.039. e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu S.Z., Al-Eryani G., Roden D.L., Junankar S., Harvey K., Andersson A., et al. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53:1334–1347. doi: 10.1038/s41588-021-00911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y., Banerjee A., Xie P., Ivanov A.A., Uddin A., Jiao Q., et al. Pharmacological suppression of the OTUD4-CD73 proteolytic axis revives antitumor immunity against immune-suppressive breast cancers. J Clin Invest. 2024 doi: 10.1172/JCI176390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rui Wu W.G., Qiu Xinyao, Wang Shicheng, Sui Chengjun, Lian Qiuyu, Wu Jianmin, et al. Comprehensive analysis of spatial architecture in primary liver cancer. Sci Adv. 2021:1–19. doi: 10.1126/sciadv.abg3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toshida K., Itoh S., Iseda N., Tomiyama T., Yoshiya S., Toshima T., et al. Impact of ACSL4 on the prognosis of hepatocellular carcinoma: Association with cancer-associated fibroblasts and the tumour immune microenvironment. Liver Int: J Int Assoc Study Liver. 2024;44:1011–1023. doi: 10.1111/liv.15839. [DOI] [PubMed] [Google Scholar]
- 24.Darmanis S., Sloan S.A., Croote D., Mignardi M., Chernikova S., Samghababi P., et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Rep. 2017;21:1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui L., Xu L., Wang G., Wen J., Luo L., Zhao H., et al. STAT3-PTTG11 abrogation inhibits proliferation and induces apoptosis in malignant glioma cells. Oncol Lett. 2020;20:6. doi: 10.3892/ol.2020.11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui L., Ren T., Zhao H., Chen S., Zheng M., Gao X., et al. Suppression of PTTG1 inhibits cell angiogenesis, migration and invasion in glioma cells. Med Oncol. 2020;37:73. doi: 10.1007/s12032-020-01398-2. [DOI] [PubMed] [Google Scholar]
- 27.Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagathihalli N.S., Beesetty Y., Lee W., Washington M.K., Chen X., Lockhart A.C., et al. Novel mechanistic insights into ectodomain shedding of EGFR Ligands Amphiregulin and TGF-α: impact on gastrointestinal cancers driven by secondary bile acids. Cancer Res. 2014;74:2062–2072. doi: 10.1158/0008-5472.CAN-13-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao X., Li H., Lyu S., Zhai J., Ji Z., Zhang Z., et al. Single-cell transcriptomics reveals heterogeneous progression and EGFR activation in pancreatic adenosquamous carcinoma. Int J Biol Sci. 2021;17:2590–2605. doi: 10.7150/ijbs.58886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiyokawa J., Wakimoto H. Preclinical And Clinical Development Of Oncolytic Adenovirus For The Treatment Of Malignant Glioma. Oncolytic Virother. 2019;8:27–37. doi: 10.2147/OV.S196403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J., Zhao D., Liu Z., Liu F. Repurposing psychiatric drugs as anti-cancer agents. Cancer Lett. 2018;419:257–265. doi: 10.1016/j.canlet.2018.01.058. [DOI] [PubMed] [Google Scholar]
- 32.Wang L., Rubinstein R., Lines J.L., Wasiuk A., Ahonen C., Guo Y., et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J Exp Med. 2011;208:577–592. doi: 10.1084/jem.20100619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flies D.B., Wang S., Xu H., Chen L. Cutting edge: A monoclonal antibody specific for the programmed death-1 homolog prevents graft-versus-host disease in mouse models. J Immunol. 2011;187:1537–1541. doi: 10.4049/jimmunol.1100660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He X.L., Zhou Y., Lu H.Z., Li Q.X., Wang Z. Prognostic value of VISTA in solid tumours: a systematic review and meta-analysis. Sci Rep. 2020;10:2662. doi: 10.1038/s41598-020-59608-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flies D.B., Han X., Higuchi T., Zheng L., Sun J., Ye J.J., et al. Coinhibitory receptor PD-1H preferentially suppresses CD4(+) T cell-mediated immunity. J Clin Invest. 2014;124:1966–1975. doi: 10.1172/JCI74589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghouzlani A., Lakhdar A., Rafii S., Karkouri M., Badou A. The immune checkpoint VISTA exhibits high expression levels in human gliomas and associates with a poor prognosis. Sci Rep. 2021;11 doi: 10.1038/s41598-021-00835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L.C., Wang Y.L., He B., Zheng Y.J., Yu H.C., Liu Z.Y., et al. Expression and clinical significance of VISTA, B7-H3, and PD-L1 in glioma. Clin Immunol (Orlando, Fla) 2022;245 doi: 10.1016/j.clim.2022.109178. [DOI] [PubMed] [Google Scholar]
- 38.McGranahan T., Li G., Nagpal S. History and current state of immunotherapy in glioma and brain metastasis. Ther Adv Med Oncol. 2017;9:347–368. doi: 10.1177/1758834017693750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ransohoff R.M., Engelhardt B. The anatomical and cellular basis of immune surveillance in the central nervous system. Nat Rev Immunol. 2012;12:623–635. doi: 10.1038/nri3265. [DOI] [PubMed] [Google Scholar]
- 40.Guo Q., Shen S., Guan G., Zhu C., Zou C., Cao J., et al. Cancer cell intrinsic TIM-3 induces glioblastoma progression. iScience. 2022;25 doi: 10.1016/j.isci.2022.105329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J.E., Patel M.A., Mangraviti A., Kim E.S., Theodros D., Velarde E., et al. Combination Therapy with Anti-PD-1, Anti-TIM-3, and Focal Radiation Results in Regression of Murine Gliomas. Clin Cancer Res: J Am Assoc Cancer Res. 2017;23:124–136. doi: 10.1158/1078-0432.CCR-15-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghouzlani A., Rafii S., Karkouri M., Lakhdar A., Badou A. The promising IgSF11 immune checkpoint is highly expressed in advanced human gliomas and associates to poor prognosis. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.608609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Omuro A., Vlahovic G., Lim M., Sahebjam S., Baehring J., Cloughesy T., et al. Nivolumab with or without ipilimumab in patients with recurrent glioblastoma: results from exploratory phase I cohorts of CheckMate 143. Neuro Oncol. 2018;20:674–686. doi: 10.1093/neuonc/nox208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X., Guo G., Guan H., Yu Y., Lu J., Yu J. Challenges and potential of PD-1/PD-L1 checkpoint blockade immunotherapy for glioblastoma. J Exp Clin Cancer Res. 2019;38:87. doi: 10.1186/s13046-019-1085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim Y., Danaher P., Cimino P.J., Hurth K., Warren S., Glod J., et al. Highly Multiplexed Spatially Resolved Proteomic and Transcriptional Profiling of the Glioblastoma Microenvironment Using Archived Formalin-Fixed Paraffin-Embedded Specimens. Mod Pathol: J U S Can Acad Pathol, Inc. 2023;36 doi: 10.1016/j.modpat.2022.100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petterson S.A., Sørensen M.D., Burton M., Thomassen M., Kruse T.A., Michaelsen S.R., et al. Differential expression of checkpoint markers in the normoxic and hypoxic microenvironment of glioblastomas. Brain Pathol (Zur, Switz) 2023;33 doi: 10.1111/bpa.13111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J., Wu G., Manick B., Hernandez V., Renelt M., Erickson C., et al. VSIG-3 as a ligand of VISTA inhibits human T-cell function. Immunology. 2019;156:74–85. doi: 10.1111/imm.13001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang X.Y., Xiong Y.L., Shi X.G., Zhao Y.B., Shi A.P., Zheng K.F., et al. IGSF11 and VISTA: a pair of promising immune checkpoints in tumor immunotherapy. Biomark Res. 2022;10:49. doi: 10.1186/s40364-022-00394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thakkar D., Paliwal S., Dharmadhikari B., Guan S., Liu L., Kar S., et al. Rationally targeted anti-VISTA antibody that blockades the C-C′ loop region can reverse VISTA immune suppression and remodel the immune microenvironment to potently inhibit tumor growth in an Fc independent manner. J Immunother Cancer. 2022;10 doi: 10.1136/jitc-2021-003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu X., Qie C., Jiang J., Xie X., Chen W., Liu W., et al. M351-0056 is a novel low MW compound modulating the actions of the immune-checkpoint protein VISTA. Br J Pharmacol. 2021;178:1445–1458. doi: 10.1111/bph.15357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie X., Chen C., Chen W., Jiang J., Wang L., Li T., et al. Structural basis of VSIG3: the ligand for VISTA. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.625808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.