Summary
A quarter of marine mammals are at risk of extinction, with disease and poor habitat quality contributing to population decline. Investigation of the Major Histocompatibility Complex (MHC) provides insight into species’ capacity to respond to immune and environmental challenges. The eighteen available cetacean chromosome level genomes were used to annotate MHC Class I loci, and to reconstruct the phylogenetic relationship of the described loci. The highest number of loci was observed in the striped dolphin (Stenella coeruleoalba), while the least was observed in the pygmy sperm whale (Kogia breviceps) and rough toothed dolphin (Steno bredanensis). Of the species studied, Mysticetes had the most pseudogenes. Evolutionarily, MHC Class I diverged before the speciation of cetaceans. Yet, locus one was genomically and phylogenetically similar in many species, persisting over evolutionary time. This characterisation of MHC Class I in cetaceans lays the groundwork for future population genetics and MHC expression studies.
Subject areas: immune system evolution, evolutionary biology
Graphical abstract

Highlights
-
•
The number of MHC Class I functional loci in cetaceans ranged from one to nine
-
•
Mysticetes had the most pseudogenes of any taxa studied
-
•
MHC Class I diverged before the speciation of cetaceans
-
•
Locus one was genomically and phylogenetically similar across taxa
Immune system evolution; Evolutionary biology.
Introduction
A key region of the genome that is frequently targeted in conservation biology studies is the Major Histocompatibility Complex (MHC). MHC genes code for a transmembrane receptor that is found on the cells of all jawed vertebrates and is involved in recognising cells as ‘self’, and detecting anything foreign that enters the body. The ability of MHC receptors to recognise a plethora of pathogens depends on the structure, folding and biochemical properties of proteins translated from MHC genes.1 The MHC Class I and II genes are the most polymorphic in vertebrate genomes and play an integral role in adaptive immune function.2 There is mounting evidence that MHC is essential to consider when investigating the immune health of a population or species and for conservation management strategies.3,4,5 High heterozygosity at the MHC loci is advantageous for an individual, since it correlates with higher fitness,6,7,8 as MHC diversity not only influences immune function but also reproductive success,9,10 and mate choice.11 While this is true, diversity of the MHC is so well maintained in some species/populations that using it as an indicator of fitness may not always be applicable, as there is evidence that some small, inbred populations are still able to maintain MHC diversity.12,13
MHC is a complex gene family that is under intense selective pressure, these genes evolve by gene duplication and a process known as birth-death evolution.12,14,15,16,17,18 The rapid diversification in the MHC is driven by host-pathogen coevolution and balancing selection that maintain high MHC diversity in host populations.6,15,19 Due to intense selective pressures and rapid evolution, the structure and diversity of the MHC can differ significantly between closely related species.17 For instance, where humans and chimpanzees usually share 99% similarity across the genome, they only share 86% similarity in the MHC region.20 Further adding to the complexity, some species have groups of unlinked MHC genes or have a number of MHC genes silenced or expressed in low quantities.18 The genomic region containing the MHC genes is highly mosaic, with high diversity observed in the areas containing the MHC Class I and II genes, while the surrounding genes are highly conserved and are shared across mammals.14
MHC has been used frequently in conservation biology studies of both terrestrial and aquatic species,14,16 yet knowledge and data on the MHC of cetaceans (whales, dolphins, and porpoises) is incomplete. This is surprising as MHC in marine mammals may have unique characteristics due to their evolutionary history. Originating from terrestrial mammals that returned to the sea, they have been exposed to different types and numbers of pathogens than terrestrial species.21,22,23 Investigation of cetacean evolution via the innate immune Toll-Like Receptor (TLR) gene TLR4 revealed signatures of positive and diversifying selection during the transition from terrestrial to marine environments, as well as during the transition from semi-aquatic to full aquatic species.21 Studying MHC loci in cetacean populations is hindered by its high complexity, therefore the genomic organisation of the MHC regions needs to be characterised prior to population level studies being conducted. Moreover, marine mammals are often cryptic, and the ability and effectiveness of sampling may be difficult due to high cost and logistics.
Next-generation sequencing has revolutionised genome biology over the last decades and generated publicly available high quality genomes of several cetacean species. These genomes can be utilised to investigate the cetacean MHC regions, both at the species and population level. Previous studies have so far mostly focused on MHC Class II in cetaceans, and characterised this gene region in beluga whales (Delphinapterus leucas),24 Yangtze River dolphin/baiji (Lipotes vexillifer),25 Amazon River dolphin/boto/boutu (Inia geoffrensis),26 Indo-Pacific finless porpoise (Neophocaena phocaenoides),27,28 blue whale (Balaenoptera musculus),29 and Indo-Pacific humpback dolphin (Sousa chinensis).30 Two recent studies have provided a detailed overview of the structural organisation of MHC Class II in the genome of marine mammals31 and cetaceans.32 Both studies suggested that like terrestrial species, MHC Class II gene copies have duplicated and evolved during the radiation of cetaceans.
In contrast to MHC Class II, less is known about MHC Class I in cetaceans, with studies describing its diversity and organisation only in a few species, including the gray whale (Eschrichtius robustus),33 Hector’s dolphin (Cephalorhynchus hectori),34 North Atlantic right whale (Eubalaena glacialis),35 Yangtze finless porpoise (N. asiaeorientalis asiaeorientalis),27,36,37 and the Yangtze River dolphin/baiji.37 An early study used molecular cloning to identify the coding region of Class I in the Tursiops genus,38 but technology used by the authors did not allow the characterisation of the organisation of MHC Class I.39 A preprint study conducted by Gambón-Deza, (2020)40 used a machine learning program to define the location and structure of the MHC Class I in cetaceans, however it was unable to determine the chromosomal locations of the loci and did not discuss the evolutionary ecology of the cetacean MHC Class I. Manual annotation using Basic Local Alignment Search Tool (BLAST) searches has proven to be a useful tool for defining the structure of MHC in species, especially those assembled to the chromosome level. He et al., (2020)41 found that BLAST searches of published genomes can accurately predict number and location of MHC loci, which can be verified by long read sequencing. Studies describing the structure and organisation of these genes in cetaceans is lacking, yet necessary to enable the investigation of population genetic diversity of endangered marine mammal species.
Cetaceans are key for ecosystem health, occupying many different niches and acting as ecosystem sentinels.42,43,44 Many of these species are capable of huge migrations like humpback whales (Megaptera novaeangliae) that travel from the Antarctic to northern Australia to birth their calves,45 while others exhibit site fidelity, like the Indo-Pacific bottlenose dolphin (T. aduncus) with a very distinct home range.46 Cetaceans are exposed to a number of risk factors,47,48,49,50,51,52,53,54,55,56 with 25% of marine mammals being on the brink of extinction.57 This number could be as high as 40%, however there is a lack of data to inform about the status of underreported species.53,57 One such risk factor is habitat degradation via terrestrial runoff and eutrophication, which has been associated with viruses, pathogenic bacteria, and parasites entering marine ecosystems. This causes disease outbreaks, such as lethal infections by Toxoplasma gondii in southern sea otters (Enhydra lutris nereis).58 Also, viral outbreaks like Phocine distemper virus which caused the death of 18,000 gray seals (Halichoerus grypus) in 1988,59 and the cetacean Morbillivirus (CeMV) which affects many dolphin species60 with outbreaks in Western Australia (2009) and Queensland (2010). A population of Burrunan dolphins (T. australis) exhibiting endemism in Victoria, Australia has experienced two mass mortality events due to Fresh Water Skin Disease (FWSD) caused by intense and extended La Niña periods of rainfall.61 This environmental change resulted in reduced salinity in their habitat, breaking down the skin barrier causing ulcerative lesions leading to secondary infection, organ failure and subsequent death.61 There is both a rise in the incidences of marine mammal diseases, and simultaneously a reduction in their immune health. Population decline also leads to loss of genetic diversity through allelic drop out and inbreeding, that impairs the species’ capacity to face and adapt to environmental stressors.62 Extrinsic challenges along with reduced genetic variation increase the populations’ and species’ susceptibility to illness including infectious diseases that can compromise individual health, reduce reproductive success, and cause mass mortality of cetaceans.63
This study provides exploratory insights into the genomic organisation and evolution of the MHC genes in cetaceans by using available genomes on the National Center for Biotechnology Information (NCBI) database. The study focused on the coding sequence from exons two, three and four from MHC Class I, with exons two and three being integral components of the peptide binding region (PBR).3,64 Due to the trans-specific nature of the MHC evolution, it was foremost expected that the sequences would cluster according to loci, indicating the timing of appearance in the genome and their evolutionary lineage. To investigate this theory, the study focused on species with chromosome-level genome assemblies in NCBI to characterise the MHC Class I gene region and its evolution in Cetaceans.
Results
Genomic organisation and characterisation of MHC Class I in cetaceans
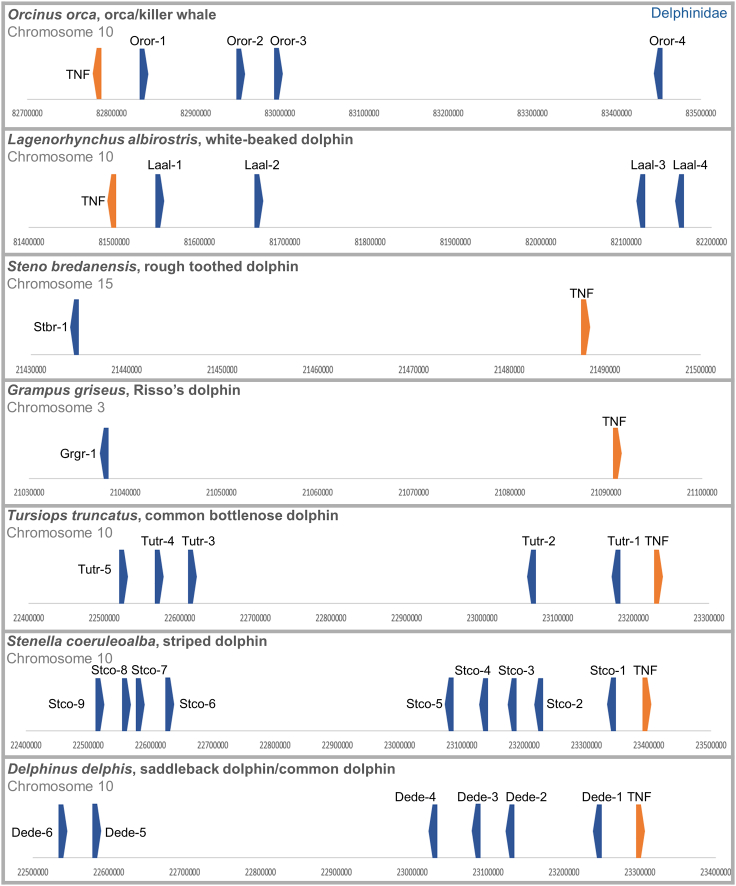
The MHC Class I loci were present on different chromosomes, with varied number of loci across genera, which is summarised in Table 1 and visualised in Figures 1, 2, and 3 (see Table 2 for species names and genome information). The loci were named according to the IPD-MHC non-human naming system,65,66 and numbered according to appearance in the genome, with respect to a single copy gene, Tumor Necrosis Factor (TNF) for orientation. For example, ‘Tutr-1’ denotes Tursiops truncatus, locus one. The pseudogenes were labeled with an asterisk to indicate that they are non-functional. All functional MHC loci appeared on the same chromosome; described hereafter as the MHC Class I region. The MHC Class I region was found on chromosome 10 in most species (10 out of 18 spp.), however it also appeared on chromosome 3 (Risso’s dolphin), 7 (North Atlantic right whale), 11 (blue whale, Rice’s whale, vaquita), 12 (gray whale), 15 (rough toothed dolphin), and 18 (sperm whale). Striped dolphin had the highest number of presumed functional loci with nine, while Risso’s dolphin and the pygmy sperm whale had the least with one locus each.
Table 1.
Number and location of loci
| Species | Common name | Abbreviation | Chromosome | Number of loci: functional (pseudo) |
|---|---|---|---|---|
| Balaenoptera acutorostrata scammoni | minke whale | Baac | 10 | 4(1) |
| 9 | (1) | |||
| Balaenoptera musculus | blue whale | Bamu | 11 | 2(2) |
| 8 | (1) | |||
| Balaenoptera ricei | rice’s whale | Bari | 11 | 3(2) |
| 8 | (1) | |||
| Delphinus delphis | saddleback dolphin (common dolphin) | Dede | 10 | 6 |
| 8 | (1) | |||
| Eschrichtius robustus | gray whale | Esro | 12 | 3(2) |
| 11 | (1) | |||
| Eubalaena glacialis | North Atlantic right whale | Eugl | 7 | 3(2) |
| 10 | (1) | |||
| Grampus griseus | Risso’s dolphin | Grgr | 3 | 1 |
| 9 | (1) | |||
| Hyperoodon ampullatus | Northern bottlenose whale | Hyam | 10 | 3 |
| 7 | (1) | |||
| Kogia breviceps | pygmy sperm whale | Kobr | 10 | 1 |
| 7 | (1) | |||
| Lagenorhynchus albirostris | white-beaked dolphin | Laal | 10 | 4 |
| 9 | (1) | |||
| Mesoplodon densirostris | Blainville’s beaked whale | Mede | 10 | 4 |
| 7 | (1) | |||
| Neophocaena sunameri | East Asian finless porpoise | Nesu | 10 | 5 |
| 8 | (1) | |||
| Orcinus orca | Orca | Oror | 10 | 4 |
| 8 | (1) | |||
| Phocoena sinus | Vaquita | Phsi | 11 | 2(1) |
| 8 | (1) | |||
| Physeter catodon | sperm whale | Phca | 18 | 2(1) |
| 16 | (1) | |||
| Stenella coeruleoalba | striped dolphin | Stco | 10 | 9 |
| Steno bredanensis | rough toothed dolphin | Stbr | 15 | 1 |
| 2 | (1) | |||
| Tursiops truncatus | common bottlenose dolphin | Tutr | 10 | 5 |
| 8 | (1) |
Functional loci are bold, pseudogenes denoted by parentheses.
Figure 1.
Genomic organisation and characterisation of MHC Class I in cetaceans
Balaenidae and Balaenopteridae MHC Class I loci. Loci were obtained via NCBI’s BLAST function and includes exons two, three and four. Loci marked with an asterisk are pseudogenes. See also Table S1 for gene lengths, premature stop codons and indels.
Figure 2.
Genomic organisation and characterisation of MHC Class I in cetaceans
Physeteridae, Kogiidae, Ziphiidae, Neophocaena and Phocoenidae MHC Class I loci. Loci were obtained via NCBI’s BLAST function and includes exons two, three and four. Loci marked with an asterisk are pseudogenes. See also Table S1 for gene lengths, premature stop codons and indels.
Figure 3.
Genomic organisation and characterisation of MHC Class I in cetaceans
Delphinidae MHC Class I loci. Loci were obtained via NCBI’s BLAST function and includes exons two, three and four. Loci marked with an asterisk are pseudogenes. See also Table S1 for gene lengths, premature stop codons and indels.
All species appeared to have an isolated pseudogene locus on a separate chromosome to the MHC Class I region. This pseudogene had a truncated exon 2 (39-43bp), and exon 3 was sometimes missing, therefore it was not included in the MHC genomic maps. Pseudogenes were defined by premature stop codons, as well as frameshift indels that resulted in premature stop codons. Mysticetes (baleen whales) appeared to have the most pseudogenes with the minke whale, blue whale, rice’s whale, gray whale, and North Atlantic right whale having one to two pseudogenes each in the MHC Class I region. Of the Odontocetes (toothed whales), sperm whale had one pseudogene in the MHC Class I region that contained a large insertion (21bp) which was not present in any other species. Locus three in Mysticetes and locus one in Physter (Bari-3∗, Bamu-3∗, Esro-3∗, Baac-3 and Phca-1∗) included a 5bp frameshift insertion, which also resulted in premature stop codons. A pseudogene was present in the Vaquita’s MHC Class I region which contained four 1bp insertions along the gene. The gene lengths, premature stop codons and indels are summarised in Table S1.
This study used a single copy gene, TNF, to orientate in the genome, and to determine synteny. This allowed the identification of several inversion events of the MHC Class I region, Figures 1, 2, and 3. For instance, during the divergence of the gray whale and the blue whale, the region was flipped (Figure 1). The relative location of the genes also allowed us to view where there may have been loss and gain of loci. In Balaenopteridae (Figure 1), for example, the species have two loci located near TNF, with two to three loci located a greater distance along the chromosome, in close association with each other. Comparably, the sperm whale (Figure 2) lacks the two loci near TNF but maintains the three loci (including a pseudogene) further along the chromosome.
Evolution and phylogenetic reconstruction of MHC Class I in cetaceans
MHC diverged before the speciation of cetaceans, evidenced by the species’ loci being scattered across the tree and, for the most part, not clustering according to species (Figure 4). For example, in clade C sequences were present from both Mysticetes (Eugl-5, Bamu-4, Bari-4) and Odontocetes (Tutr-5, Stco-8, Oror-2, Oror-4). An exception to this pattern can be seen in subclade D where loci 2–5 of the East Asian finless porpoise (Nesu-2-5) group together, along with two loci of the vaquita (Phsi-2-3∗). The grouping of the East Asian finless porpoise is a part of a larger clade (clade E) including loci from the Northern bottlenose whale (Hyam-2-3), and Blainville’s beaked whale (Mede-2-4). Therefore, despite the East Asian finless porpoise loci grouping together, there remains a trans-specific pattern of evolution overall.
Figure 4.
Evolution and phylogenetic reconstruction of MHC Class I in cetaceans
Maximum likelihood phylogenetic tree generated using aLRT statistics with branch lengths shown. Scale bar indicates evolutionary distance. Nodes denote loci, with the loci name being derived from the first two letters of the species’ scientific name and the number indicating location in the genome (Tutr-1 indicates Tursiops truncatus, locus one). Loci with an asterisk are pseudogenes. Clades discussed in the text are labeled (A–H).
Locus one from most of the species studied formed a clade (clade F), though the clade was paraphyletic, as it did not include sperm whale, pygmy sperm whale or the rough toothed dolphin (Figure 4). The clade also included locus five from the saddleback dolphin (Dede-5) and locus four from the gray whale (Esro-4), however, locus one in these two species was also present in the clade (Dede-1, Esro-1). Within clade F, Dede-5, Tutr-1 and Stco-1 cluster together (labeled subclade G). In addition, within clade F, subclade H has included six families: Balaenidae, Balaenopteridae, Ziphiidae, Phocoenidae, Neophocaena and Delphinidae that had a branching pattern wherein delphinids and porpoises diverged from Mysticetes and Ziphiidae.
The pseudogenes in Mysticetes formed a monophyletic clade, clade B (Figure 4), which included locus 3 from minke whale (Baac-3∗), gray whale (Esro-3∗), blue whale (Bamu-3∗), rice’s whale (Bari-3∗) and North Atlantic right whale (Eugl-3∗). It also included locus 1 from sperm whale (Phca-1∗), being the most ancestral node for this clade. The clade had the longest branch length of the whole tree (0.17). Another monophyletic clade, clade A, included the pseudogenes on locus two of the Mysticetes (Eugl-2∗, Bari-2∗, Bamu-2∗, Esro-2∗) as well as loci two and five from the minke whale (Baac-2, Baac-5). Interestingly, Baac-2 and Baac-5 retained function at the locus, while the Mysticetes had a pseudogene.
Discussion
Genomic mapping of MHC Class I in cetaceans
Most often, the functional loci were found on chromosome 10, however they were also located on various other chromosomes across genera (Figures 1, 2, and 3). Additional to the MHC Class I region found on a single chromosome, an isolated pseudogene was located on a separate chromosome, a pattern contrasting MHC Class II where all loci were located on the same chromosome.31,32 This study identified varied numbers of MHC Class I loci across cetacean species, from one locus (Risso’s dolphin and pygmy sperm whale) to nine (striped dolphin). This was similar to those reported by Xu et al., (2009),67 who found eight striped dolphin alleles and two alleles in the Risso’s dolphin. Indeed, previous studies of the MHC Class I genes in cetaceans have identified at least three loci in five species, including the Yangtze River dolphin, gray whale, Hector’s dolphin, Yangtze finless porpoise, and North Atlantic Right whale,27,33,34,35,36,37 often identifying 3–5 unique alleles per individual. The MHC loci in cetaceans also tends to be fewer than terrestrial ungulates, for instance, the loss of the MHC Class II DY locus in cetaceans and only one DQ locus compared to terrestrials with a variable number of DQ loci.28,31,32 Class I in bovines may be similar in size to that of humans, with at least six loci expressed.68 Fan et al., (2019)69 also found that the sperm whale had lost 73 MHC genes compared to cattle. This may be due to adaption to marine ecological niche, as well as distribution, transmission and diversity of both marine mammals and marine pathogens differing from terrestrial mammals.
Understanding the number of MHC loci is useful for conducting population genetic analysis, by providing context to what is meant by ‘high’ and ‘low’ genetic diversity. It also gives insight into the immunological evolutionary pressure on a species, even between closely related taxa. Our study identified five loci in the East Asian finless porpoise, while four loci where previously described by Ruan et al., (2016)36 in the Yangtze finless porpoise. The pattern of closely related taxa having highly diverged MHC has been reported previously20,67 and could be explained by different ecological and ethological processes influencing the tempo and mode of MHC evolution.18 Evidence suggests that although cetaceans are geographically dispersed, there is still strong influence of environment-type in their evolution and adaption.70 For instance, populations of the Tursiops genus that live close to each other, but in differing environmental conditions (i.e., enclosed bay versus open coastal), can cause distinct population differentiation due to low gene flow.71,72 There are likely multiple complex factors at play causing the divergence of MHC in cetacean species, including but not limited to, pathogenic load,6 species distribution,21 movement patterns,73 social group size,18 inter species interactions and sexual selection.11 Indeed, it is not just pathogenic stress that determines MHC copy number.74 While the diversity of MHC relies on heterozygote advantage (overdominance hypothesis) and negative frequency dependent selection (rare allele advantage),18 the mechanism and strength of selection can vary by loci. Further testing of population ecology and life history traits, considering MHC diversity and evolution would be needed to determine the factors driving MHC evolution in cetaceans.
Our study identified sequences with stop codons and indels in some species, supporting the notion that not all loci are functional. A similar pattern was observed in North Atlantic right whales by Gillett et al., (2013)35 where half of the unique sequences included stop codons and indels. It has been noted before that there may be a higher presence of pseudogenes in Mysticetes than in Odontocetes.35,75 However, one study that investigated MHC Class I genes of 11 cetacean species found no stop codons or indels in the 3–5 alleles identified per individual.67 Future studies on MHC Class I in cetaceans should consider investigating the expression of the characterised loci to determine the functional relevance of the number of loci present, as evidenced in Hector’s dolphin in which the most common allele was also the most commonly expressed.34
The rapid evolution of the MHC resulting in different number of MHC loci across species makes the characterisation of this genomic region particularly challenging. While attempts to automate loci detection via machine learning have been made,40 manual annotation of these regions can provide more detailed and accurate characterisation of these complex loci. For example, Gambón-Deza (2020)40 identified six loci in the common bottlenose dolphin, while we detected five, and they found three loci in vaquita, however failed to identify that one is a pseudogene. In addition, similar patterns were also detected by the two approaches, such as locating the same two loci in the blue whale and sperm whale. The results obtained via differing methods highlights how manual annotation can finetune genome annotations, and that use of long-read sequencing may be necessary to characterise regions of the genome that are complex and evolving rapidly. Furthermore, these results also highlight the importance of detailed structural characterisation of the MHC gene region so that MHC markers can be used accurately and meaningfully in population level studies.
Phylogenetic reconstruction of MHC Class I in cetaceans
It is well documented that the MHC diverged before the speciation of cetaceans,12,26,27,29,33,35,67 and this was further supported by our study. The presence of trans species allelism is indicative of selection acting on the MHC loci, whereby the divergence of species is delayed (with respect to the MHC), and alleles are maintained across taxa.18 That being said, advantageous non-synonymous mutations and rare alleles may undergo positive selection. Locus one is both phylogenetically and structurally similar across cetacean species. For this reason, it is likely an ortholog between species, potentially having ecological relevance for most, with similar alleles being maintained by selection. For example, if there is a pathogen present in the species’ environment, alleles that favor detection of that pathogen may be retained, a known underlying factor in the evolution of MHC.12,26,37,76 MHC alleles can persist for at least 40 million years, if maintained by balancing selection,13,15,77 in fact, Xu et al., (2009)67 had estimated that MHC Class I polymorphism in cetaceans arose 56.9 MYA (±24.82). The phylogenetic relationships presented here support this hypothesis, showing multiple families contain similar alleles on locus one, despite the divergence of Mysticetes and Odontocetes approximately 40 MYA.78,79,80,81,82 Previous studies have compared the evolutionary relationship of MHC in cetaceans to terrestrial mammals with clades that group according to loci and placed distantly related taxa within these lineages.27,36,40,83 However, when only looking at the cetaceans, the fact that the gene divergence pre-dates the species divergence, as well as the rapid diversification, adaptive radiation and wide habitat ranges makes clustering by loci unclear. Similar patterns have been observed in Class II phylogeny also, where the divergence of the MHC genes in the Tursiops genus occurred well before the first emergence of dolphins.76 Nevertheless, other studies, such as Ruan et al., (2016)36 showed that the cetacean loci formed sister groups to terrestrial artiodactyls, providing evidence that the cetacean loci are paralogous of the terrestrial species.
We did, however, see clustering of the East Asian finless porpoise loci 2–5, along with vaquita loci 2–3∗, subclade D, Figure 4. A previous study by Xu et al., (2009)67 also detected distinct clustering of the Yangtze finless porpoise, when comparing to other cetaceans and terrestrial ungulates, and Gillett et al., (2013)35 observed strong clade formation of porpoise and dolphin alleles only. The porpoise species described in these studies have distinct home ranges, with small populations existing in close proximity to anthropogenic influences.84,85,86,87 It may be that there are unique and stronger selective pressures acting on these species, causing their distinction from other cetaceans. Interestingly, the species in clade E face quite different selective pressures, with the East Asian finless porpoise and the vaquita having limited home ranges, preferring warm, shallow coastal habitats.88,89 Conversely, the Northern bottlenose whale and Blainville’s beaked whale are offshore, deep diving animals.90,91 The pattern observed here may be due to speciation, since Phocoenidae and Ziphiidae are sister taxa, falling between Balaenids and Delphinids. What we see here contrasts with some hypotheses that immune gene diversity can be driven by habitat type and proximity to land.76 There has previously been evidence of geographical isolation of beaked whales via an equatorial barrier70 which may contribute to their divergence from other species. Gray whales, a predominantly offshore species, have also been found to have low polymorphism at Class I loci,83 which may be due to less exposure to a variety of pathogens (or due to adaptive response to a particular fitness limiting pathogen). On the other hand, high MHC Class II diversity was observed in Yangtze River dolphins that otherwise have low genetic diversity,8,25 potentially owing to increased pathogenic stress as a species residing in a highly polluted region. There has been cause to suggest that proximity to land and exposure to anthropogenic-induced pathogenic stress could influence the immune health and evolution of cetaceans and other marine mammals.21,44,92,93,94,95,96 The pattern observed in this study, and ecological pathogen exposure hypothesis may be further explored by increasing sample size, including more species with small ranges in coastal, inshore habitats.
Of note, the pygmy sperm whale and sperm whale are unusual in their MHC organisation and evolution. The pygmy sperm whale had only one MHC locus, while the sperm whale was the only species to have MHC loci located on chromosome 18. Both species, were absent from clade F, which contained most species’ locus one. The sperm whale’s locus one included two insertions and premature stop codons, clustering with the Myticetes locus three, all of which were pseudogenes. The A 6Mb MHC region was identified by Fan et al., (2019)69 also on chromosome 18 in the sperm whale, including Class I, II and III genes. The unusual organisation of MHC genes in the sperm whale genome compared to other cetaceans may originate from karyotype differences across species (sperm whales having two less chromosomes than most species97), and/or from the sperm whale being the only surviving species of toothed whales in the genus Physeter.79 Low numbers of MHC loci in both the sperm whale and pygmy sperm whale could be of conservation concern, since we know that high heterozygosity at the MHC loci can provide a selective advantage.7
Cetacean species are more at risk now than they ever have been with the rise of emerging pathogens and immune stressors caused anthropogenic impacts. Over the last 40 years, disease outbreaks among marine mammals have increased due to eutrophication and terrestrial pathogens spreading to the ocean.63 Exposure to freshwater runoff is linked to skin diseases leading to secondary infection and mortality.61 Certainly, the increase in cetacean disease and exposure to immune stressors brings into question the ability of cetacean species to adapt to these challenges. MHC is a useful indicator of population fitness, especially when compared with neutral genetic markers.3 With the rise of disease and reduction in habitat quality for many cetacean populations, genomic mapping of the MHC and its evolution advances our knowledge on the adaption and potential immune risks that species face. For instance, will species with low MHC copy number who are exposed to a high number of pathogens and other immune stressors be able to cope with under immune stress or when faced with a new pathogen? It is therefore crucial to gain insight into the complexity, diversity, and adaptability of the MHC for each species.
This paper is the first to characterise the genomic location and organisation of the MHC Class I genes across multiple cetacean species, information that is essential for future population and conservation genetics studies on members of this infraorder. We found varied numbers and locations of MHC Class I genes across cetacean species, and further confirmed that the evolution of MHC is complex, rapid and differs even between closely related species. Future investigation should focus on the factors that drive the evolution of MHC in Cetacea, with a particular focus on habitat and proximity to terrestrial runoff. Moreover, expression analysis of MHC loci would be beneficial to understand the functionality of multiple Class I genes in cetaceans.
Limitations of the study
The emergence of NGS technologies has meant that high-quality genomes are freely available for analysis and allowed us to conduct this study. However, due to the extreme complexity of MHC gene family, only genome sequences that were generated with longread technologies can provide reliable source for gene and loci annotations. Since high quality genomes were only available from 18 species, more species would be ideal to be included in the analysis. Species from a range of niches, and information on the different pathogen communities could assist in determining the factors driving MHC evolution in cetaceans. As well as this, we were unable to identify genuine locus functionality, which would need to be confirmed with expression analysis, which was out of the scope of this study.
Glossary
Allele: alternative versions of a gene that may produce distinguishable phenotypic effects.
Cetacea/cetaceans: whales, dolphins, and porpoises.
Locus/loci: a specific place along the length of a chromosome where a given gene is located (plural, loci).
Major Histocompatibility Complex (MHC): trans-membrane receptor that functions in antigen presentation and pathogen recognition.
Paralog: homologous genes that are found in the same genome because of gene duplication.
Paraphyly: in phylogenetics, an incomplete grouping of descendants.
Monophyly: in phylogenetics, all descendants from a common ancestor form a group/clade.
Mysticetes: whales that lack teeth, having baleen instead.
Odontocetes: toothed whales.
Trans-specific: genetic variants’ origin predates speciation event(s).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Eubalaena glacialis, North Atlantic right whale reference genome mEubGla1.hap1 | GenBank | GCA_028571275.1 |
| Balaenoptera acutorostrata scammony, minke whale reference genome mBalAcu1.1 | GenBank | GCA_949987535.1 |
| Balaenoptera musculus, blue whale reference genome mBalMus1.pri.v3 | GenBank | GCA_009873245.3 |
| Balaenoptera ricei, rice’s whale reference genome mBalRic1.hap2 | GenBank | GCA_028023285.1 |
| Eschrichtius robustu, gray whale reference genome mEscRob2.pri | GenBank | GCA_028021215.1 |
| Delphinus delphis, saddleback dolphin (common dolphin) reference genome ASM3006286v1 | GenBank | GCA_030062865.1 |
| Grampus griseus, Risso’s dolphin reference genome Grampus_griseus_HiC | GenBank | GCA_028646425.1 |
| Lagenorhynchus albirostris, white-beaked dolphin reference genome mLagAlb1.1 | GenBank | GCA_949774975.1 |
| Orcinus orca, orca/killer whale reference genome mOrcOrc1.1 | GenBank | GCA_937001465.1 |
| Stenella coeruleoalba, striped dolphin reference genome mSteCoe1.1 | GenBank | GCA_951394435.1 |
| Steno bredanensis, rough toothed dolphin reference genome Steno_bredanensis_HiC | GenBank | GCA_028646385.1 |
| Tursiops truncates, common bottlenose dolphin reference genome mTurTru1.mat.Y | GenBank | GCF_011762595.1 |
| Kogia breviceps, pygmy sperm whale reference genome mKogBre1 haplotype 1 | GenBank | GCA_026419965.1 |
| Neophocaena sunameri, East Asian finless porpoise reference genome ASM2622585v1 | GenBank | GCA_026225855.1 |
| Phocoena sinus, vaquita reference genome mPhoSin1.pri | GenBank | GCA_008692025.1 |
| Physeter catodon, sperm whale reference genome ASM283717v5 | GenBank | GCA_002837175.5 |
| Hyperoodon ampullatus, Northern bottlenose whale reference genome mHypAmp2.1 | GenBank | GCA_949752795.1 |
| Mesoplodon densirostris, Blainville’s beaked whale reference genome mMesDen1_primary_haplotype | GenBank | GCA_025265405.1 |
| Software and algorithms | ||
| Basic Local Alignment Search Tool (BLAST) | Altschul et al., (1990)98 | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| Geneious Prime 2021.2.2 | Drummond et al., (2010)99 | https://www.geneious.com/ |
| Multiple Sequence Comparison by Log-Expectation (MUSCLE) alignment algorithm | Edgar, R.C., (2004)100 | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC390337/ |
| PhyML 3.3.20180621 | Guindon et al., (2010)101 | http://www.atgc-montpellier.fr/phyml/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and fulfilled by the lead contact, Beata Ujvari (beata.ujvari@deakin.edu.au).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
This paper analyses existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
Experimental model and study participant details
This article does not contain any studies with all the experimental models (animals, human subjects, plants, microbe strains, cell lines, primary cell cultures) performed by any of the authors. See Table 2 for species and genomic information. There is no expected bias in the results based on sex or gender.
Table 2.
Genomes used and assembly information, including number of BLAST hits obtained from NCBI
| Species | Common name | Abbreviation | Family | Sample collection location | Assembly name | Assembly accession | Date | Coverage | Assembly method | Sequencing technology | Number of hits |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Eubalaena glacialis | North Atlantic right whale | Eugl | Balaenidae | USA: Amelia Island, Florida | mEubGla1.hap1 | GCA_028571275.1 | 9/02/2023 | 32.0x | Hifiasm + Hi-C phasing v. 0.16.1 + galaxy2; Bionano Solve v. 3.7.0 + galaxy0; salsa v. 2.3 + galaxy3 | PacBio Sequel II HiFi; Bionano DLS; 3D DNA Hi-C | 28 |
| Balaenoptera acutorostrata scammoni | minke whale | Baac | Balaenopteridae | South Korea | mBalAcu1.1 | GCA_949987535.1 | 1/05/2023 | 30x | Various | PacBio, Arima2 | 23 |
| Balaenoptera musculus | blue whale | Bamu | Balaenopteridae | USA: Pacific Ocean, Santa Barbara | mBalMus1.pri.v3 | GCA_009873245.3 | 2/10/2020 | 51.16x | FALCON v. 1.0.2; FALCON-Unzip v. 1.0.2; purge_haplotigs v. bitbucket 7.10.2018; scaff10x v. git 4.28.2018; Bionano Solve DLS v. 3.2.1; Salsa2 HiC v. 2.0; Arrow polishing and gap filling v. 5.1.0.26412; longranger align v. 2.2.2; freebayes v. 1.2.0; gEVAL manual curation v. 2019-10-25; VGP standard assembly pipeline v. 1.5; Additional Purge_dup and Polish v. 2020-05-28 | PacBio Sequel I; Illumina NovaSeq; 10X Genomics chromium; Dovetail Genomics HiC | 23 |
| Balaenoptera ricei | rice’s whale | Bari | Balaenopteridae | USA: Gulf of Mexico, San Destin, Florida | mBalRic1.hap2 | GCA_028023285.1 | 30/01/2023 | 30.0x | Hifiasm + Hi-C phasing v. 0.16.1 + galaxy3; Bionano Solve v. 3.7.0 + galaxy0; yahs v. 1.2a + galaxy1 | PacBio Sequel II HiFi | 27 |
| Eschrichtius robustus | gray whale | Esro | Balaenopteridae | USA: North Pacific Ocean, near Crescent City, California | mEscRob2.pri | GCA_028021215.1 | 31/01/2023 | 29.61x | HiFiasm v. 0.16.1 + galaxy3; yahs v. 1.2a + galaxy2 | PacBio Sequel II HiFi; 3D-DNA Hi-C | 25 |
| Delphinus delphis | saddleback dolphin (common dolphin) | Dede | Delphinidae | NA | ASM3006286v1 | GCA_030062865.1 | 24/05/2023 | 32.6x | FALCON v. 0.3; SOAPdenovo v. 2.04 | Illumina NovaSeq; PacBio Sequel | 33 |
| Grampus griseus | Risso’s dolphin | Grgr | Delphinidae | NA | Grampus_griseus_HiC | GCA_028646425.1 | 16/02/2023 | 131x | 3D-DNA v. 2020; JBAT v. 2020 (see dnazoo.org/methods) | Illumina NovaSeq 6000 | 9 |
| Lagenorhynchus albirostris | white-beaked dolphin | Laal | Delphinidae | UK: East Lothian, Morrisons Haven, Scotland | mLagAlb1.1 | GCA_949774975.1 | 8/04/2023 | 33x | Various | PacBio, Arima2 | 25 |
| Orcinus orca | orca | Oror | Delphinidae | Netherlands: Hardewijk Dolphinarium North Atlantic Ocean type |
mOrcOrc1.1 | GCA_937001465.1 | 3/05/2022 | 34x | NA | NA | 26 |
| Stenella coeruleoalba | striped dolphin | Stco | Delphinidae | UK: Highland, Ardmair, Scotland | mSteCoe1.1 | GCA_951394435.1 | 16/06/2023 | 35x | Various | PacBio, Arima2 | 49 |
| Steno bredanensis | rough toothed dolphin | Stbr | Delphinidae | NA | Steno_bredanensis_HiC | GCA_028646385.1 | 16/02/2023 | 95x | 3D-DNA v. 2020; JBAT v. 2020 (see dnazoo.org/methods) | Illumina NovaSeq 6000 | 10 |
| Tursiops truncatus | common bottlenose dolphin | Tutr | Delphinidae | USA: Baltimore, Maryland | mTurTru1.mat.Y | GCF_011762595.1 | 27/03/2020 | 63.7x | TrioCanu v. 1.8; purge_dups v. 1.0.0; Scaff 10x v. 4.1.0; Bionano solve v. 3.2.1_04122018; Salsa2 HiC v. 2.2; Arrow polishing and gap filling v. smrtlink_6.0.0.47841; Freebayes v. 1.3.1; gEVAL manual curation v. 2020-01-15; VGP trio assembly pipeline v. 1.6 | PacBio Sequel I CLR; Illumina NovaSeq; Arima Genomics Hi-C; Bionano Genomics DLS | 28 |
| Kogia breviceps | pygmy sperm whale | Kobr | Kogiidae | USA: La Jolla, California | mKogBre1 haplotype 1 | GCA_026419965.1 | 27/11/2022 | 23.0x | Hifiasm + Hi-C phasing v. 0.16.1 (r375) + galaxy1; Bionano solve v. 3.7; salsa v. 2.3 | NA | 14 |
| Neophocaena sunameri | East Asian finless porpoise | Nesu | Neophocaena | China: Yellow Sea, near Lianyungang City | ASM2622585v1 | GCA_026225855.1 | 18/11/2022 | 62.0x | hifiasm v. JULY-2022 | PacBio | 32 |
| Phocoena sinus | vaquita | Phsi | Phocoenidae | Mexico: San Felipe, Baja California | mPhoSin1.pri | GCA_008692025.1 | 26/09/2019 | 67.26x | FALCON v. 5.1.1; FALCON-Unzip v. 1.0.2; Arrow smrtanalysis Pacbio polishing v. 6.0.0.47841; purge_haplotigs v. 1.0.4; scaff10X v. 4.1; Bionano solve DLS v. 3.3_10252018; Salsa HiC v. 2.2; Arrow smrtanalysis Pacbio polishing & gap filling v. 6.0.0.47841; longranger align v. 2.2.2; freebayes Illumina polishing v. 1.2.0; gEVAL manual curation v. 2019-07-23; VGP assembly pipeline individual v. 1.5 | PacBio Sequel I; Illumina NovaSeq; Arima Genomics Hi-C; Bionano Genomics DLE-1 | 19 |
| Physeter catodon | sperm whale | Phca | Physeteridae | NA | ASM283717v5 | GCA_002837175.5 | 28/02/2023 | 248x | SOAPdenovo v. SEP-2017; ARCS pipeline v. SEP-2017; Supernova v. SEP-2017 | BGISEQ-500 | 19 |
| Hyperoodon ampullatus | Northern bottlenose whale | Hyam | Ziphiidae | Canada: Nova Scotia, Halifax Gully Marine Protected Area | mHypAmp2.1 | GCA_949752795.1 | 8/04/2023 | 28x | Various | PacBio, Arima2 | 19 |
| Mesoplodon densirostris | Blainville’s beaked whale | Mede | Ziphiidae | USA: Melbourne Beach, Florida | mMesDen1_primary_haplotype | GCA_025265405.1 | 19/09/2022 | 43.5x | Hifiasm v. 0.16.1+galaxy1; purge_dups v. 1.2.5+galaxy3; Bionano solve v. 3.7.0+galaxy0; Salsa v. 2.3+galaxy2 | NA | 24 |
Only genomes with chromosome level assembly were used. Common and Latin names listed are based on records in NCBI GenBank.
Method details
Sequences were obtained from NCBI by using the BLAST98 search function from chromosome-level genomes of 18 cetacean species, across eight families (refer to Table 2 for scientific names and detailed information on the species genomes used in this study). A previously described 704bp MHC Class I sequence (Genbank: EU024810.1) from a Hector’s dolphin, obtained from cDNA was used to BLAST the genomes, searching for somewhat similar sequences. Using Geneious Prime version 2021.2.299 BLAST hits from exons 2, 3 and 4 were concatenated to form the full-length target sequence. BLAST searches resulted in several hits within the given genome, and these hits were included as they represent potential MHC loci. However, if the hit didn’t contain all three target exons, was too short (<50bp) or had an identity <70%, it was excluded from further analysis. As such, one locus was identified on a different chromosome, separate from other loci. This locus had a truncated exon 2 (39-43bp) and exon 3 was sometimes missing, therefore it was excluded from further analysis. A single copy gene, TNF, was used to orientate loci within the genome. BLAST hit data is recorded in Table S2.
Sequences were aligned using Multiple Sequence Comparison by Log-Expectation (MUSCLE) algorithm.100 The alignment was also translated into reading frame one to view functionally relevant amino acid sequences. The nucleotide alignment was used to build a Maximum-Likelihood (ML) tree using PhyML 3.3.20180621.101 The tree was constructed using the TN93 substitution model and aLRT statistics, with a fixed proportion of invariable sites at zero, an estimated transition/transversion ratio and four substitution categories. Cattle was chosen as the outgroup for the cetacean evolutionary tree as it is part of the Artiodactyl order, sister taxa to cetaceans and due to the likelihood that MHC diverged before the speciation of cetaceans.31
Quantification and statistical analysis
This study defined ‘n’ as number of species (18). Information on the alignment and phylogenetic parameters can be found in the method details. These approaches did not require a formal statistical analysis.
Acknowledgments
Author contributions
Conceptualisation, G.A.D., K.R., B.U.; Methodology, G.A.D, K.R, B.U.; Formal Analysis, G.A.D.; Investigation, G.A.D.; Data Curation, G.A.D.; Visualisation, G.A.D.; Writing – Original Draft, G.A.D.; Writing – Review & Editing, K.R., A.O., M.T.S, B.U.; Supervision, K.R., B.U.; Project Administration, K.R., B.U.
Declaration of interests
The authors declare no competing interests.
Published: March 27, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109590.
Supplemental information
References
- 1.Madden D.R. The 3-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 2.Janeway C.J., Travers P., M W. Garland Science; 2001. The Major Histocompatibility Complex and its Functions. [Google Scholar]
- 3.Manlik O., Krützen M., Kopps A.M., Mann J., Bejder L., Allen S.J., Frère C., Connor R.C., Sherwin W.B. Is MHC diversity a better marker for conservation than neutral genetic diversity? A case study of two contrasting dolphin populations. Ecol. Evol. 2019;9:6986–6998. doi: 10.1002/ece3.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ujvari B., Belov K. Major histocompatibility complex (MHC) markers in conservation biology. Int. J. Mol. Sci. 2011;12:5168–5186. doi: 10.3390/ijms12085168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front. Zool. 2005;2:16. doi: 10.1186/1742-9994-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borghans J.A.M., Beltman J.B., De Boer R.J. MHC polymorphism under host-pathogen coevolution. Immunogenetics. 2004;55:732–739. doi: 10.1007/s00251-003-0630-5. [DOI] [PubMed] [Google Scholar]
- 7.Penn D.J., Damjanovich K., Potts W.K. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl. Acad. Sci. USA. 2002;99:11260–11264. doi: 10.1073/pnas.162006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedrick P.W. Pathogen resistance and genetic variation at MHC loci. Evolution. 2002;56:1902–1908. doi: 10.1111/j.0014-3820.2002.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 9.Kalbe M., Eizaguirre C., Dankert I., Reusch T.B.H., Sommerfeld R.D., Wegner K.M., Milinski M. Lifetime reproductive success is maximized with optimal major histocompatibility complex diversity. Proc. Biol. Sci. 2009;276:925–934. doi: 10.1098/rspb.2008.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas M.L., Harger J.H., Wagener D.K., Rabin B.S., Gill T.J. HLA sharing and spontaneous abortion in humans. Am. J. Obstet. Gynecol. 1985;151:1053–1058. doi: 10.1016/0002-9378(85)90379-5. [DOI] [PubMed] [Google Scholar]
- 11.Eizaguirre C., Yeates S.E., Lenz T.L., Kalbe M., Milinski M. MHC-based mate choice combines good genes and maintenance of MHC polymorphism. Mol. Ecol. 2009;18:3316–3329. doi: 10.1111/j.1365-294X.2009.04243.x. [DOI] [PubMed] [Google Scholar]
- 12.Villanueva-Noriega M.J., Baker C.S., Medrano-González L. Evolution of the MHC-DQB exon 2 in marine and terrestrial mammals. Immunogenetics. 2013;65:47–61. doi: 10.1007/s00251-012-0647-8. [DOI] [PubMed] [Google Scholar]
- 13.Aguilar A., Roemer G., Debenham S., Binns M., Garcelon D., Wayne R.K. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proc. Natl. Acad. Sci. USA. 2004;101:3490–3494. doi: 10.1073/pnas.0306582101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumánovics A., Takada T., Lindahl K.F. Genomic organization of the mammalian MHC. Annu. Rev. Immunol. 2003;21:629–657. doi: 10.1146/annurev.immunol.21.090501.080116. [DOI] [PubMed] [Google Scholar]
- 15.Hughes A.L., Yeager M. Natural selection and the evolutionary history of major histocompatibility complex loci. Front. Biosci. 1998;3:d509–d516. doi: 10.2741/a298. [DOI] [PubMed] [Google Scholar]
- 16.Bernatchez L., Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J. Evol. Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- 17.Kelley J., Walter L., Trowsdale J. Comparative genomics of major histocompatibility complexes. Immunogenetics. 2005;56:683–695. doi: 10.1007/s00251-004-0717-7. [DOI] [PubMed] [Google Scholar]
- 18.Piertney S.B., Oliver M.K. The evolutionary ecology of the major histocompatibility complex. Heredity. 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. [DOI] [PubMed] [Google Scholar]
- 19.Van Oosterhout C., Joyce D.A., Cummings S.M., Blais J., Barson N.J., Ramnarine I.W., Mohammed R.S., Persad N., Cable J. Balancing selection, random genetic drift, and genetic variation at the major histocompatibility complex in two wild populations of guppies (Poecilia reticulata) Evolution. 2006;60:2562–2574. doi: 10.1554/06-286.1. [DOI] [PubMed] [Google Scholar]
- 20.Anzai T., Shiina T., Kimura N., Yanagiya K., Kohara S., Shigenari A., Yamagata T., Kulski J.K., Naruse T.K., Fujimori Y., et al. Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc. Natl. Acad. Sci. USA. 2003;100:7708–7713. doi: 10.1073/pnas.1230533100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen T., Xu S., Wang X., Yu W., Zhou K., Yang G. Adaptive evolution and functional constraint at TLR4 during the secondary aquatic adaptation and diversification of cetaceans. BMC Evol. Biol. 2012;12:39. doi: 10.1186/1471-2148-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mccallum H., Kuris A., Harvell C., Lafferty K., Smith G., Porter J. Does terrestrial epidemiology apply to marine systems? Trends Ecol. Evol. 2004;19:585–591. doi: 10.1016/j.tree.2004.08.009. [DOI] [Google Scholar]
- 23.Suttle C.A. Marine viruses — major players in the global ecosystem. Nat. Rev. Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 24.Murray B.W., Malik S., White B.N. Sequence variation at the major histocompatibility complex locus DQ beta in beluga whales (Delphinapterus leucas) Mol. Biol. Evol. 1995;12:582–593. doi: 10.1093/oxfordjournals.molbev.a040238. [DOI] [PubMed] [Google Scholar]
- 25.Yang G., Yan J., Zhou K., Wei F. Sequence variation and gene duplication at MHC DQB Loci of Baiji (Lipotes vexillifer), a Chinese River dolphin. J. Hered. 2005;96:310–317. doi: 10.1093/jhered/esi055. [DOI] [PubMed] [Google Scholar]
- 26.Martínez-Agüero M., Flores-Ramírez S., Ruiz-García M. First report of major histocompatibility complex class II loci from the Amazon pink river dolphin (genus Inia) Genet. Mol. Res. 2006;5:421–431. [PubMed] [Google Scholar]
- 27.Xu S., Sun P., Zhou K., Yang G. Sequence variability at three MHC loci of finless porpoises (Neophocaena phocaenoides) Immunogenetics. 2007;59:581–592. doi: 10.1007/s00251-007-0223-9. [DOI] [PubMed] [Google Scholar]
- 28.Ruan R., Ruan J., Wan X.L., Zheng Y., Chen M.M., Zheng J.S., Wang D. Organization and characteristics of the major histocompatibility complex class II region in the Yangtze finless porpoise (Neophocaena asiaeorientalis asiaeorientalis) Sci. Rep. 2016;6 doi: 10.1038/srep22471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreno-Santillán D.D., Lacey E.A., Gendron D., Ortega J. Genetic variation at exon 2 of the MHC class II DQB locus in blue whale (Balaenoptera musculus) from the Gulf of California. PLoS One. 2016;11 doi: 10.1371/journal.pone.0141296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X., Lin W., Zhou R., Gui D., Yu X., Wu Y. Low major histocompatibility complex class II variation in the endangered Indo-Pacific humpback dolphin (Sousa chinensis): Inferences about the role of balancing selection. J. Hered. 2016;107:143–152. doi: 10.1093/jhered/esv138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Sá A.L.A., Breaux B., Burlamaqui T.C.T., Deiss T.C., Sena L., Criscitiello M.F., Schneider M.P.C. The marine mammal class II major histocompatibility complex organization. Front. Immunol. 2019;10:696. doi: 10.3389/fimmu.2019.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z., Sun X., Chen M., Li L., Ren W., Xu S., Yang G. Genomic organization and phylogeny of MHC class II loci in cetaceans. J. Hered. 2019;110:332–339. doi: 10.1093/jhered/esz005. [DOI] [PubMed] [Google Scholar]
- 33.Flores-Ramirez S., Urban-Ramirez J., Miller R.D. Major histocompatibility complex class I loci from the gray whale (Eschrichtius robustus) J. Hered. 2000;91:279–282. doi: 10.1093/jhered/91.4.279. [DOI] [PubMed] [Google Scholar]
- 34.Heimeier D., Baker C.S., Russell K., Duignan P.J., Hutt A., Stone G.S. Confirmed expression of MHC class I and class II genes in the New Zealand endemic Hector's dolphin (Cephalorhynchus hectori) Mar. Mamm. Sci. 2009;25:68–90. doi: 10.1111/j.1748-7692.2008.00244.x. [DOI] [Google Scholar]
- 35.Gillett R.M., Murray B.W., White B.N. Characterization of class I– and class II–Like major histocompatibility complex loci in pedigrees of North Atlantic Right Whales. J. Hered. 2014;105:188–202. doi: 10.1093/jhered/est095. [DOI] [PubMed] [Google Scholar]
- 36.Ruan R., Wan X.-L., Zheng Y., Zheng J.-S., Wang D. Assembly and characterization of the MHC class I region of the Yangtze finless porpoise (Neophocaena asiaeorientalis asiaeorientalis) Immunogenetics. 2016;68:77–82. doi: 10.1007/s00251-015-0885-7. [DOI] [PubMed] [Google Scholar]
- 37.Xu S., Chen B., Zhou K., Yang G. High similarity at three MHC loci between the baiji and finless porpoise: Trans-species or convergent evolution? Mol. Phylogenet. Evol. 2008;47:36–44. doi: 10.1016/j.ympev.2007.05.026. [DOI] [PubMed] [Google Scholar]
- 38.Shirai K., Sakai T., Oike T. Molecular cloning of bottle-nosed dolphin (Tursiops truncatus) MHC class I cDNA. J. Vet. Med. Sci. 1998;60:1093–1096. doi: 10.1292/jvms.60.1093. [DOI] [PubMed] [Google Scholar]
- 39.Manlik O. University of New South Wales; 2016. Fitness and major histocompatibility complex diversity of two bottlenose dolphin populations. Doctor of Philosophy. [Google Scholar]
- 40.Gambón-Deza F. Immunoglobulins, MHC and T-cell receptors genes in cetaceans. bioRxiv. 2020;2 doi: 10.1101/2020.10.24.353342. Preprint at. [DOI] [Google Scholar]
- 41.He K., Minias P., Dunn P.O. Long-read genome assemblies reveal extraordinary variation in the number and structure of MHC loci in birds. Genome Biol. Evol. 2021;13 doi: 10.1093/gbe/evaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simeone C.A., Gulland F.M.D., Norris T., Rowles T.K. A systematic review of changes in marine mammal health in North America, 1972-2012: The need for a novel integrated approach. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor J.S., Hart L.B., Adams J. Skin lesion prevalence of estuarine common bottlenose dolphins (Tursiops truncatus) in North Carolina, with comparisons to other east coast study sites. Mar. Mamm. Sci. 2021;37:127–141. doi: 10.1111/mms.12731. [DOI] [Google Scholar]
- 44.Bossart G.D. Marine mammals as sentinel species for oceans and human health. Vet. Pathol. 2011;48:676–690. doi: 10.1177/0300985810388525. [DOI] [PubMed] [Google Scholar]
- 45.Jenner K.C.S., Jenner M.-N.M., McCabe K.A. Geographical and temporal movements of humpback whales in Western Australian waters. APPEA J. Aust. Pet. Prod. Explor. Assoc. 2001;41:749–765. doi: 10.1071/AJ00044. [DOI] [Google Scholar]
- 46.Braulik G., Natoli A., Kiszka J., Parra G., Plön S., Smith B. 2019. Tursiops aduncus. IUCN Red List Assess. [DOI] [Google Scholar]
- 47.MacLeod C.D. Global climate change, range changes and potential implications for the conservation of marine cetaceans: a review and synthesis. Endanger. Species Res. 2009;7:125–136. doi: 10.3354/esr00197. [DOI] [Google Scholar]
- 48.Simmonds M.P., Isaac S.J. The impacts of climate change on marine mammals: early signs of significant problems. Oryx. 2007;41:19–26. doi: 10.1017/S0030605307001524. [DOI] [Google Scholar]
- 49.Monk A., Charlton-Robb K., Buddhadasa S., Thompson R.M. Comparison of mercury contamination in live and dead dolphins from a newly described species, Tursiops australis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waerebeek K.V., Baker A., Félix F., Gedamke J., Iñiguez M., Sanino G., Secchi E., Sutaria D., Helden A.v., Wang Y. Vessel collisions with small cetaceans worldwide and with large whales in the Southern Hemisphere, an initial assessment. Lat. Am. J. Aquatic Mamm. 2007;6 doi: 10.5597/lajam00109. [DOI] [Google Scholar]
- 51.Wisniewska D.M., Johnson M., Teilmann J., Siebert U., Galatius A., Dietz R., Madsen P.T. High rates of vessel noise disrupt foraging in wild harbour porpoises (Phocoena phocoena) Proc. Biol. Sci. 2018;285 doi: 10.1098/rspb.2017.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Read A.J., Drinker P., Northridge S. Bycatch of marine mammals in U.S. and global fisheries. Conserv. Biol. 2006;20:163–169. doi: 10.1111/j.1523-1739.2006.00338.x. [DOI] [PubMed] [Google Scholar]
- 53.Avila I.C., Kaschner K., Dormann C.F. Current global risks to marine mammals: taking stock of the threats. Biol. Conserv. 2018;221:44–58. doi: 10.1016/j.biocon.2018.02.021. [DOI] [Google Scholar]
- 54.Chapin F.S., 3rd, Zavaleta E.S., Eviner V.T., Naylor R.L., Vitousek P.M., Reynolds H.L., Hooper D.U., Lavorel S., Sala O.E., Hobbie S.E., et al. Consequences of changing biodiversity. Nature. 2000;405:234–242. doi: 10.1038/35012241. [DOI] [PubMed] [Google Scholar]
- 55.Kaschner K., Tittensor D.P., Ready J., Gerrodette T., Worm B. Current and future patterns of global marine mammal biodiversity. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lotze H.K., Lenihan H.S., Bourque B.J., Bradbury R.H., Cooke R.G., Kay M.C., Kidwell S.M., Kirby M.X., Peterson C.H., Jackson J.B.C. Depletion, degradation, and recovery potential of estuaries and coastal seas. Science. 2006;312:1806–1809. doi: 10.1126/science.1128035. [DOI] [PubMed] [Google Scholar]
- 57.Davidson A.D., Boyer A.G., Kim H., Pompa-Mansilla S., Hamilton M.J., Costa D.P., Ceballos G., Brown J.H. Drivers and hotspots of extinction risk in marine mammals. Proc. Natl. Acad. Sci. USA. 2012;109:3395–3400. doi: 10.1073/pnas.1121469109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller M.A., Gardner I.A., Kreuder C., Paradies D.M., Worcester K.R., Jessup D.A., Dodd E., Harris M.D., Ames J.A., Packham A.E., Conrad P.A. Coastal freshwater runoff is a risk factor for Toxoplasma gondii infection of southern sea otters (Enhydra lutris nereis) Int. J. Parasitol. 2002;32:997–1006. doi: 10.1016/S0020-7519(02)00069-3. [DOI] [PubMed] [Google Scholar]
- 59.Duignan P.J., Van Bressem M.-F., Baker J.D., Barbieri M., Colegrove K.M., De Guise S., De Swart R.L., Di Guardo G., Dobson A., Duprex W.P., et al. Phocine distemper virus: Current knowledge and future directions. Viruses. 2014;6:5093–5134. doi: 10.3390/v6125093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Bressem M.F., Duignan P.J., Banyard A., Barbieri M., Colegrove K.M., De Guise S., Di Guardo G., Dobson A., Domingo M., Fauquier D., et al. Cetacean morbillivirus: current knowledge and future directions. Viruses. 2014;6:5145–5181. doi: 10.3390/v6125145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duignan P.J., Stephens N.S., Robb K. Fresh Water Skin Disease in Dolphins: A case definition based on pathology and environmental factors in Australia. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-78858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chapman J.R., Nakagawa S., Coltman D.W., Slate J., Sheldon B.C. A quantitative review of heterozygosity–fitness correlations in animal populations. Mol. Ecol. 2009;18:2746–2765. doi: 10.1111/j.1365-294X.2009.04247.x. [DOI] [PubMed] [Google Scholar]
- 63.Gulland F.M.D., Hall A.J. Is marine mammal health deteriorating? Trends in the global reporting of marine mammal disease. EcoHealth. 2007;4:135–150. doi: 10.1007/s10393-007-0097-1. [DOI] [Google Scholar]
- 64.Babik W. Methods for MHC genotyping in non-model vertebrates. Mol. Ecol. Resour. 2010;10:237–251. doi: 10.1111/j.1755-0998.2009.02788.x. [DOI] [PubMed] [Google Scholar]
- 65.Maccari G., Robinson J., Bontrop R.E., Otting N., de Groot N.G., Ho C.-S., Ballingall K.T., Marsh S.G.E., Hammond J.A. IPD-MHC: nomenclature requirements for the non-human major histocompatibility complex in the next-generation sequencing era. Immunogenetics. 2018;70:619–623. doi: 10.1007/s00251-018-1072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Groot N.G., Otting N., Robinson J., Blancher A., Lafont B.A.P., Marsh S.G.E., O'Connor D.H., Shiina T., Walter L., Watkins D.I., Bontrop R.E. Nomenclature report on the major histocompatibility complex genes and alleles of Great Ape, Old and New World monkey species. Immunogenetics. 2012;64:615–631. doi: 10.1007/s00251-012-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu S.X., Ren W.H., Li S.Z., Wei F.W., Zhou K.Y., Yang G. Sequence polymorphism and evolution of three cetacean MHC genes. J. Mol. Evol. 2009;69:260–275. doi: 10.1007/s00239-009-9272-z. [DOI] [PubMed] [Google Scholar]
- 68.Ellis S. The cattle major histocompatibility complex: is it unique? Vet. Immunol. Immunopathol. 2004;102:1–8. doi: 10.1016/j.vetimm.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 69.Fan G., Zhang Y., Liu X., Wang J., Sun Z., Sun S., Zhang H., Chen J., Lv M., Han K., et al. The first chromosome-level genome for a marine mammal as a resource to study ecology and evolution. Mol. Ecol. Resour. 2019;19:944–956. doi: 10.1111/1755-0998.13003. [DOI] [PubMed] [Google Scholar]
- 70.Holt B.G., Marx F.G., Fritz S.A., Lessard J.-P., Rahbek C. Evolutionary diversification in the marine realm: a global case study with marine mammals. Frontiers of Biogeography. 2020;12 doi: 10.21425/F5FBG45184. [DOI] [Google Scholar]
- 71.Möller L.M., Wiszniewski J., Allen S.J., Beheregaray L.B. Habitat type promotes rapid and extremely localised genetic differentiation in dolphins. Mar. Freshw. Res. 2007;58:640–648. doi: 10.1071/MF06218. [DOI] [Google Scholar]
- 72.Charlton-Robb K., Taylor A.C., McKechnie S.W. Population genetic structure of the Burrunan dolphin (Tursiops australis) in coastal waters of South-Eastern Australia: conservation implications. Conserv. Genet. 2015;16:195–207. doi: 10.1007/s10592-014-0652-6. [DOI] [Google Scholar]
- 73.Minias P., Whittingham L.A., Dunn P.O. Coloniality and migration are related to selection on MHC genes in birds. Evolution. 2017;71:432–441. doi: 10.1111/evo.13142. [DOI] [PubMed] [Google Scholar]
- 74.Bentkowski P., Radwan J. Evolution of major histocompatibility complex gene copy number. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1007015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baker C.S., Vant M.D., Dalebout M.L., Lento G.M., O’Brien S.J., Yuhki N. Diversity and duplication of DQB and DRB-like genes of the MHC in baleen whales (suborder: Mysticeti) Immunogenetics. 2006;58:283–296. doi: 10.1007/s00251-006-0080-y. [DOI] [PubMed] [Google Scholar]
- 76.Yang W.-C., Hu J.-m., Chou L.-S. Phylogenetic analyses of MHC class II genes in bottlenose dolphins and their terrestrial relatives reveal pathogen-driven directional selection. Zool. Stud. 2010;49:132–151. [Google Scholar]
- 77.Garrigan D., Hedrick P.W. Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution. 2003;57:1707–1722. doi: 10.1111/j.0014-3820.2003.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 78.McGowen M.R., Tsagkogeorga G., Álvarez-Carretero S., dos Reis M., Struebig M., Deaville R., Jepson P.D., Jarman S., Polanowski A., Morin P.A., Rossiter S.J. Phylogenomic resolution of the cetacean tree of life using target sequence capture. Syst. Biol. 2020;69:479–501. doi: 10.1093/sysbio/syz068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cabrera A.A., Bérubé M., Lopes X.M., Louis M., Oosting T., Rey-Iglesia A., Rivera-León V.E., Székely D., Lorenzen E.D., Palsbøll P.J. A genetic perspective on cetacean evolution. Annu. Rev. Ecol. Evol. Syst. 2021;52:131–151. doi: 10.1146/annurev-ecolsys-012021-105003. [DOI] [Google Scholar]
- 80.Thewissen J.G.M., Cooper L.N., George J.C., Bajpai S. From land to water: the origin of whales, dolphins, and porpoises. Evo. Edu. Outreach. 2009;2:272–288. doi: 10.1007/s12052-009-0135-2. [DOI] [Google Scholar]
- 81.Thewissen J.G.M., Williams E.M. The early radiations of Cetacea (mammalia): Evolutionary pattern and developmental correlations. Annu. Rev. Ecol. Systemat. 2002;33:73–90. doi: 10.1146/annurev.ecolsys.33.020602.095426. [DOI] [Google Scholar]
- 82.Marx F.G., Fordyce R.E. Baleen boom and bust: a synthesis of mysticete phylogeny, diversity and disparity. R. Soc. Open Sci. 2015;2 doi: 10.1098/rsos.140434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Flores-Ramirez S., Miller R.D., Urban-Ramirez J. Major histocompatibility complex I polymorphism in a cetacean: The gray whale (Eschrichtius robustus) Mar. Mamm. Sci. 2004;20:262–273. doi: 10.1111/j.1748-7692.2004.tb01155.x. [DOI] [Google Scholar]
- 84.Tian J., Gan Z., Sanganyado E., Lu Z., Wu J., Han J., Liu W. Tissue distribution and health risk of trace elements in East Asian finless porpoises. Environ. Pollut. 2021;290 doi: 10.1016/j.envpol.2021.118007. [DOI] [PubMed] [Google Scholar]
- 85.Cheng Z., Pine M.K., Li Y., Zuo T., Niu M., Wan X., Zhao X., Wang K., Wang J. Using local ecological knowledge to determine ecological status and threats of the East Asian finless porpoise, Neophocaena asiaeorientalis sunameri, in south Bohai Sea, China. Ocean Coast Manag. 2021;203 doi: 10.1016/j.ocecoaman.2021.105516. [DOI] [Google Scholar]
- 86.Xiong X., Chen X., Zhang K., Mei Z., Hao Y., Zheng J., Wu C., Wang K., Ruan Y., Lam P.K.S., Wang D. Microplastics in the intestinal tracts of East Asian finless porpoises (Neophocaena asiaeorientalis sunameri) from Yellow Sea and Bohai Sea of China. Mar. Pollut. Bull. 2018;136:55–60. doi: 10.1016/j.marpolbul.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 87.D'agrosa C., Lennert-Cody C.E., Vidal O. Vaquita bycatch in Mexico's artisanal gillnet fisheries: Driving a small population to extinction. Conserv. Biol. 2000;14:1110–1119. doi: 10.1046/j.1523-1739.2000.98191.x. [DOI] [Google Scholar]
- 88.Zhou X., Guang X., Sun D., Xu S., Li M., Seim I., Jie W., Yang L., Zhu Q., Xu J., et al. Population genomics of finless porpoises reveal an incipient cetacean species adapted to freshwater. Nat. Commun. 2018;9:1276. doi: 10.1038/s41467-018-03722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rojas-Bracho L., Reeves R.R., Jaramillo-Legorreta A. Conservation of the vaquita Phocoena sinus. Mamm Rev. 2006;36:179–216. doi: 10.1111/j.1365-2907.2006.00088.x. [DOI] [Google Scholar]
- 90.Hooker S.K., Baird R.W. Deep-diving behaviour of the northern bottlenose whale, Hyperoodon ampullatus (Cetacea: Ziphiidae) Proc. Roy. Soc. Lond. B. 1999;266:671–676. doi: 10.1098/rspb.1999.0688. [DOI] [Google Scholar]
- 91.Aguilar de Soto N., Madsen P.T., Tyack P., Arranz P., Marrero J., Fais A., Revelli E., Johnson M. No shallow talk: Cryptic strategy in the vocal communication of Blainville's beaked whales. Mar. Mamm. Sci. 2012;28:E75–E92. doi: 10.1111/j.1748-7692.2011.00495.x. [DOI] [Google Scholar]
- 92.Van Bressem M.F., Van Waerebeek K., Raga J.A. A review of virus infections of cataceans and the potential impact of morbilliviruses, poxviruses and papillomaviruses on host population dynamics. Dis. Aquat. Org. 1999;38:53–65. doi: 10.3354/dao038053. [DOI] [PubMed] [Google Scholar]
- 93.Van Bressem M.F., Raga J.A., Di Guardo G., Jepson P.D., Duignan P.J., Siebert U., Barrett T., Santos M.C.d.O., Moreno I.B., Siciliano S., et al. Emerging infectious diseases in cetaceans worldwide and the possible role of environmental stressors. Dis. Aquat. Org. 2009;86:143–157. doi: 10.3354/dao02101. [DOI] [PubMed] [Google Scholar]
- 94.Harvell C.D., Kim K., Burkholder J.M., Colwell R.R., Epstein P.R., Grimes D.J., Hofmann E.E., Lipp E.K., Osterhaus A.D., Overstreet R.M., et al. Emerging marine diseases - climate links and anthropogenic factors. Science. 1999;285:1505–1510. doi: 10.1126/science.285.5433.1505. [DOI] [PubMed] [Google Scholar]
- 95.Meager J.J., Limpus C. Mortality of inshore marine mammals in eastern Australia is predicted by freshwater discharge and air temperature. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koch M., da Silva V., Bracarense A., Domit C. Environmental Aspects and Diseases Related to Immunosuppression in Cetaceans: A Concise Review. Ciencias Agrarias. 2018;39:2897–2918. doi: 10.5433/1679-0359.2018v39n6p2897. [DOI] [Google Scholar]
- 97.Árnason U. Banding studies on the gray and sperm whale karyotypes. Hereditas. 1981;95:277–281. doi: 10.1111/j.1601-5223.1981.tb01418.x. [DOI] [PubMed] [Google Scholar]
- 98.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 99.Drummond A., Ashton B., Buxton S., Cheung M., Cooper A., Duran C., Field M., Heled J., Kearse M., Markowitz S., et al. Geneious version 2021.2.2. 2010. http://www.geneious.com
- 100.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper analyses existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.