Summary
We present a method of in vitro/in vivo protein detection by pairing CRISPR-Cas9 genome editing with the NanoBiT system. We describe steps for cell culturing, in vitro CRISPR-Cas9 ribonucleoprotein delivery, cell monitoring, efficiency assessments, and edit analysis through HiBiT assays. We then detail procedures to determine edit specificity through genomic DNA analysis, small interfering RNA reverse transfection, and HiBiT blotting. This protocol is simple to execute and multifunctional, and it enables high-throughput screens on endogenous proteins to be conducted with ease.
Subject areas: Health Sciences, High Throughput Screening, Molecular Biology, Gene Expression, CRISPR, Protein Biochemistry, Biotechnology and bioengineering
Graphical abstract

Highlights
-
•
Design of CRISPR-Cas9 reagents for inclusion of a HiBiT tag into a protein of interest
-
•
Instruction for execution/verification of in vitro genomic editing via electroporation
-
•
Steps for monitoring endogenous protein expression using HiBiT-related assays
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
We present a method of in vitro/in vivo protein detection by pairing CRISPR-Cas9 genome editing with the NanoBiT system. We describe steps for cell culturing, in vitro CRISPR-Cas9 ribonucleoprotein delivery, cell monitoring, efficiency assessments, and edit analysis through HiBiT assays. We then detail procedures to determine edit specificity through genomic DNA analysis, small interfering RNA reverse transfection, and HiBiT blotting. This protocol is simple to execute and multifunctional, and it enables high-throughput screens on endogenous proteins to be conducted with ease.
Before you begin
Biochemical detection methods such as western blotting and enzyme-linked immunosorbent assays (ELISAs) have been successfully applied to monitor thousands of individual proteins over the past decades. Yet, thousands more proteins remain challenging to detect and monitor due to low levels of endogenous protein expression, fast protein turnover, or poor antibody availability. Moreover, reliance on protein overexpression mediated through plasmid or viral means can cause elevated stress response activation, non-physiological protein complex stoichiometries, and inefficient client protein folding. To circumvent these challenges, we describe pairing the technologies of CRISPR-Cas9 genome editing with the Nano Luciferase Binary Technology (NanoBiT; LgBiT+HiBiT) luciferase complementation system.
Nano luciferase (NanoLuc) is a small (19.3 kDa), monomeric enzyme best known for its application as a bioluminescent reporter.1 Its high stability and ATP independence has made it one of the most used proteins for biological reporting.2 Compared to conventional reporters, such as green fluorescent protein (GFP), firefly luciferase, or even small full-length luciferases such as Gaussia luciferase,3 NanoLuc has shown exceptional sensitivity and versatility, making it ideal for use in multiple biological studies.4,5,6,7
In an effort to expand the applications of this technology, NanoLuc has since been modified into a structural complementation reporter system known as NanoBiT,8 which consists of two components: LgBiT, a 159 amino acid, 18 kDa subunit and HiBiT, an 11 amino acid, 1.3 kDa high affinity (KD = 700 pM) subunit. Due to their affinity for each other, LgBiT and HiBiT spontaneously associate within cells or during an assay to form full length NanoBiT, which produces a bright and stable glow-like luminescence when supplemented with furimazine and assay buffer. The HiBiT subunit is specifically important in physiological protein studies as its relatively small size (similar to an epitope tag) makes it possible to efficiently insert into genomic DNA using CRISPR gene editing.9,10 This property enables detection and monitoring of proteins at endogenous levels. LgBiT can be either produced within a HiBiT cell line (and spontaneously reconstitute to form NanoBiT), or be provided in trans during the NanoBiT assay, allowing for flexible complementation options. Ultimately, the formation of the NanoBiT luciferase produces bright and quantitative luminescence proportional to the amount of HiBiT tagged protein present.
The primary goal of this STAR Protocols paper is to guide prospective users in utilizing HiBiT tagging as a method of endogenous protein monitoring for in vitro studies. This method can be used in a variety of different cell lines and on a large number of target proteins. The ease and high sensitivity of the NanoBiT assay makes this method favorable for endogenous protein investigations and studies involving bioluminescence imaging,10 protein-protein interaction studies,8 monitoring protein degradation in real-time,11 and high throughput screening (HTS),5 such as compound and genetic screens. We have decided to use our HiBiT tagging of the cholesterol ester transfer protein (CETP), a protein associated with age-related macular degeneration (AMD)12,13,14,15 and atherosclerosis,16 in HepG2 cells as an example workflow for this method and the subsequent analysis/validation of CRISPR editing.
Experimental design considerations
Selecting a cell line
Effective gene editing and endogenous protein analysis are heavily dependent on a chosen cell line. First, prior to initiating HiBiT editing, it is ideal to pick a cell line that is predicted to express a particular protein of interest (POI) and is biologically relevant to the given project. This can be accomplished empirically, though literature searches, or potentially using the Human Protein Atlas (https://www.proteinatlas.org). Where possible, we recommend carrying out a pilot experiment assessing POI expression in a given cell line prior to editing as this may avoid future difficulties in evaluating seemingly negative results (i.e., no NanoBiT signal arising from edited cell lines). Additional considerations for the cell line chosen to edit should also include ploidy and whether the cell line is known to have active homology directed DNA repair pathways.
Second, to minimize off-target editing resulting from sustained expression of Cas9 and a guide RNA (gRNA), we recommend performing CRISPR editing by delivering transient ribonucleoprotein (RNP) via electroporation. Accordingly, it is important to select a cell line that can be electroporated easily and efficiently. Since every cell line responds to electroporation differently, we recommend starting with literature references for previously established protocols and parameters associated with your cell line. A useful reference is the Neon Transfection System Cell Line Data and Transfection Parameters website (https://www.thermofisher.com/us/en/home/life-science/cell-culture/transfection/neon-transfection-system/neon-transfection-system-cell-line-data.html), which has suggested parameters for over 140 cells lines at the time of publication. For cell lines with little to no previously established protocols, we strongly suggest a thorough, small scale optimization exploring a variety of parameters using a plasmid encoding for a fluorescent protein to determine which are the best for electroporation efficiency and cell viability (see Figure 1 for examples of different electroporation parameters for HepG2 cells).
Note: As we describe a protocol that utilizes double strand breaks to subsequently insert the HiBiT tag, it is a possibility that indel formation could occur at edited loci or in highly similar genomic regions (provided as a list by IDT when a particular crRNA is selected, see below). Indel formation can be evaluated by a T7E1 assay or by next-generation sequencing.
Figure 1.
Example whole-well Celigo images 1-day post-electroporation of HepG2 cells demonstrating successful transfection of plasmid using a variety of parameters
Seventy thousand HepG2 cells (per well) were electroporated with 250 ng (per well) pEGFP-N1 using the method described in the ‘In vitro electroporation of CRISPR-Cas9 RNP’ section. One day later, bright-field and green fluorescence (ex: 483 nm/em: 536 nm, integration time of 50,000 μs) images were taken from unelectroporated wells and wells electroporated with plasmid under the indicated electroporation conditions. While all the electroporation conditions used yielded observable transfection of HepG2 cells, we found that 1050 V, 30 ms, 2 pulses gave the best combination of transfection and sustained viability. These parameters were used for the remaining experiments going forward. Scale bar = 2 mm.
An alternative method for introduction of Cas9 RNP is by lipofection using reagents such as CRISPRMAX.17,18 While electroporation has been routinely used for difficult-to-transfect cells, such as primary cells, a disadvantage of electroporation is that it cannot be scaled like lipofection can be, and it requires specialized, costly instrumentation (i.e., an electroporation system). Lipofection, on the other hand, can be scaled to virtually any size, and does not require instrumentation other than what is commonly found in a wet lab (vortex, centrifuge, etc.). Nonetheless, we use electroporation throughout this protocol.
Third, we recommend taking into consideration the downstream analyses to be carried out on the cell line. For example, for studies that require single colony clones to be grown from the edited culture, it is necessary to pick a cell line that grows well (and ideally quickly) as single cells. Similarly, experiments geared towards high-throughput screening of the edited cells should pick a cell line suited to tolerate any manipulation required during screening (i.e., tolerant of media changes, etc).
Early consideration of these applications is crucial for developing an efficient experiment that minimizes the necessity to troubleshoot. Finally, it is imperative to understand that genomic editing and electroporation is variable based on a variety of parameters. Because of this, we recommend that edited cells showing no signs of HiBiT signal go through the procedure of edit analysis prior to making conclusions. Thorough analysis of the edit will provide more information on the potential cause of low/no signal or editing failure and provide a foundation for further experimental development. In some cases, it may be necessary to change cell lines to achieve effective editing and ultimate production of your HiBiT POI for downstream applications.
Design of gene-specific reagents for CRISPR-Cas9 editing
Prior to editing cells, one must first design and purchase the proper CRISPR-Cas9 reagents for effective and accurate editing including a CRISPR RNA (crRNA) proximal to desired HiBiT insertion site within the genomic DNA (gDNA), and a single-stranded DNA oligo nucleotide (ssODN) sequence for homology-directed repair (HDR). Other CRISPR reagents, such as tracrRNA (which anneals to the designed crRNA) or the Staphylococcus pyogenes (S.p.) Cas9 enzyme, are general reagents not specific to editing site and can be purchased without any design requirements (although each providing company may have optimized versions of each of these reagents). An example design for HiBiT insertion into CETP is shown in Figure 2.
-
1.
Once the target gene has been identified, determination of the HiBiT tag placement is important. The goal should be to insert the tag in a place that will be i) functionally-tolerated within the POI, ii) in an area with crRNA sequences that minimize off target effects, and iii) in a location easily accessible for LgBiT to bind. For these reasons, we recommend prioritization of either N-terminal or C-terminal editing. In order to obtain the genomic DNA sequence, we recommend using Benchling (https://benchling.com) which can provide easy-to-use visualization of gene exons, introns and protein sequence simultaneously across a variety of laboratory model species (e.g., human, mouse, etc.).
Note: While we have found Benchling to be reliable source for accurate genomic DNA sequences, there is always the possibility that a chosen cell line may have synonymous or non-synonymous mutations in the DNA region selected for editing. Thus, it may be worth investing time to empirically verify the sequence of the targeted gDNA region prior to editing.
CRITICAL: If one desires to place an N-terminal HiBiT tag on a secreted POI, it is important to insert it after the signal sequence such that upon processing, since the HiBiT tag will still be attached to the processed POI instead of being cleaved off. SignalP19 online software (https://services.healthtech.dtu.dk/services/SignalP-5.0/) can help identify predicted signal sequences if they are not known, and a FLAG sequence (DYKDDDDK) may be necessary to force signal sequence cleavage20 and to avoid potential cleavage of the HiBiT tag if located close to the signal sequence cleavage site. Additional online software such as Phobius21 (https://phobius.sbc.su.se/) can be used to cross-verify signal sequence cleavage prediction using SignalP.
Note: If there is concern that the HiBiT tag, however small, could interfere with the function of the edited POI, it could be possible to change the termini edited (N- vs. C-, or C- vs. N-), or to evaluate potential functional changes using transient overexpression of a similarly designed HiBiT fusion using plasmids prior to attempting gene editing. However, a functional assay for the POI must exist, and one must also take the time to design and possibly synthesize genes for insertion into an appropriate plasmid.
-
2.When identifying the Cas9 cut site for the insertion of the HiBiT tag itself, it is important to consider the protospacer adjacent motif (PAM) site. PAM sites are small regions of DNA, approximately 2–6 base pairs in length, that are located directly downstream of the Cas9 cut site. The Cas9 nuclease’s priority is to recognize a PAM site, which will guide the unravelling and cutting of the DNA, typically around 3 or 4 base pairs upstream of the PAM.22
 CRITICAL: Therefore, it is important to both identify a PAM site near the location of editing (typically a 2–5 bp window surrounding desired cut site) as well as choose the appropriate nuclease, which, in our case, is the S.p. Cas9 endonuclease (double strand DNA cutting).
CRITICAL: Therefore, it is important to both identify a PAM site near the location of editing (typically a 2–5 bp window surrounding desired cut site) as well as choose the appropriate nuclease, which, in our case, is the S.p. Cas9 endonuclease (double strand DNA cutting).-
a.Cas proteins originating from different species can recognize divergent PAM sites, allowing users the ability to edit almost any DNA regardless of the PAM site present. We recommend using S.p. Cas9, as the recombinant protein is commercially available from multiple companies (we use Integrated DNA Technologies (IDT) Alt-R S.p. Cas9 Nuclease V3) and its guidance and activity is well understood.23 The PAM sequence recognized by this enzyme is 5′-NGG-3′ where N can be any nucleotide base.
-
a.
-
3.After determining the cut site and locating the PAM, development of the appropriate crRNAs can take place. It is our recommendation to order crRNAs from commercial companies, like IDT, that have the appropriate software to design and create custom crRNAs. Many of these companies have large banks of predesigned crRNA that have been validated and are able to provide relative on-target and off-target information to aid in the design process. We use the IDT website to identify and rank crRNAs from the ∼100 bp region surrounding our desired HiBiT insertion site (https://www.idtdna.com/site/order/designtool/index/CRISPR_CUSTOM). It is our recommendation that prioritization of crRNA designs begin with the validated suggestions. The best crRNA are ones that are predesigned, validated, and fall close to the cut site. Secondarily, in IDT’s ranking system, higher numbers for both on-target and off-target scores indicate a (theoretically) better crRNA. Since prediction software is not flawless, we recommend purchasing at least two crRNAs to test initially in order to increase the likelihood of editing success. See Figure 3 as an example of this set-based variability.Note: crRNA/tracrRNA duplexes were used because of the universal modularity of the tracrRNA to complex with S.p. Cas9 crRNAs. That is to say, that one can purchase a large amount of tracrRNA and use it to form crRNA/tracrRNA duplexes for many separate editing strategies. Alternatively, one could purchase a short guide RNA (sgRNA), but at an additional overall cost per editing reaction due to extra RNA synthesis steps.Note: CETP crRNA/tracrRNA set 2 was used for subsequent CETP editing experiments.
-
a.For edits that cannot be achieved using predesigned crRNA suggestions from IDT, the main priority when picking crRNA’s should be the crRNA location in relation to the PAM and cut site.
-
a.
-
4.Development of the ssODN is custom to the desired insertion and is comprised of two main segments: the homology arms and the insertion sequence. Identification of the homology arms is solely based on where the insertion should take place. Homology arms are identified as the exact gDNA sequence immediately downstream and upstream of the cut site. These homology arms should be ∼50 base pairs long and may include only exon sequences, intron sequences, or a combination of both. The insertion sequence itself lies between the identified homology arms and consists of the 33-base pair, 11 amino acid HiBiT tag (DNA sequence: GTGAGCGGCTGGCGGCTGTTCAAGAAGATTAGC, protein sequence: VSGWRLFKKIS).Note: Use of the HiBiT tag is proprietary to the Promega Corporation and appropriate agreements must be completed prior to use of this sequence.
-
a.Depending on where the HiBiT tag is inserted, it may be important to use a valine and serine (VS) linker adjacent to the HiBiT sequence, and/or inclusion of sequences that may ‘protect’ the HiBiT sequence from cleavage (by the signal peptidase, for example) such as a FLAG tag (see above). Inclusion of a FLAG tag has the added benefit that it also allows for a secondary detection method, should the HiBiT assay fail, and can be used in for other downstream protein analyses.
-
b.To prevent the ssODN repair template from being cleaved by the Cas9 endonuclease, it is imperative that the designed ssODN does not contain significant portions (or key elements) of the crRNA sequence or PAM site. In some instances, the crRNA + PAM site will be completely separated upon insertion of the HiBiT sequence. If this is the case, then no additional modifications to the ssODN need to be made. Alternatively, one can use silent mutagenesis to eliminate the PAM site in the ssODN. If this is not an option due to the positioning of the PAM relative to codons, one can make multiple (3–4) silent mutations in the crRNA binding area to minimize possible Cas9 cutting.
-
c.While HiBiT editing will result in insertion of at least 33 bp in the gDNA, which, depending on the size of the DNA post PCR amplification, may be able to be resolved on a high percentage agarose gel, there is also the possibility of researchers incorporating new (or removing existing) restriction enzyme sequences located in the ssODN using silent mutation introduction. This approach would allow for restriction enzyme-based confirmation of successful editing.
-
a.
Figure 2.
Schematic design of our CETP HiBiT editing strategy in human cells
Presented is a C-terminal HiBiT tagging immediately prior to the CETP stop codon located in exon 16. Since the insertion of the VS 2×FLAG VS HiBiT tag occurs within the gRNA sequence, successful introduction of this insert via ssODN substantially disrupts the gRNA recognition sequence. Therefore, mutation of the PAM site is not necessary to prevent Cas9 cutting of the ssODN. Note: the gRNA indicated in this figure originates from ‘set 2’ in Figure 3.
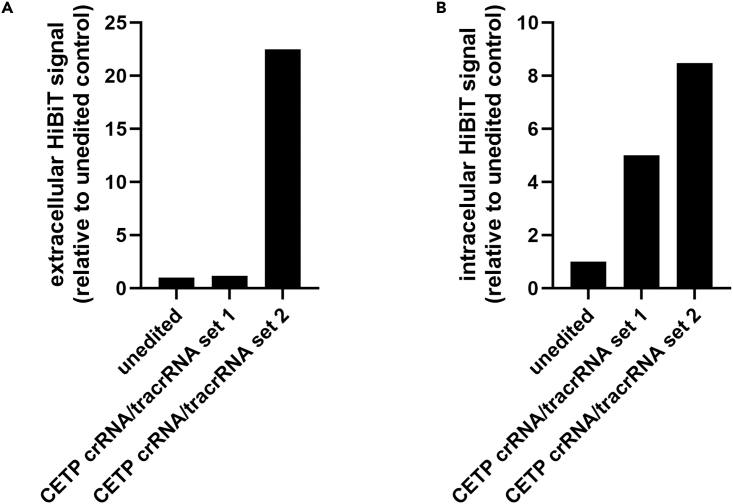
Figure 3.
Example data from two sets of crRNA/tracrRNA duplexes + ssODN both designed to insert a C-terminal HiBiT tag on CETP
(A) Intracellular or (B) secreted CETP HiBiT signal obtained ∼2 weeks post initial electroporation. Secreted luminescent signal (performed as described in the ‘HiBiT assay analysis of edits’ section using the Lytic Detection System) was normalized to the control sample (wild-type, edited cells) to compare relative HiBiT expression levels. In this example, CETP was edited in HepG2 cells using one of two sets of crRNA/tracrRNA respectively. Edit set 1 shows little to no HiBiT signal relative to the control, indicating a potentially failed edit. However, edit set 2 has much higher signal response representing what results of positive edits should look like, if not a more obvious difference. As these are example data that were taken at a select time point, n = 1.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Tris base (white crystals or crystalline powder/molecular biology), Fisher BioReagents | Thermo Fisher Scientific | Cat# BP152-500 |
| Sodium chloride, ACS, ≥99%, Ultrapure, Thermo Scientific Chemicals | Thermo Fisher Scientific | Cat# J21618.36 |
| Tween 20 | MilliporeSigma | Cat# P1379 |
| 10× TBE buffer | Research Products International | Cat# 30UA80 |
| 2-Mercaptoethanol (BME) | MilliporeSigma | Cat# M6250 |
| Hank’s balanced salt solution | Corning | Cat# MT21022CV |
| Trypsin-EDTA (0.25%), phenol red | Thermo Fisher Scientific | Cat# 25200056 |
| Nuclease-free duplex buffer | Integrated DNA Technologies | Cat# 11-05-01-12 |
| Alt-R S.p. Cas9 nuclease V3, 100 μg | Integrated DNA Technologies | Cat# 1081058 |
| Alt-R Cas9 electroporation enhancer | Integrated DNA Technologies | Cat# 1075915 |
| Alt-R HDR enhancer V2 | Integrated DNA Technologies | Cat# 10007910 |
| Q5 high-fidelity 2× master mix | New England Biolabs | Cat# M0492S |
| GelGreen nucleic acid stain | MilliporeSigma | Cat# SCT125 |
| Gel loading dye, purple (6×) | New England Biolabs | Cat# B7024S |
| 100 bp DNA ladder | New England Biolabs | Cat# N3231S |
| siCETP | Thermo Fisher Scientific | Cat# s2933 |
| Silencer siRNA positive and negative controls | Thermo Fisher Scientific | Cat# AM4611 |
| siGLO Lamin A/C Control siRNA (human) | Horizon Discovery | Cat# D-001620-02-05 |
| Opti-MEM I reduced serum medium | Thermo Fisher Scientific | Cat# 31985062 |
| DharmaFECT 4 transfection reagent | Horizon Discovery | Cat# T-2004-03 |
| Halt protease inhibitor cocktail (100×) | Thermo Fisher Scientific | Cat# 78429 |
| Benzonase | MilliporeSigma | Cat# 9025-65-4 |
| RIPA lysis and extraction buffer | Thermo Fisher Scientific | Cat# 89901 |
| Laemmli SDS-sample buffer (4×) | Boston BioProducts | Cat# BP-110NR |
| Critical commercial assays | ||
| Nano-Glo HiBiT lytic detection system | Promega | Cat# N3030 |
| Nano-Glo HiBiT blotting system | Promega | Cat# N2410 |
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat# 23225 |
| Experimental models: Cell lines | ||
| Hep G2 cells | American Type Culture Collection | Cat# HB-8065 |
| Oligonucleotides | ||
| Alt-R CRISPR-Cas9 crRNA | Integrated DNA Technologies | N/A |
| Alt-R CRISPR-Cas9 tracrRNA, 5 nmol | Integrated DNA Technologies | Cat# 1072532 |
| Custom 25 nmol single-stranded oligodeoxyribonucleotide (ssODN) | Integrated DNA Technologies | Varies |
| Custom DNA oligos for gDNA amplification | MilliporeSigma | Varies |
| Software and algorithms | ||
| crRNA identification and ssODN synthesis | Integrated DNA Technologies (online) | https://www.idtdna.com/pages/products/crispr-genome-editing/alt-r-crispr-cas9-system |
| Genomic DNA analysis | Benchling (online) | https://www.benchling.com/ |
| Sanger sequencing | Eurofins Genomics | https://www.eurofinsus.com/genomic-services/ |
| DNA sequencing alignment software | SnapGene | https://www.snapgene.com/ |
| Other | ||
| Invitrogen Neon NxT electroporation system | Thermo Fisher Scientific | Cat# NEON1 |
| Invitrogen Neon NxT electroporation system 10-μL Kit | Thermo Fisher Scientific | Cat# N1025 |
| VWR thermal Shake Touch thermoshaker with 1.5 mL block | Avantor | Cat# 89232-908 |
| Thermo Scientific Sorvall Legend Micro 17 microcentrifuge | Thermo Fisher Scientific | Cat# 75-002-431 |
| Celigo image cytometer | Nexcelom | N/A |
| Heracell 150i GP CO₂ incubator | Thermo Fisher Scientific | Cat# 51032872 |
| Thermo Scientific 120 V digital microplate shaker | Thermo Fisher Scientific | Cat# S81139 |
| GloMax Discover microplate reader | Promega | Cat# GM3000 |
| Thermo Scientific NanoDrop OneC Microvolume UV-Vis spectrophotometer | Thermo Fisher Scientific | Cat# 13-400-519 |
| SimpliAmp thermal cycler | Thermo Fisher Scientific | Cat# A24811 |
| Owl EasyCast B2 mini gel electrophoresis systems | Thermo Fisher Scientific | Cat# B2-BP |
| PowerPac universal power supply | Bio-Rad | Cat# 1645070 |
| Safe Imager 2.0 Blue-Light transilluminator | Thermo Fisher Scientific | Cat# G6600 |
| iBlot 2 Gel transfer device | Thermo Fisher Scientific | Cat# IB21001 |
| iBlot Transfer stack, nitrocellulose, regular size | Thermo Fisher Scientific | Cat# IB301001 |
| Odyssey Fc imaging system | LI-COR | N/A |
| 10 cm tissue culture-treated dishes | Corning | Cat# 430167 |
| DMEM high glucose (4.5 g/L) | Thermo Fisher Scientific | Cat# 11965092 |
| Antibiotic free culture media (∼50 mL, contains glutamine, 10% FBS) | N/A | N/A |
| Fetal bovine serum | Omega Scientific | Cat# FB-02 |
| Hausser Scientific Levy hemacytometer chamber set | Thermo Fisher Scientific | Cat# 02-671-55A |
| Costar 24-well clear TC-treated multiple well plates | Corning | Cat# 3524 |
| 96-well black plate, non-treated surface | Thermo Fisher Scientific | Cat# 237108 |
| Monarch Genomic DNA Purification Kit | New England Biolabs | Cat# T3010S |
| Snap Cap low-retention microcentrifuge tubes | Thermo Fisher Scientific | Cat# 3434PK |
| Parafilm M wrapping film | Thermo Fisher Scientific | Cat# S37440 |
| QIAquick Gel Extraction Kit | QIAGEN | Cat# 28704 |
| Novex Tris-glycine mini protein gels, 4%–20% | Thermo Fisher Scientific | Cat# XP04205BOX |
| Razor blades | N/A | N/A |
| pEGFP-N1 | Takara | Discontinued, suggested replacement: pAcGFP-F vector (Takara Cat# 632511) |
| pCMV-GFP – GFP plasmid control (or similar) | Addgene | Plasmid 11153 |
Materials and equipment
Tris-Buffered Saline (TBS), 10× Stock Solution, pH 7.6
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris Base | 200 mM | 24 g |
| NaCl | 1500 mM | 88 g |
| ddH2O | N/A | Adjust to final volume of 1 L (∼ 900 mL) |
| Total | 10× | 1 L |
Store at 4 for 3 months.
CRITICAL: Adjust to final pH of 7.6 using 12 N HCl prior to bringing up to volume with ddH2O.
Tris-Buffered Saline with 0.1% Tween 20 (TBST) 1× Solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 10× TBS Stock Solution | 1× | 100 mL |
| Tween 20 Detergent | 0.1% w/v | 1 mL |
| ddH2O | N/A | Adjust to final volume of 1 L (∼900 mL) |
| Total | 1× |
Store at 4 for 3 months.
Step-by-step method details
Cell culturing
Prior to editing, cells should be routinely maintained and subcultured according to their specific requirements. The cells that we have chosen to edit in this protocol, HepG2 (ATCC HB-8065), should be 70% confluent in a 10 cm plate on the day of electroporation.
In vitro CRISPR-Cas9 RNP delivery via electroporation
Timing: 4–7 days, longer for culturing cells to scale or generating single colony clones
-
1.Day 1: Editing.Note: This protocol is written for sample preparation of 2 control wells of unelectroporated cells, 2 control wells of a GFP plasmid control, and 2 wells of edited cells in a 24 well plate using the 10 L Neon Neon NxT Transfection tips. These experiments can be scaled up as necessary. Alternatively, for much larger experiments, one can use 100 L Neon tips. Editing can also be performed using previous versions of the Neon Transfection system with slight modification, including using “R” buffer instead of Genome Editing Buffer, and using 3 mL of “E” buffer to fill the electroporation cassette. For the sake of simplicity, we have just focused on using the Neon NxT Transfection system. All editing reagents used for HiBiT-tagging CETP are listed in Table 1.
-
a.Dissolve the crRNAs in TE or nuclease-free molecular biology grade water to a final concentration of 100 μM.
-
b.Dissolve the tracrRNA in TE or nuclease-free molecular biology grade water to a final concentration of 100 μM.
-
c.Prepare the crRNA/tracrRNA duplex in a biosafety cabinet:
-
i.1.5 μL of 100 μM crRNA.
-
ii.1.5 μL of 100 μM tracrRNA.
-
iii.3.25 μL duplex buffer.
-
i.
-
d.Heat the solution at 95°C for 5 min and then allow to cool to RT slowly to anneal the crRNA and tracrRNA (−0.5°C/min or within the heat block while it is turned off).
-
e.Spin down the crRNA/tracrRNA duplex at max speed on a microcentrifuge for ∼10 s.
-
f.Prepare the ribonucleoprotein (RNP) in a culture hood by combining:
-
i.5 μL crRNA/tracrRNA duplex.
-
ii.3.66 μL Genome Editing resuspension buffer.
-
iii.1.33 μL Cas9 enzyme.
 CRITICAL: Cas9 should be added last and slowly, while stirring the solution with the pipette tip.Note: The use of Cas9 directly labeled with GFP (e.g., IDT catalog# 1081060) can serve as an alternative editing enzyme that has the additional benefit of downstream FACS sorting for single colony isolation or population enrichment. Alternatively, it may be possible to enrich for HiBiT edited populations using HiBiT immunoreagents, depending on the POI edited.
CRITICAL: Cas9 should be added last and slowly, while stirring the solution with the pipette tip.Note: The use of Cas9 directly labeled with GFP (e.g., IDT catalog# 1081060) can serve as an alternative editing enzyme that has the additional benefit of downstream FACS sorting for single colony isolation or population enrichment. Alternatively, it may be possible to enrich for HiBiT edited populations using HiBiT immunoreagents, depending on the POI edited.
-
i.
-
g.Flick to mix and allow the solution to incubate in the culture hood at RT for 15 min while preparing the cells.
-
h.To prepare cells from a 10 cm dish, aspirate the media and wash with 5 mL Hank’s Balanced Salt Solution (HBSS).
-
i.Aspirate the HBSS and add 2 mL Trypsin-EDTA (0.25%).
-
i.Gently rock the plate to disperse the trypsin and incubate the plate at 37°C until all cells are in suspension, typically 3–5 min.
-
ii.Verify thorough trypsinization under a microscope prior to neutralizing.
-
i.
-
j.Quench the trypsin with 8 mL antibiotic free culture media and resuspend thoroughly.
-
k.Count the cells on a hemocytometer and aliquot the desired number of cells into an Eppendorf tube.
-
i.While every cell line will vary slightly in the desired seeding density, we have found a density of 70,000 cells per well to be the most universally applicable. Therefore, we would recommend aliquoting 420,000 cells total for 6 wells of a 24-well plate.
-
i.
-
l.Spin down the cells in a centrifuge at 1,000 x g for 3–5 min at RT.
-
m.While the cells are spinning down, turn on the Neon NxT Transfection system and load the electroporation parameters. Starting parameters can be found in the NxT software, or on the Life Technologies website (https://www.thermofisher.com/us/en/home/life-science/cell-culture/transfection/neon-transfection-system/neon-transfection-system-cell-line-data.html). For HepG2 cells, we have used 1050 V, 30 ms, 2 pulses successfully.Note: Every cell line will vary slightly in the exact electroporation parameters used (pulse voltage, pulse length, pulse number, etc.). Thermo Fisher Scientific provides optimized electroporation conditions for the Neon NxT Transfection system based on the cell line. We recommend following the parameters listed if the cell line of interest is included in their database. In the case of our HepG2 edits, our optimization experiments showed 1050 V, 30 ms, 2 pulses to be the best parameters for electroporation and subsequent cell viability. However, we have found that parameters (i.e., 1200 V, 50 ms, 1 pulse, 1230 V, 20 ms, 3 pulses) can also be used (Figure 1). If no parameters are suggested, 1400 V, 20 ms, 2 pulses tends to be universally effective. We recommend using these parameters in the first electroporation attempt, especially if no database information on the cell line of interest is available.
-
n.Prepare a 24 well plate with 500 μL of warmed antibiotic free culture media in each experimental well.
-
o.Once the cells have been spun down, aspirate the media from the tube and resuspend the pellet in 37.5 μL Genome Editing resuspension buffer.
-
i.For every 140,000 cells (or two wells of a 24 wells), use 12.5 μL of Genome Editing resuspension buffer.
-
ii.It is important to minimize the amount of time cells spend in Genome Editing resuspension buffer. We recommend resuspension as close to the electroporation point as possible.
-
i.
-
p.Prepare the RNP/ssODN/cell suspension mix (25 μL) by combining:
-
i.1.25 μL electroporation enhancer.
-
ii.1.25 μL ssODN (dissolved previously to 100 μM in TE or nuclease-free molecular biology grade water).
-
iii.12.5 μL cell suspension.
-
iv.10 μL RNP mixture (described above).Note: it is important to avoid making bubbles within the electroporation mixture as this will lead to arcing (i.e., a flash) during electroporation. Sample arcing can drastically decrease editing efficiency and cell viability. For this reason, we recommend preparing all edit-focused sample mixtures (i.e., those containing RNPs, ssODNs, and cell suspensions) prior to preparing control samples.Note: The final molar ratio of crRNA/tracrRNA:Cas9:donor DNA template using this method is: 4.8:3.28:5 μM. This can serve as a starting point for future experiments/optimizations.
-
i.
-
q.Prepare the GFP control sample (for 2 wells of a 24 well plate) by combining:
-
i.500 ng GFP plasmid (250 ng/well, e.g., pEGFP-N1, or pCMV GFP).
-
ii.1.25 μL electroporation enhancer.
-
iii.12.5 μL cell suspension.
-
iv.Bring to a final volume of 25 μL with Genome Editing resuspension buffer.Optional: one can use GFP-tagged Cas9 (e.g., Alt-R S.p. Cas9-GFP, Catalog #10008100) to visualize electroporation efficiency (and possibly use for sorting, see above), although this signal is typically not bright and is transient.
-
i.
-
r.Prepare the transfection tube by adding 2 mL of electrolyte solution (“E” solution that comes with the Neon Transfection kit) and place in the chamber. This tube will click into place.
-
i.These tubes can be used up to 10 times granted that all the same reagents are being used. When using different ssODNs, RNPs, etc. it is imperative to use fresh E buffer for each new composition.
-
i.
-
s.Install the Neon 10 μL tip onto the provided pipette.
-
i.Push the pipette plunger down on the provided pipette until the clasps emerge at the base of the tip.
-
ii.Press the pipette firmly into a Neon NxT Transfection pipette tip until it clicks.
-
iii.Release the plunger so the clamps grasp the tip.
-
iv.Remove the tip from the provided box (now attached to the pipette) and inspect for proper fit.
 CRITICAL: There should be no space between the top of the plastic tip and the base of the metal pipette. Depressing and releasing the plunger should also result in movement of the gold tip within the plastic sheath.Note: In the instance that the pipette tip is not properly on the pipette, replace the pipette tip in the box by depressing the plunger completely and pressing down firmly until the tip is released. Follow the previously described steps to try and attach the tip again.Note: Each tip can be used for 2 electroporations containing the same reagents before being disposed. When using different ssODNs, RNPs, etc. it is imperative to use a new tip.
CRITICAL: There should be no space between the top of the plastic tip and the base of the metal pipette. Depressing and releasing the plunger should also result in movement of the gold tip within the plastic sheath.Note: In the instance that the pipette tip is not properly on the pipette, replace the pipette tip in the box by depressing the plunger completely and pressing down firmly until the tip is released. Follow the previously described steps to try and attach the tip again.Note: Each tip can be used for 2 electroporations containing the same reagents before being disposed. When using different ssODNs, RNPs, etc. it is imperative to use a new tip.
-
i.
-
t.Gently resuspend the RNP/ssODN/cell mixture by flicking, careful to avoid bubbles.
-
u.Pipette 10 μL of the solution into the electroporation tip on the provided pipette.
-
i.Depress the plunger until the very tip of the gold pipette is exposed at the bottom of the sheath.
 CRITICAL: Do not depress too far or the tip will be released.
CRITICAL: Do not depress too far or the tip will be released. -
ii.Insert the pipette tip into solution and slowly release the plunger to pick up the sample, being cautious to avoid air bubbles.
-
iii.Inspect the sample within the tip for any air bubbles.
 CRITICAL: If there are air bubbles in the tip, return the sample to the tube and try collecting the sample again.
CRITICAL: If there are air bubbles in the tip, return the sample to the tube and try collecting the sample again. CRITICAL: If air bubbles persist, make sure the pipette tip has been properly installed. If the pipette tip is properly installed, add 1–3 μL of Genome Editing resuspension buffer to the sample and try collection again.
CRITICAL: If air bubbles persist, make sure the pipette tip has been properly installed. If the pipette tip is properly installed, add 1–3 μL of Genome Editing resuspension buffer to the sample and try collection again.
-
i.
-
v.Place the pipette in the electroporation tube containing E buffer.
-
w.Gently press down until it clicks into place.
-
x.Confirm that the desired parameters are selected in the Neon menu.
-
y.Click start on the Neon NxT Transfection system to begin electroporation.
-
i.Watch the tip containing the sample for bright flashes.Note: These flashes are indicative of arcing and the samples that show arcing should be noted.
-
i.
-
z.Pipette the 10 μL of electroporated sample into one well of a 24 well plate prepared with antibiotic free culture media.
-
aa.Gently rock the plate to disperse the cells and repeat electroporation for all edited samples and GFP control samples.
 CRITICAL: Plate untransfected cells as a control by adding 5 μL of the cell suspension per well.Optional: If you would like to increase the chances of homology directed repair (HDR), add HDR enhancer solution to the wells containing electroporated cells to a final concentration of 1 μM per well. Rock gently to disperse.Optional: HDR enhancers such as SCR7 (e.g., STEMCELL Technologies Cat# SCR7) can also be considered here.
CRITICAL: Plate untransfected cells as a control by adding 5 μL of the cell suspension per well.Optional: If you would like to increase the chances of homology directed repair (HDR), add HDR enhancer solution to the wells containing electroporated cells to a final concentration of 1 μM per well. Rock gently to disperse.Optional: HDR enhancers such as SCR7 (e.g., STEMCELL Technologies Cat# SCR7) can also be considered here. -
ab.Incubate the cells at 37°C for 48–72 h.
 CRITICAL: Cells should be monitored daily to assess cell attachment, growth, and viability. Approximately 24 h after transfection, cells can be imaged for GFP plasmid transfection to determine electroporation efficiency. We recommend using the Celigo Image Cytometer by Nexcelom for cell imaging (or a similar instrument). Imaging with this instrument will provide whole-well information that can be easily tracked across time. However, a standard fluorescence microscope would also work for assessing electroporation efficiency and cell viability.
CRITICAL: Cells should be monitored daily to assess cell attachment, growth, and viability. Approximately 24 h after transfection, cells can be imaged for GFP plasmid transfection to determine electroporation efficiency. We recommend using the Celigo Image Cytometer by Nexcelom for cell imaging (or a similar instrument). Imaging with this instrument will provide whole-well information that can be easily tracked across time. However, a standard fluorescence microscope would also work for assessing electroporation efficiency and cell viability.
-
a.
-
2.Day 2: Cell Monitoring and Efficiency Assessment (troubleshooting 1).
-
a.Remove cell plate from the incubator and take to the instrument.
-
b.Turn on the computer and Celigo instrument. Open the software and log into the appropriate account.
-
c.Click “Create New Scan”.
-
d.Select the culture plate specifications from the drop-down list.
-
i.If using the recommended 24 well plates from the key resources table, select “24-well Corning 3524 Plate”.
-
i.
-
e.Appropriately name the experiment and click “Load Plate”. The Celigo will eject the tray to hold your culture plate.
-
f.Load the plate with the lid on.
-
g.Once the plate has been placed in the tray, click “Next” on the pop up on the computer and the instrument will take in the tray and automatically switch screens.
-
h.From the drop-down list labeled “Please Select…” in the top left of the screen, select “Plate Reader” and “Whole Well 1 + 2”.Note: This will allow for imaging with 2 different channels. We recommend imaging all wells of the plate in “Brightfield” and “Green 483/536” settings. This will allow for comparison of cell densities, dispersion, and GFP expression. Within the GFP control wells themselves, comparison of GFP expressing and non-expressing cells will determine electroporation efficiency.
-
i.Under the section titled “Channel”, select “Channel 1”.
-
j.Under “Configuration”, the default illumination setting will be set to “Far Red 632/692”. Change this setting to “Brightfield”.Note: The main portion of the screen will show a preview of the selected well shown in the plate map on the right of the screen. We recommend changing the viewing setting to “Live” using the “Camera Control” buttons. This will allow for more accurate focusing while setting up imaging parameters.
-
k.Under “Motion Control” settings, select “Focus Setup” and a small pop-up window will appear with options to help focus the image.
-
l.Select “Register Auto” and allow the instrument to find the appropriate focus settings. This concludes the setup for Channel 1 imaging.
-
m.Select “Channel 2” from the “Channel” settings box.
-
i.Change this setting to “Green 483/536” and the main portion of the screen will show a preview of the selected well shown in the plate map on the right.Note: You can change the previewed well by clicking the dot on the plate map corresponding to a well containing the GFP control. Allow a few seconds for the instrument to move the plate and reset the preview image. Once the GFP well is selected, the well preview will be automatically updated on the screen.
-
i.
-
n.Under “Configuration” settings, change the “Exposure Time (μs)” to adjust to the level of GFP expression present in the well.Note: We recommend choosing an exposure that allows for a detailed image of individual cells expressing GFP without overexposing and showing non-expressing cells. We have found that the exposure setting typically lies between 10,000 and 35,000, however this can vary.
-
o.Once the exposure settings have been set, click “Find Focus” under “Motion Control” settings and allow the instrument a few seconds to adjust. This will set the focus for Channel 2.
-
p.Select “Set Offset” to save the focus settings specific to Channel 2 separate from those determined for Channel 1.
-
q.In the plate map on the right of the screen, change the settings from “Navigation” to “Selection” and select the appropriate wells for imaging.Note: Selected wells will be yellow and unselected wells will be gray.
-
r.At the bottom right of the screen, select “Start Scan” and allow the instrument time to go through and image the wells.
-
i.Once a well has been imaged for both channels, the well in the plate map will turn green and is ready for preview.
-
ii.To preview, select the green well from the plate map.
-
iii.Change the viewing settings to see the entire well by selecting “Well” and then “Well” again. This will change the image on the screen to show the entirety of the well that was imaged.
-
iv.The default settings will show the selected well with both channels overlaid. This can be manipulated using the “Channel 1” and “Channel 2” buttons under “Camera Controls”.
-
v.Use the mouse to zoom in and out of the preview image.
-
vi.In GFP expressing wells, we recommend zooming into the well until you can identify individual cells. Overlay channels 1 and 2 to assess GFP expressing and non-expressing cells.
-
vii.Without changing the field of view settings, you can move between the GFP expressing wells in the plate map and compare the exact same field of view. Between the 2 GFP control wells, estimate the editing efficiency of the electroporation.
-
viii.For example, in a field of view containing 10 cells, if 8 cells are expressing GFP and 2 are not, we can estimate the editing efficiency to be 80%.
-
i.
-
s.Once all of the selected wells have been imaged and turned green, select “Start Analysis” at the bottom right of the screen. This will take you to the analysis page where you can compare the densities, intensities, and well images all on one screen.Note: While this information can be valuable, we have found that the numbers produced in the analysis step may be unreliable for the purposes of this experiment. Therefore, we recommend prioritizing the estimation of efficiency from the image preview screen and using the analysis data as secondary verification if necessary.
-
t.At the top right of the screen, select “Export All Images” to save the images outside of the instrument software.
-
i.In the pop up, select the box next to “Stitch Images” and change the pixel number according to how sharp the image needs to be.Note: The lower the pixel number, the more detailed the image but the longer exporting will take and the larger the final file will be. We recommend 2–5 pixels for clear, detailed images that can be used for validation while also maintaining a manageable file size.
-
ii.Export data.
-
i.
-
u.At the top of the screen, select the eject icon to eject the plate tray from the instrument.
-
v.Remove the culture plate and select “Next” on the pop up on the screen. The instrument will take the tray back in.
-
w.Log out of the system and be sure to transfer any exported files as appropriate.
-
x.Replace the culture plate containing the cells back in the incubator.Note: While we describe a protocol to image whole wells using a Celigo instrument, one can also use a standard inverted fluorescence microscope to also image eGFP signal and estimate transfection efficiency. We would recommend taking images with low magnification (4–10×) under fluorescence and using bright field to quantify as many cells as possible within each well.
 CRITICAL: Two or three days after transfection, cell media can be changed back to full growth media containing antibiotics. We recommend leaving cells alone for the full 72 h after transfection to increase editing efficiency and cell recovery. However, if cell viability is decreasing, it may be beneficial to change media closer to 48 h post-transfection.
CRITICAL: Two or three days after transfection, cell media can be changed back to full growth media containing antibiotics. We recommend leaving cells alone for the full 72 h after transfection to increase editing efficiency and cell recovery. However, if cell viability is decreasing, it may be beneficial to change media closer to 48 h post-transfection.
-
a.
-
3.Day 3 or 4 (48–72 h post-transfection): Changing Media.
-
a.Remove cell plate from the incubator and place in a culture hood.
-
b.Aspirate the media from the wells and wash with 250 μL HBSS.
-
c.Aspirate the HBSS and replace with 500 μL of full culture media (containing antibiotics).
-
d.Return cell plate in the incubator and allow to continue to grow. Begin subculturing as normal.
-
a.
Table 1.
Sequences of gRNA, ssODN, and gDNA amplification primers used to generate and validate CETP HiBiT editing
| HiBiT edit | gRNA | PAM | ssODN | Exon 16 primers |
|---|---|---|---|---|
| CETP VS 2×FLAG VS HiBiT (human) |
GAGACTTCTAGCTCAAGCTC (set 2 from Figure 3; on-target score: 83, off-target score: 49) | TGG | ACTTTGGCTTCCCTGAGCACCTGCTGGTGGATTT CCTCCAGAGCTTGAGCGTGAGCGATTACAAGGA TGACGACGATAAGGATTACAAGGATGACGACG ATAAGGTGAGCGTGAGCGGCTGGCGGCTGTTC AAGAAGATTAGCTAGAAGTCTCCAAGGAGGTC GGGATGGGGCTTGTAGCAGAAGGCAAGCAC |
Forward: TACCAGCTTGGCTCCCTCCTGGT Reverse: GCTTGCCTTCTGCTACAAGCCCCAT |
HiBiT assay analysis of edits
Timing: 10–30 min
Analysis of electroporated cells can be done in various ways. For editing confirmation, we recommend starting with HiBiT assays since it is a simple and quick method. For secreted proteins that have been edited, we recommend running extracellular HiBiT assays starting 48 h after transfection. We have found that editing is typically not easily detectable until closer to the 72 h mark, or even after media has been changed back to full culture media. However, this is likely dependent on the amount of HiBiT tagged protein produced by a cell and the editing efficiency. We recommend to continue to take 24 h time point assays for data collection. True HiBiT signal should accumulate in the media over time. For proteins that are not secreted, analysis by intracellular HiBiT assays is recommended but should only be done once the electroporated samples have had time to grow and be expanded as this assay will lyse the sample for analysis.
-
4.Secreted HiBiT Assay (troubleshooting 2).
 CRITICAL: This protocol is written for 1 HiBiT assay in correspondence to analysis of 1 sample. This can be scaled up to the desired number of assays. The purpose of this assay is to evaluate HiBiT signal associated with appropriately tagged proteins that have been secreted into the media. Since we remove an aliquot of media from the edited cells, we can use the NanoGlo HiBiT Lytic Detection System, which is cheaper than the Extracellular Detection version. However, if one would like to measure HiBiT amounts within a well containing cells (i.e., media is not transferred to a new plate), the NanoGlo HiBiT Extracellular Detection System should be used to prevent cell lysis. More information about these systems can be found on the Promega website (https://www.promega.com/resources/technologies/hibit-protein-tagging-system/).Optional: One has the option of performing live-cell HiBiT assays (i.e., Promega catalog# N2570, N2580) if desired. Our protocol is outlined for a Promega GloMax Discover plate reader, but can be similarly adapted to other plate readers.
CRITICAL: This protocol is written for 1 HiBiT assay in correspondence to analysis of 1 sample. This can be scaled up to the desired number of assays. The purpose of this assay is to evaluate HiBiT signal associated with appropriately tagged proteins that have been secreted into the media. Since we remove an aliquot of media from the edited cells, we can use the NanoGlo HiBiT Lytic Detection System, which is cheaper than the Extracellular Detection version. However, if one would like to measure HiBiT amounts within a well containing cells (i.e., media is not transferred to a new plate), the NanoGlo HiBiT Extracellular Detection System should be used to prevent cell lysis. More information about these systems can be found on the Promega website (https://www.promega.com/resources/technologies/hibit-protein-tagging-system/).Optional: One has the option of performing live-cell HiBiT assays (i.e., Promega catalog# N2570, N2580) if desired. Our protocol is outlined for a Promega GloMax Discover plate reader, but can be similarly adapted to other plate readers.-
a.Thaw HiBiT Lytic Buffer (100 mL) provided in the Nano-Glo HiBiT Lytic Detection System kit.
-
i.This can be done by leaving it to thaw for a few hours at 25°C or for at least 10 h at 4°C.
 CRITICAL: Do not thaw above 25°C as this buffer contains labile dithiothreitol (DTT).Note: We recommend aliquoting the lytic buffer post-thaw. Stock aliquots can be stored at −20°C for long term storage and in our experience, can endure up to 10 freeze thaw cycles with no obvious change in performance. Working aliquots can be kept at 4°C for up to 1 year, or at RT for up to 3 months. We recommend keeping working aliquots at RT as we have found that equilibration of this buffer at RT leads to less variability in assay readouts.
CRITICAL: Do not thaw above 25°C as this buffer contains labile dithiothreitol (DTT).Note: We recommend aliquoting the lytic buffer post-thaw. Stock aliquots can be stored at −20°C for long term storage and in our experience, can endure up to 10 freeze thaw cycles with no obvious change in performance. Working aliquots can be kept at 4°C for up to 1 year, or at RT for up to 3 months. We recommend keeping working aliquots at RT as we have found that equilibration of this buffer at RT leads to less variability in assay readouts.
-
i.
-
b.Add 50 μL HiBiT Lytic Buffer at RT to an Eppendorf tube.
 CRITICAL: A single assay requires 25 μL of HiBiT Lytic Buffer, but values should be scaled up to account for pipetting error. For larger assays, we recommend scaling up an extra 10% to account for pipette error (e.g., for 20 samples, prepare enough HiBiT assay solution for 22 samples, etc.).
CRITICAL: A single assay requires 25 μL of HiBiT Lytic Buffer, but values should be scaled up to account for pipetting error. For larger assays, we recommend scaling up an extra 10% to account for pipette error (e.g., for 20 samples, prepare enough HiBiT assay solution for 22 samples, etc.). -
c.Add 0.5 μL HiBiT LgBiT protein to HiBiT Lytic Buffer/Substrate mixture (1:100 ratio).
-
d.Add 1 μL HiBiT Lytic Substrate to the HiBiT Lytic Buffer (1:50 ratio).Note: The resulting mixture is a 2× HiBiT assay solution.
-
e.Mix the 2× HiBiT assay solution by pipette resuspension or gently flicking to mix.
 CRITICAL: Do not vortex the mixture, as vortexing can cause bubbles that can negatively impact the assay variability.
CRITICAL: Do not vortex the mixture, as vortexing can cause bubbles that can negatively impact the assay variability. -
f.Add 25 μL of the 2× HiBiT assay solution to a black 96 well plate.Note: A white 96 well plate can also be used that will increase the resultant signal, but will also increase background luminescence readings.
-
g.Place the assay plate in a culture hood along with the cell plate to be assayed.
-
h.Remove the lid of the cell plate and tilt the plate towards you slightly to gather the media at the base of the well.
-
i.Remove 25 μL of the media from the well without touching the adherent cells.
-
j.Plate the 25 μL of media directly on top of the 2× HiBiT assay solution in the black 96 well plate and gently resuspend.
-
k.Return the cell plate in the incubator.
-
l.Shake the 96 well assay plate on a plate shaker at 400 rpm for 5 min.
-
m.While the sample is shaking, turn on the Promega GloMax Discover plate reader.
-
n.Select “Luminescence”.
-
o.Select the wells to be analyzed.
-
i.Blue squares correspond to selected wells.
-
ii.Integration time can vary; however, we use 2 s/well.
-
i.
-
p.Select the door button in the top right to eject the tray.
-
q.After 5 min of shaking, remove the lid from the 96 well assay plate and load into the instrument.
-
r.Select the door button again to take in the tray.
-
s.Select “Start” at the bottom right of the screen to begin instrument initialization.
-
t.A pop-up window will appear. Appropriately title your scan here and select “Start”. The instrument will begin acquisition and data will appear on the screen as the readings are taken.
 CRITICAL: It is important to include controls when reading luminescence values in order to determine background. When analyzing electroporated samples, we recommend including at least one assay of the unelectroporated control that was plated to give context to the readouts. Typically, we see background readings anywhere from 25–200 arbitrary luminescence units (ALU) for media alone or media from unedited cells.
CRITICAL: It is important to include controls when reading luminescence values in order to determine background. When analyzing electroporated samples, we recommend including at least one assay of the unelectroporated control that was plated to give context to the readouts. Typically, we see background readings anywhere from 25–200 arbitrary luminescence units (ALU) for media alone or media from unedited cells. -
u.After acquisition, save and transfer the data as necessary. Data analysis should include averaging and normalization as appropriate. Signal from electroporated samples should be normalized to unelectroporated controls to determine whether editing took place or not. Recall that luminescence is proportional to the amount of protein that has been HiBiT tagged.
-
a.
-
5.Intracellular HiBiT Assay (troubleshooting 3).
 CRITICAL: This protocol is written for 1 HiBiT assay in correspondence to analysis of 1 sample in a 24 well plate. This can be scaled up or down depending on the sample size and number of assays. The purpose of this assay is to evaluate HiBiT signal associated with appropriately tagged proteins that have been expressed within cultured cells. We recommend this type of evaluation only after growing up and expanding edited cells to an appropriate volume, as this assay will lyse the cells for intracellular analysis.
CRITICAL: This protocol is written for 1 HiBiT assay in correspondence to analysis of 1 sample in a 24 well plate. This can be scaled up or down depending on the sample size and number of assays. The purpose of this assay is to evaluate HiBiT signal associated with appropriately tagged proteins that have been expressed within cultured cells. We recommend this type of evaluation only after growing up and expanding edited cells to an appropriate volume, as this assay will lyse the cells for intracellular analysis.-
a.Add 50 μL HiBiT Lytic Buffer at RT to an Eppendorf tube.
-
i.A single assay requires 25 μL of HiBiT Lytic Buffer, but values should be scaled up to account for pipetting error.Note: For larger assays, we recommend scaling up an extra 10% to account for pipette error (i.e. for 20 samples, prepare enough HiBiT assay solution for 22 samples, etc.).
-
i.
-
b.Add 1 μL HiBiT Lytic Substrate to the HiBiT Lytic Buffer (1:50 ratio).
-
c.Add 0.5 μL HiBiT LgBiT Protein to HiBiT Lytic Buffer/Substrate mixture (1:100 ratio).Note: The resulting mixture is a 2× HiBiT assay solution.
-
d.Place the Eppendorf tube containing the mixture in a culture hood and dilute in a 1:1 ratio with HBSS to produce a 1× working solution.
-
e.Remove the cell plate to be assayed and place in the culture hood.
-
f.Aspirate the media from the well to analyze.
-
g.Wash the cells with 250 μL HBSS.
-
h.Aspirate the HBSS.
-
i.Add 50 μL (for a 24 well plate) of the 1× HiBiT assay solution directly to the adherent cells in the well.
 CRITICAL: For culture plates of various sizes, add 10% of the well volume of the 1× HiBiT assay solution. This may require additional scaling up when preparing the 2× solution when working with plates larger than 24 wells, so plan volumes accordingly.
CRITICAL: For culture plates of various sizes, add 10% of the well volume of the 1× HiBiT assay solution. This may require additional scaling up when preparing the 2× solution when working with plates larger than 24 wells, so plan volumes accordingly. -
j.Shake the 24 well assay plate on a multiplate shaker at 400 rpm for 5 min.
-
k.While the sample is shaking, turn on the Promega GloMax Discover plate reader.
-
l.Select “Luminescence”.
-
m.Select the wells to be analyzed. Blue squares correspond to selected wells.
-
n.After 5 min of shaking, transfer 40 μL of the lysed solution from the culture plate to a well in a black 96 well assay plate.
 CRITICAL: For plates of various sizes, it is recommended to normalize the volume transferred from the culture plate to the assay plate to be 80% of volume of HiBiT solution (in this case 40 out of 50 total μL) added to the wells. This will help avoid pipette error and normalize samples for easier comparison.
CRITICAL: For plates of various sizes, it is recommended to normalize the volume transferred from the culture plate to the assay plate to be 80% of volume of HiBiT solution (in this case 40 out of 50 total μL) added to the wells. This will help avoid pipette error and normalize samples for easier comparison. -
o.Select the door button in the top right to eject the tray.
-
p.Load the 96 well assay plate onto the tray.
-
q.Select the door button again to take in the tray.
-
r.Select “Start” at the bottom right of the screen to begin instrument initialization.
-
s.A pop-up window will appear.
-
i.Appropriately title your scan here and select “Start”. The instrument will begin acquisition and data will appear on the screen as the readings are taken.
-
i.
-
t.After acquisition, save and transfer the data as necessary.
 CRITICAL: Data analysis should include averaging and normalization as appropriate. Signal from electroporated samples should be normalized to unelectroporated controls to determine whether editing took place or not. Further analysis can be done by comparing intracellular assay results to extracellular assay results for a bigger picture of the nature of the HiBiT tagged protein.
CRITICAL: Data analysis should include averaging and normalization as appropriate. Signal from electroporated samples should be normalized to unelectroporated controls to determine whether editing took place or not. Further analysis can be done by comparing intracellular assay results to extracellular assay results for a bigger picture of the nature of the HiBiT tagged protein.
-
a.
gDNA analysis of edits
Timing: 4–6 h, longer if extracting DNA and sending for Sanger sequencing
Extracellular and intracellular HiBiT assays can be used to quickly assess whether HiBiT tagging of proteins may or may not have taken place. While a positive signal usually indicates that editing has occurred, a lack of HiBiT signal does not necessarily mean that no editing has taken place. In this latter scenario, it is possible that editing has taken place, but that the editing is inefficient, or that the protein is not being produced, or that it is being produced at very low levels. The electroporation protocol listed above prepares samples in duplicate with the intention of potentially using one well for gDNA analysis and keeping one to be cultured further. The purpose of this portion of the protocol is to harvest and analyze gDNA editing of a target gene from edited cells to verify HiBiT editing.
Note: This protocol is written for analysis of 1 well of a 24 well plate but can be scaled up or down accordingly. Example data of gDNA amplification and analysis from CETP HiBiT-edited HepG2 cells is found in Figure 4.
CRITICAL: It is very important to also include an unedited control well of cells for appropriate comparisons.
-
6.Cell Harvesting.
-
a.Remove the plate containing potentially edited cells from the incubator and place the plate in the culture hood.
-
b.Aspirate the media from the desired well.
-
c.Wash the well with 250 μL of HBSS.
-
d.Aspirate the HBSS.
-
e.Add 200 μL Trypsin-EDTA (0.25%).
-
f.Replace the plate in the incubator.
-
i.Allow adequate time for the cells to trypsinize, typically 3–5 min.
-
ii.Verify trypsinization under a microscope before proceeding.
-
i.
-
g.Neutralize the well with 800 μL of full culture media.
-
h.Resuspend the 1 mL volume within the well using a pipette. This will help to detach any cells that may still be slightly adherent as well as homogenize the sample.
-
i.Transfer the entire sample volume (1 mL) to an Eppendorf tube.
-
j.Spin the sample down at 1,000 × g for 3–5 min to pellet the cells.
-
k.Aspirate the supernatant being mindful not to disturb the pellet.Optional: Add 500 μL HBSS to the pellet to wash.
-
l.Spin the sample down at 3,000 rpm for 3–5 min to pellet the cells again.
-
m.Aspirate the HBSS. The cells have now been harvested as a pellet and are ready for gDNA extraction.Note: If gDNA extraction will not be performed immediately, the pellet can be frozen at −20°C until ready. For longer term storage of the pellet, keep frozen at −80°C.
-
a.
-
7.gDNA Extraction.
-
a.Extract the gDNA from the cell pellet.Note: We recommend using the Monarch Genomic DNA Purification kit and following the protocol provided by New England Biolabs. Alternatively, a simple alkaline extraction has been used successfully to extract gDNA also.24Note: We recommend following the lower volume elution (35 μL) directions for increased gDNA concentration using the Monarch Genomic DNA Purification kit. We also recommend elution with molecular biology grade water instead of the provided elution buffer.
 CRITICAL: if using water to elute, make sure water is also used as the blank for any DNA quantification.
CRITICAL: if using water to elute, make sure water is also used as the blank for any DNA quantification. -
b.Quantify the DNA elution. We recommend using Thermo Scientific NanoDrop OneC Microvolume UV-Vis Spectrophotometer for quantification of double stranded DNA.
-
i.Turn on the instrument.
-
ii.Select “dsDNA” to quantify double stranded DNA samples (e.g., gDNA extractions).
-
iii.Lift the arm and load 2 μL of blank elution solution (either water or elution buffer) to the center of the bottom pedestal.
-
iv.Carefully lower the arm and allow the instrument to read the blank.
-
v.Lift the arm and clean the pedestal.
-
vi.Load 2 μL of sample onto the center of the bottom pedestal.
-
vii.Carefully lower the arm and allow the instrument to read the sample. The measured concentration (in ng/μL) will appear on the screen.
-
viii.Record the concentration.
-
ix.Lift the arm, clean the pedestal, and repeat steps ‘vi’ through ‘viii’ until all samples have been read.
-
x.Click “End Experiment” at the bottom right of the screen.
-
xi.Click “End” on the pop-up screen.
-
xii.Clean both pedestals on the instrument, lower the arm back to resting position, and turn off the instrument.
-
i.
-
c.DNA samples can be kept at 4°C for short term storage or −20°C for longer term storage.
-
a.
-
8.PCR.
-
a.For preparation of a 20 L reaction, combine the following reagents in a PCR tube:
-
i.10 μL Q5 Master Mix (2×).
-
ii.1 μL forward gDNA primer, 1 μL reverse gDNA primer.Note: CETP Exon 16 forward primer: TACCAGCTTGGCTCCCTCCTGGTNote: CETP Exon 16 reverse primer: GCTTGCCTTCTGCTACAAGCCCCAT
 CRITICAL: Primers can be designed in any number of ways. We use Benchling to identify amplification areas surrounding our HiBiT insertion site using primers with a melting temperature of ∼62°C–65°C and 25–35 bp in length. Typically amplicons should be 150–300 bp. This size will allow one to detect insertion of the HiBiT tag (and any other accompanying sequence, such as a VS linker and FLAG tag(s).Optional: The VS linker is a starting point for linker addition, but may need to be changed in terms of composition and/or length depending on the context of each edited POI and how well tolerated the HiBiT insertion is.20 ng gDNA extracted from cell pellet.
CRITICAL: Primers can be designed in any number of ways. We use Benchling to identify amplification areas surrounding our HiBiT insertion site using primers with a melting temperature of ∼62°C–65°C and 25–35 bp in length. Typically amplicons should be 150–300 bp. This size will allow one to detect insertion of the HiBiT tag (and any other accompanying sequence, such as a VS linker and FLAG tag(s).Optional: The VS linker is a starting point for linker addition, but may need to be changed in terms of composition and/or length depending on the context of each edited POI and how well tolerated the HiBiT insertion is.20 ng gDNA extracted from cell pellet.
-
i.
-
b.Bring to volume with molecular biology grade water.Note: The amount of gDNA can vary by PCR. However, we recommend normalizing the amount of gDNA across samples for easier comparison. Furthermore, we recommend that the volume of gDNA does not exceed 10% of the reaction volume so that the PCR can be effectively run. For verification of editing, it is typically not necessary to run a PCR reaction volume greater than 30 μL.
-
c.Turn on the MiniAmp Thermal Cycler.
-
d.Set up the PCR according to the following table:
PCR step Temperature Time Cycles Initialization 98°C 30 s 1× Denaturation 98°C 10 s 30–35× Annealing Varies by primer set, 63°C for CETP 30 s Elongation 72°C 1 min Final Elongation 72°C 2 min Hold 4°C - 1×
-
a.
-
9.Gel Electrophoresis and Imaging (troubleshooting 4, 5, and 6).
-
a.Prepare a 1.8% agarose gel.
-
i.Weigh 0.9 g of agarose in a weigh boat and transfer to a 250 mL flask.
-
ii.Measure out 50 mL 1× TBE in a graduated cylinder and add to the 250 mL flask containing the agarose.
-
iii.Gently swirl to begin dissolving the agarose.
-
iv.Stuff the opening of the flask with Kim Wipes to prevent evaporation.
-
v.Microwave the flask for 70 s.
-
i.
-
b.Prepare a small gel form by taping off the open ends of the gel holder and adding a comb.
-
c.Once the agarose has been heated, remove carefully from the microwave using a heat protectant glove.
-
i.Discard the Kim Wipes.
-
ii.Visually inspect the gel solution to confirm that all the agarose has been dissolved in the 1× TBE.
-
i.
-
d.Place the flask on the bench and allow it to cool slightly so that it is warm, but not hot to the touch.
-
e.Add 2 μL Gel Green dye to the gel solution and swirl to distribute.
-
f.Once the gel solution has begun to cool, pour into the prepared gel form.
-
g.Use a small pipette tip to pop or move any bubbles to the edges of the gel.
-
i.Allow the gel to cool completely before proceeding (typically, minimum of 20 min).
-
i.
-
h.Once the gel has completely set, remove the tape and comb from the gel and place in a gel rig.
-
i.Fill the rig with 1× TBE running buffer until the wells of the gel are completely covered.
-
j.Add Purple Gel Loading Dye (6×) to PCR products for a final concentration of 1×. For a 20 μL reaction volume, add 4 μL of Purple Gel Loading Dye (6×).
-
k.Load 4 μL of 100 bp ladder in lane 1 of the gel.
-
l.Load the PCR products onto the gel.
-
m.Connect the gel rig to a power source and run at 140 V for 30–60 min.
-
i.Image the gel. We recommend using the LI-COR Odyssey FC imager.
-
ii.Remove gel from the gel rig and gently place on a LI-COR Odyssey FC imaging tray.
-
iii.On the LI-COR Odyssey FC imager, select the eject button and load the imaging tray.
-
iv.Press the eject button again to retract the tray.
-
v.Log into the LI-COR software and connect to the FC imager.
-
vi.Image the gel using the 600 channel and a 30 s exposure time.
-
i.
-
n.Verify that the DNA amplicon size is as expected for the primers used in the PCR.
-
o.Determine whether editing took place or not.
-
i.If editing did not take place, it is expected that the amplicon of the edited sample would exactly match that of the wild-type (or unedited) control.
-
ii.If editing did take place, the gDNA would include an insertion segment that is not present in the unedited wild-type sample. This would show up as a higher band on the gel corresponding to the increased number of base pairs equal to that of the insertion sequence length.
-
i.
-
p.Calculate editing efficiency.
-
i.In the LI-COR software, click the “Analysis” tab at the top of the screen.
-
ii.Select “Draw shape” and outline the bands present in the gel.
 CRITICAL: It is important to be cautious of the shape dimensions as this can impact the software’s determination of background. The space between the inner and outer squares will determine background. Make sure none of the bands lie within this region. The band should be fully enclosed within the inner square of the shape.
CRITICAL: It is important to be cautious of the shape dimensions as this can impact the software’s determination of background. The space between the inner and outer squares will determine background. Make sure none of the bands lie within this region. The band should be fully enclosed within the inner square of the shape. -
iii.The software will determine relative readings of band intensity based on the drawn shapes. Use these intensities to determine editing efficiency by dividing the insertion band’s intensity by the total intensity within that lane.
-
i.
-
q.If an edited band is present, proceed with gel excision, DNA extraction, and Sanger Sequencing to verify the proper insertion.
-
a.
-
10.Gel Excision and Extraction.
-
a.Place the gel on a blue light transilluminator.
-
b.Using the protective screen or glasses, view the gel and identify the band(s) corresponding to edited samples.
-
c.Using a fresh razor blade, excise the band of interest by cutting completely through the gel around the band.
 CRITICAL: Be careful not to cut through the band itself. However, it is recommended to cut as closely to the band as possible.
CRITICAL: Be careful not to cut through the band itself. However, it is recommended to cut as closely to the band as possible. CRITICAL: Do not use the same razor blade for different samples as this could cross contaminate DNA samples.
CRITICAL: Do not use the same razor blade for different samples as this could cross contaminate DNA samples. CRITICAL: Dispose of the razor blades in a “sharps only” disposal after one use.
CRITICAL: Dispose of the razor blades in a “sharps only” disposal after one use. -
d.Confirm that the correct band has been excised and transfer the excision into a new Eppendorf tube.
-
i.This gel can be immediately used for DNA extraction or stored for DNA extraction at a later date. Gel excisions can be stored at 4°C for short term or −20°C for longer term. It is recommended to wrap the Eppendorf tube with Parafilm prior to storage to avoid drying out.
-
i.
-
e.Extract the DNA from the gel. We recommend using the QIAquick Gel Extraction kit.Note: This kit comes with all necessary reagents and columns (except for molecular grade ethanol, molecular grade isopropanol, and Eppendorf collection tubes), as well as an instruction manual for the step-by-step procedure.Note: The protocol recommends an additional incubation time of 2–5 min in Buffer PE before centrifuging if the samples will be used for salt sensitive applications. We do not recommend following this step as we have found it to be unnecessary and have little to no impact on sequencing results. However, we do recommend a longer final spin after PE wash (2 min instead of 1 min), and an additional 1 min at RT to allow any excess ethanol to evaporate prior to elution.Note: To elute, we recommend using a lower volume (30 μL) of molecular biology grade water. This will result in an increased DNA concentration. We also recommend following the increased incubation time during elution to increase yield.
 CRITICAL: If using water to elute, make sure water is also used as the blank for any DNA quantification.
CRITICAL: If using water to elute, make sure water is also used as the blank for any DNA quantification. -
f.Quantify the DNA concentration. We recommend using Thermo Scientific NanoDrop OneC Microvolume UV-Vis Spectrophotometer for quantification of double stranded DNA.
-
i.Turn on the instrument.
-
ii.Select “dsDNA” to quantify double stranded DNA samples (i.e., gDNA extractions).
-
iii.Lift the arm and load 2 μL of blank to the center of the bottom pedestal.
-
iv.Carefully lower the arm and allow the instrument to read the blank.
-
v.Lift the arm and clean the pedestal.
-
vi.Load 2 μL of sample onto the center of the bottom pedestal.
-
vii.Carefully lower the arm and allow the instrument to read the sample. The measured concentration (in ng/μL) will appear on the screen.
-
viii.Record the concentration.
-
ix.Lift the arm, clean the pedestal, and repeat steps 'vi' through 'viii' until all samples have been read.
-
x.Click “End Experiment” at the bottom right of the screen.
-
xi.Click “End” on the pop-up screen.
-
xii.Clean both pedestals on the instrument, lower the arm back to resting position, and turn off the instrument.
-
i.
-
g.DNA samples can be kept at 4°C for short term storage or −20°C for longer term storage.
-
a.
-
11.Sanger Sequencing (troubleshooting 7).Note: Sanger sequencing is a process that can be used to determine the exact DNA sequence within a sample. For the purposes of this experiment, Sanger sequencing allows determination of the DNA sequence within the excised gel bands corresponding to edited DNA. This provides positional information of the possible HiBiT insertion within the excised sequence. One can use this information to verify that that the insertion matches the ssODN used in electroporation as well as verify editing within the right target gene.Optional: We use Eurofins Genomics, but any Sanger Sequencing Core could do. Follow specific guidelines for your selected Core or company. Example Sanger chromatograms of unedited CETP gDNA (i.e., lower band in Figure 4) and edited CETP HiBiT gDNA (i.e., upper band in Figure 4) are provided as Figure 5.Optional: As an alternative to Sanger sequencing, one can also perform next generation sequencing (NGS), however, in our experience, this is overkill and is not necessary unless the Sanger sequencing is inconclusive.
-
a.Prepare Eppendorf tubes labeled appropriately to differentiate samples. It is important to keep track of what DNA template is in which tube, as well as which primer was used.
-
b.To each tube, add:
-
i.5 μL DNA template (typically 10–100 ng/μL).
-
ii.5 μL 10 μM primer.Note: We recommend 5 μL for DNA concentrations between 10–100 ng/μL. For DNA samples of a lower concentration, you can increase the volume of DNA template used to 8 μL, making sure to match the primer volume 1:1. This will yield a higher volume sample for sequencing that may increase the chances of better sequencing results. For higher concentrations, dilute the samples appropriately prior to preparation.
-
i.
-
c.Go to Eurofins Genomics website (https://eurofinsgenomics.com/en/home/).
-
d.Select the “Products & Services” tab.
-
e.In the drop-down menu titled “Sanger Sequencing”, select “Tube Sequencing”.
-
f.On the right of the next screen, select the blue button labeled “Tube Sequencing” with the cart icon.
-
g.This will prompt you to log into, or make, an account.
-
h.Once logged in, select the number of samples from the drop-down menu.
-
i.Select “Standard & SimpleSeq tubes” and click next.
-
j.Under “Sample Type”, specify PCR product. We do not recommend any additional services for this sequencing, but it is an available option if necessary.
-
k.Check the box that specifies premixed samples.
-
l.Select product length from the list.
-
m.Under “Template Name”, add the tube labels for all samples. Barcodes are not necessary but can be used instead if preferred.
-
n.Click “Add to Cart”.Note: If you are not using barcodes, a pop-up window will ask for confirmation before you are able to proceed.
-
o.Check out and deliver samples as appropriate.Note: Typically, sequencing results will be received the day after the samples have been picked up.
-
p.To view and analyze sequencing results, we recommend using Benchling to identify the expected gDNA sequence.
-
i.Load this sequence into SnapGene as a new DNA/RNA file.
-
i.
-
q.Add the insertion from the ssODN design to the appropriate spot within the gDNA.
-
i.Identify the homology arms from the ssODN.
-
ii.Add the insertion sequence (containing HiBiT tag) by copy/paste into the SnapGene file.
-
i.
-
r.Download and open the sequencing results from Eurofins Genomics.
-
s.Align the sequencing results to the expected sequence in SnapGene.
-
i.This can be done by clicking “View” → “Show Alignments” → “Align Imported Sequences” → select the .ab1 file.
-
i.
-
t.Analyze the alignments to verify positively edited gDNA.
 CRITICAL: After positively identifying HiBiT tagged cell lines by HiBiT assays and gDNA verification, it is important to next evaluate the specificity of the editing prior to further experiments. For example, while you may have determined that your POI has been tagged with HiBiT, it is possible that a portion of the luminescence signal that you observe during the NanoBiT assay could arise from off target HiBiT editing effects on a separate gene. For determining editing specificity, we recommend simple reverse transfection small interfering RNA (siRNA) and HiBiT blotting experiments.
CRITICAL: After positively identifying HiBiT tagged cell lines by HiBiT assays and gDNA verification, it is important to next evaluate the specificity of the editing prior to further experiments. For example, while you may have determined that your POI has been tagged with HiBiT, it is possible that a portion of the luminescence signal that you observe during the NanoBiT assay could arise from off target HiBiT editing effects on a separate gene. For determining editing specificity, we recommend simple reverse transfection small interfering RNA (siRNA) and HiBiT blotting experiments.
-
a.
Figure 4.
Example image of an agarose gel of amplified gDNA after successful CETP HiBiT editing has occurred in HepG2 cells
Whereas unedited samples show a single amplicon, amplification of HiBiT edited samples show two bands. The upper band (orange box) correlates to incorporation of the HiBiT tag (along with linkers and a 2×FLAG tag). Comparison of the relative intensity of the bands in the edited sample yields an editing efficiency of ∼49%. Both bands can be excised and sequenced for further confirmation of editing (see Figure 5).
Figure 5.
Example of Sanger sequencing results of HepG2 CETP HiBiT gDNA
The expected gDNA sequence of CETP exon 16 edited with insertion of the HiBiT sequence (VS 2×FLAG VS HiBiT) is shown for comparison at the top of this panel. Below that is unedited gDNA corresponding to the lower bands in Figure 4. At the bottom is the gDNA sequence that perfectly matches the expected insertion post editing. The consistency of the chromatogram also indicates the specificity of the editing (i.e., that the tag was placed correctly and consistently).
siRNA reverse transfection
Timing: 4–5 days
siRNA can be used to induce knockdown of specific gene transcripts. For the purposes of this experiment, siRNA can be used to determine the specificity of the HiBiT tag by knocking down the POI and monitoring the effects on HiBiT luminescence. The purpose of this protocol is to effectively run siRNA knockdowns using reverse transfection of edited cells to find any off-target effects of the HiBiT editing and determine editing specificity. Example data of an siRNA experiment that demonstrates on-target CETP HiBiT editing can be found in Figure 6.
-
12.
Day 1: Reverse Transfection in a 24 well plate.
This protocol requires 250 μL siRNA mix and 250 μL cell suspension per well. These volumes can be scaled up accordingly if larger well sizes are desired. Reverse transfections can be a quicker way to knockdown genes and can be performed on cells that are sometimes recalcitrant to standard lipofection, like ARPE-19 cells. Each siRNA should be performed in triplicate (i.e., three separate wells each). We typically use Ambion Silencer Select siRNAs, which are effective and cost efficient.-
a.For each well, sterilely prepare the siRNA master mix by combining:
-
i.250 μL Opti-MEM.
-
ii.2.133 μL Dharmafect 4.
-
i.
-
b.Aliquot the 250 μL of siRNA/Dharmafect 4 master mix into tubes.
-
c.Add 2.5 μL of 10 μM siRNA into the tubes.Note: This siRNA mix will contain 100 nM siRNA, and the final concentration after cell addition will be 50 nM.Note: We recommend including at least the following siRNAs: non-targeting (siNT, control), toxic (siTOX, transfection estimation), or fluorescent (siGLO, transfection estimation) siRNAs as experimental controls. The siTOX control should kill the cells if properly transfected. Therefore, in this well, a higher level of cell death indicates better transfection efficiency. There should be minimal or no cell death in the siNT or siGLO wells.
-
d.Vortex the siRNA/Dharmafect 4/Opti-MEM mix for 15–30 s.
-
e.Incubate the siRNA mix in the culture hood at RT for 20 min.
-
i.Cells should be prepared during this time.
-
i.
-
f.Prepare cells from a 10 cm dish as follows:
-
i.Aspirate the media.
-
ii.Wash with 5 mL HBSS.
-
iii.Aspirate the HBSS and add 2 mL Trypsin-EDTA (0.25%). Incubate at 37°C until all cells are in suspension.
-
iv.Quench with 8 mL full culturing media.
-
v.Prepare the required number of cells to a final density of ∼466,000 cells/mL.Note: The final cell density will vary by cell line, but we have found that ∼116,500 cells per well of a 24 well plate works well across multiple cell lines.
-
i.
-
g.After the 20-min incubation for the siRNA mix is finished, spin the tube briefly in the centrifuge.
-
h.Add 250 μL of siRNA mix per well in a 24 well plate.
-
i.Add 250 μL of cell suspension per well in a 24 well plate. The cell suspension should be added directly on top of the siRNA mix.
-
j.Rock the plate until well dispersed.
-
k.Incubate at 37°C for approximately 12 h.
-
a.
-
13.Day 2 (24 h knockdown):
-
a.Check siTOX/siGLO controls accordingly by imaging on the Nexcelom Celigo.Note: Cells transfected with siTOX at this stage may not be dead, but should look haggard.
-
b.Run an extracellular HiBiT assay. Change media in all wells to full culturing media.
-
a.
-
14.Day 3 (48 h knockdown):
-
a.Check siTOX controls accordingly by imaging on the Nexcelom Celigo. Cells transfected with siTOX at this stage should be clearly dying.
-
b.Run an extracellular HiBiT assay.
-
a.
-
15.Day 4 (72 h knockdown):
-
a.Check siTOX controls accordingly by imaging on the Nexcelom Celigo. Cells transfected with siTOX at this stage should be ∼80–90% dead.
-
b.Run an extracellular HiBiT assay.
-
c.Change media.
-
a.
-
16.Day 5 (96 h knockdown): troubleshooting 8.
-
a.Run an extracellular HiBiT assay.
-
b.Run an intracellular HiBiT assay.
-
a.
Figure 6.
Example data of a siRNA time course experiment using HepG2 CETP HiBiT edited cells
Edited HepG2 cells were cultured and treated as indicated in the ‘siRNA reverse transfection’ section of this protocol. The negative control is a non-targeting siRNA that does not anneal to any known mammalian sequence. siTOX is a positive control siRNA that will kill cells if incorporated during the transfection (it knocks down essential genes for cell viability), and therefore decrease the HiBiT signal with time. The experimental silencer RNA is a CETP-specific siRNA (siCETP) that should not kill the cells but should decrease HiBiT signal by decreasing CETP mRNA. Secreted CETP HiBiT data were collected at 24 h time points and normalized to siNT. As expected, silencing of CETP closely resembled the positive control, and suggests high editing specificity for the CETP gene. n = 2–3 biological replicates, mean ± S.D. presented.
HiBiT blotting
Timing: 4–6 h
A HiBiT blot is very similar to the more commonly known western blot. However, instead of primary and conjugated secondary antibodies, one uses LgBiT to recognize and bind to HiBiT on the membrane, and when substrate is added to the mix, the newly formed NanoBiT complex will produce a chemiluminescent band detectable by conventional film or chemiluminescent images. The purpose of this portion of the protocol is to resolve and analyze all HiBiT-tagged secreted or intracellular proteins from an edited line to again confirm specific HiBiT tagging of a correctly sized POI. HiBiT blotting serves as a secondary verification of the specificity of HiBiT editing as well as verification that the tagged protein molecular weight matches that of the target protein. This step can also be combined with the previous siRNA step (i.e., run siRNA samples on a HiBiT blot) as further confirmation of specificity of editing if desired.
Note: It is important to realize that the NanoBiT assay (in plate reader format) can be very sensitive and yields a wider dynamic range than HiBiT Blotting. Therefore, in some cases, such as in our example case with CETP HiBiT, luminescence levels can be detected in plate reader format, but not HiBiT blotting format. This is typically a challenge for HiBiT signals that hover in the low thousands (1,000–5,000). As noted below, this challenge can sometimes be overcome by concentration or enrichment, but is not guaranteed. While we had difficulties performing CETP HiBiT blotting due to its low expression, we nonetheless include a HiBiT blotting section for readers to follow for their specific HiBiT-tagged POI.
-
17.
Media Collection and Cell Lysis.
This protocol is written for cells cultured in a 24 well plate. Volumes can be scaled up as needed.-
a.24–72 h prior to sample collection, culture the cells in serum free or low serum culture media if your protein is secreted.
 CRITICAL: High levels of serum can be disruptive to the blotting process and can mask HiBiT-tagged proteins during the imaging process. If collecting media for secreted protein analysis, make sure to allow enough incubation time for protein to accumulate in the media, which can be confirmed prior to blotting using an extracellular/secreted HiBiT assay. Concentration with an Amicon filter may be required to enrich for your secreted protein enough to be visible by HiBiT blotting.
CRITICAL: High levels of serum can be disruptive to the blotting process and can mask HiBiT-tagged proteins during the imaging process. If collecting media for secreted protein analysis, make sure to allow enough incubation time for protein to accumulate in the media, which can be confirmed prior to blotting using an extracellular/secreted HiBiT assay. Concentration with an Amicon filter may be required to enrich for your secreted protein enough to be visible by HiBiT blotting. -
b.Remove the media and place in an Eppendorf tube. This tube contains the secreted HiBiT tagged protein sample.
-
c.Wash the culture plate with a full volume of HBSS.
-
d.In the meantime, spin down the media samples at 3,000 × g for 3 min to pellet any dead cells and cellular debris.
-
i.Transfer the supernatant to a fresh tube and store on ice.
-
i.
-
e.Aspirate the HBSS from the cell culture plate.
-
f.Add 50 μL of lysis buffer to each well.
-
i.Lysis buffer consists of 1:100 protease inhibitor and 1:1000 benzonase in RIPA buffer. We recommend preparing extra to account for pipetting error.
-
i.
-
g.Rock the cell plate for 15 min at 4°C to lyse the cells.
-
i.Verify sufficient lysis under the microscope before proceeding.
-
ii.If the lysis is insufficient, rock at 4°C for an additional 10 min, or until effective lysis can be verified.
-
i.
-
h.Transfer the lysate to a fresh Eppendorf tube.
-
i.Spin down at >12,000 x g for 10 min at 4°C to pellet cellular debris.
-
j.Transfer the supernatant to a fresh tube.
-
i.This sample will contain the soluble protein sample and should be stored on ice. Discard the pellet, which contains any cellular debris from lysis.
-
ii.Samples can be stored at −20°C for longer term, if needed.
-
i.
-
a.
-
18.BCA Standard Curve and Sample Preparation.
-
a.Run a BCA assay to determine the total protein amount within the lysate samples. For media samples, we recommend normalizing to volume instead of protein concentration.
-
i.Dilute the BSA standard with microbiology grade nuclease-free water to the final standard concentrations of 2, 1, 0.5, 0.25, 0.125 and 0 mg/mL. We recommend making ∼ 1 mL of each standard as these can be stored at RT and used for later experiments as needed.
-
ii.Using the reagents provided in the kit, create the assay master mix of reagent A and B in a 50:1 ratio. Each well of the assay will require 200 L of master mix. We recommend making extra for pipetting error.
-
iii.Add 10 μL of BSA standards to respective wells of a 96 well clear plate.
-
iv.Add 2–3.33 μL of lysate samples to respective wells.
-
v.Add 200 μL of master mix to each well on top of the sample.Note: The reaction starts as soon as the master mix is added, so it is imperative to add this last and as quickly as possible.
-
vi.Cover the plate loosely with aluminum foil and incubate at 37°C for 20 min.
-
vii.Once the 20-min incubation period is finished, quickly read the absorbance of the wells at 562 nm.
-
viii.Using the absorbance readings of the BSA standards, plot a standard curve of OD vs. BSA concentration (mg/mL). We recommend using Excel to develop the graph. Make sure to include a linear trendline, trendline equation, and the R2 value. The R2 value should be above 0.95.
-
ix.Calculate the protein concentration of your samples using the following formula: Protein Concentration (mg/mL) = ((Sample OD – b)/m)∗5.Note: ‘b’ is the trendline y intercept and m is the trendline slope. The values will be scaled up to 10 depending on the amount of sample added to the well (i.e. if 2 μL of sample was used, scale up to 10 by multiplying by 5).
-
i.
-
b.Prepare samples of normalized protein concentration diluted appropriately to 20 μL with RIPA in fresh Eppendorf tubes.Note: Ideally, samples should be made with a final volume of 20 μL containing 20–30 μg of protein. We do not recommend exceeding 24 μL samples as addition of loading dye will make the volume too large to load into the gel.
-
c.Prepare the media samples by concentrating the media. We recommend concentration of 500 μL into ∼25 μL.
-
i.Add the concentration filter to a clean tube and load 500 μL of media sample.
-
ii.Spin at >12,000 × g for 30 min.
-
iii.Remove the filter and place it upside down into a clean collection tube. Discard of the original collection tube containing flow through.
-
iv.Spin at 5,000 × g for 5 min, or until all of the concentrated sample has been collected.
-
v.Transfer 20 μL of media sample to a new tube for SDS PAGE.
-
i.
-
d.Prepare the SDS Page loading dye (4× SDS BME) according to the following:
-
i.Divide your sample volume (ideally, 20 μL) by 3 to calculate how much 4× loading dye should be added to each sample for a final concentration of 1×.
-
ii.Multiply the loading dye volume per sample by the number of samples. Scale up enough to account for pipetting error.
-
iii.Create the appropriate volume of loading dye by adding 4× reducing SDS to BME in a 30:1 ratio.
-
iv.Add the loading dye to each sample.
-
v.Boil samples for 5 min in a 95°C heat block.
-
vi.Carefully remove samples from the heat block. Avoid popping of the Eppendorf lids as this can lead to decrease in sample yield.
-
vii.Spin the samples down at >12,000 × g or 30 s–1 min.
-
i.
-
e.Prepare the SDS PAGE blank by diluting 4× reducing SDS with RIPA to a final concentration of 1×.
 CRITICAL: You will need 20 μL per well. We recommend scaling up to account for pipetting error.
CRITICAL: You will need 20 μL per well. We recommend scaling up to account for pipetting error.
-
a.
-
19.SDS PAGE.
-
a.Set up the SDS PAGE gel (4–20% Tris-Gly) in 1× Tris-Gly with SDS running buffer.
-
b.Load 0.75 μL Page Ruler Protein Plus Ladder.
-
c.Load 20 μL of each sample.
-
d.Load 20 μL blank (1x SDS PAGE buffer) in the appropriate wells.
-
i.We recommend keeping one well of blank between ladder and sample, as well as in between sample sets (i.e., separating media and lysate samples).
-
i.
-
e.Run the gel at 140 V for 80 min.
-
a.
-
20.Blot Transfer.
-
a.Once the SDS PAGE has been completely run, transfer the proteins to a nitrocellulose membrane using the iBlot 2 transfer system as follows:
-
i.Fill a 1 L beaker with deionized water and soak 1 filter paper while preparing the transfer.
-
ii.Open the iBlot transfer stack.
-
iii.Align the stack into the iBlot transfer instrument and remove the top of the stack.
 CRITICAL: There is a white insert between the top stack and nitrocellulose membrane to mark the point of separation. Be sure that the nitrocellulose membrane does not get stuck to the top of the stack.
CRITICAL: There is a white insert between the top stack and nitrocellulose membrane to mark the point of separation. Be sure that the nitrocellulose membrane does not get stuck to the top of the stack. -
iv.Remove the SDS PAGE gel from the cassette and rinse with DI water.
-
v.Dip a spatula in the running buffer and use to remove the plastic gel cover.
-
vi.Using the same spatula, cut off the gel wells and dye front. Make sure to keep the spatula damp with running buffer at all times.
-
vii.Dampen your fingers in running buffer. Carefully, pick up the SDS PAGE gel and place on top of the nitrocellulose membrane.
 CRITICAL: Be careful not to rip or tear the gel. Avoid air bubbles when placing onto the membrane.
CRITICAL: Be careful not to rip or tear the gel. Avoid air bubbles when placing onto the membrane. -
viii.Dampen a small roller in running buffer and gently roll over the gel to ensure it lays flat on the nitrocellulose membrane.
-
ix.Place the pre-soaked filter paper on top of the gel and roll with the damp roller.
-
x.Place the top of the transfer stack on top of the filter paper and roll again.
-
xi.Place the transfer absorbent pad on top of the stack, making sure to align the electrode properly in the machine. Gently roll once more with the damp roller.
-
xii.Close the iBlot system and run according to the following protocol. This process will take 7 min.
-
i.
-
b.After the proteins have been transferred to the membrane, remove it from the iBlot system, trim the membrane as appropriate, and incubate in 1× TBST.
 CRITICAL: The Tween in TBST exposes the HiBiT tag, allowing LgBiT to recognize and bind to it.
CRITICAL: The Tween in TBST exposes the HiBiT tag, allowing LgBiT to recognize and bind to it. -
c.Incubation in 1× TBST for approximately 12 h has shown the best results for HiBiT blotting. However, the incubation period can be as little as 10 min before moving on to the blot incubations with LgBiT and substrate.
-
a.
-
21.HiBiT Blot Incubations (troubleshooting 9 and 10).
-
a.Using the reagents from the NanoGlo HiBiT Blotting kit, prepare the first incubation solution by diluting the provided HiBiT blotting buffer in nuclease-free water in a 1:10 dilution.
-
i.For example, dilute 1 mL of HiBiT blotting buffer in 9 mL water.
-
i.
-
b.Add the LgBiT to the buffer solution in a 1:200 ratio. For example, add 50 μL of LgBiT to 10 mL of diluted HiBiT blotting buffer.
-
c.Remove the 1× TBST from the nitrocellulose membrane and replace it with the HiBiT blotting buffer containing LgBiT. Rock for 1 h at RT.
-
d.After 1 h, add NanoLuc Substrate (not the Lytic or Extracellular Substrate mentioned above) to the blotting solution in a 1:500 ratio.
-
i.For example, add 20 μL of substrate to 10 mL of blotting buffer. This can be done in the blotting container by tilting the container to collect all the buffer in one area and adding the substrate directly to the buffer.
-
i.
-
e.Gently rock a few times by hand to ensure distribution of the substrate. Rock for 5 min at RT.
-
f.Remove the membrane from the blotting container and image using the LI-COR Odyssey FC following the instructions provided by LI-COR.
-
i.When imaging the blot, make sure to include the 700 nm channel to view the ladder as well as the chemiluminescence channel in order to view the HiBiT luminescence.
-
i.
-
a.
Expected outcomes
Editing cells and imaging for GFP control
After electroporation, some cell death may occur, but cells should become mostly adherent 24 h after electroporation. Cell cultures should begin growing normally after the media has been changed back to normal culture media and allowed to recover for a couple of days. When imaging electroporated cells, including the GFP control, it is expected that all cell densities and distributions are comparable. Only the wells electroporated with the GFP control should express green fluorescence (unless Cas9 GFP protein was used for editing). Additionally, it is expected that the cells begin to express HiBiT anywhere from 48–72 h after electroporation. This can most easily be monitored with HiBiT assays.
Extracellular HiBiT assay
If the target protein is secreted, extracellular HiBiT assays of media from electroporated cells should show increased luminescence signal over HiBiT assays of media from unelectroporated cells. Typically, this signal will not be noticeable until 48–72 h after electroporation. It may take a day or two for secreted protein to accumulate in the media, but signal should be noticeably higher, at minimum 3×, than that of the unelectroporated control within 5 days of electroporation. As the edited cells grow, the difference between unedited cells and HiBiT edited cells should continue to increase, but eventually plateau.
Intracellular HiBiT assay
If the target protein is not secreted, extracellular HiBiT assays will not aid in determination of editing and an intracellular assay may be required. If this is the case, then one needs to plan to grow (or edit) additional well solely for the purpose of assaying for HiBiT. If the target protein is secreted, intracellular HiBiT assays can be executed to compare the amount of HiBiT tag protein that is expressed within the cell versus what is secreted. In both cases, the luminescence signal should be significantly higher than that of the unelectroporated control within 5 days of electroporation.
gDNA gel imaging
Positive edits will exhibit 2 amplified DNA bands, whereas negative edits will appear identical to controls. The lower band will correspond to that of unedited DNA and should match gDNA amplified from the unelectroporated cells. The higher band on the gel corresponds to the DNA including the HiBiT knockin. This band should not be apparent in wild type, or unelectroporated, DNA sample. This band should be higher than the unedited band by the number of base pairs present in the insertion sequence located in the ssODN. Editing efficiency as determined by a gDNA amplicon can vary depending on the locus edited, the cell line used, and the electroporation efficiency of the cells. We have observed HiBiT signal from cells that were only 15% edited with the HiBiT tag and have obtained successful HiBiT editing up to ∼88% in other cases.
Sanger Sequencing
Results of Sanger sequencing should align well with the expected DNA sequence. There should be a clear insertion sequence that matches the designed ssODN sequence. The insertion is expected to be in the location determined by the homology arms identified in the ssODN development stage. The sequence chromatogram should show clearly defined peaks and very few discrepancies in base pair identification. If Sanger sequencing fails, an alternative sequence approach would be to use next generation sequencing (NGS). However, this is costly and can take a long time to complete, unlike Sanger sequencing.
siRNA
The non-targeting siRNA control should not decrease HiBiT signal at all compared to untransfected controls, if used. The toxic siRNA control should both decrease luminescence signal and kill cells if delivered properly. Knockdown of the target protein should lead to a decrease in the HiBiT signal, similar to the values produced by the toxic control. Knockdown of other proteins not targeted in editing should not reduce the HiBiT signal and results should be comparable to the non-targeting control values. The ability of siRNA to knockdown a POI is of course dependent on the half-life of the POI, which should also be taken into account during this experiment.
HiBiT blot
For samples of concentrated media, if the target protein is secreted it is expected to see one band expressing HiBiT signal. The molecular weight of this band should match approximately that of the target protein. For samples of cell lysate, there should be only one protein band expressing HiBiT signal that also corresponds to the molecular weight of the target protein. Any unelectroporated or wild type controls should not show any HiBiT expression. If issues persist with detecting HiBiT-tagged POIs via blotting, one could use a set of different medias to determine which, if any, may yield enhanced production of your HiBiT-tagged POI (Figure 7). POIs with multiple isoforms may appear as multiple bands.
Figure 7.
Using alternative media yields different levels of CETP HiBiT production from edited HepG2 cells
CETP HiBiT HepG2 cells were seeded at a density of 350,000 cells/well of a 6 well plate for 12 h in normal growth media (full DMEM with 10% FBS, PSQ). The next day, media was replaced with the indicated media and incubated for an additional 1 or 2 days. Serum-free Opti-MEM yields the highest production of CETP HiBiT of all the media tested and could be used moving forward to increase chances of confirming the CETP HiBiT using HiBiT Blotting. As these are example data that were taken at select time points, n = 1.
Limitations
The knock-in of the HiBiT tag is dependent on the efficiency and accuracy of CRISPR-Cas9 technology. In our research, we have demonstrated the efficacy of this editing technique in adult pigment retinal epithelium (ARPE-19) cells. However, CRISPR-Cas9 editing is not always accepted the same across cell lines. This can manifest itself in various ways, such as off target effects, lack of insertion acceptance, or the absence of indel formation from Cas9. Thus, it is possible that editing will require additional optimization and characterization of the resulting cell lines (i.e., phenotyping for biomarker(s), immunolabeling or qPCR, or performing a relevant phenotypic assay such as wound healing, migration, or tube formation) not outlined in this protocol. Potential limitations such as these will need to be explored further in order for a more generalized protocol to be determined.
Troubleshooting
As with any experiment, issues can potentially arise from the contamination or mishandling of equipment, reagents, and cells. Please ensure that all materials are handled and stored properly and according to their instructions. Use of calibrated pipettes is strongly recommended as is the use of aerosol barrier tips wherever possible.
CRITICAL: It is important to ensure that HiBiT cell lines are well labeled so that they are not mixed or accidentally mistaken for a different HiBiT cell line. If you believe that you may have mixed up HiBiT cell lines, the easiest experiment to perform to confirm their identity is through an siRNA experiment.
Problem 1
Cells are dead, or not attaching, within 24 h of electroporation.
Potential solutions
-
•
We have found that the easiest fix to this problem lies within the length of time that cells sit in antibiotic free media containing electroporation enhancer, HDR enhancer, and other reagents after electroporation. It is possible that some cell lines will need full culture media sooner than others in order to survive (i.e., they may be more sensitive to the aforementioned editing reagents). In this case, we recommend changing the media no sooner than 24 h after electroporation.
Note: The sooner the media is changed and the CRISPR-Cas9 reagents are removed, the less time cells have to perform HDR, and the less effective editing will be.
-
•
This problem could be due to overwhelming the cells intake of reagents during electroporation. We recommend trying electroporation with a decreased concentration, or complete removal, of HDR enhancer to make the electroporation recovery process a little easier on the cells.
-
•
This could be due to using the wrong electroporation parameters. Some cell lines are more sensitive to this technique than others. We recommend trying different pulse voltages, pulse lengths, and number of pulses to better optimize electroporation for the target cell line. We have found that sensitive cell lines typically benefit from using the following parameters: 1050 V, 30 ms, 2 pulses. However, we recommend executing optimization by electroporating the target cell line with only GFP plasmid and changing the electroporation parameters. Parameters that result in live cell cultures that highly express GFP should be prioritized and used for further editing-based electroporations of that cell line.
-
•This could be due to using too high of a concentration of reagents during electroporation. While the recommended proportions have been shown to be the most universally accepted across cell lines, some cells will be more sensitive to the introduction of CRISPR-Cas9 reagents than others. For example, we have found that primary cells are more sensitive and require more dilute reagents. We recommend electroporating sensitive cell lines with the following values:
-
○For preparing the duplex crRNA/tracrRNA:
-
-4.4 μL crRNA at 100 M
-
-4.4 μL tracrRNA at 100 M
-
-1.2 μL duplex buffer
-
-
-
○For preparing the RNP (per 1 well):
-
-0.5 μL dilute Cas9 or Cas9-GFP
-
-Dilute the Cas9 according to the following:
-
-0.3 μL Cas9
-
-0.2 μL Genome editing buffer
-
-0.5 μL duplex crRNA/tracrRNA
-
-
-
○For preparing the RNP/ssODN/Cell suspension mix (per 1 well):
-
-1 μL RNP
-
-9 μL cell suspension
-
-1 μL dilute ssODN. Dilute from 100 μM to 20 μM prior to use
-
-1 μL dilute electroporation enhancer. Dilute from 100 μM to 20 μM prior to use
-
-2 μL Genome editing buffer
-
-
-
○For preparing the GFP control (per 1 well):
-
-250 ng of GFP plasmid
-
-9 μL cell suspension
-
-1 μL dilute electroporation enhancer
-
-Bring to a final volume of 14 μL with Genome editing buffer
-
-
-
○
Problem 2
Cells are not showing increased luminescence signal after electroporation in extracellular HiBiT assays.
Potential solutions
-
•
If the target protein is known to be secreted, the most likely cause of this problem is low, or no, editing efficiency. An intracellular HiBiT assay should be run first to see if there is HiBiT signal within the cells. No intracellular signal is indicative of low editing efficiency (see problem 3), or possibly that the cell line does not produce your POI at sufficient levels. Presence of intracellular signal is indicative of effective editing with low protein secretion (see potential solutions: problem 2).
-
•
The other likely cause of this issue is a lack of protein secretion from the target cell line. An intracellular HiBiT assay should be run to see if there is HiBiT signal within the cells corresponding to expressed protein that is not being secreted. Cell lines with high intracellular HiBiT signal but no extracellular signal indicates effective editing with no protein secretion. In this case, we recommend finding ways to induce (through differentiation, perhaps) or moving onto a new, model cell line with a higher secretion propensity of the target protein. For cell lines that do not show intracellular HiBiT signal, see problem 3.
-
•
It is also possible that depending on the edited POI, there may be a need to stabilize a labile protein (if it is rapidly targeted for degradation, for example), or induce the expression of the POI using differentiation factors. Finally, a POI may have dynamic expression that changes based on a stimulus, cellular growth status, etc. To account for this latter possibility, it is recommended to assay at different points of time during the culturing process.
Problem 3
Cells are not showing increased luminescence signal after electroporation in intracellular HiBiT assays.
Potential solutions
-
•
The most likely cause of this problem is a lack of protein expression from the target cell line. We recommend starting with gDNA analysis to determine whether effective editing took place or not before moving forward. Should the agarose gel image show a band corresponding to effective DNA editing, then the issue most likely lies within the cell’s expression of this protein and not the editing process itself. At this point, we would recommend finding ways to increase protein expression in the current cell line or editing the target protein within a new, model cell line with better protein expression. However, should the agarose gel not show a band corresponding to effective DNA editing, then the issue most likely lies within the electroporation itself. In this case, we recommend running a T7E1 assay to verify indel formation achieved by the selected crRNA/tracrRNA duplex and retrying electroporation and analysis.
-
•
It is also possible that depending on the edited POI, there may be a need to stabilize a labile protein (if it is rapidly targeted for degradation, for example), or induce the expression of the POI using differentiation factors. Finally, a POI may have dynamic expression that changes based on a stimulus, cellular growth status, etc. To account for this latter possibility, it is recommended to assay at different points of time during the culturing process.
Problem 4
Edited cell gDNA shows edited DNA but there is no HiBiT signal in extracellular and/or intracellular assays.
Potential solution
-
•This means that the CRISPR-Cas9 editing was executed effectively and resulted in an insertion within the target protein, but the expression of this protein within the target cell line is low or nonexistent. We would recommend trying the same exact editing in a new cell line that has better expression patterns for the target protein to be used as an experimental model. We recommend utilization of RNA-seq data sets as they may be useful in determining cell types that are likely to express the gene of interest, or to use other websites such as the Human Protein Atlas (https://www.proteinatlas.org).
-
○Alternatively, it is possible that the HiBiT tagging design was flawed and resulted in insertion of the HiBiT tag, but not in frame with the open reading frame. This can be determined by analysis of the Sanger sequencing chromatogram. If a mistake has been found, one will need to reengineer the ssODN repair template and perform the experiments again.
-
○
Problem 5
Edited cell gDNA does not show edited DNA but there is HiBiT signal in extracellular and/or intracellular assays.
Potential solution
-
•
This means that either the editing efficiency is very low, or that the DNA you are analyzing on the gel is not contributing to the HiBiT signal and editing took place elsewhere within the genomic DNA. First, verify the correct use of primers in the PCR. Specificity experiments, such as siRNA and HiBiT blotting, can be used to determine whether off-target editing may have taken place. However, to remedy this latter problem and edit the appropriate gene, we recommend redesigning the CRISPR-Cas9 reagents with a new editing site while prioritizing specific homology arms and guide RNA that may help reduce off target editing.
Problem 6
Extracted gDNA does not show edited DNA and there is no HiBiT signal in extracellular and/or intracellular assays.
Potential solution
-
•
This means that CRISPR-Cas9 editing was not executed effectively. We recommend optimization of electroporation parameters (pulse voltage, pulse width, and number of pulses) using only GFP plasmid. Parameters that produce cells expressing high levels of GFP should be prioritized. Some cells accept editing in different locations easier than others, so we also recommend exploration into other editing sites. For example, if you started with N-terminal editing, try designing new reagents focused on C-terminal editing. Exploration of additional crRNAs may also be warranted.
CRITICAL: It is also important to include the HDR enhancer in the editing protocol.
Problem 7
Sanger sequencing cannot be aligned.
Potential solutions
-
•
One potential cause of this issue is excision of an unclear gel band. Extraction of DNA from a smeared or very small band can lead to poor sequencing results that will not align in SnapGene. We recommend rerunning the PCR amplification using the extracted DNA from the first excised band as the DNA template. This will reamplify the potentially edited DNA that should produce a clearer band on the gel. Sanger sequencing should be run on the new band to try for better results.
-
•
If the sequencing results yield a lot of undetermined (or “N”) bases, it could be that the DNA extracted is not clear enough to sequence. In this case, we would recommend starting with PCR optimization to get a clearer gel band for excision and DNA extraction. This can be done by increasing the annealing temperature (to reduce laddering or the presence of primer dimers), increasing the amount of template DNA, increasing the amplification cycles, or simply running the gel longer to allow for better band separation. If these solutions fail, then re-harvesting of fresh gDNA sample or re-editing of cells would be recommended.
-
•
If the sequencing results produce a relatively clear sequence with very few undetermined (or “N”) bases and is not aligning to the expected DNA sequence, we recommend running the sequencing results through the National Library of Medicine Nucleotide BLAST software (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) to identify the sequence itself. If the BLAST results confirm the sequence as matching the target gene, check the alignment set up in SnapGene. If the BLASTN results show the sequence belongs to a different gene, then potential off-target HiBiT editing has taken place or PCR amplification was done for the wrong DNA sample.
-
•
Sanger sequencing is only successful with consistent editing. If the Sanger sequencing fails, it is possible that the editing was heterogeneous. An alternative would be to attempt Next Generation Sequencing (NGS) to aid in the confirmation of editing and sort out any discrepancies.
Problem 8
siRNA knockdown of the target protein does not decrease luminescence signal.
Potential solutions
-
•
If the cell line has been verified to be edited and produces good, secreted HiBiT signal, then this problem is possibly due to ineffective introduction of the siRNA. Transfection delivery should be analyzed by observing the controls. We recommend rerunning the experiment with fresh reagents, intentional focus placed on the siRNA mix preparation, and making sure to plate the cells on top of the siRNA mix and rock sufficiently. In extreme cases, other reverse transfection protocols, such as those utilizing RNAi Max, may be necessary to try. Additional protocols can be found online but may require additional optimization. We would also suggest to confirm knock down of the gene transcript by quantitative PCR should this issue arise.
-
•
If the tagged protein is not secreted, the extracellular HiBiT assay will not produce valuable data. In this case, we recommend plating 12 wells per siRNA (triplicate wells to be analyzed daily). All wells should be transfected at the same time, but every 24 h a new set of triplicate wells should be analyzed by both extracellular and intracellular HiBiT assays for data collection.
-
•
The inefficient knockdown of luminescence signal can occur when editing efficiency or HiBiT expression is very low. In these cases, it is necessary to troubleshoot the electroporation itself in order to establish a viable cell line for analysis. Cell lines should produce consistent HiBiT signal that is significantly higher than the background in order to be useful in high volume assays and compound screens.
Problem 9
HiBiT blotting results in multiple bands in imaging.
Potential solution
-
•
This result may indicate that HiBiT editing yielded off-target effects and multiple proteins are expressing HiBiT. Evaluation of the molecular weights where these bands exist on the blot is necessary to determine what proteins may be edited. It is possible to see multiple bands, but only have one edited protein if the protein exists as different splice variants or isoforms in the cell. Combine blot results with siRNA data to differentiate between potential causes. Re-editing at a new insertion site may be necessary to avoid these results.
Problem 10
HiBiT blotting results in no bands in imaging.
Potential solutions
-
•
The samples used in this analysis should consist of positively edited cell lines previously verified. Therefore, the issue most likely lies within the execution of the SDS PAGE and blotting incubations. Verify that the sample preparations, SDS PAGE, nitrocellulose membrane transfer, and incubation procedures were executed correctly. We recommend using fresh reagents that have been properly stored and maintained. It is also possible that the amount of HiBiT tagged protein is not sufficient for detection via blotting. We would recommend that one concentrate the sample if possible prior to SDS PAGE. Typically HiBiT signal of >50,000 ALU can be detected by HiBiT blotting.
-
•
A monoclonal HiBiT antibody is available from Promega (catalog #N7200) that could be used in standard western blotting for a HiBiT POI which may allow for better detection due to primary and secondary antibody-based signal amplification.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, John D. Hulleman (hulleman@umn.edu).
Technical contact
Questions about the technical specifics of performing the protocol should be directed to and will be answered by the technical contact, John D. Hulleman (hulleman@umn.edu).
Materials availability
Sequences of the reagents for the HiBiT tagging of CETP are provided within this document (Table 1). For access to the engineered HepG2 CTEP HiBiT cell line, please contact the lead contact, John D. Hulleman (hulleman@umn.edu).
Data and code availability
For access to raw data presented in this paper, please contact the lead contact, John D. Hulleman (hulleman@umn.edu). No new code was developed as part of this project.
Acknowledgments
J.D.H. is the Larson Endowed Chair for Macular Degeneration and was supported by the Fichtenbaum Charitable Trust, an endowment from the Roger and Dorothy Hirl Research Fund, and a National Eye Institute (NEI) R01 grant (EY027785). Additional support was provided by an NEI Visual Science Core grant (P30 EY030413, to the UT Southwestern Department of Ophthalmology). J.D.H. is also a member of the Promega Advanced Academic Access Program, which defrayed reagent costs. We would also like to acknowledge the Green Fellows Program at UT Dallas, supported by the Cecil and Ida Green Foundation. The graphical abstract was created with BioRender.com.
Author contributions
Conceptualization, J.D.H.; writing – original draft, K.P.L.; writing – review and editing, K.P.L. and J.D.H.
Declaration of interests
The authors declare no competing interests.
References
- 1.England C.G., Ehlerding E.B., Cai W. NanoLuc: A Small Luciferase Is Brightening Up the Field of Bioluminescence. Bioconjug. Chem. 2016;27:1175–1187. doi: 10.1021/acs.bioconjchem.6b00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boute N., Lowe P., Berger S., Malissard M., Robert A., Tesar M. NanoLuc Luciferase - A Multifunctional Tool for High Throughput Antibody Screening. Front. Pharmacol. 2016;7:27. doi: 10.3389/fphar.2016.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hulleman J.D., Brown S.J., Rosen H., Kelly J.W. A high-throughput cell-based Gaussia luciferase reporter assay for identifying modulators of fibulin-3 secretion. J. Biomol. Screen. 2013;18:647–658. doi: 10.1177/1087057112469405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loh J.M.S., Proft T. Comparison of firefly luciferase and NanoLuc luciferase for biophotonic labeling of group A Streptococcus. Biotechnol. Lett. 2014;36:829–834. doi: 10.1007/s10529-013-1423-z. [DOI] [PubMed] [Google Scholar]
- 5.DiCesare S.M., Ortega A.J., Collier G.E., Daniel S., Thompson K.N., McCoy M.K., Posner B.A., Hulleman J.D. GSK3 inhibition reduces ECM production and prevents age-related macular degeneration-like pathology. bioRxiv. 2023 doi: 10.1101/2023.12.14.571757. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boursier M.E., Levin S., Zimmerman K., Machleidt T., Hurst R., Butler B.L., Eggers C.T., Kirkland T.A., Wood K.V., Friedman Ohana R. The luminescent HiBiT peptide enables selective quantitation of G protein-coupled receptor ligand engagement and internalization in living cells. J. Biol. Chem. 2020;295:5124–5135. doi: 10.1074/jbc.RA119.011952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boursier M.E., Levin S., Hurst R., Ohana R.F. Equilibrium and Kinetic Measurements of Ligand Binding to HiBiT-tagged GPCRs on the Surface of Living Cells. Bio. Protoc. 2020;10 doi: 10.21769/BioProtoc.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon A.S., Schwinn M.K., Hall M.P., Zimmerman K., Otto P., Lubben T.H., Butler B.L., Binkowski B.F., Machleidt T., Kirkland T.A., et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016;11:400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- 9.Schwinn M.K., Machleidt T., Zimmerman K., Eggers C.T., Dixon A.S., Hurst R., Hall M.P., Encell L.P., Binkowski B.F., Wood K.V. CRISPR-Mediated Tagging of Endogenous Proteins with a Luminescent Peptide. ACS Chem. Biol. 2018;13:467–474. doi: 10.1021/acschembio.7b00549. [DOI] [PubMed] [Google Scholar]
- 10.Schwinn M.K., Steffen L.S., Zimmerman K., Wood K.V., Machleidt T. A Simple and Scalable Strategy for Analysis of Endogenous Protein Dynamics. Sci. Rep. 2020;10:8953. doi: 10.1038/s41598-020-65832-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riching K.M., Mahan S., Corona C.R., McDougall M., Vasta J.D., Robers M.B., Urh M., Daniels D.L. Quantitative Live-Cell Kinetic Degradation and Mechanistic Profiling of PROTAC Mode of Action. ACS Chem. Biol. 2018;13:2758–2770. doi: 10.1021/acschembio.8b00692. [DOI] [PubMed] [Google Scholar]
- 12.Liutkeviciene R., Vilkeviciute A., Streleckiene G., Kriauciuniene L., Chaleckis R., Deltuva V.P. Associations of cholesteryl ester transfer protein (CETP) gene variants with predisposition to age-related macular degeneration. Gene. 2017;636:30–35. doi: 10.1016/j.gene.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 13.Momozawa Y., Akiyama M., Kamatani Y., Arakawa S., Yasuda M., Yoshida S., Oshima Y., Mori R., Tanaka K., Mori K., et al. Low-frequency coding variants in CETP and CFB are associated with susceptibility of exudative age-related macular degeneration in the Japanese population. Hum. Mol. Genet. 2016;25:5027–5034. doi: 10.1093/hmg/ddw335. [DOI] [PubMed] [Google Scholar]
- 14.Cheng C.Y., Yamashiro K., Chen L.J., Ahn J., Huang L., Huang L., Cheung C.M.G., Miyake M., Cackett P.D., Yeo I.Y., et al. New loci and coding variants confer risk for age-related macular degeneration in East Asians. Nat. Commun. 2015;6:6063. doi: 10.1038/ncomms7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landowski M., Bowes Rickman C. Targeting Lipid Metabolism for the Treatment of Age-Related Macular Degeneration: Insights from Preclinical Mouse Models. J. Ocul. Pharmacol. Ther. 2022;38:3–32. doi: 10.1089/jop.2021.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Grooth G.J., Klerkx A.H.E.M., Stroes E.S.G., Stalenhoef A.F.H., Kastelein J.J.P., Kuivenhoven J.A. A review of CETP and its relation to atherosclerosis. J. Lipid Res. 2004;45:1967–1974. doi: 10.1194/jlr.R400007-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Yu X., Liang X., Xie H., Kumar S., Ravinder N., Potter J., de Mollerat du Jeu X., Chesnut J.D. Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX. Biotechnol. Lett. 2016;38:919–929. doi: 10.1007/s10529-016-2064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grätz L., Sajkowska-Kozielewicz J.J., Wesslowski J., Kinsolving J., Bridge L.J., Petzold K., Davidson G., Schulte G., Kozielewicz P. NanoBiT- and NanoBiT/BRET-based assays allow the analysis of binding kinetics of Wnt-3a to endogenous Frizzled 7 in a colorectal cancer model. Br. J. Pharmacol. 2023:1–17. doi: 10.1111/bph.16090. [DOI] [PubMed] [Google Scholar]
- 19.Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen A., Hulleman J.D. Evidence of Alternative Cystatin C Signal Sequence Cleavage Which Is Influenced by the A25T Polymorphism. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Käll L., Krogh A., Sonnhammer E.L.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gleditzsch D., Pausch P., Müller-Esparza H., Özcan A., Guo X., Bange G., Randau L. PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol. 2019;16:504–517. doi: 10.1080/15476286.2018.1504546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudbeck L., Dissing J. Rapid, simple alkaline extraction of human genomic DNA from whole blood, buccal epithelial cells, semen and forensic stains for PCR. Biotechniques. 1998;25:588–592. doi: 10.2144/98254bm09. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
For access to raw data presented in this paper, please contact the lead contact, John D. Hulleman (hulleman@umn.edu). No new code was developed as part of this project.