Abstract
We previously described that type B retrovirus-like particles released from the human mammary carcinoma cell line T47D are pseudotypes and package retroviral RNA of different origins (W. Seifarth, H. Skladny, F. Krieg-Schneider, A. Reichert, R. Hehlmann, and C. Leib-Mösch, J. Virol. 69:6408–6416, 1995). One preferentially packaged retroviral sequence, ERV-MLN, has now been used to isolate the corresponding full-length provirus from a human genomic library. The 9,315-bp proviral genome comprises a complete retroviral structure except for a 3′ long terminal repeat (LTR) truncation. A lysine tRNA primer-binding site and phylogenetic analyses assign this human endogenous retroviral element, now called HERV-K-T47D, to the HML-4 subgroup of the HERV-K superfamily. The gag, prt, pol, and env genes exhibit 40 to 60% amino acid identity to HERV-K10. HERV-K-T47D is located on human chromosome 10, with five closely related elements on chromosomes 8, 9, 15, 16, and 19 and several hundred HERV-K-T47D-related solitary LTRs dispersed over the human genome. HERV-K-T47D-related sequences are detected in the genomes of higher primates and Old World monkeys but not in those of New World monkeys. High HERV-K-T47D transcription levels were observed in human placenta tissue, whereas transcription in T47D cells was strictly steroid dependent.
Human endogenous retroviral sequences (HERVs) are inherited genomic elements with structural features of integrated retroviruses. To date, HERVs are estimated to comprise at least 1% of the human genome (for reviews, see references 14, 16, and 44). The biologically most active HERVs are members of the HERV-K superfamily (for a review, see reference 20). Members of this family are characterized by the presence of primer binding sites (PBSs) for lysine tRNA, hence the designation K. They represent about 70 to 100 elements and a large number of solitary long terminal repeats (LTRs) in the human genome. These elements are related to type A, B, and D retroviruses and have been classified by alignments of short stretches of the reverse transcriptase (RT) domain into six different groups (HML-1 to -6) (23). The members within a subgroup are more than 85% identical, whereas the intersubgroup similarity does not exceed 75%. To date, full-length proviral elements from only subgroups HML-2 and -6 have been isolated and completely sequenced.
The group HERV-K(HML-6), which is the least closely related to mouse mammary tumor virus, comprises about 30 to 40 members with 40 to 68% nucleotide sequence similarity to mouse mammary tumor virus and intracisternal type A particles of the mouse and hamster (24). In addition to proviral sequences, about 50 solitary HML-6-related LTRs are found per haploid genome.
The HERV-K(HML-2) group consists of approximately 30 members with full-length genomes, a few elements with large deletions (23, 25, 31), and an estimated 10,000 to 25,000 solitary LTRs distributed throughout the human genome (15). The prototype HERV-K(HML-2) provirus is HERV-K10, which to date is the only completely sequenced full-length provirus of this group (31). HERV-K10 and most HERV-K10-related proviruses harbor a characteristic deletion of 292 nucleotides (nt) leading to a defective genome with a polymerase gene fused to the envelope gene. However, transcripts of other HERV-K-related proviruses with uninterrupted pol and env open reading frames have been detected in human teratocarcinoma cell lines (18, 19, 42), and gag and pol gene products of HERV-K(HML-2) family members have been demonstrated to be enzymatically active (12, 28, 37). The env gene of HERV-K-IDDM, which was isolated from patients with acute-onset type I diabetes, was found to encode an endogenous superantigen (4). These studies suggest that some proviruses of the HERV-K superfamily have the potential to encode functional retroviral enzymes, possibly even with sufficient genetic information for the formation of retrovirus-like particles, which have been observed in normal human placentas, oocytes, and fetuses (9, 13, 21, 26), in both malignant and nonmalignant breast tissue samples (1, 10, 11, 27), and in germ cell tumors or cell lines derived from these tissues (17). However, there remain many questions with regard to the biological significance or function of these particles, particularly since they appear to be generated by complementation between several expressed HERVs, resulting in pseudotype particles with retroviral RNA of different types (2, 32, 38). Such packaging mechanisms could lead to unforeseen consequences in the use of retroviral vectors in gene therapy or following interspecies organ transplants.
Previously, using degenerate primers from a conserved region of retroviral pol genes (39), we repeatedly amplified three different retroviral sequences from particles released by the human mammary carcinoma-derived cell line T47D (38). One predominant sequence showed about 65% sequence identity to HERV-K10 within the RT region. By screening a human genomic library with the amplified product, we isolated a proviral pol sequence which we preliminarily termed ERV-MLN. The question was whether ERV-MLN is derived from an endogenous provirus with functional retroviral gene products, particularly with the packaging capabilities of Gag proteins. Therefore, we completely analyzed its proviral structure and genomic organization. Sequence comparisons assigned this novel HERV to the HML-4 subgroup of HERV-K elements. We also determined the chromosomal location and expression pattern of the provirus, now called HERV-K-T47D.
Classification of the HERV-K-T47D provirus.
We previously isolated and cloned a human endogenous retroviral RT-related sequence from particles released by the human breast cancer cell line T47D. This pol fragment was used to isolate from a human genomic library, as described previously, a number of hybridizing λ clones (38) that were entirely sequenced by the dideoxy chain termination method (36). Two overlapping λ clones now revealed that this element, previously termed ERV-MLN, comprises an almost full-length proviral structure with an overall length of 9,315 bp (Fig. 1). Next to the 5′ LTR (nt 1 to 943) is a putative tRNA PBS (nt 946 to 963) which, despite a 3-bp mismatch, is most closely related to the complementary sequence of the 3′ end of human lysine tRNA (CUU anticodon) (Fig. 2A). The putative PBS is identical to that found in HERV-KC4 (6) and is closely related to the PBSs of other HERV elements belonging to the HERV-K superfamily. Therefore, ERV-MLN, a human endogenous retrovirus with lysine tRNA as the most likely primer for reverse transcription originating from retrovirus-like particles released by the T47D cell line, is now referred to as HERV-K-T47D.
FIG. 1.
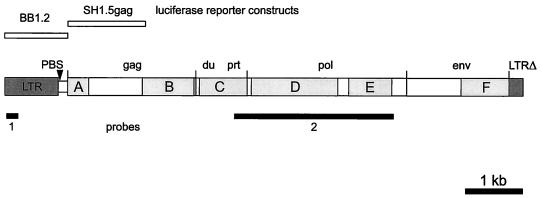
Proviral organization of HERV-K-T47D, locations of hybridization probes, and regions of amino acid identity to HERV-K10. DNA fragments used as hybridization probes are shown as black bars (LTR, probe 1, 229 bp; pol, probe 2, 2.9-kb HindIII-HindIII fragment). Six regions with amino acid identity to HERV-K10 were identified and are depicted as shaded boxes A to F. Abbreviations: du, dUTPase; prt, protease; pol, polymerase; env, envelope. HERV-K-T47D fragments (BB1.2, nt 25 to 1243, 1,218 bp; SH1.5gag, nt 1062 to 2517, 1,455 bp) employed for the construction of recombinant pBL luciferase reporter plasmids used in transient transfection experiments are shown as open bars at the top of the figure. LTRΔ indicates the truncated 3′ LTR.
FIG. 2.
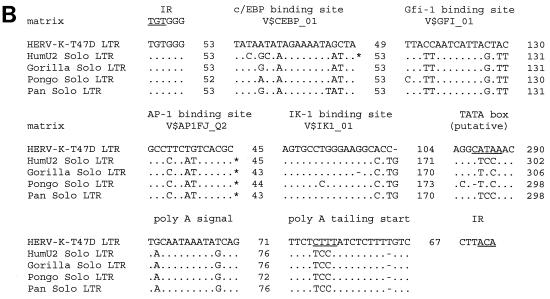
(A) Nucleotide sequence of HERV-K-T47D proviral DNA. LTRs are enclosed in brackets, and the inverted termini TGT and ACA are indicated by arrows. Transcriptional regulatory sequences, i.e., c/EBP, Gfi-1, AP-1, Ik-1, a glucocorticoid-responsive element (GRE), enhancer-like elements, a putative TATA box, a polyadenylation signal, and polyadenylation sites (CA and TA), are underlined once and labeled above. PBS and the polypurine tract (ppt) are double underlined. Sequence complementary to the 3′ end of human lysine tRNA is depicted below the PBS sequence, with lowercase letters being used for mismatches. Translated amino acid sequences with significant homology to HERV-K10 (31), shown under the nucleotide sequence in the six shaded boxes A to F, are those of gag (box A, 40% identity; box B, 49% identity), dUTPase-protease (box C, 59% identity), RT-RNase H (box D, 59% identity), integrase (box E, 59% identity), and env (box F, 58% identity). Frameshifts in the amino acid sequence are indicated with slashes; asterisks correspond to stop codons. Conserved zinc finger motifs (type CX2CX4HX4C) in the NC (box B) region are marked by underlining of the corresponding amino acids. (B) Alignment of putative regulatory elements of the HERV-K-T47D 5′ LTR with corresponding elements from solitary HERV-K-T47D-related LTRs of higher primates (33). Asterisks indicate binding sites which would not have been found with the default parameters of MathInspector (34). However, they were found when a lower threshold was used. Dots and dashes show identical and missing nucleotides, respectively. Under the binding site designations are search string variables used by the program MathInspector. IR, inverted repeat.
To further classify HERV-K-T47D within subgroups HML-1 to -6, so far characterized by a 242-bp stretch of the RT domain (23), we aligned the subgroup sequences with the corresponding HERV-K-T47D region by using the software package Gene Works (IntelliGenetics, Inc.). Sequence comparison revealed that HML-4.1 is the most closely related sequence, showing about 80% nucleotide homology. Therefore, HERV-K-T47D is the first identified full-length prototypic element of the HERV-K(HML-4) subgroup.
Genomic organization and coding regions.
Since a full-length HML-4 group provirus has not yet been identified, we used the well-characterized HERV-K10 (31), a member of the HML-2 subgroup, for alignments with a computer-assisted translation of the complete nucleotide sequence of HERV-K-T47D in order to analyze its genomic organization and identify putative reading frames. At the amino acid level, this revealed an HERV-K-T47D retroviral structure, comprising gag, prt, pol, and env genes (Fig. 2A), with six regions exhibiting significant protein similarity to HERV-K10, ranging from 40 to 60% homology (boxes A to F in Fig. 1 and 2A). Within the HERV-K-T47D gag gene, which tentatively extends from nt 1095, based on comparison with HERV-K10 and the location of a suitable methionine, to nt 3480, two regions with significant amino acid similarity to HERV-K10 were found (box A, nt 1058 to 1385, 40% homology; and box B, nt 2416 to 3308, 49% homology). As shown in Fig. 2A, box A corresponds to the amino-terminal part of the matrix protein, whereas box B comprises the carboxy-terminal half of the capsid (CA) protein and almost the entire nucleocapsid (NC) protein. These regions are separated by 1,235 nt displaying no significant nucleotide (or, hence, amino acid) identity to the corresponding region of HERV-K10, which is shorter and comprises only 627 nt. FASTA database searches based on the differing 608 nt of this sequence revealed an 89% nucleotide homology in a 203-bp overlap with a human CpG island (5). Within the NC protein of HERV-K-T47D, two Zn finger domains of the CX2CX4HX4C type were identified (nt 3114 to 3155 and nt 3237 to 3275). The first motif is defective, lacking the initial Cys, whereas the second Zn finger is intact. A third conserved HERV-K-T47D region (box C in Fig. 1 and 2A) extends from nt 3510 to 4250. It exhibits 59% amino acid identity to the corresponding region of HERV-K10 and comprises the complete retroviral dUTPase (nt 3510 to 3883) and part of the retroviral protease (nt 3884 to 4340).
Retroviral pol genes are generally the most conserved sequences among retroviruses (22). This concurs with our observation that a 2.5-kb stretch of HERV-K-T47D pol (nt 4320 to 6998) shows a 60% overall amino acid identity to HERV-K10 (Fig. 2A, boxes D and E). The RT (nt 4341 to 5184) exhibits 65% identity, including some short stretches with almost absolute identity, while the tether region (nt 5185 to 5672), which connects the RT and RNase H protein domains, is less conserved (47% amino acid identity). RNase H (nt 5673 to 6086) shows 50% identity to HERV-K10, including a common feature of retroviral RNase H proteins, the DEDD motif.
The env gene of HERV-K-T47D shows the least homology to HERV-K10, with the exception of the transmembrane domain (TM). At the amino terminus of the HERV-K-T47D TM, a region with 59% amino acid identity to HERV-K10 is found (Fig. 1 and 2A, box F, nt 8347 to 8991). Specifically, two clusters of hydrophobic amino acids (nt 8401 to 8478 and 8931 to 8970) are highly conserved (86% identity). The 3′ end of HERV-K-T47D env is followed by a polypurine tract (nt 9046 to 9059), which is a conserved motif of the retroviral env-LTR border. Despite their well-defined structure, the coding regions of HERV-K-T47D are interrupted by nonsense and frameshift mutations.
The putative LTRs of HERV-K-T47D were defined by aligning the sequences flanking the proviral coding regions at the 5′ and 3′ ends. Sequence repeats of 254 bp which differ from one another in 23 positions (91% homology) were identified. However, database searches revealed that the 943-bp region from the 5′ end of HERV-K-T47D exhibits 70% homology to a solitary retroviral LTR sequence at the human RNU2 locus on chromosome 17q21 (33). This LTR can be traced back to a complete retroviral element of 6 kb which still exists in the corresponding chromosomal locus of the baboon. During primate evolution, excision of the provirus by homologous recombination created the solitary LTR now found in the genomes of the chimpanzee, gorilla, orangutan, and human (33). This LTR is considered to be associated with the concerted evolution of the tandem array encoding U2 snRNA. Direct sequence alignment of this solitary RNU2 LTR with the ends of HERV-K-T47D revealed that its 5′ LTR is intact whereas the 3′ LTR is truncated after nt 254. The 5′ LTR is bordered by short inverted repeats (TGT…ACA) and is followed by an untranslated leader sequence of 150 bp (nt 944 to 1094) containing the PBS (nt 946 to 963). Several potential regulatory elements were identified by using the program ModelInspector (8). Putative binding sites for transcription factors C/EBP, Gfi-1, AP1, and Ik-1 were detected within the U3 region (Fig. 2A and B) by using the program MathInspector (34). These sequences were found to be conserved in solitary human and various solitary primate RNU2 LTRs (Fig. 2B). However, a putative TATA box at position 462 of HERV-K-T47D is not present in those solitary LTRs. A glucocorticoid-responsive element and two enhancer-like structures were also tentatively assigned (29). A polyadenylation signal [poly(A)] was detected in the 5′ LTR but not in the 3′ LTR, which is truncated in this region. Therefore, a poly(A) signal located either within the coding region of the provirus or within 3′ cellular flanking sequences may be used to generate HERV-K-T47D mRNA. To examine these possibilities, we screened a cDNA library from steroid-induced T47D cells with an HERV-K-T47D LTR probe (Fig. 1, probe 1) generated by PCR with forward primer CCGAGGCAAGAGACTGAAGGCAC (nt 25 to 47) and reverse primer ACTTCTCACAATGTCCCTTCAGC (nt 232 to 254). We were not able to isolate an HERV-K-T47D cDNA by this method, but we obtained several clones containing cellular sequences which are polyadenylated by solitary HERV-K-T47D-related LTRs (data not shown). Based on these clones, we identified two possible poly(A) addition sites (CA and TA) within the HERV-K-T47D LTR. The poly(A) addition site observed in the majority of cDNA clones was used to assign the R-U5 border (Fig. 2A).
Chromosomal location and evolution of HERV-K-T47D and related elements.
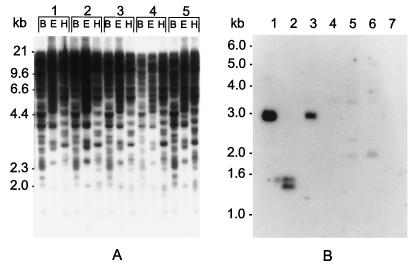
Southern blot analysis was performed under high-stringency conditions as described previously (38) to determine the copy number and chromosomal location of HERV-K-T47D and closely related sequences in the human genome. HindIII-digested DNA from a panel of 24 human-rodent monochromosomal hybrid cell lines (mapping panel no. 2; NIGMS Human Genetic Cell Repository, Camden, N.J.) was hybridized to an HERV-K-T47D pol DNA fragment (Fig. 1, probe 2). The observed banding pattern suggests that HERV-K-T47D is located on human chromosome 10. Furthermore, five related elements, probably representing other members of the HERV-K(HML-4) family, could be assigned to chromosomes 8, 9, 15, 16, and 19. Southern blot analysis of human DNA samples digested with a set of restriction enzymes revealed that in addition to those proviral sequences, several hundred solitary HERV-K-T47D LTRs may exist in the human genome (Fig. 3A). As is known from studies of HERV-K(HML-2) (15) and HERV-H elements (7), multiple solitary LTRs are a common feature of HERV families. As evolutionary relics, they reflect high-level retrotransposon activity and subsequent homologous recombination during evolution.
FIG. 3.
(A) Southern blot analysis of human genomic DNA with a probe specific for the HERV-K-T47D LTR (Fig. 1, probe 1). DNA samples (10 μg/lane) from healthy individuals (1 to 5) were restricted to completion, blotted, and hybridized under relaxed stringency conditions (5× SSC, 60°C). Restriction enzymes are abbreviated as follows: B, BamHI; E, EcoRI; and H, HindIII. Marker sizes are indicated on the left. (B) Southern blot analysis of DNA derived from Old World and New World monkeys and higher primates, using a probe specific for the HERV-K-T47D pol gene (Fig. 1, probe 2). High-molecular-weight DNA (10 μg/lane) was restricted to completion with HindIII, blotted, and hybridized under relaxed stringency conditions. The DNAs analyzed were as follows: lane 1, human; lane 2, chimpanzee; lane 3, orangutan; lane 4, Presbytis; lane 5, baboon; lane 6, rhesus monkey; and lane 7, Aotes.
To investigate HERV-K-T47D evolution, we analyzed DNAs of Old and New World monkeys and higher primates by performing Southern blot hybridization under relaxed hybridization conditions as described previously (38). High-molecular-weight DNA was digested with HindIII and probed with the 2.9-kb HERV-K-T47D pol fragment (Fig. 1, probe 2). A strong signal of the same size (2.9 kb) was detected in restricted DNA from both the human and the orangutan (Fig. 3B, lanes 1 and 3). DNA derived from the chimpanzee resulted in two smaller bands (1.3 and 1.4 kb) of similar intensity (lane 2), suggesting the presence of an additional HindIII restriction site in this element. In Old World monkeys (lanes 4 to 6), a series of weak signals differing in size (approximately 2.0, 2.4, and 3.5 kb) were detected, while DNA from the New World monkey genus Aotes (lane 7) gave no detectable hybridization signal. These data concur with previous findings indicating that most HERV elements arose early in primate evolution (for a review, see reference 16).
Transcription of HERV-K-T47D in human tissues.
Regardless of whether active, functional proteins are encoded, a crucial role of HERVs may be their ability to act as promoters of either immunologically related retroviral antigens or cellular genes or, conversely, to act as premature transcription terminators. Since the production and release of T47D particles is steroid dependent (11, 32, 38), T47D cells were treated with 10−9 M estrogen for 48 h, at which time was added 10−8 M progesterone, with subsequent incubation for 24 h (11, 30). Total RNA was prepared from steroid-treated and untreated cells in accordance with a CsCl ultracentrifugation protocol (35), separated by denaturing 1% formamide–agarose gel electrophoresis, transferred to Zeta-Probe membranes (Bio-Rad, Munich, Germany) by the vacublot procedure (Vacu-Gene XL; Pharmacia/LKB, Freiburg, Germany), and hybridized with a 32P-labeled HERV-K-T47D pol fragment (Fig. 1, probe 2) under high-stringency conditions (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1% sodium dodecyl sulfate, 5× Denhardt’s solution, and 100 μg of denatured sheared herring sperm DNA per ml for 16 h at 65°C). Expression of HERV-K-T47D was found exclusively in steroid-stimulated T47D cells (Fig. 4A), therefore correlating well with the production and release of HERV-K-T47D particles. The HERV-K-T47D transcript was about 4.5 kb, which does not match the size of a full-length or a regular spliced retroviral transcript.
FIG. 4.
HERV-K-T47D transcription in human tissues. (A) Total RNA derived from estradiol- and progesterone-induced (T47D+) or noninduced (T47D−) cells was blotted onto Hybond membranes. All filters were probed with an HERV-K-T47D pol fragment (Fig. 1, probe 2) and washed under conditions of high stringency (0.1× SSC, 65°C). (B) Multiple-tissue Northern blot with 2 μg of mRNA per lane from heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas tissues. To assess RNA quality, the blots (A and B) were rehybridized with a human APC probe and a ubiquitin probe, respectively. Marker sizes are indicated on the left.
To confirm our Northern blotting results and further analyze the observed truncated HERV-K-T47D transcript, PCR experiments were performed on T47D cDNA, using primers derived from HERV-K-T47D gag (reverse primer, CGCGGATCCTATGGCTGCAAGGATTCTAAG, nt 1358 to 1381), pol (forward primer, CGCGGATCCCTCAACAATGTCGCTCAGGCTAC, nt 4741 to 4766; reverse primer, CGCGGATCCCCAAGTAACTTTTGAAAGTC, nt 5104 to 5126), env (forward primer, CGCGGATCCGTTTAATTGCTGTTACTACAACAGC, nt 8432 to 8456), and LTR (forward primer, CGCGGATCCGCAACTTGGTGGTAGTGGTACC, nt 805 to 829) in various combinations and in combination with an oligo(dT)22 primer. T47D mRNA was reverse transcribed, using a cDNA first-strand kit (Stratagene, La Jolla, Calif.) and the target-specific reverse primers. Amplification of HERV fragments was carried out in a reaction mixture (total volume, 100 μl) containing 10 mM Tris (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, 0.25 mM each deoxynucleoside triphosphate, 2.5 U of Taq DNA polymerase (Boehringer, Mannheim, Germany), and 50 pmol of each mixed oligonucleotide primer pair. Using a Perkin-Elmer Cetus DNA thermal cycler, PCR was performed with the following cycle parameters: a hot start at 94°C for 5 min; 30 cycles of 1 min at 94°C, 2 min at 55 to 65°C, and 2 min at 72°C; and a final extension step of 7 min at 72°C. A control reaction in which the DNA template was omitted was carried out to detect any traces of contaminating genomic DNA in the solutions used.
Only amplification with gag and pol primers yielded fragments, comprising 575 and 387 bp, respectively. These PCR products were sequenced, revealing 100% sequence identity to the corresponding gag and pol sequences of HERV-K-T47D (data not shown). However, despite several trials, amplification of HERV-K-T47D env sequences by using either env-derived primers or pol and env primers in combination with oligo(dT) primers failed. Therefore, we conclude that the 4.5-kb truncated HERV-K-T47D transcript contains only gag and pol sequences and lacks a complete HERV-K-T47D env gene. The lack of a functional poly(A) signal within the 3′ LTR of HERV-K-T47D correlates with the DNA sequencing data indicating that the polyadenylation signal detected in the 5′ LTR is located in the region that is truncated in the 3′ LTR. This suggests that the 4.5-kb transcript may be generated by premature termination, using a poly(A) signal located within the coding region of the provirus, or by splicing into 3′ cellular sequences which may provide the required poly(A) site.
To investigate the transcriptional activity of HERV-K-T47D in other human tissues, Northern blot analyses were carried out with a commercial human multiple-tissue Northern blot (HMT-blot; Clontech, Palo Alto, Calif.). In all Northern blot hybridization experiments, either the adenomatous polyposis coli (APC) gene, a human tumor suppressor and housekeeping gene (kindly provided by H.-J. Butterfass, German Cancer Research Center, Heidelberg, Germany), or the human ubiquitin gene (Clontech) was used to monitor mRNA integrity. Hybridization with the HERV-K-T47D pol fragment under high-stringency conditions revealed a high level of transcription exclusively in full-term placental tissue (Fig. 4B). The signal corresponds in size to the band obtained from T47D RNA after steroid treatment (Fig. 4A). With the exception of the breast carcinoma cell line T47D, a correlation between HERV-K-T47D expression and the occurrence or progression of malignancy was not found. No HERV-K-T47D transcripts could be detected in RNA from human tumor cell lines such as melanoma (S361), lung cancer (A549), colorectal adenocarcinoma (SW480), cervix carcinoma (HeLa), Burkitt’s lymphoma (Raji), lymphoblastic leukemia (Molt-4), promyelocytic leukemia (HL-60), and chronic myeloid leukemia (K562) and in RNA from peripheral blood mononuclear cells of patients with chronic myeloid leukemia, acute lymphatic leukemia, or acute myeloid leukemia (data not shown). These results suggest that HERV-K-T47D transcription at levels detectable by Northern blot analysis is tissue specific and steroid hormone dependent.
Transcriptional activity of the 5′ LTR of HERV-K-T47D.
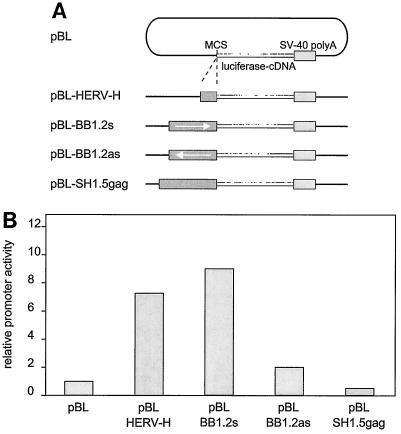
To examine the promoter activity of the putative 5′ LTR of HERV-K-T47D in T47D cells, plasmids containing the luciferase reporter gene downstream of an LTR-containing DNA fragment of HERV-K-T47D were constructed (Fig. 1, BB1.2). The BB1.2 fragment was generated by PCR from the proviral sequence by using forward primer GCGGGATCCGAGGCAAGAGACTGAAGGCAC (nt 25 to 47) and reverse primer CGCGGATCCCTCAGTTGGAAACCAAGGGC (nt 1221 to 1243). Employing the newly introduced BamHI restriction sites, BB1.2 was cloned in both the sense (pBL-BB1.2s) and the antisense (pBL-BB1.2as) directions into the multiple cloning site of the luciferase expression vector pBL (Fig. 5A) (kindly provided by Karin Butz, German Cancer Research Center, Heidelberg, Germany). As a negative control, the SH1.5gag fragment was included in the T47D cell transfection experiments. Plasmid pBL-SH1.5gag (sense direction) was constructed by cloning a 1.5-kb SpeI-HindIII DNA fragment (Fig. 1) into pBL. Amplification of this fragment was performed with forward primer GCGACTAGTTGGGCACTCAGAGTATCTCAG (nt 1062 to 1087) and reverse primer GCGAAGCTTCTGCTAAGGATTTTCGGGCGG (nt 2488 to 2517). As a positive control, a transcriptionally active HERV-H LTR was used (7). A 393-bp fragment containing the HERV-H promoter was amplified from clone H6 (kindly provided by D. Mager, Terry Fox Laboratory, Vancouver, British Columbia, Canada), using forward primer CGCGGATCCTGTCAGGCCTCTGAGCCCAA and reverse primer CGCAAGCTTATGTGAGCAACATGGCTGTT, and cloned via BamHI and HindIII restriction sites into pBL. The identity and correct insertion orientation of each construct were verified by nucleotide sequencing.
FIG. 5.
Analysis of HERV-K-T47D putative LTR promoter activity in T47D cells. (A) pBL-HERV reporter constructs used for luciferase expression assays. The putative LTR 1.2-kb PCR fragment (Fig. 1, fragment BB1.2) was cloned in the sense (pBL-BB1.2s) and the antisense (pBL-BB1.2as) orientations into the luciferase expression vector pBL. As controls, plasmid pBL-SH1.5gag with the insert (SH1.5gag, Fig. 1) and pBL-HERV-H containing the HERV-H LTR promoter of H6 (7) were similarly constructed. MCS, multiple cloning site; SV-40, simian virus 40. (B) Transient expression in T47D cells of HERV-pBL luciferase reporter constructs. T47D cells were transiently transfected according to standard procedures. The luciferase expression driven by the retroviral promoter was measured by a standardized luciferase assay and is shown as bar graphs representing relative promoter activity. All results shown are derived from triplicate experiments.
Transfection of plasmids into T47D cells was performed by the calcium phosphate precipitation method (3). The day before transfection, 3 × 105 cells were seeded per 6-cm-diameter petri dish and cultivated for 24 h at 37°C and 5% CO2. For transfection, triplicate dishes were incubated in parallel with a calcium phosphate-DNA mixture containing 0.8 pmol of reporter plasmid and 1 μg of pZ (a β-actin–luciferase construct used for internal standardization). The total amount of DNA per dish was adjusted to 6.5 μg with pBluescript SK(+). Incubation was carried out for 16 to 18 h at 37°C and 5% CO2. T47D cells were then further incubated in fresh RPMI 1640 medium for 48 h. T47D cells were treated with estradiol and then progesterone (in dimethyl sulfoxide solvent), each for 24 h, as described by Keydar et al. (11), while the control dishes received medium with dimethyl sulfoxide alone. At 48 h postincubation (37°C, 5% CO2), cells were harvested and lysates were prepared according to the recommendations of the Enhanced Luciferase Assay Kit (Berthold Detection Systems, Pforzheim, Germany). Relative HERV promoter activity was calculated as the ratio between the levels of luciferase expression of the constructs and the pBL vector. Transient expression of the constructs in T47D cells revealed that pBL-BB1.2s displayed about the same relative transcriptional activity as the active promoter of the HERV-H LTR (pBL-HERV-H). Upon steroid induction of T47D cells, an about twofold enhancement of luciferase activity was observed (data not shown). This suggests that the 5′ LTR of HERV-K-T47D contains regulatory elements that are steroid dependent and can mediate efficient transcriptional activity of HERV-K-T47D or other sequences in T47D cells.
In conclusion, our results show that HERV-K-T47D is actively transcribed in T47D cells in a steroid-dependent manner, and this active transcription is easily accounted for by the promoter activity and the presence of a number of putative transcription factor binding sequences found in the 5′ LTR. Such activity may also apply to a number of related solitary LTRs which were also detected, perhaps resulting in transcriptional activation of disease-associated antigens. However, the HERV-K-T47D-specific transcript, containing only gag and pol sequences, does not comprise a full-length proviral sequence but is presumably irregularly spliced or terminated. Since HERV-K-T47D does not have the coding capacity for full-length structural proteins, the origin of the retroviral proteins responsible for particle formation and the RT activity found associated with T47D particles (11, 32, 38) is still unclear. Particularly, the gag gene, which is essential for virus packaging and particle formation, is inactivated by stop codons and frameshifts in HERV-K-T47D. As rescue experiments with defective retroviruses lacking the gag, pol, and env open reading frames suggest (41), these activities must be provided in trans by other coding-competent HERV elements. Examples of such coding-competent HERVs are members of the HERV-K(HML-2) subgroup, transcripts of which have been detected in some human teratocarcinoma cell lines (20, 42). Since we were not able to isolate intact protein-coding HERVs from particle preparations, the packaging signals of these genomes may be defective in order to prevent the generation of replication-competent and possibly infective retroviral particles. Particularly in light of the use of retroviral vectors in gene therapy or the prospect of xenotransplantation (40, 43), identification of such HERV sequences and understanding the mechanisms and risks of generating new, infectious retroviral particles will be of major importance.
Nucleotide sequence accession number.
The complete nucleotide sequence of HERV-K-T47D has been deposited in GenBank under accession no. AF020092.
Acknowledgments
We thank Dixie Mager, Terry Fox Laboratories, Vancouver, for providing the HERV-H LTR clone H6 and C. Ross and J. Wienberg, European Cell Bank of Primates, for providing primate fibroblast cell cultures and blood. We further thank A. Arthur-Goettig for critically reading the manuscript.
This work was supported in part by the Commission of the European Union (contracts GENE-CT93-0019 and BIO4-CT95-0226), the Bayerische Forschungsstiftung (FORBIOSICH), and the Faculty of Clinical Medicine Mannheim, University of Heidelberg.
REFERENCES
- 1.Al-Sumidaie A M, Hart C A, Leinster S J, Green C D, McCarthy K. Particles with properties of retroviruses in monocytes from patients with breast cancer. Lancet. 1988;ii:5–9. doi: 10.1016/s0140-6736(88)90998-1. [DOI] [PubMed] [Google Scholar]
- 2.Boyd M T, Foley B, Brodsky I. Evidence for copurification of HERV-K-related transcripts and a reverse transcriptase activity in human platelets from patients with essential thrombocythemia. Blood. 1997;90:4022–4030. [PubMed] [Google Scholar]
- 3.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conrad B, Weissmahr R N, Böni J, Arcari R, Schüpbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90:303–313. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 5.Cross S H, Charlton J A, Nan X, Bird A P. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- 6.Dangel A W, Mendoza A R, Baker B J, Daniel C M, Carroll M C, Wu L C, Yu C Y. The dichotomous size variation of human complement C4 genes is mediated by a novel family of endogenous retroviruses, which also establishes species-specific genomic patterns among Old World primates. Immunogenetics. 1994;40:425–436. doi: 10.1007/BF00177825. [DOI] [PubMed] [Google Scholar]
- 7.Feuchter A, Mager D. Functional heterogeneity of a large family of human LTR-like promoters and enhancers. Nucleic Acids Res. 1990;18:1261–1270. doi: 10.1093/nar/18.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frech, K., J. Danescu-Mayer, and T. Werner. A novel method to develop highly specific models for regulatory units detects a new LTR in GenBank which contains a functional promoter. J. Mol. Biol., in press. [DOI] [PubMed]
- 9.Johnson P M, Lyden T W, Mwenda J M. Endogenous retroviral expression in the human placenta. Am J Reprod Immunol. 1990;23:115–120. doi: 10.1111/j.1600-0897.1990.tb00683.x. [DOI] [PubMed] [Google Scholar]
- 10.Keydar I, Chen L, Karby S, Weiss F, Delarea J, Radu M, Chaitcik S, Brenner H. Establishment and characterization of a cell line of human breast carcinoma origin. Eur J Cancer. 1979;15:659–670. doi: 10.1016/0014-2964(79)90139-7. [DOI] [PubMed] [Google Scholar]
- 11.Keydar I, Ohno T, Nayak R, Sweet R, Simoni F, Weiss F, Karby S, Mesa-Tejada R, Spiegelman S. Properties of retrovirus-like particles produced by a human breast carcinoma cell line: immunological relationship with mouse mammary tumor virus proteins. Proc Natl Acad Sci USA. 1984;81:4188–4192. doi: 10.1073/pnas.81.13.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitamura Y, Ayukawa T, Ishikawa T, Kanda T, Yoshiike K. Human endogenous retrovirus K10 encodes a functional integrase. J Virol. 1996;70:3302–3306. doi: 10.1128/jvi.70.5.3302-3306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson E, Nilsson O B, Sundstrom P, Widehn S. Morphological and microbiological signs of endogenous C-virus in human oocytes. Int J Cancer. 1981;18:551–556. doi: 10.1002/ijc.2910280504. [DOI] [PubMed] [Google Scholar]
- 14.Leib-Mösch, C., R. Brack-Werner, T. Werner, M. Bachmann, O. Faff, V. Erfle, and R. Hehlmann. 1990. Endogenous retroviral elements in human DNA. Cancer Res. 50(Suppl. 1):5636–5642. [PubMed]
- 15.Leib-Mösch C, Haltmeier M, Werner T, Geigl E-M, Brack-Werner R, Francke U, Erfle V, Hehlmann R. Genome distribution and transcription of solitary HERV-K LTRs. Genomics. 1993;18:261–269. doi: 10.1006/geno.1993.1464. [DOI] [PubMed] [Google Scholar]
- 16.Leib-Mösch C, Seifarth W. Evolution and biological significance of human retroelements. Virus Genes. 1996;11:133–145. doi: 10.1007/BF01728654. [DOI] [PubMed] [Google Scholar]
- 17.Löwer J, Wondrak E M, Kurth R. Genome analysis and reverse transcriptase activity of human teratocarcinoma-derived retroviruses. J Gen Virol. 1987;68:2807–2815. doi: 10.1099/0022-1317-68-11-2807. [DOI] [PubMed] [Google Scholar]
- 18.Löwer R, Boller K, Hasenmaier B, Korbmacher C, Müller-Lantzsch N, Löwer J, Kurth R. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Löwer R, Tönjes R R, Korbmacher C, Kurth R, Löwer J. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J Virol. 1995;69:141–149. doi: 10.1128/jvi.69.1.141-149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyden T W, Johnson P M, Mwenda J M, Rote N S. Ultrastructural characterization of endogenous retroviral particles isolated from normal human placentas. Biol Reprod. 1994;51:152–157. doi: 10.1095/biolreprod51.1.152. [DOI] [PubMed] [Google Scholar]
- 22.McClure M A, Johnson M S, Feng D-F, Doolittle R F. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc Natl Acad Sci USA. 1988;85:2469–2473. doi: 10.1073/pnas.85.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medstrand P, Blomberg J. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J Virol. 1993;67:6778–6787. doi: 10.1128/jvi.67.11.6778-6787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medstrand P, Mager D L, Yin H, Dietrich U, Blomberg J. Structure and genomic organization of a novel human endogenous retrovirus family: HERV-K(HML6) J Gen Virol. 1997;78:1731–1744. doi: 10.1099/0022-1317-78-7-1731. [DOI] [PubMed] [Google Scholar]
- 25.Meese E, Göttert E, Zang K D, Sauter M, Schommer S, Mueller-Lantzsch N. Human endogenous retroviral element K10 (HERV-K10): chromosomal localization by somatic hybrid mapping and fluorescence in situ hybridization. Cytogenet Cell Genet. 1996;72:40–42. doi: 10.1159/000134157. [DOI] [PubMed] [Google Scholar]
- 26.Mondal H, Hofschneider P H. Isolation and characterization of retrovirus-like elements from normal human fetuses. Int J Cancer. 1982;30:281–287. doi: 10.1002/ijc.2910300305. [DOI] [PubMed] [Google Scholar]
- 27.Moore R, Dixon M, Smith R, Peters G, Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J Virol. 1987;61:480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller-Lantzsch N, Sauter M, Weiskircher A, Kramer K, Best B, Buck M, Grässer F. Human endogenous retroviral element K10 (HERV-K10) encodes a full length Gag homologous 73 kD protein and a functional protease. AIDS Res Hum Retroviruses. 1993;9:343–350. doi: 10.1089/aid.1993.9.343. [DOI] [PubMed] [Google Scholar]
- 29.Ono M. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J Virol. 1986;58:937–944. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono M, Kawakami M, Ushikubo H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol. 1987;61:2059–2062. doi: 10.1128/jvi.61.6.2059-2062.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986;60:589–598. doi: 10.1128/jvi.60.2.589-598.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patience C, Simpson G R, Colletta A A, Welch H M, Weiss R A, Boyd M T. Human endogenous retrovirus expression and reverse transcriptase activity in the T47D mammary carcinoma cell line. J Virol. 1996;70:2654–2657. doi: 10.1128/jvi.70.4.2654-2657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavelitz T, Rusche L, Matera A G, Scharf J M, Weiner A M. Concerted action of the tandem array encoding primate U2 snRNA occurs in situ, without changing the cytological context of the RNU2 locus. EMBO J. 1995;14:169–177. doi: 10.1002/j.1460-2075.1995.tb06987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauter M, Schommer S, Kremmer E, Remberger K, Dölken G, Lemm I, Buck M, Best B, Neumann-Haefelin D, Mueller-Lantzch N. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J Virol. 1995;69:414–421. doi: 10.1128/jvi.69.1.414-421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seifarth W, Skladny H, Krieg-Schneider F, Reichert A, Hehlmann R, Leib-Mösch C. Retrovirus-like particles released from the human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J Virol. 1995;69:6408–6416. doi: 10.1128/jvi.69.10.6408-6416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shih A, Misra R, Rush M G. Detection of multiple, novel reverse transcriptase coding sequences in human nucleic acids: relation to primate retroviruses. J Virol. 1989;63:64–75. doi: 10.1128/jvi.63.1.64-75.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoye J P, Coffin J M. The dangers of xenotransplantation. Nat Med. 1995;1:1100. doi: 10.1038/nm1195-1100a. [DOI] [PubMed] [Google Scholar]
- 41.Tchenio T, Heidmann T. Defective retroviruses can disperse in the human genome by intracellular transposition. J Virol. 1991;65:2113–2118. doi: 10.1128/jvi.65.4.2113-2118.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tönjes R R, Limbach C, Löwer R, Kurth R. Expression of human endogenous retrovirus type K envelope glycoprotein in insect and mammalian cells. J Virol. 1997;71:2747–2756. doi: 10.1128/jvi.71.4.2747-2756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss R A. Transgenic pigs and virus adaptation. Nature. 1998;391:327–328. doi: 10.1038/34772. [DOI] [PubMed] [Google Scholar]
- 44.Wilkinson D A, Mager D L, Leong J C. Endogenous human retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 3. New York, N.Y: Plenum Press; 1994. pp. 465–535. [Google Scholar]